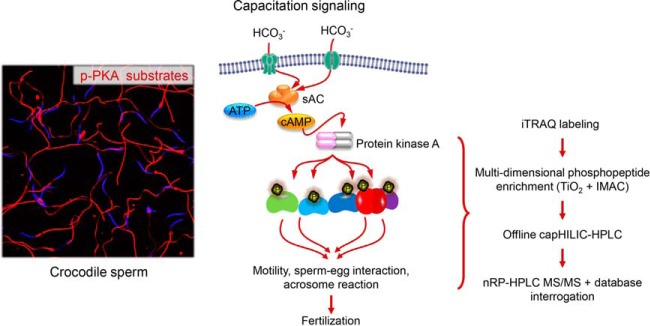
Quantitative proteomic and phosphoproteomic analyses of Australian saltwater crocodile spermatozoa reveals that these cells respond to capacitation stimuli by mounting a cAMP-mediated signaling cascade that is analogous to that of spermatozoa from the mammalian lineage. Similarly, the phosphorylated proteins responsible for driving the functional maturation of crocodile spermatozoa share substantial evolutionary overlap with those documented in mammalian spermatozoa.
Keywords: Phosphoproteome, Phosphorylation, Cell biology*, Developmental biology*, Physiology*, Post-translational modifications*, Protein Identification*, Protein Modification*, Serine/Threonine Kinases*, Signal Transduction*, capacitation, crocodile, Crocodylus porosus, sperm
Graphical Abstract
Highlights
Comparative and quantitative phosphoproteomics of Australian saltwater crocodile spermatozoa.
Capacitation stimuli elicit a cAMP-mediated signaling cascade in crocodile spermatozoa.
Mechanistic insights into conservation of sperm activation pathways.
Abstract
Competition to achieve paternity has contributed to the development of a multitude of elaborate male reproductive strategies. In one of the most well-studied examples, the spermatozoa of all mammalian species must undergo a series of physiological changes, termed capacitation, in the female reproductive tract before realizing their potential to fertilize an ovum. However, the evolutionary origin and adaptive advantage afforded by capacitation remains obscure. Here, we report the use of comparative and quantitative proteomics to explore the biological significance of capacitation in an ancient reptilian species, the Australian saltwater crocodile (Crocodylus porosus,). Our data reveal that exposure of crocodile spermatozoa to capacitation stimuli elicits a cascade of physiological responses that are analogous to those implicated in the functional activation of their mammalian counterparts. Indeed, among a total of 1119 proteins identified in this study, we detected 126 that were differentially phosphorylated (± 1.2 fold-change) in capacitated versus, noncapacitated crocodile spermatozoa. Notably, this subset of phosphorylated proteins shared substantial evolutionary overlap with those documented in mammalian spermatozoa, and included key elements of signal transduction, metabolic and cellular remodeling pathways. Unlike mammalian sperm, however, we noted a distinct bias for differential phosphorylation of serine (as opposed to tyrosine) residues, with this amino acid featuring as the target for ∼80% of all changes detected in capacitated spermatozoa. Overall, these results indicate that the phenomenon of sperm capacitation is unlikely to be restricted to mammals and provide a framework for understanding the molecular changes in sperm physiology necessary for fertilization.
The molecular processes leading to fertilization remain among the key unresolved questions in the field of reproductive biology. Based on studies of the mammalian lineage, it is widely accepted that terminally differentiated spermatozoa develop the capacity to fertilize an ovum during sequential phases of post-testicular maturation as they pass through the male (epididymis) (1) and female reproductive tracts (2). The latter of these is termed capacitation and is defined as a time-dependent process during which spermatozoa experience a suite of biochemical and biophysical changes that collectively endow them with the ability to recognize and fertilize an ovum (2, 3). A distinctive characteristic of this phase of functional maturation is that it occurs in the complete absence of nuclear gene transcription and de novo, protein synthesis. Instead, the regulation of capacitation rests almost exclusively with convergent signaling cascades that transduce extracellular signals to effect extensive post-translational modification of the intrinsic sperm proteome (4). In this context, differential protein phosphorylation has emerged as a dominant molecular switch, which regulates sperm-oocyte recognition and adhesion, the ability to undergo acrosomal exocytosis, and the propagation of an altered pattern of movement referred to as hyperactivation (5).
A curiosity of the capacitation cascade is its evolutionary origin and the adaptive advantage that is afforded by such an elaborate form of post-testicular sperm maturation. Indeed, studies of the spermatozoa of sub-therian vertebrate species such as those of the aves, have failed to document a process equivalent to capacitation (6–8); with spermatozoa of fowls and turkeys appearing refractory to the need for any such physiological changes during their residence in the female reproductive system (6, 8). Similarly, in the quail it has been shown that most testicular sperm can bind to a perivitelline membrane and acrosome react with no additional advantage being afforded by exposure to capacitation stimuli (9). Although it has been suggested that reptilian spermatozoa also experience minimal post-testicular maturation, this paradigm has recently been challenged by functional analysis of ejaculated spermatozoa from the Australian saltwater crocodile (Crocodylus porosus,) (10). In this context, exposure to capacitation stimuli, which elevate intracellular levels of cyclic AMP (cAMP), promoted a significant enhancement of the motility profile recorded in crocodile spermatozoa. Notably, dilution in capacitation medium also enhances the post-thaw survival of cryopreserved crocodile spermatozoa (11). Conversely, crocodile spermatozoa are rendered quiescent upon incubation in bicarbonate-free media formulated to suppress the capacitation of eutherian spermatozoa (10). We contend that such changes may reflect physiological demands imposed by the transferal of sperm storage responsibilities from the male to the female reproductive tract, and the attendant need to alternatively silence and reactivate spermatozoa to enhance their longevity and fertilization competence, respectively. Nevertheless, the mechanistic basis of these opposing responses remain obscure, as does the identity of the proteins implicated in their regulation.
Here, we have used mass spectrometry-based proteomics to generate a comprehensive protein inventory of mature crocodile spermatozoa and subsequently explore signatures of capacitation via quantitative phosphoproteomic profiling strategies. Our data confirm that the phosphorylation status of the crocodile sperm proteome is substantially modified in response to capacitation stimuli, thus refuting the tenet that this phenomenon is restricted to the mammalian lineage and providing a framework for understanding the molecular changes in sperm physiology necessary for fertilization.
EXPERIMENTAL PROCEDURES
Chemicals and Reagents
Unless otherwise specified, chemical reagents were obtained from Sigma (St. Louis, MO). Anti-phosphoserine (P5747), phosphothreonine (P6623), phosphotyrosine (P5964), flotillin1 (F1180), CABYR (SAB2107035), and tubulin (T5168) antibodies were purchased from Sigma. Anti-phospho (Ser/Thr) PKA substrate antibodies (9621) was from Cell Signaling (Danvers, MA). Anti-ZPBP2 (H00124626-B01) antibody was from Abnova (Taipei City, Taiwan). Anti-DNM3 (14737–1-AP) and SPATC1 (25861–1-AP) antibodies were from ProteinTech (Rosemont, IL). Anti-ZPBP1 (S1587) antibody was from Epitomics (Burlingame, CA). Anti-ACR (sc-46284) and ACRBP (sc-109379) were from Santa Cruz Biotechnology (Dallas, TX). Anti-AKAP4 (4BDX-1602) antibody was from 4BioDx (Lille, France). Anti-rabbit IgG-horseradish peroxidase (HRP) was purchased from Merck Millipore (Billerica, MA), and anti-mouse IgG and anti-goat IgG HRP were from Santa Cruz Biotechnology. All fluorescently labeled (Alexa Fluor) secondary antibodies were from Thermo Fisher Scientific (Waltman, MA). Fluorescein isothiocyanate (FITC) conjugated Pisum sativum, agglutinin (PSA) (FL-1051) was from Vector Laboratories (Burlingame, CA).
Animals and Semen Collection
The study was undertaken at Koorana Crocodile Farm, QLD, Australia with the approval of the University of Queensland Animal Ethics Committee (SAS/361/10) and Queensland Government Scientific Purposes Permit (WISP09374911). Semen used throughout this study was collected by digital massage (12) from mature (>3.0 m) saltwater crocodiles during the breeding season (November 2014, 2015).
Sperm Capacitation
The in vitro, capacitation of crocodile spermatozoa was achieved via elevation of intracellular cAMP levels as previously described (13, 14). In brief, raw semen samples were diluted 1:4 into one of two modified formulations of Biggers, Whitten and Whittingham (BWW)1 medium (15), namely: (1) noncapacitating BWW media control (NC) [comprising 120 mm NaCl, 4.6 mm KCl, 1.7 mm CaCl2.2H2O, 1.2 mm KH2PO4, 1.2 mm MgSO4.7H2O, 5.6 mm d-glucose, 0.27 mm sodium pyruvate, 44 mm sodium lactate, 5 U/ml penicillin, 5 mg/ml streptomycin and 20 mm HEPES buffer and 3 mg/ml BSA (pH 7.4, osmolality of 300mOsm/kg)], or (2) capacitating BWW (CAP), an equivalent formulation to that of NC BWW, with additional supplementation of 25 mm NaHCO3, a phosphodiesterase inhibitor (pentoxifylline, 1 mm) and a membrane permeable cAMP analogue (dibutyryl cyclic AMP, dbcAMP, 1 mm). Following dilution, an aliquot of sperm was immediately assessed for viability, motility characteristics, acrosomal integrity, and phosphorylation status. The remainder of the sample was incubated at 30 °C for 120 min. At the completion of incubation, sperm suspensions were prepared for mass spectrometry analysis as described below. To substantiate capacitation-like changes in crocodile spermatozoa, a sub-population of these cells were assessed for phosphorylation of serine, threonine and tyrosine residues. Similarly, the spermatozoa were also assayed for overall levels of phospho-PKA substrates.
Comparative and Quantitative Sperm Proteome and Phosphoproteome Analysis
Preparations of crocodile spermatozoa (noncapacitated and capacitated) were subjected to membrane protein enrichment by dissolving in 100 μl of ice-cold 0.1 m Na2CO3 supplemented with protease (Sigma) and phosphatase inhibitors (Roche, Complete EDTA free). These suspensions were subjected to probe tip sonication at 4 °C for 3 × 10 s intervals before incubation for 1 h at 4 °C. Soluble proteins were isolated from membrane proteins by ultracentrifugation (100,000 × g, for 90 min at 4 °C) (16). Membrane-enriched pellets and soluble proteins were dissolved in urea (6 m urea, 2 m thiourea), reduced using 10 mm DTT (30 min, room temperature), alkylated using 20 mm iodoacetamide (30 min, room temperature, in the dark), and subsequently digested with 0.05 activity units of Lys-C endoproteinase (Wako, Osaka, Japan) for 3 h at 37 °C. After Lys-C digestion, the solution was diluted in 0.75 m urea, 0.25 m thiourea with TEAB, and digested using 2% w/w trypsin (specificity for positively charged lysine and arginine side chains; Promega, Madison, WI) overnight at 37 °C in 500 mm triethylammonium bicarbonate (TEAB) and centrifuged at 14,000 × g, for 30 min at 4 °C (17, 18). Peptides were desalted and cleaned up using a modified StageTip microcolumn (19). Quantitative fluorescent peptide quantification (Qubit Assay; Thermo Fisher Scientific) was employed and 100 μg of each peptide sample was labeled using isobaric tag based methods (20), according to manufacturer's specifications (iTRAQ; SCIEX, Framingham, MA). Digestion and isobaric tag labeling efficiency was determined by NanoLC-MS/MS (described below). Samples were then mixed in 1:1 ratio and phosphopeptides enriched using a multidimensional strategy employing TiO(2) pre-enrichment step followed by separate multi- and mono-phosphorylated peptides post-fractionation using a sequential elution from immobilized metal affinity chromatography (21). Nonmodified peptides were then subjected to offline hydrophilic interaction liquid chromatography (HILIC) before high resolution LC-MS/MS (18).
Tandem Mass Spectrometry (NanoLC-MS/MS) Quantitative Analyses
NanoLC-MS/MS, was performed using a Dionex UltiMate 3000RSLC nanoflow HPLC system (Thermo Fisher Scientific). Membrane and soluble mono- and multiphosphorylated peptides, and nonmodified peptides (seven membrane enriched fractions and seven soluble fractions) were suspended in buffer A (0.1% formic acid) and directly loaded onto an Acclaim PepMap100 C18 75 μm × 20 mm trap column (Thermo Fisher Scientific) for pre-concentration and online desalting. Separation was then achieved using an EASY-Spray PepMap C18 75 μm × 500 mm column (Thermo Fisher Scientific), employing a linear gradient from 2 to 32% acetonitrile at 300 nl/min over 120 min. Q-Exactive Plus MS System (Thermo Fisher Scientific) was operated in full MS/data dependent acquisition MS/MS mode (data-dependent acquisition). The Orbitrap mass analyzer was used at a resolution of 70 000, to acquire full MS with an m,/z, range of 390–1400, incorporating a target automatic gain control value of 1e6 and maximum fill times of 50 ms. The 20 most intense multiply charged precursors were selected for higher-energy collision dissociation fragmentation with a normalized collisional energy of 32. MS/MS fragments were measured at an Orbitrap resolution of 35,000 using an automatic gain control target of 2e5 and maximum fill times of 110 ms (18). Fragmentation data were converted to peak lists using Xcalibur (Thermo Fisher Scientific), and the HCD data were searched using Proteome Discoverer version 2.1 (Thermo Fisher Scientific) against the Archosauria, crown group comprising birds and crocodilians within the UniprotKB database (Archosauria,, downloaded on the 31st January of 2018, 622,090 sequences). Mass tolerances in MS and MS/MS modes were 10 ppm and 0.02 Da, respectively; trypsin was designated as the digestion enzyme, and up to two missed cleavages were allowed. S-carbamidomethylation of cysteine residues was designated as a fixed modification, as was modification of lysines and peptide N termini with the isobaric iTRAQ 8-plex label. Variable modifications considered were: acetylation of lysine, oxidation of methionine and phosphorylation of serine, threonine and tyrosine residues. Results from searches of membrane-enriched and soluble fractions were merged and interrogation of the corresponding reversed database was also performed to evaluate the false discovery rate (FDR) of peptide identification using Percolator based on q-values, which were estimated from the target-decoy search approach. To filter out target peptide spectrum matches (target-PSMs) over the decoy-PSMs, a fixed false discovery rate (FDR) of 1% was set at the peptide level (17). The dataset (Dataset S1) analyzed here have been deposited in the Mass Spectrometry Interactive Virtual Environment (MassIVE) database and are publicly accessible at: https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=8acd6725da734f6f89bbd64460d03686.
SDS-PAGE and Immunoblotting
After incubation, crocodile spermatozoa were pelleted (400 × g,, 1 min), washed in NC BWW media and re-centrifuged (400 × g,, 1 min). The sperm pellet was re-suspended in SDS extraction buffer (0.375 m Tris pH 6.8, 2% (w/v) SDS, 10% (w/v) sucrose, protease inhibitor mixture), incubated at 100 °C for 5 min and equivalent amounts of protein (10 μg) were separated by SDS-PAGE (22). Gels were either stained with silver reagent or transferred onto nitrocellulose membranes (Hybond C-extra; GE Healthcare, Buckinghamshire, England, UK) (23). Membranes were blocked for 1 h in Tris buffered saline (TBS) containing 5% w/v skim milk powder. After rinsing with TBS containing 0.1% v/v Tween-20 (TBST), membranes were sequentially incubated with appropriate primary antibody at 4 °C overnight and its corresponding HRP-conjugated secondary antibody for 1 h. Following three washes in TBST, labeled proteins were detected using enhanced chemiluminescence reagents (GE Healthcare).
Immunocytochemistry
Spermatozoa were fixed in 4% paraformaldehyde, washed three times with 0.05 m glycine in PBS and then applied to poly-l-lysine coated glass coverslips. The cells were permeabilized with 0.2% Triton X-100 and blocked in 3% BSA/PBS for 1 h. Coverslips were then washed in PBS and incubated in a humidified chamber with appropriate primary and secondary antibodies (1 h at 37 °C). Coverslips were washed (3 × 5 min) in filtered PBS after each antibody incubation, before mounting in antifade reagent comprising 10% Mowiol 4–88 (Merck Millipore), 30% glycerol and 2.5% 1,4-diazobicyclo-(2.2.2)-octane (DABCO) in 0.2 m Tris (pH 8.5). Sperm cells were then examined by confocal microscopy (Olympus, Nagano, Japan).
Duolink Proximity Ligation Assay
In situ, proximity ligation assays (PLA) were conducted according to manufacturer's instructions (OLINK Biosciences, Uppsala, Sweden; as described by (24)) using combinations of either anti-CABYR and anti-phosphoserine or anti-SPATC1 and anti-phosphoserine antibodies. Coverslips were mounted as previously described for immunocytochemistry and visualized by fluorescence microscopy (Carl Zeiss, Sydney, NSW, Australia). If target proteins resided within a maximum distance of 40 nm, this reaction resulted in the production of a discrete fluorescent foci that appeared as red spots (25). PLA fluorescence was quantified for 200 cells per slide.
Experimental Design and Statistical Rationale
All MS analyses were performed in duplicate using ejaculated semen samples collected from different crocodiles (n, = 2). Similarly, immunoblotting analyses to confirm the phospho-serine, -threonine, -tyrosine, and -PKA status of crocodile spermatozoa were conducted on the same two semen samples (n, = 2). However, immunocytochemical, PLA, and functional experiments to substantiate crocodile sperm proteomic data were performed in triplicate using individual biological samples differing from those employed for MS analyses (n, = 3). Where appropriate, graphical data are presented as means ± S.E. (n, = 3), with statistical significance being determined by analysis of variance (ANOVA).
RESULTS
Global Proteomic Analysis of Crocodile Spermatozoa
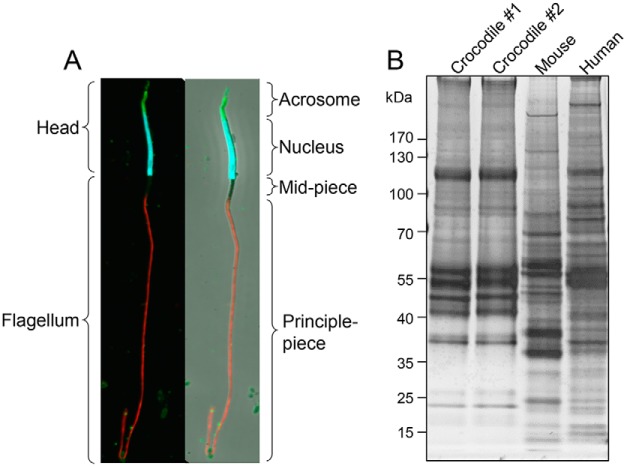
Before analysis, the quality of spermatozoa in each ejaculate was assessed via immunolabeling with markers of acrosomal (PSA, green) and nuclear integrity (DAPI, blue) and counterstaining of the flagellum with anti-tubulin antibodies (red) (Fig. 1A,). A portion of the sperm sample from each animal was subjected to standard cell lysis and the proteins resolved by SDS-PAGE to confirm broadly equivalent proteomic profiles, which differed substantially from equivalent lysates of mouse and human spermatozoa (Fig. 1B,). The complexity of the remaining samples was reduced via fractionation into membrane-enriched and soluble cell lysates; both of which were subjected to mass spectrometry analysis. Notwithstanding the incomplete annotation of the crocodile genome, and the attendant need for sequence alignment to be performed against the Archosauria, crown group, our experimental strategy identified a complex proteomic signature comprising a total of 1119 proteins (supplemental Table S1). Among these proteins, an average of 4.6 peptide matches (encompassing 3.4 unique peptide matches) were generated per protein; representing an average peptide coverage of 11% per protein (supplemental Table S1).
Fig. 1.
Assessment of crocodile sperm samples. A,, The integrity of crocodile spermatozoa isolated from ejaculated semen samples were assessed via immunolabeling of cells with acrosomal (FITC-conjugated PSA, green), nuclear (DAPI, blue) and flagellar (tubulin, red) markers. B,, A portion of the spermatozoa from each of two animals were subjected to standard cell lysis before extracted proteins were resolved by SDS-PAGE and silver stained to confirm broadly equivalent proteomic profiles. For comparative purposes, crocodile sperm lysates were resolved alongside equivalent lysates prepared from mouse and human spermatozoa.
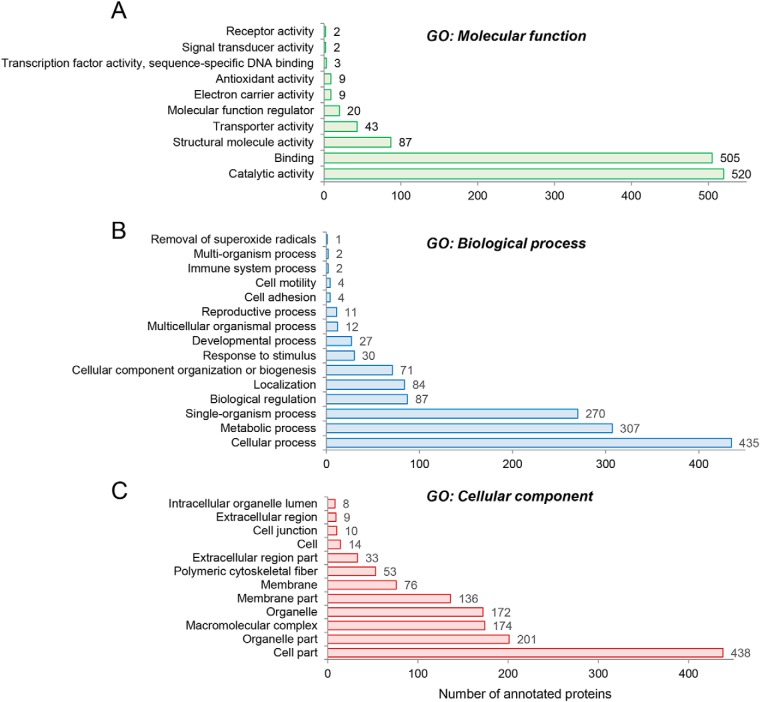
Provisional interrogation of this global crocodile sperm proteome on the basis of Gene Ontology (GO) classification (26) returned dominant terms of catalytic activity, binding, structural molecule activity, and transporter activity among the top GO molecular function categories when ranked on the basis of number of annotated proteins (Fig. 2A,, supplemental Table S2). Notable enrichment was also identified in the broad GO biological process categories of cellular process, metabolic process, single organism process, biological regulation and localization (Fig. 2B,, supplemental Table S2). Additional categories of direct relevance to sperm physiology/function included: developmental process, reproductive process, cell motility and cell adhesion (Fig. 2B,, supplemental Table S2). The dominant GO cellular compartments represented in the crocodile sperm proteome included that of the cell, organelle, macromolecular complex, and membrane, with some 438, 201, 174, and 136 proteins mapping to each of these respective categories (Fig. 2C,, supplemental Table S2).
Fig. 2.
GO annotation of crocodile sperm proteome. The complete inventory of 1119 identified proteins was subjected to provisional Gene Ontology (GO) classification using the universal protein knowledgebase (UniProtKB) functional annotation tools (26). Proteins were curated on the basis of (A,) GO molecular function, (B,) GO biological process, and (C,) GO cellular compartment.
Our application of isobaric peptide labeling afforded the opportunity to investigate the relative abundance of each protein in opposing populations of noncapacitated and capacitated spermatozoa. As might be expected of a cell that lacks the capacity to engage in protein synthesis, the majority (∼90%) of crocodile sperm proteins were detected at equivalent levels irrespective of capacitation status. Among the remaining proteins, we documented an apparent reduction in the abundance of 53 proteins and conversely, an apparent increase in abundance of 71 proteins (supplemental Table S2); possibly reflecting differential partitioning and/or translocation between intracellular domains upon exposure to capacitation stimuli, thus influencing the efficiency of their extraction.
Conservation of the Crocodile Sperm Proteome
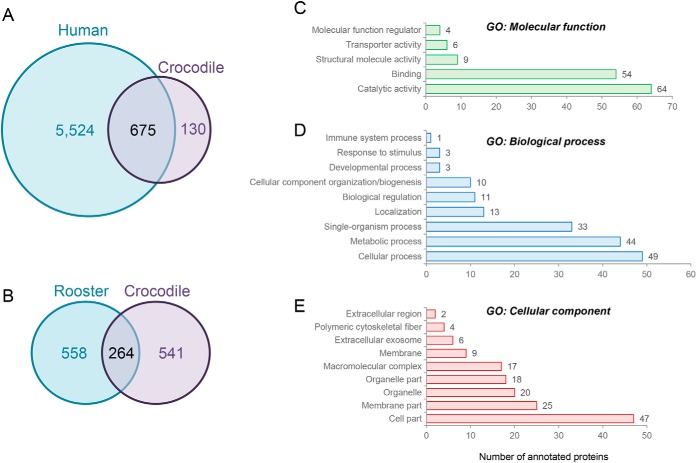
To begin to explore the extent of conservation between the crocodile sperm proteome and that of more widely studied mammalian species, we elected to survey the published proteome compiled for human spermatozoa; a comprehensive dataset comprising 6199 of the predicted ∼7500 that constitute this cell type (27). To facilitate this comparison, all Archosauria, UniProt accession numbers were manually converted into accession numbers equating to their human homologues (supplemental Table S1). During this annotation, 232 proteins were unable to be assigned accession numbers because of ambiguous nomenclature (e.g., proteins remaining as uncharacterized or failure to unequivocally determine the relevant protein isoform) and an additional 82 potentially redundant protein identifications were detected among our initial crocodile proteomic inventory (e.g., apparently equivalent proteins that have been assigned different gene names within the Archosauria, database). Of the remaining 805 proteins, homologues of 675 (84%) were identified in the compiled human sperm proteome (supplemental Table S1; Fig. 3A,). Although we remain cognizant that neither sperm proteomic database is yet to be fully annotated, functional classification of the 130 putatively nonconserved crocodile sperm proteins revealed notable enrichment in the molecular function category of catalytic activity (Fig. 3C,) and the biological process of metabolism (Fig. 3D,). Moreover, a substantial number of these proteins mapped to the membrane domain (Fig. 3E,), suggestive of differing specialization of the surface and possibly the metabolic characteristics of these cells. In expanding this analysis to include comparison with the published rooster sperm proteome comprising 822 proteins (28), we were only able to identify homologues of 264 crocodile sperm proteins (supplemental Table S1; Fig. 3B,). However, owing to the modest annotation of these datasets we caution against interpreting these data as evidence for the divergence of the sperm proteomes in these species.
Fig. 3.
Conservation of the crocodile sperm proteome and annotation of proteins not curated within the compiled human sperm proteome. A,, B,, Venn diagrams illustrating the conservation of crocodile sperm proteins with those reported for (A,) human (27) and (B,) rooster spermatozoa (28). C–E,, Gene Ontology (GO) analysis was performed to assess the functional classification of the 130 crocodile sperm proteins that are not currently annotated within the compiled human sperm proteome. For this purpose, proteins were curated on the basis of (C,) GO molecular function, (D,) GO biological process, and (E,) GO cellular compartment using the universal protein knowledgebase (UniProtKB) functional annotation tools (26).
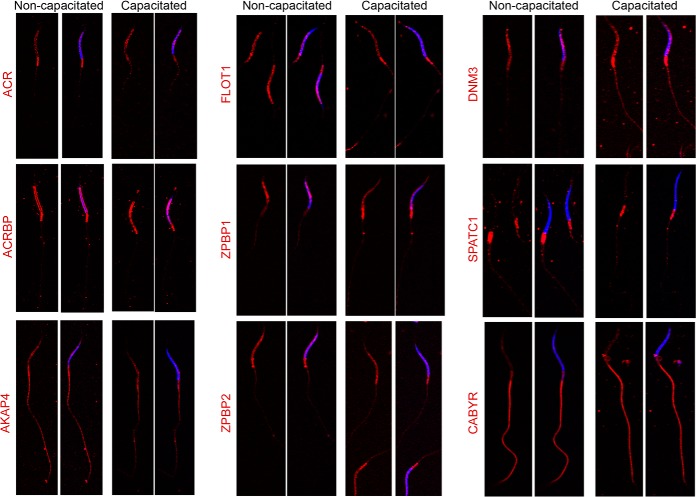
To validate these in silico, comparisons, nine candidate proteins with well characterized roles in the spermatozoa of eutherian species were assessed for their presence and localization in crocodile spermatozoa. This analysis confirmed the presence of each of the nine targeted proteins in crocodile spermatozoa and demonstrated labeling patterns consistent with those expected based on studies of eutherian spermatozoa [i.e., sperm head: acrosin (ACR), acrosin binding protein (ACRBP), zona pellucida binding protein 1 (ZPBP1) and 2 (ZPBP2), flotillin 1 (FLOT1), dynamin 3 (DNM3); sperm flagellum: protein kinase A anchoring protein 4 (AKAP4), calcium binding tyrosine phosphorylation regulated protein (CABYR), and spermatogenesis and centriole associated 1 (formerly speriolin; SPATC1)] (Fig. 4). Moreover, we failed to document any form of overt capacitation-associated change in protein abundance or distribution between subcellular domains (Fig. 4). One possible exception was that of acrosin, a highly conserved proteinase that dominates the profile of acrosomal proteins found in mammalian spermatozoa. Thus, although acrosin was present throughout the peri-nuclear domain of both noncapacitated and capacitated crocodile spermatozoa, this localization pattern was accompanied by a particularly intense foci of posterior head labeling in noncapacitated cells. The loss of this labeling in capacitated cells coincided with a 1.77-fold increase in acrosin abundance in the membrane-enriched sperm fraction, and a reciprocal 1.56-fold decrease in the soluble fraction (supplemental Table S1).
Fig. 4.
Validation of the conservation of crocodile sperm proteins. To validate our proteomic data, 9 proteins were targeted for assessment of their presence and localization in crocodile spermatozoa. These candidates included proteins known to reside in either the sperm head [acrosin (ACR), acrosin binding protein (ACRBP), flotillin 1 (FLOT1), zona pellucida binding protein 1 (ZPBP1) and 2 (ZPBP2), and dynamin 3 (DNM3)] or flagellum [protein kinase A anchoring protein 4 (AKAP4), calcium binding tyrosine phosphorylation regulated protein (CABYR), and spermatogenesis and centriole associated 1 (formerly speriolin; SPATC1)]. For this purpose, populations of noncapacitated and capacitated crocodile spermatozoa were fixed and permeabilized before labeling with appropriate antibodies (red). The cells were then counterstained with DAPI (blue) and examined by confocal microscopy. These experiments were replicated on independent samples from three different crocodiles, and representative labeling patterns are shown. Scale bar, 5 μm.
Analysis of Capacitation-associated Protein Phosphorylation of Crocodile Spermatozoa
In seeking to refine our analysis of proteins potentially involved in the regulation of crocodile sperm function, we performed phosphopeptide-enrichment in tandem with isobaric-tag based labeling to identify signatures of capacitation-associated signaling within these cells. Before performing this analysis, sperm lysates were subjected to immunolabeling with anti-phospho-serine, -threonine, -tyrosine, and -PKA antibodies, confirming that capacitation stimuli elicited equivalent responses in the spermatozoa of crocodiles used in this study (n, = 2; Fig. 5) and those we have reported in previous studies (n, = 5; (10)). Importantly, this consistency also extended to the relative abundance of phosphorylated peptides detected in each biological replicate (supplemental Table S3).
Fig. 5.
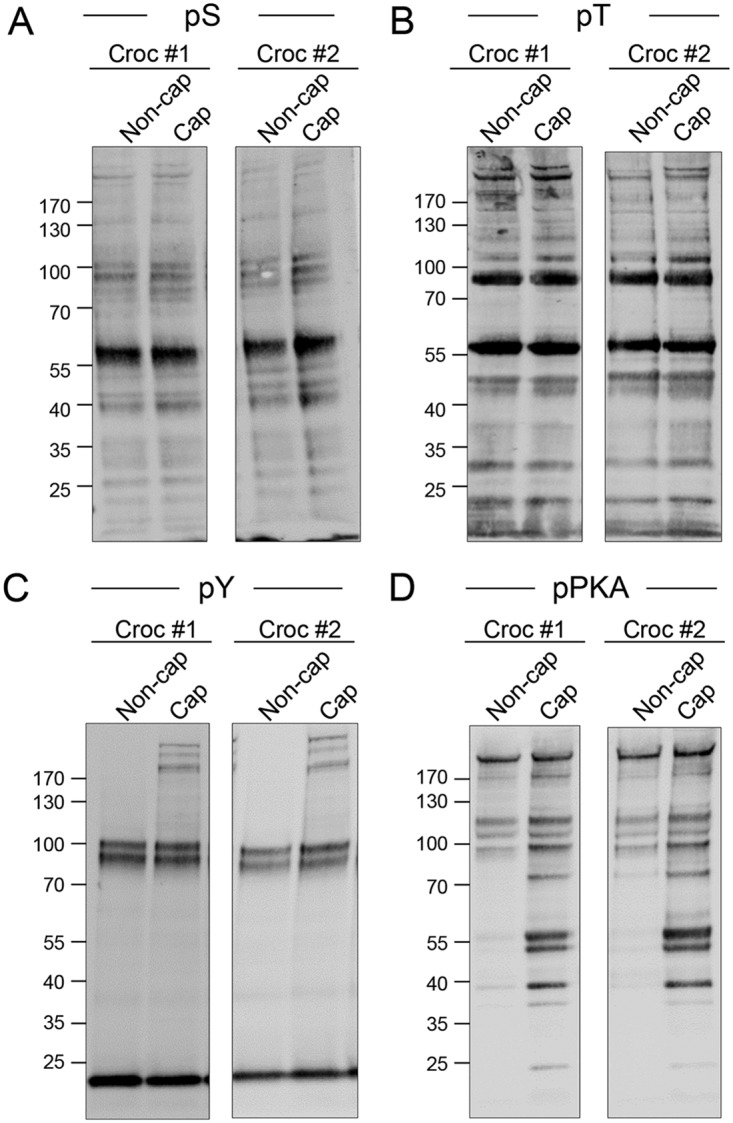
Assessment of equivalent physiological response to capacitation stimuli. To confirm that capacitation stimuli elicited equivalent responses in the spermatozoa of both crocodiles used in the phospho-enrichment aspect of this study, cell lysates were prepared from noncapacitated (Non-cap) and capacitated (Cap) populations of crocodile spermatozoa. Lysates were resolved by SDS-PAGE before being subjected to immunoblotting with (A,) anti-phosphoserine (pS), (B,) anti-phosphothreonine (pT), (C,) anti-phosphotyrosine (PY) or (D,) anti-phospho-PKA substrate (pPKA) antibodies.
Using a threshold of ± 1.2 fold-change, we identified 174/269 (∼65%; supplemental Table S3) phosphopeptides that experienced differential phosphorylation in capacitated versus, noncapacitated spermatozoa (Table I). Among these peptides, 31 were characterized by the presence of multiple phosphorylation sites and similarly, several additional candidates mapped to proteins possessing multiple phosphopeptides. Thus, 22 proteins were identified as being targeted for multiple (de)phosphorylation events, with prominent examples including: fibrous sheath CABYR protein, centrosomal protein of isoform B, phosphodiesterase, outer dense fiber 2 and sulfotransferase family cytosolic 1B member 1 isoform C, each with as many as 17, 6, 5, 3, and 3 differentially phosphorylated residues, respectively (Table I). Taking these events into consideration, a total of 126 unique proteins were identified as being differentially phosphorylated in capacitated versus, noncapacitated spermatozoa (Table I).
Table I. Differential phosphorylation of peptides in capacitated versus non-capacitated crocodile spermatozoa.
| Change (peptides) | Phosphorylated peptide | Phosphorylated residue(s) | Protein name | Uniprot ID | Fold change (Cap/Non-cap) |
|
|---|---|---|---|---|---|---|
| Membrane-enriched | Soluble | |||||
| MEMBRANE-ENRICHED ONLY | ||||||
| Increased (19) | [R].DSHFEKAGTSQSSR.[S] | pS | A-kinase anchor protein 4 | A0A151M6A1 | 1.87 | – |
| [R].SEHALQSEQR.[K] | pS | Coiled-coil domain-containing protein 40 | A0A151NKJ4 | 1.71 | – | |
| [K].AQQTYSLISLGGETWINR.[R] | 2 × pS | Fibrous sheath CABYR-binding protein | A0A151PDZ5 | 1.28 | – | |
| [R].RGSQQPTGQESR.[R] | pS | Fibrous sheath CABYR-binding protein | A0A151PDZ5 | 2.02 | – | |
| [R].RTGKVMASHSQQTR.[E] | pS | Fibrous sheath CABYR-binding protein | A0A151PDZ5 | 2.49 | – | |
| [R].SEEYAEQLTVQLAEKDSYVAEALSTLESWR.[S] | pS | Outer dense fiber protein 2 (Fragment) | A0A087RGV2 | 2.99 | – | |
| [K].TLTDVVSPGPCYFVDPHVSR.[F] | pS | Outer dense fiber protein 3-like protein 1 | A0A151M9U6 | 1.79 | – | |
| [K].STGQNDGDEPQSAEHIESR.[T] | pS | Perilipin | A0A091HQZ3 | 1.29 | – | |
| [R].RGSADQGYPAR.[R] | pS | Phosphatidylinositol 4,5-bisphosphate 5-phosphatase A isoform A | A0A151NDF4 | 1.66 | – | |
| [K].DWEDDSDEDLSNFDR.[F] | pS | Prostaglandin E synthase 3 | A0A151PBS0 | 1.52 | – | |
| [R].RASVCAEAYNPDEEEDDAESR.[I] | pS | Protein kinase cAMP-dependent type II regulatory subunit beta | H0YY13 | 1.31 | – | |
| [R].RNSSISGTATPR.[H] | pS | RNA-binding protein MEX3D | A0A151NGQ2 | 1.37 | – | |
| [R].SRSPSPLPLR.[S] | pS | Spermatogenesis associated 18 | U3I5S0 | 1.36 | – | |
| [R].SRSPSPLPLR.[S] | pS | Spermatogenesis associated 18 | U3I5S0 | 1.26 | – | |
| [R].LQAEAPVQTSGFDLLERLSPLSQTESQTQR.[L] | pT/S | Sulfotransferase family cytosolic 1B member 1 isoform C | A0A151MJ41 | 1.38 | – | |
| [R].NIYNPPESNASVIMDYNEEGQLLQTAFLGTSR.[R] | pS + pY | Teneurin transmembrane protein 3 | U3KAS7 | 1.40 | – | |
| [K].RAHTPTPGIYMGRPTHSGGGGGGGAGR.[R] | pT | Transformer 2 alpha homolog | F1NPM7 | 1.22 | – | |
| [R].RRSPSPYYSR.[GY] | pS | Transformer 2 alpha homolog | F1NPM7 | 1.40 | – | |
| [R].QASTDAGTAGALTPQHVR.[A] | pS | Yorkie isoform X1 | A0A0Q3MCH6 | 1.21 | – | |
| Decreased (28) | [K].CKSQFTITPGSEQIR.[A] | pS | Aconitate hydratase, mitochondrial (Aconitase) | A0A093TJW6 | –1.35 | – |
| [K].EVNQQSSNRNEFCHER.[G] | pS | A-kinase anchor protein 7 | A0A151P6K4 | –1.32 | – | |
| [R].SLAEASGSRPAGTR.[N] | pS | A-kinase anchor protein 9 | A0A151M6A1 | –1.30 | – | |
| [K].AKTSPDAFIQLALQLAHYR.[D] | pT/S/Y | Carnitine palmitoyltransferase 1A | U3J8S7 | –1.31 | – | |
| [R].TPEELDDSDFETEDFDVR.[S] | pS | Catenin alpha-2 isoform A | A0A151NQU8 | –1.20 | – | |
| [R].SLSPSPPPSR.[Y] | pS | Centrosomal protein of | A0A151N2J7 | –1.22 | – | |
| [R].SSSPLSSTLRSPSHSPER.[A] | pS | Centrosomal protein of isoform B | A0A151PAH0 | –1.28 | – | |
| [R].RLQEQLLNVASEDDITTSR.[K] | pS | Centrosome and spindle pole-associated protein 1-like | A0A151MN72 | –1.30 | – | |
| [K].QGISNTVPSGELLPSPGVLR.[L] | pS | Cilia-and flagella-associated protein 57 | A0A151MKT8 | –1.61 | – | |
| [R].ALAAMEQDSPVQR.[I] | pS | Cilia-and flagella-associated protein 58 | A0A151NVA7 | –1.45 | – | |
| [R].QDPSPTVSSEGVGAR.[A] | pS | Diphthine methyltransferase isoform B | A0A151MF01 | –1.25 | – | |
| [R].SAATSGAGSTTSGVVSGSLGSR.[E] | pS | E3 ubiquitin-protein ligase HUWE2 | A0A151NCW7 | –1.47 | – | |
| [R].GSGTASDDEFENLR.[I] | pS | E3 ubiquitin-protein ligase HUWE3 | A0A151NCW7 | –1.46 | – | |
| [K].AQQTYSLISLGGETWINRR.[T] | pS | Fibrous sheath CABYR-binding protein | A0A151PDZ5 | –1.23 | – | |
| [K].VMASHSQQTRESWIQEFR.[V] | pS/T | Fibrous sheath CABYR-binding protein | A0A151PDZ5 | –1.33 | – | |
| [K].AEEPAPVPVEKPPEPAMSELTVGINGFGRIGR.[L] | pT | Glyceraldehyde-3-phosphate dehydrogenase | A0A151M7J4 | –1.22 | – | |
| [R].QDSFPDENHLSR.[K] | pS | Intraflagellar transport 43-like protein isoform B | A0A151MLI4 | –1.41 | – | |
| [R].ADSSDTDLEIEDAER.[S] | pS | Leucine-rich repeat-containing protein 74A isoform A | A0A151MLE8 | –1.27 | – | |
| [K].APASPPPTSGLWTTQR.[D] | pS | Long-chain-fatty-acid–CoA ligase ACSBG1 | A0A151NWJ6 | –1.31 | – | |
| [R].VDHGAEIITQSPGR.[S] | pS | Microtubule-associated protein | A0A151N9F1 | –1.23 | – | |
| [R].IRTPEQIPSPVNTYLTEEDLFHR.[K] | pT + pS | MYCBP-associated protein isoform B | A0A151N2T7 | –1.79 | – | |
| [R].EVDNLTLTPSDSQDDVR.[S] | pS | Pericentrin isoform A | A0A151NAF1 | –1.43 | – | |
| [K].TSPETSGIFSGEDFPIIR.[F] | pT/S | Regulator of G-protein signaling protein-like isoform B | A0A151MIB2 | –1.46 | – | |
| [R].SASGLLEGLSPLVSEQDLSTIQPLIR.[Y] | pS + pS/T | TAO kinase 2 (Fragment) | A0A151NQE0 | –1.40 | – | |
| [K].TQYASAESQR.[S] | pS | Tektin-1 | A0A151N5N4 | –1.38 | – | |
| [R].SASHQIR.[Q] | pS | Tektin-2 isoform B | A0A151PGB9 | –2.87 | – | |
| [R].SSGASSSSLNLIR.[W] | pS | Testis anion transporter 1 (Solute carrier family 26 member 8) | A0A151N8J2 | –1.40 | – | |
| [K].RLQYVQSELR.[L] | pS | Uncharacterized protein | A0A151P2B6 | –1.67 | – | |
| SOLUBLE ONLY | ||||||
| Increased (33) | [K].QDAANLYHHK.[H] | pY | Axin-1 | A0A093PEG4 | – | 5.58 |
| [R].RTSMGGTQQQFVEGVR.[M] | pS | Catenin beta 1 | G1NGP2 | – | 1.37 | |
| [R].RHGLSTSSLR.[AD] | pS | Centrosomal protein of 135 kDa (Fragment) | A0A091F4F0 | – | 1.33 | |
| [R].SSSPLSSTLRSPSHSPER.[A] | pS | Centrosomal protein of isoform B | A0A151PAH0 | – | 1.54 | |
| [R].RASPRPSSSISFRPAAER.[A] | pS | Coiled-coil domain-containing protein 136-like | A0A151M3U3 | – | 1.34 | |
| [R].HADHGALTLGSGSATTR.[L] | pT/S | E3 ubiquitin-protein ligase HUWE1 | A0A151NCW7 | – | 1.26 | |
| [R].SAATSGAG[-TTSGVVSGSLGSR.[E] | pS | E3 ubiquitin-protein ligase HUWE2 | A0A151NCW7 | – | 1.31 | |
| [K].GQQTSLIWSR.[KQ] | pS | Fibrous sheath CABYR-binding protein | A0A151PDZ5 | – | 1.20 | |
| [K].VMASHSQQTRESWIQEFR.[V] | pS | Fibrous sheath CABYR-binding protein | A0A151PDZ5 | – | 3.91 | |
| [K].VMASHSQQTRESWIQEFR.[V] | pS/T | Fibrous sheath CABYR-binding protein | A0A151PDZ5 | – | 1.94 | |
| [K].FGNLQIEESR.[R] | pS | Fibrous sheath-interacting protein 3 | A0A151MNU1 | – | 1.66 | |
| [R].EINQSETNVTNEIIR.[T] | pS | Fibrous sheath-interacting protein 4/5 | A0A151MNU1 | – | 2.35 | |
| [R].GEPNVSYICSR.[Y] | pY | Glycogen synthase kinase 3 beta | A0A151MD62 | – | 1.48 | |
| [RK].NAEPEQSHSNTSTLTER.[E] | pS | GTPase-activating Rap/RanGAP domain-like 1 protein transcript variant 4 | G0ZS69 | – | 1.45 | |
| [K].LFPIGSSTSSIQGDHPQGR.[R] | pS | Lamin tail domain-containing protein 2 isoform A | A0A151NXE0 | – | 1.24 | |
| [R].GRYDSQVALR.[G] | pS | Myeloid leukemia factor 3 | A0A151NB71 | – | 1.26 | |
| [R].HGESAWNLENR.[F] | pS | Phosphoglycerate mutase (EC 5.4.2.11) (EC 5.4.2.4) | A0A0Q3PMK9 | – | 1.49 | |
| [R].THNGESVSYLFSHVPL.[-] | pS | Phosphoribosyl pyrophosphate synthetase 2 | U3I8U7 | – | 1.66 | |
| [R].RFSEGTSADR.[E] | pS | Proteasome 26S subunit, ATPase 6 | F1NCS8 | – | 1.50 | |
| [R].YHGHSMSDPGVSYR.[T] | pS | Pyruvate dehydrogenase E1 component subunit alpha | A0A151N056 | – | 1.55 | |
| [K].IGPQQPSGVAPGAGSR.[H] | pS | Ras-related protein Rab-2B | A0A151P513 | – | 1.46 | |
| [R].RNSAPVSVSAVR.[T] | pS | Rho GTPase-activating protein 31 | A0A0Q3MRE9 | – | 1.27 | |
| [K].SASSISLFASR.[E] | pS | Sperm-tail PG-rich repeat-containing protein 2 isoform C | A0A151NMF5 | – | 1.44 | |
| [R].SASGLLEGLSPLVSEQDLSTIQPLIR.[Y] | 2 × pS + pS/T | TAO kinase 2 (Fragment) | A0A151NQE0 | – | 2.45 | |
| [K].LHEVALNTGPDSSCGLATAGFR.[T] | pS | Tektin-4 | A0A151N2D8 | – | 1.67 | |
| [K].GQSIHLLNGR.[K] | pS | Transmembrane protein 41A | A0A151NH62 | – | 1.55 | |
| [K].FWEVISDEHGIDIAGNYYGGASLQLER.[I] | pS | Tubulin beta chain | A0A0Q3U5V4 | – | 1.65 | |
| [K].FWEVISDEHGIDPSGNYVGDSDLQLER.[I] | pS/Y | Tubulin beta-4 chain (Beta-tubulin class-III) | P09652 | – | 1.20 | |
| [K].HAFSLHQLQNDIR.[I] | pS | Ubiquitinyl hydrolase 1 (EC 3.4.19.12) | A0A151MCT8 | – | 1.47 | |
| [R].NSFQNVLEPDITR.[V] | pS | Uncharacterized protein | A0A151P8I0 | – | 1.29 | |
| [R].VHSFLSSCLPHR.[K] | pS | Uncharacterized protein | A0A151MIN4 | – | 1.88 | |
| [K].STDESPYTPPSDSQR.[M] | pT | WD repeat-containing protein 17 isoform C | A0A151PB29 | – | 1.36 | |
| [R].DTVISLSDVQVR.[R] | pS | WD repeat-containing protein 78 | A0A151NJC1 | – | 1.42 | |
| Decreased (18) | [K].AQQTYSLISLGGETWINRR.[T] | 2 × pS | Fibrous sheath CABYR-binding protein | A0A151PDZ5 | – | −1.27 |
| [K].IESEGGLQLLQRIYQLRK.[D] | pY | Protein SERAC1 (Fragment) | A0A091W9Y3 | – | −1.27 | |
| [K].SESMGNTSPRR.[S] | pS | Lamin tail domain-containing protein 2 isoform A | A0A151NXE0 | – | −2.71 | |
| [K].SETSAFGAPSQNSLGAVSNAETQR.[R] | 2 × pS | Phosphodiesterase (EC 3.1.4.-) | A0A151LZS8 | – | −1.20 | |
| [K].YISLCRSEHALQSEQR.[K] | pS/Y | Coiled-coil domain-containing protein 41 | A0A151NKJ4 | – | −1.33 | |
| [R].AGLSQLCDSSDEEQQDTQPGPR.[E] | pS | Nucleolar protein with MIF4G domain 1 | U3JWQ2 | – | −1.44 | |
| [R].EPSLHNVEELPPSR.[R] | pS | Fibroblast growth factor (FGF) | A0A151P6U2 | – | −1.35 | |
| [R].HMFYHDLQVRPEDHALLMSDPPLSPTTNR.[E] | 2 × pS | Actin-like protein 9 | A0A151NXK5 | – | −1.28 | |
| [R].LQYVQSELR.[L] | pS | Uncharacterized protein | A0A151P2B6 | – | −1.23 | |
| [R].LSPLSQTESQTQR.[L] | pS | Sulfotransferase family cytosolic 1B member 1 isoform C | A0A151MJ41 | – | −1.32 | |
| [R].QVQDTQQLLER.[A] | pT | Laminin subunit gamma-2 (Fragment) | A0A091Q153 | – | −1.91 | |
| [R].SQSSAQFLSGDQEPWAFR.[G] | pS | Meningioma expressed antigen 5 (hyaluronidase) | H0ZI42 | – | −1.22 | |
| [R].SSGASSSSLNLIR.[W] | pS | Testis anion transporter 1 (Solute carrier family 26 member 8) | A0A151N8J2 | – | −1.33 | |
| [R].VISQEAIGLQSR.[H] | 2 × pS | Fibrous sheath CABYR-binding protein | A0A151PDZ5 | – | −1.43 | |
| [R].VISQEAIGLQSR.[H] | pS | Fibrous sheath CABYR-binding protein | A0A151PDZ5 | – | −1.59 | |
| [R].YDSQVALR.[G] | pS | Myeloid leukemia factor 6 | A0A151NB71 | – | −1.60 | |
| [R].YHGHSMSDPGISYR.[T] | pS | Pyruvate dehydrogenase E1 component subunit alpha (EC 1.2.4.1) | A0A1D5PEH3 | – | −1.63 | |
| [R].TGKVMASHSQQTR.[E] | pS | Fibrous sheath CABYR-binding protein | A0A151PDZ5 | – | −3.18 | |
| CHANGED IN BOTH MEMBRANE-ENRICHED AND SOLUBLE FRACTIONS | ||||||
| Increased in both fractions (25) | [K].DGSGQHVDVSPTSQR.[L] | pS | Aconitate hydratase, mitochondrial (Aconitase) (EC 4.2.1.-) | A0A1D5NWW1 | 1.31 | 1.28 |
| [R].ASSPGYIDSPTYSR.[Q] | pS | Actin binding LIM protein family member 3 | U3K2G6 | 1.40 | 1.53 | |
| [K].RNIQQYNSFVSLSV.[-] | pS | cAMP-dependent protein kinase type I-alpha regulatory subunit | R0L5G8 | 1.86 | 1.78 | |
| [K].CSPSGHLNTQPHYR.[L] | pS | Centrosomal protein of isoform B | A0A151P2B4 | 1.57 | 2.34 | |
| [R].RLYGGSQSSR.[K] | pS | Doublecortin domain-containing protein 2C isoform A | A0A151N849 | 2.94 | 1.73 | |
| [R].RSPPPSPSTQR.[R] | pS | Dynamin 3 | A0A151NTM6 | 1.73 | 1.23 | |
| [R].SHHAAGAAPAPTPAAR.[A] | pS | E3 ubiquitin-protein ligase HUWE1 | A0A151NPY6 | 2.06 | 1.20 | |
| [R].ASSTTPEHDATR.[S] | pS | EF-hand calcium-binding domain-containing protein 3 isoform C | A0A151PBR5 | 1.67 | 1.61 | |
| [K].RLSSIGAENTEENRR.[W] | pS | Fructose-bisphosphate aldolase (EC 4.1.2.13) | A0A151MLN4 | 1.32 | 1.31 | |
| [R].HCGGSHTITYPYR.[H] | pS | Glutamine-rich protein 2 | A0A151N352 | 1.39 | 1.61 | |
| [R].SASLVEESR.[I] | pS | Hydrocephalus-inducing protein-like protein isoform B | A0A151MSN2 | 1.21 | 1.23 | |
| [R].DGHSSVEDAR.[A] | pS | Interferon-stimulated exonuclease-like 2 | A0A151P910 | 1.28 | 1.30 | |
| [K].SESMGNTSPR.[R] | pS | Lamin tail domain-containing protein 2 isoform A | A0A151NXE0 | 1.91 | 1.49 | |
| [R].LLLKPHIQSXEDLQLILELLEK.[M] | pS | Malignant fibrous histiocytoma-amplified sequence 1 | A0A0Q3PZZ6 | 1.79 | 3.14 | |
| [R].TASNEHLTR.[A] | pS | MICOS complex subunit | A0A151NPV1 | 2.23 | 1.26 | |
| [R].LTNASRHAHLVAR.[F] | pS | Mitochondria-eating protein isoform A | A0A151PA77 | 1.34 | 1.30 | |
| [K].STDAQLQEEAAR.[T] | pS | Phosphodiesterase (EC 3.1.4.-) | A0A151LZS8 | 1.53 | 1.21 | |
| [K].SETSAFGAPSQNSLGAVSNAETQR.[R] | pS | Phosphodiesterase (EC 3.1.4.-) | A0A151LZS8 | 1.33 | 1.33 | |
| [K].RNSFGSCQDR.[N] | pS | Protein pitchfork | A0A151NBQ8 | 1.48 | 1.90 | |
| [K].EHLQTRTPEPVEGR.[K] | pT | Radial spoke head 3-like protein | A0A151M2H0 | 1.23 | 1.28 | |
| [R].NLGSINTELQDVQR.[I] | pS | SEC22 homolog B, vesicle trafficking protein (gene/pseudogene) | G1MZU9 | 1.59 | 2.43 | |
| [K].RASGQAFELILSPR.[S] | pS | Stathmin isoform A | A0A151MMG4 | 1.55 | 2.58 | |
| [K].RAHTPTPGIYMGRPTHSGGGGGGGAGR.[R] | 2 × pT | Transformer 2 alpha homolog | F1NPM7 | 1.27 | 1.40 | |
| [K].FWEVISDEHGIDIAGNYRGAAPLQLER.[I] | pY | Tubulin beta chain (Fragment) | A0A093GRG0 | 1.22 | 1.51 | |
| [R].HVLHDAY.[-] | pY | Uncharacterized protein | A0A151MFN7 | 1.28 | 1.59 | |
| Decreased in both fractions (9) | [R].RVSVCAEAFNPDEEEEDTEPR.[V] | pS | cAMP-dependent protein kinase type II-alpha regulatory subunit | A0A151MKS7 | −1.78 | −1.44 |
| [R].HGLSTSSLR.[AD] | pS | Centrosomal protein of 135 kDa (Fragment) | A0A091F4F0 | −1.69 | −1.42 | |
| [K].LNQAQQTDSNLSVYKR.[K] | 2 × pS | Fibrous sheath CABYR-binding protein | A0A151PDZ5 | −1.20 | −1.67 | |
| [K].LGLGIDEDEVTAEVLGAAAADEIPPLEGDEDTSR.[M] | pT/S | Heat shock cognate protein HSP 90-beta (Fragment) | A0A091RXG0 | −1.38 | −1.29 | |
| [RK].HVLIAEVIFTNIGGAATAVGDPPNVIIVSK.[QR] | pS | P protein | A0A151NX71 | −1.37 | −1.29 | |
| [R].YYSPGYSEALLER.[V] | pS | Sperm-associated antigen 6 | A0A151PCW8 | −1.35 | −1.52 | |
| [R].ATNELDQVPSPELGSEGIFYR.[H] | pS | Sperm-associated antigen 8 isoform A | A0A151MTE8 | −1.87 | −1.74 | |
| [R].EAVCGSPASARSAGNATVLAFSR.[C] | pS | WD repeat-containing protein 16 (Fragment) | A0A099YQN6 | −1.22 | −1.47 | |
| [R].RDTVISLSDVQVR.[R] | pS | WD repeat-containing protein 80/81 | A0A151NJC1 | −2.31 | −1.20 | |
| RECIPROCAL CHANGE IN MEMBRANE-ENRICHED AND SOLUBLE FRACTIONS | ||||||
| Increased in soluble and decreased in membrane-enriched (31) | [K].GYSVGDILQEVMR.[Y] | pS | A-kinase anchor protein 10/11 | A0A151M6A1; A0A151ND25 | −1.43 | 2.02 |
| [K].MAQNSDTSLK.[S] | pS | A-kinase anchor protein 5 | A0A151M6A1 | −1.42 | 1.93 | |
| [K].STEILEAMVR.[R] | pS | A-kinase anchor protein 8 | A0A151M6A1 | −1.22 | 1.41 | |
| [KR].NLQAVVQTPGGR.[KR] | pT | Centrosomal protein of 135 kDa (Fragment) | A0A091F4F0; A0A151PAH0 | −1.75 | 1.70 | |
| [K].GTSEVRVTTTVTTR.[G] | pT/S | Centrosomal protein of isoform B | A0A151P2B4 | −1.37 | 1.67 | |
| [R].LVLALDGGRSHDIISLESR.[S] | pS | Centrosomal protein of isoform B | A0A151PAH0 | −2.39 | 1.50 | |
| [R].SHDIISLESR.[S] | pS | Centrosomal protein of isoform B | A0A151PAH0 | −3.22 | 1.65 | |
| [R].DLILGNSETDQSR.[S] | pS | Coiled-coil domain-containing protein 63 | A0A151NSE3 | −2.04 | 2.19 | |
| [R].DKKYKPTWHCIVGR.[N] | pY | Dynein light chain 1, cytoplasmic (Fragment) | A0A094KGE2 | −1.20 | 1.42 | |
| [K].GQQTSLIWSR.[KQ] | pT/S | Fibrous sheath CABYR-binding protein | A0A151PDZ5 | −1.25 | 1.22 | |
| [K].VMASHSQQTRESWIQEFR.[V] | 2 × pS | Fibrous sheath CABYR-binding protein | A0A151PDZ5 | −1.30 | 1.38 | |
| [K].VMASHSQQTRESWIQEFR.[V] | pS | Fibrous sheath CABYR-binding protein | A0A151PDZ5 | −1.69 | 2.25 | |
| [K].LTQHIRSLEELR.[N] | pS | Golgin subfamily B member 1 isoform B | A0A151PCT3 | −1.52 | 1.94 | |
| [K].HTGPNSPDTANDGFVR.[L] | pS | Heterogeneous nuclear ribonucleoprotein H1-like protein | Q6WNG8 | −1.23 | 1.35 | |
| [K].GVHTAMSALSVAPTR.[A] | pS | MICOS complex subunit MIC13/14 | A0A151P0P1 | −1.34 | 1.35 | |
| [K].NFDELSINPDAHTFR.[S] | pS | Myeloid leukemia factor 5 | A0A151NB71 | −1.32 | 1.27 | |
| [K].EELNEVTQELAESEHENTLLR.[R] | pT/S | Outer dense fiber protein 2 | E1BSP2 | −1.93 | 1.25 | |
| [R].SEEYAQQLTVQLAEKDSYVAEALSTLESWR.[S] | pS/T/Y | Outer dense fiber protein 2 (Fragment) | A0A091SXF2 | −1.53 | 1.35 | |
| [K].HMTSSDINTLTR.[Q] | pS | Outer dense fiber protein 4 | A0A151MFS7 | −1.73 | 1.36 | |
| [K].MLDLETQLSR.[NIST] | pS | Outer dense fiber protein 5 | A0A151MFS7 | −2.40 | 1.20 | |
| [R].SIVHAVQAGIFVER.[M] | pS | Phosphodiesterase (EC 3.1.4.-) | A0A151LZS8 | −2.15 | 1.53 | |
| [R].YNHSHDQLVLTGSSDSR.[V] | pS | Protein TSSC1 | A0A151N7Y4 | −1.20 | 1.21 | |
| [R].ASSTTPEHDATR.[S] | pS | EF-hand calcium-binding domain-containing protein 3 isoform C | A0A151PBR5 | 1.67 | 1.61 | |
| [R].HDSETEDMLWDDLLHGPECR.[S] | pS | Putative homeodomain transcription factor 1 isoform B | A0A151NHA5 | −1.39 | 1.24 | |
| [R].DQGQGHIDTSPFSSVVR.[A] | pS | Septin-4/5/6 | A0A151NTM3 | −1.55 | 1.53 | |
| [R].AVGYAATAVTLSR.[L] | pS/T/Y | SH3 domain and tetratricopeptide repeat-containing protein 2 | A0A091IYK3 | −1.39 | 1.45 | |
| [K].HSLSSVLNR.[S] | pS | Spermatogenesis-associated protein 6 isoform A | A0A151M4J1 | −1.35 | 2.75 | |
| [R].RVEHPSPSGDLPTWCTPR.[E] | pS | Spermatogenesis-associated protein 6 isoform A | A0A151M4J1 | −1.38 | 1.36 | |
| [R].LSPLSQTESQTQR.[L] | 2 × pS | Sulfotransferase family cytosolic 1B member 1 isoform C | A0A151MJ41 | −2.15 | 1.46 | |
| [K].VQRAVGMLSNTTAIAEAWAR.[L] | pT + pS/T | Tubulin alpha chain (Fragment) | A0A093K505 | −1.26 | 1.23 | |
| [K].VLHEAGFNIEDSSSETNK.[S] | pS/T | Uncharacterized protein | A0A151NBB1 | −1.31 | 1.26 | |
| [K].VLLSHSNNLSNIR.[A] | pS | Uncharacterized protein | A0A151MFN7 | −1.44 | 1.51 | |
| Increased in membrane-enriched and decreased in soluble (10) | [R].SVELKTAKPIDPSKTDPTVLLFVESQYSQLGQDIIAILESSR.[F] | pS + pT | Bifunctional heparan sulfate N-deacetylase/N-sulfotransferase 4 | A0A091G6U2 | 2.35 | −2.46 |
| [R].IGHHSTSDDSSAYR.[S] | pS | Branched-chain alpha-keto acid dehydrogenase E1-alpha subunit | Q98UJ8 | 1.90 | −1.24 | |
| [R].RGSQQPTGQESR.[R] | pS | Fibrous sheath CABYR-binding protein | A0A151PDZ5 | 2.01 | −1.49 | |
| [R].HLGIDISPEGR.[A] | pS | Hydrocephalus-inducing protein-like protein isoform B | A0A151MSN2 | 1.30 | −1.21 | |
| [R].SRSMSPVLSR.[R] | pS | Mitochondria-eating protein isoform A | A0A151PA77 | 2.66 | −1.28 | |
| [R].IVSAQSLAEDDVE.[-] | pS | Mitochondrial import receptor subunit TOM20-like protein | A0A151M2X9 | 1.47 | −1.78 | |
| [KR].LLDTEDELSDIQSDSVPLEVR.[D] | pT | Phosphodiesterase (EC 3.1.4.-) | A0A151LZS8 | 2.00 | −1.31 | |
| [R].AKEHLQTRTPEPVEGR.[K] | pT | Radial spoke head 3-like protein | A0A151M2H0 | 1.76 | −1.46 | |
| [R].GFGSEEGSR.[A] | pS | RNA-binding protein 8A | A0A091EDJ7 | 1.60 | −1.20 | |
| [R].TTERPMSVRDSIQPGLGPR.[D] | pS | Speriolin | A0A151P497 | 2.00 | −1.23 | |
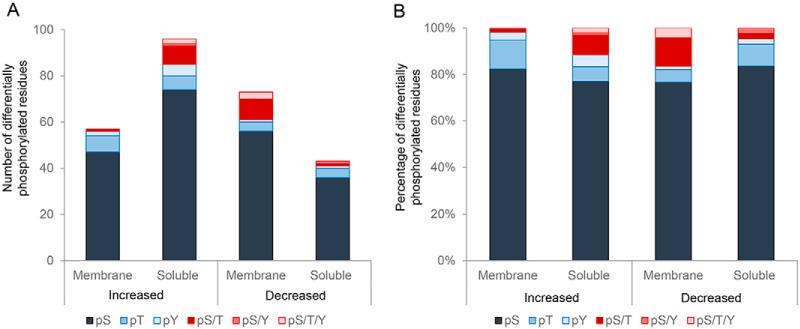
Among these phosphorylation events, a notable bias was detected for serine residues (Fig. 6A,, Table I), with this amino acid featuring as the target for ∼80% of all differentially phosphorylated sites (Fig. 6B,, Table I). Thereafter, threonine was the next most common phospho-target, with only relatively few tyrosine residues being identified as differentially phosphorylated in our analysis (Fig. 6).
Fig. 6.
Assessment of differentially phosphorylated peptides. The (A,) total number and (B,) proportion of phospho-serine (pS), -threonine (pT), and -tyrosine residues (pY) was determined among those peptides experiencing differential phosphorylation (i.e., ± 1.2 fold change) in populations of noncapacitated versus, capacitated crocodile spermatozoa. pS/T, pS/Y, pS/T/Y = ambiguous phosphorylation of either: a serine or threonine residue, a serine or tyrosine residue, or a serine, threonine, or tyrosine residue, respectively.
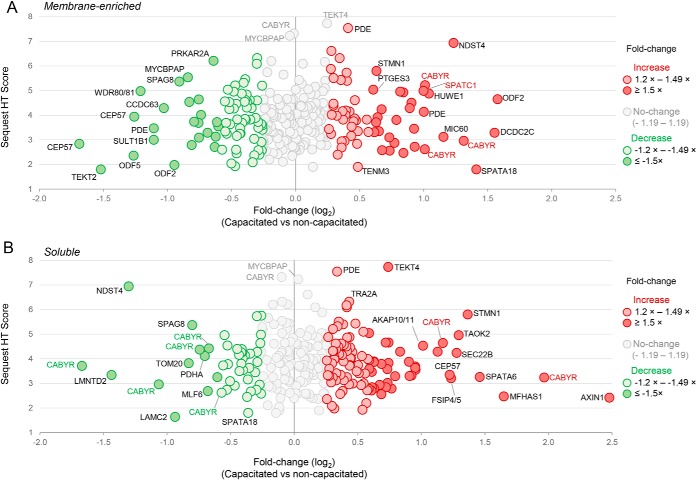
Overall, we noted a trend for proportionally more peptides experiencing increased, as opposed to reduced, phosphorylation in capacitated spermatozoa versus, that of their noncapacitated counterparts (77 versus, 55, respectively; Table I). The former of these capacitation-associated changes included several phosphopeptides that more than doubled their basal levels documented in noncapacitated spermatozoa (Fig. 7, Table I). Among the most dominant of these were peptides mapping to CABYR, a protein that was common to both membrane-enriched and soluble sperm fractions; and featured phosphopeptides that showed reduced phosphorylation in capacitated spermatozoa (Fig. 7, Table I). Alternatively, phosphopeptides mapping to proteins such as CEP57, TEKT2, ODF5, LMNTD2, and NDST1 were characterized by an apparent halving of their abundance in capacitated cells (Fig. 7, Table I).
Fig. 7.
Plots depicting fold changes associated with differentially phosphorylated peptides. Plots were constructed to demonstrate the fold change (x, axis; depicted as log2 fold change) and overall SequestHT Score (y, axis) of phosphorylated peptides in (A,) membrane-enriched and (B,) soluble lysates extracted from noncapacitated and capacitated crocodile spermatozoa. For the purpose of this analysis, a threshold of at least a ± 1.2 fold change in reporter ion intensity was implemented to identify differentially phosphorylated peptides. The identity of the parent protein from which a portion of the peptides originate has been annotated.
Although the quantitative changes documented among most of these peptides are likely attributed to genuine changes in phosphorylation status driven by capacitation-associated signaling cascades, we cannot equivocally conclude that all such changes are strictly tied to these events. Rather, some 41 phosphopeptides were identified that displayed reciprocal profiles of accumulation into different sub-cellular fractions depending on the capacitation status of the spermatozoa from which they were extracted (Table I), thus supporting our previous supposition that their parent proteins may have undergone capacitation-associated translocation between different sub-cellular domains.
Gene Ontology Analysis of Differentially Phosphorylated Proteins
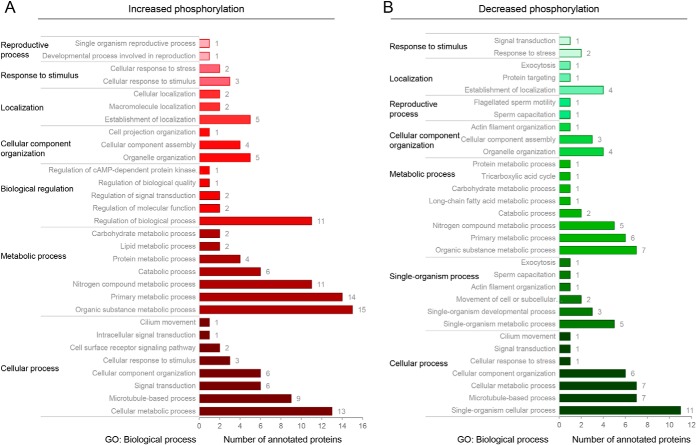
The majority of proteins that experienced increased phosphorylation during capacitation mapped to the dominant GO biological process categories of cellular process, metabolic process, and biological regulation (Fig. 8A,), with additional sub-categories of direct relevance to sperm capacitation including: cellular response to stimulus, regulation of signal transduction, cell surface receptor signaling pathway, regulation of cAMP-dependent protein kinase (i.e., PKA), cilium movement, and reproductive process (Fig. 8A,). Interestingly, the opposing subset of crocodile sperm proteins that experienced reduced phosphorylation following induction of capacitation mapped to similar biological process categories (i.e., cellular process, metabolic process, capacitation, flagellated sperm motility, response to stress, and signal transduction) suggesting that regulation of sperm activity is tightly coupled to the opposing action of cellular kinases and phosphatases.
Fig. 8.
GO annotation of crocodile sperm proteins harboring peptides that experienced differential capacitation-associated phosphorylation. Gene Ontology (GO) analysis was performed to assess the functional classification of all peptides that experienced a ± 1.2 fold change in reporter ion intensity in noncapacitated versus, capacitated crocodile spermatozoa. Proteins were curated on the basis of GO biological process using the universal protein knowledgebase (UniProtKB) functional annotation tools (26) and whether they underwent (A,) increased or (B,) decreased phosphorylation.
Validation of Phosphorylated Proteins
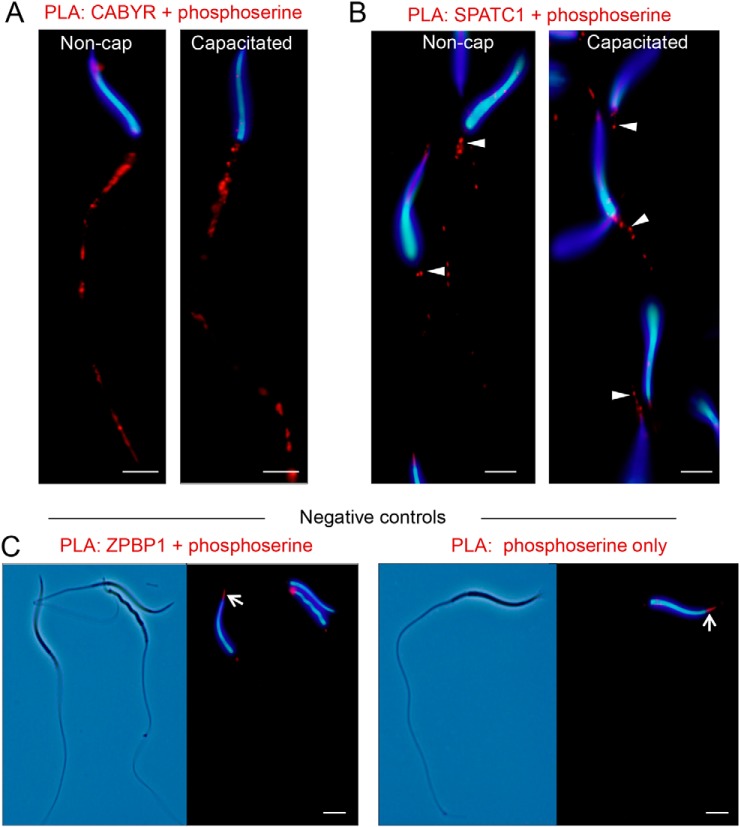
To begin to validate the phosphorylation of crocodile sperm proteins, we employed a proximity ligation assay with paired anti-phosphoserine and anti-CABYR or anti-SPATC1 antibodies. This strategy confirmed that CABYR was targeted for serine phosphorylation throughout the mid and principal-piece of the crocodile sperm flagellum (Fig. 9A,); cellular domains consistent with those that harbor the CABYR protein itself (Fig. 3). Equivalent results were also obtained for PLA labeling of phosphoserine/SPATC1 albeit restricted to the sperm neck coinciding with the distribution of this centriole associated protein. The specificity of PLA labeling was supported by the absence of fluorescence, save for a discrete foci of weak staining in the anterior region of the sperm head, in negative controls featuring the use of anti-phosphoserine antibodies alone or in combination with a nonphosphorylated target (i.e., ZPBP1).
Fig. 9.
Confirmation of CABYR and SPATC1 as targets for serine phosphorylation in crocodile spermatozoa. Proximity ligation assays (PLA) were employed to confirm that representative proteins, CABYR and SPATC1, were substrates for phosphorylation in noncapacitated (Non-cap) and capacitated crocodile spermatozoa. This assay results in the production of punctate red fluorescent signals when target antigens of interest, i.e., (A,) CABYR and phosphoserine residues or (B,) SPATC1 and phosphoserine residues reside within a maximum of 40 nm from each other. These experiments were replicated on independent samples from three different crocodiles, and representative PLA labeling patterns are shown. Arrowheads in (B,) indicate PLA labeling of the neck of the flagellum. C,, Negative controls included the labeling of spermatozoa with paired antibodies against phosphoserine and ZPBP1, a protein that was not identified as a substrate for serine phosphorylation, and the omission of one of the primary antibodies from the initial incubation (i.e., phosphoserine only). As anticipated, neither of these negative controls generated positive PLA labeling of the sperm flagellum. They did however, result in discrete, nonspecific PLA labeling in the anterior region of the sperm head (arrows). Scale bar, 5 μm.
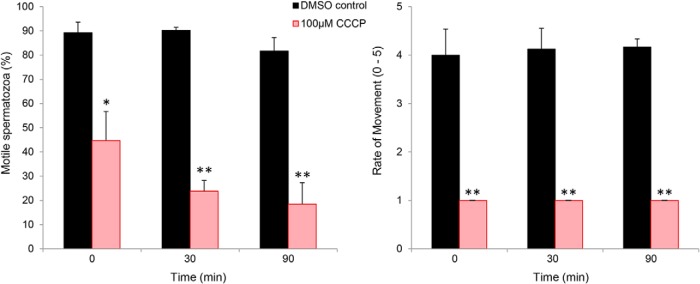
In view of the demonstration that a significant proportion of differentially phosphorylated proteins corresponded to those housed in the sperm flagellum and mapped to aerobic metabolic pathways (e.g., fructose-bisphosphate aldolase, glyceraldehyde-3-phosphate dehydrogenase, phosphoglycerate mutase, pyruvate dehydrogenase, aconitase, long-chain-fatty-acid-CoA ligase, carnitine palmitoyltransferase), we sought to determine whether oxidative phosphorylation does support the enhanced motility profile of crocodile spermatozoa that occurs in response to capacitation stimuli (10). For this purpose, populations of capacitating crocodile spermatozoa were co-incubated with carbonyl cyanide m-chlorophenyl hydrazone (CCCP), a chemical uncoupler of oxidative phosphorylation. As noted in Fig. 10, the application of CCCP lead to a significant decrease in sperm motility, which was manifest in the form of a reduced percentage of motile spermatozoa (Fig. 10A,) as well as a pronounced reduction in the overall rate of movement among these cells (Fig. 10B,). This suppression of sperm motility occurred independent of an attendant loss of vitality, which remained above 80% in all treatment groups. Furthermore, lipidomic profiling of noncapacitated versus, capacitated crocodile sperm membranes revealed a significant, 3-fold reduction in the abundance of palmitoleic acid (16:1, n,-9)(n, = 3; 2.33 ± 0.67 versus, 0.78 ± 0.50; p, < 0.02). Notably, the loss of palmitoleic acid appeared selective such that the levels of an additional 17 phospholipid fatty acid substrates remained essentially unchanged in response to capacitation (data not shown).
Fig. 10.
Assessment of the impact of the mitochondrial uncoupling agent, CCCP on crocodile sperm motility. To determine the contribution of oxidative phosphorylation to supporting the enhanced motility parameters elicited in response to capacitation stimuli, capacitating populations of crocodile spermatozoa were co-incubated CCCP (a chemical uncoupler of oxidative phosphorylation) or the appropriate DMSO vehicle control. Spermatozoa were sampled from each treatment group immediately after introduction of CCCP (t = 0) and at 30 and 60 min intervals and assessed for (A,) overall percentage of motile cells and (B,) the rate of sperm movement using criteria defined by Barth (49). These experiments were replicated on independent samples from three different crocodiles, and data are presented as mean ± S.E. * p, < 0.05, ** p, < 0.01.
DISCUSSION
It is well recognized the spermatozoa of all mammalian species only acquire functional maturity as they are conveyed through the male and female reproductive tracts. Despite decades of research however, the evolutionary origin, and adaptive advantage, of these elaborate forms of post-testicular maturation remain obscure. Here, we have exploited quantitative proteomics coupled with phosphopeptide-enrichment strategies to explore the crocodile sperm proteome and identify signatures of post-translational modification associated with the functional activation of these cells. Our data confirm that the phosphorylation status of the crocodile sperm proteome is substantially modified in response to stimuli formulated to elevate intracellular levels of the second messenger cAMP; thus supporting the necessity for capacitation-like changes in promoting the fertility of these cells. Moreover, we have established that the enhanced motility profile of capacitated crocodile spermatozoa is likely fueled by aerobic metabolism of selective membrane fatty acid substrates.
Although spermatozoa have now been successfully recovered from several reptilian species, systematic attempts to modulate the physiology of these cells are rare and global proteomic analyses are currently lacking. Thus, in completing the first comprehensive proteomic assessment of reptilian spermatozoa, we have been able to initiate comparative analyses with the curated proteomes from representative mammalian (human, (27)) and avian (rooster, (28)) spermatozoa with a view to furthering our understanding of this cell's complex biological machinery. These analyses confirmed the presence of some 84% of the identified crocodile spermatozoa proteins within the human sperm proteome; a level of conservation that suggests the core proteomic architecture of spermatozoa from these distantly related vertebrate species are broadly comparable. Working within the limitations imposed by incomplete coverage, functional classification of crocodile sperm proteins that are not currently annotated in the human sperm proteome revealed enrichment in the molecular function category of catalytic activity, and the biological process of metabolism. In addition, a substantial number of these proteins mapped to the membrane domain. Based on these data, we infer specialization of the surface, and possibly the metabolic characteristics, of crocodile spermatozoa.
The former explanation is consistent with evidence that the plasma membrane of crocodile spermatozoa displays exceptionally high tolerance to anisotonic osmotic stress (29). Indeed, crocodile spermatozoa retain high levels of plasma membrane integrity (>50%) during exposure to osmotic excursions of between 25–1523 mOsm kg−1 (29). Such characteristics are perhaps a physiological necessity owing to the potential for these cells to encounter dilution into fresh, or brackish, water following ejaculation into the cloacal chamber of the female. Alternatively, a high tolerance to anisotonic media could be linked to sperm storage in this species, as it appears to be in microbats (30). Although, the preservation of plasma membrane integrity in the face of extreme osmotic challenge undoubtedly reflects its lipid architecture, such properties may be augmented by the synergistic action of ion transport and drug efflux proteins in the lipid bilayer. An interesting example of one such protein is that of the testis anion transporter 1 (SLC26A8), an anion exchanger that mediates chloride, sulfate and oxalate transport and has been postulated to fulfil critical functions in the male germ line (31). Noteworthy in the context of our study, SLC26A8 has been implicated in the formation of a molecular complex involved in the regulation of chloride and bicarbonate ions fluxes during induction of sperm capacitation (32). Thus, an increased understanding of the functional relationships between the proteomic composition of the crocodile sperm membrane and their ability to survive osmotic excursions may ultimately help inform protocols to address the emerging need for the successful cryopreservation of crocodile spermatozoa (11).
Consistent with energy-production being a key attribute in the support of motility needed for spermatozoa to ascend the female reproductive tract and achieve fertilization, metabolic enzymes were identified as one of the dominant functional categories represented among the crocodile sperm proteome. Notably, enzymes mapping to glycolysis, oxidative phosphorylation and lipid metabolism were each highly enriched in the crocodile sperm proteome, with those of the latter category including proteins implicated in lipid catabolism, modification, and transport. Among these proteins we identified carnitine palmitoyl transferase 1 (CPT1A), an enzyme that catalyzes the rate-limiting reaction of beta-oxidation of fatty acids (33), and one that experienced among the highest fold changes (2.27 increase) of accumulation into the detergent labile (soluble) fraction of capacitated sperm lysates. From its position in the outer mitochondrial membrane, CPT1 catalyzes the formation of long-chain acylcarnitines from their respective CoA esters and thus commits them to β-oxidation within the mitochondrial matrix (33). It follows that pharmacological inhibition of CPT1A reduces flux through β-oxidation and, in species such as the horse, this manifests in the form of compromised sperm motility (34). The fact that this response occurs independently of any attendant loss of vitality, has been taken as evidence that stallion spermatozoa are able to effectively use endogenous fatty acids as an energy substrate to support motility (34). Although we have not yet had the opportunity to test this hypothesis directly in crocodile spermatozoa, we did secure several lines of correlative evidence that these cells use a similar metabolic strategy. Thus, capacitated crocodile spermatozoa experienced a selective depletion of palmitoleic acid [a monounsaturated fatty acid substrate known to enhance the motility profile of spermatozoa from species as diverse as sheep and fowls (35, 36)], as well as a significant reduction in motility following the uncoupling of oxidative phosphorylation. Moreover, we identified CPT1A as a substrate for differential phosphorylation in noncapacitated versus, capacitated crocodile spermatozoa. This finding takes on added significance in view of the demonstration that CPT1 catalytic activity can be selectively modulated by a mechanism of cAMP-dependent phosphorylation/dephosphorylation in somatic cells (37–39). It is therefore conceivable that the differential phosphorylation of CPT1A witnessed in crocodile spermatozoa may serve as a physiological switch to divert their metabolism either toward, or away from, fatty acid oxidation. Because fatty acid metabolism is conducive to long-term sustained release of energy, this strategy could assist with prolonging in vivo, sperm storage before ovulation (40, 41), while also proving advantageous in the context of enabling crocodile spermatozoa to negotiate the many meters of female reproductive tract before arriving at the site of fertilization (41).
Beyond its putative impact on CPT1A activity, elevation of intracellular cAMP also elicited the phosphorylation of numerous alternative substrates implicated in sperm motility initiation and maintenance. Notably, these proteins included peptides mapping to the alpha and beta regulatory subunits of protein kinase A (PKA), a promiscuous cAMP-dependent serine/threonine kinase. In eutherian spermatozoa, PKA is widely acknowledged as the central hub of the canonical capacitation cascade owing to its ability to integrate cAMP signaling with the downstream tyrosine kinase signaling pathways that underpin the functional activation of the cell (2). Consistent with data from our own immunolocalization studies (10), PKA primarily resides in the axoneme, a structure that forms an integral part of the motility apparatus of the sperm flagellum. Indeed, PKA is effectively anchored within this specific subcellular location by interaction between a docking domain present in the enzyme's regulatory subunit and that of scaffolding proteins of the protein kinase A anchoring protein (AKAP) family (42, 43); multiple members of which also displayed differential phosphorylation in capacitating crocodile spermatozoa (i.e., AKAP4, AKAP5, AKAP8, AKAP10/11). This sequestration of PKA ensures that the enzyme is juxtaposed with its relevant axonemal protein targets, while simultaneously segregating its activity to prevent indiscriminate phosphorylation of alternative substrates.
These data are entirely consistent with the demonstration that the bulk of the crocodile sperm proteins identified as undergoing differential capacitation-associated phosphorylation were those harbored within the sperm flagellum; with prominent examples including fibrous sheath CABYR-binding protein, outer dense fiber proteins (ODF2, ODF3, ODF4, ODF5), cilia-and flagella-associated proteins (CFAP57, CFAP58), fibrous sheath-interacting proteins (FSIP3, FSIP4/5), microtubule-associated protein, tubulins (TUBA, TUBB), and dynein (DYNLL1). They also accord with our previous observations of a rapid and sustained increase in the rate of motility as being among the principle changes witnessed in capacitating crocodile spermatozoa (10). Although the conservation of phospho-substrates documented above suggests conservation of the core activation pathways employed by reptilian and eutherian spermatozoa, it is also apparent that downstream signaling events show some degree of divergence. Thus, unlike eutherian sperm capacitation in which tyrosine phosphorylation appears to exert overriding control (4), we identified comparatively few tyrosine phosphorylated peptides in capacitated crocodile spermatozoa. Such findings agree with our previous immunoblotting studies in which we also documented only relatively subtle changes in tyrosine phosphorylation status, save for a small subset of very high molecular weight proteins (10). With the increased resolution afforded by the MS strategy employed herein, we have now affirmed the identity of at least one of these proteins as dynein; a microtubule-dependent force-generating ATPase that plays a pivotal role in axonemal microtubule sliding and hence the propagation of sustained flagellum beating (44).
Although the identification of relatively few phosphotyrosine substrates represents a departure from the widely accepted models of eutherian sperm capacitation, our findings do more closely approximate those experienced in somatic cells wherein, phosphorylation of serine, threonine and tyrosine amino acids occurs at an estimated ratio of 1000:100:1 (45). In seeking to reconcile these apparently incongruous results, it is perhaps noteworthy that a subset of the serine/threonine substrates identified herein are instead regulated by tyrosine phosphorylation in the spermatozoa of mammalian species, thus raising the possibility of lineage specific expansion of the role of tyrosine kinases in the spermatozoa of higher vertebrates. Illustrative of this phenomenon, we identified the fibrous sheath calcium-binding tyrosine phosphorylation-regulated protein (CABYR) as comprising as many as 17 differentially phosphorylated peptides, not one of which features a phospho-tyrosine residue. As its name suggests, this represents a marked departure from the homologue characterized in mouse (46, 47) and human spermatozoa (48), the former of which harbors as many as seven potential tyrosine phosphorylation motifs that are subject to extensive phosphorylation during in vitro, capacitation (46). Characterization of the adaptive significance of such changes remains as an intriguing focus for future research.
In summary, we have exploited an advanced proteomic platform to improve our understanding of sperm biology in a model reptilian species, the Australian saltwater crocodile. Through the identification of recognized hallmarks of the capacitation cascade, our collective data affirm the hypothesis that crocodile sperm do engage a network of signaling pathways, centering on PKA activity, to promote their functional activation (10). In doing so, these data challenge the widely promulgated view that post-testicular sperm maturation is limited to the mammalian lineage.
DATA AVAILABILITY
The data set (supplemental Dataset S1) analyzed here have been deposited in the Mass Spectrometry Interactive Virtual Environment (MassIVE) database (Project ID: MassIVE MSV000082258), and are publicly accessible at: https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=8acd6725da734f6f89bbd64460d03686.
Supplementary Material
Acknowledgments
We thank the staff at Koorana Crocodile Farm, and in particular John Lever and Robbie McLeod, for assistance with collection of crocodile semen. We are also grateful for the technical assistance of Tamara Keeley and Ed Qualischefski.
Footnotes
 This article contains supplemental Tables.
This article contains supplemental Tables.
1 The abbreviations used are:
- BWW
- Biggers, Whitten, and Whittingham medium
- ACR
- Acrosin
- AKAP4
- A-kinase anchoring protein 4
- CABYR
- Calcium binding tyrosine phosphorylation regulated protein
- cAMP
- cyclic adenosine monophosphate
- CCCP
- carbonyl cyanide m-chlorophenyl hydrazone
- DDA
- data dependent acquisition
- DNM
- Dynamin
- FDR
- false discovery rate
- FITC
- Fluorescein isothiocyanate
- FLOT1
- Flotillin 1
- GO
- gene ontology
- HILIC
- hydrophilic interaction chromatography
- HRP
- Horseradish peroxidase
- LC-MS/MS
- liquid chromatography-tandem mass spectrometry
- PKA
- protein kinase A
- PLA
- proximity ligation assay
- PSA
- Pisum sativum, agglutinin
- Ser
- Serine
- SPATC1
- Spermatogenesis and centriole associated 1
- Thr
- Threonine
- Tyr
- Tyrosine
- TEAB
- ttriethylammonium bicarbonate
- ZPBP1
- Zona pellucida binding protein 1
- ZPBP2
- Zona pellucida binding protein 2.
REFERENCES
- 1. Zhou W., De Iuliis G. N., Dun M. D., and Nixon B. (2018) Characteristics of the epididymal luminal environment responsible for sperm maturation and storage. Front. Endocrinol. 9, 59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aitken R. J., and Nixon B. (2013) Sperm capacitation: a distant landscape glimpsed but unexplored. Mol. Hum. Reprod. 19, 785–793 [DOI] [PubMed] [Google Scholar]
- 3. Baker M. A., Nixon B., Naumovski N., and Aitken R. J. (2012) Proteomic insights into the maturation and capacitation of mammalian spermatozoa. Syst. Biol. Reprod. Med. 58, 211–217 [DOI] [PubMed] [Google Scholar]
- 4. Gervasi M. G., and Visconti P. E. (2016) Chang's meaning of capacitation: A molecular perspective. Mol. Reprod. Dev. 83, 860–874 [DOI] [PubMed] [Google Scholar]
- 5. Stival C., Puga Molina Ldel C., Paudel B., Buffone M. G., Visconti P. E., and Krapf D. (2016) Sperm capacitation and acrosome reaction in mammalian sperm. Adv. Anat. Embryol. Cell Biol. 220, 93–106 [DOI] [PubMed] [Google Scholar]
- 6. Howarth B., Jr. (1970) An examination for sperm capacitation in the fowl. Biol. Reprod. 3, 338–341 [DOI] [PubMed] [Google Scholar]
- 7. Howarth B., Jr. (1983) Fertilizing ability of cock spermatozoa from the testis epididymis and vas deferens following intramagnal insemination. Biol. Reprod. 28, 586–590 [DOI] [PubMed] [Google Scholar]
- 8. Howarth B. Jr., and Palmer M. B. (1972) An examination of the need for sperm capacitation in the turkey, Meleagris gallopavo,. J. Reprod. Fertil. 28, 443–445 [DOI] [PubMed] [Google Scholar]
- 9. Nixon B., Ewen K. A., Krivanek K. M., Clulow J., Kidd G., Ecroyd H., and Jones R. C. (2014) Post-testicular sperm maturation and identification of an epididymal protein in the Japanese quail (Coturnix coturnix japonica,). Reproduction 147, 265–277 [DOI] [PubMed] [Google Scholar]
- 10. Nixon B., Anderson A. L., Smith N. D., McLeod R., and Johnston S. D. (2016) The Australian saltwater crocodile (Crocodylus porosus) provides evidence that the capacitation of spermatozoa may extend beyond the mammalian lineage. Proc. Biol. Sci. 283, 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Johnston S. D., Qualischefski E., Cooper J., McLeod R., Lever J., Nixon B., Anderson A. L., Hobbs R., Gosalvez J., Lopez-Fernandez C., and Keeley T. (2017) Cryopreservation of saltwater crocodile (Crocodylus porosus) spermatozoa. Reprod. Fertil. Dev. 29, 2235–2244 [DOI] [PubMed] [Google Scholar]
- 12. Johnston S. D., Lever J., McLeod R., Oishi M., Qualischefski E., Omanga C., Leitner M., Price R., Barker L., Kamaue K., Gaughan J., and D'Occhio M. (2014) Semen collection and seminal characteristics of the Australian saltwater crocodile (Crocodylus porosus,). Aquaculture 422–423, 25–35 [Google Scholar]
- 13. Asquith K. L., Baleato R. M., McLaughlin E. A., Nixon B., and Aitken R. J. (2004) Tyrosine phosphorylation activates surface chaperones facilitating sperm-zona recognition. J. Cell Sci. 117, 3645–3657 [DOI] [PubMed] [Google Scholar]
- 14. Mitchell L. A., Nixon B., and Aitken R. J. (2007) Analysis of chaperone proteins associated with human spermatozoa during capacitation. Mol. Hum. Reprod. 13, 605–613 [DOI] [PubMed] [Google Scholar]
- 15. Biggers J. D., Whitten W. K., and Whittingham D. G. (1971) The culture of mouse embryos in vitro. In: Daniel J. C., ed. Methods in Mammalian Embryology, pp. 86–116, Freeman Press, San Francisco, CA [Google Scholar]
- 16. Fujiki Y., Hubbard A. L., Fowler S., and Lazarow P. B. (1982) Isolation of intracellular membranes by means of sodium carbonate treatment: application to endoplasmic reticulum. J. Cell Biol. 93, 97–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dun M. D., Chalkley R. J., Faulkner S., Keene S., Avery-Kiejda K. A., Scott R. J., Falkenby L. G., Cairns M. J., Larsen M. R., Bradshaw R. A., and Hondermarck H. (2015) Proteotranscriptomic Profiling of 231-BR Breast Cancer Cells: Identification of Potential Biomarkers and Therapeutic Targets for Brain Metastasis. Mol. Cell. Proteomics 14, 2316–2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Degryse S., de Bock C. E., Demeyer S., Govaerts I., Bornschein S., Verbeke D., Jacobs K., Binos S., Skerrett-Byrne D. A., Murray H. C., Verrills N. M., Van Vlierberghe P., Cools J., and Dun M. D. (2017) Mutant JAK3 phosphoproteomic profiling predicts synergism between JAK3 inhibitors and MEK/BCL2 inhibitors for the treatment of T-cell acute lymphoblastic leukemia. Leukemia 32, 788–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Larsen M. R., Cordwell S. J., and Roepstorff P. (2002) Graphite powder as an alternative or supplement to reversed-phase material for desalting and concentration of peptide mixtures before matrix-assisted laser desorption/ionization-mass spectrometry. Proteomics 2, 1277–1287 [DOI] [PubMed] [Google Scholar]
- 20. Ross P. L., Huang Y. N., Marchese J. N., Williamson B., Parker K., Hattan S., Khainovski N., Pillai S., Dey S., Daniels S., Purkayastha S., Juhasz P., Martin S., Bartlet-Jones M., He F., Jacobson A., and Pappin D. J. (2004) Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169 [DOI] [PubMed] [Google Scholar]
- 21. Engholm-Keller K., Birck P., Storling J., Pociot F., Mandrup-Poulsen T., and Larsen M. R. (2012) TiSH–a robust and sensitive global phosphoproteomics strategy employing a combination of TiO2, SIMAC, and HILIC. J. Proteomics 75, 5749–5761 [DOI] [PubMed] [Google Scholar]
- 22. Laemmli U. K. (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685 [DOI] [PubMed] [Google Scholar]
- 23. Towbin H., Staehelin T., and Gordon J. (1979) Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc. Natl. Acad. Sci. U.S.A. 76, 4350–4354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dun M. D., Smith N. D., Baker M. A., Lin M., Aitken R. J., and Nixon B. (2011) The chaperonin containing TCP1 complex (CCT/TRiC) is involved in mediating sperm-oocyte interaction. J. Biol. Chem. 286, 36875–36887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Soderberg O., Gullberg M., Jarvius M., Ridderstrale K., Leuchowius K. J., Jarvius J., Wester K., Hydbring P., Bahram F., Larsson L. G., and Landegren U. (2006) Direct observation of individual endogenous protein complexes in situ, by proximity ligation. Nat. Methods 3, 995–1000 [DOI] [PubMed] [Google Scholar]
- 26. Consortium T. U. (2017) UniProt: the universal protein knowledgebase. Nucleic Acids Res. 45, D158–D169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Amaral A., Castillo J., Ramalho-Santos J., and Oliva R. (2014) The combined human sperm proteome: cellular pathways and implications for basic and clinical science. Hum. Reprod. Update 20, 40–62 [DOI] [PubMed] [Google Scholar]
- 28. Labas V., Grasseau I., Cahier K., Gargaros A., Harichaux G., Teixeira-Gomes A. P., Alves S., Bourin M., Gerard N., and Blesbois E. (2015) Qualitative and quantitative peptidomic and proteomic approaches to phenotyping chicken semen. J. Proteomics 112, 313–335 [DOI] [PubMed] [Google Scholar]
- 29. Johnston S. D., Lever J., McLeod R., Qualischefski E., Brabazon S., Walton S., and Collins S. N. (2014) Extension, osmotic tolerance and cryopreservation of saltwater crocodile (Crocodylus porosus,) spermatozoa. Aquaculture 426–427, 213–221 [Google Scholar]
- 30. Crichton E. G. (2000) Sperm storage and fertilization. In: Crichton E. G., and Krutzsch P. H., eds. Reproductive Biology of Bats, pp. 295–320, Academic Press, London [Google Scholar]
- 31. Toure A., Lhuillier P., Gossen J. A., Kuil C. W., Lhote D., Jegou B., Escalier D., and Gacon G. (2007) The testis anion transporter 1 (Slc26a8) is required for sperm terminal differentiation and male fertility in the mouse. Hum. Mol. Genet. 16, 1783–1793 [DOI] [PubMed] [Google Scholar]
- 32. Rode B., Dirami T., Bakouh N., Rizk-Rabin M., Norez C., Lhuillier P., Lores P., Jollivet M., Melin P., Zvetkova I., Bienvenu T., Becq F., Planelles G., Edelman A., Gacon G., and Toure A. (2012) The testis anion transporter TAT1 (SLC26A8) physically and functionally interacts with the cystic fibrosis transmembrane conductance regulator channel: a potential role during sperm capacitation. Hum. Mol. Genet. 21, 1287–1298 [DOI] [PubMed] [Google Scholar]
- 33. Lee K., Kerner J., and Hoppel C. L. (2011) Mitochondrial carnitine palmitoyltransferase 1a (CPT1a) is part of an outer membrane fatty acid transfer complex. J. Biol. Chem. 286, 25655–25662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Swegen A., Curry B. J., Gibb Z., Lambourne S. R., Smith N. D., and Aitken R. J. (2015) Investigation of the stallion sperm proteome by mass spectrometry. Reproduction 149, 235–244 [DOI] [PubMed] [Google Scholar]
- 35. Eslami M., Ghasemiyan H., and Zadeh Hashem E. (2017) Semen supplementation with palmitoleic acid promotes kinematics, microscopic and antioxidative parameters of ram spermatozoa during liquid storage. Reprod. Domest. Anim. 52, 49–59 [DOI] [PubMed] [Google Scholar]
- 36. Rad H. M., Eslami M., and Ghanie A. (2016) Palmitoleate enhances quality of rooster semen during chilled storage. Anim. Reprod. Sci. 165, 38–45 [DOI] [PubMed] [Google Scholar]
- 37. Harano Y., Kashiwagi A., Kojima H., Suzuki M., Hashimoto T., and Shigeta Y. (1985) Phosphorylation of carnitine palmitoyltransferase and activation by glucagon in isolated rat hepatocytes. FEBS Lett. 188, 267–272 [DOI] [PubMed] [Google Scholar]
- 38. Pegorier J. P., Garcia-Garcia M. V., Prip-Buus C., Duee P. H., Kohl C., and Girard J. (1989) Induction of ketogenesis and fatty acid oxidation by glucagon and cyclic AMP in cultured hepatocytes from rabbit fetuses. Evidence for a decreased sensitivity of carnitine palmitoyltransferase I to malonyl-CoA inhibition after glucagon or cyclic AMP treatment. Biochem. J. 264, 93–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guzman M., and Geelen M. J. (1992) Activity of carnitine palmitoyltransferase in mitochondrial outer membranes and peroxisomes in digitonin-permeabilized hepatocytes. Selective modulation of mitochondrial enzyme activity by okadaic acid. Biochem. J. 287 (Pt 2), 487–492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Davenport M. (1995) Evidence of possible sperm storage in the caiman, Paleosuchus palpebrosus,. Herpetol. Rev. 26, 14–15 [Google Scholar]
- 41. Gist D. H., Bagwill A., Lance V., Sever D. M., and Elsey R. M. (2008) Sperm storage in the oviduct of the American alligator. J Exp Zool A Ecol Genet Physiol 309, 581–587 [DOI] [PubMed] [Google Scholar]
- 42. Wong W., and Scott J. D. (2004) AKAP signalling complexes: focal points in space and time. Nat. Rev. Mol. Cell Biol. 5, 959–970 [DOI] [PubMed] [Google Scholar]
- 43. Luconi M., Carloni V., Marra F., Ferruzzi P., Forti G., and Baldi E. (2004) Increased phosphorylation of AKAP by inhibition of phosphatidylinositol 3-kinase enhances human sperm motility through tail recruitment of protein kinase A. J. Cell Sci. 117, 1235–1246 [DOI] [PubMed] [Google Scholar]
- 44. Loreng T. D., and Smith E. F. (2017) The Central Apparatus of Cilia and Eukaryotic Flagella. Cold Spring Harb. Perspect. Biol. 9, a028118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Raggiaschi R., Gotta S., and Terstappen G. C. (2005) Phosphoproteome analysis. Biosci. Rep. 25, 33–44 [DOI] [PubMed] [Google Scholar]
- 46. Naaby-Hansen S., Mandal A., Wolkowicz M. J., Sen B., Westbrook V. A., Shetty J., Coonrod S. A., Klotz K. L., Kim Y. H., Bush L. A., Flickinger C. J., and Herr J. C. (2002) CABYR, a novel calcium-binding tyrosine phosphorylation-regulated fibrous sheath protein involved in capacitation. Dev. Biol. 242, 236–254 [DOI] [PubMed] [Google Scholar]
- 47. Li Y. F., He W., Kim Y. H., Mandal A., Digilio L., Klotz K., Flickinger C. J., and Herr J. C. (2010) CABYR isoforms expressed in late steps of spermiogenesis bind with AKAPs and ropporin in mouse sperm fibrous sheath. Reprod. Biol. Endocrinol. 8, 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li Y. F., He W., Mandal A., Kim Y. H., Digilio L., Klotz K., Flickinger C. J., Herr J. C., and Herr J. C. (2011) CABYR binds to AKAP3 and Ropporin in the human sperm fibrous sheath. Asian J. Androl. 13, 266–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Barth A. D. (1995) Evaluation of frozen bovine semen by the veterinary practitioner. Proc. Bovine Short Course, pp. 105–110 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data set (supplemental Dataset S1) analyzed here have been deposited in the Mass Spectrometry Interactive Virtual Environment (MassIVE) database (Project ID: MassIVE MSV000082258), and are publicly accessible at: https://massive.ucsd.edu/ProteoSAFe/dataset.jsp?task=8acd6725da734f6f89bbd64460d03686.