Abstract
Human embryonic stem cells subjected to a one-time uniaxial stretch for as short as 30-min on a flexible substrate coated with Matrigel experienced rapid and irreversible nuclear-to-cytoplasmic translocation of NANOG and OCT4, but not Sox2. Translocations were directed by intracellular transmission of biophysical signals from cell surface integrins to nuclear CRM1 and were independent of exogenous soluble factors. On E-CADHERIN-coated substrates, presumably with minimal integrin engagement, mechanical strain-induced rapid nuclear-to-cytoplasmic translocation of the three transcription factors. These findings might provide fundamental insights into early developmental processes and may facilitate mechanotransduction-mediated bioengineering approaches to influencing stem cell fate determination.
Keywords: Mechanotransduction, hESCs, Nanog, Oct4, CRM1, FAK, cytoplasmic translocation, stem cells
Insight, innovation, integration.
How do mechanical stimuli, sensed through different types of cell adhesions, alter embryonic stem cell fate? This paper employs a stretchable device coated with different cell adhesion molecules to apply stretch to human embryonic stem cells in a manner that might mimic differentiation of cell lineages present at the blastocyst stage of development. The work reveals that stretch induces an unexpectedly rapid export of a subset of pluripotent transcription factors from the nucleus to the cytoplasm before significant decreases in overall levels. Interestingly, the subset of transcription factors exported differs between cells stretched on Matrigel- versus E-CADHERIN-coated devices. These results have physiological relevance to embryonic development where some cells experience mechanical forces predominantly through cell–cell contact while other cells also experience cell–ECM interactions leading to different cell-fate specification. These insights might enhance our understanding of early development and may also guide bioengineering approaches where stretching is used to accelerate or regulate the direction of differentiation.
INTRODUCTION
Human pluripotent stem cells (PSCs) including human embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) are a promising resource for regenerative medicine. Pluripotency, described as the ability of a stem cell to generate all of the cell types of an organism [1], is regulated by a core group of transcription factors—NANOG, OCT4, and SOX2—expressed in pluripotent stem cells and in cells located in the inner cell mass of the blastocyst stage embryo [2–4]. These transcription factors have fundamental roles in early development and are required for the propagation of undifferentiated ESCs in culture [5]. OCT4 and NANOG are essential regulators of early development and ESC identity, and have distinct roles maintaining pluripotency since disruption of OCT4 or NANOG regulatory circuitry results in the differentiation of inner cell mass and ESCs to trophectoderm and extra embryonic endoderm, respectively [1, 5, 6]. Therefore, a better understanding of the mechanisms that alter expression of the pluripotency-related transcription factors and PSC fate may help comprehending early embryo development processes [7, 8].
Embryonic development is a dynamic, self-organizational process involving cell division, cell fate decisions, and morphogenic-patterning events coordinated through physical and soluble signals [9, 10]. Biochemical signals known as morphogens modulate cell activities within the developing embryo [9, 11]. However, emerging evidence increasingly suggests that physical factors can dictate cellular fates in response to alterations within the internal and external environment during embryogenesis [10]. Cells experience extrinsic tensile-compressive forces from neighboring cells and they exert intrinsic forces to the extracellular matrix (ECM) through several mechanisms, including acto-myosin contractility and cytoskeletal assembly [12]. Advanced understanding of the mechanisms by which human ESCs respond to local forces will inform knowledge of development and lineage determination.
During pre-implantation development, blastomeres experience mechanical forces that alter cell fates within the embryo. Maintenance of stem cell pluripotency requires the activity of the transcription factors, NANOG, OCT4, and SOX2. During differentiation, the levels of all three factors decrease, but do so in different orders depending on the direction of differentiation. What could be the role of mechanical forces in regulating the timing of changes in levels of these three key transcription factors? We hypothesized that mechanical forces transmitted through different combinations of cell surface receptors may play a role. Upon performing experiments, we found that mechanical strain applied to human ESCs for as short as 30-min induces rapid and irreversible nuclear-to-cytoplasmic translocation of NANOG and OCT4, but not SOX2.
On Matrigel-coated substrates, these translocations are directed by intracellular transmission of biophysical signals from cell surface integrins to nuclear CRM1 and is not reliant on exogenous soluble factors. Interestingly, on E-CADHERIN-coated substrates, presumably with minimal integrin engagement, mechanical strain induces rapid nuclear-to-cytoplasmic translocation of the three transcription factors. These findings provide fundamental insights into early developmental processes and may facilitate mechanotransduction-mediated bioengineering approaches to influencing stem cell fate determination.
RESULTS AND DISCUSSION
Due to physical and ethical concerns, it is challenging to test and determine how mechanical forces alter PSC fate during human development. Previous work utilizing cultured ESCs in vitro has provided interesting results that aim to fill this gap, but has also led to apparently contradictory outcomes, where, for example, mechanical stretch has led to either maintenance of pluripotency [13, 14] or the promotion of differentiation [15, 16]. Due to methodological differences among studies such as the type of external mechanical stimuli, the length of stimulus (hours, days), and supplementation of growth factors and small inhibitors (TGFβ, Smad2/3) administered to cells, it is not clear what parameters promote differentiation versus maintenance of pluripotency [13, 14]. For example, continuous application of a 10% equibiaxial cyclic strain at frequencies above 6 cycles/min for 12 days maintains pluripotency through action of the TGFβ/Activin/Nodal signaling pathway [13, 14]. In contrast, 15% applied strains at 12 cycles/min for 12 h reduces pluripotency gene expression via activation of Rho/ROCK and subsequent decrease of AKT phosphorylation [15]. It has also been shown that a 17.5 Pa force applied to cells via RGD-coated microbeads at 18 cycles/min for 1 h reduced OCT4 expression [16]. Furthermore, matrix stiffnesses in the presence of external growth factors lead human ESCs into mesoderm differentiation [17]. A recent study has shown applying 100% continuous-stretching to human PSCs (pulse width of 2 h for 4 h periods) under neural induction medium condition phosphorylated myosin and actin filaments and enhanced cytoskeletal activity [18]. In this study, we examined the effects of a non-cyclical 10% uniaxial strain based on the rationale that during gastrulation, strain is more of a one-time stretch rather than a cyclical stretch-relax event that repeats every few seconds [19].
To investigate the role of mechanical forces in human ESCs, we applied a 10% uniaxial stretch and held the cells in that stretched state thereafter (Figs. 1A and S1, see online supplementary material for a color version of this figure). From preliminary tests with different uniaxial strains (0%, 5%, 10%, 15%, and 20%) applied to human ESCs, we observed translocation of transcription factors by strain conditions above 10%. However, at 20% strain, human ESCs detached from the substrate surface and failed to thrive (Fig. S2A, see online supplementary material for a color version of this figure). Hence, we used the 10% strain condition to investigate the effects of mechanical forces on human ESCs.
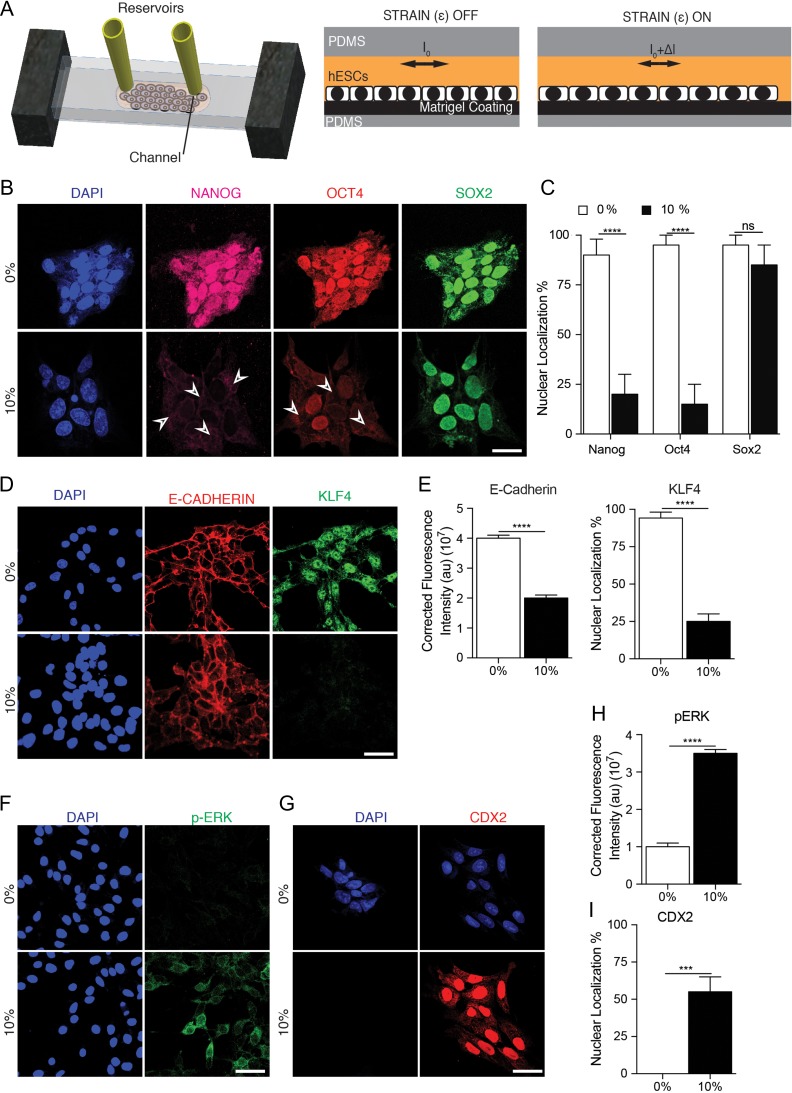
Figure 1.
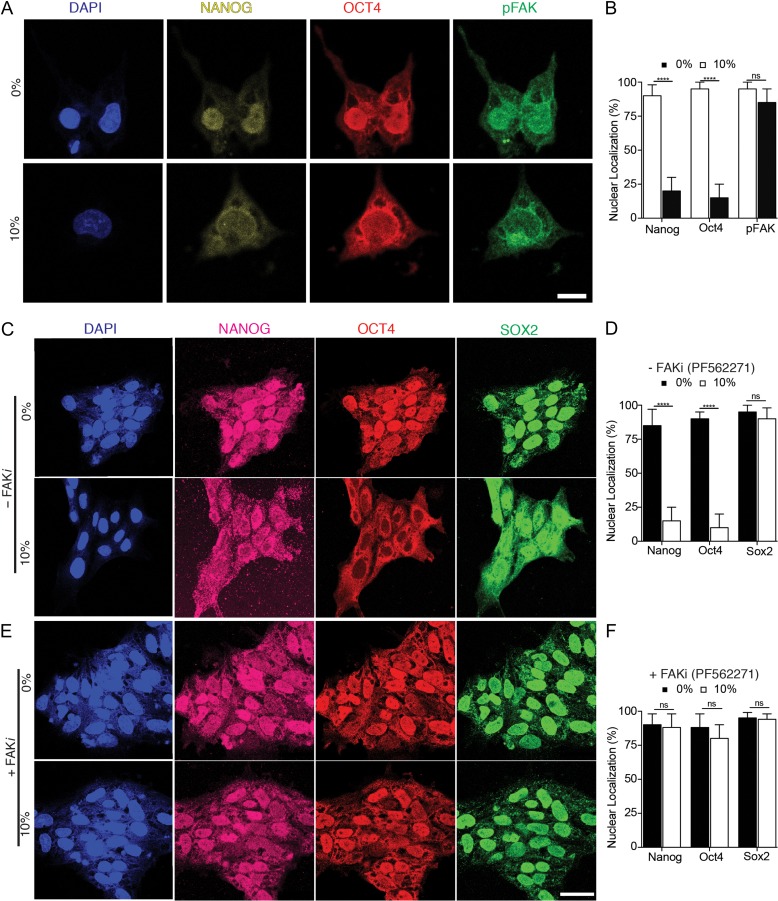
Uniaxial mechanical force exerted on human ESC. (A) Schematic of chip to exert 10% uniaxial strain to human ESCs. Side view with and without stretch applied illustrates how distortion of flexible substrates results in stretch of adherent human ESCs. Top view depicts reservoirs where cells are loaded and supplied with medium. (B) Representative micrographs showing the expression of NANOG, OCT4, and SOX2 in human ESC colonies during 2 h of no strain (0%) and 10% strain applied via uniaxial forces. DAPI stained nuclei. Scale bar = 50 μm. (C) Percentages of cells with nuclear localization of indicated transcription factor in each condition. (D) Representative images of E-CADHERIN and KLF4 expression during 2 h of 10% stretching. (E) Corrected fluorescence intensity of E-CADHERIN and percentage of nuclear localization of KLF4 in each conditions (F) Representative micrographs showing phosphorylation of ERK in human ESCs under 10% strain, and (H) its corrected fluorescence intensity measurements. (G) Representative micrographs showing expression of CDX2 in cells under 10% strain, and (I) its quantification from three independent replicates. Unpaired t test P values <0.05 (*), <0.01 (**), <0.001(***), n.s.: not significant.
Mechanical stimulation led to a surprisingly fast-induced translocation of NANOG and OCT4 from the nucleus to the cytoplasm that was observed within 30 min (Fig. S2, see online supplementary material for a color version of this figure) and continuing beyond 2 h (Fig. 1B and C). In contrast, SOX2 remained in the nucleus regardless of stretch application to the cells attached to Matrigel-coated substrates. It has been reported that ERK-mediated phosphorylation initiates the exportation of KLF4 from the nucleus to the cytoplasm [20] and reduction in E-cadherin expression in differentiated human ESCs [17, 21]. Therefore, we determined the extent to which KLF4 and E-CADHERIN expression decreased after uniaxial stretching (Fig. 1D and E). Because phosphoryled ERK (pERK) plays a role during differentiation of PSCs [20], we examined whether mechanical strain induces changes of ERK in human ESCs. We detected pERK in the cytoplasm of human ESCs subjected to external uniaxial strain in comparison to untreated controls (Fig. 1F and H). Importantly, the culture of human ESCs in a non-strain condition for an additional 24 h after being subjected to 2 h of applied strain demonstrated that the translocation of transcription factors was irreversible, NANOG was degraded and OCT4 remained in the cytoplasm, while SOX2 remained in the nucleus (Fig. S2, see online supplementary material for a color version of this figure).
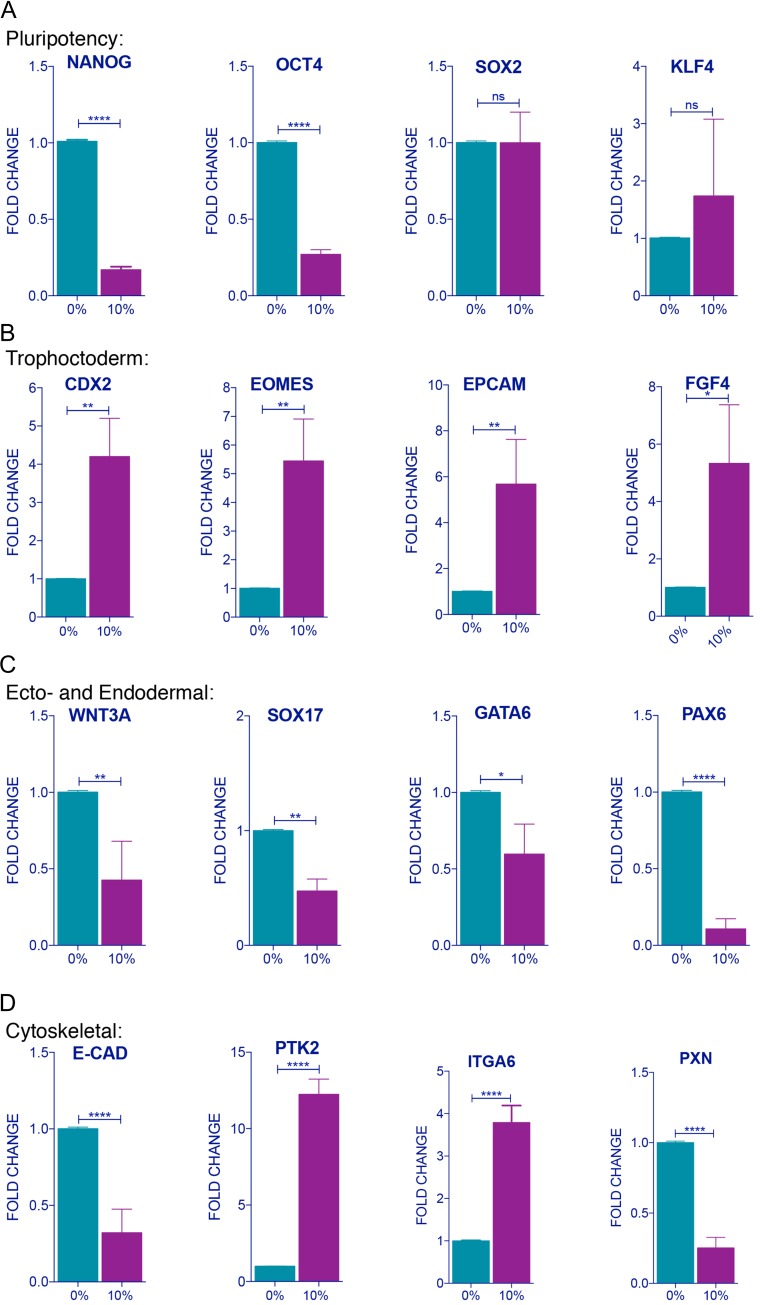
Interestingly, mechanical strain in human ESCs elevated the translation of CDX2, a transcription factor antagonist of OCT4 [22] and a determinant of trophectoderm lineage [23] (Fig. 1G and I). Gene expression analysis also further confirmed that 10% stretching for 2 h increased trophectoderm markers—CDX2, EOMES, EpCAM, and FGF4, while expression of ecto- and endodermal markers—WNT3A, SOX17, GATA6, and PAX6—decreased (Fig. 2B). No change was observed in SOX2 and KLF4, while the RNA expression of NANOG and OCT4 decreased (Fig. 2A). In addition, the expression of E-CADHERIN and PAXILLIN decreased, while PTK2 (FAK) and ITGA6 (Integrin alpha 6) expression were elevated after 10% mechanical stretching was applied compared to non-stretched controls (Fig. 2D). These effects at the RNA and protein levels were a direct effect of mechanical forces and not due to exogenous chemical cues, as cells were cultured in the same culture medium (Materials and Methods).
Figure 2.
Molecular characterization and identification of 10% strain applied human ESCs for 2 h via qRT-PCR analysis for temporal expression of (A) pluripotency (NANOG, OCT4, SOX2, and KLF4), (B) trophoctoderm markers (CDX2, EOMES, EpCAM, and FGF4), (C) ecto- and endodermal lineage differentiation (WNT3A, SOX17, GATA6, and PAX6), and (D) cytoskeletal, focal adhesion kinase (Ptk2) and PXN (Paxillin), Integrin alpha 6 (ITGA6) and E-CADHERIN with and without 10% strain. All quantification from three independent replicates. Unpaired t test P values <0.05 (*), <0.01 (**), <0.001(***), n.s.: not significant.
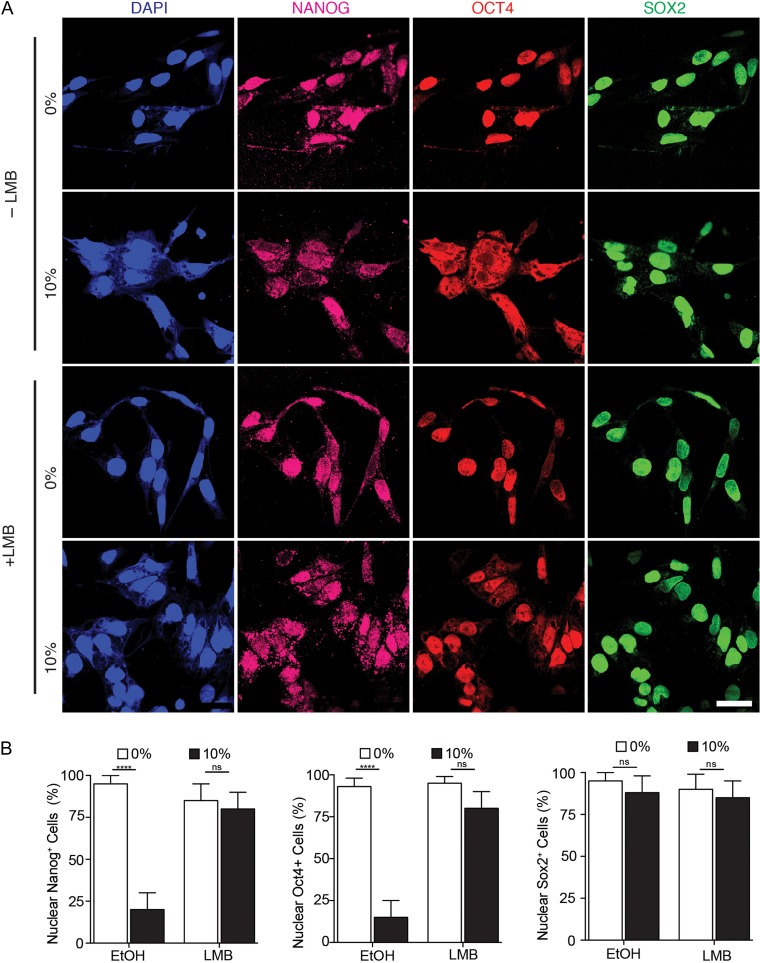
OCT4 is transported through nuclear pores by the nuclear export protein CRM1 (also known as XPO1) [20, 24] and the nuclear import protein, importin α [25–27]. Therefore, we investigated the involvement of CRM1 in the translocation of pluripotency-related transcription factors in human ESCs during uniaxial forces. In the presence of Leptomysin B (LMB), a chemical inhibitor of CRM1, NANOG and OCT4 remained in the nucleus in contrast to their cytoplasmic localization in the control group (Fig. 3). This suggests that mechanical stretching activates CRM1 in human ESCs.
Figure 3.
CRM1 inhibitor, Leptomyosin (LMB), abolished the effect of mechanical strain in pluripotent transcription factors. (A) Representative micrographs showing translocation of NANOG and OCT4 to the cytoplasm of human ESCs under mechanical strain during 2 h in the absence of LMB inhibitor but treated with ethanol (EtOH) as diluent. (B) Pre-treatment of human ESCs with LMB inhibitor abolished the shuttling mechanism of transcription factors during 2 h of 10% strain forces. (C) Percentage of cells with nuclear localization of NANOG, OCT4, and SOX2 within 2 h upon mechanical strain with and without LMB. Scale bars equal to 50 μm. All quantifications were from at least three independent experiments with two replicates per experiments. Unpaired t test P values <0.05 (*), <0.01(**), <0.001 (***). n.s.: not significant.
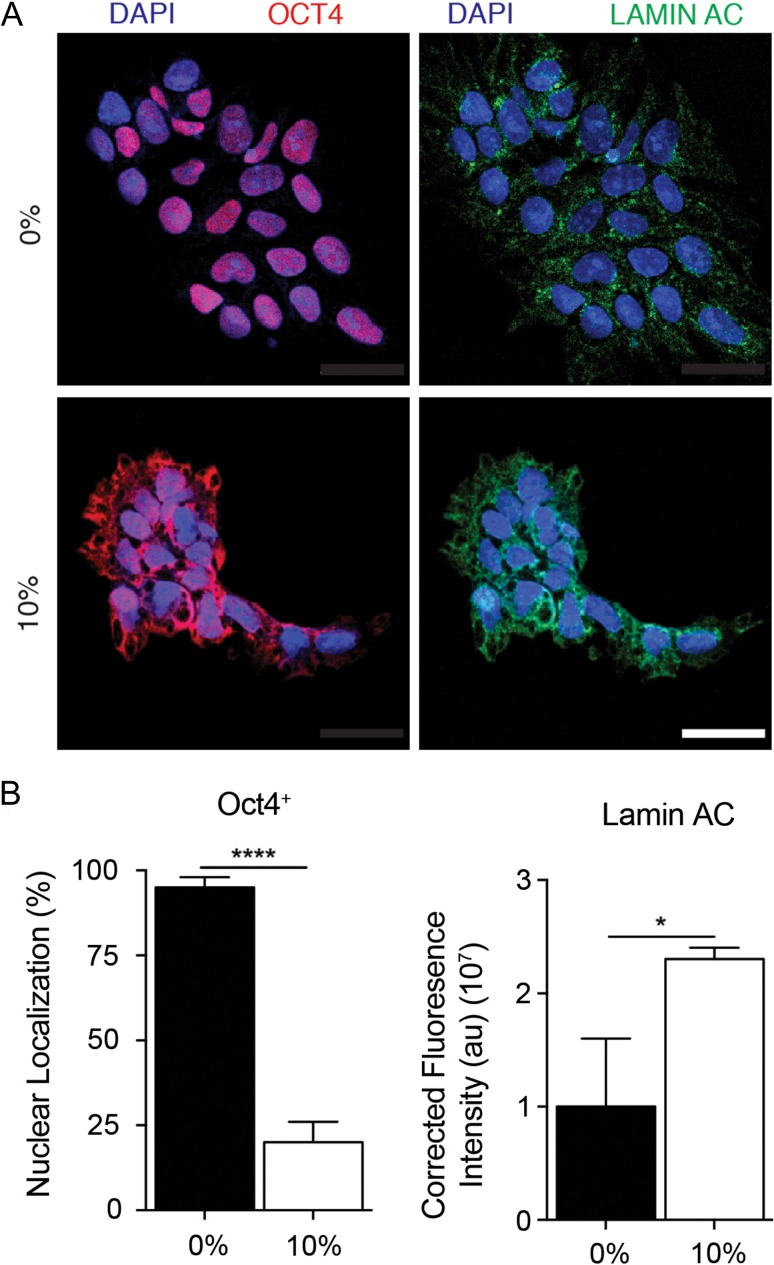
To determine what cellular machinery might be involved in sensing the micro-environmental forces exerted by uniaxial stretching, we investigated nuclear lamins, which sense extracellular forces through the cytoskeleton [28]. LAMIN A/C in particular plays a central role in the organization of the nuclear structure and gene function, and it is activated during human ESC differentiation [29]. Our results demonstrate that LAMIN A/C expression increased in response to mechanical strain, concurrent with the translocation of pluripotency-related-factors to the cytoplasm (Fig. 4). These results suggest the involvement of mechanosensitive protein(s) mediating the activation of CRM1 and LAMINA/C in the translation of NANOG and OCT4. Thus, we analyzed the phosphorylation of focal adhesion kinase (FAK), a mechanotransducive protein that previously was shown to be associated with translocation of OCT4 to the cytoplasm and with differentiation of human ESCs [30]. Uniaxial force application to human ESCs induced FAK phosphorylation (Fig. 4A) concurrent with NANOG and OCT4 translocation to the cytoplasm (Fig. 5A and B). These results were consistent with our previous report showing the translocation of OCT4 to the cytoplasm in response to FAK phosphorylation, induced by Mn+2-mediated activation or integrin β1 activation in human ESCs [30] (Fig. S3A and B, see online supplementary material for a color version of this figure). Pre-treatment of human ESCs with the chemical FAK inhibitor, PF562271, before uniaxial stretching prevented the cytoplasmic translocation of NANOG and OCT4 (Fig. 5C–F). Thus, uniaxial strain-mediated effects on human ESCs cultured on Matrigel-coated substrates involve FAK activation to transmit the extracellular information from outside to inside the cell.
Figure 4.
Effect of uniaxial mechanical force on mechanotransductive molecular components. (A) Representative micrographs showing expression of Lamin A/C and OCT4 in cells during 2 h without (0% strain) and with (10% strain) mechanical strain. DAPI stained nuclei. Scale bar = 50 μm. Unpaired t test P values < 0.05 (*), >0.01 (**), <0.001(***), n.s.: not significant. (B) Quantification of fluorescence for each protein after 10% uniaxial strain.
Figure 5.
FAK translocate to cytoplasm after strain forces are applied and prevention of FAK phosphorylation eliminates the effects of mechanical strain shuttling pluripotent transcription factors from nucleus to cytoplasm. (A) NANOG, OCT4, and pFAK shuttle to cytoplasm after mechanical strain exerted, (B) NANOG and OCT4 shuttled to cytoplasm under the mechanical strain within 2 h in the absence of PF562271, chemical inhibitor of FAK phosphorylation (treated with DMSO, as diluent); while their localization of these markers remains in the nuclei within 2 h under 10% strain in the presence of PF562271, (C and D) percentage of cells with nuclear localization of NANOG, OCT4, and SOX2 within 2 h upon mechanical strain with and without PF562271. Scale bars equal to 50 μm. All quantifications were from at least three independent experiments with two replicates per experiments. Unpaired t test P values <0.05 (*), <0.01(**), <0.001 (***), n.s.: not significant.
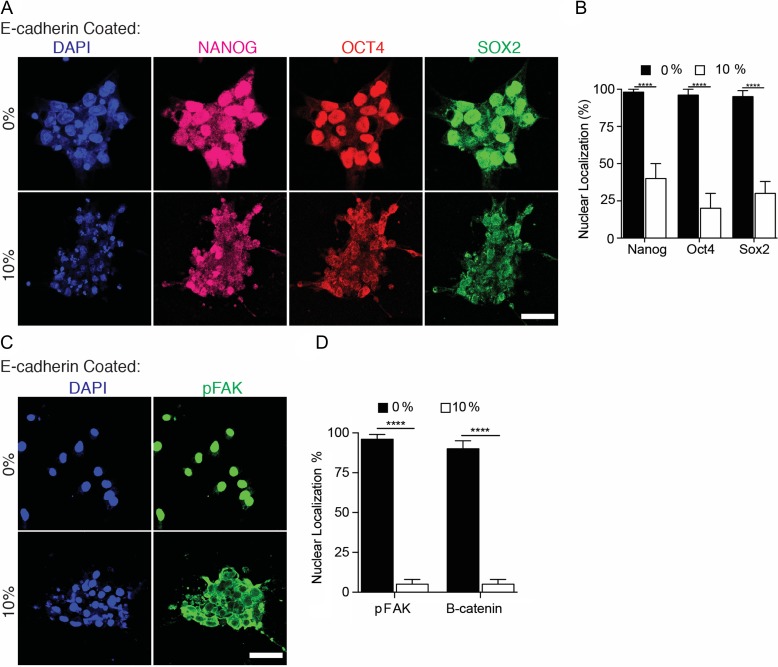
Similar to early developmental events, our stretching experiments might activate mechanisms that sense changes in cell–cell adhesion [31, 32] and cell–ECM interactions [33, 34]. To identify the effects of cell–cell interactions with less involvement of cell–ECM interactions, we performed experiments with substrates coated with E-CADHERIN. In the absence of mechanical strain, NANOG, OCT4, and SOX2 remained in the nucleus. However, upon mechanical straining of human ESCs cultured on E-cadherin, the three transcription factors translocated to the cytoplasm within 2 h (Fig. 6A and B). Further, upon application of mechanical strain, FAK became phosphorylated and localized in the cytoplasm, while in the control group, FAK was localized in the nucleus (Fig. 6C and D). The translocation of the three transcription factors to the cytoplasm contrasted with results observed on Matrigel-coated chips, where SOX2 remained nuclear, while NANOG and OCT4 translocated to the cytoplasm. This suggests that strain sensed through integrin-mediated and E-CADHERIN-mediated mechanisms have different effects on human ESCs. Mechanical cues on cell–ECM interaction are well known to be involved in mechanotransduction via integrins and focal adhesions [35, 36], however, pathway(s) of strain-induced differentiation through cell–cell interaction remains elusive. Our results and methods open new avenues for exploring the distinct role of mechanical cues from cell–cell interactions vs. cell–ECM interactions.
Figure 6.
Effect of uniaxial mechanical strain on E-cadherin-coated substrates. (A) Representative micrographs showing translocating to cytoplasm of NANOG, OCT4, and SOX2 on E-CADHERIN-coated PDMS chip within 2 h of mechanical strain. (B) Graph indicating percentage of cell with nuclear localization of NANOG, OCT4, and SOX2 within 2 h upon mechanical strain on cell cultured on E-CADHERIN-coated chips, compared to the control group. (C) Representative micrographs showing shuttled to cytoplasm of pFAK within 2 h of mechanical strain. (D) Graph indicating percentage of cell with nuclear localization of pFAK within 2 h upon mechanical strain on cell cultured on E-CADHERIN-coated chips, compared to the non-stretched group. Scale bars equal to 50 μm. All quantifications were from at least three independent experiments with two replicates per experiments. Unpaired t test P values <0.05 (*), <0.01(**), <0.001 (***). n.s.: not significant.
CONCLUSION
Human ESCs are pluripotent cells that are derived from the inner cell mass of the mammalian blastocyst, and they are capable of self-renewal in vitro under certain conditions [37, 38]. It is generally accepted that ESC differentiation is accompanied by decreased expression of NANOG, OCT4, and SOX2. Regulation of differentiation, however, is more complex and the precise balance of transcription factors, NANOG, OCT4, and SOX2, plays essential roles in the regulation of human ESC pluripotency. For example, not only down-regulation, but overexpression of any of these factors can also lead to a loss of maintenance of pluripotency and initiation of differentiation. Specifically, overexpression of SOX2 prompts neuroectodermal specification [39], while overexpression/ down-regulation of OCT4 and NANOG supports mesodermal differentiation [2, 40].
The delicate interactions between pluripotency related transcription factors suggest that changes in their subcellular localization may also play a role in exit from self-renewal [20, 41], and in the early stages of differentiation. Previous reports of translocation of pluripotency-related transcription factors are limited to a recent report showing that mutations in ERK phosphorylation in mouse ESCs lead to KLF4 and NANOG translocation from the nucleus to the cytoplasm, followed by subsequent differentiation [20].
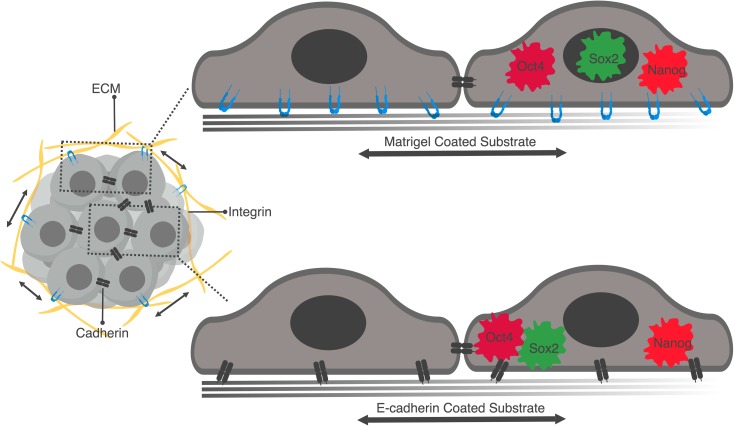
Here, we demonstrate that that the pluripotency circuit can be altered with a one-time uniaxial stretch in the absence of any genetic manipulation or use of exogenously added growth factors or cytokines with visible effects as early as 30 min after stretching. Interestingly, inhibition of translocation of NANOG and OCT4, through blocking of CRM1, prevented differentiation of human ESCs. Moreover, our data suggest that mechanotransducive signals through focal adhesions propagate rapidly to phosphorylate FAK and promote LAMIN A/C expression and activity of CRM1, affecting the localization and function of pluripotency-related transcription factors (Fig. 7). Our result contradict a report that shows cyclic mechanical strain maintains pluripotency [13, 14], while it supports another report that demonstrates mechanical strain inhibits pluripotency gene expression by inhibiting AKT activation [15]. It is worth noting that unlike these previous studies that use cyclic strain, we applied a one-time sustained stretch.
Figure 7.
Schematic illustration of mechanotransductive signals activated in hESCs by uniaxial mechanical strain.
Cell–cell adhesion transmits physical forces and influences the dynamics of tissue formation by enabling cellular processes including tissue remodeling, migration, and growth during early development [31, 42–44]. Interestingly, we found that uniaxial stretch translocated NANOG and OCT4 on Matrigel-coated substrates, while in addition to those two, SOX2 also translocated to the cytoplasm on E-CADHERIN-coated substrate. These results show that translocation of pluripotency factors from nucleus to cytoplasm in human ESCs is mechanosensitive and it is impacted by forces transduce through cell–ECM and cell–cell interactions.
Overall, these results confirm that biologic responses to mechanical stress may be propagated faster than biochemical cues [45]. These mechanotransductive processes on human ESCs are dependent on cell–matrix and cell–cell interactions and might be indicative of early events of embryo development, as observed here with the upregulation of trophectoderm related markers. Understanding how mechanical cues alter stem cell fate will provide us key insights into comprehending early development and also will contribute to the use of stem cells in regenerative medicine and biotechnology. For example, in stem cell differentiation applications, it is a standard practice to add different activators and inhibitor molecules at different timepoints to mimic physiological development. Our results suggest that timed application of one-time stretch to human ESCs may be a practical and powerful biomimetic cue to guide stem cell differentiation.
MATERIALS AND METHODS
Microfabrication and PDMS device preparation
PDMS devices were fabricated using standard soft lithographic techniques [46]. Briefly, a rectangular pattern (13 mm × 3 mm) was designed using a commercial computer-aided design software, AutoCAD (Autodesk, Inc.) and printed on a transparent film (CAD Art, Inc.). With the transparent film mask, a SU-8 master structure with a thickness of 200 μm was developed on a silicon wafer. A 10:1 mixture of a PDMS prepolymer base (Sylard 184, Dow Corning) and a curing agent was deposited on the silicon mold and cured overnight at 60°C flat oven. Glass slides were salinized overnight, and then a 10:1 mixture of a PDMS prepolymer base and a curing agent was deposited on the salinized glass slide, then spin coated at 650 Rpm for 40 s to obtain 100 mm thick membrane and cured overnight at 60°C flat oven. After peeling the finished PDMS from the mold, two holes at each end of the pattern were punched on the PDMS. The peeled PDMS slabs and membrane were rendered hydrophilic by exposure to oxygen plasma (SPI Supplies, West Chester, PA) for 5 min and then the slabs were bonded to a PDMS membrane, then two tip reservoirs were connected to each hole. Shortly afterwards, channels were filled with PBS to maintain their hydrophilic state and devices were sterilized with UV light for 30 min.
ECM coating on channel
Microchannels were coated with hESC qualified Matrigel (Corning) and recombinant human E-cadherin protein (R&D Systems).
Matrigel coating: One aliquot of Matrigel (112 ul) was diluted in 10 mL of cold DMEM/F12 to coat channels. 50 uL of Matrigel/DMEM/F12 solution was added to channels and incubated at room temperature for at least 2 h before use. To remove excess Matrigel, the channel system was washed three times with PBS.
E-cadherin coating: Company’s protocol was followed for human recombinant E-cadherin protein coating. Briefly, human recombinant E-cadherin protein was reconstituted at 250 μg/ml in PBS with Ca++ and Mg++ and then added to microchannels for incubation at 37°C for 90 min. To remove the excess E-cadherin, the channel system was washed three times with PBS with Ca++ and Mg++.
hESC culture
Human embryonic stem cells (hESCs) Lines H1, H9 (NIH code: WA09; WiCell Research Institute, Madison, WI), and CHB10 (NIH Code: NIHhESC-09-0009, Children’s Hospital Corporation) were cultured on the synthetic surface PMEDSAH as described previously [47] with human-cell-conditioned medium (HCCM, MTI-Global Stem, Gaithersburg, MD, http://www.mti-globalstem.com) supplemented with 5 ng/ml of human recombinant basic fibroblast growth factor (FGF2; InvitrogenTM, Carlsbad, CA, http://www.invitrogen.com), and 1%antibiotic–antimycotic (Gibco). The hPSC culture medium was replaced every other day and all cell culture was performed in designated incubators at 37°C in 5% CO2 and high humidity. Differentiated cells were mechanically removed using a sterile pulled-glass pipet under a stereomicroscope (LeicaMZ9.5, Leica Microsystems Inc., Buffalo Grove, IL).
hESC seeding in the channel
Hundred microliter of HCCM with roughly 10 undifferentiated hESC’s clusters (100–150 mm) were loaded into the channel from one end of the device for attachment overnight at 37°C in 5% CO2 and high humidity.
Stretching
After observing cell attachment in the channel, hESC colonies were subjected to uniaxial strain using a home-made strainer (S.T. Japan USA LLC, FL, USA) (Fig. S1, see online supplementary material for a colur version of this figure) [48]. The PDMS device was stretched by an applied strain of 10%. The applied strain was further confirmed by a caliper rule. When the uniaxial strain is applied to the PDMS device, a compressive strain will be formed simultaneously due to Poisson’s effect. Also, non-uniform mechanical strain will be applied to the cells depending on their locations in the device, resulting in different translocation profiles. To solve such problems, we took images of the human ESCs consistently at the center of the channels and used them for the analysis.
Inhibition and activation studies
Inhibition: PF562271 (Sigma-Aldrich) was added to the culture media at 1 μM an hour before stretching the cells to inhibit focal adhesion kinase phosphorylation.
To block nuclear export signal via interaction with a cysteine residue in the CRM1 (exportin1), LMB (Exportin1 inhibitor) was added to the media at 100 nM an hour before applying mechanical strain.
Activation : To activate integrin, human ESCs were treated with 0.5 mM manganese chloride (MnCI2) (Sigma, Cat#7773-01-5) for 24 h.
Cell immunocytochemistry analysis
hESCs in microchannel were washed with PBS for 5 min and aspirated out the supernant and added 1 ml of Z-Fix solution (Anatech LTD: cat# 170) for 10 min at RT shaking. Next, cells were washed 3× with 1 ml of PBS for 15 min each at RT. This was followed by sequential incubation with unmasking solution (PBS, 2 N HCL, 0.5% TritonX) for 15 min, its removal, quenching solution (TBS, 0.1% sodium borohydride) for 15 min, its removal, and permeabilization solution (PBS, 0.02% TritonX) for 15 min. Then, blocking solution (5% BSA in 1× PBS) was added for 1 h. Microchannels were then incubated in primary antibodies overnight at 4°C shaking. Channels were then washed with 1× PBS three times for 15 min at RT while shaking. We then incubated in secondary antibodies covered in foil for at least 1 h at RT while shaking. Afterwards, channels were washed twice in PBS for 20 min, incubated in DAPI solution for 20 min and washed in 1× PBS for 20 min. We used Nikon Ti Eclipse Confocal Microscope, 20× and 60× magnification lenses, with water to capture images with or without 3× digital zoom, ¼ frames per second, 512 × 512 image capture, 1.2 Airy Units, 2× line averaging, appropriate voltage and power settings optimized per antibody. No modification was done, except image sizing reduction, rotation, or gray scale change for figure preparation. All original and unaltered blots are found in the Supplementary figures.
All antibodies were used as following with a working volume of 1 ml in 5% BSA in PBS, unless noted otherwise.
Primary antibodies for immunofluorescence in Table 1.
Table 1.
List of primary antibodies.
| Antibody name | Company | Catalog number | Dilutions |
|---|---|---|---|
| OCT4 | SantaCruz | #sc8629 | 1:500 |
| NANOG | Abcam | #ab62724 | 1:100 |
| SOX2 | Millipore | #ab56603 | 1:500 |
| PFAK (TYR397) | Invitrogen | #700255 | 1:200 |
| CDX2 | SantaCruz | #393572 | 1:500 |
| KLF4 | ABCAM | #ab129473 | 1:200 |
| E-CADHERIN | R&D Systems | #AF748 | 1:500 |
| INTEGRIN ALPHA 6 | SantaCruz | #374057 | 1:500 |
| EOMES | Cell Signaling | #81493S | 1:200 |
| EPCAM | Cell Signaling | #2929S | 1:300 |
| pERK | Cell Signaling | #9101 | 1:250 |
Secondary antibodies for immunofluorescence: All antibodies were used at a concentration of 1:1500 with a working volume of 1.5 ml in 5% BSA in PBS. DAPI stain was used for DNA. Donkey anti-Rabbit IgG secondary antibody, Alexa Fluor® 488 (TFS: cat# A-21207), donkey anti-Goat IgG secondary antibody, Alexa Fluor® 594 (TFS: cat# A-11058), and donkey anti-Mouse IgG secondary antibody, Alexa Fluor® 647 (TFS: cat# R37114).
Image analysis
Image J was used to quantify fluorescent intensity [49, 50]. Briefly, after selecting the cell of interest using any of the polygon drawing/selection tools, ‘set measurement’ was selected from the Analyze menu by selecting area, integrated density, and mean gray value. Then, ‘Measure’ was selected from the analyze menu, and a region next to the cell that has no fluorescence was selected for background. This step was repeated for each single cell in the field and then the all data were analyzed on excel, and this formula was followed for the corrected total cell fluorescence (CTCF):
Extraction and purification of total RNA
Plates were washed with PBS and 1000 μl of Trizol Reagent (Invitrogen, Carlsbad, CA) was added to the plates, and RNAs were collected after vigorous pipetting. Two hundred microliter of chloroform was added to this solution followed by centrifugation (13 000g 15 min). Aqueous phase containing RNA was separated and 500 μl isopropanol was added and stored at 200 °C at least overnight. Then, the manufacturer’s RNA clean-up protocol, RNeasy Mini-Kit (Qiagen, Valencia, CA), with the optional On-column DNAse treatment was followed. RNA quality and concentration were checked using a Synergy NEO HTS Multi-Mode Microplate Reader (BioTek Instruments, Winooski, VT).
Reverse-transcription PCR (RT-PCR) analysis
Reverse transcription from 2.5 μg of total RNA in a 20 μl reaction into cDNA was performed using SuperScript™ VILO™ Master Mix (ThermoFisher Cat#11755050). The synthesis of first-stranded cDNA was carried out in the PCR tube after combining SuperScript VILO, RNA, and DEPC-treated water, in the first cycle at 25°C for 10 min, incubating at 42°C for 60 min, and terminating the reaction at 85°C for 5 min. Quantitative PCR was performed triplicate for each sample using TaqMan probes (Applied Biosystems) and TaqMan Universal PCR Master Mix (Applied Biosystems) on 7900 HT Fast Real Time PCR system (Applied Biosystems). Relative quantification of Nanog, Oct4, Sox2, FAK, Snai1, T, ITGAV, and Pax6 gene expression data was normalized to the GAPDH expression and calculated using the 2−ΔΔCT expression level [51].
A list of primers used in qRT-PCR are given in Table 2. All primers were purchased from ThermoFisher Life Technologies.
Table 2.
Primer sets used in qRT-PCR
| Gene symbol | Assay ID | UniGene ID |
|---|---|---|
| NANOG | Hs02387400_g1 | Hs.635882 |
| POU5F1 (OCT 3/4) | Hs03005111_g1 (FAM-MGB) | Hs.249184 |
| SOX2 | Hs01053049 (FAM-MGB) | Hs.518438 |
| FAK (PTK2) | Hs03657683 | Hs.395482 |
| CDX2 | Hs01078080_m1 | Hs.127383 |
| E-CADHERIN | Hs01023894_m1 | Hs.461086 |
| PXN | Hs01104424_m1 | Hs.446336 |
| EOMES | Hs00172872_m1 | Hs.591663 |
| FGF4 | Hs00173564_m1 | Hs.1755 |
| EPCAM | Hs00901885_m1 | Hs.542050 |
| WNT3A | Hs00263977_m1 | Hs.336930 |
| SOX17 | Hs00751752_s1 | Hs.98367 |
| GATA6 | Hs00232018_m1 | Hs.514746 |
| PAX6 | Hs00240871_m1 | Hs.446336 |
| ITGA6 | Hs01041011_m1 | Hs.133397 |
| KLF4 | Hs00358836_m1 | Hs.376206 |
| GAPDH | Hs02786624_g1 | Hs.544577 |
Statistical analysis
Results are presented as mean ± SEM. Unpaired two-tailed Student’s t test was performed for comparisons, and a P value < 0.05 was considered statistically significant.GraphPad Prism 7 was used to analyze the data.
Supplementary Material
Funding
The National Institute of Dental and Craniofacial Research (1F30DE026048-01 and R01-DE016530 to P.H.K), the National Institutes of Health (R01 EB019436 to C.X.D.), and the National Institute of Allergy and Infectious Diseases (AI 116482 to S.T.) supported these studies. TT was supported by the Republic of Turkey's Ministry of National Education (MoNE) Fellowship.
Fig. 7 in this manuscript is created using BioRender (https://biorender.io/).
Authors' contribution
T.T. led the designing of experiments, performed all studies and associated analysis, and wrote the manuscript. B.C.K. designed the chip. L.G.V-D., C.X.D., S.T., and P.H.K. oversaw the study, and contributed to interpretation of data and writing of the manuscript. T.T., L.G.V-D., S.T., and P.H.K. participated in the final editing of the manuscript.
Conflict of interest statement
None declared.
REFERENCES
- 1. Nichols J, Zevnik B, Anastassiadis K et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell 1998;95:379–91. [DOI] [PubMed] [Google Scholar]
- 2. Niwa H, Miyazaki J, Smith AG. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet 2000;24:372–76. [DOI] [PubMed] [Google Scholar]
- 3. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006;126:663–76. [DOI] [PubMed] [Google Scholar]
- 4. Parfitt DE, Shen MM. From blastocyst to gastrula: gene regulatory networks of embryonic stem cells and early mouse embryogenesis. Philos Trans R Soc Lond B Biol Sci 2014;369:20130542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chambers I, Colby D, Robertson M et al. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 2003;113:643–55. [DOI] [PubMed] [Google Scholar]
- 6. Mitsui K, Tokuzawa Y, Itoh H et al. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 2003;113:631–42. [DOI] [PubMed] [Google Scholar]
- 7. Odorico JS, Kaufman DS, Thomson JA. Multilineage differentiation from human embryonic stem cell lines. Stem Cells 2001;19:193–204. [DOI] [PubMed] [Google Scholar]
- 8. Yu J, Vodyanik MA, Smuga-Otto K et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007;318:1917–20. [DOI] [PubMed] [Google Scholar]
- 9. Christian JL. Morphogen gradients in development: from form to function. Wiley Interdiscip Rev Dev Biol 2012;1:3–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. LeGoff L, Lecuit T. Mechanical forces and growth in animal tissues. Cold Spring Harb Perspect Biol 2015;8:a019232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stathopoulos A, Iber D. Studies of morphogens: keep calm and carry on. Development 2013;140:4119–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lecuit T, Lenne PF. Cell surface mechanics and the control of cell shape, tissue patterns and morphogenesis. Nat Rev Mol Cell Biol 2007;8:633–44. [DOI] [PubMed] [Google Scholar]
- 13. Saha S, Ji L, de Pablo JJ et al. Inhibition of human embryonic stem cell differentiation by mechanical strain. J Cell Physiol 2006;206:126–37. [DOI] [PubMed] [Google Scholar]
- 14. Saha S, Ji L, de Pablo JJ et al. TGFbeta/Activin/Nodal pathway in inhibition of human embryonic stem cell differentiation by mechanical strain. Biophys J 2008;94:4123–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Teramura T, Takehara T, Onodera Y et al. Mechanical stimulation of cyclic tensile strain induces reduction of pluripotent related gene expressions via activation of Rho/ROCK and subsequent decreasing of AKT phosphorylation in human induced pluripotent stem cells. Biochem Biophys Res Commun 2012;417:836–41. [DOI] [PubMed] [Google Scholar]
- 16. Chowdhury F, Na S, Li D et al. Material properties of the cell dictate stress-induced spreading and differentiation in embryonic stem cells. Nat Mater 2010;9:82–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Przybyla L, Lakins JN, Weaver VM. Tissue mechanics orchestrate Wnt-dependent human embryonic stem cell differentiation. Cell Stem Cell 2016;19:462–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xue X, Sun Y, Resto-Irizarry AM et al. Mechanics-guided embryonic patterning of neuroectoderm tissue from human pluripotent stem cells. Nat Mater 2018;17:633–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Zhang HT, Hiiragi T. Symmetry breaking in the mammalian embryo. Annu Rev Cell Dev Biol 2018;34:405–26. [DOI] [PubMed] [Google Scholar]
- 20. Dhaliwal NK, Miri K, Davidson S et al. KLF4 nuclear export requires ERK activation and initiates exit from naive pluripotency. Stem Cell Reports 2018;10:1308–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Topal T, Hong X, Xue X et al. Acoustic tweezing cytometry induces rapid initiation of human embryonic stem cell differentiation. Sci Rep 2018;8:12977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mammoto A, Mammoto T, Ingber DE. Mechanosensitive mechanisms in transcriptional regulation. J Cell Sci 2012;125:3061–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Strumpf D, Mao CA, Yamanaka Y et al. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development 2005;132:2093–02. [DOI] [PubMed] [Google Scholar]
- 24. Giritharan G, Ilic D, Gormley M et al. Human embryonic stem cells derived from embryos at different stages of development share similar transcription profiles. PLoS One 2011;6:e26570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Pan G, Qin B, Liu N et al. Identification of a nuclear localization signal in OCT4 and generation of a dominant negative mutant by its ablation. J Biol Chem 2004;279:37013–20. [DOI] [PubMed] [Google Scholar]
- 26. Okuyama T, Yamagishi R, Shimada J et al. Structural and mechanistic insights into nuclear transport and delivery of the critical pluripotency factor Oct4 to DNA. J Biomol Struct Dyn 2018;36:767–78. [DOI] [PubMed] [Google Scholar]
- 27. Yasuhara N, Shibazaki N, Tanaka S et al. Triggering neural differentiation of ES cells by subtype switching of importin-alpha. Nat Cell Biol 2007;9:72–9. [DOI] [PubMed] [Google Scholar]
- 28. Osmanagic-Myers S, Dechat T, Foisner R. Lamins at the crossroads of mechanosignaling. Genes Dev 2015;29:225–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Constantinescu D, Gray HL, Sammak PJ et al. Lamin A/C expression is a marker of mouse and human embryonic stem cell differentiation. Stem Cells 2006;24:177–85. [DOI] [PubMed] [Google Scholar]
- 30. Villa-Diaz LG, Kim JK, Laperle A et al. Inhibition of focal adhesion kinase signaling by integrin alpha6beta1 supports human pluripotent stem cell self-renewal. Stem Cells 2016;34:1753–64. [DOI] [PubMed] [Google Scholar]
- 31. Weber GF, Bjerke MA, DeSimone DW. A mechanoresponsive cadherin–keratin complex directs polarized protrusive behavior and collective cell migration. Dev Cell 2012;22:104–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Simoes Sde M, Mainieri A, Zallen JA. Rho GTPase and Shroom direct planar polarized actomyosin contractility during convergent extension. J Cell Biol 2014;204:575–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pines M, Das R, Ellis SJ et al. Mechanical force regulates integrin turnover in Drosophila in vivo. Nat Cell Biol 2012;14:935–43. [DOI] [PubMed] [Google Scholar]
- 34. Crawford BD, Henry CA, Clason TA et al. Activity and distribution of paxillin, focal adhesion kinase, and cadherin indicate cooperative roles during zebrafish morphogenesis. Mol Biol Cell 2003;14:3065–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Moore SW, Roca-Cusachs P, Sheetz MP. Stretchy proteins on stretchy substrates: the important elements of integrin-mediated rigidity sensing. Dev Cell 2010;19:194–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Schwartz MA, DeSimone DW. Cell adhesion receptors in mechanotransduction. Curr Opin Cell Biol 2008;20:551–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pera MF, Reubinoff B, Trounson A. Human embryonic stem cells. J Cell Sci 2000;113:5–10. [DOI] [PubMed] [Google Scholar]
- 38. Donovan PJ, Gearhart J. The end of the beginning for pluripotent stem cells. Nature 2001;414:92–7. [DOI] [PubMed] [Google Scholar]
- 39. Kopp JL, Ormsbee BD, Desler M et al. Small increases in the level of Sox2 trigger the differentiation of mouse embryonic stem cells. Stem Cells 2008;26:903–11. [DOI] [PubMed] [Google Scholar]
- 40. Loh YH, Wu Q, Chew JL et al. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet 2006;38:431–40. [DOI] [PubMed] [Google Scholar]
- 41. Oka M, Moriyama T, Asally M et al. Differential role for transcription factor Oct4 nucleocytoplasmic dynamics in somatic cell reprogramming and self-renewal of embryonic stem cells. J Biol Chem 2013;288:15085–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Heisenberg CP, Bellaiche Y. Forces in tissue morphogenesis and patterning. Cell 2013;153:948–62. [DOI] [PubMed] [Google Scholar]
- 43. Leckband DE, le Duc Q, Wang N et al. Mechanotransduction at cadherin-mediated adhesions. Curr Opin Cell Biol 2011;23:523–30. [DOI] [PubMed] [Google Scholar]
- 44. Budnar S, Yap AS. A mechanobiological perspective on cadherins and the actin-myosin cytoskeleton. F1000Prime Rep 2013;5:35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Maitre JL, Turlier H, Illukkumbura R et al. Asymmetric division of contractile domains couples cell positioning and fate specification. Nature 2016;536:344–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Whitesides GM, Ostuni E, Takayama S et al. Soft lithography in biology and biochemistry. Annu Rev Biomed Eng 2001;3:335–73. [DOI] [PubMed] [Google Scholar]
- 47. Villa-Diaz LG, Nandivada H, Ding J et al. Synthetic polymer coatings for long-term growth of human embryonic stem cells. Nat Biotechnol 2010;28:581–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kim BC, Moraes C, Huang J et al. Fracture-based fabrication of normally closed, adjustable, and fully reversible microscale fluidic channels. Small 2014;10:4020–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McCloy RA, Rogers S, Caldon CE et al. Partial inhibition of Cdk1 in G 2 phase overrides the SAC and decouples mitotic events. Cell Cycle 2014;13:1400–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Burgess A, Vigneron S, Brioudes E et al. Loss of human greatwall results in G2 arrest and multiple mitotic defects due to deregulation of the cyclin B-Cdc2/PP2A balance. Proc Natl Acad Sci USA 2010;107:12564–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 2008;3:1101–8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.