Abstract
Long interspersed nuclear element 1 (LINE-1 or L1) is the dominant retrotransposon in mammalian genomes. L1 encodes two proteins ORF1p and ORF2p that are required for retrotransposition. ORF2p functions as the replicase. ORF1p is a coiled coil-mediated trimeric, high affinity RNA binding protein that packages its full- length coding transcript into an ORF2p-containing ribonucleoprotein (RNP) complex, the retrotransposition intermediate. ORF1p also is a nucleic acid chaperone that presumably facilitates the proposed nucleic acid remodeling steps involved in retrotransposition. Although detailed mechanistic understanding of ORF1p function in this process is lacking, recent studies showed that the rate at which ORF1p can form stable nucleic acid-bound oligomers in vitro is positively correlated with formation of an active L1 RNP as assayed in vivo using a cell culture-based retrotransposition assay. This rate was sensitive to minor amino acid changes in the coiled coil domain, which had no effect on nucleic acid chaperone activity. Additional studies linking the complex nucleic acid binding properties to the conformational changes of the protein are needed to understand how ORF1p facilitates retrotransposition.
1. Introduction
The long interspersed nuclear element 1 (LINE1, L1) non-LTR retrotransposon is an autonomously replicating genomic parasite that has been amplifying and evolving in mammalian genomes for >100 Myr and now constitutes 20% or more of certain mammalian genomes [1–3]. In addition, L1 can also copy non-L1 transcripts into genomic DNA, and as a consequence L1 activity has generated upwards of 40% of the mass of many mammalian genomes [4,5]. Only a subset of L1 elements actively retrotranspose in the mammalian genomes [6–9]. Currently, 80–100 L1 copies that belong to a single subfamily Ta are active in the human genome [10]while ~3000 L1 copies of 3 subfamilies, TF, A and GF, are active in the mouse genome [11–13]. Replication and evolution continue in humans [14–16], and L1 activity creates genetic diversity and various genetic alterations [17–20]. Given its detrimental [21–23] and potential catastrophic effects [24,25], explaining its persistence as well as determining a mechanistic understanding of its replication remain formidable biological issues.
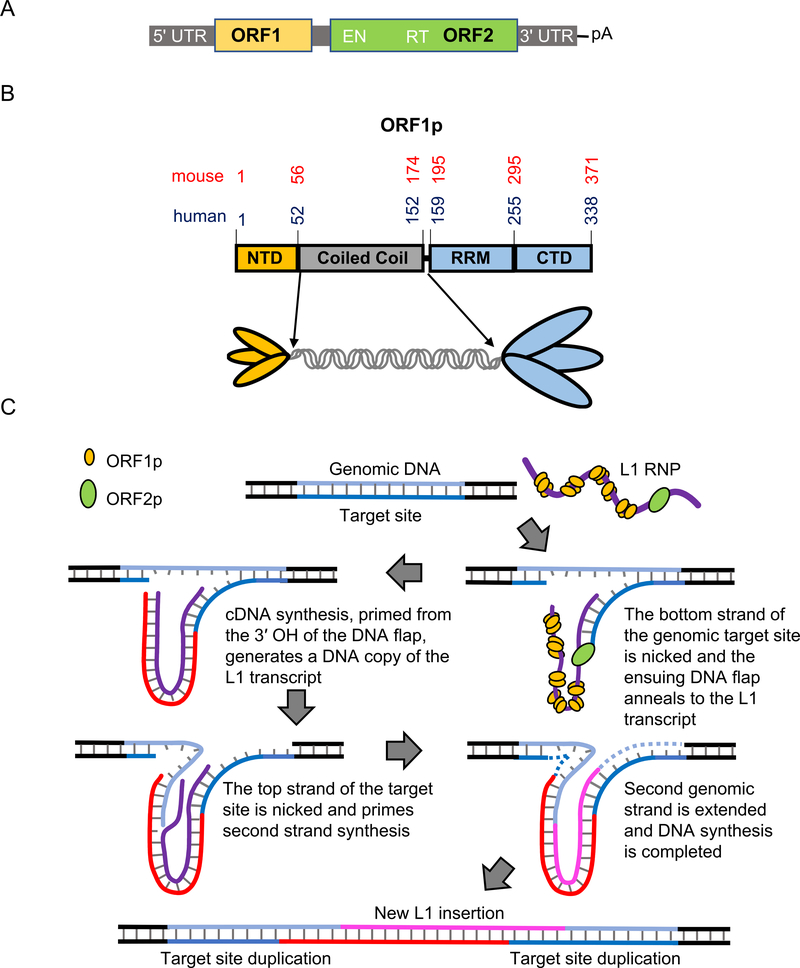
Mammalian L1 elements are 6–7 kb in length, contain a regulatory 5′ UTR, two long open reading frames that encode two proteins ORF1p and ORF2p, and a 3′ UTR (Fig. 1A). Both ORF1p (Fig. 1B) and ORF2p are required for retrotransposition [26]. The two proteins associate with their encoding transcript to form a ribonucleoprotein complex (RNP), which mediates retrotransposition [27–30]. L1 replicates by reverse transcription of its transcript. While this is analogous to retroviruses and retroviral-like retrotransposons, L1 replicates by the dramatically different process of target-site-primed reverse transcription, or TPRT [31,32] (Fig. 1C). In this mechanism reverse transcription of the L1 RNA transcript is primed from a 3′ hydroxyl at the genomic insertion site [33]. Highly conserved endonuclease and reverse transcriptase domains in ORF2p indicate its replicase function in TPRT [26,34,35]. In contrast, ORF1p lacks any known enzymatic domains, although it does contain highly conserved non-canonical RNA binding domains and phosphorylation sites [36,37], that are required for retrotransposition activity.
Fig. 1.
A) L1 domain structure. The domain organization of a typical full-length human L1 element. Approximate positions (not to scale) of the 5′ untranslated region (5′ UTR), ORF1, ORF2 (which includes endonuclease (EN) and reverse transcriptase (RT)), 3′ UTR, and the poly A tail are noted. B) Domain structure and trimeric depiction of ORF1p. The amino acid positions of the N-terminal domain (NTD), coiled coil domain (CC); RNA recognition motif (RRM) and C-terminal domain (CTD) for mouse [13,40,47,55] and human [36,44,46] ORF1p are denoted in red and blue text, respectively. C) Schematic of the proposed steps in TPRT [3,31,32]. Target site of the genomic DNA is depicted in blue shades and L1 RNA in purple. The first DNA strand at the target site is cleaved by ORF2p EN. The L1 RNA is annealed to the cleaved site and reverse transcribed by ORF2p RT to synthesize the first L1 cDNA (red). The second genomic target site strand is cleaved, and it primes the L1 cDNA to synthesize the second L1 DNA strand (magenta). Subsequent DNA synthesis required for completion is denoted in dashed lines. Consequently, target site duplications (TSD) are produced at the flanks of the newly synthesized L1 element. ORF1p may mediate strand exchange reactions required to anneal the primers and/or facilitate nucleic acid arrangements during reverse transcription.
Human and mouse ORF1 encode a ~40 kDa protein and early studies showed that they could be isolated as L1 RNP particles associated with their encoding L1 RNA [30,38]. As presented in detail below, subsequent studies showed that the 40 kDa monomer forms a coiled coil-mediated trimer that binds nucleic acids with high affinity (Fig. 1B) [38,39]. In vitro studies using bulk biochemical and single molecule assays showed that human and mouse ORF1p (from here on referring to the trimer) act as nucleic acid chaperones, presumably during TPRT [32,40–43], however, at which steps and by what means is unknown. In addition, both human and mouse ORF1p can polymerize in the presence or absence of nucleic acids [43–45]. Recent structural and theoretical studies on human and mouse ORF1p addressed its complex nucleic acid binding properties [46–49], yet we do not understand how they contribute to the molecular mechanism of L1 retrotransposition. We now review the current understanding of ORF1p in the context of those nucleic acid binding properties that had been obtained with the purified protein. Therefore, we will not be discussing the extensive and informative cell biological literature on the structure and properties of the L1RNP and its possible interaction with host factors as exemplified by two recently published papers [50,51].
2. ORF1p structure
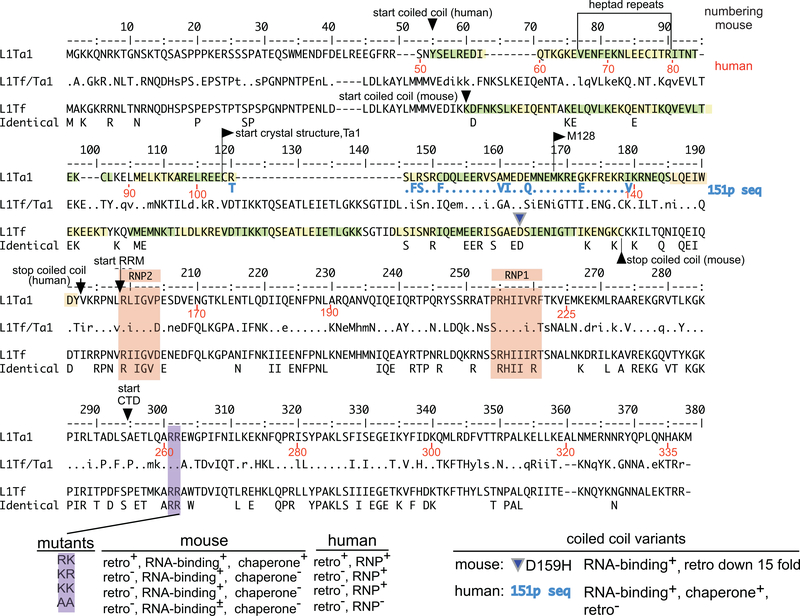
The amino- and carboxy-terminal halves of mammalian ORF1p evolved under dramatically different evolutionary constraints. The carboxy terminal half is highly conserved (Fig. 2) and contains a non-canonical RNA recognition motif (RRM) and a distinct C-terminal domain (CTD) (Fig. 1B). Residues in the carboxy-terminal half mediate high affinity RNA binding and nucleic acid chaperone activity [32,39–42,44,52]. In contrast, the sequence of the amino-terminal half can be highly variable, although a coiled coil motif is conserved throughout vertebrate evolution [21,53,54]. Direct visualization of mouse ORF1p using atomic force microscopy revealed an elongated dumbbell-like structure, consistent with its trimeric conformation [55] mediated by the coiled coil [44,46,55], which is parallel in nature and stabilized by additional inter-chain hydrogen bonds [46]. Molecular structures for the human C-terminal half [36,46] and the mouse CTD [47] were solved. The RRM domain of human ORF1p is structured with a typical βαββαβ fold and contains non-canonical RNP1 and RNP2 sequences (Fig. 2) with aromatic side chains that could mediate base-stacking or hydrophobic interactions with nucleic acid substrates. It also contains two highly conserved salt bridges (E165-R215 and E169-R202) that are formed between two loops, L(β1-α1) and L (β2–β3) [36,46]. The L (β2-β3) loop is intrinsically disordered [46] and contains the two T/SP motifs (T203P204, and T213P214) out of the four (S18P19, S27P28 in the NTD) highly conserved proline-directed protein kinase (PDPK) targets in ORF1p. T or S phosphorylatable residues at these respective sites are required for L1 retrotransposition [37,56].
Fig. 2. Alignment of mouse and human ORF1p sequences.
Consensus sequences of the mouse L1Tf family [13] and the human Ta1 subfamily of the currently active L1Pa1 [15] family were aligned. The middle L1T1f/Ta1 sequences shows the comparison between the aligned elements. A dot indicates identity, dashes indicate gaps, and lower case indicates conservative amino acid differences. The bottom entry (Identical) shows the 100% identical positions. The numbers in black refer to the L1Tf, and those in red, the L1Ta1 sequence. The beginning and end of the coiled coil domain, and the starts of the RNA recognition motif (RRM) and C-terminal domain (CTD) are indicated. The coordinates of the RRM and CTD were taken from references [36,47]. The alignment is also annotated with the following information: The beginning of truncated M128 [44], which is largely a monomer at ≥20 °C; the location of a natural variant in the mouse coiled coil domain (D159H, using the L1Tf numbering with an offset of plus 4 in the mouse alignment only to account for the 4 position gap introduced into the alignment of L1Tf between positions 40 and 45) [41,26,38,46] paired mutations in two adjacent highly conserved arginine residues of the CTD, in human ([26,29]) and in mouse [42]. Heptad repeats are highlighted in green and yellow. RNP1 and RNP2 sequences are highlighted in light brown [36]. Regarding the “phenotype” of these mutations: retro means retrotransposition as measured in a cell culture based retrotransposition assay [26]; RNP means the presence of ORF1p in RNP particles isolated from in vivo assays; RNA binding means as performed with purified ORF1p protein in vitro; chaperone means chaperone activity (as described in [42]).
A crystal structure of human ORF1p, which lacked most of its N-terminal half, yielded three distinct orientations[46] consistent with three structural states for the CTD: resting, lifting, and twisting. The π-stacking interaction between Y282 and R155 was shown to primarily influence the dynamics of the CTD. A lifting motion of the CTD was proposed to open the RRM–CTD cleft and expose the basic residues for nucleic acid binding. Subsequent twisting or rotational motions would then wrap the nucleic acid, such that a continuous strand would occupy binding sites at each ORF1p monomer. The resting position of the CTD render the basic patch residues of the coiled coil domain accessible, which could allow further nucleic acid binding. This binding mechanism is speculative, however, because the structure did not contain nucleic acid. To support the proposed binding mechanism, several mutants were generated and probed in NA binding and retrotransposition studies. For instance, the highly conserved R261 (Fig. 2), at which substitutions eliminated retrotransposition both in human [26,29] and mouse ORF1p [42], was predicted from the structure to inhibit RNA binding, which was also confirmed by size-exclusion chromatography. Other mutations, such as R220A, abolished retrotransposition but did not inhibit RNA binding. The effect of this and other similar mutations was proposed to be due to a defect in ORF1p flexibility and dynamics based on the structure.
However, it is important to keep in mind that, as mentioned above, the crystal structure was derived from a protein that lacks the NTD and slightly more than half of the coiled coil domain, both of which contain structures critical to ORF1p function. The intrinsically disordered NTD contains highly conserved PDPK phosphorylation sites [37] and the coiled coil facilitates rapid polymerization of the trimers on single-stranded NA [43], two functions that are vital for L1 retrotransposition. Furthermore, the structural conformation of ORF1p may significantly vary in the presence of nucleic acids. Therefore, although the ORF1p C-terminal half structure suggests that flexible inter-domain orientations are functionally important, the exact details of the structure-function correlation may differ for full length protein. Nevertheless, it is clear that multiple domains in ORF1p simultaneously interact with nucleic acids and are also capable of mediating inter-protein interactions [44] (discussed in the next section, see Fig. 3). This results in a complex set of possible configurations, which in turn appear to depend strongly on solution conditions. An understanding of these configurations is essential to determine how ORF1p facilitates different stages of retrotransposition.
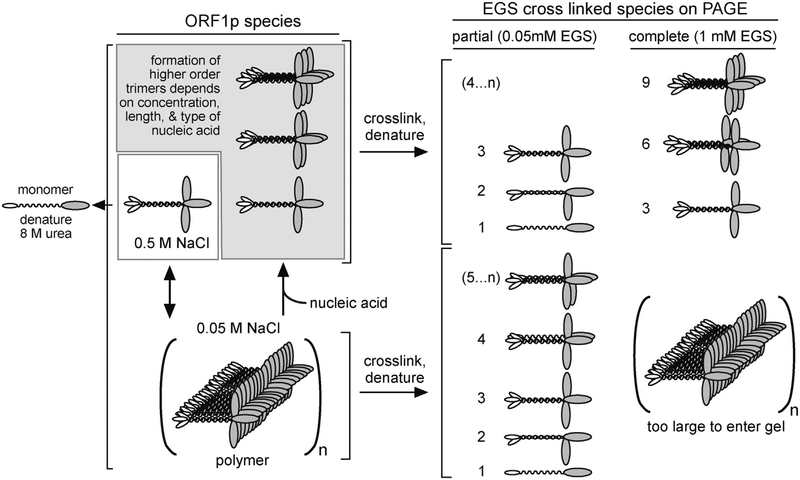
Fig. 3. ORF1p polymerization.
Schematic representation of possible ORF1p species and their cross-linked products as observed in Callahan et al. [44]. The numbers to the left of the cartoons of the cross-linked species indicate their monomer content. 4..n and 5..n indicate higher orders of multimers beyond 3 or 4, respectively. Higher orders of ORF1p multimers of trimers were observed in crosslinking experiments with 1 mM EGS, in the presence of oligonucleotides at 0.05 M NaCl.
3. Nucleic acid binding properties of ORF1p
Several early studies demonstrated co-localization of ORF1p and L1 RNA in L1 RNPs. For example, L1 RNA of L1RNP isolated from mouse embryonal carcinoma cells was efficiently cross-linked to protein by a brief exposure to ultraviolet light, indicating that ORF1p and L1 RNA were in close contact [57]. Similarly, high concentrations of monovalent cations and RNase dissociated L1 RNPs isolated from human teratocarcinoma cell lines, again indicating co-localization between the protein and L1 RNA [38,58]. Co-localization of ORF1p with L1 RNA was further confirmed when HeLa cells were transfected with an L1 that contained an epitope tagged ORF1p [29]. Subsequent quantitative binding studies of ORF1p to NA showed that ORF1p binds without sequence specificity to both RNA and DNA [41–44,52,59,60].
Electrophoretic mobility shift assays showed that mouse L1 ORF1p binds RNA with positive cooperativity, providing early evidence of protein-protein interactions between the polypeptides [61]. The nature of protein-protein interactions was extensively studied in Callahan et al. [44] with human ORF1p purified from insect cells (Fig. 3). In this study, the cross-linking reagent EGS, which interacts with primary amines (e.g.,ε-amino group of lysine), was used to capture ORF1p interaction products, prior to treatment with 8 M urea, which fully denatures non-crosslinked ORF1p into its constituent components, i.e., monomers, partial trimers, and multiples of trimers (Fig. 3). In 0.5 M NaCl ORF1p is a soluble trimer but is unable to bind nucleic acids. Optimal nucleic acid binding occurs at 50 mM NaCl, but in the absence of nucleic acids, ORF1p trimers rapidly form massive polymers that precipitate from solution. When cross linked with 1 mM EGS the cross-linked polymers are too large to enter a 6% denaturing polyacrylamide (PAGE) gel (Fig. 3). Addition of equimolar oligonucleotides (N ≥ 20) immediately resolves the ORF1p polymers to trimers or multimers of trimers (depending on the length of the oligonucleotide, (Fig. 3)). Therefore, ORF1p remains active in its polymeric conformation, its presumed state in the L1RNP. Partial crosslinking occurs with limiting EGS (0.05 mM) and generates a ladder of products starting at the size of the ~40 kDa monomer and multimers thereof (Fig. 3). Carrying out these experiments with an amino-terminal truncated ORF1p (M128, Fig. 2) [44], which contains just 3.5 heptads of the 14-heptad coiled coil and is largely a monomer at ≥20 °C, showed that the amino acid residues that mediate inter-trimer interactions reside in the conserved C-terminal half of the protein.
The ability to form large oligomers in the absence of nucleic acid may provide a mechanism to prevent the diffusion of the newly synthesized protein into the cytoplasm before binding to L1 RNA. However, the relationship between ORF1p translation from the L1 transcript and formation of the L1 RNP with the L1 transcript remains obscure as any such large clusters must dissociate in order to optimally coat the transcript. ORF1p binds RNA and ssDNA with similar affinity, but with an ~8-fold lower affinity to a perfectly matched duplex. In contrast, ORF1p binds mismatched duplexes with about the same affinity as ssDNA, and, in fact, stabilizes it from dissociation [44]. This occurs at near equimolar or a 10-fold higher molar excess of ORF1p/duplex but at sufficient molar excess, ORF1p dissociated the duplex. The monomeric form of ORF1p (i.e., N-terminal truncated M128p, Fig. 2) bound nucleic acids with substantially lower affinities, in comparison to the trimer, and could not protect a mismatched duplex from dissociation [44], even though M128p contains the intact RRM and CTD domains critical for both high affinity nucleic acid binding and NA chaperone activity [36,42,45,47]. These results indicate that intra-trimeric interactions play vital roles for the mechanism of nucleic acid binding by ORF1p.
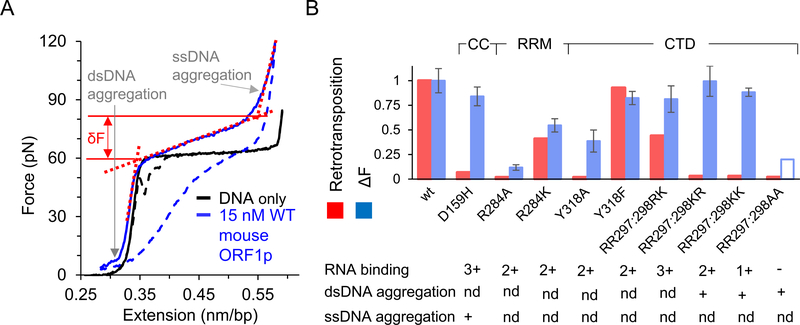
Single molecule DNA stretching studies have been used to quantitatively characterize the affinity of mouse ORF1p for DNA. Fig. 4 shows a diagram of a typical single molecule DNA stretching experiment while Fig. 5A shows a DNA stretching curve in the presence and absence of wild type mouse ORF1p. The width of the transition, shown as F in Fig. 5A, demonstrates the strong effect of ORF1p on DNA conformation. Transition width is positively correlated with protein binding to DNA, so the width as a function of concentration was used to determine the DNA binding affinity by fitting to a binding isotherm [41]. Here the protein concentration dependence of the change in the DNA force-extension profile is used to determine the DNA binding affinity. Below we will discuss the biophysical interpretation of the change in transition width in relation to nucleic acid chaperone activity. The DNA binding affinity measurements demonstrate that mouse ORF1p binding to DNA is in the range of 10 nM in near-physiological solution conditions (50–100mM Na+). However, many ORF1p mutants did not alter binding affinity but strongly affected retrotransposition [41,42]. Thus, more complex interactions beyond simple nucleic acid binding determine the effect of mouse ORF1p on retrotransposition, as will be discussed below in the section on nucleic acid chaperone activity
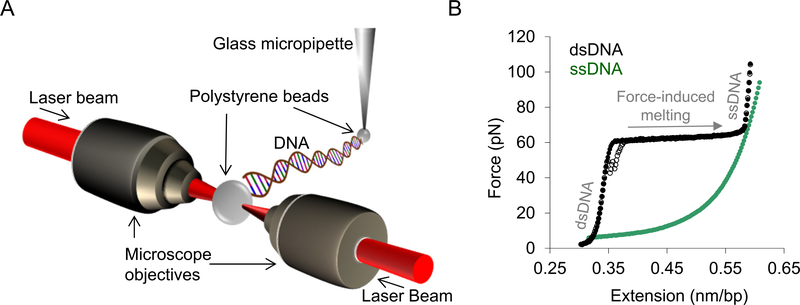
Fig. 4. Single molecule DNA stretching.
A) Schematic depiction of an optical tweezers system. A single molecule of DNA (48.5 kbp) is attached by its biotinylated ends to streptavidin-coated polystyrene beads. One bead is immobilized by a glass micropipette attached to a flow cell while the other is held in an optical trap. The optical trap is created by converging two counter-propagating laser beams to overlap in space using microscope objectives. By moving the glass micropipette attached to the flow cell, the tethered DNA molecule is stretched, and the force exerted on the DNA is measured as a function of extension in the presence and absence of protein. B) Solid and empty black circles (also the solid and dashed black lines in Figs. 5A and 6 A) represent the stretch and return curves of a bare dsDNA molecule. The green circles represent the force-extension curve of an ssDNA molecule. The distinct regimes of the dsDNA force-extension profile are denoted in grey text. At forces ≪~60 pN the DNA molecule is primarily double-stranded. The plateau at ~60 pN represents a sharp overstretching transition, where very little additional force causes the DNA molecule to stretch to almost twice its contour length. This is due to force-induced melting of the DNA molecule and this transition is referred to as the helix-coil transition [73]. The force range over which this transition occurs is defined as the helix-coil transition width (or transition width). In the salt conditions used in these studies, the stretched dsDNA molecule is mostly single-stranded at forces higher than the helix-coil transition. The stretch and return cycles of the bare DNA molecule are almost reversible and immediate re-annealing of the duplex is observed when the stretched DNA molecule is returned to zero-force (empty black circles).
Fig. 5. Single molecule DNA stretching results from mouse ORF1p mutational analyses [41,42,59].
Solid and dashed blue lines are the stretch and return curves of dsDNA in the presence of 15 nM wild type mouse ORF1p. ΔF is the relative increase in the helix-coil transition width due to bound protein, where ΔF = δF–δF0. Here, δF0 (~4pN) and δF are the helix-coil transition widths in the absence and presence of protein, respectively. δF (shown as the force difference between the solid red lines) is determined by the intersection of the dashed red lines on the figure, which represent ds- and ss- DNA regimes of the ORF1p-DNA complex. Thus, ΔF is a measure of the relative increase in force (due to bound protein) required in the cooperative conversion of ds- to ss- DNA, which was shown to be positively correlated with the nucleic acid chaperone capabilities of the bound protein as described in the text. B) ΔF at 15 or 20 nM protein concentration, as measured in single molecule DNA stretching assays (blue), and retrotransposition activity in cultured in vivo assays (red) for ORF1p mutants, presented after normalizing with the corresponding values for the wild type protein. The transition width for the RR297:298AA mutant at these concentrations was not reported due to its lower binding affinity. However, it was shown to saturate at a 5-fold higher concentration than what was used for wild type protein. The empty bar is to denote that the transition width is likely much smaller at these concentrations in comparison with the wild type protein. Extensive aggregation of single- or double- stranded DNA in comparison with the wildtype protein, are denoted with a plus sign. While aggregation is important to facilitate DNA interactions and can increase chaperone activity, extensive aggregate formation can also slow down DNA interaction kinetics, which may in turn inhibit chaperone activity [79]. The ability to bind RNA as observed in the bulk solution assays is also reported. The plus sign denotes comparable binding affinity as wild type and these mutants are ranked according to their relative binding affinities, with (−) the lowest and (3+) the highest affinity. ‘nd’ represents ‘not determined’.
4. Nucleic acid chaperone activity of ORF1p
The L1 retrotransposon, like retroviruses, replicates by reverse transcription of an RNA intermediate. Reverse transcription of retroviruses is facilitated by the nucleic acid chaperone activity of its nucleocapsid (NC) proteins. This appears to be achieved by lowering the energy barrier of rearranging nucleic acids to achieve maximal base complementarity [62]. Therefore, in vitro, a nucleic acid chaperone will primarily mediate nucleic acid annealing and strand exchange reactions. NC proteins from multiple retroviruses and retroelements [63–66] act as nucleic acid chaperones and contain the CCHC zinc finger motif, critical for retroviral replication processes [67,68]. In the case of HIV-1 NC, nucleic acid chaperone activity is positively correlated with retroviral infectivity [69,70]. ORF1p of I factor, a LINE-like element of Drosophila melanogaster [71], also behaves as a nucleic acid chaperone [72]. Although some retrotransposons such as Ty3 [66] contain an NC protein, L1 does not. Thus, as mammalian L1 ORF1p does not contain a CCHC motif it likely does not function by the same mechanism as NC proteins. Nevertheless, both mouse and human L1 ORF1p were shown to be nucleic acid chaperones [32,43].
To address the mechanism of ORF1p chaperone activity mutational analyses on mouse ORF1p were performed to correlate in vitro nucleic acid binding with retrotransposition. As discussed above, DNA binding affinities for several mutants was determined by single molecule DNA stretching assays using helix-coil transition width as a function of ORF1p concentration. The helix-coil transition in the absence of protein is very narrow (δF is small), due to cooperative conversion of dsDNA to ssDNA, i.e., the requirement for near simultaneous melting of multiple base pairs. Therefore, it was suggested that when an NA-binding protein increases the transition width (transition is less cooperative) it does so by lowering the energetic barrier to rearrangement of nucleic acid secondary structure, thereby promoting melting of small numbers of base pairs [67,68]. Thus, measurement of the transition width reflects both DNA binding affinity and its chaperone activity; i.e., that a greater transition width represents greater chaperone capabilities. When fully bound by protein, mouse ORF1p mutants altered the transition width to different degrees, and the extent to which the proteins increased the transition width at saturated protein binding appeared to correlate with retrotransposition activity. This result is shown in Fig. 5B, which shows the transition width at high protein concentration, near saturated binding, for several mutants [73]. As described above, HIV-NC protein dramatically increased the transition width [67,68,74,75] as also observed with ORF1p. Thus, it was suggested that the mutations reduced ORF1p nucleic acid chaperone activity, which was responsible for reduced retrotransposition [42]. However, the exact mechanism by which this dramatic increase in helix-coil transition occurs is not clear. A similar effect is also observed in the presence of DNA intercalators such as ethidium [76] and ruthenium complexes [77,78], which prevent DNA strand separation. Therefore, the observed increase in the helix-coil transition width in the presence of HIV-1 NC was subsequently attributed to the simultaneous destabilization of the duplex while maintaining protein-mediated interactions between the two DNA strands [65,69,70]. Below we discuss the ORF1p mutations that altered both retrotransposition and their interaction with DNA as measured by single molecule DNA stretching. The mutations are shown in Fig. 2 and the results from stretching studies are summarized in Fig. 5B.
Substitutions in the highly conserved R284 (Fig. 2, note the +4 offset in the mouse ORF1p residue positions as described in the legend to Fig. 2) in the RRM and RR297–298 and Y318 in the CTD as well as D159 in the coiled coil of mouse ORF1p (Fig. 2) were analyzed by DNA stretching [41,42,59]. Alanine substitutions at 284 and 318 (R284A and Y318A) eliminated retrotransposition, while substitutions that conserved charge or hydrophobicity (R284K and Y318F) were active in retrotransposition. In contrast, KR297–298, KK297–298 or AA297–298 substitutions at RR297–298 eliminated retrotransposition while the RK297–298 mutation remained active, so simply preserving the charge is not always sufficient to retain retrotransposition activity. A [26,29] D159H substitution at the coiled coil lowered the retrotransposition frequency by a factor of ~15.
As shown in Fig. 5B, many of the mutants show a direct correlation between reduced transition width and reduced retrotransposition relative to wild type ORF1p (R284A, R284K, Y318A, Y318F, R298K), suggesting that the reduced retrotransposition is due to inhibition of ORF1p chaperone activity. However, there are some outliers, which show similar transition widths to wild type ORF1p but are defective in retrotransposition (D159H, RR297:298KR, RR297:298KK). These outliers required further analysis to understand why retrotransposition was inhibited, however we note that D159H is a coiled coil mutant and in Section 5, we explore the effect of coiled coil mutants of human ORF1p and their effect on retrotransposition.
To further understand the interactions between these additional ORF1p mutants and DNA, the full stretching curve was analyzed. In addition to an increase in transition width, extensive dsDNA aggregation by dsDNA binding proteins can increase the force required to reach the dsDNA contour length (RR297–298 KR, RR297–298KK, and RR297–298AA in Fig. 5B) [59]. In a recent study, the dsDNA aggregation by NC proteins from SIV and HIV-1 were quantitatively measured and used to differentiate the nucleic acid chaperone activity of these two proteins [63]. Strong dsDNA aggregation and wild-type transition widths was observed for RR297–298KR and RR297–298KK. While this shows that these proteins behave differently than wild type ORF1p, we do not know why retrotransposition is inhibited. Although dsDNA aggregation is a strong component of nucleic acid chaperone activity, aggregation can also inhibit chaperone activity if it slows protein-nucleic acid interaction kinetics [79]. The reduced retrotransposition for RR297–298AA was attributed to weak RNA binding. In comparison, all of the other mutants studied showed RNA binding affinity similar to wild type ORF1p. Finally, while D159H showed a wild type transition width and wild type levels of dsDNA aggregation, it was defective in retrotransposition. However, upon analysis of the full stretching curve, it was found that the defective activity of the protein is correlated with its extensive ssDNA aggregation. The DNA observed at extensions beyond 0.55 nm/bp is mostly single-stranded (Fig. 4B) in the salt conditions (50–100mM Na+) used in the studies discussed here[80–82]. Therefore, a relative decrease in ssDNA extension after the helix-coil transition in the presence of protein is indicative of ssDNA aggregation due to bound protein. This reduced retrotransposition activity resulting from ssDNA aggregation may also be attributed to a reduction in protein-DNA interaction kinetics [41,79].Thus, all of the mutants described here that exhibited wild type nucleic acid binding but were defective in retrotransposition showed a difference in ssDNA aggregation, dsDNA aggregation, or helix-coil transition properties in single molecule stretching experiments. These differences in turn may be related to nucleic acid chaperone activity. However, the mechanisms behind these changes were not determined in wild type and mutant mouse ORF1p studies. Therefore, understanding the reason for the observed behavior will require quantitative DNA interaction kinetics studies and characterization of ORF1p oligomerization states as discussed below [43,44].
5. ORF1p oligomerization and single molecule DNA binding kinetics
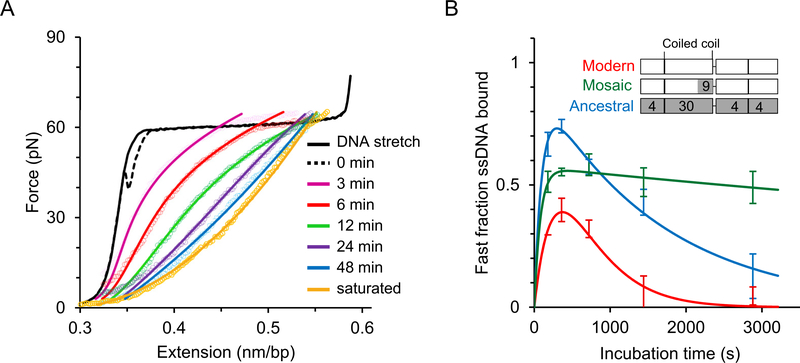
Single molecule DNA stretching assays were also carried out on human ORF1p to determine its DNA interaction kinetics in the context of its previously measured oligomerization properties [43,44]. A single DNA molecule was captured in the optical tweezers instrument and stretched to high forces beyond the helix-coil transition, such that most of the DNA molecule was single-stranded. The DNA molecule was held at this force in the presence of ORF1p for different incubation times, as shown in Fig. 6A. After subsequent relaxation to lower forces, the resulting length of DNA strongly depended on the time of incubation, indicating that ORF1p formed stably bound oligomers on ssDNA, with more stably bound (longer) oligomers at longer incubation time [43]. Three ORF1p variants were compared; modern human ORF1p, its resuscitated primate ancestor and a mosaic variant of the modern protein in which 9 of the 30 substitutions in the coiled coil domain retained their ancestral state (Fig. 2 and inset of Fig. 6B). Although these proteins formed equally stable trimers and behaved indistinguishably in bulk binding and in vitro nucleic chaperone assays, the mosaic 151p was completely inactive in an in vivo retrotransposition assay (Fig. 7). Here, the rate of ORF1p oligomerization on ssDNA was determined (Fig. 6). During incubation, stable oligomers formed on the ssDNA molecule held between polystyrene beads in solution in the optical tweezers. After a fixed time, the fraction of stable oligomers was determined. The results showed that both retrotransposition-competent ORF1p proteins (i.e, the modern human protein and its resuscitated ancestral counterpart) rapidly formed stably-bound oligomers on ssDNA. In contrast, the retrotransposition-defective mosaic ORF1p formed such stably bound oligomers at a rate at least 10-fold slower (Fig. 6B, green curve). This property is a function of the coiled coil sequence, presumably by positioning the carboxy-terminal halves ORF1p in a way that is conducive to the inter-trimer contacts that mediate oligomerization. Other than trimer formation per se (which is necessary but clearly is not sufficient for retrotransposition), this study revealed the first functional role for the coiled coil in L1 retrotransposition.
Fig. 6. Single molecule studies of human ORF1p [43].
A) In contrast to the studies described in Fig. 5, here, the DNA molecule is overstretched above the helix-coil transition up to the ssDNA regime before incubating it with the protein, in order to minimize dsDNA binding effects and exclusively investigate ssDNA-ORF1p binding kinetics. Solid and dashed line represent the stretch-return cycle of a bare dsDNA, with the return curve also referred to as 0 min incubation. The colored empty circles are the return curves of a DNA molecule after incubating with 2 nM protein at ~70 pN for different time periods. The gold curve listed as saturated represents the maximum shift in extension towards ssDNA, obtained at long incubation times and greater than 15 nM protein concentration. The ssDNA-bound protein during the incubation prevents reannealing and thereby shift the relaxation curve after protein incubation towards the ssDNA curve (see green circles, Fig. 4B), allowing one to quantitatively probe the ssDNA fraction bound as a function of time. The solid lines represent the fitted curves as described elsewhere [43]. ORF1p rapidly binds ssDNA. However, it transforms relatively slowly into stable oligomers. Less stable proteins (presumably un-transformed trimers) dissociate during the return cycle and the ssDNA fraction bound by such proteins is defined as the fast fraction in this study. The fast fraction decreases with incubation time as they transform into more stable oligomers on ssDNA. B) Fast fraction as a function of time for modern human (111p, from L1Pa1 family), its resuscitated ancestral primate (555p, from L1Pa5 family) and a mosaic ORF1p (151p) in which 9 residues in the coiled coil are replaced with the corresponding ancestral residues, as shown in the inset (also see Fig. 2 where these residues are indicated in teal). Domain boundaries in the inset correspond to Fig. 1B and white and grey shades represent the amino acid residues of the modern and ancestral ORF1p, respectively. The number of amino acid substitutions relative to the modern ORF1p is denoted in the relevant domains in the other two ORF1p variants. The measured fast fraction is modeled as a sum of increasing and decreasing exponential functions (solid lines) and red, blue and green represent modern, ancestral, and mosaic proteins, respectively. The fast fraction rapidly saturates for all three variants, indicating rapid protein binding to ssDNA. However, this fraction decreases with increasing incubation time as the proteins form more stable oligomers. Therefore, the rate at which the fast fraction decreases is proportional to the rate of stable oligomerization of protein on ssDNA. The retrotransposition-incompetent mosaic ORF1p was at least 10-fold slower in forming stable oligomers in comparison with the active human and primate protein variants.
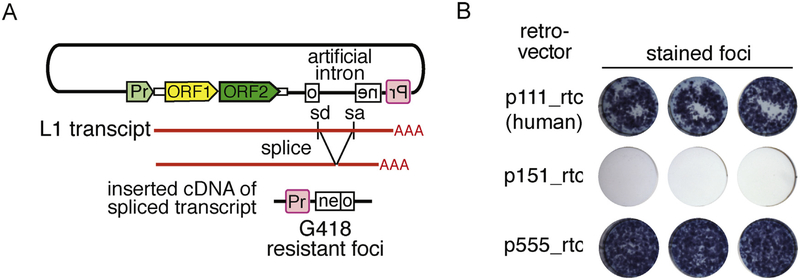
Fig. 7. Retrotransposition assay.
A) Organization of an L1 vector in a typical retrotransposition assay. The L1 vector contains an antisense copy of the neo gene disrupted by an intron in the sense orientation. sd and sa are the splice donor and splice acceptor sites, respectively. The intron of the transcribed L1 vector will be spliced and contain the antisense copy of the neo gene. Retrotransposition competent elements will support subsequent cDNA synthesis of this transcript at a DNA target site and ultimately the insertion of an active copy of the neo gene, which when expressed from its promoter (Pr, in red) generates colonies of G418 resistant cells or foci. B) An example of stained foci generated from the retrotransposition assay described in (A) [43]. p111_rtc, p151_rtc and p555_rtc are the L1 vectors containing the modern human, mosaic and the resuscitated primate ORF1 sequences (see inset of Fig. 6B), respectively.
6. Conclusions and future directions
Although ORF1p is known to be critical for L1 retrotransposition in mouse and human systems, the mechanism by which it facilitates retrotransposition is not yet understood. To provide a clear mechanism, it will be necessary to fully characterize its structure function relationship and correlate its in vitro activities with its function in the cell. For mouse ORF1p, several studies have identified specific mutations that determine the locations of regions that are critical for nucleic acid chaperone activity and for retrotransposition. However, studies of mouse ORF1p have not yet characterized its oligomerization properties, although aggregation of DNA was observed in many single molecule experiments. This indicates that oligomerization and polymerization, as observed for human ORF1p, likely also occur for mouse ORF1p. It would be interesting to characterize these properties for wild type and mutant mouse ORF1p using similar methods for comparison.
The recent experiments on human ORF1p demonstrate a wide variety of protein-protein and protein-nucleic acid interactions that likely govern ORF1p function in the cell. It is important to quantify the behavior of human ORF1p under multiple solution conditions to understand the dynamics of protein oligomerization. Although protein oligomerization is difficult to study, it is now clear that oligomerization kinetics are a determinant of retrotransposition activity. Therefore, future studies should attempt to determine the relationship between structure and human ORF1p function while including supramolecular structures formed during oligomerization. Not only are these large structures themselves important, but the process of assembly of the structures is clearly also critical for protein function.
Acknowledgements
This work was supported by the Intramural Program of the National Institute of Diabetes, Digestive and Kidney Diseases; National Institutes of Health [HNK6H7 A.V.F]; National Institutes of Health [R01GM072462 to M.C.W.]; National Science Foundation [MCB-1243883 to M.C.W.].
References
- [1].Furano AV, The biological properties and evolutionary dynamics of mammalian LINE-1 retrotransposons, Prog. Nucleic Acid Res. Mol. Biol 64 (2000) 255–294. [DOI] [PubMed] [Google Scholar]
- [2].Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, et al. , Initial sequencing and analysis of the human genome, Nature 409 (6822) (2001) 860–921. [DOI] [PubMed] [Google Scholar]
- [3].Kazazian HH Jr., Moran JV, Mobile DNA in health and disease, N. Engl. J. Med 377 (4) (2017) 361–370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Esnault C, Maestre J, Heidmann T, Human LINE retrotransposons generate processed pseudogenes, Nat. Genet 24 (4) (2000) 363–367. [DOI] [PubMed] [Google Scholar]
- [5].Wei W, Gilbert N, Ooi SL, Lawler JF, Ostertag EM, Kazazian HH, Boeke JD, Moran JV, Human L1 retrotransposition: cis preference versus trans complementation, Mol. Cell. Biol 21 (4) (2001) 1429–1439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Moran JV, Gilbert N, Mammalian LINE-1 retrotransposons and related elements, in: Craig NL, Craigie R, Gellert M, Lambowitz AM (Eds.), Mobile DNA II, American Society of Microbiology Press, Washington, DC, 2002, pp. 836–869. [Google Scholar]
- [7].Martin SL, Characterization of a Line-1 Cdna that originated from Rna present in Ribonucleoprotein-particles–implications for the structure of an active-mouse Line-1, Gene 153 (2) (1995) 261–266. [DOI] [PubMed] [Google Scholar]
- [8].Schichman SA, Severynse DM, Edgell MH, Hutchison CA 3rd, Strand-specific LINE-1 transcription in mouse F9 cells originates from the youngest phylogenetic subgroup of LINE-1 elements, J. Mol. Biol 224 (3) (1992) 559–574. [DOI] [PubMed] [Google Scholar]
- [9].Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH Jr., Hot L1s account for the bulk of retrotransposition in the human population, Proc. Natl. Acad. Sci. U. S. A 100 (9) (2003) 5280–5285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Sassaman DM, Dombroski BA, Moran JV, Kimberland ML, Naas TP, DeBerardinis RJ, Gabriel A, Swergold GD, Kazazian HH Jr., Many human L1 elements are capable of retrotransposition, Nat. Genet 16 (1) (1997) 37–43. [DOI] [PubMed] [Google Scholar]
- [11].Goodier JL, Ostertag EM, Du K, Kazazian HH, A novel active L1 retrotransposon subfamily in the mouse (vol 11, pg 1677, 2001), Genome Res. 11 (11) (2001) 1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Kingsmore SF, Giros B, Suh D, Bieniarz M, Caron MG, Seldin MF, Glycine receptor beta-subunit gene mutation in spastic mouse associated with LINE-1 element insertion, Nat. Genet 7 (2) (1994) 136–141. [DOI] [PubMed] [Google Scholar]
- [13].Naas TP, DeBerardinis RJ, Moran JV, Ostertag EM, Kingsmore SF, Seldin MF, Hayashizaki Y, Martin SL, Kazazian HH, An actively retrotransposing, novel subfamily of mouse L1 elements, EMBO J. 17 (2) (1998) 590–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Skowronski J, Fanning TG, Singer MF, Unit-length line-1 transcripts in human teratocarcinoma cells, Mol. Cell. Biol 8 (4) (1988) 1385–1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Boissinot S, Chevret P, Furano AV, L1 (LINE-1) retrotransposon evolution and amplification in recent human history, Mol. Biol. Evol 17 (6) (2000) 915–928. [DOI] [PubMed] [Google Scholar]
- [16].Boissinot S, Entezam A, Young L, Munson PJ, Furano AV, The insertional history of an active family of L1 retrotransposons in humans, Genome Res. 14 (7) (2004) 1221–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Chen JM, Stenson PD, Cooper DN, Ferec C, A systematic analysis of LINE-1 endonuclease-dependent retrotranspositional events causing human genetic disease, Hum. Genet 117 (5) (2005) 411–427. [DOI] [PubMed] [Google Scholar]
- [18].Beck CR, Garcia-Perez JL, Badge RM, Moran JV, LINE-1 elements in structural variation and disease, Annu. Rev. Genom. Hum. Genet 12 (2011) 187–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Huang CR, Schneider AM, Lu Y, Niranjan T, Shen P, et al. , Mobile interspersed repeats are major structural variants in the human genome, Cell 141 (7) (2010) 1171–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, Van Meir EG, Vertino PM, Devine SE, Natural mutagenesis of human genomes by endogenous retrotransposons, Cell 141 (7) (2010) 1253–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Boissinot S, Furano AV, Adaptive evolution in LINE-1 retrotransposons, Mol. Biol. Evol 18 (12) (2001) 2186–2194. [DOI] [PubMed] [Google Scholar]
- [22].Boissinot S, Davis J, Entezam A, Petrov D, Furano AV, Fitness cost of LINE-1 (L1) activity in humans, Proc. Natl. Acad. Sci. U. S. A 103 (25) (2006) 9590–9594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Myers S, Bottolo L, Freeman C, McVean G, Donnelly P, A fine-scale map of recombination rates and hotspots across the human genome, Science 310 (5746) (2005) 321–324. [DOI] [PubMed] [Google Scholar]
- [24].Bourc’his D, Bestor TH, Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L, Nature 431 (7004) (2004) 96–99. [DOI] [PubMed] [Google Scholar]
- [25].Soper SFC, van der Heijden GW, Hardiman TC, Goodheart M, Martin SL, de Boer P, Bortvin A, Mouse maelstrom, a component of Nuage, is essential for Spermatogenesis and transposon repression in meiosis, Dev. Cell 15 (2) (2008) 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH Jr., High frequency retrotransposition in cultured mammalian cells, Cell 87 (5) (1996) 917–927. [DOI] [PubMed] [Google Scholar]
- [27].Doucet AJ, Hulme AE, Sahinovic E, Kulpa DA, Moldovan JB, et al. , Characterization of LINE-1 ribonucleoprotein particles, PLoS Genet. 6 (10) (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kulpa DA, Moran JV, Cis-preferential LINE-1 reverse transcriptase activity in ribonucleoprotein particles, Nat. Struct. Mol. Biol 13 (7) (2006) 655–660. [DOI] [PubMed] [Google Scholar]
- [29].Kulpa DA, Moran JV, Ribonucleoprotein particle formation is necessary but not sufficient for LINE-1 retrotransposition, Hum. Mol. Genet 14 (21) (2005) 3237–3248. [DOI] [PubMed] [Google Scholar]
- [30].Martin SL, Ribonucleoprotein particles with LINE-1 RNA in mouse embryonal carcinoma cells, Mol. Cell. Biol 11 (9) (1991) 4804–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Luan DD, Korman MH, Jakubczak JL, Eickbush TH, Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition, Cell 72 (4) (1993) 595–605. [DOI] [PubMed] [Google Scholar]
- [32].Martin SL, Bushman FD, Nucleic acid chaperone activity of the ORF1 protein from the mouse LINE-1 retrotransposon, Mol. Cell. Biol 21 (2) (2001) 467–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Eickbush TH, Malik HS, Origins and evolution of retrotransposons, in: Craig NL, Craigie R, Gellert M, Lambowitz AM (Eds.), Mobile DNA II, American Society of Microbiology Press, Washington, DC, 2002, pp. 1111–1144. [Google Scholar]
- [34].Feng Q, Moran JV, Kazazian HH Jr., Boeke JD, Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition, Cell 87 (5) (1996) 905–916. [DOI] [PubMed] [Google Scholar]
- [35].Mathias SL, Scott AF, Kazazian HHJ, Boeke JD, Gabriel A, Reverse transcriptase encoded by a human transposable element, Science 254 (5039) (1991) 1808–1810. [DOI] [PubMed] [Google Scholar]
- [36].Khazina E, Weichenrieder O, Non-LTR retrotransposons encode noncanonical RRM domains in their first open reading frame, Proc. Natl. Acad. Sci. U. S. A 106 (3) (2009) 731–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Cook PR, Jones CE, Furano AV, Phosphorylation of ORF1p is required for L1 retrotransposition, Proc. Natl. Acad. Sci. U. S. A 112 (14) (2015) 4298–4303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Hohjoh H, Singer MF, Cytoplasmic ribonucleoprotein complexes containing human LINE-1 protein and RNA, EMBO J 15 (3) (1996) 630–639. [PMC free article] [PubMed] [Google Scholar]
- [39].Martin SL, The ORF1 protein encoded by LINE-1: structure and function during L1 retrotransposition, J. Biomed. Biotechnol 2006 (1) (2006) 45621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Martin SL, Nucleic acid chaperone properties of ORF1p from the non-LTR retrotransposon, LINE-1, RNA Biol. 7 (6) (2010) 706–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Martin SL, Bushman D, Wang F, Li PW, Walker A, Cummiskey J, Branciforte D, Williams MC, A single amino acid substitution in ORF1 dramatically decreases L1 retrotransposition and provides insight into nucleic acid chaperone activity, Nucleic Acids Res. 36 (18) (2008) 5845–5854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Martin SL, Cruceanu M, Branciforte D, Wai-Lun Li P, Kwok SC, Hodges RS, Williams MC, LINE-1 retrotransposition requires the nucleic acid chaperone activity of the ORF1 protein, J. Mol. Biol 348 (3) (2005) 549–561. [DOI] [PubMed] [Google Scholar]
- [43].Naufer MN, Callahan KE, Cook PR, Perez-Gonzalez CE, Williams MC, Furano AV, L1 retrotransposition requires rapid ORF1p oligomerization, a novel coiled coil-dependent property conserved despite extensive remodeling, Nucleic Acids Res. 44 (1) (2016) 281–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Callahan KE, Hickman AB, Jones CE, Ghirlando R, Furano AV, Polymerization and nucleic acid-binding properties of human L1 ORF1 protein, Nucleic Acids Res. 40 (2) (2012) 813–827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Basame S, Wai-lun Li P, Howard G, Branciforte D, Keller D, Martin SL, Spatial assembly and RNA binding stoichiometry of a LINE-1 protein essential for retrotransposition, J. Mol. Biol 357 (2) (2006) 351–357. [DOI] [PubMed] [Google Scholar]
- [46].Khazina E, Truffault V, Buttner R, Schmidt S, Coles M, Weichenrieder O, Trimeric structure and flexibility of the L1ORF1 protein in human L1 retrotransposition, Nat. Struct. Mol. Biol 18 (9) (2011) 1006–1014. [DOI] [PubMed] [Google Scholar]
- [47].Januszyk K, Li P.W.-l., Villareal V, Branciforte D, Wu H, et al. , Identification and solution structure of a highly conserved C-terminal domain within ORF1p required for retrotransposition of long interspersed nuclear element-1, J. Biol. Chem 282 (34) (2007) 24893–24904. [DOI] [PubMed] [Google Scholar]
- [48].Muthukumaran R, Sangeetha B, Amutha R, Conformational analysis on the wild type and mutated forms of human ORF1p: a molecular dynamics study, Mol. Biosyst 11 (7) (2015) 1987–1999. [DOI] [PubMed] [Google Scholar]
- [49].Rajagopalan M, Balasubramanian S, Ramaswamy A, Insights into the RNA binding mechanism of human L1-ORF1p: a molecular dynamics study, Mol. Biosyst 13 (9) (2017) 1728–1743. [DOI] [PubMed] [Google Scholar]
- [50].Taylor MS, Altukhov I, Molloy KR, Mita P, Jiang H, et al. , Dissection of affinity captured LINE-1 macromolecular complexes, Elife 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Mita P, Wudzinska A, Sun X, Andrade J, Nayak S, et al. , LINE-1 protein localization and functional dynamics during the cell cycle, Elife 7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kolosha VO, Martin SL, High-affinity, non-sequence-specific RNA binding by the open reading frame 1 (ORF1) protein from long interspersed nuclear element 1 (LINE-1), J. Biol. Chem 278 (10) (2003) 8112–8117. [DOI] [PubMed] [Google Scholar]
- [53].Sookdeo A, Hepp CM, McClure MA, Boissinot S, Revisiting the evolution of mouse LINE-1 in the genomic era, Mob. DNA 4 (1) (2013) 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Boissinot S, Sookdeo A, The evolution of LINE-1 in vertebrates, Genome Biol. Evol 8 (12) (2016) 3485–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Martin SL, Branciforte D, Keller D, Bain DL, Trimeric structure for an essential protein in L1 retrotransposition, Proc. Natl. Acad. Sci. U. S. A 100 (24) (2003) 13815–13820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Furano AV, Cook PR, The challenge of ORF1p phosphorylation: Effects on L1 activity and its host, Mob. Genet. Elem 6 (1) (2016) e1119927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Saxton JA, Martin SL, Recombination between subtypes creates a mosaic lineage of LINE-1 that is expressed and actively retrotransposing in the mouse genome, J. Mol. Biol 280 (4) (1998) 611–622. [DOI] [PubMed] [Google Scholar]
- [58].Hohjoh H, Singer MF, Ribonuclease and high salt sensitivity of the ribonucleoprotein complex formed by the human LINE-1 retrotransposon, J. Mol. Biol 271 (1) (1997) 7–12. [DOI] [PubMed] [Google Scholar]
- [59].Evans JD, Peddigari S, Chaurasiya KR, Williams MC, Martin SL, Paired mutations abolish and restore the balanced annealing and melting activities of ORF1p that are required for LINE-1 retrotransposition, Nucleic Acids Res. 39 (13) (2011) 5611–5621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Martin SL, Li J, Weisz JA, Deletion analysis defines distinct functional domains for protein-protein and nucleic acid interactions in the ORF1 protein of mouse LINE-1, J. Mol. Biol 304 (1) (2000) 11–20. [DOI] [PubMed] [Google Scholar]
- [61].Kolosha VO, Martin SL, In vitro properties of the first ORF protein from mouse LINE-1 support its role in ribonucleoprotein particle formation during retrotransposition, Proc. Natl. Acad. Sci. U. S. A 94 (19) (1997) 10155–10160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Levin JG, Guo J, Rouzina I, Musier-Forsyth K, Nucleic acid chaperone activity of HIV-1 nucleocapsid protein: critical role in reverse transcription and molecular mechanism, Prog. Nucleic Acid Res. Mol. Biol 80 (2005) 217–286. [DOI] [PubMed] [Google Scholar]
- [63].Post K, Olson ED, Naufer MN, Gorelick RJ, Rouzina I, Williams MC, Musier-Forsyth K, Levin JG, Mechanistic differences between HIV-1 and SIV nucleocapsid proteins and cross-species HIV-1 genomic RNA recognition, Retrovirology 13 (1) (2016) 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Qualley DF, Stewart-Maynard KM, Wang F, Mitra M, Gorelick RJ, Rouzina I, Williams MC, Musier-Forsyth K, C-terminal domain modulates the nucleic acid chaperone activity of human T-cell leukemia virus type 1 nucleocapsid protein via an electrostatic mechanism, J. Biol. Chem 285 (1) (2010) 295–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Wu H, Wang W, Naiyer N, Fichtenbaum E, Qualley DF, et al. , Single aromatic residue location alters nucleic acid binding and chaperone function of FIV nucleocapsid protein, Virus Res. 193 (2014) 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Chaurasiya KR, Geertsema H, Cristofari G, Darlix JL, Williams MC, A single zinc finger optimizes the DNA interactions of the nucleocapsid protein of the yeast retrotransposon Ty3, Nucleic Acids Res. 40 (2) (2012) 751–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Williams MC, Rouzina I, Wenner JR, Gorelick RJ, Musier-Forsyth K, Bloomfield VA, Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching, Proc. Natl. Acad. Sci. U. S. A 98 (11) (2001) 6121–6126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Williams MC, Gorelick RJ, Musier-Forsyth K, Specific zinc-finger architecture required for HIV-1 nucleocapsid protein’s nucleic acid chaperone function, Proc. Natl. Acad. Sci. U. S. A 99 (13) (2002) 8614–8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Wu H, Mitra M, McCauley MJ, Thomas JA, Rouzina I, Musier-Forsyth K, Williams MC, Gorelick RJ, Aromatic residue mutations reveal direct correlation between HIV-1 nucleocapsid protein’s nucleic acid chaperone activity and retroviral replication, Virus Res. 171 (2) (2013) 263–277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Wu H, Mitra M, Naufer MN, McCauley MJ, Gorelick RJ, Rouzina I, Musier-Forsyth K, Williams MC, Differential contribution of basic residues to HIV-1 nucleocapsid protein’s nucleic acid chaperone function and retroviral replication, Nucleic Acids Res. 42 (4) (2014) 2525–2537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Bucheton A, I transposable elements and I-R hybrid dysgenesis in Drosophila, Trends Genet. 6 (1) (1990) 16–21. [DOI] [PubMed] [Google Scholar]
- [72].Dawson A, Hartswood E, Paterson T, Finnegan DJ, A LINE-like transposable element in Drosophila, the I factor, encodes a protein with properties similar to those of retroviral nucleocapsids, EMBO J. 16 (14) (1997) 4448–4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Rouzina I, Bloomfield VA, Force-induced melting of the DNA double helix 1, Thermodyn. Anal. Biophys. J 80 (2) (2001) 882–893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Williams MC, Rouzina I, Bloomfield VA, Thermodynamics of DNA interactions from single molecule stretching experiments, Acc. Chem. Res 35 (3) (2002) 159–166. [DOI] [PubMed] [Google Scholar]
- [75].Wu H, Rouzina I, Williams MC, Single-molecule stretching studies of RNA chaperones, RNA Biol. 7 (6) (2010) 712–723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Vladescu ID, McCauley MJ, Rouzina I, Williams MC, Mapping the phase diagram of single DNA molecule force-induced melting in the presence of ethidium, Phys. Rev. Lett 95 (15) (2005), 158102. [DOI] [PubMed] [Google Scholar]
- [77].Vladescu ID, McCauley MJ, Nunez ME, Rouzina I, Williams MC, Quantifying force-dependent and zero-force DNA intercalation by single-molecule stretching, Nat. Methods 4 (6) (2007) 517–522. [DOI] [PubMed] [Google Scholar]
- [78].Almaqwashi AA, Paramanathan T, Rouzina I, Williams MC, Mechanisms of small molecule-DNA interactions probed by single-molecule force spectroscopy, Nucleic Acids Res. 44 (9) (2016) 3971–3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Cruceanu M, Gorelick RJ, Musier-Forsyth K, Rouzina I, Williams MC, Rapid kinetics of protein-nucleic acid interaction is a major component of HIV-1 nucleocapsid protein’s nucleic acid chaperone function, J. Mol. Biol 363 (5) (2006) 867–877. [DOI] [PubMed] [Google Scholar]
- [80].King GA, Gross P, Bockelmann U, Modesti M, Wuite GJ, Peterman EJ, Revealing the competition between peeled ssDNA, melting bubbles, and S-DNA during DNA overstretching using fluorescence microscopy, Proc. Natl. Acad. Sci. U. S. A 110 (10) (2013) 3859–3864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Zhang X, Chen H, Le S, Rouzina I, Doyle PS, Yan J, Revealing the competition between peeled ssDNA, melting bubbles, and S-DNA during DNA overstretching by single-molecule calorimetry, Proc. Natl. Acad. Sci. U. S. A 110 (10) (2013) 3865–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Chaurasiya KR, Paramanathan T, McCauley MJ, Williams MC, Biophysical characterization of DNA binding from single molecule force measurements, Phys. Life Rev 7 (3) (2010) 299–341. [DOI] [PMC free article] [PubMed] [Google Scholar]