Abstract
To carry out effective genome-wide association studies, information about linkage disequilibrium (LD) is essential. Here, we used medium-density SNP chips to provide estimates of LD in native Tunisian cattle. The two measures of LD that were used, mean r2 and D’, decreased from 0.26 to 0.05 and from 0.73 to 0.40, respectively, when the distance between markers increased from less than 20 Kb to 200 Kb. The decay in LD over physical distance occurred at a faster rate than that reported for European and other indigenous breeds, and reached background levels at less than 500 Kb distance. This is consistent with the absence of strong selective pressure within the Tunisian population and suggests that, in order to be effective, any potential genome-wide association mapping studies will need to use chips with higher marker density. An analysis of effective population size (Ne) based on LD data showed a decline in past Ne, with a sudden drop starting about eight generations ago. This finding, combined with the high levels of recent inbreeding revealed by runs of homozygosity (ROH) analysis, indicate that this population is endangered and may be in urgent need of a conservation plan that includes a well-designed genetic management program.
Keywords: Tunisian cattle, conservation, linkage disequilibrium, effective population size, single nucleotide polymorphism
Introduction
Native Tunisian cattle are small animals that are well adapted to the harsh environmental conditions that characterize their rearing area, such as reduced food resources, elevated temperatures during the hot season, and an abundance of parasites and pathogens. They are currently endangered by the result of extensive crossbreeding with imported breeds (Djemali and Kayouli, 2003). Local cattle in Tunisia belong to three main ecotypes, defined according to coat color. These are the Blonde du Cap Bon (BLCAP) (white), the Brune de l’Atlas Grise (BRATG) (grey), and the Brune de l’Atlas Fauve (BRATF) (tawny). Native Tunisian cattle have not been subjected to artificial selection, and are in need of breeding programs to improve their production performance. Nowadays, the availability of high-throughput genotyping technologies makes it possible to use genetic markers to map genes that are responsible for economically important traits. Such information can then be incorporated into breeding programs. Before one can efficiently map these genes, it is, however, necessary to first determine the marker density that is compatible with the distances across which linkage disequilibrium (LD) extends in the population of interest. LD refers to the non-random association of alleles at different loci in gametes. LD patterns vary substantially among populations and can arise both through physical linkage between loci and, more generally, through a variety of evolutionary forces that structure a genome, such as genetic drift, admixture, and natural selection. By characterizing the pattern of LD within a population, we are able to determine the amount of markers needed to optimize the mapping resolution of genetic variants related to economic traits. Furthermore, LD is used routinely for the study of the demographic history of populations in many species, such as humans (Reich et al., 2001; McEvoy et al., 2011), cattle (Khatkar et al., 2008; Porto-Neto et al., 2014), and sheep (McRae et al., 2002; Al-Mamun et al., 2015). It can also be used to infer recent and historical effective population size (Ne). Ne is defined as the size of an “ideal” population – a hypothetical population that is panmictic, large enough that sampling error is negligible (thereby preventing genetic drift), with an equal sex ratio, and without migration, mutation, or selection – that would experience the effects of inbreeding or drift to the same degree as observed in the actual breeding population (Falconer and Mackay, 1996). Ne is an important parameter for monitoring the genetic health of endangered populations as it reflects the risk of extinction that is posed by genetic drift (which drives the loss of alleles in endangered populations). Here, we characterized LD in the genome of local Tunisian cattle by analyzing 87 individuals using the Illumina BovineSNP50 chip assay. We then used LD estimates to infer changes in the past effective population size of native Tunisian cattle over time. In addition, to get an overview of population inbreeding, we present a genome-wide characterization of runs of homozygosity (ROH) in the Tunisian individuals.
Materials and Methods
Animal sampling
We identified for sampling 40 individuals of indigenous Tunisian cattle; these originated from both the northwestern and central parts of Tunisia, which are two of the main regions in the country where local cattle breeds are found. Animal sampling was carried out by a multi-institutional team representing the Livestock and Pasture Office (OEP), the Office of Silvopastoral Development of the North-West (ODESYPANO), the National Gene Bank (BNG), and the National Institute of Agronomic Research of Tunisia (INRAT).
Since most native cattle have been extensively crossed with imported breeds, significant efforts were made to select purebred individuals. Selection was performed based on morphological criteria and targeted isolated mountainous regions where no historical artificial insemination activity was recorded. Our sample included 25 BRATF and 15 BRATG individuals. Blood samples were collected in EDTA Vacutainer tubes under procedures approved by the Tunisian Veterinary Authority.
DNA extraction and genotyping assays
DNA was extracted using the phenol-chloroform protocol (Cheng et al., 1995). DNA quantity and quality were evaluated using a Nanodrop instrument, and DNA samples were then genotyped on the BovineSNP50 BeadChip Ver. 2 (Illumina, San Diego, CA, USA) at the Labogena core facility (Jouy-en-Josas, France) using standard operating procedures, as recommended by the manufacturer (http://www.illumina.com).
Marker quality control and selection
Overall, we obtained genotyping data from all 40 animals for 52,274 SNPs. Quality control of these data was performed using PLINK software v1.9 (Purcell et al., 2007). Samples with a genotyping rate of <90% across autosomal SNPs and markers with a call rate <90% were discarded. Using these criteria, we excluded one BRATF individual and 2,232 SNPs from further analysis. An exact test for Hardy-Weinberg equilibrium (p <0.01) was then carried out on the remaining SNPs using PLINK, which resulted in the retention of 49,362 SNPs for subsequent analyses. We combined these newly generated genotypes with those already available for 48 individuals of Tunisian cattle: 18 BRATF, 15 BRATG, and 15 BLCAP (Ben Jemaa et al., 2015). Thus, our final data set was composed of 48,458 SNPs genotyped for 87 Tunisian individuals.
To evaluate the effect of minor allele frequencies (MAFs) on LD, we estimated the LD score using three different MAF thresholds: 0.01 (MAF 0.01), 0.05 (MAF 0.05), and 0.1 (MAF 0.1). The application of these three thresholds resulted in the retention of 42,699, 38,078, and 33,146 SNPs, respectively. In order to facilitate comparisons among the three datasets, we reduced the size of the MAF 0.01 and MAF 0.05 datasets to 33,333 and 33,509 SNPs, respectively.Marker coverage and SNP density per chromosome are indicated for each MAF threshold in Table S1 (67.2KB, pdf) , and the distribution of allele frequencies for each MAF cutoff value is illustrated in Figure S1 (53.5KB, pdf) .
Detection of runs of homozygosity
To assess levels of inbreeding in our 87 individuals, we examined our genetic data for runs of homozygosity (ROH). These were detected in sliding windows of 20 SNPs using PLINK. No more than five missing calls and one heterozygous SNP were allowed in each window. The minimum length of a ROH segment was set to 1 Mb, and a ROH was declared if it contained at least 20 SNPs. The minimum required SNP density was one SNP per 200 kb and the maximum gap allowed between any two consecutive SNPs was 1000 kb. ROHs were classified into seven categories based on length (1 to 4.999 Mb, 5 to 9.999 Mb, 10 to 14.999 Mb, 15 to 19.999 Mb, 20 to 24.999 Mb, 25 to 49.999 Mb, >50 Mb).
Estimation of linkage disequilibrium
We used the genotyping data from these 87 individuals to estimate linkage disequilibrium in our sample. For this, we used Haploview v4.2 (Barrett et al., 2005) to generate two standard descriptive LD parameters, D’ and r2, for all syntenic SNP pairs that were less than 50 Mb apart. All values of r2 between syntenic markers were corrected for sample size using the following equation:
where n is the number of haplotypes in the sample (Villa-Angulo et al., 2009). To better understand how LD estimates were related to physical distances between syntenic marker pairs, we used R software (https://www.r-project.org/) to analyze LD decay over the following windows of genetic distance (Kb): 0-19; 20-39; 40-59; 60-99; 100-199; 200-499; 500-999; 1,000-1,999; 2,000-4999; 5,000-9,999; 10,000-11,999; 12,000-14,999; 15,000-16,999; 17,000-19,999; 20,000-29,999; 30,000-50,000. Average r2 and D’ values were determined and plotted for all combined autosomes for each distance window and for each MAF cutoff value. LD parameters and the percentage of SNP pairs with r2 values >0.2 or 0.8 were also calculated between adjacent SNPs within each chromosome. We further determined the amount of background linkage disequilibrium by calculating r2 and D’ for non-syntenic SNPs. This was done by randomly selecting 1,700 SNPs (~5% of the markers) distributed over all chromosomes.
Estimation of effective population size
We used SNeP software (Barbato et al., 2015) to estimate Ne and its change over time using the observed extent of LD calculated for the three MAF thresholds. SNeP estimates the historic effective population size based on the following equation (Corbin et al., 2012).
where N T(t) is the effective population size estimated t generations ago, f(ct) is the mapping function used to estimate the recombination rate (ct) t generations ago, r2adj is the LD estimation, adjusted for sampling bias (r2adj=r2-(1/2N), where N is the population sample size), and “a” is a constant that accounts for mutation. Inter-marker distances used in our NT(t) estimates ranged from 0.02 to 50 Mb. We also set a to 1 when f(ct) was calculated using the Sved and Feldman (1973) 0 approximation: f(ct) = d(1-d/2), where d is the linkage distance between SNPs, which is proportional to the physical distance, δ (δ=10-8*d).
Results
Detection of ROHs in the Tunisian cattle genome
A total of 3,313 ROHs were identified in the 87 Tunisian cattle individuals. The average number of ROHs per individual was 38 ± 12.6 (range: 16 to 81). Most ROHs (85%) were between 1 and 5 Mb long. Chromosome BTA01 had the highest number of ROHs (305), while BTA28 had the least (39) (Figure S2 (47.3KB, pdf) ). Overall, the number of ROHs per chromosome tended to decrease with chromosome length (correlation between chromosome length and the number of ROHs was ~92%). Among the 87 individuals, 36 (~41%) had a total sum of ROHs >100 Mb, while 15 individuals (~ 17%) had a total sum of ROHs >250 Mb (Figure S3 (46.4KB, pdf) ). Among these, 14 individuals (~93%) had at least one ROH that was more than 25 Mb long (Table S2 (23.4KB, pdf) ). Approximately 5.4%, on average, of the entire genome of Tunisian cattle was covered by ROH segments. Chromosome BTA25 showed the highest coverage by ROHs (12.4%) while BTA01 had the least (1.6%) (Figure S2 (47.3KB, pdf) ).
Characterization of Linkage Disequilibrium
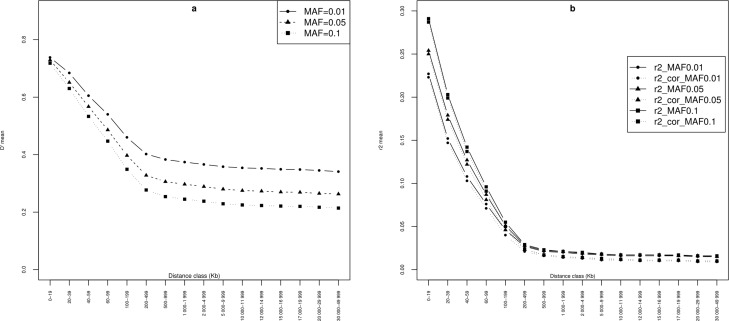
The extent of LD was first evaluated for all syntenic SNP pairs for all chromosomes combined. Table 1 shows the values estimated for the decay of r2, r2 corrected for sample size, and D’ for all 29 autosomes; this information is displayed graphically in Figure 1. Correction for sample size resulted in a slight decrease (~7% on average) in r2 estimates for markers that were less than 500 Kb apart. Overall, LD parameters declined rapidly over short inter-marker distances. For example, the estimated r2 values averaged over the three MAF thresholds decreased from 0.26 for syntenic SNPs less than 20 Kb apart to 0.05 for those that were less than 200 Kb apart. However, LD decay was slower over longer physical distances. For example, mean D’ dropped from 0.33 for SNPs separated by 200-499 Kb to 0.28 for those that were more than 30 Mb apart.
Table 1. Average D’ and r2 estimates by distance class.
| Distance_class (Kb) | Number of pairs | Average D’ (sd) | Average r2 (sd) | Average r2 corrected (sd) | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF0.01 | MAF0.05 | MAF0.1 | MAF0.01 | MAF0.05 | MAF0.1 | MAF0.01 | MAF0.05 | MAF0.1 | MAF0.01 | MAF0.05 | MAF0.1 | |
| 0-19 | 546 | 551 | 580 | 0.738(0.34) | 0.729(0.33) | 0.718(0.33) | 0.227(0.3) | 0.254(0.29) | 0.291(0.31) | 0.223(0.299) | 0.250(0.29) | 0.287(0.30) |
| 20-39 | 11 888 | 12 264 | 12 074 | 0.684(0.35) | 0.651(0.34) | 0.630(0.34) | 0.152(0.22) | 0.179(0.24) | 0.203(0.25) | 0.147(0.226) | 0.174(0.23) | 0.199(0.25) |
| 40-59 | 9 556 | 9 738 | 9 571 | 0.605(0.35) | 0.567(0.34) | 0.533(0.34) | 0.108(0.18) | 0.127(0.19)) | 0.142(0.2) | 0.103(0.179) | 0.122(0.19) | 0.137(0.19) |
| 60-99 | 19 319 | 19 759 | 19 398 | 0.540(0.36) | 0.486(0.34) | 0.447(0.32) | 0.076(0.14) | 0.087(0.14) | 0.096(0.15) | 0.071(0.137) | 0.081(0.14) | 0.091(0.15) |
| 100-199 | 47 353 | 48 233 | 47 311 | 0.460(0.35) | 0.397(0.31) | 0.349(0.28) | 0.046(0.09) | 0.051(0.09 | 0.055(0.1) | 0.040(0.090) | 0.046(0.09) | 0.050(0.09) |
| 200-499 | 139 280 | 141 276 | 138 233 | 0.402(0.34) | 0.328(0.29 | 0.277(0.24) | 0.027(0.05) | 0.028(0.05) | 0.029(0.05) | 0.021(0.047) | 0.023(0.04) | 0.024(0.04) |
| 500-999 | 228 259 | 231 215 | 226 852 | 0.383(0.33 | 0.306(0.28) | 0.254(0.23) | 0.021(0.03) | 0.022(0.03) | 0.023(0.03 | 0.016(0.033) | 0.017(0.03) | 0.017(0.03) |
| 1 000-1 999 | 447 448 | 453 482 | 442 903 | 0.374(0.33) | 0.297(0.27) | 0.245(0.22) | 0.020(0.03) | 0.021(0.03) | 0.021(0.03) | 0.014(0.029) | 0.015(0.02) | 0.015(0.02) |
| 2 000-4 999 | 1 290 593 | 1 303 873 | 1 274 310 | 0.366(0.33) | 0.289(0.27) | 0.238(0.22) | 0.018(0.03) | 0.019(0.03) | 0.020(0.03) | 0.013(0.027) | 0.014(0.02) | 0.014(0.02) |
| 5 000-9 999 | 2 010 370 | 2 027 558 | 1 982 159 | 0.358(0.33) | 0.280(0.27) | 0.229(0.21) | 0.017(0.02) | 0.018(0.02) | 0.018(0.02) | 0.011(0.025) | 0.012(0.02) | 0.013(0.02) |
| 10 000-11 999 | 760 396 | 765 970 | 749 864 | 0.354(0.33) | 0.275(0.26) | 0.225(0.21) | 0.016(0.02) | 0.017(0.02) | 0.017(0.02) | 0.011(0.023) | 0.012(0.02) | 0.012(0.02) |
| 12 000-14 999 | 1 100 143 | 1 106 024 | 1 081 484 | 0.352(0.33) | 0.273(0.26) | 0.223(0.21 | 0.016(0.02) | 0.017(0.02) | 0.017(0.02) | 0.010(0.023) | 0.011(0.02) | 0.011(0.02) |
| 15 000-16 999 | 708 390 | 711 382 | 695 392 | 0.349(0.33) | 0.270(0.26) | 0.221(0.21) | 0.016(0.02) | 0.017(0.02) | 0.017(0.02) | 0.010(0.022) | 0.011(0.02) | 0.011(0.02) |
| 17 000-19 999 | 1 027 451 | 1 033 865 | 1 011 227 | 0.348(0.33) | 0.269(0.26) | 0.220(0.21) | 0.016(0.02) | 0.016(0.02 | 0.017(0.02) | 0.010(0.022) | 0.011(0.02) | 0.011(0.02) |
| 20 000-29 999 | 3 100 869 | 3 115 808 | 3 048 049 | 0.345(0.33) | 0.265(0.26) | 0.217(0.21) | 0.015(0.02) | 0.016(0.02) | 0.016(0.02) | 0.009(0.022) | 0.010(0.02) | 0.010(0.02) |
| 30 000-49 999 | 4 710 189 | 4 739 668 | 4 629 739 | 0.341(0.33) | 0.263(0.26) | 0.214(0.2) | 0.015(0.02) | 0.015(0.02) | 0.016(0.02) | 0.009(0.021) | 0.010(0.02) | 0.010(0.02) |
| Non-syntenic SNPs | 1 444 151 | 1 444 151 | 1 444 151 | 0.342(0.32) | 0.258(0.25) | 0.214(0.2) | 0.0145(0.02) | 0.015(0.02) | 0.015(0.02) | 0.008(0.020) | 0.009(0.02) | 0.009(0.02) |
Figure 1. Average LD decay pooled over all autosomes for three different minor allele frequency (MAF) thresholds in the Tunisian cattle population: (a) D’ , (b) r2 (as estimated by Haploview) and r2 corrected for sample size (r2_cor).
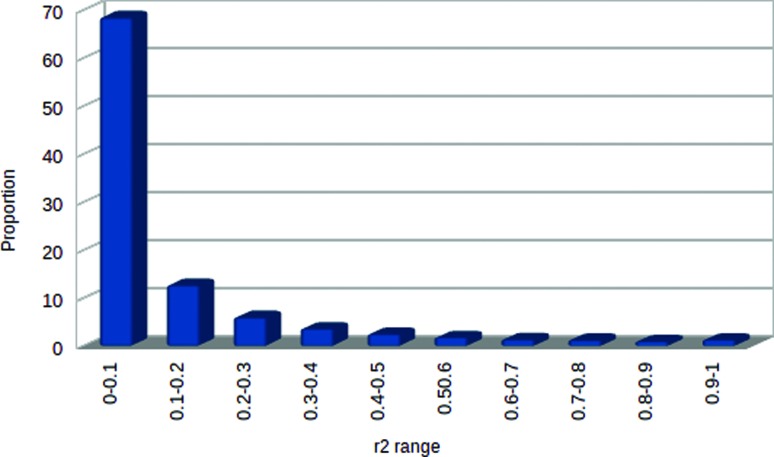
The effect of MAF on the extent of LD among syntenic SNPs was studied using three different MAF thresholds: 1, 5, and 10% (Figure 1 and Table 1). Overall, we analyzed 17,056,201 SNP pairs for MAF=0.01, 17,164,817 for MAF=0.05, and 16,813,297 SNP pairs for MAF=0.1. It seemed that the MAF had a strong effect on mean r2 over short physical distances (≤ 200 Kb), while its effect on D’ was more pronounced over long distances (Figure 1 and Table 1). For instance, within the range 0-19 Kb, mean r2 increased by 28% passing from MAF=0.01 to MAF=0.1, while the increase was only 8% within the 500-999 Kb interval. The range of D’ and r2 values for SNP pairs more than 500 Kb apart were similar to those observed for non-syntenic markers, for which around 79% of D’ values were lower than 0.4 and 94% of r2 values were lower than 0.05 (Figure S4 (69.6KB, pdf) ). The distribution of r2 values between adjacent SNP pairs, combined over all autosomes, is shown in Figure 2. Most adjacent markers (~80%) had an average r2 value <0.2 while only 2% were in strong or complete LD (0.8 <r2 <1). Mean genomic distance between these marker pairs was 73.4 Kb and 35.4 Kb, respectively (data not shown). The mean value of r2 between adjacent SNP pairs pooled over all autosomes was 0.132 ± 0.19. Some variations in the LD values between adjacent SNPs were observed within chromosomes (Table 2), with BTA 19, 23, 25, 26, 28, and 29 having the lowest LD values and BTA 1, 6, and 8 having the highest ones. Per-chromosome values ranged from 0.106 ± 0.17 for BTA28 to 0.158 ± 0.228 for BTA06 (r2) and from 0.494 ± 0.33 for BTA23 to 0.158 ± 0.22 for BTA06 (D’). Unsurprisingly, chromosomes with the lowest LD values tended to have the lowest proportion of marker pairs with strong and moderate LD.
Figure 2. Distribution of r2 values between adjacent SNP pairs pooled over all autosomes.
Table 2. Mean r2 and D’ between adjacent SNPs on each chromosome in Tunisian native cattle and percentage of marker pairs with r2 >0.2 or 0.8.
| BTA | Intermarker average distance (Kb) | Average D’ ± sd | Average r2 ± sd | % r2 >0.2 | % r2 >0.8 |
|---|---|---|---|---|---|
| 1 | 71.47 ± 64.51 | 0.583 ± 0.35 | 0.153 ± 0.21 | 21.61 | 2.82 |
| 2 | 76.82 ± 78.74 | 0.573 ± 0.35 | 0.143 ± 0.20 | 19.81 | 2.64 |
| 3 | 74.60 ± 71.88 | 0.574 ± 0.35 | 0.141 ± 0.20 | 20.17 | 2.22 |
| 4 | 75.29 ± 64.98 | 0.572 ± 0.35 | 0.134 ± 0.20 | 18.57 | 2.27 |
| 5 | 86.16 ± 86.86 | 0.553 ± 0.35 | 0.127 ± 0.19 | 17.44 | 2.25 |
| 6 | 71.51 ± 79.12 | 0.583 ± 0.35 | 0.158 ± 0.22 | 21.93 | 3.40 |
| 7 | 77.68 ± 97.71 | 0.564 ± 0.35 | 0.144 ± 0.20 | 19.82 | 2.68 |
| 8 | 73.45 ± 63.08 | 0.579 ± 0.35 | 0.147 ± 0.21 | 20.47 | 2.65 |
| 9 | 81.52 ± 79.84 | 0.57 ± 0.35 | 0.141 ± 0.20 | 18.22 | 3.02 |
| 10 | 74.76 ± 110.10 | 0.555 ± 0.35 | 0.140 ± 0.20 | 19.74 | 2.89 |
| 11 | 75.18 ± 72.59 | 0.552 ± 0.35 | 0.143 ± 0.21 | 19.81 | 3.24 |
| 12 | 83.46 ± 135.71 | 0.551 ± 0.35 | 0.127 ± 0.19 | 18.27 | 1.74 |
| 13 | 72.71 ± 65.71 | 0.563 ± 0.35 | 0.131 ± 0.18 | 17.87 | 1.77 |
| 14 | 71.59 ± 66.72 | 0.563 ± 0.35 | 0.142 ± 0.20 | 19.50 | 2.44 |
| 15 | 77.33 ± 70.82 | 0.558 ± 0.34 | 0.130 ± 0.19 | 17.71 | 2.08 |
| 16 | 74.79 ± 78.75 | 0.565 ± 0.35 | 0.139 ± 0.20 | 20.40 | 2.05 |
| 17 | 73.13 ± 77.88 | 0.547 ± 0.35 | 0.128 ± 0.19 | 18.00 | 1.95 |
| 18 | 78.10 ± 81.84 | 0.549 ± 0.35 | 0.136 ± 0.20 | 19.66 | 2.26 |
| 19 | 69.24 ± 63.14 | 0.529 ± 0.35 | 0.116 ± 0.18 | 16.63 | 1.34 |
| 20 | 70.29 ± 60.83 | 0.559 ± 0.35 | 0.134 ± 0.20 | 18.19 | 2.29 |
| 21 | 77.96 ± 86.18 | 0.554 ± 0.35 | 0.139 ± 0.19 | 21.37 | 1.76 |
| 22 | 73.13 ± 64.58 | 0.549 ± 0.35 | 0.126 ± 0.19 | 18.06 | 1.76 |
| 23 | 73.63 ± 75.66 | 0.494 ± 0.34 | 0.108 ± 0.17 | 14.46 | 1.23 |
| 24 | 74.13 ± 65.68 | 0.541 ± 0.35 | 0.129 ± 0.19 | 16.70 | 2.22 |
| 25 | 68.39 ± 56.42 | 0.506 ± 0.34 | 0.122 ± 0.17 | 17.58 | 1.01 |
| 26 | 72.89 ± 61.70 | 0.521 ± 0.35 | 0.109 ± 0.17 | 13.47 | 1.38 |
| 27 | 71.65 ± 71.04 | 0.529 ± 0.35 | 0.122 ± 0.19 | 16.38 | 1.53 |
| 28 | 72.90 ± 62.37 | 0.51 ± 0.34 | 0.106 ± 0.17 | 13.72 | 1.37 |
| 29 | 76.08 ± 78.60 | 0.526 ± 0.35 | 0.110 ± 0.17 | 14.02 | 0.99 |
| Mean | 74.82 ± 75.62 | 0.551 ± 0.35 | 0.132 ± 0.19 | 18.26 | 2.11 |
Effective population size
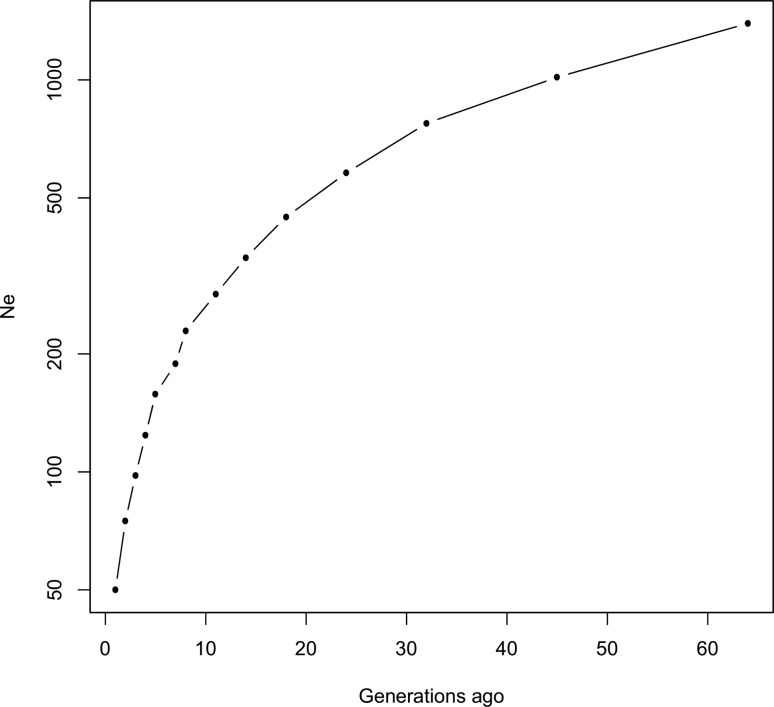
Average effective population size over the past 64 generations was estimated from the mean r2 values for the 29 bovine autosomes using the three MAF thresholds (Table 3). Figure 3 illustrates an example for MAF 0.1. Similar Ne estimates were found between the MAF 0.1 and the MAF 0.05 datasets. However, between these two datasets and the MAF 0.01 dataset, we observed discrepancies that increased over generations. In general, we observed a continual decrease in Ne across generations, with the average value falling from 1410 to 51 after 64 generations. It appeared that Ne declined slowly between 64 and 32 generations ago, and then this decline underwent a sudden acceleration from eight generations ago to the present, when Ne dropped from an average of 232 to an average of 191 (Table 3).
Table 3. Average estimated effective population size truncated at 64 generations for three different MAF thresholds.
| Generations ago | Ne_MAF0.1 | Ne_MAF0.05 | Ne_MAF0.01 | Mean Ne |
|---|---|---|---|---|
| 1 | 50 | 51 | 52 | 51 |
| 2 | 75 | 75 | 78 | 76 |
| 3 | 98 | 98 | 101 | 99 |
| 4 | 124 | 124 | 127 | 125 |
| 5 | 158 | 159 | 163 | 160 |
| 7 | 189 | 190 | 195 | 191 |
| 8 | 229 | 230 | 237 | 232 |
| 11 | 284 | 284 | 292 | 287 |
| 14 | 352 | 354 | 366 | 357 |
| 18 | 447 | 450 | 463 | 453 |
| 24 | 579 | 577 | 597 | 584 |
| 32 | 775 | 770 | 799 | 781 |
| 45 | 1016 | 1023 | 1053 | 1031 |
| 64 | 1393 | 1397 | 1440 | 1410 |
Figure 3. Average-estimated effective population size in the Tunisian cattle population, plotted against average generations in the past, truncated at 64 generations. Estimated effective population size was plotted on a log scale.
Discussion
In the present study, we first identified runs of homozygosity in a sample of 87 Tunisian cattle individuals. Characterization of ROHs, particularly their size and frequency, provides information about relatedness within a population. Our findings showed that more than one sixth of the individuals in our sample had a total sum of ROHs >250 Mb, with almost all individuals having at least one ROH >25 Mb. Because long ROH fragments are indicators of recent inbreeding, we can hypothesize that consanguineous mating is quite common in the local livestock farming system. This is particularly problematic as long autozygous ROHs are generally associated with an increased incidence of strongly deleterious mutations as well as mildly deleterious variants (Szpiech et al., 2013). However, the mean number of ROHs per individual observed for native Tunisian cattle (38 ± 12.6) was low compared to that found in other European cattle breeds. For instance, Ferencakovic et al. (2013) reported that Brown Swiss, Tyrol Grey, and Pinzgauer cattle had, on average, 94.76 ± 14.55, 70.86 ± 9.51, and 59.96 ± 9.91 ROHs per individual, respectively. This most likely reflects differences in the history of breed formation between European breeds and Tunisian cattle, with the former originating from a more recent bottleneck and subjected to a more pronounced selection pressure.
Next, we aimed to assess the extent of genome-wide LD and to infer the effective population size in the native Tunisian cattle population using medium-density SNP chips and two of the most popular metrics for quantifying LD: D’ and r2. Our results revealed a gradual decline in these two parameters over increasing inter-marker distances, with a steeper rate of decay for r2 than for D’. Furthermore, we observed an inflation in estimates of D’ (which was more noticeable over long inter-marker distances) with a decrease in the MAF threshold used. This finding has previously been reported in other studies (e.g., Khatkar et al., 2008; Bohmanova et al., 2010; Espigolan et al., 2013) and might be explained by the fact that, in the formula for D’, the denominator is set to the minimal product of SNP allele frequencies and will be lower when the difference between allele frequencies is higher, thus leading to over-estimation of D’. Khatkar et al. (2008) reported that the observed inflation in D’ estimates with an increase in the proportion of rare alleles is mainly due to the fact that rare alleles are, in general, younger than common alleles, and hence may still be in LD. Instead, mean values of r2 dropped noticeably as the MAF threshold was lowered at all inter-marker distances.
Correction for sample size resulted in only a small change in r2 estimates at short inter-marker distances, indicating that 87 individuals were sufficient to estimate r2 with acceptable reliability. However, this number may be insufficient for a correct estimate of D’, because small samples may fail to sample rare fourth gametes which, therefore, can inflate D’ (Khatkar et al., 2008). Indeed, Bohmanova et al. (2010) reported that a minimal sample size of 55 animals was required for accurate estimation of r2, while 444 animals were required for D’, and other studies have likewise reported that D’ is more sensitive to sample size variations than r2 (e.g., Ardlie et al., 2002; Du et al., 2007). We therefore considered our estimates of r2 to be more reliable than the D’ estimates.
In order to further reduce the effect of sampling bias on r2 estimates in our study, it would be reasonable to consider only values obtained using a MAF threshold of 0.1. This is because LD around common alleles (those with a moderate-to-high frequency) can be measured with a modest sample size (80 ± 100 chromosomes) to a precision within 10 ± 20% of the asymptotic limit (Reich et al., 2001). Around 74% of the SNPs in the MAF 0.1 dataset had a moderate-to-high MAF (>0.2) (Figure S1 (53.5KB, pdf) ), making the r2 estimates that were based on this dataset robust over short inter-marker distances.
Differences in LD were observed between chromosomes and might result from variations in chromosome length, with longer chromosomes having higher LD than shorter ones. Indeed, we found a high correlation (~78%) between r2 value and chromosome length. In agreement with our results, Stapley et al. (2010) found that LD extended further on the macrochromosomes than on microchromosomes in the zebra finch genome. Another possible explanation for inter-chromosomal variations in LD might be the difference in total sum of ROHs observed between high- and low-LD chromosomes (Table S3 (13.5KB, pdf) ). This hypothesis is corroborated by the high positive correlation (~82%) found between LD and total sum of ROHs per chromosome. Similarly, studies of LD in humans have reported that ROHs are more common in regions with high linkage disequilibrium and a low recombination rate (e.g., Gibson et al., 2006).
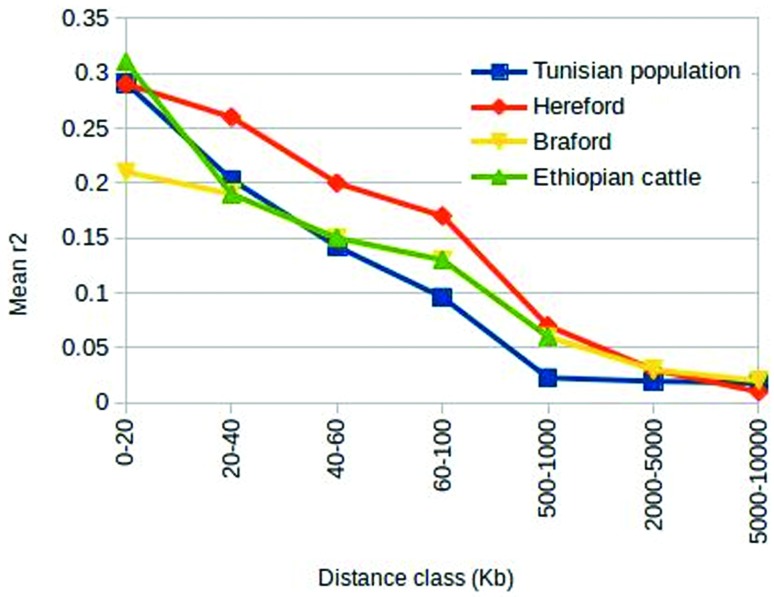
Next, we compared our results with those obtained from other taurine breeds using a similar SNP panel. We found that LD decay in the Tunisian population was faster than that reported by Biegelmeyer et al. (2016) for a sample of 391 Braford and 2079 Hereford cattle, as well as that published by Edea et al. (2014) for Ethiopian local cattle (Figure 4). Qanbari et al. (2010) found that, in German Holstein cattle, mean r2 decreased from 0.30 between marker pairs separated by less than 25 Kb to 0.12 for those separated by 100-200 Kb; this decrease was slower than that observed in our sample (0.29 to 0.05 for Tunisian native cattle over the same change in physical distance). Instead, in reporting LD estimates for a sample of 887 North American Holstein bulls, Bohmanova et al. (2010) found that mean r2 for markers 40-60 kb apart was 0.20, which is higher than that found in our study (0.14) for the same inter-marker distance. However, it is difficult to compare our results directly with these previous studies due to differences in the scale of sampling, given that the level of LD decreases with increasing sample size (Hill and Robertson, 1968). When we restricted our comparisons to studies with smaller sample sizes, we found that the mean r2 value for adjacent markers in the present study (pooled over all autosomes; mean = 0.132, range: 0.106 to 0.158) was lower than those reported by Flury et al. (2010) for 128 indigenous Swiss cattle (mean r2 = 0.24, range: 0.19 to 0.26), by Mastrangelo et al. (2014) for 76 Italian Modicana individuals (mean r2 = 0.200, range: 0.168 to 0.234), and by Beghain et al. (2013) for 30 Blonde d’Aquitaine bulls (mean r2 = 0.205, range: 0.172 to 0.241). In summary, although our results are based on a smaller sample size, they paint a coherent picture: the lower level of genome-wide LD and its faster decay in Tunisian cattle compared to other breeds is consistent with the absence of strong selective pressure and with the exclusive use of natural reproductive methods within this population (unlike Holstein, Hereford, or Braford breeds). This hypothesis is further corroborated by the fact that mean r2 values between adjacent SNPs for the artificially unselected Italian breed Cinisara were similar to those reported in our study (mean r2 = 0.168, range: 0.138 to 0.184; Mastrangelo et al., 2014).
Figure 4. Decay in linkage disequilibrium (r2) over increasing inter-marker distance in Tunisian, Hereford, Braford, and local Ethiopian cattle.
We used average r2 estimates (combined over all autosomes) at different inter-marker distances to infer past effective population sizes of the local Tunisian cattle population. Short inter-marker distances reflect the effective population size over historical periods of time while long inter-marker distances give information on population size in the immediate past (Hill, 1981). Using a MAF threshold of 0.01 tends to increasingly overestimate Ne across generations, while a MAF threshold of 0.05 is less subject to erroneous estimates of effective population size. Our study revealed that the Tunisian cattle population has undergone a sudden, substantial drop in Ne, starting from eight generations ago. If we assume a generation time for local cattle of 5 years, this period corresponds to the mid-1970s, and indeed, it was reported that the numbers of local and cross-bred cattle in Tunisia fell by 32% between 1975 and 1990 because of the massive introduction of European breeds (Kayouli, 2001). In addition, it is worth noting that, due to the small sample size in our study, it is likely that the values we found for recent Ne underestimate the true Ne in the whole native Tunisian cattle population; according to Corbin et al. (2012), when Ne is assumed to be variable, a severe underestimation occurs in recent Ne estimates. Therefore, the inferences for recent Ne in the present study should be considered cautiously due to the sampling bias. Taken together, the pattern of ROHs and the low levels of long-range LD found in the present study, combined with the relatively high levels of genetic diversity reported in a previous study (Ben Jemaa et al., 2015) may indicate that native Tunisian cattle have not been subjected to strong genetic drift. We thus hypothesize that there has been a significant drop in the effective population size of Tunisian cattle over successive generations, but with higher real Ne values than those found in our study (especially regarding recent Ne estimates).
The fact that the recent effective population size that was inferred here for local Tunisian cattle was small implies that this population is endangered. According to Franklin (1980), populations with Ne <50 are considered to be at immediate risk of extinction. Other authors have gone even further, suggesting that a threshold of Ne = 50 is too small to ensure long-term population survival. For instance, Meuwissen (2009) reported that a threshold of Ne = 100 would be necessary to ensure the long-term viability of an animal population. This is because such small populations are exposed to loss of genetic variation through high levels of both inbreeding and genetic drift, which can substantially increase the extinction probability of populations in changing environments.
In conclusion, this work presents the first assessment of ROH patterns and LD distribution at the genome-wide level in the native Tunisian cattle population. Compared to European breeds, we found that Tunisian cattle had less LD, and that it decayed faster with physical distance between markers. Therefore, a higher SNP density might be necessary to carry out effective genome-wide association mapping within this population. We provide evidence of a rapid decline in the effective population size in Tunisian cattle, associated with high levels of recent inbreeding within a significant proportion of individuals. These two observations indicate an urgent need to establish a conservation plan that includes a well-designed genetic management program for this population.
Acknowledgments
The authors would like to thank the participating farmers, the General Directorate of Veterinary Services of Tunisia, the Tunisian Livestock and Pasture Office (OEP), the Tunisian Office of Silvopastoral Development of the North-West (ODESYPANO), and the National Bank of Genes of Tunisia for their invaluable help. Genotyping assays were funded by INRA, the International Foundation for Science (IFS grant B/5478), and by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Science, ICT & Future Planning (2013K2A4A1044519).
Supplementary material
The following online material is available for this article:
Footnotes
Associate Editor: Alexandre Rodrigues Caetano
Conflict of interest
The authors declare that they have no conflicts of interest.
Author contributions
SBJ conceived and designed the study, NT and SM performed the blood sampling, ER made DNA extraction and quality control, SBJ and MB analyzed the data, DR managed genotyping step, SBJ, MB and DR wrote the manuscript, all authors read and approved the final version.
References
- Al-Mamun HA, Clark SA, Kwan P, Gondro C. Genome-wide linkage disequilibrium and genetic diversity in five populations of Australian domestic sheep. Genet Sel Evol. 2015;47:90. doi: 10.1186/s12711-015-0169-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Al-Mamun HA, Clark SA, Kwan P and Gondro C (2015) Genome-wide linkage disequilibrium and genetic diversity in five populations of Australian domestic sheep. Genet Sel Evol 47:90. [DOI] [PMC free article] [PubMed]
- Ardlie KG, Kruglyak L, Seielstad M. Patterns of linkage disequilibrium in the human genome. Nat Rev Genet. 2002;3:299–309. doi: 10.1038/nrg777. [DOI] [PubMed] [Google Scholar]; Ardlie KG, Kruglyak L and Seielstad M (2002) Patterns of linkage disequilibrium in the human genome. Nat Rev Genet 3:299–309. [DOI] [PubMed]
- Barbato M, Orozco-terWengel P, Tapio M, Bruford MW. SNeP: A tool to estimate trends in recent effective population size trajectories using genome-wide SNP data. Front Genet. 2015;6:109. doi: 10.3389/fgene.2015.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]; Barbato M, Orozco-terWengel P, Tapio M and Bruford MW (2015) SNeP: A tool to estimate trends in recent effective population size trajectories using genome-wide SNP data. Front Genet 6:109. [DOI] [PMC free article] [PubMed]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]; Barrett JC, Fry B, Maller J and Daly MJ (2005) Haploview: Analysis and visualization of LD and haplotype maps. Bioinformatics 21:263–265. [DOI] [PubMed]
- Beghain J, Boitard S, Weiss B, Boussaha M, Gut I, Rocha D. Genome-wide linkage disequilibrium in the Blonde d’Aquitaine cattle breed. J Anim Breed Genet. 2013;130:294–302. doi: 10.1111/j.1439-0388.2012.01020.x. [DOI] [PubMed] [Google Scholar]; Beghain J, Boitard S, Weiss B, Boussaha M, Gut I and Rocha D (2013) Genome-wide linkage disequilibrium in the Blonde d’Aquitaine cattle breed. J Anim Breed Genet 130:294–302. [DOI] [PubMed]
- Ben Jemaa S, Boussaha M, Ben Mehdi M, Lee JH, Lee SH. Genome-wide insights into population structure and genetic history of Tunisian local cattle using the illumina bovinesnp50 beadchip. BMC Genomics. 2015;16:677. doi: 10.1186/s12864-015-1638-6. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ben Jemaa S, Boussaha M, Ben Mehdi M, Lee JH and Lee SH (2015) Genome-wide insights into population structure and genetic history of Tunisian local cattle using the illumina bovinesnp50 beadchip. BMC Genomics 16:677. [DOI] [PMC free article] [PubMed]
- Biegelmeyer P, Gulias-Gomes CC, Caetano AR, Steibel JP, Cardoso FF. Linkage disequilibrium, persistence of phase and effective population size estimates in Hereford and Braford cattle. BMC Genetics. 2016;17:32. doi: 10.1186/s12863-016-0339-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; Biegelmeyer P, Gulias-Gomes CC, Caetano AR, Steibel JP and Cardoso FF (2016) Linkage disequilibrium, persistence of phase and effective population size estimates in Hereford and Braford cattle. BMC Genetics 17:32. [DOI] [PMC free article] [PubMed]
- Bohmanova J, Sargolzaei M, Schenkel FS. Characteristics of linkage disequilibrium in North American Holsteins. BMC Genomics. 2010;11:421. doi: 10.1186/1471-2164-11-421. [DOI] [PMC free article] [PubMed] [Google Scholar]; Bohmanova J, Sargolzaei M and Schenkel FS (2010) Characteristics of linkage disequilibrium in North American Holsteins. BMC Genomics 11:421. [DOI] [PMC free article] [PubMed]
- Cheng S, Chen Y, Monforte JA, Higuchi R, Van Houten B. Template integrity is essential for PCR amplification of 20- to 30-kb sequences from genomic DNA. PCR Methods Appl. 1995;4:294–298. doi: 10.1101/gr.4.5.294. [DOI] [PubMed] [Google Scholar]; Cheng S, Chen Y, Monforte JA, Higuchi R and Van Houten B (1995) Template integrity is essential for PCR amplification of 20- to 30-kb sequences from genomic DNA. PCR Methods Appl 4:294–298. [DOI] [PubMed]
- Corbin LJ, Liu AYH, Bishop SC, Woolliams JA. Estimation of historical effective population size using linkage disequilibria with marker data. J Anim Breed Genet. 2012;129:257–270. doi: 10.1111/j.1439-0388.2012.01003.x. [DOI] [PubMed] [Google Scholar]; Corbin LJ, Liu AYH, Bishop SC and Woolliams JA (2012) Estimation of historical effective population size using linkage disequilibria with marker data. J Anim Breed Genet 129:257–270. [DOI] [PubMed]
- Djemali M, Kayouli C. L’élevage laitier en Tunisie. In: Proceedings of the joint EAAP-CIHEAM-FAO Symposium on Prospects for a Sustainable Dairy Sector in the Mediterranean. Hammamet, Tunisia: 2003. pp. 96–105. [Google Scholar]; Djemali M and Kayouli C (2003) L’élevage laitier en Tunisie. In: Proceedings of the joint EAAP-CIHEAM-FAO Symposium on Prospects for a Sustainable Dairy Sector in the Mediterranean, Hammamet, Tunisia, pp 96-105.
- Du FX, Clutter AC, Lohuis MM. Characterizing linkage disequilibrium in pig populations. Int J Biol Sci. 2007;3:166–178. doi: 10.7150/ijbs.3.166. [DOI] [PMC free article] [PubMed] [Google Scholar]; Du FX, Clutter AC and Lohuis MM (2007) Characterizing linkage disequilibrium in pig populations. Int J Biol Sci 3:166–178. [DOI] [PMC free article] [PubMed]
- Edea Z, Dadi H, Kim SW, Park JH, Shin GH, Dessie T, Kim KS. Linkage disequilibrium and genomic scan to detect selective loci in cattle populations adapted to different ecological conditions in Ethiopia. J Anim Breed Genet. 2014;131:358–366. doi: 10.1111/jbg.12083. [DOI] [PubMed] [Google Scholar]; Edea Z, Dadi H, Kim SW, Park JH, Shin GH, Dessie T and Kim KS (2014) Linkage disequilibrium and genomic scan to detect selective loci in cattle populations adapted to different ecological conditions in Ethiopia. J Anim Breed Genet 131:358–366. [DOI] [PubMed]
- Espigolan R, Baldi F, Boligon AA, Souza FR, Gordo DG, Tonussi RL, Cardoso DF, Oliveira HN, Tonhati H, Sargolzaei M, et al. Study of whole genome linkage disequilibrium in Nellore cattle. BMC Genomics. 2013;14:305. doi: 10.1186/1471-2164-14-305. [DOI] [PMC free article] [PubMed] [Google Scholar]; Espigolan R, Baldi F, Boligon AA, Souza FR, Gordo DG, Tonussi RL, Cardoso DF, Oliveira HN, Tonhati H, Sargolzaei M et al. (2013) Study of whole genome linkage disequilibrium in Nellore cattle. BMC Genomics 14:305. [DOI] [PMC free article] [PubMed]
- Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. 4th edition. Longmans Green; Harlow: 1996. p. 480. [Google Scholar]; Falconer DS and Mackay TFC (1996) Introduction to Quantitative Genetics. 4th edition. Longmans Green, Harlow, 480 pp.
- Ferencakovic M, Sölkner J, Curik I. Estimating autozygosity from high-throughput information: effects of SNP density and genotyping errors. Genet Sel Evol. 2013;45:42. doi: 10.1186/1297-9686-45-42. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ferencakovic M, Sölkner J and Curik I (2013) Estimating autozygosity from high-throughput information: effects of SNP density and genotyping errors. Genet Sel Evol 45:42 [DOI] [PMC free article] [PubMed]
- Flury C, Tapio M, Sonstegard T, Drögemüller C, Leeb T, Simianer H, Hanotte O, Rieder S. Effective population size of an indigenous Swiss cattle breed estimated from linkage disequilibrium. J Anim Breed Genet. 2010;127:339–347. doi: 10.1111/j.1439-0388.2010.00862.x. [DOI] [PubMed] [Google Scholar]; Flury C, Tapio M, Sonstegard T, Drögemüller C, Leeb T, Simianer H, Hanotte O and Rieder S (2010) Effective population size of an indigenous Swiss cattle breed estimated from linkage disequilibrium. J Anim Breed Genet 127:339–347. [DOI] [PubMed]
- Franklin IR. Evolutionary change in small populations. In: Soule ME, Wilcox BA, editors. Conservation Biology: An Evolutionary-Ecological Perspective. Sinauer Associates; Sunderland: 1980. pp. 135–140. [Google Scholar]; Franklin IR (1980) Evolutionary change in small populations. In: Soule ME and Wilcox BA (eds) Conservation Biology: An Evolutionary-Ecological Perspective. Sinauer Associates, Sunderland, pp 135–140.
- Gibson J, Morton NE, Collins A. Extended tracts of homozygosity in outbred human populations. Hum Mol Genet. 2006;15:789–795. doi: 10.1093/hmg/ddi493. [DOI] [PubMed] [Google Scholar]; Gibson J, Morton NE and Collins A (2006) Extended tracts of homozygosity in outbred human populations. Hum Mol Genet 15:789–795. [DOI] [PubMed]
- Hill WG. Estimation of effective population size from data on linkage disequilibrium. Genet Res. 1981;38:209–216. [Google Scholar]; Hill WG (1981) Estimation of effective population size from data on linkage disequilibrium. Genet Res 38:209–216.
- Hill WG, Robertson A. Linkage disequilibrium in finite populations. Theor Appl Genet. 1968;38:226–231. doi: 10.1007/BF01245622. [DOI] [PubMed] [Google Scholar]; Hill WG and Robertson A (1968) Linkage disequilibrium in finite populations. Theor Appl Genet 38:226–231. [DOI] [PubMed]
- Kayouli C. Country Pasture/Forage Resource Profiles in Tunisia. 2001. https://docplayer.net/21479393-Country-pasture-forage-resource-profiles-tunisia-by-chedly-kayouli.html.; Kayouli C (2001) Country Pasture/Forage Resource Profiles in Tunisia, https://docplayer.net/21479393-Country-pasture-forage-resource-profiles-tunisia-by-chedly-kayouli.html.
- Khatkar MS, Nicholas FW, Collins AR, Zenger KR, Cavanagh JA, Barris W, Schnabel RD, Taylor JF, Raadsma HW. Extent of genome-wide linkage disequilibrium in Australian Holstein-Friesian cattle based on a high-density SNP panel. BMC Genomics. 2008;9:187. doi: 10.1186/1471-2164-9-187. [DOI] [PMC free article] [PubMed] [Google Scholar]; Khatkar MS, Nicholas FW, Collins AR, Zenger KR, Cavanagh JA, Barris W, Schnabel RD, Taylor JF and Raadsma HW (2008) Extent of genome-wide linkage disequilibrium in Australian Holstein-Friesian cattle based on a high-density SNP panel. BMC Genomics 9:187. [DOI] [PMC free article] [PubMed]
- Mastrangelo S, Saura M, Tolone M, Salces-Ortiz J, Di Gerlando R, Bertolini B, Fontanesi L, Sardina MT, Serrano M, Portolano B. The genome-wide structure of two economically important indigenous Sicilian cattle breeds. J. Anim. Sci. 2014;92:4833–4842. doi: 10.2527/jas.2014-7898. [DOI] [PubMed] [Google Scholar]; Mastrangelo S, Saura M, Tolone M, Salces-Ortiz J, Di Gerlando R, Bertolini B, Fontanesi L, Sardina MT, Serrano M and Portolano B (2014) The genome-wide structure of two economically important indigenous Sicilian cattle breeds. J. Anim. Sci 92:4833–4842. [DOI] [PubMed]
- McEvoy BP, Powell JE, Goddard ME, Visscher PM. Human population dispersal “Out of Africa” estimated from linkage disequilibrium and allele frequencies of SNPs. Genome Res. 2011;21:821–829. doi: 10.1101/gr.119636.110. [DOI] [PMC free article] [PubMed] [Google Scholar]; McEvoy BP, Powell JE, Goddard ME and Visscher PM (2011) Human population dispersal “Out of Africa” estimated from linkage disequilibrium and allele frequencies of SNPs. Genome Res 21:821–829. [DOI] [PMC free article] [PubMed]
- McRae AF, McEwan JC, Dodds KG, Wilson T, Crawford AM, Slate J. Linkage disequilibrium in domestic sheep. Genetics. 2002;160:1113–1122. doi: 10.1093/genetics/160.3.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]; McRae AF, McEwan JC, Dodds KG, Wilson T, Crawford AM and Slate J (2002) Linkage disequilibrium in domestic sheep. Genetics 160:1113–1122. [DOI] [PMC free article] [PubMed]
- Meuwissen T. Genetic management of small populations: A review. Acta Agric Scand Sect Anim Sci. 2009;59:71–79. [Google Scholar]; Meuwissen T (2009) Genetic management of small populations: A review. Acta Agric Scand Sect Anim Sci 59:71–79.
- Porto-Neto LR, Kijas JW, Reverter A. The extent of linkage disequilibrium in beef cattle breeds using high-density SNP genotypes. Genet Sel Evol. 2014;46:22. doi: 10.1186/1297-9686-46-22. [DOI] [PMC free article] [PubMed] [Google Scholar]; Porto-Neto LR, Kijas JW and Reverter A (2014) The extent of linkage disequilibrium in beef cattle breeds using high-density SNP genotypes. Genet Sel Evol 46:22. [DOI] [PMC free article] [PubMed]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, deBakker PIW, Daly MJ, et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]; Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, deBakker PIW, Daly MJ et al. (2007) PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. [DOI] [PMC free article] [PubMed]
- Qanbari S, Pimentel ECG, Tetens J, Thaller G, Lichtner P, Sharifi AR, Simianer H. The pattern of linkage disequilibrium in German Holstein cattle. Anim Genet. 2010;41:346–356. doi: 10.1111/j.1365-2052.2009.02011.x. [DOI] [PubMed] [Google Scholar]; Qanbari S, Pimentel ECG, Tetens J, Thaller G, Lichtner P, Sharifi AR and Simianer H (2010) The pattern of linkage disequilibrium in German Holstein cattle. Anim Genet 41:346–356. [DOI] [PubMed]
- Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, Lavery T, Kouyoumjian R, Farhadian SF, Ward R, et al. Linkage disequilibrium in the human genome. Nature. 2001;411:199–204. doi: 10.1038/35075590. [DOI] [PubMed] [Google Scholar]; Reich DE, Cargill M, Bolk S, Ireland J, Sabeti PC, Richter DJ, Lavery T, Kouyoumjian R, Farhadian SF, Ward R et al. (2001) Linkage disequilibrium in the human genome. Nature 411:199–204. [DOI] [PubMed]
- Szpiech ZA, Xu J, Pemberton TJ, Peng W, Zöllner S, Rosenberg NA, Li JZ. Long runs of homozygosity are enriched for deleterious variation. Am J Hum Genet. 2013;93:90–102. doi: 10.1016/j.ajhg.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]; Szpiech ZA, Xu J, Pemberton TJ, Peng W, Zöllner S, Rosenberg NA and Li JZ (2013) Long runs of homozygosity are enriched for deleterious variation. Am J Hum Genet 93:90–102. [DOI] [PMC free article] [PubMed]
- Stapley J, Birkhead TR, Burke T, Slate J. Pronounced inter- and intrachromosomal variation in linkage disequilibrium across the zebra finch genome. Genome Res. 2010;20:496–502. doi: 10.1101/gr.102095.109. [DOI] [PMC free article] [PubMed] [Google Scholar]; Stapley J, Birkhead TR, Burke T and Slate J (2010) Pronounced inter- and intrachromosomal variation in linkage disequilibrium across the zebra finch genome. Genome Res 20:496-502. [DOI] [PMC free article] [PubMed]
- Sved JA, Feldman MW. Correlation and probability methods for one and two loci. Theor Popul Biol. 1973;4:129–132. doi: 10.1016/0040-5809(73)90008-7. [DOI] [PubMed] [Google Scholar]; Sved JA and Feldman MW (1973) Correlation and probability methods for one and two loci. Theor Popul Biol 4:129–132. [DOI] [PubMed]
- Villa-Angulo R, Matukumalli LK, Gill CA, Choi J, Van Tassell CP, Grefenstette JJ. High-resolution haplotype block structure in the cattle genome. BMC Genetics. 2009;10:19. doi: 10.1186/1471-2156-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]; Villa-Angulo R, Matukumalli LK, Gill CA, Choi J, Van Tassell CP and Grefenstette JJ (2009) High-resolution haplotype block structure in the cattle genome. BMC Genetics 10:19. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.