Abstract
Autosomal recessive nonsyndromic hearing loss (ARNSHL) is a genetically heterogeneous neurosensory disorder, usually characterized by congenital or prelingual hearing loss. We report a Han Chinese male, born to consanguineous parents, presenting with nonsyndromic sensorineural hearing loss, whose clinical phenotype was also consistent with auditory neuropathy spectrum disorder (ANSD). After exome sequencing, a gap junction protein beta 2 gene (GJB2) c.235delC variant in the homozygous state was detected in the patient. Both parents were heterozygous for this variant, as documented by Sanger sequencing. The known pathogenic GJB2 c.235delC variant was not detected in 200 healthy controls. It is predicted to be a disease-causing alteration by generating a truncated protein p.(L79Cfs*3), disturbing the appropriate folding and/or oligomerization of connexins and leading to defective gap junction channels. This study shows that the association of homozygosity of the GJB2 c.235delC variant with ARNSHL and ANSD in a patient.
Keywords: Auditory neuropathy spectrum disorder, exome sequencing, hearing loss, GJB2 gene, GJB2 c.235delC variant
Autosomal recessive nonsyndromic hearing loss (ARNSHL) is a genetically heterogeneous neurosensory disorder, usually characterized by congenital or prelingual hearing loss, and not accompanied by other clinical features (Xia et al., 2015; Meena and Ayub, 2017). ARNSHL accounts for 45-52.5% of cases of inherited hearing loss, which occurs in approximately 1/1000-2000 newborns (Duman and Tekin, 2012; Duan et al., 2015; Xia et al., 2015). Individuals with ARNSHL usually present difficulty in language development and social interactions.
Since variants in the gap junction protein beta 2 gene (GJB2) were first identified as causative of ARNSHL in 1997 (Kelsell et al., 1997), to date (January, 2019), at least pathogenic variants in other 72 genes have been causally associated with ARNSHL according to the Hereditary Hearing Loss Homepage (https://hereditaryhearingloss.org).
The extreme genetic heterogeneity of nonsyndromic hearing loss makes the use of regular Sanger sequencing to identify its genetic cause very challenging, and exome sequencing has been recommended as a cost-effective alternative strategy (Xia et al., 2016). In the present study, homozygosity for a GJB2 variant was detected by exome sequencing, as causative of autosomal recessive hearing loss in a Han Chinese male presenting auditory neuropathy spectrum disorder (ANSD).
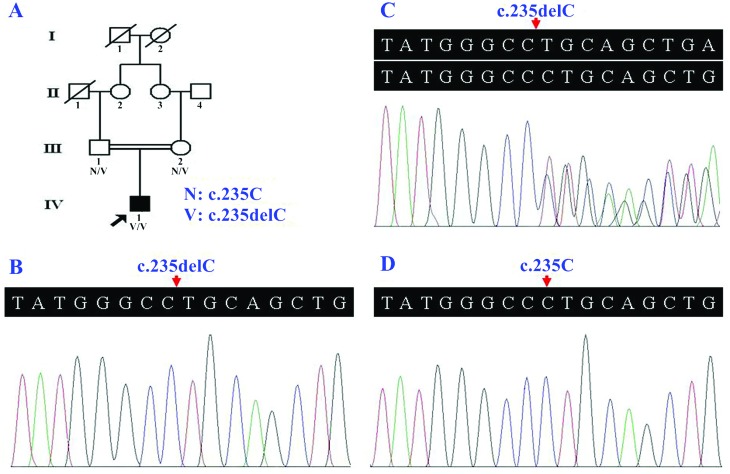
Three members of a Han Chinese family from Hunan, including two normal-hearing first-cousin parents (III:1 and III:2, Figure 1A) and a patient (IV:1, a 27-year-old male), took part in this study. Bilateral prelingual hearing impairment was diagnosed in his first year of life, but neither hearing aids nor cochlear implantation was offered during his childhood. Two hundred unrelated subjects (female/male: 100/100, aged 27.0 ± 6.8 years) without hearing impairments were recruited as healthy controls. Clinical and audiological evaluations were performed on all participants at the Third Xiangya Hospital of Central South University, Changsha, China. Peripheral blood samples were obtained from all participants, and genomic DNA was extracted using a saturated phenol-chloroform extraction method (Yuan et al., 2015). The present study was reviewed and approved by the Institutional Review Board of the Third Xiangya Hospital, Central South University (Changsha, China), in accordance with the Declaration of Helsinki. Written informed consent forms were provided by all participants.
Figure 1. Pedigree of the Han Chinese individual with nonsyndromic hearing loss, and the GJB2 Sanger sequencing electropherograms. (A) The patient was born to first-cousin normal-hearing parents. (B) Homozygosity for the GJB2 c.235delC variant in the individual with hearing loss (IV:1). (C) The heterozygosity for the GJB2 c.235delC variant in the normal-hearing father (III:1). (D) The GJB2 sequence in a normal control.
A series of auditory evaluations, including pure tone audiometry (PTA), tympanometry, acoustic reflex (AR) thresholds, auditory brainstem responses (ABR), transient evoked otoacoustic emission (TEOAE), and distortion product otoacoustic emission (DPOAE) were performed. Potential inner ear congenital malformations were evaluated with magnetic resonance imaging (MRI). Audiometric thresholds were evaluated at frequencies 250, 500, 1000, 2000, 4000, and 8000 Hz by PTA. Hearing acuity is considered ‘normal’ at a threshold within 25 decibels (dB), and the degree of hearing loss was classed as mild (26-40 dB), moderate (41-60 dB), severe (61-80 dB), or profound (> 81 dB) (Asghari et al., 2017). TEOAE and DPOAE were tested using GN otometrics-Madsen capellaTM. Fast-Screen mode and 80 dB hearing level were set for TEOAE examination. DP1, 65 dB hearing level for f1, and 55 dB hearing level for f2 were set for DPOAE.
Three micrograms of the patient’s genomic DNA was used for exome sequencing. It was first sonically sheared and then enriched, hybridized, and captured by the Agilent SureSelect Human All Exon V4 kit at BGI-Shenzhen (Shenzhen, China), according to the manufacturer’s protocol. The library with the targeted exome was sequenced using the Illumina HiSeq 2000 platform. The mean sequencing depth was 101.78, and 99.43% of the targeted exome was covered. Single nucleotide polymorphisms (SNPs) and insertions/deletions (InDels) were detected. Alleles with a frequency > 0.5% in the following databases were screened out based on the SNPs database (dbSNP version 138), 1000 genomes project (1000 genomes release phase 3), HapMap project (2010-08_phase II + III), Exome Sequencing Project 6500 (ESP6500) (Zheng et al., 2016), Exome Aggregation Consortium, and an in-house exome database of BGI. The functional effects of non-synonymous SNPs in the coding regions were predicted by Sorting Intolerant from Tolerant (SIFT, http://sift.jcvi.org/) and Polymorphism Phenotyping version 2 (PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/).
Sanger sequencing was performed to identify whether candidate variants co-segregated with ARNSHL phenotype in the family, using an ABI3500 sequencer (Applied Biosystems, Foster City, CA, USA) (Zheng et al., 2016). Primer sequences for the pathogenic variant in the GJB2 gene were designed and synthesized as follows: forward, 5’-TCGCATTATGATCCTCGTTG-3’ and reverse, 5’-CTCCCCCTTGATGAACTTCC-3’. The function effects of possible candidate variants were further predicted using MutationTaster (http://www.mutationtaster.org/).
The patient’s audiological evaluation revealed profound bilateral sensorineural hearing loss, a type A tympanometric curve, and absent AR and ABR. TEOAE and DPOAE were absent in the patient’s left ear. TEOAE and low amplitude DPOAE at 4000 or 8000 Hz were elicited in the patient’s right ear. MRI showed no anomaly in the patient’s inner ears. The patient’s clinical phenotype was also consistent with ANSD, a disorder of the auditory pathway characterized by the absence of ABR and the presence of OAE (Manchaiah et al., 2011). PTA of his parents showed normal hearing level.
Exome sequencing generated 104,662 SNPs and 16,813 InDels. After screening out common and nonpathogenic variants, homozygosity for the c.235delC variant (rs80338943, a known pathogenic variant, NM_004004.5) in the GJB2 gene was found, and there were no other potentially causative variants for hearing loss.
Homozygosity for the c.235delC variant in the GJB2 gene was confirmed in the patient by Sanger sequencing (Figure 1B). His parents were found to be heterozygous for this variant (Figure 1C). The GJB2 c.235delC variant was not detected in the 200 healthy controls (Figure 1D), and it is predicted to be disease-causing by MutationTaster, resulting in a shift in the reading frame at codon 79 and a premature stop codon at codon 81, p.(L79Cfs*3).
Variants in the GJB2 gene are the primary cause of ARNSHL and responsible for 5-43% of nonsyndromic hearing loss in different ethnicities (Kenneson et al., 2002; Duan et al., 2015). Presently at least 400 pathogenic variants in the GJB2 gene are known on the basis of the Human Gene Mutation Database (http://www.hgmd.cf.ac.uk/ac/index.php). Mutation spectrum and frequency in the GJB2 gene vary with ethnicity (Zheng et al., 2015).
In this study, by exome sequencing, a homozygous GJB2 c.235delC variant, known to be pathogenic (Dai et al., 2015), was found in an individual with hearing loss, inherited from first-cousin normal-hearing heterozygous parents. Variants in other causative genes for hearing loss were excluded. Exome sequencing is a powerful strategy for accurate diagnosis of ARNSHL or ANSD, a highly genetically heterogeneous disorder.
The GJB2 gene encodes connexin 26, a gap-junction protein, expressed in the human and rat cochlear cells (Kelsell et al., 1997). Connexin 26 consists of an N-terminal helix, four transmembrane helices (TM1-4), two extracellular loops (E1 and E2), a cytoplasmic loop (CL), and a C-terminus (Maeda et al., 2009). The protein is involved in recycling potassium ions (Kelsell et al., 1997), ATP release, intercellular signaling, hearing function regulation (Zhao et al., 2005), cochlear development, and active cochlear amplification (Chen et al., 2014). Connexin 26 knockout mice displayed congenital hearing loss and cochlear developmental disorders (Chen et al., 2014). Conditional knockout mice showed severe hearing loss and DPOAE reductions (Zhu et al., 2013).
The c.235delC variant in the GJB2 gene, predicted to produce a truncated protein, was reported in different populations, especially in East Asia (Dai et al., 2015; Taniguchi et al., 2015). The GJB2 c.235delC variant involving the TM2 domain is predicted to be a disease-causing alteration by MutationTaster. It generates a truncated protein p.(L79Cfs*3) missing important functional segments, including CL, TM3, E2, TM4, and C-terminal segments. The glutamine (p.Q80) in the TM2 segment of connexin 26 interacts with arginine (p.R32) in the TM1 segment, thus the variant may interfere with the interplay between the two TM domains, disturb the appropriate folding and/or oligomerization of connexins, and lead to defective gap junction channels (Maeda et al., 2009).
GJB2 variants have been reported in 7.5% of patients with ANSD (Carvalho et al., 2016). Our patient was diagnosed with ANSD due to the presence of right ear OAE, but the absence of ABR, and this is the first report of the GJB2 c.235delC variant in connection with ANSD. ANSD may result from an abnormality in the inner hair cells (IHC), in the synapse between IHC and auditory nerve, or in the auditory nerve itself (Starr et al., 1996). Connexin 26 is expressed in the cochlear basement membrane on which the hair cells lie (Kelsell et al., 1997). Connexin 26 expression contributes to IHC functional maturation (Johnson et al., 2017), thus GJB2-associated ANSD may be caused by immature IHCs.
In conclusion, our study shows a novel association of homozygosity for the c.235delC variant in the GJB2 gene with the phenotypes of ARNSHL and ANSD.
Acknowledgments
The work was supported by grants from National Key Research and Development Program of China (2016YFC1306604); National Natural Science Foundation of China (81670216); Natural Science Foundation of Hunan Province, China (2016JJ2166 and 2017JJ3469); the Foster Key Subject of the Third Xiangya Hospital of Central South University, China (Clinical Laboratory Diagnostics); the New Xiangya Talent Project of the Third Xiangya Hospital of Central South University, China (20150301); National-level College Students’ Innovative Training Plan Program, China (201710533227 and 201810533244).
Footnotes
Associate Editor: Angela M. Vianna-Morgante
Conflicts of interest
The authors declare that there is no conflict of interest.
Author contributions
HX and HD conceived and designed the study. HX, XH, HBX, YAZ, LG, ZY, JL and HD conducted the experiments. HX, XH and HD analyzed the data. HX and HD wrote the manuscript. All authors read and approved the final version.
References
- Asghari A, Farhadi M, Daneshi A, Khabazkhoob M, Mohazzab-Torabi S, Jalessi M, Emamjomeh H. The prevalence of hearing impairment by age and gender in a population-based study. Iran J Public Health. 2017;46:1237–1246. [PMC free article] [PubMed] [Google Scholar]; Asghari A, Farhadi M, Daneshi A, Khabazkhoob M, Mohazzab-Torabi S, Jalessi M and Emamjomeh H (2017) The prevalence of hearing impairment by age and gender in a population-based study. Iran J Public Health 46:1237-1246. [PMC free article] [PubMed]
- Chen J, Chen J, Zhu Y, Liang C, Zhao HB. Deafness induced by Connexin 26 (GJB2) deficiency is not determined by endocochlear potential (EP) reduction but is associated with cochlear developmental disorders. Biochem Biophys Res Commun. 2014;448:28–32. doi: 10.1016/j.bbrc.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]; Chen J, Chen J, Zhu Y, Liang C and Zhao HB (2014) Deafness induced by Connexin 26 (GJB2) deficiency is not determined by endocochlear potential (EP) reduction but is associated with cochlear developmental disorders. Biochem Biophys Res Commun 448:28-32. [DOI] [PMC free article] [PubMed]
- Dai ZY, Sun BC, Huang SS, Yuan YY, Zhu YH, Su Y, Dai P. Correlation analysis of phenotype and genotype of GJB2 in patients with non-syndromic hearing loss in China. Gene. 2015;570:272–276. doi: 10.1016/j.gene.2015.06.038. [DOI] [PubMed] [Google Scholar]; Dai ZY, Sun BC, Huang SS, Yuan YY, Zhu YH, Su Y and Dai P (2015) Correlation analysis of phenotype and genotype of GJB2 in patients with non-syndromic hearing loss in China. Gene 570:272-276. [DOI] [PubMed]
- de Carvalho GM, Ramos PZ, Castilho AM, Guimarães AC, Sartorato EL. Relationship between patients with clinical auditory neuropathy spectrum disorder and mutations in Gjb2 gene. Open Neurol J. 2016;10:127–135. doi: 10.2174/1874205X01610010127. [DOI] [PMC free article] [PubMed] [Google Scholar]; de Carvalho GM, Ramos PZ, Castilho AM, Guimarães AC and Sartorato EL (2016) Relationship between patients with clinical auditory neuropathy spectrum disorder and mutations in Gjb2 gene. Open Neurol J 10:127-135. [DOI] [PMC free article] [PubMed]
- Duan SH, Zhu YM, Wang YL, Guo YF. Common molecular etiology of nonsyndromic hearing loss in 484 patients of 3 ethnicities in northwest China. Acta Otolaryngol. 2015;135:586–591. doi: 10.3109/00016489.2015.1006334. [DOI] [PubMed] [Google Scholar]; Duan SH, Zhu YM, Wang YL and Guo YF (2015) Common molecular etiology of nonsyndromic hearing loss in 484 patients of 3 ethnicities in northwest China. Acta Otolaryngol 135:586-591. [DOI] [PubMed]
- Duman D, Tekin M. Autosomal recessive nonsyndromic deafness genes: a review. Front Biosci. 2012;17:2213–2236. doi: 10.2741/4046. [DOI] [PMC free article] [PubMed] [Google Scholar]; Duman D and Tekin M (2012) Autosomal recessive nonsyndromic deafness genes: a review. Front Biosci 17: 2213-2236. [DOI] [PMC free article] [PubMed]
- Johnson SL, Ceriani F, Houston O, Polishchuk R, Polishchuk E, Crispino G, Zorzi V, Mammano F, Marcotti W. Connexin-mediated signaling in nonsensory cells is crucial for the development of sensory inner hair cells in the mouse cochlea. J Neurosci. 2017;37:258–268. doi: 10.1523/JNEUROSCI.2251-16.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]; Johnson SL, Ceriani F, Houston O, Polishchuk R, Polishchuk E, Crispino G, Zorzi V, Mammano F and Marcotti W (2017) Connexin-mediated signaling in nonsensory cells is crucial for the development of sensory inner hair cells in the mouse cochlea. J Neurosci 37:258-268 [DOI] [PMC free article] [PubMed]
- Kelsell DP, Dunlop J, Stevens HP, Lench NJ, Liang JN, Parry G, Mueller RF, Leigh IM. Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature. 1997;387:80–83. doi: 10.1038/387080a0. [DOI] [PubMed] [Google Scholar]; Kelsell DP, Dunlop J, Stevens HP, Lench NJ, Liang JN, Parry G, Mueller RF and Leigh IM (1997) Connexin 26 mutations in hereditary non-syndromic sensorineural deafness. Nature 387:80-83. [DOI] [PubMed]
- Kenneson A, Van Naarden Braun K, Boyle C. GJB2 (connexin 26) variants and nonsyndromic sensorineural hearing loss: A HuGE review. Genet Med. 2002;4:258–274. doi: 10.1097/00125817-200207000-00004. [DOI] [PubMed] [Google Scholar]; Kenneson A, Van Naarden Braun K and Boyle C (2002) GJB2 (connexin 26) variants and nonsyndromic sensorineural hearing loss: A HuGE review. Genet Med 4:258-274. [DOI] [PubMed]
- Maeda S, Nakagawa S, Suga M, Yamashita E, Oshima A, Fujiyoshi Y, Tsukihara T. Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature. 2009;458:597–602. doi: 10.1038/nature07869. [DOI] [PubMed] [Google Scholar]; Maeda S, Nakagawa S, Suga M, Yamashita E, Oshima A, Fujiyoshi Y and Tsukihara T (2009) Structure of the connexin 26 gap junction channel at 3.5 A resolution. Nature 458:597-602. [DOI] [PubMed]
- Manchaiah VK, Zhao F, Danesh AA, Duprey R. The genetic basis of auditory neuropathy spectrum disorder (ANSD) Int J Pediatr Otorhinolaryngol. 2011;75:151–158. doi: 10.1016/j.ijporl.2010.11.023. [DOI] [PubMed] [Google Scholar]; Manchaiah VK, Zhao F, Danesh AA and Duprey R (2011) The genetic basis of auditory neuropathy spectrum disorder (ANSD). Int J Pediatr Otorhinolaryngol 75:151-158. [DOI] [PubMed]
- Meena R, Ayub M. Genetics of human hereditary hearing impairment. J Ayub Med Coll Abbottabad. 2017;29:671–676. [PubMed] [Google Scholar]; Meena R and Ayub M (2017) Genetics of human hereditary hearing impairment. J Ayub Med Coll Abbottabad 29:671-676. [PubMed]
- Starr A, Picton TW, Sininger Y, Hood LJ, Berlin CI. Auditory neuropathy. Brain. 1996;119:741–753. doi: 10.1093/brain/119.3.741. [DOI] [PubMed] [Google Scholar]; Starr A, Picton TW, Sininger Y, Hood LJ and Berlin CI (1996) Auditory neuropathy. Brain 119:741-753. [DOI] [PubMed]
- Taniguchi M, Matsuo H, Shimizu S, Nakayama A, Suzuki K, Hamajima N, Shinomiya N, Nishio S, Kosugi S, Usami S, et al. Carrier frequency of the GJB2 mutations that cause hereditary hearing loss in the Japanese population. J Hum Genet. 2015;60:613–617. doi: 10.1038/jhg.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]; Taniguchi M, Matsuo H, Shimizu S, Nakayama A, Suzuki K, Hamajima N, Shinomiya N, Nishio S, Kosugi S, Usami S et al. (2015) Carrier frequency of the GJB2 mutations that cause hereditary hearing loss in the Japanese population. J Hum Genet 60:613-617. [DOI] [PMC free article] [PubMed]
- Xia H, Huang X, Guo Y, Hu P, He G, Deng X, Xu H, Yang Z, Deng H. Identification of a novel MYO15A mutation in a Chinese family with autosomal recessive nonsyndromic hearing loss. PLoS One. 2015;10:e0136306. doi: 10.1371/journal.pone.0136306. [DOI] [PMC free article] [PubMed] [Google Scholar]; Xia H, Huang X, Guo Y, Hu P, He G, Deng X, Xu H, Yang Z and Deng H (2015) Identification of a novel MYO15A mutation in a Chinese family with autosomal recessive nonsyndromic hearing loss. PLoS One 10:e0136306. [DOI] [PMC free article] [PubMed]
- Xia H, Xu H, Deng X, Yuan L, Xiong W, Yang Z, Deng H. Compound heterozygous GJB2 mutations associated to a consanguineous Han family with autosomal recessive non-syndromic hearing loss. Acta Otolaryngol. 2016;136:782–785. doi: 10.3109/00016489.2016.1157727. [DOI] [PubMed] [Google Scholar]; Xia H, Xu H, Deng X, Yuan L, Xiong W, Yang Z and Deng H (2016) Compound heterozygous GJB2 mutations associated to a consanguineous Han family with autosomal recessive non-syndromic hearing loss. Acta Otolaryngol 136:782-785. [DOI] [PubMed]
- Yuan L, Deng X, Song Z, Yang Z, Ni B, Chen Y, Deng H. Genetic analysis of the RAB39B gene in Chinese Han patients with Parkinson’s disease. Neurobiol Aging. 2015;36:2907.e11-2. doi: 10.1016/j.neurobiolaging.2015.06.019. [DOI] [PubMed] [Google Scholar]; Yuan L, Deng X, Song Z, Yang Z, Ni B, Chen Y and Deng H (2015) Genetic analysis of the RAB39B gene in Chinese Han patients with Parkinson’s disease. Neurobiol Aging 36:2907.e11-2. [DOI] [PubMed]
- Zhao HB, Yu N, Fleming CR. Gap junctional hemichannel-mediated ATP release and hearing controls in the inner ear. Proc Natl Acad Sci U S A. 2005;102:18724–18729. doi: 10.1073/pnas.0506481102. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhao HB, Yu N and Fleming CR (2005) Gap junctional hemichannel-mediated ATP release and hearing controls in the inner ear. Proc Natl Acad Sci U S A 102:18724-18729. [DOI] [PMC free article] [PubMed]
- Zheng J, Ying Z, Cai Z, Sun D, He Z, Gao Y, Zhang T, Zhu Y, Chen Y, Guan MX. GJB2 mutation spectrum and genotype-phenotype correlation in 1067 Han Chinese subjects with non-syndromic hearing loss. PLoS One. 2015;10:e0128691. doi: 10.1371/journal.pone.0128691. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zheng J, Ying Z, Cai Z, Sun D, He Z, Gao Y, Zhang T, Zhu Y, Chen Y and Guan MX (2015) GJB2 mutation spectrum and genotype-phenotype correlation in 1067 Han Chinese subjects with non-syndromic hearing loss. PLoS One 10:e0128691. [DOI] [PMC free article] [PubMed]
- Zheng W, Zhang J, Deng X, Xiao J, Yuan L, Yang Y, Guan L, Song Z, Yang Z, Deng H. Identification of a premature termination mutation in the proline-rich transmembrane protein 2 gene in a Chinese family with febrile seizures. Mol Neurobiol. 2016;53:835–841. doi: 10.1007/s12035-014-9047-4. [DOI] [PubMed] [Google Scholar]; Zheng W, Zhang J, Deng X, Xiao J, Yuan L, Yang Y, Guan L, Song Z, Yang Z and Deng H (2016) Identification of a premature termination mutation in the proline-rich transmembrane protein 2 gene in a Chinese family with febrile seizures. Mol Neurobiol 53:835-841. [DOI] [PubMed]
- Zhu Y, Liang C, Chen J, Zong L, Chen GD, Zhao HB. Active cochlear amplification is dependent on supporting cell gap junctions. Nat Commun. 2013;4:1786. doi: 10.1038/ncomms2806. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhu Y, Liang C, Chen J, Zong L, Chen GD and Zhao HB (2013) Active cochlear amplification is dependent on supporting cell gap junctions. Nat Commun 4:1786. [DOI] [PMC free article] [PubMed]