An expanded understanding of mosquito reproductive biology may assist efforts to control wild mosquito populations by manipulating reproduction. Given that molecular mechanisms of seminal fluid and sperm function are poorly understood, we have catalogued proteins that compose the ejaculate in Aedes aegypti,—the vector of dengue, Zika, and other viruses. The sperm and seminal fluid proteomes characterized herein represent a valuable tool for investigations of mosquito biology and the development of molecular targets by which mosquito populations may be suppressed.
Keywords: Gene Expression*, Omics, Tandem Mass Spectrometry, RNA SEQ, Label-free quantification, Accessory gland, Fertility, Mating, Reproduction, Testis
Graphical Abstract
Highlights
First high throughput mosquito sperm proteome, with 870 identified proteins.
Nearly four-fold expansion of Ae. aegypti, seminal fluid proteome, with 280 proteins.
Transcriptomics illuminate post-mating male accessory gland gene expression.
Refined genetic architecture of male reproduction in a human disease vector.
Abstract
The yellow fever mosquito, Aedes aegypti,, transmits several viruses causative of serious diseases, including dengue, Zika, and chikungunya. Some proposed efforts to control this vector involve manipulating reproduction to suppress wild populations or to replace them with disease-resistant mosquitoes. The design of such strategies requires an intimate knowledge of reproductive processes, yet our basic understanding of reproductive genetics in this vector remains largely incomplete. To accelerate future investigations, we have comprehensively catalogued sperm and seminal fluid proteins (SFPs) transferred to females in the ejaculate using tandem mass spectrometry. By excluding female-derived proteins using an isotopic labeling approach, we identified 870 sperm proteins and 280 SFPs. Functional composition analysis revealed parallels with known aspects of sperm biology and SFP function in other insects. To corroborate our proteome characterization, we also generated transcriptomes for testes and the male accessory glands—the primary contributors to Ae. aegypti, sperm and seminal fluid, respectively. Differential gene expression of accessory glands from virgin and mated males suggests that transcripts encoding proteins involved in protein translation are upregulated post-mating. Several SFP transcripts were also modulated after mating, but >90% remained unchanged. Finally, a significant enrichment of SFPs was observed on chromosome 1, which harbors the male sex determining locus in this species. Our study provides a comprehensive proteomic and transcriptomic characterization of ejaculate production and composition and thus provides a foundation for future investigations of Ae. aegypti, reproductive biology, from functional analysis of individual proteins to broader examination of reproductive processes.
The mosquito, Aedes aegypti,, is the most important vector of arboviruses globally, transmitting viruses that cause dengue (1), Zika (2), chikungunya (3), and yellow fever (4). Consequently, Ae. aegypti, places a severe strain on public health infrastructure around the world (5). Despite decades of effort to control mosquito populations, Ae. aegypti, continues to contribute to human disease epidemics. New and improved control strategies are needed to prevent future outbreaks and mitigate disease burden.
Some promising control strategies under development target reproduction to suppress mosquito populations. For example, sterilized males can be released to suppress populations by impairing reproduction by their wild mates (6, 7). Manipulating reproductive phenotypes may also provide a means of driving disease-refractory traits into a population (reviewed in 8). One such strategy employs the intracellular bacterium Wolbachia,, which, when introduced into Ae. aegypti,, induces cytoplasmic incompatibility that allows the bacterium to spread in a population, potentially to fixation (9). Cytoplasmic incompatibility causes sperm of males with Wolbachia, to be incompatible with uninfected females' eggs, whereas Wolbachia-,positive females can reproduce with any male, regardless of infection status (reviewed in 10), giving Wolbachia,-positive individuals a fitness advantage over their uninfected counterparts. This bacterium also blocks or reduces transmission of several viruses, including dengue (11) and Zika (12). Consequently, introduction of novel Wolbachia, infections into vector populations is being explored as a transmission reducing strategy.
Designing mosquito control strategies that target reproduction requires an intimate knowledge of the underlying cellular and molecular mechanisms. Yet, only a few functions of proteins involved in mosquito reproduction have been described to date. For example, in Ae. aegypti, seminal fluid proteins (SFPs)1 induce several physiological and behavioral changes in females, including refractoriness to future mating (13–18), stimulation of oogenesis (19), enhanced survival (18), and the ability to fertilize eggs (20). However, the molecular identity of active SFP components for this species remains elusive. Seminal fluid initiates sperm motility via the action of proteases in many insects (silkworm (21); water strider (22); Culex, mosquito (23)), but the precise sperm proteins on which seminal fluid acts in Ae. aegypti, have not been identified. Similarly, sperm-associated odorant receptors may control motility in Ae. aegypti,, although the exact function and ligands of these receptors are unknown (24). Finally, the mechanism by which Wolbachia, induces cytoplasmic incompatibility has not been described in Ae. aegypti,, but Wolbachia, proteins contained in sperm are hypothesized to be involved (25, 26).
Identification of sperm proteins and SFPs that are transferred to females during copulation is an important objective to enable future investigations into specific reproductive processes. Components of the transferred ejaculate include sperm and seminal fluid—both of which play vital roles in mosquito reproduction. An Ae. aegypti, seminal fluid proteome was first reported by Sirot et al., (27) based on mass spectrometry analyses. That study described 93 putative SFPs transferred during mating. Although not a primary focus of their work, they also identified 101 putative sperm proteins. Later work identified more than twice as many SFPs from Ae. albopictus, using similar methodology (28). Proteome complexity of other insects' seminal fluid (reviewed in 29, 30, 31) and sperm (32–34) suggests that more proteins remain to be identified in the Ae. aegypti, ejaculate. Here, we used tandem mass spectrometry (MS/MS) with greater sensitivity and the recently revised and expanded genome to build on the foundational work of Sirot et al., by identifying constituent proteins of both Ae. aegypti, sperm and seminal fluid. Importantly, significantly increased coverage of the sperm proteome allowed more accurate differentiation between seminal fluid and sperm proteins in the mixed ejaculate sample. We also profiled the transcriptomes of the male accessory glands (MAG; before and after mating) and testes, the major source tissues for SFPs and sperm proteins, respectively. Our proteomic characterization represents a nearly 4-fold expansion of putative SFPs and a more than 8-fold expansion in the Ae. aegypti, sperm proteome. Our results yield insights into the molecular function, genome organization, regulation, and evolution of sperm proteins and SFPs in this important disease vector. Ultimately, these proteomes provide a basis for future studies of Ae. aegypti, reproduction and, potentially, a catalogue of molecular targets for the development of novel mosquito control methods.
EXPERIMENTAL PROCEDURES
Rearing
Mosquitoes were derived from a laboratory colony of Ae. aegypti, that was established from individuals collected in Bangkok, Thailand, in 2011 and supplemented with wild caught mosquitoes every 2 to 3 years. Mosquitoes were reared as described previously by Degner and Harrington (35). Briefly, eggs were hatched under vacuum pressure, and a day later 200 first instar larvae were transferred to rearing trays of 1 L deionized water with four Cichlid Gold fish food pellets (Hikari, Himeji, Japan) as diet (except in the case of 15N-labeled females; see below). Pupae were separated by sex based on size, allowed to eclose individually in separate test tubes, and adults were transferred into single-sex cages with 10% sucrose provided ad libitum,.
Sperm Isolation
Males used for sperm protein sample preparation were aged between 5 and 8 days post-eclosion (dpe). Sperm were isolated from the seminal vesicles of each male to ensure a relatively homogenous population of mature sperm which have completed spermiogenesis. Seminal vesicle sperm likely also include proteins that could have been deposited on the sperm coat by the vas deferentia or seminal vesicle after sperm leave the testes (Ndiaye et al., 1997). Males were dissected in physiological saline (133 mm NaCl, 2.63 mm KCl, 9.75 mm Na2HPO4, 3 mm KH2PO4, 2 mm CaCl2, adjusted to pH 6.9; hereafter “saline”). The seminal vesicle was isolated from other tissue and consecutively transferred to two clean droplets of saline to remove any adherent fat body or other debris. Clean dissecting tools were used to transfer the seminal vesicle to a final droplet of saline where it was ruptured to release sperm. Sperm suspended in saline were transferred to a microcentrifuge tube on ice, and pooled sperm samples were flash frozen in liquid nitrogen every 2 h.
Two biological replicates included sperm combined from 400 and 470 randomly selected males, respectively. Pooled samples were centrifuged at 25,000 × g, for 10 min at 4 °C. The supernatant was removed, leaving 18 μl saline with the pellet. An equal volume of 2x Laemmli buffer + 5% β-mercaptoethanol was added to the pellet, and samples were solubilized by sonicating for 30 s, boiling for 15 min, and re-sonicating for 30 s. To remove any particulate debris, samples were spun down at 10,000 × g, at 4 °C for 10 min, and the supernatant was placed in a fresh tube. Protein was quantitated using a 1:5 dilution of the sample using the EZQ assay (Thermo Fisher Scientific, Waltham, MA). Protein used for subsequent mass spectrometry was standardized across biological replicates (16 μg).
Transferred Ejaculate Isolation
To identify SFPs that are unequivocally transferred to the female, we used a reverse-labeling technique pioneered in Drosophila, by Findlay et al., (36) and adapted to Ae. aegypti, by Sirot et al., (27). Females labeled with 15N were mated to unlabeled males, allowing the identification of only transferred ejaculate proteins via MS/MS. Males were reared as described above. As larvae, females were labeled with 15N using the rearing methodology of Sirot et al., (27). Briefly, a prototrophic yeast strain (D273–10B) was grown in media whose only nitrogen source was 15N ammonium sulfate (Cambridge Isotope Laboratories; Cambridge, MA). Yeast were grown to saturation, pelleted, and resuspended in PBS to a final volume of one sixteenth of the growth media. A few drops of yeast slurry were provided to newly hatched first instar larvae after vacuum hatching. One day after hatching, 200 larvae were placed in a rearing tray with 200 ml water from a previous cohort of 15N-yeast-reared mosquitoes (to seed their rearing environment with beneficial microbiota) and 800 ml of deionized water. Larvae were fed 4 ml of labeled yeast slurry a day after hatching, and again at 4 d after hatching. Pupae were isolated into individual tubes as described above, and females were put into an 8 L bucket cage and provided sucrose solution ad libitum,. Sucrose was replaced every 2 d to preclude the introduction of unlabeled nitrogen via microbial contamination.
At 4 to 5 dpe, matings between labeled females and unlabeled males were observed as in Degner and Harrington (35). Because mosquito seminal fluid is known to contain proteases (27), mosquitoes were dissected in saline with protease inhibitors (cOmplete Mini Protease Inhibitor Mixture; Sigma Aldrich, St. Louis, MO). In contrast to Sirot et al., (27), we dissected only bursae (and not spermathecae). Adjoining tissue associated with the gonotreme (vaginal lips) was left in place and acted as a tissue barrier that prevented escape of seminal fluid into the surrounding saline. We cannot rule out the possibility that some seminal proteins may be processed, modified, or trafficked outside of the bursa in the short time between mating and dissection. To minimize these processes, we immediately placed females on ice after mating and dissected them within 3 min of mating. Upon excision, bursae (n, = 35–43 per replicate) were transferred to 33 μl 1× Laemmli buffer diluted from a 2x stock solution with saline, with 2.5% β-mercaptoethanol and 1x protease inhibitors. Samples were sonicated on high with a Bioruptor UCD-200 (Diagenode, Liège, Belgium) for 30 s, boiled for 10 min, sonicated for 30 s, and centrifuged at 10,000 × g, at 4 °C for 10 min. The supernatant was removed; 30 μl were frozen at −80 °C, and 2 μl were diluted 1:3 with buffer for protein quantitation using the EZQ assay. In parallel, we also prepared samples of bursae from virgin, labeled females from each cohort to assess the efficiency of 15N-labeling. Mated and virgin bursae samples contained 18 and 8 μg of protein, respectively.
Tandem Mass Spectrometry Analysis
Solubilized proteins were separated on a 1-dimensional SDS-PAGE gel and split into 6 fractions, with two biological replicates run in parallel (supplemental Fig. S1). Gel fractions were cut into 1 mm cubes, washed, and dehydrated (37). Subsequently, proteins were reduced with dithiothreitol and alkylated with iodoacetamide (38). Gel pieces were subsequently digested in 50 μl trypsin in 50 mm ammonium bicarbonate, 10% acetonitrile (20 ng/μl) at 37 °C for 16 h. Resultant peptides were extracted with two washes of 50% acetonitrile, 5% formic acid, and one with 90% acetonitrile, 5% formic acid. Extracts from each wash were pooled, lyophilized, reconstituted in 0.5% formic acid, and subjected to nanoLC-ESI-MS/MS analysis using an Orbitrap Fusion Tribrid mass spectrometer (Thermo-Fisher Scientific, San Jose, CA) equipped with nanospray Flex Ion Source, and coupled with a Dionex UltiMate 3000RSLC nano system (Thermo, Sunnyvale, CA). Peptide samples were injected onto a PepMap C-18 RP nano trap column (5 μm, 100 μm i.d. × 20 mm, Dionex) with nanoViper fittings at 20 μl/min flow rate for desalting. Samples were then separated on a PepMap C-18 RP nano column (2 μm, 75 μm × 15 cm) at 35 °C, followed by elution on a 90 min gradient of 5% to 35% acetonitrile in 0.1% formic acid at 300 nL/min. Finally, a 5 min ramping to 90% acetonitrile in 0.1% formic acid and a 5 min hold at this eluent completed each run cycle. Between cycles, the column was re-equilibrated for 25 min using 0.1% formic acid. The Orbitrap Fusion was run in positive spray ion mode with spray voltage set at 1.6 kV and a source temperature at 275 °C. External calibration for FT, IT, and quadrupole mass analyzers was performed. In data-dependent acquisition analysis, the instrument was operated using FT mass analyzer in MS scan to select precursor ions followed by 3 s “Top Speed” data-dependent CID ion trap MS/MS scans at 1.6 m,/z, quadrupole isolation for precursor peptides with multiple charged ions above a threshold ion count of 10,000 and normalized collision energy of 30%. MS survey scans at a resolving power of 120,000 (fwhm at m,/z, 200), for the mass range of m,/z, 375–1575. Dynamic exclusion parameters were set at repeat count 1 with a 20 s repeat duration, an exclusion list size of 500, and 40 s of exclusion duration with ±10 ppm exclusion mass width. The activation time was 10 ms for CID analysis. All data were acquired under Xcalibur 3.0 operation software. All post-quantitation sample preparation was conducted at the Cornell Biotechnology Resource Center.
Peptide Identification and Protein Annotation
RAW data were converted to mzML format using msconvert from the Trans-Proteomic Pipeline (TPP v5.0 (Typhoon) rev 0; (39)), using the default peak filtering parameters. The processed data from each MS/MS run was analyzed by X!Tandem (40) and Comet (41) against the Ae. aegypti, L5.0 protein database (GCF_002204515.2; 42). Only the longest protein isoform of each gene was included in the search database, resulting in a database of 14,626 proteins. For X!Tandem, a fragment ion mass tolerance of 0.40 Da and a parent ion tolerance of 10.0 PPM were used. For Comet, a fragment bin tolerance of 1.0005 with a 0.4 offset and a parent ion tolerance of 10.0 PPM were used. Iodoacetamide derivative of cysteine was specified as a fixed modification, whereas oxidation of methionine and deamidation of glutamine and asparagine were specified as variable modifications. Peptides were allowed up to two missed trypsin cleavage sites. All downstream analyses were conducted using the Trans-Proteomic Pipeline (TPP v5.0 (Typhoon) rev 0; 39). False Discovery Rates (FDRs) for each tissue (sperm or ejaculate) were estimated with a randomized decoy database using PeptideProphet (43), employing accurate mass binning model and the nonparametric negative distribution model. X!Tandem and Comet PeptideProphet results were merged using iProphet (44), to provide more robust peptide identification. Peptide identifications were accepted if they could be established at greater than 95.0% iProphet probability, and protein assignations were accepted if they could be established at greater than 99.0% probability. Proteins that contained identical peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy parsimony principles. Proteins were required to be identified by two or more unique peptide spectral matches to be included.
Verification of Labeling Efficiency
For ejaculate samples, protein from virgin female controls was run on the same gel as protein from their mated counterparts, and labeling efficiency was verified on a representative fraction in each cohort's virgin sample (supplemental Fig. S1). No peptides or proteins were identified using our statistical criteria in the labeled virgin female samples when searched using standard, unlabeled mass parameters. Thus, whole female labeling was complete in relation to MS/MS peptide and protein identification and precluded the identification of female proteins from mated female bursae.
Protein Quantitation
Protein quantitation was conducted using the semi-quantitative spectral counting approach implemented by the APEX Quantitative Proteomics Tool (45). The 50 proteins with the highest protein identification probabilities (as determined by iProphet) were utilized as the training dataset. The 35 physicochemical properties available in the APEX tool were used for prediction of peptide detection/nondetection in the construction of a training dataset file. Protein probabilities (Oi,) were computed using the Random Forest classifier algorithm trained with the dataset generated in the previous step. APEX protein abundances per sample were calculated using the protXML file generated by ProteinProphet.
Experimental Design and Statistical Rationale
To ensure the reproducibility of protein identifications, two biological replicates of each tissue were analyzed, with each replicate prepared from independent cohorts of mosquitoes. The reproducibility of protein identification was high for both tissues (see results). We therefore combined both replicates to increase sensitivity and proteome coverage (see above). To control for the possibility that unlabeled female-derived proteins were identified in our ejaculate samples, we assessed labeling efficiency by conducting mass spectrometry on virgin bursae alone, using a representative gel slice from each biological replicate (supplemental Fig. S1). False Discovery Rates (FDRs) were estimated with a randomized decoy database using PeptideProphet (43), employing accurate mass binning model and the nonparametric negative distribution model. For differential mRNA expression, GO, and KEGG pathway analyses, FDR correction was performed by applying the Benjamini-Hochberg method on the calculated p, values (46).
Transcriptome Analysis of Testes and Male Accessory Glands
Testes were harvested from males at 1 dpe and transferred to TRIzol. Because mature sperm are actively produced at this age (47), and spermatogenesis is at its peak (48, 49), testes at this age likely contain most transcripts that contribute to the testicular sperm proteome. Male accessory glands (MAG) including the connecting ejaculatory duct were dissected from virgin males aged 6 and 8 dpe, and care was taken to remove as much of the adjoining seminal vesicle as possible with a minutien pin. We also analyzed MAG from mated males at the same age. Previous work has demonstrated that Ae. aegypti, males become depleted after mating with three to five females in succession, and seminal fluid is slowly regenerated over 48 h (50). In our study, we provided males with four virgin females for a period of 8 h (beginning 2 h after simulated dawn) to allow for male seminal fluid depletion. On average, each male mated with more than three females in this period (as determined by dissection of females' spermathecae). Males were dissected in saline 16 h after their mates had been removed. We generated four biological replicates from independent cohorts for each treatment, and each replicate contained combined tissue from 20–40 (testes) or 40–60 (MAG) males. Total RNA was extracted from each sample in Trizol following manufacturer's instructions (Invitrogen, Carlsbad, CA). Poly-A mRNA was isolated and cDNA libraries were prepared using the QuantSeq 3′ mRNA-Seq Library Prep Kit FWD for Illumina (Lexogen, Vienna, Austria). Amplified cDNA products were run on an AATI Fragment Analyzer (Advanced Analytical Technologies, Inc.; Hialeah, FL) so that the cDNA was of enough concentration for sequencing. Library concentrations were balanced using digital PCR (51), and each of the 12 uniquely barcoded samples were sequenced in one lane using the Illumina HiSeq 2500 platform with 100 bp reads. All sequencing was conducted at the Cornell Biotechnology Resources Center.
For further analysis of the transcriptomes, we included additional, publicly available data to evaluate tissue-biased gene expression. These include gonadectomized male carcass (SRP075464; 52) and a virgin female reproductive tract sample (SRP068996; 53). Raw RNA-seq reads were processed by trimming the first 10 bases from the 3′ position, followed by quality trimming of both ends to a minimum quality Phred score of 20 (Sickle v.1.210; 54). Processed reads were then mapped to the Ae. aegypti, genome (VectorBase release L5.1; 42) using Hisat2 (v.2.1.0; 55) with default parameters, and transcript abundance was estimated for each sample with StringTie (v.1.3.4; 56). Raw counts for each sample were extracted from the StringTie abundance estimates using the auxiliary “prepDE.py” script provided on the StringTie website (https://ccb.jhu.edu/software/stringtie/). Signal peptides in the translated transcriptome were predicted in silico, using a local installation of SignalP (v.4.1; 57).
We used raw counts from the RNA-seq samples to (1) classify genes based on tissue-biased expression in the MAG and testes, and (2) identify genes differentially expressed between virgin and mated MAG (based on transcripts per million; TPM). Count matrices were filtered to remove low abundance transcripts (counts per million < 5). First, we compared expression levels of testes or virgin MAG (in the present study) to levels in three other tissues: gonadectomized male carcass (52), virgin female reproductive tract (53), and virgin MAG (compared with testes) or testes (compared with virgin MAG). We classified genes as testes- or MAG-biased if they had >2-fold higher transcript abundance compared with other samples at a minimum FDR cutoff of 0.05 (edgeR v.3.23.2; 58). We identified differentially expressed genes between virgin and mated MAGs as having >2-fold abundance difference at an FDR cutoff of 0.01. Using the same differential expression criteria, we also re-analyzed data from female reproductive tracts of virgin and just-mated females (53) to identify transcripts putatively transferred to females in the ejaculate using current annotated gene models. Finally, we assessed tissue-biased mRNA expression of putative SFPs and sperm proteins using a Wilcoxon signed rank test with continuity correction to test whether the mean log2 ratio of MAG/testis mRNA expression significantly deviated from zero.
Chromosomal Distribution of Male Reproductive Genes
To evaluate the chromosomal distribution of SFPs, sperm proteins and MAG/testes-biased genes, we calculated the expected number of genes for each class on each chromosome, assuming a random distribution of genes across the genome. We then multiplied the total number of genes within each class by the expected proportion for each chromosome (based on the proportion of total genes on that chromosome) to establish an observed/expected ratio. We also calculated this ratio for a 123 Mb region on chromosome 1 that surrounds the sex determining locus and has low rates of recombination (59). A Chi-square test (df, = 1) for each gene class per chromosome was used to test for biased representation.
Orthology Relationships and Functional Enrichment Analysis
Protein orthology was assessed using a local installation of OrthoDB with default SWIFT and clustering parameters (56). Briefly, best reciprocal hits (BRHs) were first identified using an all-versus,-all approach via the algorithm SWIPE (60), and clusters were built progressively with e-value cutoffs of 1E-3 for triangulating BRHs and 1E-6 for pair-only BRHs. One-to-one, one-to-many, and many-to-many relationships were included in subsequent analyses. Protein sequences for Ae. aegypti, and Ae. albopictus, were retrieved from NCBI (GCF_002204515.2 and GCF_001876365.2, respectively), and Drosophila melanogaster, protein sequences were retrieved from FlyBase (r6.18). Protein sequences for each species were filtered to retain only the longest isoform for each gene. Sperm proteins and SFPs for Ae. albopictus, and D. melanogaster, were based on previous mass spectrometry-based proteomic studies (28, 32, 36). Because SFP identifications by Boes et al., (28) were made using a de novo, transcriptome (the Ae. albopictus, genome had not yet been released), we converted the original SFP identifications to their current accession numbers by BLASTing assembled transcripts from Boes et al., (28) to the assembled Ae. albopictus, genome (61). All Gene Ontology (GO) analyses were conducted using GOseq (62), with gene lengths derived from the longest transcript of each assembled StringTie gene. GO terms were extracted from BLAST results against the SwissProt database (www.uniprot.org). KEGG pathway analysis was performed with clusterProfiler (63) using the Ae. aegypti, KEGG database.
RESULTS
Sperm and Ejaculate Proteome Characterization
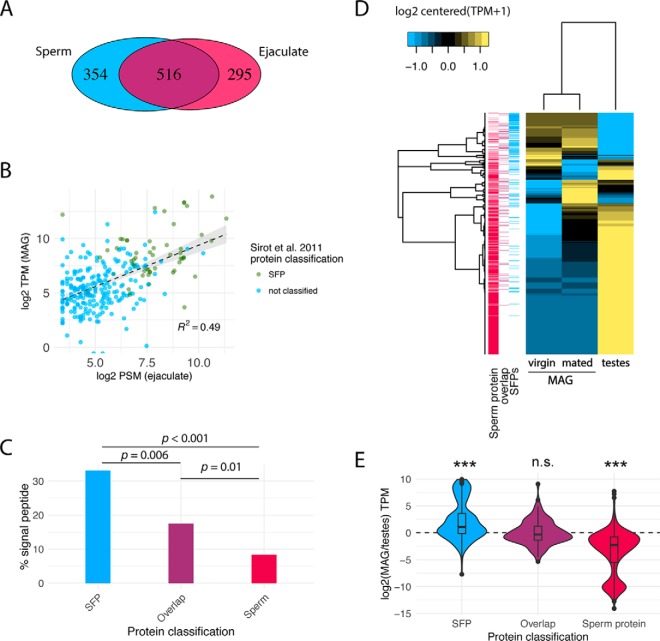
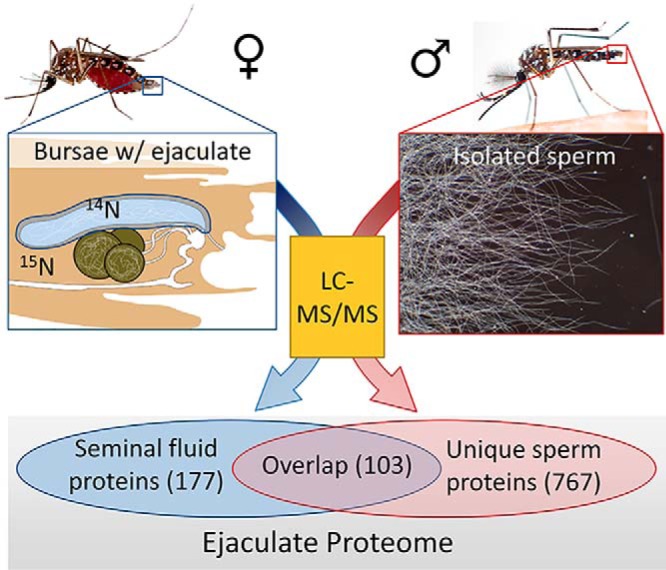
To identify Ae. aegypti, proteins transferred by males to females during mating and to distinguish sperm and nonsperm seminal fluid components, we used MS/MS to analyze proteins from (1) purified sperm isolated from male seminal vesicles and (2) whole ejaculate from bursae of mated females. For our analysis of the whole ejaculate, females were labeled with heavy nitrogen to preclude detection of female-contributed proteins. Labeling efficiency was determined to be complete regarding peptide and protein identification (see Experimental Procedures). Two replicates were analyzed per sample. High levels of reproducibility were observed, including 85% of sperm proteins and 78% of ejaculate proteins identified in both biological replicates. Similar levels of inter-replicate consistency have been reported for sperm proteomes of other organisms (34). To maximize protein identification, we combined spectra across biological replicates for our final proteome determination, resulting in 54,894 peptide spectral matches (PSMs) for sperm, and 30,801 for the ejaculate (supplemental Table S1). The nearly 2-fold disparity in PSMs is because of the contribution of labeled female proteins in the ejaculate sample. In total, 870 and 811 proteins with at least 2 unique PSMs were identified in sperm and ejaculate, respectively (supplemental Table S2). Sperm proteins were identified by an average of 11.4 unique PSMs and 62.8 total PSMs per protein. Ejaculate proteins were identified with an average of 8.0 unique PSMs and 37.8 total PSMs. As was expected given the substantial contribution of sperm cells to ejaculate composition, extensive overlap was observed between sperm and ejaculate proteomes; 516 proteins were detected in both samples, whereas 354 proteins were only identified in sperm and 295 proteins were uniquely detected in the ejaculate (Fig. 1A,).
Fig. 1.
Characteristics of the Ae. aegypti, sperm and ejaculate proteomes. A,, Venn diagram representing the number of proteins identified by ≥2 unique peptides using LC-MS/MS in sperm (red) and ejaculate (blue) proteomes. B,, Scatter plot of the ejaculate protein (log2PSM) and virgin MAG mRNA (log2TPM) abundances of the 177 high-confidence SFPs and 103 sperm/SFP overlap proteins. Proteins classified as SFPs in the Sirot et al., study are highlighted in green and those not identified in Sirot et al., (27) are highlighted in blue. Dotted line is a linear model fit, and gray shading represents the 95% confidence interval. C,, Percentage of proteins containing a predicted signal peptide sequence among SFPs, sperm, and sperm/SFP overlap protein classes; all proportions are significantly different from each other (Chi-square, p, < 0.05). D,, Heatmap representing the mRNA abundance profile of male reproductive classes identified in this study. Cladograms on the left and top represent Pearson cluster grouping of genes and samples, respectively. Annotation bars on the left indicate the protein classification for each gene. E,, Violin plots displaying the fold-change (log2) distribution of mRNA abundance between MAG and testes samples for each male reproductive protein class. Width of violins represents the number of proteins at any given TPM ratio; boxes represent inner quartiles and outliers drawn using the Tukey method. Asterisks indicate groups that are significantly different from each other (Wilcoxon Signed rank test; *** p, < 0.001).
The primary goal of this study was to use MS/MS with higher sensitivity and accuracy to expand upon the prior characterization of Ae. aegypti, SFPs and sperm by Sirot et al., (27). They identified 74 SFPs that mapped to the recently refined Ae. aegypti, genome (42); some of the 93 SFPs described by Sirot et al., (27) do not map to the new genome or are now part of larger, fused gene models. Of these, we identified 60 (81%) in our ejaculate sample, 32 of which were also identified in our purified sperm sample. It is noteworthy that we detected an additional 5 SFPs from Sirot et al., (27), but these were not included as SFPs because they did not meet our two unique peptide inclusion criteria. As such our proteomic characterization expands the previous Ae. aegypti, SFP characterization.
Refined Seminal Fluid Protein Classification
Because the ejaculate is a complex mixture of sperm and seminal fluid, we applied stringent inclusion criteria to our ejaculate samples to refine the delineation between SFPs and sperm proteins. We first removed 10 proteins involved in protein translation (i.e., ribosomal proteins, translation initiation factors, and elongation factors; supplemental Table S2) from the list of putative SFPs. These proteins exhibit ubiquitous patterns of expression, including both MAG and testes, and are unlikely to be bona fide, secreted SFPs. Although we cannot rule out that they are secreted SFPs, their presence in our samples may also be the result of holocrine secretion in the accessory glands, inclusion in apocrine vesicles (64, 65), or transfer of MAG cells to the female, as has been described in D. melanogaster, (66). Next, to reduce the possible inclusion of sperm proteins that were absent in our sperm proteome but present in the ejaculate (perhaps because of low abundance), we defined “high confidence SFPs” as proteins with a minimum of 6 total PSMs in the ejaculate, 2 unique PSMs, and not present in our sperm proteome; this resulted in 177 high-confidence SFPs (supplemental Table S2).
Previous analyses of insect sperm proteomes have consistently identified proteins generally considered to be SFPs (i.e., highly expressed in the MAG and believed to be secreted molecules transferred to females as nonsperm components (32, 67, 68)). To identify proteins predominantly produced by the MAG, but identified in both our sperm and ejaculate sample, we used a 2.5-fold greater protein abundance threshold in the ejaculate relative to sperm. This resulted in the identification of 103 additional putative SFPs, which we label as “sperm/SFP overlap” (supplemental Table S2), including 53 with 5-fold greater protein abundance in the ejaculate relative to sperm. In total, this resulted in a combined SFP proteome of 280 proteins.
To better understand the relationship between SFPs identified in the current study and by Sirot et al., (27), we examined protein abundance and transcript levels of both protein sets (50 of the 74 SFPs identified by those authors are present in our SFP proteome). This revealed that SFPs identified by Sirot et al., (27) were significantly more abundant than the remainder of our 280 SFPs (Fig. 1B; Wilcoxon rank sum test; W, = 3786, p, < 0.001), consistent with the higher sensitivity and coverage of the MS/MS methods utilized in this study. Proteins identified by Sirot et al., (27) also exhibited significantly higher levels of MAG expression relative to SFPs identified in this study (Fig. 1B; Wilcoxon rank sum test; W, = 3005, p, < 0.001). We therefore conclude that the sensitivity of our SFP characterization resulted in the addition of a greater number of low abundance SFPs.
To evaluate our SFP characterization, we explored three types of analyses. First, we determined the presence of predicted signal peptides in identified proteins—a hallmark of secreted proteins. Among our high-confidence SFPs, ∼33% contained signal peptides, compared with only ∼9% of sperm proteins (Chi-square = 69.2, p, < 0.001). Additionally, ∼17% of the sperm/SFP overlap proteins had a predicted signal peptide—also significantly more than the sperm proteome (Chi-square = 6.47, p, = 0.01), and less than high-confidence SFPs (Chi-square = 7.7, p, = 0.006; Fig. 1C,). Second, we used RNA-seq to characterize the transcriptomes of the MAG and testis, the predominant source tissues of proteins in our analysis. As expected, SFPs (Wilcoxon Signed rank test; p, < 0.001) and sperm proteins (Wilcoxon Signed rank test; p, < 0.001) exhibit MAG- and testis-biased expression, respectively, whereas the sperm/SFP overlap proteins exhibit less tissue-biased expression overall (Wilcoxon Signed rank test; p, = 0.36; Fig. 1D–E,). Fewer than 1% of all identified proteins were not present in the transcriptome of their predicted source tissue (9 sperm proteins, 2 high-confidence SFPs, and 1 sperm/SFP overlap protein). Third, we assessed the amount of high-confidence SFP protein abundance variation explained by variation in MAG expression levels. This revealed a significant correlation between protein and transcript abundance (R2, = 0.49, F, = 88.24, p, < 0.001; Fig. 1B,). In conjunction, these results support our proteomic characterization of SFPs produced in the MAG, including proteins present in both sperm and ejaculate samples but highly enriched in the ejaculate proteome.
Functional Enrichment in Sperm and Seminal Fluid Proteomes
Gene ontology (GO) categories enriched in the Ae. aegypti, sperm proteome are largely similar to those found in other insects' sperm (Table I, supplemental Table S3; 32, 34, 67, 68). Proteins associated with mitochondria and the axoneme were the most enriched cellular components in the sperm proteome. Proteasome components were also enriched. Over-represented biological processes in sperm include nucleotide biosynthesis, metabolic processes related to the tricarboxylic acid cycle, and proteins regulating ciliar function. Nucleotide binding, ion transport, and oxidoreductase activity were enriched molecular functions in sperm. KEGG pathways enriched in the sperm proteome were dominated by those involved in metabolism, including carbon metabolism (e.g., pyruvate and butyrate metabolism, the TCA cycle, and oxidative phosphorylation) as well as the metabolism of several classes of amino acids (supplemental Table S4).
Table I. Gene Ontology analysis of sperm proteins, high-confidence SFPs, and sperm/SFP overlap.
List of significant terms is abbreviated to exclude redundancy and to focus on terms discussed in text. For exhaustive list, see Table S3. FDR; false discovery rate.
| Data set | Ontology | GO Term | Proteins (total in category) | FDR |
|---|---|---|---|---|
| Sperm Proteome | Biological Process | Organonitrogen compound metabolic process | 102 (978) | 5.3E-11 |
| Carboxylic acid metabolic process | 100 (553) | 1.9E-23 | ||
| Electron transport chain | 13 (55) | 6.5E-05 | ||
| Cellular Component | Mitochondrion | 150 (834) | 1.2E-45 | |
| Microtubule | 36 (210) | 1.3E-07 | ||
| Cilium | 32 (154) | 1.7E-08 | ||
| Proteasome complex | 31 (47) | 2.1E-25 | ||
| Dynein complex | 19 (40) | 1.7E-11 | ||
| Molecular Function | Nucleoside phosphate binding | 187 (1629) | 5.7E-20 | |
| Hydrolase activity | 163 (2153) | 4.7E-04 | ||
| Oxidoreductase activity | 103 (839) | 2.0E-13 | ||
| Threonine-type endopeptidase activity | 14 (15) | 8.3E-16 | ||
| High-confidence SFP proteome | Biological Process | Organonitrogen compound metabolic process | 30 (978) | 0.009 |
| Carboxylic acid metabolic process | 21 (553) | 0.033 | ||
| Hexose metabolic process | 10 (79) | 7.5E-04 | ||
| Cellular Component | Extracellular region part | 33 (1116) | 0.029 | |
| Molecular Function | Hydrolase activity | 62 (2153) | 0.000 | |
| Nucleoside phosphate binding | 43 (1629) | 0.040 | ||
| Peptidase activity | 27 (739) | 0.006 | ||
| Sperm/SFP overlap | Biological Process | Carboxylic acid metabolic process | 29 (553) | 7.2E-11 |
| Vesicle-mediated transport | 15 (536) | 0.017 | ||
| Protein folding | 10 (113) | 2.6E-05 | ||
| ATP hydrolysis coupled proton transport | 8 (25) | 4.3E-08 | ||
| Oocyte microtubule cytoskeleton polarization | 3 (10) | 0.014 | ||
| Molecular Function | Nucleoside phosphate binding | 39 (1629) | 3.4E-06 |
In our high-confidence SFP proteome, extracellular structure was significantly enriched among cellular component categories, further supporting the accuracy of our SFP identification. Over-represented biological processes include proteolysis and both carbohydrate and amide metabolism. Significantly enriched molecular functions include hydrolase activity and peptidase activity. This observation is consistent with the widespread presence of proteolytic enzymes and regulators in SFPs of other insects (reviewed in 69). The sperm/SFP overlap proteome shared several enriched categories with both the sperm and high-confidence SFP proteomes, including carboxylic acid metabolic processes and nucleotide binding. Several additional GO terms emerged in this protein set as well, including ATP hydrolysis-coupled proton transport, vesicle-mediated transport, and protein folding, the latter of which was previously reported by Sirot et al., ((70); Table I, supplemental Table S3). Enriched KEGG groups in the combined seminal fluid (high-confidence SFP and sperm/SFP overlap) proteomes included those related to phagosomes and lysosomes, as well as pathways related to carbon metabolism and gluconeogenesis.
Orthology With Sperm Proteins and SFPs In Other Species
We next examined orthology of sperm proteins and SFPs in two different species: D. melanogaster,, given its well-characterized sperm proteome and SFPs (32, 36), and Ae. albopictus,, which is the closest species to Ae. aegypti, with characterized SFPs (28). Orthology was determined between the complete genome of all three species, and then orthologs of Ae. aegypti, SFPs and sperm proteins also classified as SFPs or sperm proteins in the other species were identified. We focused solely on SFPs in the comparison with Ae. albopictus,, because a thorough sperm proteome is lacking for this species.
Overall, ∼87 and ∼98% of proteins in the Ae. aegypti, genome have an ortholog (either as one-to-one or one-to-many relationships) in D. melanogaster, and Ae. albopictus,, respectively. Among proteins unique to the Ae. aegypti, sperm proteome, 760 (99%) have an ortholog in D. melanogaster,, and 451 (59%) of these are also found in the D. melanogaster, sperm proteome (supplemental Table S2; 32, 68). Out of the 280 Ae. aegypti, SFPs characterized in our study (including 177 high-confidence and 103 sperm/SFP overlap proteins), 275 (98%) have orthologs in D. melanogaster,. Of these, only 11 have also been identified as D. melanogaster, SFPs (4%; supplemental Table S2). Proteins that contribute to the seminal fluid proteome have therefore diverged extensively during Dipteran evolution. Orthologs were identified in the Ae. albopictus, genome for 275 (98%) of our SFPs. Of the Ae. albopictus, SFPs identified to date (28), 86 (43%) were classified as SFPs in our study (supplemental Table S2).
MAG and Testis Transcriptome Characterization and Differential Expression
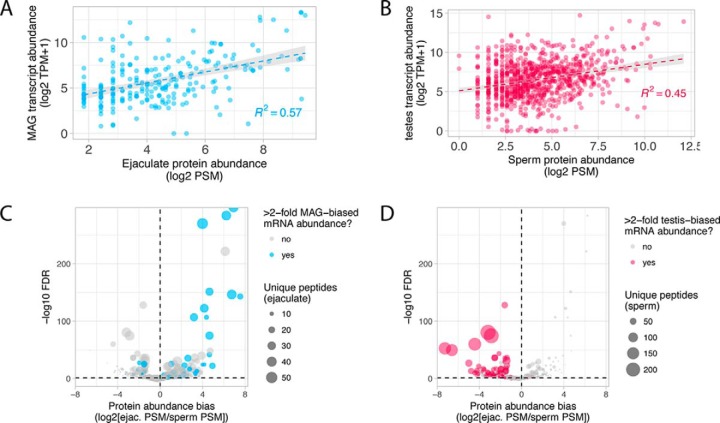
We used short-read RNA sequencing to examine gene expression in testes and MAGs to (1) identify transcripts with tissue-biased expression, (2) compare transcript and protein abundance, (3) assess the transfer of male RNAs to females during mating and (4) characterize the effect of mating on MAG gene expression. In both MAG and testes, we found a significant correlation between transcript abundance and protein abundance for high confidence SFPs and sperm proteins (SFPs: R2 = 0.57, F, = 110.4, p, < 0.001; sperm: R2 = 0.45, F, = 223.8, p, < 0.001; Fig. 2A,–2B,). As such, variation in transcript abundance explains a substantial amount of protein variation in both of our samples. In total, ∼11,000 and ∼7000 genes had detectable mRNA expression in the testes and MAG, respectively. However, only a subset of these exhibit >2-fold expression bias in testes or MAG compared with other tissues (testes: 2863; MAGs: 1485). We examined the association between tissue-biased mRNA expression and differential protein abundance for proteins that were detected in both ejaculate and sperm samples. These results demonstrate that proteins with significant protein abundance differences between sperm and ejaculate samples also tend to show >2-fold tissue-biased mRNA expression (Fig. 2C, and 2D,), further supporting our SFP classification criteria (see above). However, we note that this relationship is less faithful for lower abundance proteins.
Fig. 2.
Protein and mRNA abundance relationships. (A,) and (B,) Scatterplots of normalized mRNA and protein abundance for MAG transcripts and ejaculate-specific proteins (A,) and testis transcripts and sperm proteins (B,). The line and gray shading represent a linear model fit with its 95% confidence interval respectively. Coefficients of determination (R2,) are indicated. (C,) and (D,) Volcano plots of protein abundance differences for all proteins detected in both the ejaculate and sperm samples. Proteins that show >2-fold mRNA expression-bias in MAG or testes tissue are indicated in blue (MAG-biased) or red (testes-biased), and the size of each point corresponds to the number of unique peptides detected for each protein.
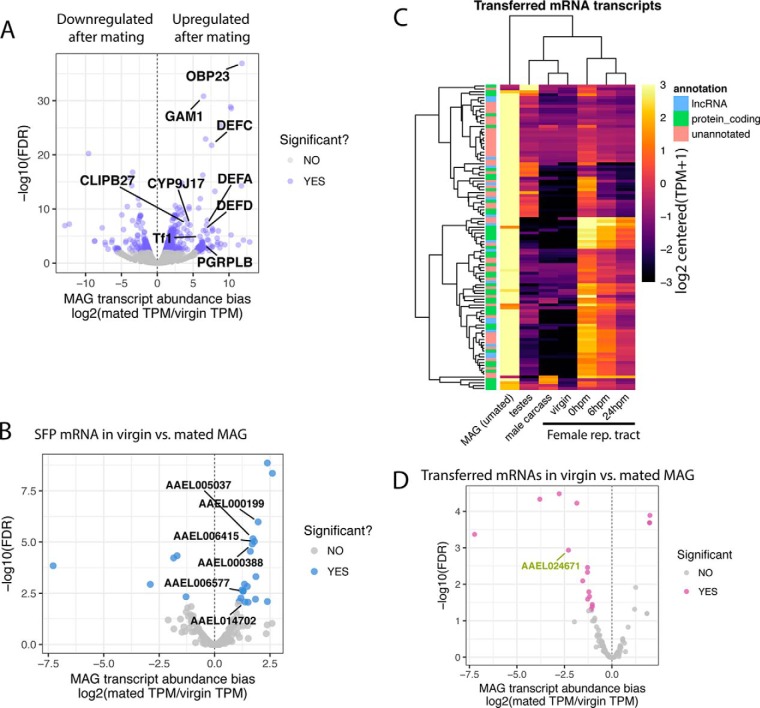
Males regenerate seminal fluid over the course of 48 h after depleting their reserves by repeated insemination (50). Although transcripts that are upregulated or abundant during this period of replenishment should not be assumed to be SFPs, we nonetheless reasoned that MAG transcriptional regulation after mating might inform our understanding of pathways required to produce depleted SFPs. Differential expression analysis of virgin and mated MAGs' transcriptomes revealed a significant bias toward gene upregulation in mated males, with 320 transcripts that are upregulated and 126 that are downregulated after mating (binomial test; p, < 0.001; Fig. 3A,). In contrast to downregulated transcripts—which were not enriched for any functional category—upregulated transcripts were enriched for several GO categories, many of which are consistent with this tissue's primary function of producing secreted proteins (Table II). For example, amino acid metabolism and aminoacyl-tRNA ligase activity were found to be enriched. Enrichment of proteins involved in ubiquitin-dependent protein catabolism and those making up components of the proteasome suggests that there may be extensive protein recycling as seminal fluid is regenerated. Furthermore, the over-representation of proteins associated with endoplasmic reticulum targeting and signal peptide processing suggests that many seminal proteins are targeted for post-translational processing and export from the cell. We also note that a variety of immune-related genes are upregulated in MAGs after mating, including a family of Defensin, antimicrobial genes (Fig. 3A,). Lastly, a similar pattern was observed among our characterized SFPs, of which 19 were upregulated after mating and only five that decreased in abundance (binomial test; p, < 0.001; Fig. 3B,). Interestingly, upregulated SFPs included six different cytoplasmic tRNA ligases and a catalytic subunit of a signal peptidase responsible for post-translation removal of secretion signal peptides, consistent with the importance of protein production and secretion in the mated MAG (see Discussion).
Fig. 3.
Differential expression between virgin and mated MAGs and abundance of transferred mRNAs. A,, Volcano plot of the 446 differentially abundant RNAs between virgin and mated MAGs. Immune-related genes upregulated after mating are highlighted. B,, Volcano plot displaying differential expression of SFP-encoding RNAs between virgin and mated MAGs. tRNA ligase transcripts that are upregulated after mating are highlighted. C,, Heatmap of the 106 transferred RNAs in the MAG and testes samples, female reproductive tract samples (53), and a gonadectomized male carcass sample (52). The annotation classification of each transcript is indicated on the left. D,, Volcano plot of the 106 putatively transferred mRNAs and their differential expression status between virgin and mated MAGs. The only lncRNA that shows reduced abundance after mating is indicated.
Table II. Gene Ontology analysis of upregulated genes in MAGs after mating.
| Ontology | GO Term | Proteins (total in category) | FDR |
|---|---|---|---|
| Biological Process | Protein targeting to ER | 9 (17) | 1.88E-07 |
| Ubiquitin-dependent protein catabolic process | 21 (197) | 5.70E-06 | |
| Response to stress | 56 (1321) | 8.11E-04 | |
| Cellular amino acid metabolic process | 19 (246) | 0.001 | |
| Signal peptide processing | 4 (7) | 0.004 | |
| Molecular Function | Aminoacyl-tRNA ligase activity | 12 (52) | 2.46E-06 |
| Threonine-type endopeptidase activity | 7 (15) | 1.36E-05 | |
| Cellular Component | proteasome complex | 17 (47) | 1.18E-12 |
| Endoplasmic reticulum part | 33 (660) | 0.005 | |
| Small nucleolar ribonucleoprotein complex | 5 (18) | 0.015 | |
| Organelle inner membrane | 18 (288) | 0.031 | |
| Nucleolus | 20 (345) | 0.035 |
Paternal mRNA Transfer During Mating
Previously, Alfonso-Parra et al., (53) demonstrated that males transfer mRNA to females in the ejaculate. Using the newly annotated genome (42), we re-analyzed data from those experiments and identified 106 transcripts, including 41 RNAs that encoded proteins and 17 long noncoding RNAs, that are putatively transferred to females. Identification was based on >2-fold increase in transcript abundance in females immediately after mating, followed by a subsequent decrease in transcript abundance. The remainder of the identified transcripts (48 of 106) are currently unannotated. Using the transcriptomic data in the current study, we determined that the majority of these transcripts have MAG-biased expression, with only two transcripts exhibiting testis-biased expression (Fig. 3C). The MAG therefore appears to be a primary source of RNA transferred to females in the ejaculate. In total, 27 proteins encoded by transferred mRNA transcripts were identified in our proteomes, including 22 in the high-confidence SFP proteome, three in the sperm/SFP overlap proteome, and five in the sperm proteome. Interestingly, the putatively transferred transcripts whose products were present in our seminal fluid proteome encode highly abundant proteins that were on average six times more plentiful than the remainder of the seminal fluid proteins. Lastly, transferred transcripts exhibited a general trend toward downregulation in the MAGs of mated males compared with MAGs of virgins, including 13 transferred transcripts that were significantly downregulated after mating between virgin and mated MAGs (Fig. 3C,); this pattern may be because of their transfer without replenishment in the MAG by the time of dissection.
SFPs are Enriched on Chromosome 1
Sex chromosomes present exclusively in males, such as the mammalian and insect Y chromosome, are highly enriched for genes with male-biased function, including many critical to spermatogenesis and sperm function (32, 71, 72). Although Ae. aegypti, lack heteromorphic sex chromosomes, chromosome 1 harbors a region of robust linkage disequilibrium surrounding the recently characterized male sex determination locus, Nix, (59, 73). To assess the enrichment of male-biased genes on this chromosome we examined the physical distribution of SFPs, sperm proteins, and MAG/testes-biased genes by chromosome. This revealed a ∼1.5-fold enrichment of SFPs on chromosome 1 (62 observed, 39 expected; Chi-square = 12.4, p, < 0.001) and ∼1.2-fold enrichment of sperm proteins on chromosome 2 (377 observed, 316 expected; Chi-square = 11.6, p, < 0.001), whereas MAG- and testis-biased transcripts showed no such enrichment on any chromosome (all Chi-square < 3.84, p, > 0.05; Fig. 4). Despite the strong enrichment of SFPs on chromosome 1, we did not observe an overrepresentation of SFPs in the linked region in the vicinity of the male determining locus, Nix, (15 observed, 16 expected; Chi-square = 0.08, p, = 0.8). Nonuniform physical distribution of sperm proteins across autosomes has also been observed in Drosophila, (68). Although the functional significance of sperm proteins' clustering on chromosome 2 remains to be determined, it is possible that this pattern facilitates co-expression during spermiogenesis, which is characterized by progressive genome silencing during the histone-to-protamine repackaging transition.
Fig. 4.
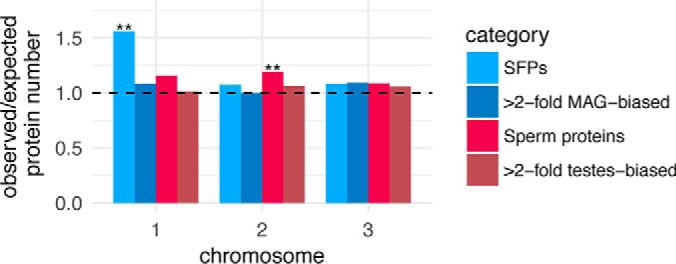
Chromosomal distribution of genes encoding SFPs, sperm proteins, or transcripts with MAG-biased expression or testis-biased expression. The y, axis represents the ratio of observed/expected number of genes for each of the chromosomes in Ae. aegypti,, and the dashed line represents the expectation under no enrichment/depletion. Asterisks indicate significant enrichment (Chi-square; **p, < 0.01).
DISCUSSION
A rapidly expanding body of evidence supports the critical roles of seminal fluid proteins (SFPs) in a wide array of reproductive phenotypes (reviewed in 29). Although this has been most extensively investigated in Drosophila,, seminal peptides and proteins are also associated with post-mating behavioral and physiological responses in mosquitoes such as Ae. aegypti, (15, 16, 18). The primary goals of this study were to comprehensively catalogue male proteins transferred to Ae. aegypti, females during insemination and establish a reliable methodology for delineating between sperm proteins and SFPs. To accomplish this, we (1) conducted an in-depth proteomic characterization of sperm, (2) used a whole-female labeling approach to identify unlabeled male proteins transferred by the male during insemination and (3) characterized the transcriptomes of the testis and male accessory gland (MAG). Importantly, we note that the whole-female labeling approach has been employed previously in Ae. aegypti, but the assignment of proteins as SFPs was limited by the lack of information regarding proteins found in sperm. Thus, distinctions between sperm proteins and SFPs were previously difficult to achieve. In the present study, we have re-assigned 39 proteins identified by Sirot et al.,: 23 previously identified SFPs and 4 sperm proteins as putative components of both sperm and seminal fluid, 10 previously identified SFPs as putative sperm proteins, and 2 previously identified sperm proteins reassigned as high-confidence SFPs. We acknowledge that our MS/MS-based approach still includes some inherent uncertainty, and that this should be kept in mind when interpreting our classifications. It is also noteworthy that advances in MS/MS sensitivity and accuracy have resulted in far greater power of detection in our study, and our analysis has also benefited tremendously from the recent resequencing and reannotation of the Ae. aegypti, genome (42). Our proteomic characterization resulted in a nearly 4-fold expansion of the current Ae. aegypti, seminal fluid proteome, and an 8-fold expansion of identified proteins in sperm. Together, this Ae. aegypti, “ejaculatome” provides a foundation for future molecular studies of mosquito reproduction and associated applications to control mosquito populations (see below).
Proteome Characteristics Independently Validate Identification
Our work differs from previous SFP characterization studies (27, 28, 36) in that our classification was supported by a detailed knowledge of sperm proteome composition. Nonetheless, several independent validation approaches were helpful in assessing the quality of our proteomic characterization. For example, we quantified the proportion of proteins with predicted secretion signals and analyzed transcriptome profiles in testes and MAGs. As would be predicted, SFPs identified in this study possessed a significantly higher proportion of predicted secretion signals than sperm proteins. Although only 33% possessed predicted secretion signals in our high-confidence SFPs, this proportion is consistent with what has been reported in seminal fluid of the grasshopper Melanoplus sanguinipes, (74). The large proportion of proteins that lacked this signal may be because much seminal fluid secretion in Ae. aegypti, has been reported to occur through both apocrine and holocrine mechanisms (64, 65). The genes encoding high-confidence SFPs were also, on average, highly specific or biased toward expression in the MAG. RNAs encoding over 99% of the identified proteins were also represented in their target tissues' transcriptomes, adding further validation to their identification as seminal fluid and sperm proteins. Those 11 proteins with no expression in their target tissue may represent proteins that were produced outside of the testes or accessory glands (e.g., in adjoining tissues such as the vas deferentia or trafficked into these organs) or transcripts that were not expressed at the time of tissue dissection.
Analysis of the functional composition of our proteomes revealed that they were closely aligned with the results of previous sperm (32–34, 68) and SFP studies in insects (30, 36). For example, our expanded sperm proteome was highly enriched for proteins related to flagellar structure, including microtubules, dynein complexes, and ciliar components, and proteins likely associated with the mitochondrial derivatives, which are a predominant structure in mosquito sperm (75, 76) and that of other insects. Consistent with what was described by Sirot et al., (27), as well as in other insects (reviewed in 29, 30, 36) and humans (77), proteases were highly enriched among our high-confidence SFPs, supporting the likely accuracy of our expanded characterization (reviewed in 69). The observed enrichment of vesicle-mediated transport proteins is also consistent with the fact that Ae. aegypti, seminal fluid is in part produced by apocrine secretion (64). Additionally, exosomes and other vesicles are believed to play a role in a variety of post-insemination cellular interactions. For example, vesicles transferred in Drosophila, seminal fluid have been reported to fuse with sperm and interact with the female reproductive tract (78), exosomes of the mouse epididymis have recently been implicated in the control of sperm RNA stores (79), and the abundance of exosome markers in avian SFPs has led to speculation about vesicle-mediated mechanisms in post-testicular sperm maturation (80). Therefore, the accuracy of our expanded proteomic characterization of sperm and SFP proteomes is corroborated by several independent lines of evidence.
It is important to note that, despite the application of stringent proteomic thresholds, some proteins could not be definitively assigned as either sperm protein or SFP. Previous studies in Drosophila, and Lepidoptera have consistently identified known SFPs (such as Drosophila, Acp36DE) at appreciable abundance levels in sperm that have yet to be combined with MAG secretions (32, 67, 68). Our identification of a relatively large protein set that is highly MAG-biased in expression but also present in sperm further suggests that the incorporation of “SFPs” during testicular sperm maturation occurs and is worthy of additional functional investigation. Although Drosophila, expression profiles in the testis and accessory gland are quite distinct, many SFPs exhibit low levels of co-expression in the testis (Dorus, unpublished data). Our transcriptomic analyses here further support such patterns of co-expression. As such, dichotomous distinctions between sperm proteins and SFPs may be an oversimplification of a more nuanced relationship between these reproductive systems. We acknowledge this ambiguity in our classification of MAG-biased proteins that were also identified in our sperm proteome. We also note that our sperm purification method could have allowed the inclusion of seminal fluid proteins (that is, nonsperm ejaculate proteins) that are produced in the seminal vesicle, vas deferentia, or testes. Similarly, the possibility exists that some male accessory gland proteins may migrate into the seminal vesicle. Although the contribution of such proteins to the sperm proteome, if any, is likely small, we cannot rule out this uncertainty.
We also note that despite our expanded proteomic coverage, several proteins that we anticipated to be identified were absent. The most notable case was Head Peptide-1, a seminal fluid peptide which has been shown to be transferred in the ejaculate (81) and has been reported to induce short term monogamy in the female after mating (15). Head Peptide-1, like many SFPs, undergoes extensive post-translational modification and may therefore be challenging to identify bioinformatically without a priori, knowledge of the biochemical composition of the proteolytic products (such as in the case of the well-studied Drosophila, Sex Peptide; 82). Another example was adipokinetic hormone (AAEL011996), which did not meet our two unique peptide inclusion threshold, although we did identify five copies of one peptide from its precursor protein that was also identified in Ae. albopictus, seminal fluid (28). This protein has been postulated to contribute to sperm protection from oxidative stress (83) and the regulation of feeding behavior (84) in other insects. We suggest that the complexity of proteolytic pathways, governed both by male and female interacting proteins, is a major barrier in the use of shotgun proteomics to study SFP identity and function in the female reproductive tract. Conducting similar analyses with an alternative digestive enzyme or de novo, peptide sequencing may allow the detection of additional proteins whose tryptic products could not be identified using standard database searches. Searching for proteins in the supernatant of our sperm samples may also identify soluble proteins that associate with the surface of sperm, as has been shown with sex peptide in D. melanogaster, (85). Finally, future investigations would also likely benefit from the inclusion of a targeted proteomic approach (reviewed in 86). Such approaches require an a priori, list of candidate peptides; in Ae. aegypti,, the neuropeptides and protein hormones catalogued by Predel et al., (87) represent a useful pool of potentially important molecules.
Evolution of Male Reproductive Proteomes
Male reproductive proteins, including SFPs, are consistently among the fastest evolving classes of protein (reviewed in 88). Although initially a goal of our study, conducting a robust analysis of the molecular evolution of proteins identified in this study was limited by the availability of genomic resources appropriate for both inter- and intraspecific tests of positive selection. Obtaining high quality genomic data for different populations of Ae. aegypti, has proven difficult, given the genome's repetitive nature (42, 89). Further, this mosquito's ability to move globally as diapausing eggs has allowed for frequent mixing and a complex population structure (90, 91). The development of appropriate population level genetic data for the analysis of recent selective sweeps should be a priority in Ae. aegypti,, as it has been in Anopheles gambiae, (92, 93). Furthermore, given the extent of molecular divergence between Ae. aegypti, and Ae. albopictus, (28), the development of genomic resources for a more closely related outgroup to Ae. aegypti, will assist in understanding evolutionary patterns at the gene level. Despite these limitations, our analysis of orthology did reveal that the suite of proteins contributing to seminal fluid, but not sperm, has diverged substantially from other Dipterans. Although sperm proteins and SFPs possess levels of orthology to the Drosophila, genome that are comparable to the genome as a whole, only 59 and 4% of orthology was observed when comparing the Ae. aegypti, sperm and SFP proteomes (respectively) with those of Drosophila, (32, 36). Although some of this disparity may be attributed to differences in overall proteome size and coverage, such a stark contrast is nonetheless compelling evidence of tissue-specific evolutionary patterns. Orthology between Ae. aegypti, SFPs and Ae. albopictus, SFPs (28), whereas more extensive (43%), was still lower than orthology between the sperm proteomes of Ae. aegypti, and D. melanogaster,—two distantly related Dipterans. These general patterns of orthology among Ae. aegypti, Ae. albopictus,, and D. melanogaster, are consistent with those described by Boes et al., (28), although the absolute level of orthology between studies varies considerably because of methodological differences. The patterns we observe suggest a process of “turn-over” in seminal fluid proteomes, whereby overall protein composition diverges rapidly even when there is evidence for conservation with regard to overarching molecular functions represented in seminal fluid. For example, a priori, expectations about Gene Ontology enrichment were met for both Ae. aegypti, sperm (e.g., cilium and mitochondrial proteins) and SFPs (extracellular localization and hydrolase activity), despite overall SFP divergence. SFPs are a pronounced target of selection and have been discussed as a driver of sexual conflict (reviewed in 94), and thus they are expected to rapidly diverge. By contrast, we note that strong conservation of sperm proteins exists across distant taxa, with different insect orders displaying 25% orthology between sperm proteomes (34), and even D. melanogaster, and mammals with 20% sperm proteome orthology (32). The overall lack of conservation in seminal fluid proteomes makes comparing the roles of specific SFPs across species difficult, but conserved molecular functions among SFPs will nevertheless allow the wealth of knowledge in Drosophila, to be leveraged toward an understanding of SFP function in nonmodel insects.
Unlike other mosquitoes with heteromorphic sex chromosomes, Culicine mosquitoes (e.g. Aedes and Culex,) harbor male-determining loci on undifferentiated, homomorphic sex chromosomes (95). Theory predicts the evolution of heteromorphic sex chromosomes following the acquisition of a sex determining locus, suppression of recombination, and expansion of the nonrecombining region. It remains unclear why homomorphic sex chromosomes appear to be retained in some taxa (96, 97). One proposed mechanism to mediate the selective effect of sexually antagonistic alleles on the promotion of recombination suppression is the establishment of efficient sex-biased expression (98). Although previously lacking, the significant enrichment of SFPs on chromosome 1 is the first evidence in support of this hypothesis in Ae. aegypti., This trend was restricted to SFPs and was not observed for genes solely over-expressed in the MAG or testis. It is intriguing to speculate that this distinction between SFPs and other male reproductive genes might be because of the prevalence (and selective strength) of sexually antagonistic alleles specifically among SFPs, which may favor their localization on chromosome 1. This is consistent with their putative role as drivers of sexual conflict (reviewed in 94), including the mediation of female post-mating responses such as sexual receptivity and longevity (14, 18).
Functional Relevance of Abundant Sperm Proteins and SFPs
Our sperm and SFP proteomes exhibited skewed abundance distribution with the top ten most abundant proteins comprising 25 and 17% of protein composition in sperm and SFPs, respectively. Interestingly, the most abundant sperm protein, cytosol aminopeptidase (AAEL006975), accounted for more than 7.4% of all protein and two other cytosol aminopeptidases were in the top ten most abundant proteins (AAEL000108, AAEL023987). These three proteins are orthologs of the eight sperm-leucyl aminopeptidases (S-LAPs) in Drosophila, with similar expression patterns, including ∼1000-fold higher expression in testes than in MAG, ∼50 times more transcript in whole male carcasses than gonadectomized carcasses (99), and upregulation during later stages of spermatogenesis (52). S-LAP orthologs constitute a significant proportion of the protein composition of Drosophila, (100) and Lepidoptera sperm (34, 67). Little is known about the specific function of S-LAPs, although it has been postulated that they may serve a structural function given the inferred loss of enzymatic capacity of several S-LAPs during Drosophila, evolution (100). Additionally, a Y-linked S-LAP in D. pseudoobscura, has been implicated in a cryptic meiotic drive system, where suppression of this locus results in aberrant spermatogenesis and a higher proportion of X-bearing sperm (101). It will be of great interest to establish the specific function of these proteins in Ae. aegypti, sperm, given their high abundance and expression patterns during spermiogenesis. Furthermore, the proteins and transcripts involved in spermatogenesis described in this study may assist in the identification of other genes involved in meiotic drive systems (reviewed in 102), which have been proposed as potential genetic means to reduce wild populations through the induction of sex ratio biases (103).
Although no SFP was as abundant as cytosol aminopeptidase in sperm, the top ten most abundant SFPs ranged from 1.2–2.6% of the protein in our ejaculate sample. l-asparaginase (AAEL002796) was the most abundant SFP (61% more abundant than the next protein) and the tenth most abundant mRNA transcript in the MAG out of over 11,000 transcripts. Although the relevance of the abundance of this enzyme is currently unclear, it may relate to several other notable observations. First, transcript AAEL020035, whose protein product is comprised of ∼60% asparagine residues, is the single most abundant MAG transcript and was also, by far, the most abundant putatively transferred transcript. (We did not identify AAEL020035 in our SFP proteome but note that it results in few identifiable peptides because of its extreme amino acid composition). Second, asparagine tRNA-ligase (AAEL006577) was two times more abundant in seminal fluid than any other tRNA-ligase. Third, asparagine tRNA-ligase was upregulated in the MAG after mating and was the most abundant tRNA-ligase transcript. Together, these suggest that MAGs are well-equipped to produce ample protein with a strong asparagine amino acid bias. Finally, two other enzymes, aspartate transaminase (AAEL002399) and citrate synthase (AAEL004297), are abundantly present in seminal fluid and could convert aspartate produced by asparaginase to oxaloacetate and citric acid, respectively. Although it is premature to draw any firm conclusions based on these observations alone, it is intriguing to speculate that the SFP proteome has the capacity to conduct gluconeogenesis (of asparagine and potentially other amino acids) and that this may feed into to the citric acid cycle. This hypothesis is supported by the results of our KEGG analysis, in which carbon metabolism, gluconeogenesis, and alanine, aspartate, and glutamate metabolism were enriched in the seminal fluid proteome. The citric acid cycle is believed to be functional in mammalian sperm (reviewed in 104), and our KEGG analysis reveals an enrichment of citric acid cycle enzymes in sperm but not seminal fluid. Whether metabolite precursors to the citric acid cycle are transported from seminal fluid to sperm remains to be determined.
The most abundant seminal fluid proteins also exhibited a strong enrichment for function in protein cleavage. Proteins with protease, dipeptidase, and aminopeptidase activity represent 12 of the top 28 most abundant proteins present in the seminal fluid proteome. Proteolytic functions have been described previously in the seminal fluid of Ae. aegypti, (27), Ae. albopictus, (28), Cx. quinquefasciatus, (105), and several nonmosquito taxa (21, 22, reviewed in 69, 106), and are a common function of many insects' seminal fluid. Based on studies in other insects, functions of these enzymes may include the activation of sperm motility or the cleavage of propeptides into their active forms (107). Our seminal fluid proteome also contains abundant enzymes that catabolize smaller substrates, such as amino acids and carbohydrates. Taken together, the enzymatic mixture in seminal fluid may be well equipped to break down many of the molecules they contain. Seminal fluid proteins were also enriched for proteins involved in maintaining proton and redox homeostasis. We identified several proteins contributing to V-type proton ATPases, which use ATP to regulate pH via proton transport. Maintaining an optimal pH in seminal fluid may allow for efficient sperm motility (reviewed in 108). Regulating pH may also create an ideal environment in which enzymatic reactions occur, either in organelles such as phagosomes and lysosomes (whose constituents were enriched in our KEGG analyses), in sperm, or in the extracellular environment. Seminal fluid also contained several proteins that function to neutralize free radicals, such as catalase (AAEL013407), peroxidase (AAEL013171), and several dehydrogenases. Regulating the physiochemical environment in seminal fluid is likely critical for the function and protection of sperm before their storage in the female's long term storage organs (spermathecae).
Ejaculate RNAs Transferred to Females
There has been much conjecture about the importance of spermatozoal RNA to fertility (109), and recent work has confirmed that the regulation of sperm ncRNA stores in the mammalian epididymis is necessary for proper embryogenesis (110). Little is known about the function of spermatozoal RNAs in insects, although they have been demonstrated to have substantial functional coherence, including an overwhelming enrichment of loci involved in translation (111). New data in this study allowed us to probe for patterns in previously described transcripts that are putatively transferred to females during mating. A total of 106 transcripts were identified, including both coding and noncoding transcripts, and most of these exhibit high levels of expression in the MAG. Based on our SFP proteome, most of the protein coding transcripts are also translated at high levels. Their high expression in the MAG suggests that they may simply hitchhike into seminal fluid with other secreted molecules. Alternatively, as has been demonstrated in Drosophila,, they could be transferred in intact MAG cells (66), or via vesicles derived from the MAG (78). Interestingly these vesicles, which may carry RNA cargo including miRNAs, fuse with sperm and have the capacity to interact with the female reproductive tract. Some male-derived transcripts are detectable in the female for up to 24 h post-mating (50), and it has been postulated that they could be used by females in some capacity (112). In Ae. aegypti,, both vesicles and RNAs are transferred in the ejaculate to the female, but their fate and function have not been investigated. Whether they impact the female or her future offspring is an intriguing, and potentially important, line of future investigation.
Mosquito Control and Future Directions
Understanding the molecular architecture of Ae. aegypti, reproduction holds great potential for vector control strategies. Mosquito reproduction is an ideal control target to reduce vector populations and the burden of disease transmission. The most direct application of this study will be the identification of modulators of female reproductive behavior. Mosquito SFPs induce behavioral responses that prevent female remating (14, 16, 17), including short term mating refractory behavior (15). To date, the molecule(s) responsible for long term refractoriness has yet to be identified. Given the strength and duration of responses to low SFP “doses” (14), identification of the responsible proteins will provide powerful tools for manipulating female reproduction in a species-specific manner. In addition, such knowledge may provide a molecular metric by which the quality of males in modified mosquito release strategies (such as those employing sterile or Wolbachia,-infected males; reviewed in 113) may be monitored and optimized. Functional analysis of specific sperm proteins and SFPs may yield insights into processes such as sperm motility and activation (21–23), sperm storage (114), and sperm-egg recognition (115). Very few studies have explored these processes in Ae. aegypti, (reviewed in 116). A mechanistic understanding of complex post-copulatory male-by-female interactions is critical to genetically modified mosquito release strategies that manipulate reproduction. Our detailed characterization of the male contributions to these interactions should serve as the foundation for the design and improvement of vector control strategies that limit the transmission of arboviruses that cause serious human illness and mortality.
Data Availability
Mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD010293 and 10.6019/PXD010293. The R analysis pipeline used for transcriptomic analysis is available as part of this project's GitHub repository (https://github.com/YazBraimah/Aegypti.ejaculatomics). Raw RNA-seq reads from this study can be obtained from the Sequence Read Archive (SRA) Accession # SRP158536. RNA-seq data from Sutton et al,. (52) and Alfonso-Parra et al., (53) can be accessed at SRA Accession # SRP075464 and SRA Accession # SRP068996, respectively.
Supplementary Material
Acknowledgments
We thank Sylvie Pitcher, Sheng Zhang, Jen Grenier, Peter Schweitzer, and the staff at the Cornell Biotechnology Resource Center for technical support, and Laura Sirot for experimental guidance and feedback. This content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
* This study was supported by NIH/NIAID grant R01AI095491 to MFW and LCH, NIH/NICHD grant R21HD088910 to S.D. and MFW, a Cornell Graduate School fellowship to ECD, and a Cornell Entomology Department Griswold grant to ECD and LCH. YHAB was supported by NIH/NICHD grant R01HD059060 to MFW and Andrew G. Clark. RNA-seq data and mass spectrometry data were made possible by NIH grants 1S10OD010693-01 and 1S10OD017992-01, respectively. Proteomics computing was supported by NSF grant OAC-1541396/ACI-1541396 to Eric Sedore of the Syracuse University Information Technology Services. Mosquito images in Graphical Abstract used with permission, © 2016 David Felix Duneau.
 This article contains supplemental material.
This article contains supplemental material.
1 The abbreviations used are:
- SFP
- seminal fluid protein
- MS/MS
- tandem mass spectrometry
- APEX
- absolute protein expression
- FDR
- false discovery rate
- PSM
- peptide spectral match
- MAG
- male accessory gland
- dpe
- days post eclosion
- TPM
- transcripts per million
- GO
- gene ontology
- S-LAP
- Sperm leucyl-aminopeptidase.
REFERENCES
- 1. Bhatt S., Gething P. W., Brady O. J., Messina J. P., Farlow A. W., Moyes C. L., Drake J. M., Brownstein J. S., Hoen A. G., Sankoh O., Myers M. F., George D. B., Jaenisch T., Wint G. R. W., Simmons C. P., Scott T. W., Farrar J. J., and Hay S. I. (2013) The global distribution and burden of dengue. Nature 496, 504–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chouin-Carneiro T., Vega-Rua A., Vazeille M., Yebakima A., Girod R., Goindin D., Dupont-Rouzeyrol M., Lourenco-de-Oliveira R., and Failloux A. B. (2016) Differential susceptibilities of Aedes aegypti, and Aedes albopictus, from the Americas to Zika virus. PLoS Negl. Trop. Dis. 10, e0004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pialoux G., Gauzere B. A., Jaureguiberry S., and Strobel M. (2007) Chikungunya, an epidemic arbovirosis. Lancet Infect. Dis. 7, 319–327 [DOI] [PubMed] [Google Scholar]
- 4. Monath T. P., and Vasconcelos P. F. (2015) Yellow fever. J. Clin. Virol. 64, 160–173 [DOI] [PubMed] [Google Scholar]
- 5. Fredericks A. C., and Fernandez-Sesma A. (2014) The burden of dengue and chikungunya worldwide: implications for the southern United States and California. Ann. Glob. Health 80, 466–475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Klassen W., and Curtis C. F. (2005) History of the Sterile Insect Technique. In: Dyck V. A., Hendrichs J., and Robinson A. S., eds. Sterile Insect Technique: Principles and Practice in Area-Wide Integrated Pest Management, pp. 3–36, Springer Netherlands, Dordrecht [Google Scholar]
- 7. Carvalho D. O., McKemey A. R., Garziera L., Lacroix R., Donnelly C. A., Alphey L., Malavasi A., and Capurro M. L. (2015) Suppression of a field population of Aedes aegypti, in Brazil by sustained release of transgenic male mosquitoes. PLoS Negl. Trop. Dis. 9, e0003864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Macias V. M., Ohm J. R., and Rasgon J. L. (2017) Gene drive for mosquito control: where did it come from and where are we headed? Int. J. Env. Res. Public Health 14, pii: E1006. doi: 10.3390/ijerph14091006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hoffmann A. A., Montgomery B. L., Popovici J., Iturbe-Ormaetxe I., Johnson P. H., Muzzi F., Greenfield M., Durkan M., Leong Y. S., Dong Y., Cook H., Axford J., Callahan A. G., Kenny N., Omodei C., McGraw E. A., Ryan P. A., Ritchie S. A., Turelli M., and O'Neill S. L. (2011) Successful establishment of Wolbachia, in Aedes, populations to suppress dengue transmission. Nature 476, 454–457 [DOI] [PubMed] [Google Scholar]
- 10. LePage D., and Bordenstein S. R. (2013) Wolbachia,: Can we save lives with a great pandemic? Trends Parasitol. 29, 385–393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Walker T., Johnson P. H., Moreira L. A., Iturbe-Ormaetxe I., Frentiu F. D., McMeniman C. J., Leong Y. S., Dong Y., Axford J., Kriesner P., Lloyd A. L., Ritchie S. A., O'Neill S. L., and Hoffmann A. A. (2011) The wMel Wolbachia, strain blocks dengue and invades caged Aedes aegypti, populations. Nature 476, 450–453 [DOI] [PubMed] [Google Scholar]
- 12. Dutra H. L., Rocha M. N., Dias F. B., Mansur S. B., Caragata E. P., and Moreira L. A. (2016) Wolbachia, blocks currently circulating Zika virus isolates in Brazilian Aedes aegypti, mosquitoes. Cell Host Microbe 19, 771–774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gwadz R. W., Craig G. B. Jr, and Hickey W. A. (1971) Female sexual behavior as the mechanism rendering Aedes aegypti, refractory to insemination. Biol. Bull. 140, 201–214 [DOI] [PubMed] [Google Scholar]
- 14. Helinski M. E., Deewatthanawong P., Sirot L. K., Wolfner M. F., and Harrington L. C. (2012) Duration and dose-dependency of female sexual receptivity responses to seminal fluid proteins in Aedes albopictus, and Ae. aegypti, mosquitoes. J. Insect Physiol. 58, 1307–1313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Duvall L. B., Basrur N. S., Molina H., McMeniman C. J., and Vosshall L. B. (2017) A peptide signaling system that rapidly enforces paternity in the Aedes aegypti, mosquito. Curr. Biol. 27, 3734–3742 e3735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fuchs M. S., Craig G. B., and Despommier D. D. (1969) The protein nature of the substance inducing female monogamy in Aedes aegypti,. J. Insect Physiol. 15, 701–709 [Google Scholar]
- 17. Fuchs M. S., Craig G. B. Jr, and Hiss E. A. (1968) The biochemical basis of female monogamy in mosquitoes. I. Extraction of the active principle from Aedes aegypti,. Life Sci. 7, 835–839 [DOI] [PubMed] [Google Scholar]
- 18. Villarreal S. M., Pitcher S., Helinski M. E. H., Johnson L., Wolfner M. F., and Harrington L. C. (2018) Male contributions during mating increase female survival in the disease vector mosquito Aedes aegypti,. J. Insect Physiol. 108, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Klowden M. J., and Chambers G. M. (1991) Male accessory gland substances activate egg development in nutritionally stressed Aedes aegypti, mosquitoes. J. Insect Physiol. 37, 721–726 [Google Scholar]
- 20. Adlakha V., and Pillai M. K. (1975) Involvement of male accessory gland substance in the fertility of mosquitoes. J. Insect Physiol. 21, 1453–1455 [DOI] [PubMed] [Google Scholar]
- 21. Nagaoka S., Kato K., Takata Y., and Kamei K. (2012) Identification of the sperm-activating factor initiatorin, a prostatic endopeptidase of the silkworm, Bombyx mori,. Insect Biochem. Mol. Biol. 42, 571–582 [DOI] [PubMed] [Google Scholar]
- 22. Miyata H., Thaler C. D., Haimo L. T., and Cardullo R. A. (2012) Protease activation and the signal transduction pathway regulating motility in sperm from the water strider Aquarius remigis,. Cytoskeleton 69, 207–220 [DOI] [PubMed] [Google Scholar]
- 23. Thaler C. D., Miyata H., Haimo L. T., and Cardullo R. A. (2013) Waveform generation is controlled by phosphorylation and swimming direction is controlled by Ca2+ in sperm from the mosquito Culex quinquefasciatus,. Biol. Reprod. 89, 135. [DOI] [PubMed] [Google Scholar]
- 24. Pitts R. J., Liu C., Zhou X., Malpartida J. C., and Zwiebel L. J. (2014) Odorant receptor-mediated sperm activation in disease vector mosquitoes. Proc. Natl. Acad. Sci. U.S.A. 111, 2566–2571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Beckmann J. F., and Fallon A. M. (2013) Detection of the Wolbachia protein WPIP0282 in mosquito spermathecae: implications for cytoplasmic incompatibility. Insect Biochem. Mol. Biol. 43, 867–878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Beckmann J. F., Ronau J. A., and Hochstrasser M. (2017) A Wolbachia, deubiquitylating enzyme induces cytoplasmic incompatibility. Nat. Microbiol. 2, 17007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Sirot L. K., Hardstone M. C., Helinski M. E. H., Ribeiro J. M. C., Kimura M., Deewatthanawong P., Wolfner M. F., and Harrington L. C. (2011) Towards a semen proteome of the dengue vector mosquito: protein identification and potential functions. PLoS Negl. Trop. Dis. 5, e989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Boes K. E., Ribeiro J. M. C., Wong A., Harrington L. C., Wolfner M. F., and Sirot L. K. (2014) Identification and characterization of seminal fluid proteins in the Asian tiger mosquito, Aedes albopictus,. PLoS Negl. Trop. Dis. 8, e2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Avila F. W., Sirot L. K., LaFlamme B. A., Rubinstein C. D., and Wolfner M. F. (2011) Insect seminal fluid proteins: identification and function. Annu. Rev. Entomol. 56, 21–40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Baer B., Zareie R., Paynter E., Poland V., and Millar A. H. (2012) Seminal fluid proteins differ in abundance between genetic lineages of honeybees. J. Proteomics 75, 5646–5653 [DOI] [PubMed] [Google Scholar]
- 31. Sepil I., Hopkins B. R., Dean R., Thezenas M. L., Charles P. D., Konietzny R., Fischer R., Kessler B. M., and Wigby S. (2018) Quantitative proteomics identification of seminal fluid proteins in male Drosophila melanogaster,. Molecular & Cellular Proteomics, 10.1074/mcp.RA118.000831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wasbrough E. R., Dorus S., Hester S., Howard-Murkin J., Lilley K., Wilkin E., Polpitiya A., Petritis K., and Karr T. L. (2010) The Drosophila melanogaster, sperm proteome-II (DmSP-II). J. Proteomics 73, 2171–2185 [DOI] [PubMed] [Google Scholar]
- 33. Zareie R., Eubel H., Millar A. H., and Baer B. (2013) Long-term survival of high quality sperm: insights into the sperm proteome of the honeybee Apis mellifera,. J. Proteome Res. 12, 5180–5188 [DOI] [PubMed] [Google Scholar]
- 34. Whittington E., Zhao Q., Borziak K., Walters J. R., and Dorus S. (2015) Characterisation of the Manduca sexta, sperm proteome: Genetic novelty underlying sperm composition in Lepidoptera. Insect Biochem. Mol. Biol. 62, 183–193 [DOI] [PubMed] [Google Scholar]
- 35. Degner E. C., and Harrington L. C. (2016) Polyandry depends on postmating time interval in the dengue vector Aedes aegypti,. Am. J. Trop. Med. Hyg. 94, 780–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Findlay G. D., Yi X., Maccoss M. J., and Swanson W. J. (2008) Proteomics reveals novel Drosophila, seminal fluid proteins transferred at mating. PLos Biol. 6, e178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Yang Y., Thannhauser T. W., Li L., and Zhang S. (2007) Development of an integrated approach for evaluation of 2-D gel image analysis: Impact of multiple proteins in single spots on comparative proteomics in conventional 2-D gel/MALDI workflow. Electrophoresis 28, 2080–2094 [DOI] [PubMed] [Google Scholar]
- 38. Shevchenko A., Wilm M., Vorm O., and Mann M. (1996) Mass spectrometric sequencing of proteins from silver stained polyacrylamide gels. Anal. Chem. 68, 850–858 [DOI] [PubMed] [Google Scholar]
- 39. Deutsch E. W., Mendoza L., Shteynberg D., Slagel J., Sun Z., and Moritz R. L. (2015) Trans-Proteomic Pipeline, a standardized data processing pipeline for large-scale reproducible proteomics informatics. Proteomics Clin. Appl. 9, 745–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Craig R., and Beavis R. C. (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20, 1466–1467 [DOI] [PubMed] [Google Scholar]
- 41. Eng J. K., Jahan T. A., and Hoopmann M. R. (2013) Comet: an open-source MS/MS sequence database search tool. Proteomics 13, 22–24 [DOI] [PubMed] [Google Scholar]
- 42. Matthews B. J., Dudchenko O., Kingan S., Koren S., Antoshechkin I., Crawford J. E., Glassford W. J., Herre M., Redmond S. N., Rose N. H., Weedall G. D., Wu Y., Batra S. S., Brito-Sierra C. A., Buckingham S. D., Campbell C. L., Chan S., Cox E., Evans B. R., Fansiri T., Filipovic I., Fontaine A., Gloria-Soria A., Hall R., Joardar V. S., Jones A. K., Kay R. G. G., Kodali V., Lee J., Lycett G. J., Mitchell S. N., Muehling J., Omer A., Partridge F. A., Peluso P., Aiden A. P., Ramasamy V., Rasic G., Roy S., Saavedra-Rodriguez K., Sharan S., Sharma A., Smith M., Turner J., Weakley A. M., Zhao Z., Akbari O. S., Black W. C., Cao H., Darby A. C., Hill C., Johnston J. S., Murphy T. D., Raikhel A. S., Sattelle D. B., Sharakhov I. V., White B. J., Zhao L., Aiden E. L., Mann R. S., Lambrechts L., Powell J. R., Sharakhova M. V., Tu Z., Robertson H. M., McBride C. S., Hastie A. R., Korlach J., Neafsey D. E., Phillippy M., and Vosshall L. B. (2018) Improved reference genome of Aedes aegypti, informs arbovirus vector control. Nature 563, 501–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Keller A., Nesvizhskii A. I., Kolker E., and Aebersold R. (2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem. 74, 5383–5392 [DOI] [PubMed] [Google Scholar]
- 44. Shteynberg D., Deutsch E. W., Lam H., Eng J. K., Sun Z., Tasman N., Mendoza L., Moritz R. L., Aebersold R., and Nesvizhskii A. I. (2011) iProphet: multi-level integrative analysis of shotgun proteomic data improves peptide and protein identification rates and error estimates. Mol. Cell. Proteomics 10, M111.007690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Braisted J. C., Kuntumalla S., Vogel C., Marcotte E. M., Rodrigues A. R., Wang R., Huang S. T., Ferlanti E. S., Saeed A. I., Fleischmann R. D., Peterson S. N., and Pieper R. (2008) The APEX Quantitative Proteomics Tool: generating protein quantitation estimates from LC-MS/MS proteomics results. BMC Bioinformatics 9, 529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Benjamini Y., and Hochberg Y. (1995) Controlling the false discovery rate - a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. B Met. 57, 289–300 [Google Scholar]
- 47. Clements A. N. (1992) The Biology of Mosquitoes: Development, Nutrition, and Reproduction, New York, CABI Publishing [Google Scholar]
- 48. Hatala A. J., Harrington L. C., and Degner E. C. (2018) Age and body size influence sperm quantity in male Aedes albopictus, (Diptera: Culicidae) mosquitoes. J. Med. Entomol. 55, 1051–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ponlawat A., and Harrington L. C. (2007) Age and body size influence male sperm capacity of the dengue vector Aedes aegypti, (Diptera : Culicidae). J. Med. Entomol. 44, 422–426 [DOI] [PubMed] [Google Scholar]
- 50. Alfonso-Parra C., Avila F. W., Deewatthanawong P., Sirot L. K., Wolfner M. F., and Harrington L. C. (2014) Synthesis, depletion and cell-type expression of a protein from the male accessory glands of the dengue vector mosquito Aedes aegypti,. J. Insect Physiol. 70, 117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Morley A. A. (2014) Digital PCR: A brief history. Biomol. Detect. Quantif. 1, 1–2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sutton E. R., Yu Y., Shimeld S. M., White-Cooper H., and Alphey A. L. (2016) Identification of genes for engineering the male germline of Aedes aegypti, and Ceratitis capitata,. BMC Genomics 17, 948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Alfonso-Parra C., Ahmed-Braimah Y. H., Degner E. C., Avila F. W., Villarreal S. M., Pleiss J. A., Wolfner M. F., and Harrington L. C. (2016) Mating-induced transcriptome changes in the reproductive tract of female Aedes aegypti,. PLoS Negl. Trop. Dis. 10, e0004451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Joshi N. A., and Fass J. N. (2011) Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files. 1.33 Ed. [Google Scholar]
- 55. Kim D., Langmead B., and Salzberg S. L. (2015) HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pertea M., Kim D., Pertea G. M., Leek J. T., and Salzberg S. L. (2016) Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 11, 1650–1667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Nielsen H. (2017) Predicting secretory proteins with SignalP. Methods Mol. Biol. 1611, 59–73 [DOI] [PubMed] [Google Scholar]
- 58. Robinson M. D., McCarthy D. J., and Smyth G. K. (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Fontaine A., Filipovic I., Fansiri T., Hoffmann A. A., Cheng C., Kirkpatrick M., Rasic G., and Lambrechts L. (2017) Extensive genetic differentiation between homomorphic sex chromosomes in the mosquito vector, Aedes aegypti,. Genome Biol. Evol. 9, 2322–2335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Rognes T. (2011) Faster Smith-Waterman database searches with inter-sequence SIMD parallelisation. BMC Bioinformatics 12, 221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Chen X. G., Jiang X., Gu J., Xu M., Wu Y., Deng Y., Zhang C., Bonizzoni M., Dermauw W., Vontas J., Armbruster P., Huang X., Yang Y., Zhang H., He W., Peng H., Liu Y., Wu K., Chen J., Lirakis M., Topalis P., Van Leeuwen T., Hall A. B., Jiang X., Thorpe C., Mueller R. L., Sun C., Waterhouse R. M., Yan G., Tu Z. J., Fang X., and James A. A. (2015) Genome sequence of the Asian Tiger mosquito, Aedes albopictus, reveals insights into its biology, genetics, and evolution. Proc. Natl. Acad. Sci. U.S.A. 112, E5907–E5915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Young M. D., Wakefield M. J., Smyth G. K., and Oshlack A. (2010) Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yu G. C., Wang L. G., Han Y. Y., and He Q. Y. (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: J. Integrative Biol. 16, 284–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Ramalingam S. (1983) Secretion in the male accessory glands of Aedes aegypti, (L.) (Diptera: Culicidae). Int. J. Insect Morphol. Embryol. 12, 87–96 [Google Scholar]
- 65. Dapples C. C., Foster W. A., and Lea A. O. (1974) Ultrastructure of the accessory gland of the male mosquito, Aedes aegypti (L.) (Diptera: Culicidae). Int. J. Insect Morphol. Embryol. 3, 279–291 [Google Scholar]
- 66. Leiblich A., Marsden L., Gandy C., Corrigan L., Jenkins R., Hamdy F., and Wilson C. (2012) Bone morphogenetic protein- and mating-dependent secretory cell growth and migration in the Drosophila, accessory gland. Proc. Natl. Acad. Sci. U.S.A. 109, 19292–19297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Whittington E., Forsythe D., Borziak K., Karr T. L., Walters J. R., and Dorus S. (2017) Contrasting patterns of evolutionary constraint and novelty revealed by comparative sperm proteomic analysis in Lepidoptera. BMC Genomics 18, 931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Dorus S., Busby S. A., Gerike U., Shabanowitz J., Hunt D. F., and Karr T. L. (2006) Genomic and functional evolution of the Drosophila melanogaster, sperm proteome. Nat. Genet. 38, 1440–1445 [DOI] [PubMed] [Google Scholar]
- 69. Laflamme B. A., and Wolfner M. F. (2013) Identification and function of proteolysis regulators in seminal fluid. Mol. Reprod. Dev. 80, 80–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Sirot L. K., Poulson R. L., McKenna M. C., Girnary H., Wolfner M. F., and Harrington L. C. (2008) Identity and transfer of male reproductive gland proteins of the dengue vector mosquito, Aedes aegypti: potential tools for control of female feeding and reproduction. Insect Biochem. Mol. Biol. 38, 176–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Carvalho A. B., Dobo B. A., Vibranovski M. D., and Clark A. G. (2001) Identification of five new genes on the Y chromosome of Drosophila melanogaster,. Proc. Natl. Acad. Sci. U.S.A. 98, 13225–13230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Lahn B. T., and Page D. C. (1997) Functional coherence of the human Y chromosome. Science 278, 675–680 [DOI] [PubMed] [Google Scholar]
- 73. Hall A. B., Basu S., Jiang X., Qi Y., Timoshevskiy V. A., Biedler J. K., Sharakhova M. V., Elahi R., Anderson M. A., Chen X. G., Sharakhov I. V., Adelman Z. N., and Tu Z. (2015) Sex determination. A male-determining factor in the mosquito Aedes aegypti,. Science 348, 1268–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Bonilla M. L., Todd C., Erlandson M., and Andres J. (2015) Combining RNA-seq and proteomic profiling to identify seminal fluid proteins in the migratory grasshopper Melanoplus sanguinipes, (F). BMC Genomics 16, 1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bao S. N., and de Souza W. (1993) Ultrastructural and cytochemical studies of the spermatid and spermatozoon of Culex quinquefasciatus, (Culicidae). J. Submicrosc. Cytol. Pathol. 25, 213–222 [PubMed] [Google Scholar]
- 76. Ndiaye M., Mattei X., and Thiaw O. T. (1997) Maturation of mosquito spermatozoa during their transit throughout the male and female reproductive systems. Tissue Cell 29, 675–678 [DOI] [PubMed] [Google Scholar]
- 77. Pilch B., and Mann M. (2006) Large-scale and high-confidence proteomic analysis of human seminal plasma. Genome Biol. 7, R40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Corrigan L., Redhai S., Leiblich A., Fan S. J., Perera S. M., Patel R., Gandy C., Wainwright S. M., Morris J. F., Hamdy F., Goberdhan D. C., and Wilson C. (2014) BMP-regulated exosomes from Drosophila, male reproductive glands reprogram female behavior. J. Cell Biol. 206, 671–688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Sharma U., Sun F., Conine C. C., Reichholf B., Kukreja S., Herzog V. A., Ameres S. L., and Rando O. J. (2018) Small RNAs are trafficked from the epididymis to developing mammalian sperm. Dev. Cell 46, 481–494.e486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Borziak K., Alvarez-Fernandez A., T L. K., Pizzari T., and Dorus S. (2016) The seminal fluid proteome of the polyandrous red junglefowl offers insights into the molecular basis of fertility, reproductive ageing and domestication. Sci. Rep. 6, 35864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Naccarati C., Audsley N., Keen J. N., Kim J. H., Howell G. J., Kim Y. J., and Isaac R. E. (2012) The host-seeking inhibitory peptide, Aea-HP-1, is made in the male accessory gland and transferred to the female during copulation. Peptides 34, 150–157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Chen P. S., Stumm-Zollinger E., Aigaki T., Balmer J., Bienz M., and Bohlen P. (1988) A male accessory gland peptide that regulates reproductive behavior of female D. melanogaster,. Cell 54, 291–298 [DOI] [PubMed] [Google Scholar]
- 83. Bednarova A., Kodrik D., and Krishnan N. (2013) Adipokinetic hormone exerts its anti-oxidative effects using a conserved signal-transduction mechanism involving both PKC and cAMP by mobilizing extra- and intracellular Ca2+ stores. Comp. Biochem. Physiol., C: Toxicol. Pharmacol. 158, 142–149 [DOI] [PubMed] [Google Scholar]
- 84. Konuma T., Morooka N., Nagasawa H., and Nagata S. (2012) Knockdown of the adipokinetic hormone receptor increases feeding frequency in the two-spotted cricket Gryllus bimaculatus,. Endocrinology 153, 3111–3122 [DOI] [PubMed] [Google Scholar]
- 85. Peng J., Chen S., Busser S., Liu H., Honegger T., and Kubli E. (2005) Gradual release of sperm bound sex-peptide controls female postmating behavior in Drosophila,. Curr. Biol. 15, 207–213 [DOI] [PubMed] [Google Scholar]
- 86. Borras E., and Sabido E. (2017) What is targeted proteomics? A concise revision of targeted acquisition and targeted data analysis in mass spectrometry. Proteomics 17 [DOI] [PubMed] [Google Scholar]
- 87. Predel R., Neupert S., Garczynski S. F., Crim J. W., Brown M. R., Russell W. K., Kahnt J., Russell D. H., and Nachman R. J. (2010) Neuropeptidomics of the mosquito Aedes aegypti,. J. Proteome Res. 9, 2006–2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Swanson W. J., and Vacquier V. D. (2002) The rapid evolution of reproductive proteins. Nat. Rev. Genet. 3, 137–144 [DOI] [PubMed] [Google Scholar]
- 89. Nene V., Wortman J. R., Lawson D., Haas B., Kodira C., Tu Z. J., Loftus B., Xi Z. Y., Megy K., Grabherr M., Ren Q. H., Zdobnov E. M., Lobo N. F., Campbell K. S., Brown S. E., Bonaldo M. F., Zhu J. S., Sinkins S. P., Hogenkamp D. G., Amedeo P., Arensburger P., Atkinson P. W., Bidwell S., Biedler J., Birney E., Bruggner R. V., Costas J., Coy M. R., Crabtree J., Crawford M., deBruyn B., DeCaprio D., Eiglmeier K., Eisenstadt E., El-Dorry H., Gelbart W. M., Gomes S. L., Hammond M., Hannick L. I., Hogan J. R., Holmes M. H., Jaffe D., Johnston J. S., Kennedy R. C., Koo H., Kravitz S., Kriventseva E. V., Kulp D., LaButti K., Lee E., Li S., Lovin D. D., Mao C. H., Mauceli E., Menck C. F. M., Miller J. R., Montgomery P., Mori A., Nascimento A. L., Naveira H. F., Nusbaum C., O'Leary S., Orvis J., Pertea M., Quesneville H., Reidenbach K. R., Rogers Y. H., Roth C. W., Schneider J. R., Schatz M., Shumway M., Stanke M., Stinson E. O., Tubio J. M. C., VanZee J. P., Verjovski-Almeida S., Werner D., White O., Wyder S., Zeng Q. D., Zhao Q., Zhao Y. M., Hill C. A., Raikhel A. S., Soares M. B., Knudson D. L., Lee N. H., Galagan J., Salzberg S. L., Paulsen I. T., Dimopoulos G., Collins F. H., Birren B., Fraser-Liggett C. M., and Severson D. W. (2007) Genome sequence of Aedes aegypti,, a major arbovirus vector. Science 316, 1718–1723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Moore M., Sylla M., Goss L., Burugu M. W., Sang R., Kamau L. W., Kenya E. U., Bosio C., Munoz Mde L., Sharakova M., and Black W. C. (2013) Dual African origins of global Aedes aegypti, s.l. populations revealed by mitochondrial DNA. PLoS Negl. Trop. Dis. 7, e2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Shi Q. M., Zhang H. D., Wang G., Guo X. X., Xing D., Dong Y. D., Xiao L., Gao J., Liu Q. M., Sun A. J., Li C. X., and Zhao T. Y. (2017) The genetic diversity and population structure of domestic Aedes aegypti, (Diptera: Culicidae) in Yunnan Province, southwestern China. Parasit. Vectors 10, 292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lawniczak M. K., Emrich S. J., Holloway A. K., Regier A. P., Olson M., White B., Redmond S., Fulton L., Appelbaum E., Godfrey J., Farmer C., Chinwalla A., Yang S. P., Minx P., Nelson J., Kyung K., Walenz B. P., Garcia-Hernandez E., Aguiar M., Viswanathan L. D., Rogers Y. H., Strausberg R. L., Saski C. A., Lawson D., Collins F. H., Kafatos F. C., Christophides G. K., Clifton S. W., Kirkness E. F., and Besansky N. J. (2010) Widespread divergence between incipient Anopheles gambiae, species revealed by whole genome sequences. Science 330, 512–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Reidenbach K. R., Neafsey D. E., Costantini C., Sagnon N., Simard F., Ragland G. J., Egan S. P., Feder J. L., Muskavitch M. A., and Besansky N. J. (2012) Patterns of genomic differentiation between ecologically differentiated M and S forms of Anopheles gambiae, in West and Central Africa. Genome Biol. Evol. 4, 1202–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sirot L. K., Wong A., Chapman T., and Wolfner M. F. (2014) Sexual conflict and seminal fluid proteins: a dynamic landscape of sexual interactions. Cold Spring Harb. Perspect. Biol. 7, a017533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Toups M. A., and Hahn M. W. (2010) Retrogenes reveal the direction of sex-chromosome evolution in mosquitoes. Genetics 186, 763–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Charlesworth B. (1996) The evolution of chromosomal sex determination and dosage compensation. Curr. Biol. 6, 149–162 [DOI] [PubMed] [Google Scholar]
- 97. Charlesworth B., and Charlesworth D. (2000) The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond., Ser. B: Biol. Sci. 355, 1563–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Vicoso B., Kaiser V. B., and Bachtrog D. (2013) Sex-biased gene expression at homomorphic sex chromosomes in emus and its implication for sex chromosome evolution. Proc. Natl. Acad. Sci. U.S.A. 110, 6453–6458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Akbari O. S., Antoshechkin I., Amrhein H., Williams B., Diloreto R., Sandler J., and Hay B. A. (2013) The developmental transcriptome of the mosquito Aedes aegypti,, an invasive species and major arbovirus vector. G3-Genes Genom. Genet. 3, 1493–1509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Dorus S., Wilkin E. C., and Karr T. L. (2011) Expansion and functional diversification of a leucyl aminopeptidase family that encodes the major protein constituents of Drosophila, sperm. BMC Genomics 12, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Ellison C., Leonard C., Landeen E., Gibilisco L., Phadnis N., and Bachtrog D. (2018) Rampant cryptic sex chromosome drive in Drosophila,. bioRxiv [Google Scholar]
- 102. Lindholm A. K., Dyer K. A., Firman R. C., Fishman L., Forstmeier W., Holman L., Johannesson H., Knief U., Kokko H., Larracuente A. M., Manser A., Montchamp-Moreau C., Petrosyan V. G., Pomiankowski A., Presgraves D. C., Safronova L. D., Sutter A., Unckless R. L., Verspoor R. L., Wedell N., Wilkinson G. S., and Price T. A. R. (2016) The ecology and evolutionary dynamics of meiotic drive. Trends Ecol. Evol. 31, 315–326 [DOI] [PubMed] [Google Scholar]
- 103. Hammond A. M., and Galizi R. (2017) Gene drives to fight malaria: current state and future directions. Pathog. Glob. Health 111, 412–423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Visconti P. E. (2012) Sperm bioenergetics in a nutshell. Biol. Reprod. 87, 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Stephens K., Cardullo R. A., and Thaler C. D. (2018) Culex pipiens sperm motility is initiated by a trypsin-like protease from male accessory glands. Mol. Reprod. Dev. 85, 440–448 [DOI] [PubMed] [Google Scholar]
- 106. Baer B., Heazlewood J. L., Taylor N. L., Eubel H., and Millar A. H. (2009) The seminal fluid proteome of the honeybee Apis mellifera,. Proteomics 9, 2085–2097 [DOI] [PubMed] [Google Scholar]
- 107. Rhea J. M., Wegener C., and Bender M. (2010) The proprotein convertase encoded by amontillado (amon,) is required in Drosophila, corpora cardiaca endocrine cells producing the glucose regulatory hormone AKH. PLoS Genet. 6, e1000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Werner M., and Simmons L. W. (2008) Insect sperm motility. Biol. Rev. Camb. Philos. Soc. 83, 191–208 [DOI] [PubMed] [Google Scholar]
- 109. Miller D., and Ostermeier G. C. (2006) Spermatozoal RNA: why is it there and what does it do? Gynecol. Obstet. Fertil. 34, 840–846 [DOI] [PubMed] [Google Scholar]
- 110. Conine C. C., Sun F., Song L., Rivera-Perez J. A., and Rando O. J. (2018) Small RNAs gained during epididymal transit of sperm are essential for embryonic development in mice. Dev. Cell 46, 470–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Fischer B. E., Wasbrough E., Meadows L. A., Randlet O., Dorus S., Karr T. L., and Russell S. (2012) Conserved properties of Drosophila, and human spermatozoal mRNA repertoires. Proc. R. Soc. Lond., Ser. B: Biol. Sci. 279, 2636–2644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Bono J. M., Matzkin L. M., Kelleher E. S., and Markow T. A. (2011) Postmating transcriptional changes in reproductive tracts of con- and heterospecifically mated Drosophila mojavensis, females. Proc. Natl. Acad. Sci. U.S.A. 108, 7878–7883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Lees R. S., Gilles J. R., Hendrichs J., Vreysen M. J., and Bourtzis K. (2015) Back to the future: the sterile insect technique against mosquito disease vectors. Curr. Opin. Insect Sci. 10, 156–162 [DOI] [PubMed] [Google Scholar]
- 114. Avila F. W., Ravi Ram K., Bloch Qazi M. C., and Wolfner M. F. (2010) Sex peptide is required for the efficient release of stored sperm in mated Drosophila, females. Genetics 186, 595–600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Perotti M. E., Cattaneo F., Pasini M. E., Verni F., and Hackstein J. H. P. (2001) Male sterile mutant casanova, gives clues to mechanisms of sperm-egg interactions in Drosophila melanogaster,. Mol. Reprod. Dev. 60, 248–259 [DOI] [PubMed] [Google Scholar]
- 116. Degner E. C., and Harrington L. C. (2016) A mosquito sperm's journey from male ejaculate to egg: mechanisms, molecules, and methods for exploration. Mol. Reprod. Dev. 83, 897–911 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository with the dataset identifier PXD010293 and 10.6019/PXD010293. The R analysis pipeline used for transcriptomic analysis is available as part of this project's GitHub repository (https://github.com/YazBraimah/Aegypti.ejaculatomics). Raw RNA-seq reads from this study can be obtained from the Sequence Read Archive (SRA) Accession # SRP158536. RNA-seq data from Sutton et al,. (52) and Alfonso-Parra et al., (53) can be accessed at SRA Accession # SRP075464 and SRA Accession # SRP068996, respectively.