ABSTRACT
Visceral leishmaniasis (VL) is a life threatening disease in which a variety of cytokines regulating the immune responses can determine its outcome. As based on their region in the gene, some single nucleotide polymorphisms (SNP) can influence the expression of their corresponding proteins, this study aimed to investigate the association between SNP in the IL-10, IL-12, IFN-γ genes and susceptibility to VL. The study was carried out on 120 patients with VL, 67 patients’ families (family group), and 102 healthy individuals with positive leishmanin skin test as positive control group. SNPs in IL-10 (−592, −819, −1082), IL-12 (+1188) were analyzed using PCR-RFLP and allele specific polymerase chain reaction (ASPCR) was used to analyze SNPs in IFN-γ (+874 A/T). The results showed that at position +874 of IFN-γ, AT genotype was significantly more frequent in patients than that in families and controls, but TT genotype was significantly more frequent in families than in patients. Distributions of IFN-γ alleles were not significantly different between the study groups. As for IL-12 and IL-10 genotypes and alleles, no significant difference was observed between the groups. Although a strong linkage disequilibrium was observed between alleles −592, −819 and −1082 of IL-10, distributions of the most common haplotypes and haplogenotypes reconstructed from IL-10 alleles were not significantly different between the study groups.
It could be suggested that heritage of AT genotype at position +874 of IFN-γ may predispose and TT genotype can resist individual to VL in an endemic area in the southwest of Iran.
KEYWORDS: Visceral leishmaniasis, IL-10, IL-12, IFN-γ
Introduction
Visceral leishmaniasis (VL), generally known as Kala azar, is characterized by irregular fever, enlargement of both the liver and the spleen (hepatosplenomegaly), lymphadenopathy, pancytopenia, progressive anemia, weight loss and hypergammaglobulinemia with hypo albunemia [1]. The incidence of VL has been evaluated at least 0.2–0.4 million worldwide [2] and around 100–300 cases were reported in Iran, annually [3]. Outcomes and manifestations of VL principally depends on the type of host immune responses in which T helper (h) 1 cytokines, inducing cell-mediated immunity (CMI), are protective and conversely Th2 secreted cytokines by promoting humoral responses, confer susceptibility to the disease. In this processes, cytokines act as the regulators of the immune responses and play an important role throughout leishmania infection. Among them, some cytokines such as gamma interferon (IFN-γ), interleukin (IL)-12 and IL-10 play a more significant role in determining the complications of VL. IL-12 is a critical mediator of cytotoxic activity of natural killer (NK) cells and it can drive Th0 cells to Th1 [4]. In addition, it has been shown that IL-12 stimulates the production of IFN-γ by T and NK cells, which consequently activates macrophages [4]. Activated macrophages produce nitric oxide (NO), a potent mediator of killing intracellular parasites [5]. In contrast, IL-10 is known as an anti-inflammatory cytokine produced by Th2 cells as well as other cells such as M2 macrophages while T regulatory cells are the most important cells that produce IL-10 cytokine. This cytokine inhibits the production of IL-12 and NO induced by IFN-γ and it can also down-regulate the effector mechanisms of Th1 [6]. Kane et al. revealed that the severity of VL is associated with up regulated level of IL-10 [7]. Maspi et al, reviewed pro-and anti-inflammatory cytokine cytokines in the immunoprotection and immunopathology of cutaneous leishmaniasis [8]. Previous studies showed that the production of cytokines could be influenced by the single nucleotide polymorphisms (SNPs) in their genes. Among them, Al-Bashier et al. observed that SNPs in IFN-γ (+874 T/A) but not IL-10 (−1082G/A) have an association with VL [9]. Furthermore, it was shown that C/T genotype rather than allele at position −819 of IL-10 was significantly more frequent in VL patients [10]. Also, another study revealed that genotypes and alleles of IL-12B (−1188) are not significantly different between VL patients and controls [11]. The data indicate that cytokine polymorphism in different populations may influence the susceptibility and/or resistance to infectious diseases, which served as the impetus for the present study which aimed to determine the predisposing effects of gene variants of the most critical cytokines such as IL-10, IL-12 and IFN-γ on VL.
Materials and methods
Study population
This study was carried out on 120 patients with VL (66 males (55%), mean age range 4.8 ± 11.6 years and 54 females (45%), mean age range 3.3 ± 3.4), 67 siblings of the patients’ families consisting of (45 males (67%), mean age range 28.3 ± 14.4 and 22 females (33%), mean age range 26.5 ± 14.1 years) who were roommates with the patients but with negative Leishmania skin test (LST) results and did not present any clinical manifestations of VL (family group). Meanwhile, 102 asymptomatic individuals (68 males (67%), mean age range 26.3 ± 14.7 years and 34 females (33%), mean age range 26.6 ± 14.6 years) who were living in the same endemic area (L. infantum is endemic) with a positive Leishmania skin test, were enrolled in the study as the positive control group. Leishmanin for the skin test was prepared from the Pasteur Institute of Iran. Of diluted solution, 0.1 mL was injected intradermally into the extensor surface of both forearms and after 48 h, induration more than 5 mm was accepted as positive leishmania skin test. All patients were registered at pediatric infectious disease ward of Namazi hospital, affiliated with Shiraz University of Medical Sciences, Shiraz, Iran. VL was diagnosed based on clinical manifestations (i.e. anemia, hepatosplenomegally and fever), and staining smear of bone marrow aspirates and/or indirect fluorescent antibody (IFA) ≥ 1/128 for Leishmania parasite. The study was carried out under the supervision of the Research Ethics Committee of Shiraz University of Medical Sciences. Three ml blood samples were taken from all the study groups following the obtained informed written consents from children’s guardians and DNA was extracted from all the blood samples using salting out method, as previously described [12].
Evaluation of snps in the cytokine genes
Polymerase chain reaction-restriction fragment length polymorphism (PCR-RFLP)
Gene variants in the IL-10, IL-12 were analyzed using PCR-RFLP method. PCR reactions were set up using sets of primers for −592 A/C, 819 T/C and −1082 A/G positions of IL-10 gene and +1188 A/C of IL-12. Each PCR cocktail contained 250 ng of genomic DNA, 1x PCR buffer, 0.5 μM specific primers set (Primm, Italy), 0.5 units Taq DNA polymerase (CinnaGen, Iran), 0.2 mM dNTPs mix (CinnaGen, Iran), and specific concentration of MgCl2 (Table 1). PCRs were performed by a thermocycler set (5530 Mastercylcler, Eppendorf, Germany) under the following thermal conditions: for IL-10, a denaturation step for 2 min at 94°C, followed by 35 cycles of a denaturation for 30 sec. at 94°C, annealing for 45 sec. at specific temperatures (Table 1), extension for 60 sec. at 72°C and finally an extension step for 10 min. at 72°C and for IL-12 the condition was: denaturation step at 95ºC for 3 min, followed by 35 cycles of a denaturation at 95ºC for 30 sec., annealing for 20 sec. at specific temperatures (Table 1), extension at 72ºC for 30 sec and final extension at 72ºC for 2 min. The products were digested by specific restriction enzymes (Fermentas, Lithuania) (Table 1). Finally, the digested products were separated on a 3% agarose NA gel (Amersham Bioscience AB, Sweden) and studied on UV transilluminator after being stained by ethidium bromide (CinnaGen- Iran). The sequences of the primers, annealing temperatures, MgCl2 concentrations, restriction enzymes and DNA product sizes after digestion are shown in Table 1.
Table 1.
The sequence of primers and PCR conditions for determining the SNPs in IL-10, IL-12 and IFN-γ.
| PCR primers | AT (°C)◊ | MgCl2 (mM) | RE○ | AS (bp) ☼ | |
|---|---|---|---|---|---|
| IL-10 (−592) | R: GGTGAGCACTACCTGACTAGC F: CCTAGGTCACAGTGACGTGG |
63 | 5.5 | RsaI | A:236 + 176 T:412 |
| IL-10 (−819) | F: TCATTCTATGTGCTGGAGATGG R: TGGGGGAAGTGGGTAAGAGT |
60 | 3.2 | MaeIII | C:125 + 84 T:209 |
| IL-10 (−1082) | F: CTCGCTGCAACCCAACTGGC R:TCTTACCTATCCCTACTTCC |
61 | 2 | MnI I | G:106 + 33 A:139 |
| IL-12(+1188) | F: TCAGACACATTAACCTTGCA R: ACCTGCCAATGACCACATTA |
55 | 2 | Taq I | A:300 C:166 + 134 |
| IFN – γ (+874) | Antisense: TCAACAAAGCTGATACTCCA Sense+874T: TTCTTACAACACAAAATCAAATCT Sense+874A: TTCTTACAACACAAAATCAAATCA |
62 | 3.5 | – | 264 |
| BG● | F: ACACAACTGTGTTCACTAGC R: CAACTTCATCCACGTTCACC |
62 | 3.5 | – | 110 |
◊Annealing temperature, ○Restriction Enzyme, ☼Amplicon size, ●β-globin (Internal control)
The underlined letters represent difference between sense primers for determining IFN-γ SNP at position +874
Allele specific polymerase chain reaction (ASPCR)
Single nucleotide polymorphism at position +874 (A/T) IFN-γ gene was examined using ASPCR [13]. To check successful PCR amplification, human β-globin was used as an internal control. PCR reactions were set up using two specific sets of primers, which amplified a 264 bp sequence of IFN-γ gene and a 110 bp sequence of human β-globin gene. DNA was amplified in a 10 μl reaction. Each PCR cocktail contained 1x PCR buffer, 3 mM MgCl2, 200 µM dNTPs mix, 0.5 units Taq DNA polymerase (all from CinnaGen, Iran), 0.5 μM specific primers set of IFN-γ, 0.2 μM specific primer sets of internal control (Primm, Italy) and 250 ng of genomic DNA. PCRs were performed under the following thermal conditions: an initial denaturation step for 6 min. at 98°C followed by 10 cycles of denaturation for 30 sec. at 96°C, annealing for 50 sec. at 60°C, extension for 40 sec. at 72°C and second step 20 cycles of denaturation at 96°C for 30 sec., annealing at 56°C for 40 sec. and extension at 72°C for 50 sec. The amplified products were monitored by electrophoresis on 2% agarose containing ethidium bromide.
Statistical analysis
Allele and genotype frequencies were estimated by direct gene counting. Data were analysed using Chi Square test with the level of significance set at less than 0.05 by EPI info 2000 and SPSS software version 20. Arlequin software package version 3.1 was used to estimate the haplotype and haplogenotype frequencies and Hardy–Weinberg equilibrium. Furthermore, linkage disequilibrium (LD) measures, P value and D’, were estimated by LD2SNPing program V 2.0 (http://www.bio.kuas.edu.tw/LD2SNPing).
Results
All genotype SNPs were in Hardy Weinberg equilibrium. Allele and genotype frequencies of IL-10, IL-12 and IFN-γ in the above-mentioned study groups are demonstrated in Table 2. As shown, at position +874 of IFN-γ, the frequency of AT genotype was significantly higher in patient group, compared with that in family and control groups, and and in contrast, the frequency of TT genotype was significantly lower in patient, compared with that in family group. Analysis of allele and genotype frequencies in IL-10 and IL-12 was not significantly different between the studied groups.
Table 2.
Frequencies of alleles and genotypes of IL-10, IFN-γ and IL-12 in VL patients, their families and controls.
| Genotypes alleles | Patient n (%) | Family n (%) | Control N (%) | P1 | P2 | PS1 (%) | PS2 (%) |
|---|---|---|---|---|---|---|---|
|
IL-10 (−592) Genotypes |
|||||||
| AA | 11 (9.17) | 10 (14.92) | 11 (10.78) | 0.231 | 0.687 | 20 | 6 |
| AC | 45 (37.5) | 23 (34.33) | 32 (31.37) | 0.665 | 0.339 | 6 | 16 |
| CC | 64 (53.33) | 34 (50.75) | 59 (57.85) | 0.734 | 0.5 | 5 | 10 |
| Alleles | 0.395 | 0.733 | 13 | 5 | |||
| A | 67 (27.92) | 43 (32.09) | 54 (26.47) | ||||
| C | 173 (72.08) | 91 (67.91) | 150 (73.53) | ||||
|
IL-10 (−819) Genotypes |
|||||||
| TT | 11(9.17) | 10 (14.92) | 11(10.78) | 0.231 | 0.687 | 20 | 6 |
| TC | 45 (37.5) | 23 (34.33) | 32 (31.37) | 0.665 | 0.339 | 6 | 16 |
| CC | 64 (53.33) | 34 (50.75) | 59 (57.85) | 0.734 | 0.5 | 5 | 10 |
| Alleles | 0.395 | 0.733 | 13 | 5 | |||
| T | 67 (27.92) | 43 (32.09) | 54 (26.47) | ||||
| C | 173 (72.08) | 91 (67.91) | 150 (73.53) | ||||
|
IL-10(−1082) Genotypes |
|||||||
| AA | 60 (50.0) | 34 (50.75) | 50 (49.02) | 0.922 | 0.88 | 3 | 3 |
| AG | 44 (36.67) | 21 (31.34) | 33 (32.35) | 0.463 | 0.5 | 11 | 10 |
| GG | 16 (13.33) | 12 (17.91) | 19 (18.63) | 0.4 | 0.28 | 13 | 19 |
| Alleles | |||||||
| A | 164 (68.33) | 89 (66.42) | 113 (61.41) | 0.704 | 0.137 | 6 | 33 |
| G | 76 (31.67) | 45 (33.58) | 71 (38.59) | ||||
|
IFN-γ (+874) Genotypes |
|||||||
| AA | 20 (16.39) | 12 (19.05) | 23 (22.33) | 0.651 | 0.259 | 6 | 20 |
| AT | 75 (61.48) | 27 (42.86) | 48 (46.6) | 0.015 | 0.025 | 68 | 61 |
| TT | 27 (22.13) | 24 (38.09) | 32 (31.07) | 0.021 | 0.128 | 60 | 33 |
| Alleles | |||||||
| A | 115 (47.13) | 51 (40.48) | 94 (45.63) | 0.222 | 0.750 | 23 | 5 |
| T | 129 (52.87) | 75 (59.52) | 112 (54.37) | ||||
|
IL-12(+1188) Genotypes |
|||||||
| AA | 27 (22.1) | 12 (19) | 27(25) | 0.62 | 0.581 | 7 | 7 |
| AC | 39 (31.9) | 25 (39.7) | 44(41) | 0.29 | 0.15 | 18 | 29 |
| CC | 56 (46.0) | 26 (41.3) | 36 (34) | 0.54 | 0.059 | 9 | 45 |
| Alleles | |||||||
| A | 93 (38.1) | 49 (38.9) | 98 (45.8) | 0.88 | 0.096 | 4 | 38 |
| T | 151 (61.9) | 77 (61.1) | 116 (54.2) |
Each P value is the result of comparing a row with the corresponding sum of related rows.
P stands for P value indicating differences between patients and family (P1), patients and controls (P2).
PS stands for power study which evaluates the power of study comparing patients and family (PS1) as well as patients and control (PS2).
When IL-10 haplotype was reconstructed from its different alleles, a strong linkage was observed between their alleles (Figure 1). However, the comparison of most frequent IL-10 haplotypes showed no significant difference between the studied groups (Table 3). The LD measures, D΄ and P value are shown in Figure 1.
Figure 1.
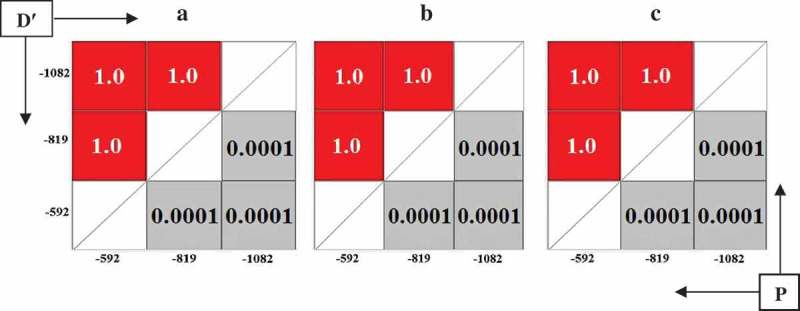
LD plots show Dʹ and P value of linkage between IL-10 alleles in VL patients (a), their families (b) and controls (c).
Table 3.
Distributions of the most common IL-10 haplotypes and haplogenotypes in VL patients, their families and controls.
| Haplotypes (−592, −819, −1082) | Patient n (%) | Family n (%) | Control n (%) | P1 | P2 | PS1 (%) | PS2 (%) |
|---|---|---|---|---|---|---|---|
| Haplotype | |||||||
| ACC | 95 (39.58) | 47 (35.07) | 79 (38.73) | 0.388 | 0.853 | 14 | 4 |
| GCC | 77 (32.08) | 43 (32.09) | 69 (33.82) | 0.999 | 0.697 | 3 | 6 |
| ATA | 68 (28.34) | 44 (32.84) | 56 (27.45) | 0.361 | 0.836 | 14 | 4 |
| Haplogenotype | |||||||
| ACC/GCC | 25 (20.83) | 12 (17.91) | 19 (18.63) | 0.63 | 0.681 | 7 | 6 |
| ACC/ACC | 23 (19.17) | 11 (16.42) | 21 (20.59) | 0.64 | 0.791 | 7 | 4 |
| ACC/ATA | 24(20) | 13 (19.4) | 18 (17.64) | 0.921 | 0.655 | 3 | 7 |
| GCC/GCC | 16 (13.33) | 11 (16.42) | 18(17.65) | 0.564 | 0.373 | 8 | 14 |
| ATA/GCC | 20 (16.67) | 9 (13.43) | 14(13.73) | 0.558 | 0.544 | 9 | 9 |
| ATA/ATA | 12 (10) | 11 (16.42) | 12 (11.76) | 0.2 | 0.673 | 23 | 6 |
Each P value is the result of comparing a row with the corresponding sum of related rows.
P stands for P value indicating differences between patients and family (P1), patients and controls (P2).
PS stands for power study which evaluates the power of study comparing patients and family (PS1) as well as patients and control (PS2).
Discussion
Genetic elements are known as the most probable important factor influencing the outcome of parasite and host combat. Given genetic variants can determine the levels of their corresponding proteins, the relationship of various SNPs in the cytokines genes predisposing Iranians to different infectious diseases was investigated previously [14–17]. Taking into account the importance of cytokines such as IL-10, IL-12 and IFN-γ in the pathogenesis of Leishmania infection and also the higher levels of these cytokines in the VL patients sera [18], the present study was undertaken to investigate the relationship between gene variants of these cytokines and susceptibility or resistance to VL.
Comparing the frequencies of IL-10, IL-12 and IFN-γ alleles showed no significant difference between VL patients, their families and controls within an endemic area in the southwest of Fars province in Iran. In addition, the frequency of haplotypes reconstructed from IL-10 alleles was not significantly different between the studied groups. In agreement with our results, Hajilooi et al. reported that the distributions of alleles A and G at position −1082 and alleles C and T at position −819 of IL-10 gene were comparable between VL patients and controls but they showed that AG and CT genotypes at position −1082 and −819, respectively, were more frequent in patients than in controls [3]. Moreover, Mishra et al. reported a variant of IL-10 (g.5311A) which was significantly associated with VL in a region of India [19]. Although IL-10 is known as an anti-inflammatory cytokine and its upregulated level has been reported in different studies during the acute phase of VL [20,21], the discrepancies between different studies from different areas of the worlds may imply that IL-10 SNPs could not explain simply its role in the VL pathogenesis and other cytokines corresponding to both innate and acquired immune responses may be involved.
To the best of our knowledge, we did not find any study in Iran evaluating the association of IL-12 gene polymorphism and VL but a study by Zahra’a A. et al. in Iraq demonstrated that neither genotypes nor alleles of IL-12B at position −1188 were different between the Iraqi VL patients and controls [11].
As for IFN-γ, we observed that AT genotype at position +874 was significantly more prevalent in patient group, compared with that in family group (P = 0.015) and conversely, the TT genotype was significantly more frequent in family group than that in the patients. It was shown that TT genotypes at position +874 of IFN-γ was strongly associated with higher circulating IFN-γ while AT and AA genotypes are related to its lower level [13]. Therefore, it seems that the individuals carrying TT genotype by producing a higher level of IFN-γ may be more resistant to VL, compared with those who inherit the low producer genotypes AT and AA. IFN-γ is an inflammatory cytokine inducing cell mediated immunity (CMI) which can control the intracellular microorganisms, especially Leishmania parasite [2] through activation of both innate and adaptive immunity and it can eradicate the parasite by the production of oxygen intermediate and reactive nitrogen species [22]. Accordingly, in the previous studies it was revealed that down-regulation of IFN-γ and IL-12 and the enormous production of immune suppressive cytokines such as IL-10, can lead to the attenuation of Th1 response and consequently susceptibility to VL [18,23]. Given the role of IFN-γ in the defense against intracellular microorganisms, the effect of its genotypes was investigated in several infectious diseases in which CMI plays the most important defensive mechanism. The association of low producing alleles of IFN-γ was reported with chronicity of hepatitis B virus [24]. Also, in parvovirus B19 infected individuals, the presence of T allele in the IFN-γ was reported to be associated with the production of anti-parvovirus antibody [24,25]. In a new meta-analysis study, the author reported that the IFNG +874 T/A (rs2430561) polymorphism is potentially associated with susceptibility to tuberculosis and may be used as a predictive biomarker [26].
In conclusion, our study indicated that among the critical cytokines influencing the outcome of VL disease such as IL-10, IL-12 and IFN-γ, the presence of AT genotype at position +874 of IFN-γ could be considered as one of the predictive factors which make individuals susceptible to visceral leishmaniasis in a sample of Iranian population from the endemic area in the southwest of Iran.
Funding Statement
This study was financially supported by a Grant No. 95-10 from Prof. Alborzi Clinical Microbiology Research Center affiliated with Shiraz University of Medical Sciences.
Acknowledgments
Our thanks go to Hassan Khajehei for language editing of the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
References
- [1].van Griensven J, Balasegaram M, Meheus F, et al. Combination therapy for visceral leishmaniasis. Lancet Infect Dis. 2010;10(3):184–194. [DOI] [PubMed] [Google Scholar]
- [2].Faleiro RJ, Kumar R, Hafner LM, et al. Immune regulation during chronic visceral leishmaniasis. PLoS Negl Trop Dis. 2014;8(7):e2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hajilooi M, Ahmadi A, Lotfi P, et al. Is the polymorphism at position-1082 of IL-10 gene associated with visceral leishmaniasis? Iran J Public Health. 2014;43(8):1107. [PMC free article] [PubMed] [Google Scholar]
- [4].Lin W-W Karin M.A cytokine-mediated link between innate immunity, inflammation, and cancer. J Clin Invest. 2007;117(5):1175–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Tripathi P, Tripathi P, Kashyap L, et al. The role of nitric oxide in inflammatory reactions. FEMS Immunol Med Microbiol. 2007;51(3):443–452. [DOI] [PubMed] [Google Scholar]
- [6].Gautam S, Kumar R, Maurya R, et al. IL-10 neutralization promotes parasite clearance in splenic aspirate cells from patients with visceral leishmaniasis. J Infect Dis. 2011;204(7):1134–1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Kane MM, Mosser DM. The role of IL-10 in promoting disease progression in leishmaniasis. J Immunol. 2001;166(2):1141–1147. [DOI] [PubMed] [Google Scholar]
- [8].Maspi N, Abdoli A, Ghaffarifar F. Pro-and anti-inflammatory cytokines in cutaneous leishmaniasis: a review. Pathog Glob Health. 2016;110(6):247–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Al-Bashier NM. Impact of IFN-(+ 874T/A) and IL-10 (−1082G/A) on the susceptibility to Visceral Leishmaniasis. Int J Curr Microbiol App Sci. 2014;3(3):662–667. [Google Scholar]
- [10].Hajilooi M, Sardarian K, Dadmanesh M, et al. Is the IL-10− 819 Polymorphism Associated with Visceral Leishmaniasis? Inflammation. 2013;36(6):1513–1518. [DOI] [PubMed] [Google Scholar]
- [11].Zahra’a A, Ad’hiah AH, Idan EM. Interleukin-12B gene polymorphism and visceral Leishmaniasis in Iraqi patients. J Biol Agri Healthcare. 2015;5:16. [Google Scholar]
- [12].Miller S, Dykes D, Polesky H. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16(3):1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Pravica V, Perrey C, Stevens A, et al. A single nucleotide polymorphism in the first intron of the human IFN-γ gene:: absolute correlation with a polymorphic CA microsatellite marker of high IFN-γ production. Hum Immunol. 2000;61(9):863–866. [DOI] [PubMed] [Google Scholar]
- [14].Rasouli M, Kiany S. Association of interferon-gamma and interleukin-4 gene polymorphisms with susceptibility to brucellosis in Iranian patients. Cytokine. 2007;38(1):49–53. [DOI] [PubMed] [Google Scholar]
- [15].Rasouli M, Kiany S, Moravej A, et al. Interleukin-12 and tumor necrosis factor-β gene polymorphisms as genetic susceptibility factors for brucellosis in Iranian patients. Iran Red Crescent Med J. 2010;2010(3):266–271. [Google Scholar]
- [16].Moravej A, Rasouli M, Kalani M, et al. IL-1beta (−511T/C) gene polymorphism not IL-1beta (+3953T/C) and LT-alpha (+252A/G) gene variants confers susceptibility to visceral leishmaniasis. Mol Biol Rep. 2012;39(6):6907–6914. doi:10.1007/s11033-012-1517-z. PubMed PMID: 22311026; eng. [DOI] [PubMed] [Google Scholar]
- [17].Rasouli M, Kalani M, Moravej A, et al. Interleukin-18 single nucleotide polymorphisms contribute to the susceptibility to brucellosis in Iranian patients. Cytokine. 2011;54(3):272–276. doi:10.1016/j.cyto.2011.02.011. PubMed PMID: 21393015; eng. [DOI] [PubMed] [Google Scholar]
- [18].Khoshdel A, Alborzi A, Rosouli M, et al. Increased levels of IL-10, IL-12, and IFN-in patients with visceral leishmaniasis. Braz J Infect Dis. 2009;13(1):44–46. [DOI] [PubMed] [Google Scholar]
- [19].Mishra A, Nizamuddin S, Arekatla G, et al. IL10 variant g. 5311A is associated with visceral leishmaniasis in Indian population. PLoS One. 2015;10(5):e0124559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Nylen S, Sacks D. Interleukin-10 and the pathogenesis of human visceral leishmaniasis. Trends Immunol. 2007September;28(9):378–384. doi:10.1016/j.it.2007.07.004. PubMed PMID: 17689290; eng [DOI] [PubMed] [Google Scholar]
- [21].Bhattacharya P, Ghosh S, Ejazi SA, et al. Induction of IL-10 and TGFβ from CD4+ CD25+ FoxP3+ T cells correlates with parasite load in Indian kala-azar patients infected with Leishmania donovani. PLoS Negl Trop Dis. 2016;10(2):e0004422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bhattacharya P, Dey R, Dagur PK, et al. Genetically modified live attenuated Leishmania donovani parasites induce innate immunity through classical activation of macrophages that direct the Th1 response in mice. Infect Immun. 2015;83(10):3800–3815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Singh OP, Sundar S. Immunotherapy and targeted therapies in treatment of visceral leishmaniasis: current status and future prospects. Front Immunol. 2014;5:296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Ben-Ari Z, Mor E, Papo O, et al. Cytokine gene polymorphisms in patients infected with hepatitis B virus. Am J Gastroenterol. 2003;98(1):144–150. [DOI] [PubMed] [Google Scholar]
- [25].Kerr J, McCoy M, Burke B, et al. Cytokine gene polymorphisms associated with symptomatic parvovirus B19 infection. J Clin Pathol. 2003;56(10):725–727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Wei Z, Wenhao S, Yuanyuan M, et al. A single nucleotide polymorphism in the interferon-γ gene (IFNG+ 874 T/A) is associated with susceptibility to tuberculosis. Oncotarget. 2017;8(31):50415. [DOI] [PMC free article] [PubMed] [Google Scholar]


