Abstract
In 1989 Normark and co-workers reported on fibrous surface structures called curli on strains of E. coli that were suspected of causing bovine mastitis. Subsequent work by many groups has revealed an elegant and highly regulated curli biogenesis pathway also referred to as Type VIII secretion system. Curli biogenesis is governed by two divergently transcribed operons, csgBAC and csgDEFG. The csgBAC operon encodes the structural subunits of curli, CsgA and CsgB, along with a chaperone-like protein, CsgC. The csgDEFG operon encodes the accessory proteins required for efficient transcription, secretion and assembly of the curli fiber. CsgA and CsgB are secreted as largely unstructured proteins, and transition to β-rich structures that aggregate into regular fibers at the cell surface. Since both these proteins have been shown to be amyloidogenic in nature, the correct spatiotemporal synthesis of the curli fiber is of paramount importance for proper functioning and viability. Gram-negative bacteria have evolved an elegant machinery for the safe handling, secretion and extracellular assembly of these amyloidogenic proteins.
Curli are extracellular proteinaceous fibers made by Gram-negative bacteria. Curli specific genes (csg) are primarily found in Proteobacteria and Bacteroidetes (1–3). The main function of curli fibers is associated with a sedimentary lifestyle and multicellular behavior in biofilms, as they form scaffolds that provide adhesive and structural support to the community (4–8). In certain pathogenic bacteria curli have also been implicated in host colonization, innate response activation and cell invasion (9–13).
Curli appear as rigid, 2-5 nm thick coiled fibers that entangle into dense congregates that surround the cell (Figure 1A, B) (1, 7, 14). Curli fibers form via a nucleation dependent self-assembly process and they adopt an amyloid fold as their native structure (15, 16). Curli are the product of a dedicated secretion-assembly pathway known as the nucleation-precipitation pathway, or Type VIII secretion system (T8SS) (3, 17, 18). In E. coli, seven curli-specific genes (csg) are clustered in two divergently described operons csgBAC and csgDEFG (17) (Figure 1C), where CsgA and B are structural components of the fiber (1, 16), CsgC is a periplasmic chaperone (19), CsgE, CsgF and CsgG form the secretion-assembly machinery (20–22) and CsgD is a transcriptional activator of the csgBAC operon (23). We will review curli biogenesis and the T8SS using the E. coli components as a representative system.
Figure 1. Curli composition and structure.
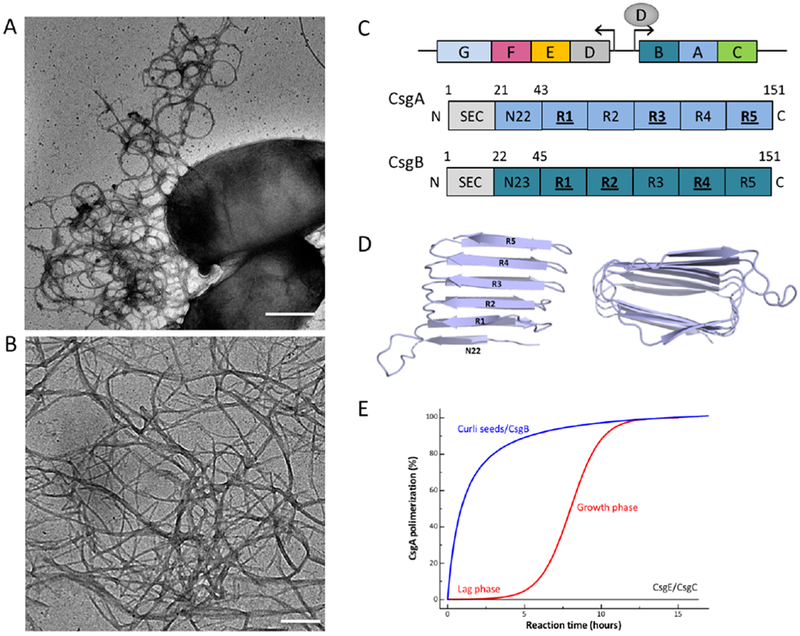
(A, B) TEM micrographs of individual E. coli cells producing curli fibers (A), and curli-like fibers grown in vitro from purified CsgA (B); scale bars: 500 nm or 200 nm, respectively. (C) Schematic organisation of the csgDEFG and csgBAC curli gene clusters and architecture of the curli subunits CsgA (blue) and CsgB (dark blue). Subunits comprise a N-terminal signal sequence (SEC) that is cleaved upon export into the periplasm. The mature proteins contain curlin pseudorepeat regions (N22, R1-R5) that guide substrate specificity in the secretion pathway and form the amyloidogenic core of the curli subunits. Repeats that efficiently self-polymerize in vitro are underscored. (D) Theoretical model of CsgA predicted based on amino acid covariation analysis (42). The predictions point to a right- or left-handed (shown) β-helix made up from stacked curlin repeats (labeled R1-R5) (E) Representation of typical in vitro CsgA polymerization profiles in absence (red) or presence (blue) of preformed fibers or the CsgB nucleator. In the presence of CsgE (1:1 ratio) or CsgC (1:500 ratio), no CsgA polymerization is observed (black curve).
The secretion-assembly machinery
In Gram-negative bacteria, assembly of surface appendages requires the passage of two lipid bilayers, the cytoplasmic or inner membrane and the outer membrane. The curli subunit proteins CsgA and CsgB cross the inner membrane via the Sec general secretory pathway, after which CsgE, CsgF and CsgG orchestrate translocation of CsgA and CsgB across the outer membrane (20–22) where they assemble into curli polymers (21). CsgG is a 262-residue lipoprotein that forms the curli translocation channel in the outer membrane (20). It forms a nonameric complex with a 36-stranded β-barrel that inserts into the outer membrane and is connected to the periplasm via a cage-like vestibule of approximately 35 Å inner diameter (24, 25) (Figure 2). The transmembrane and periplasmic domains are separated by a 9 Å wide channel constriction that would be compatible with the passage of unfolded polypeptides. Single channel current recordings and in vivo bile-salt sensitivity assays show a constitutively open channel, which can be gated in presence of the periplasmic accessory factor CsgE (24). The latter is a soluble 107-residue protein that can oligomerize into a nonameric cap-like structure that binds the periplasmic entry to the CsgG secretion channel (24) (Figure 2). Although at elevated concentrations CsgG can facilitate a non-selective leakage of periplasmic polypeptides, under native conditions secretion is specific for curli subunits and requires CsgE (22). In vitro, CsgE inhibits CsgA self-assembly when present at a 1:1 stoichiometric ratio, demonstrating it can directly interact with curli subunits. The prevailing model is for CsgE to act as specificity factor by binding periplasmic CsgA and targeting it to the CsgG secretion channel. In vitro the CsgE:CsgG complex is in a reversible equilibrium (24), suggesting CsgE may cycle between a periplasmic and CsgG-bound form. The NMR structure of the CsgE monomer shows a compact alpha/beta sandwich protein with flexible C-terminal tail (26) (Figure 2). Two regions of CsgE involved in the CsgE-CsgA interaction were determined: a head comprising a positively charged patch centered around R47 and a stem comprising a negatively charged patch containing E31 and E85 (27). R47 was found to mediate an indispensable charge-charge interaction with CsgA, while mutations in the negatively charged neck region retained CsgA binding capacity, but impact CsgE-dependent secretion of CsgA. The exact binding epitopes in CsgA are currently unknown, although the first pseudo-repeat in the curli subunits (in E. coli CsgA referred to as “N22”) is sufficient to target polypeptides for secretion through the curli pathway (20, 22). Secretion of non-native sequences was found to have a size-constraint in case of folded fusion proteins, which is likely to reflect the width of the CsgG constriction (28). The dimensions of the channel constriction suggest the native substrate navigates the CsgG channel in an extended conformation (24, 25). In agreement with this, in strains with curli assembly defects extracellular CsgA is found as an unfolded chain (16, 29). The transition of secreted CsgA to the amyloid state requires the activity of CsgF and the nucleator subunit CsgB (15, 21, 29–31).
Figure 2. Integrated model for curli subunit secretion.
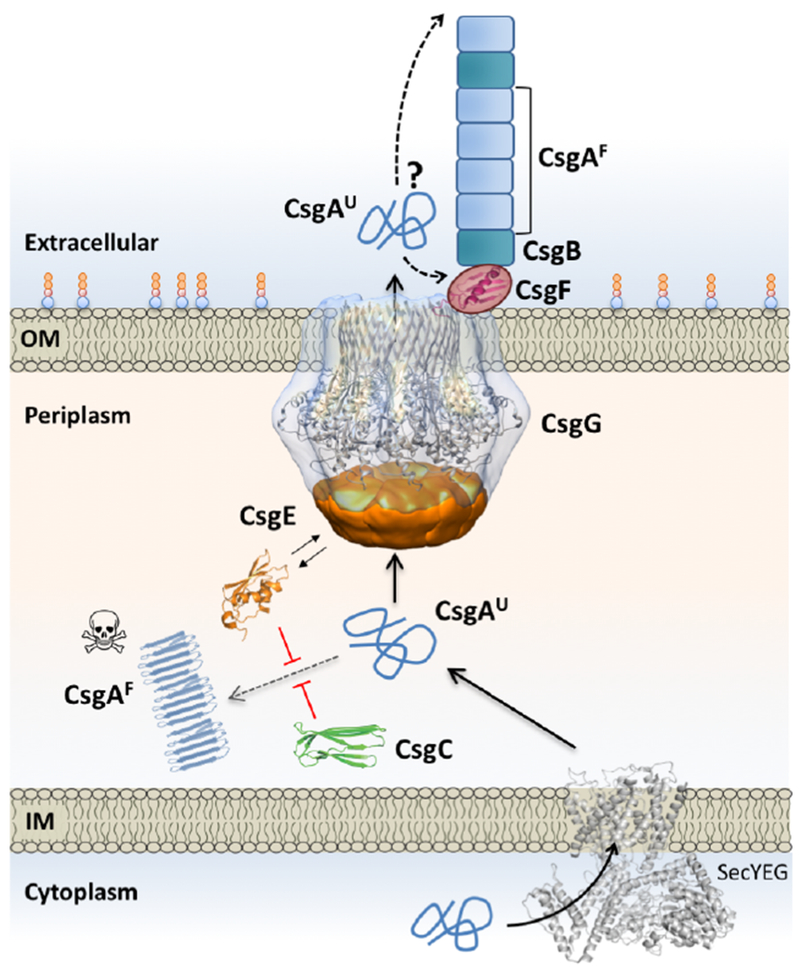
Curli subunits enter the periplasm via the SecYEG translocon, from where they progress to the cell surface as unfolded polypeptides via the curli transporter CsgG. Premature folding and polymerization of CsgA in the periplasm (right dotted line) is inhibited by CsgE and CsgC. CsgE binds and targets subunits to the secretion channel, whilst CsgC provides a safeguard against runaway polymerization, likely by the binding and neutralization of early assembly intermediates and/or nascent fibers. CsgG forms a nonameric complex that acts as a peptide diffusion channel and cooperates with the periplasmic factor CsgE, which binds the channel and forms a capping structure to the secretion complex. Recruitment and (partial) enclosure of CsgA in the secretion complex is proposed to create an entropy gradient over the channel that favors CsgA’s outward diffusion as an unfolded, soluble polypeptide. Once secreted, curli fiber formation and elongation is templated by CsgB, in a CsgF-dependent manner. CsgF is likely to be in contact or close proximity to the CsgG channel. The exact role of CsgF and whether fibers extend from the proximal or distal end (dashed arrows) is presently unknown. Abbreviations: IM, inner membrane; OM, outer membrane.
CsgF is a 119-residue protein that is found to be surface-exposed in a CsgG-dependent manner, and may be speculated to be in close proximity or direct contact with the CsgG channel (Figure 2) (21). Structural data obtained by NMR spectroscopy revealed the CsgF monomer consists of three independent elements: an N-terminal unstructured region, a 21-residue α-helix and a C-terminal antiparallel β-sheet made of 4 strands (32). The absence of tertiary structure and the presence of exposed hydrophobic surfaces may correspond to putative interaction sides with CsgG or the secretion substrates CsgA and CsgB. Although CsgF appears to be dispensable for secretion of curli subunits, it is essential for curli fiber formation (21). When csgF is deleted, CsgB is no longer associated with the cell surface and is not able to exert its nucleating function. These observations implicate CsgF as a coupling factor between CsgA secretion and extracellular polymerization into curli fibers by coordinating or chaperoning the nucleating activity of the CsgB minor subunit (21).
A striking conundrum in curli assembly is the mode of protein translocation across the outer membrane. Protein transport is an energy-dependent process, where the driving force is generally drawn from ATP or GTP hydrolysis, protein synthesis, or an electrochemical potential such as the protein-motive force (33). However, the periplasmic space is devoid of known hydrolysable energy carriers and the semi-porous outer membrane does not support the build-up of solute or ion gradients. In Gram-negative secretion pathways this hurdle is overcome by means of cell envelope-spanning complexes such as type I, II, III and IV secretion systems (34), or by drawing energy from substrate folding and assembly such as seen for the chaperone-usher assembly and type V secretion pathways (35, 36). Available structural data indicate CsgG operates as a peptide diffusion channel (24, 25). The height of the CsgG constriction is compatible with the simultaneous binding of 4-5 residues along the length of an extended polypeptide chain, so that passage of the full protein implies stepwise Brownian diffusion along the length of the polypeptide chain. The rectifying force that ensures a net forward Brownian diffusion is presently unclear. The transition from the extended pre-assembly state to the amyloid state upon secretion and incorporation into curli fibers may provide such driving force. However, in csgB deletion mutants, CsgA does not assemble into curli and can be found as disordered polypeptide in the cell medium, indicating fiber assembly is not a prerequisite for secretion (21, 29). In an alternative hypothesis secretion is driven by an entropy gradient across the outer membrane and the channel, in a process that involves the secretion factor CsgE (24, 37). It is speculated that the local high concentration and conformational confinement of curli subunits in the CsgG vestibule would raise an entropic free-energy gradient over the translocation channel. Under physiological conditions, both the entropy potential of CsgA capture at the translocation channel and the folding energy released from the disorder-order transition of subunits incorporating into surface-associated fibers may cooperate to ensure forward diffusion across the CsgG channel.
Fiber components and self-assembly
Curli fibers are linear, non-covalent polymers of the major and minor subunits CsgA and CsgB, respectively (1, 16). The major curlin subunit, CsgA is a 151 amino acid long peptide with five imperfect repeat units, R1-R5 (Figure 1C). The repeating units in CsgA are predicted to form a β-strand-loop-β-strand motifs that constitute the core of the β-sheet rich curli amyloid fiber (3, 16, 38–42) (Figure 1D). In vitro, CsgA amyloid formation can be monitored using the amyloid binding dye thioflavin-T (ThT) (43). CsgA amyloid aggregation displays a canonical sigmodal polymerization kinetics typical for amyloid proteins with a lag phase, a growth phase and a plateau phase (44) (Figure 1E). The ‘lag phase’ represents the time required for the build-up of enough primary nuclei to lead to the rapid incorporation of remaining free monomers into the nascent fibers during the ‘growth’ phase (38, 44, 45). Recent in vitro studies suggest that the nucleation of CsgA into the amyloid template is a single-step process and does not pass through one or more oligomeric aggregated states as often seen in pathological amyloids (45). The phase transitions of the curli repeats from an unfolded to a folded β-strand-loop-β-strand motif, in the CsgA subunit itself or in a minimal oligomer such as a CsgA dimer, serves as the “nucleus” in this model (45). Once formed, CsgA fibers display distinctive structural and biophysical properties that are shared by all amyloids (16, 44, 45).
CsgA is secreted by the T8SS to the cell surface as an unstructured protein and will remain in a non-amyloid form unless it interacts with the CsgB ‘nucleator’ protein. CsgB shares 30% sequence identity with CsgA and is also predicted to form five imperfect β-strand-loop-β-strand repeats, R1-R5 (31) (Figure 1C). The C-terminal R5 repeat of CsgB has been shown to be necessary for anchoring CsgB to the cell surface (31, 46). While the precise mechanism of in vivo nucleation has not been elucidated it is tempting to postulate that the R1-R4 repeats of membrane anchored CsgB assume a β-solenoid structure that provides a template for recognition by either R1 or R5 of CsgA (38, 41). The R1 and R5 repeats in CsgA are suggested to mediate interaction with CsgB, but also self-association by providing a template for secreted CsgA monomers to add on to the growing fiber tip (38, 47). A distinctive characteristic of curli assembly is that CsgA and CsgB can be independently secreted as unstructured subunits, which can then self-assemble into extracellular fibers when they contact each other on the cell surface (15, 29).
Interestingly, the R2, R3 and R4 repeating units of CsgA contain conserved Asp and Gly ‘gatekeeper’ residues (48). The positions of these gatekeeper residues are conserved in most CsgA homologs and certain gatekeeper residues modulate the amyloidogenic nature of CsgA by inhibiting its intrinsic aggregation propensity (48). The gatekeeper residues allow CsgA to remain in an unstructured form before being secreted to the cell surface and nucleated by CsgB (15, 31, 38, 46, 48). Interestingly, a CsgA mutant called CsgA* that lacks the gatekeeper residues polymerized in vivo in the absence of CsgB. However, the expression of CsgA* is toxic to cells, underlying the importance of controlled amyloid formation afforded by the T8SS system (48). In this respect, the type VIII (T8SS) evolved an intricate ability to maintain amyloidogenic proteins unstructured until secretion across the outer membrane.
Pathway control and chemical inhibition
An intriguing question in functional amyloid assembly pathways such as curli biogenesis is the avoidance of cytotoxic effects that are associated with amyloid formation in human and animal protein aggregation disorders. In addition to elaborate regulation at the transcriptional level (3, 17, 49–53), several adaptations within the pathway components ensure safe secretion of the amyloidogenic subunits and controlled fibril formation at the cell surface. A recent study found that unlike most pathological amyloid depositions, curli nucleation is a single-step process that does not involve intermediary oligomeric aggregates, which are often considered the more toxic species (45, 54). This suggest curli subunits are evolutionary optimised to have a sharp and direct transition from an intrinsically disordered conformation, to the relatively inert amyloid conformation. In addition, by the provision of the major subunit CsgA and the minor subunit CsgB, the pathway has segregated efficient fiber elongation and fiber nucleation, providing spatiotemporal control over curli deposition. On top of that, the T8SS evolved a further safeguard to quell premature CsgA amyloid formation by having a dedicated periplasmic chaperone (19). CsgC serves as a potent chaperone-like protein to prevent the runaway polymerization of CsgA in the periplasmic space (19) (Figure 2). In vitro CsgC inhibits CsgA polymerization down to substoichiometric molar ratios of 1:500 (19). The exact mechanism by which CsgC inhibits CsgA aggregation is not completely understood. The observed substoichiometric molar ratio may suggest that CsgC transiently interacts with soluble oligomeric pool of CsgA to prevent seed formation and subsequent polymerization of CsgA into amyloid fibers (19, 55). Another study, however, found that CsgC does not bind unfolded CsgA and instead acts at the growth poles of nuclei and curli fibers (45). Either way, a conserved patch of positively charged residues on the surface exposed β-strand (β4-β5 edge) of CsgC appears to mediate an electrostatic interaction between CsgA and CsgC that drives the inhibitory effect (55).
Finally, because functional amyloids like curli are important for biofilms formation and also have been shown to be responsible for host colonization, virulence and eliciting host immune response (45), chemical inhibitors of curli assembly have been developed (56, 57). A number of small molecule inhibitors effectively inhibit curli formation at low micromolar to nanomolar concentrations, although these have yet to be evaluated in in vivo disease models. In recent years, at least 10 distinct bacterial amyloid systems have been described (13). Amongst these, the Pseudomonas Fap pathway is noticeable in encompassing a dedicated secretion-assembly pathway (58). Although non-homologous to the Type VIII secretion pathway, Fap assembly involves a dedicated outer membrane channel (FapF) and a major, polymerizing (FapC) and minor, nucleating fiber subunit (FapB), reminiscent of curli assembly (59, 60).
Acknowledgements:
HR and NVG acknowledge funding by VIB and ERC through consolidator grant BAS-SBBT (grant no: 649082). MRC acknowledges generous support from NIH GM118651 and AI137535 and for helpful discussions from members of the Chapman lab.
References:
- 1.Olsen A, Jonsson A, Normark S. 1989. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338:652–655. [DOI] [PubMed] [Google Scholar]
- 2.Dueholm MS, Albertsen M, Otzen D, Nielsen PH. 2012. Curli functional amyloid systems are phylogenetically widespread and display large diversity in operon and protein structure. PLoS One 7:e51274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnhart MM, Chapman MR. 2006. Curli biogenesis and function. Annu Rev Microbiol 60:131–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collinson SK, Doig PC, Doran JL, Clouthier S, Trust TJ, Kay WW. 1993. Thin, aggregative fimbriae mediate binding of Salmonella enteritidis to fibronectin. J Bacteriol 175:12–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Römling U, Sierralta WD, Eriksson K, Normark S. 1998. Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol Microbiol 28:249–264. [DOI] [PubMed] [Google Scholar]
- 6.Kikuchi T, Mizunoe Y, Takade A, Naito S, Yoshida S. 2005. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol Immunol 49:875–884. [DOI] [PubMed] [Google Scholar]
- 7.Hung C, Zhou Y, Pinkner JS, Dodson KW, Crowley JR, Heuser J, Chapman MR, Hadjifrangiskou M, Henderson JP, Hultgren SJ. 2013. Escherichia coli biofilms have an organized and complex extracellular matrix structure. MBio 4:e00645–00613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hufnagel DA, Depas WH, Chapman MR. 2015. The Biology of the Escherichia coli Extracellular Matrix. Microbiol Spectr 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herwald H, Morgelin M, Olsen A, Rhen M, Dahlback B, Muller-Esterl W, Bjorck L. 1998. Activation of the contact-phase system on bacterial surfaces--a clue to serious complications in infectious diseases. Nat Med 4:298–302. [DOI] [PubMed] [Google Scholar]
- 10.Gophna U, Barlev M, Seijffers R, Oelschlager TA, Hacker J, Ron EZ. 2001. Curli Fibers Mediate Internalization of Escherichia coli by Eukaryotic Cells. Infect Immun 69:2659–2665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tükel Ç, Raffatellu M, Humphries AD, Wilson RP, Andrews-Polymenis HL, Gull T, Figueiredo JF, Wong MH, Michelsen KS, Akçelik M, Adams LG, Bäumler AJ. 2005. CsgA is a pathogen-associated molecular pattern of Salmonella enterica serotype Typhimurium that is recognized by Toll-like receptor 2. Mol Microbiol 58:289–304. [DOI] [PubMed] [Google Scholar]
- 12.Tükel Ç, Wilson RP, Nishimori JH, Pezeshki M, Chromy BA, Bäumler AJ. 2009. Responses to Amyloids of Microbial and Host Origin Are Mediated through Toll-like Receptor 2. Cell Host & Microbe 6:45–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gerven NV, Verren SEVD, Reiter DM, Remaut H. 2018. The Role of Functional Amyloids in Bacterial Virulence. J Mol Biol 430:3657–3684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Serra DO, Klauck G, Hengge R. 2015. Vertical stratification of matrix production is essential for physical integrity and architecture of macrocolony biofilms of Escherichia coli. Environ Microbiol 17:5073–5088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bian Z, Normark S. 1997. Nucleator function of CsgB for the assembly of adhesive surface organelles in Escherichia coli. EMBO J 16:5827–5836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chapman MR, Robinson LS, Pinkner JS, Roth R, Heuser J, Hammar M, Normark S, Hultgren SJ. 2002. Role of Escherichia coli curli operons in directing amyloid fiber formation. Science 295:851–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hammar M, Arnqvist A, Bian Z, Olsen A, Normark S. 1995. Expression of two csg operons is required for production of fibronectin- and congo red-binding curli polymers in Escherichia coli K-12. Mol Microbiol 18:661–670. [DOI] [PubMed] [Google Scholar]
- 18.Gerven NV, Klein RD, Hultgren SJ, Remaut H. 2015. Bacterial Amyloid Formation : Structural Insights into Curli Biogensis. Trends Microbiol. 23:693–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Evans Margery L, Chorell E, Taylor Jonathan D, Åden J, Götheson A, Li F, Koch M, Sefer L, Matthews Steve J, Wittung-Stafshede P, Almqvist F, Chapman Matthew R. 2015. The Bacterial Curli System Possesses a Potent and Selective Inhibitor of Amyloid Formation. Mol Cell 57:445–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robinson LS, Ashman EM, Hultgren SJ, Chapman MR. 2006. Secretion of curli fibre subunits is mediated by the outer membrane-localized CsgG protein. Mol Microbiol 59:870–881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nenninger AA, Robinson LS, Hultgren SJ. 2009. Localized and efficient curli nucleation requires the chaperone-like amyloid assembly protein CsgF. Proc Natl Acad Sci U S A 106:900–905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nenninger AA, Robinson LS, Hammer ND, Epstein EA, Badtke MP, Hultgren SJ, Chapman MR. 2011. CsgE is a curli secretion specificity factor that prevents amyloid fibre aggregation. Mol Microbiol 81:486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Prigent-Combaret C, Brombacher E, Vidal O, Ambert A, Lejeune P, Landini P, Dorel C. 2001. Complex Regulatory Network Controls Initial Adhesion and Biofilm Formation in Escherichia coli via Regulation of the csgD Gene. J Bacteriol 183:7213–7223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goyal P, Krasteva PV, Van Gerven N, Gubellini F, Van den Broeck I, Troupiotis-Tsailaki A, Jonckheere W, Pehau-Arnaudet G, Pinkner JS, Chapman MR, Hultgren SJ, Howorka S, Fronzes R, Remaut H. 2014. Structural and mechanistic insights into the bacterial amyloid secretion channel CsgG. Nature 516:250–253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao B, Zhao Y, Kou Y, Ni D, Zhang XC, Huang Y. 2014. Structure of the nonameric bacterial amyloid secretion channel. Proc Natl Acad Sci U S A 111:E5439–5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shu Q, Krezel AM, Cusumano ZT, Pinkner JS, Klein R, Hultgren SJ, Frieden C. 2016. Solution NMR structure of CsgE: Structural insights into a chaperone and regulator protein important for functional amyloid formation. Proc Natl Acad Sci U S A 113:7130–7135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Klein RD, Shu Q, Cusumano ZT, Nagamatsu K, Gualberto NC, Lynch AJL, Wu C, Wang W, Jain N, Pinkner JS, Amarasinghe GK, Hultgren SJ, Frieden C, Chapman MR. 2018. Structure-Function Analysis of the Curli Accessory Protein CsgE Defines Surfaces Essential for Coordinating Amyloid Fiber Formation. mBio 9: pii e01349–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Gerven N, Goyal P, Vandenbussche G, De Kerpel M, Jonckheere W, De Greve H, Remaut H. 2014. Secretion and functional display of fusion proteins through the curli biogenesis pathway. Mol Microbiol 91: 1022–1035. [DOI] [PubMed] [Google Scholar]
- 29.Hammar M, Bian Z, Normark S. 1996. Nucleator-dependent intercellular assembly of adhesive curli organelles in Escherichia coli. Proc Natl Acad Sci U S A 93:6562–6566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hammer ND, McGuffie BA, Zhou YZ, Badtke MP, Reinke AA, Brannstrom K, Gestwicki JE, Olofsson A, Almqvist F, Chapman MR. 2012. The C-Terminal Repeating Units of CsgB Direct Bacterial Functional Amyloid Nucleation. J Mol Biol 422:376–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hammer ND, Schmidt JC, Chapman MR. 2007. The curli nucleator protein, CsgB, contains an amyloidogenic domain that directs CsgA polymerization. Proc Natl Acad Sci U S A 104:12494–12499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schubeis T, Spehr J, Viereck J, Köpping L, Nagaraj M, Ahmed M, Ritter C. 2018. Structural and functional characterization of the Curli adaptor protein CsgF. FEBS Letters 592:1020–1029. [DOI] [PubMed] [Google Scholar]
- 33.Wickner W, Schekman R. 2005. Protein Translocation Across Biological Membranes. Science 310:1452–1456. [DOI] [PubMed] [Google Scholar]
- 34.Costa TRD, Felisberto-Rodrigues C, Meir A, Prevost MS, Redzej A, Trokter M, Waksman G. 2015. Secretion systems in Gram-negative bacteria: structural and mechanistic insights. Nat Rev Microbiol 13:343. [DOI] [PubMed] [Google Scholar]
- 35.Zavialov AV, Tischenko VM, Fooks LJ, Brandsdal BO, Aqvist J, Zav’yalov VP, Macintyre S, Knight SD. 2005. Resolving the energy paradox of chaperone/usher-mediated fibre assembly. Biochem J 389:685–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bernstein HD. 2015. Looks can be deceiving: recent insights into the mechanism of protein secretion by the autotransporter pathway. Mol Microbiol 97:205–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van den Broeck I, Goyal P, Remaut H. 2015. Insights in peptide diffusion channels from the bacterial amyloid secretor CsgG. Channels 9:65–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang X, Hammer ND, Chapman MR. 2008. The molecular basis of functional bacterial amyloid polymerization and nucleation. J Biol Chem 283:21530–21539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Debenedictis EP. 2017. Structural predictions for curli amyloid fibril subunits CsgA and CsgB RSC Adv 7:48102–48112. [Google Scholar]
- 40.Shewmaker F, Mcglinchey RP, Thurber KR, Mcphie P, Dyda F, Tycko R, Wickner RB. 2009. The Functional Curli Amyloid Is Not Based on In-register Parallel β-Sheet Structure J Biol Chem 284:25065–25076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Louros NN, Bolas GMP, Tsiolaki PL, Hamodrakas SJ, Iconomidou VA. 2016. Intrinsic aggregation propensity of the CsgB nucleator protein is crucial for curli fiber formation. J Struct Biol 195:179–189. [DOI] [PubMed] [Google Scholar]
- 42.Tian P, Boomsma W, Wang Y, Otzen DE, Jensen MH, Lindorff-Larsen K. 2015. Structure of a Functional Amyloid Protein Subunit Computed Using Sequence Variation. J Am Chem Soc 137:22–25. [DOI] [PubMed] [Google Scholar]
- 43.Biancalana M, Koide S. 2010. Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochim Biophys Acta 1804:1405–1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Arosio P, Knowles TPJ, Linse S. 2015. On the lag phase in amyloid fibril formation. Phys Chem Chem Phys 17:7606–7618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sleutel M, Broeck IVD, Gerven NV, Feuillie C, Jonckheere W, Valotteau C, Dufrêne YF, Remaut H. 2017. Nucleation and growth of a bacterial functional amyloid at single-fiber resolution. Nat Chem Biol. 13:902–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hammer ND, Mcguffie BA, Zhou Y, Badtke MP, Reinke AA, Brännström K, Gestwicki JE, Olofsson A, Almqvist F, Chapman MR. 2012. The C-Terminal Repeating Units of CsgB Direct Bacterial Functional Amyloid Nucleation. J Mol Biol 422:376–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang X, Chapman MR. 2008. Sequence Determinants of Bacterial Amyloid Formation. J Mol Biol 380:570–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang X, Zhou Y, Ren J-J, Hammer ND, Chapman MR. 2010. Gatekeeper residues in the major curlin subunit modulate bacterial amyloid fiber biogenesis. Proc Natl Acad Sci U S A 107:163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arnqvist A, Olsen A, Normark S. 1994. sigma-S-dependent growth-phase induction of the csgBA promoter in Escherichia coli can be achieved in vivo by sigma-70 in the absence of the nucieoid-associated protein H-NS. Mol Microbiol 13:1021–1032. [DOI] [PubMed] [Google Scholar]
- 50.Brown PK, Dozois CM, Nickerson CA, Zuppardo A, Terlonge J, Curtiss R. 2001. MlrA, a novel regulator of curli (AgF) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar Typhimurium. Mol Microbiol 41:349–363. [DOI] [PubMed] [Google Scholar]
- 51.Gerstel U, Park C, Römling U. 2003. Complex regulation of csgD promoter activity by global regulatory proteins. Mol Microbiol 49:639–654. [DOI] [PubMed] [Google Scholar]
- 52.Gerstel U, Römling U. 2003. The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res Microbiol 154:659–667. [DOI] [PubMed] [Google Scholar]
- 53.Olsen A, Arnqvist A, Hammar M, Sukupoli S, Normark S. 1993. The rpoS sigma factor relieved H-NS mediated transcriptional reprssion of csgA the subunit gene for fibronectin binding curli of Escherichia coli. Mol Microbiol 7:523–536. [DOI] [PubMed] [Google Scholar]
- 54.Chiti F, Dobson CM. 2017. Protein Misfolding, Amyloid Formation, and Human Disease: A Summary of Progress Over the Last Decade. Annu Rev of Biochem 86:27–68. [DOI] [PubMed] [Google Scholar]
- 55.Taylor JD, Hawthorne WJ, Lo J, Dear A, Jain N, Meisl G, Andreasen M, Fletcher C, Koch M, Darvill N, Scull N, Escalera-Maurer A, Sefer L, Wenman R, Lambert S, Jean J, Xu Y, Turner B, Kazarian SG, Chapman MR, Bubeck D, de Simone A, Knowles TPJ, Matthews SJ. 2016. Electrostatically-guided inhibition of Curli amyloid nucleation by the CsgC-like family of chaperones. Sci Rep 6:24656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cegelski L, Pinkner JS, Hammer ND, Cusumano CK, Hung CS, Chorell E, Aberg V, Walker JN, Seed PC, Almqvist F, Chapman MR, Hultgren SJ. 2009. Small-molecule inhibitors target Escherichia coli amyloid biogenesis and biofilm formation. Nat Chem Biol 5:913–919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Andersson EK, Bengtsson C, Evans ML, Chorell E, Sellstedt M, Lindgren AEG, Hufnagel DA, Bhattacharya M, Tessier PM, Wittung-stafshede P, Almqvist F, Chapman MR. 2013. Article Modulation of Curli Assembly and Pellicle Biofilm Formation by Chemical and Protein Chaperones. Chem Biol 20:1245–1254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dueholm MS, Petersen SV, Sonderkaer M, Larsen P, Christiansen G, Hein KL, Enghild JJ, Nielsen JL, Nielsen KL, Nielsen PH, Otzen DE. 2010. Functional amyloid in Pseudomonas. Mol Microbiol 77:1009–1020. [DOI] [PubMed] [Google Scholar]
- 59.Dueholm MS, Sondergaard MT, Nilsson M, Christiansen G, Stensballe A, Overgaard MT, Givskov M, Tolker-Nielsen T, Otzen DE, Nielsen PH. 2013. Expression of Fap amyloids in Pseudomonas aeruginosa, P. fluorescens, and P. putida results in aggregation and increased biofilm formation. Microbiologyopen 2:365–382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rouse SL, Stylianou F, Wu HYG, Berry JL, Sewell L, Morgan RML, Sauerwein AC, Matthews S. 2018. The FapF Amyloid Secretion Transporter Possesses an Atypical Asymmetric Coiled Coil. J Mol Biol 430:3863–3871. [DOI] [PMC free article] [PubMed] [Google Scholar]


