Abstract
Highly social insects are characterized by caste dimorphism, with distinct size differences of reproductive organs between fertile queens and the more or less sterile workers. An abundance of nutrition or instruction via diet-specific compounds has been proposed as explanations for the nutrition-driven queen and worker polyphenism. Here, we further explored these models in the honeybee (Apis mellifera) using worker nutrition rearing and a novel mutational screening approach using the clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9) method. The worker nutrition-driven size reduction of reproductive organs was restricted to the female sex, suggesting input from the sex determination pathway. Genetic screens on the sex determination genes in genetic females for size polyphenism revealed that doublesex (dsx) mutants display size-reduced reproductive organs irrespective of the sexual morphology of the organ tissue. In contrast, feminizer (fem) mutants lost the response to worker nutrition-driven size control. The first morphological worker mutants in honeybees demonstrate that the response to nutrition relies on a genetic program that is switched “ON” by the fem gene. Thus, the genetic instruction provided by the fem gene provides an entry point to genetically dissect the underlying processes that implement the size polyphenism.
In honeybees, nutrition drives dimorphic size development of reproductive organs in fertile queens and sterile workers. A study using the first induced morphological mutants in honeybees demonstrates that this developmental plasticity requires a genetic program that is switched on by the “feminizer” gene.
Author summary
In honeybees, nutrition drives dimorphic size development of reproductive organs in fertile queens and sterile workers. The first induced morphological mutants in honeybees demonstrate that this developmental plasticity requires a genetic program that is switched “ON” by the feminizer (fem) gene.
Introduction
Highly social insects are characterized by caste dimorphism, with morphologically and physiologically distinct reproductive queens and more or less sterile workers [1–3]. In honeybees, the development of two distinct phenotypes is controlled by different nutrition, and it is a prominent example of developmental plasticity and polyphenism [4, 5].
One major concern for the study of caste development involves explaining how a usually sterile worker and a queen that lays up to 2,000 eggs per day develop from different diet and feeding regimens [4, 6, 7]. Worker-destined larvae receive restricted amounts of a reduced sugar content diet (worker jelly [WJ]), while queen-destined larvae receive large quantities of a sugar-rich diet (royal jelly [RJ]) [8–11]. WJ and RJ drive the development of female larvae in two distinct morphs. Workers have a five-day longer developmental time, lower body mass, two small ovaries containing few ovarioles, and mid- and hind-leg structures adapted for pollen collection and transport. Queens have a five-day shorter developmental time, larger body mass, and two large ovaries that contain many more ovarioles, and they lack the pollen collection structures on the legs.
Two types of models have been proposed to explain how diets and feeding regimens mediate worker/queen development. The Nutrition/Growth model suggests that queen/worker development is driven by the amount of food and balance of nutrition [7, 11, 12], which modulate a developmental program. Queen-destined larvae have abundant nutrition, and organ growth is only limited by the intrinsic program. Worker-destined larvae have a shortage of nutrition that restricts growth and influences metabolic parameters accordingly. In contrast, the Instruction model proposes that the RJ has a compound (or compounds) that instruct the development of queens [13–15]. In support of the Instruction model, research over the past decades has attempted to identify a single compound from RJ [12, 14] that can determine queen development.
A recent study provided evidence that the protein royalactin has queen-determining activity [15]. However, follow-up experiments in another laboratory were unable to repeat these results [7], questioning the existence of a single determinant for queen development [4]. Gradually increasing the sugar levels of WJ and altering the composition of RJ-containing diets produced workers, intercastes, and eventually queens [9–11, 16], but it failed to rear only queens. The more continuous caste characteristics resulting from different feeding regimes [17] have been proposed in support of the Nutrition/Growth model. The RJ and the WJ produce different reaction norms of the general developmental program that determines the caste polyphenism. An alternative explanation is that the essential higher sugar levels for queen-destined larvae are a secondary effect and reflect the higher energy requirements for the faster and larger-growing queen organs of an otherwise instructed queen program. The rearing of larvae at day 5 in queenless colonies yielded bees with ovariole numbers that were discontinuous (either more worker or queen-like distributed), while other queen and worker traits were either absent or present in a noncorrelated fashion [18], suggesting two distinct states of the developmental program and the possible existence of regulatory switches [19].
One possible mechanism by which nutrients are sensed by bee larvae is the insulin/IGF signaling (IIS) and target of rapamycin (TOR) pathways, which link the abundance of nutrition with worker and queen differential gene expression [20–23]. Indeed, nutritional input can also influence growth and metabolic programs via the IIS and TOR pathways in mammals and other insects [24–26]. However, whether regulation of the IIS and TOR pathways drives caste differentiation or whether the regulation is a response to the activation of a queen developmental program is currently unknown. Consistent with the faster and larger growth of queens, gene expression studies have revealed the up-regulation of physiometabolic genes in queens, reflecting their higher metabolic rate [27, 28]. Chromatin modifications and DNA methylation analyses have indicated distinct epigenetic states in worker- and queen-destined larvae, suggesting another level of regulatory control associated with caste-specific gene expression [29–31].
Here, we explored whether nutrition is the only factor directing size polyphenism and whether further genetic instruction from the sex determination pathway is required. To do so, we introduced a method to screen mutations directly in worker bees using the clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9 (CRISPR/Cas9) technique.
Results
Worker nutrition is not a general driver for the reduced size of reproductive organs
According to the Nutrition/Growth model, nutrition is the only driver of reduced reproductive organ size, the most prominent trait in caste development. Males, like queens, receive high amounts of sugar during larval development [32] and develop large reproductive organs unlike sterile worker bees. Gradually increasing the sugar levels of WJ produces intercaste development [9, 10, 16]. Hence, if a shortage of nutrition in the worker diet (and reduced sugar levels) is the only driving component, we would expect that this diet would also mediate the size reduction of reproductive organs in males.
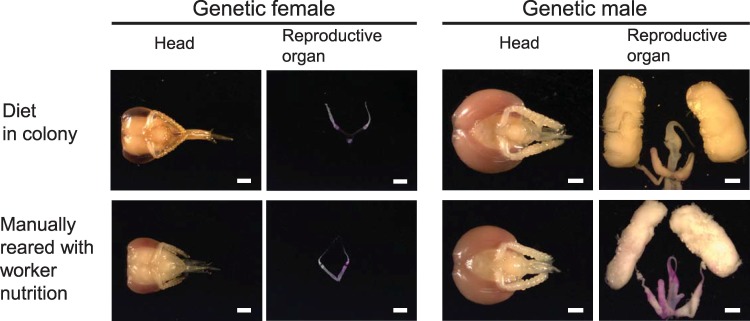
We manually reared genetic females and males on worker nutrition [16, 33] and compared their phenotypes with those of workers and genetic males reared in the colony (Fig 1 and S1 and S2 Tables). The reproductive organs of genetic female bees raised on worker nutrition either inside the colony (n = 14) or manually outside (n = 15) were equivalent in size (Fisher’s exact test, df = 1, P = 1). In both laboratory- and colony-reared genetic females, there were few ovarioles, and the size of each ovary was small compared with the size of the heads (Fig 1 and S1 Table). This contrasts with the large ovaries of the female larvae fed a queen diet in the hive (queens alone cannot be consistently reared under laboratory conditions [7]; see Fig 4A and 4B as an example of a queen phenotype). This result indicates that our manual feeding regime mirrors the effect of a worker diet in the hive [16, 33]. To examine whether only the balance and amount of nutrition (low amount of sugar) determine small reproductive organs, we reared genetic male larvae on worker nutrition in the laboratory and compared these with males that received high amounts of sugar in the colony [32]. Genetic males that were reared on the worker nutrition diet had large male reproductive organs (Fig 1 and S2 Table). They were equivalent in size (n = 20) to the males obtained from the colony (n = 8) that were reared on drone nutrition (Fisher’s exact test, df = 1, P = 1). These results indicate that worker nutrition (and a shortage of sugar) is not the only requirement for the size polyphenism, suggesting input from the sex determination pathway.
Fig 1. Reproductive organ and head phenotypes of females and males reared on worker nutrition in the laboratory and in the colony.
Scale bar = 1 mm.
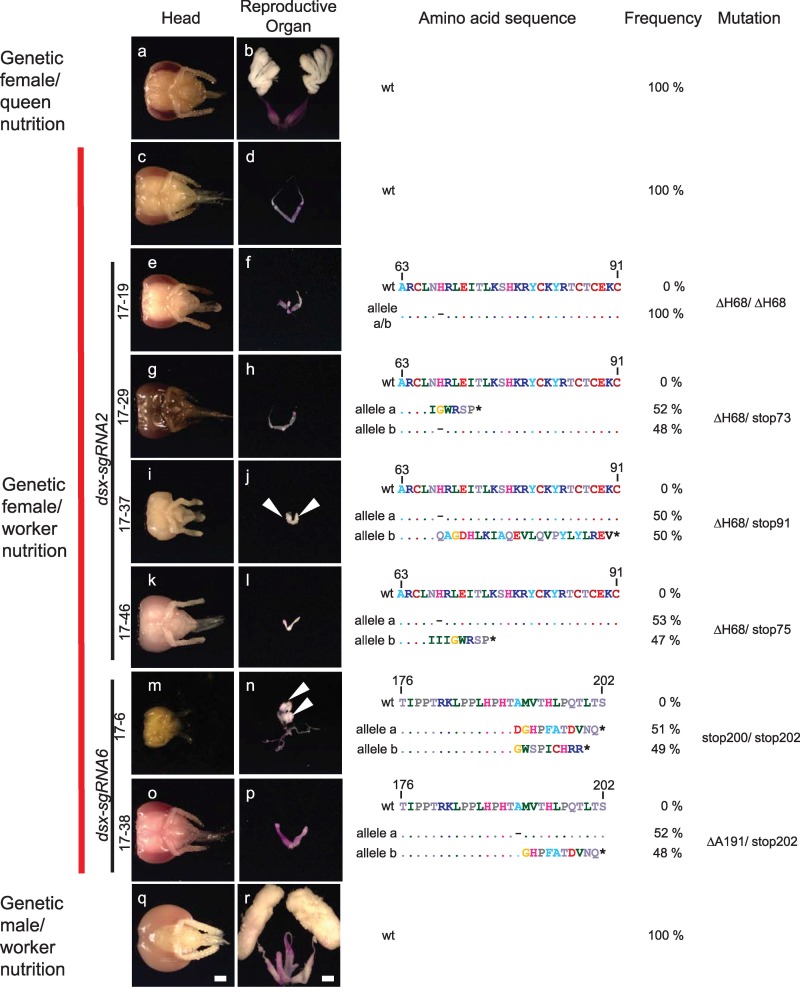
Fig 4. Size polyphenism of the reproductive organs in genetic female double mutants for the dsx gene.
Pictures of the head and internal reproductive organs of mutated and WT control bees are shown on the left, while the genotypes at the dsx locus with the deduced amino acid sequences are displayed on the right. Mutated and control genetic females and males were reared on worker nutrition. Queens were reared on the queen diet in a colony (we cannot mimic queen rearing in the laboratory). The WT amino acid sequence is shown above the detected alleles for comparison. (a, b) WT genetic female reared on queen nutrition (RJ) in the colony. (c, d) WT genetic females manually reared on worker nutrition. (e–l) Genetic females reared on worker nutrition that were double mutants for dsx via the dsx-sgRNA6 (note that a small part of the worker bee head 17–39 [picture i] is missing due to the dissection process). (m–p) Genetic females reared on worker nutrition that were double mutants for dsx via the dsx-sgRNA2. (q, r) Genetic males manually reared on worker nutrition. Organs were stained with aceto-orcein (reddish coloring) to facilitate the dissection process. Testis tissues are marked with arrows. Scale bar, 1 mm. Dashes in the sequence indicate deletions, and stars illustrate early translational stop codons. RJ, royal jelly; WT, wild type.
Somatic mutational screening in reared bees
We next established a method that enables the mutational screening of sex-determining genes directly in worker bees using the CRISPR/Cas9 method [34–36]. Following traditional mutant approaches, we would need to produce mutant queens and drones that need to be crossed to generate double-mutant worker bees. If we could mutate all nuclei in the embryo, we would be able to directly rear mutated worker bees without maintaining colonies and performing crossings. To examine whether we could mutate worker bees entirely using the CRISPR/Cas9 method, we tested different embryonic injection conditions. To determine the robustness of this approach, we studied at least two sites for three genes, the doublesex (dsx), fruitless (fru), and loc552773 genes (S1 Fig). Only the dsx gene was used later on for phenotyping. We injected into the anterior embryos of very young female embryos (0 to 1.5 hours after egg deposition) [37]. We tested a set of single guide RNAs (sgRNAs; S3 Table) at different concentrations and observed that we repeatedly mutated each injected embryo.
The fragment length (FL) and sequence analyses of the amplicons in larval stage 1 larvae revealed that up to 100% of the fru and dsx and 60% of the loc552773 target embryos were mutated (Tables 1 and S4 and S5 and Fig 2). The wild-type (WT) allele was consistently not detected in 30 of the 39 mutated larvae (77%), suggesting that all nuclei (to the level of detection) and both alleles in the larvae were mutated (generating double mutants). More than two mutated sequence variants were detected in a single larva (3%), while singly mutated sequences together with the WT allele were detected in 8 larvae (20%) (S4 and S5 Tables). Indels occurred most frequently between the 5 bp to 1 bp range, with 44% of mutations being deletions and 20% resulting in insertions (S5 and S6 Tables). All mutations occurred at the designated target site. Therefore, our results on the adjustments demonstrate that nearly 80% of the injected embryos had mutations on both alleles (double mutants) affecting the bee entirely (absence of mosaicism). This high proportion enabled us to screen for mutant effects of the sex-determining genes directly in the injected bees.
Table 1. Frequency of the mutated honeybee larvae based on FL analyses at single base-pair resolution of the amplicons.
| Treatment | pg of Cas9 mRNA per embryo |
pg of sgRNA per embryo |
No. of surviving embryos 24 h after injection | No. (%) of hatched L1 larvae | No. of genotyped larvae | No. of larvae with length variant1 | Efficiency of mutagenesis2 |
|---|---|---|---|---|---|---|---|
| fru-sgRNA1 | 800 | 29.2 | 105 | 10 (10%) | 8 | 2 | 20% |
| fru-sgRNA2 | 400 | 14.6 | 467 | 72 (15%) | 7 | 6 | 86% |
| fru-sgRNA1 | 240 | 8.8 | 78 | 2 (3%) | 2 | 2 | 100% |
| fru-sgRNA4 | 400 | 14.6 | 125 | 3 (2%) | 3 | 3 | 100% |
| fru-sgRNA5 | 400 | 14.6 | 98 | 10 (10%) | 10 | 10 | 100% |
| loc-sgRNA13 | 400 | 14.6 | 93 | 7 (8%) | 5 | 3 | 60% |
| loc-sgRNA2 | 400 | 14.6 | 102 | 32 (31%) | 28 | 1 | 4% |
| dsx-sgRNA1 | 400 | 5.5 | 52 | 1 (2%) | 1 | 1 | 100% |
| dsx-sgRNA1 | 400 | 3.7 | 93 | 5 (5%) | 4 | 1 | 25% |
| dsx-sgRNA2 | 400 | 5.5 | 178 | 2 (1%) | 2 | 2 | 100% |
| dsx-sgRNA2 | 400 | 3.7 | 89 | 5 (6%) | 5 | 5 | 100% |
| dsx-sgRNA2 | 400 | 0.7 | 82 | 21 (26%) | 19 | 3 | 16% |
| H2O | - | - | 48 | 27 (56%) | 11 | 0 | 0% |
| Uninjected | - | - | 65 | 55 (85%) | 19 | 0 | 0% |
1Fragments differed in length compared with fragments isolated from 7 nontreated (WT) larvae.
2Relative ratio of the number of mutant larvae to the number of all larvae.
3Targeted the gene loc552773.
Abbreviations: Cas9, CRISPR-associated protein 9; FL, fragment length; pg, picogram; sgRNA, single guide RNA; WT, wild type.
Fig 2. Examples of FL and nucleotide sequence analyses of the targeted genomic sites of single bees using the efficient CRISPR/Cas9 method.
FL analysis is presented on the left, and the nucleotide sequences are presented on the right for single bees. Examples of WT alleles and mutated sequences are shown. The cleavage site of the Cas9 protein is indicated with arrows. The PAM site (the essential targeting component for CRISPR/Cas9) is underlined in the nucleotide sequence. Dashes indicate deletions. CRISPR/Cas9, clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9; FL, fragment length; mut, mutated sequences; PAM, Protospacer adjacent motif; WT, wild type.
The feminizer gene is required for small size polyphenism
To examine whether the feminizer (fem) gene is required for small size polyphenism, we mutated the gene in genetic females and reared them with worker nutrition. The fem gene instructs female development and maintains the female signal during development, as revealed from fem interference RNA (RNAi) knockdown and mosaic studies using a non–worker-specific diet for bee rearing [19, 38]. The Fem protein is encoded by female-specific spliced fem transcripts but not the male spliced variant, which harbors an early stop codon [19] (Fig 3A). The female splicing of fem is directed by the complementary sex determiner (csd) gene when the genotype is heterozygous (Fig 3A) [39]. If the fem gene is required for small size polyphenism, we would expect that worker nutrition cannot drive size reduction when fem is inactive. If the fem gene is dispensable, worker nutrition would drive size reduction even when the fem gene is inactive. We induced mutations at two target sites in the first half of the female open reading frame (ORF) of the fem gene with fem-sgRNA1 and fem-sgRNA2 (S1 and S2 Figs) and reared genetic females with worker nutrition to larval stage 5. Fifteen percent of the reared and injected genetic females (heterozygous for the csd gene; S7 Table) were double mutants for nonsense mutations as revealed from the sequenced amplicons (S8 Table and S2 Fig). These double mutants (n = 4) had large gonads (Fig 3B and 3D) compared with the small gonads of WT genetic females reared on worker nutrition (n = 38, Fisher’s exact test, df = 1, P < 0.001, S9 Table). The large gonads in the mutants were of the male type. They consisted of packed layers of multiple testioles of the same size as those of the males reared on worker nutrition (Fig 3B) and those of the males in the colony (Fig 1). The female fem mutants lost the female dsx transcript and only displayed the male dsx transcript (Fig 3C), demonstrating that the mutant bees entirely switched in their development from female to male identity. These results indicate that fem is required for size polyphenism or that size polyphenism relies on the intrinsic program of the female differentiating tissue induced by fem.
Fig 3. Size polyphenism of gonads in genetic females at larval stage 5 that were double mutants for the fem gene.
(a) Model of the known components of the sex-determining pathway in honeybees with nutritional differences in females. (b) Gonad development at larval stage 5. (Right) A pair of large gonads (male type) from fem sgRNA2-treated genetic females reared on worker nutrition. The gonads display densely packed layers of folded testioles, similar to those observed in haploid males (WT males). (Left) Pairs of small gonads (female type) from WT workers and genetic female bees reared on worker nutrition. A WT large queen ovary from a queen reared in a colony on queen nutrition. A large WT testis of a haploid male manually reared on worker nutrition. (c) Male dsx (dsxM) and female dsx (dsxF) transcripts in mutated genetic females with male phenotypes (fem-sgRNA1 or fem-sgRNA2). Male and female transcripts were separately amplified by RT-PCR [64], and the male and female fragments of each single bee were resolved via agarose gel electrophoresis. Numbers indicate different control and mutated bees. (d) Deduced amino acid sequences from sequenced amplicons of the fem gene at the designated CRISPR/Cas9 cleavage sites for the four worker nutrition-reared genetic female larvae with large gonads of the male type. Stars indicate premature translation stop codons. Numbers indicate different mutated bees. Scale bars, 1 mm. CRISPR/Cas9, clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9; dsxF, female dsx; dsxM, male dsx; RT-PCR, reverse transcription PCR; sgRNA, single guide RNA; WT, wild type.
dsx is dispensable for small size polyphenism
To examine the role of female dsx on size polyphenism of the reproductive organ, we mutated the dsx gene in genetic females and reared them on worker nutrition. If dsx is dispensable, we would expect small size polyphenism even when dsx activity is compromised. In Drosophila melanogaster, the dsx gene essentially controls, beside the reproductive organs, all aspects of somatic sexual differentiation [40, 41], and it controls at least reproductive organ development in other insects that belong to different insect orders, including hymenopteran insects [42–45]. The dsx transcripts in honeybees are sex-specifically spliced by the presence of the Fem protein in females and the absence of the Fem protein in males [19] (Fig 3A). The sexual splice variants encode a transcription factor with an intertwined zinc-containing DNA binding (DM) domain and male- and female-specific termini at the carboxyl end [46–50]. We mutated the dsx gene at two target sites in the non–sex-specific expressed N-terminal portion. dsx-sgRNA2 targeted the DM domain, whereas dsx-sgRNA6 targeted a downstream region in exon 3 (S1 Fig). The treated genetic females were reared on worker nutrition and were examined for morphological changes of the reproductive organ and head. Genotyping of the mutated bees with morphological changes via next-generation sequencing (NGS) of the amplicons revealed that they were regularly double mutants with an approximate ratio of 1:1, suggesting that the mutations belong to the two chromosomes of the diploid set. If we detected more than two sequence variants per bee, we excluded these bees from further phenotype analysis as they were genetic mosaics (e.g., a mosaic of differently mutated cells). Eleven (17%) of the adult or pupal bees had intersex morphology in the reproductive organs compared with the WT genetic females (S10 Table). No effect was observed for the heads. The following mutations were the most common ones in the genetic females: (i) different nonsense mutations that introduced new stop codons at various positions in exons 2 and 3, (ii) deletions of amino acids in the DM domain mainly the histidine codon at amino acid position 68 (ΔH68), and (iii) deletion of the alanine codon (ΔA191) at amino acid position 191 (Fig 4 with the deduced amino acid sequences and S3 Fig with the detected nucleotide sequences). The ΔH68 mutation removes a histidine of the DM domain that is essential for the zinc binding and DM domain functions [47, 51] and that is conserved between vertebrates and invertebrates (S4 Fig). The intersex reproductive organs were all of the same small size (n = 11) as the worker reproductive organs in WT genetic females that were manually reared on worker nutrition (n = 17, Table 2, Fisher’s exact test, df = 1, P = 1). The small intersex reproductive organs displayed either male gonads with poorly or non–sex-specifically differentiated duct systems (n = 4), as observed in stop200/stop202 and ΔH68/stop91 genetic females (arrows in Figs 4 and S5). The potentially earlier developmental stage of some of these mutant bees cannot explain why these male-like gonads are so small because the distinct size differences of male and worker gonads are also present at earlier pupal stages (S6 Fig). In other cases, the reproductive organs were underdeveloped (n = 7), and the oviducts were consistently misshaped while the ovarioles were repeatedly missing, as identified in ΔH68/ΔH68, ΔH68/stop73, ΔH68/stop75, and ΔA191/stop202 genetic females (Figs 4 and S5). The heads of the mutant genetic females with intersex reproductive organs were all of worker type (n = 11, Fig 4 and S10 Table), suggesting that dsx is not required for sexual development of the head. The results of the consistently small, intersex reproductive organs with varying degrees of masculinization suggest that dsx is not required for size polyphenism.
Table 2. The size of the intersex reproductive organs in genetic females double mutant for dsx and reared on worker nutrition.
|
Sex |
Nutrition |
Genotype |
Reproductive organ |
Numbers |
Size of reproductive organa | |
|---|---|---|---|---|---|---|
| <2.5 mm; <0.7 times the size of the head width |
>6 mm; >1.2 times the size of the head width |
|||||
|
Genetic female |
Manually reared on worker nutrition |
dsx double mutants | Intersex | 11 | 11 (100%) | 0 (0%) |
| WT | Worker | 17 | 17 (100%) | 0 (0%) | ||
| Queen diet in colony | WT | Queen | 3 | 0 (0%) | 3 (100%) | |
| Genetic male | Manually reared on worker nutrition | WT | Male | 16 | 0 (0%) | 16 (100%) |
aLength between the fused left and right part of the reproductive organ to its end in the sagittal plane.
Abbreviation: WT, wild type.
Discussion
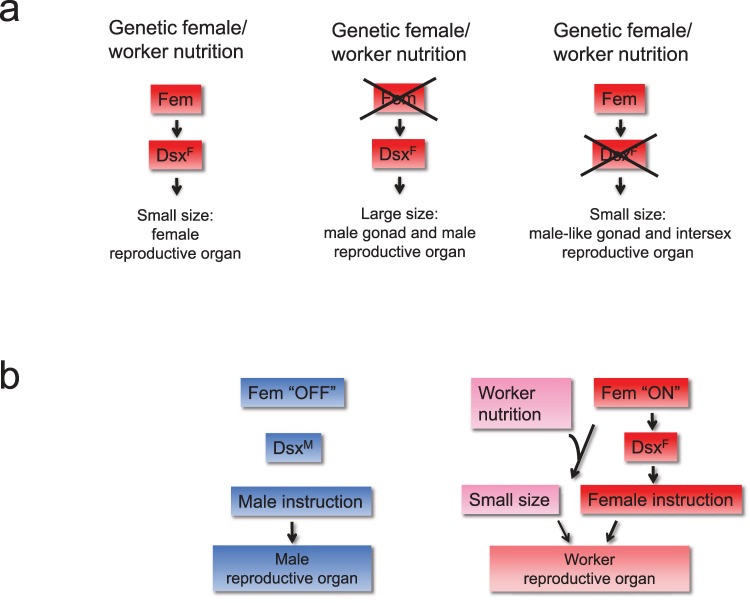
Caste polyphenism in honeybees is determined by different nutrition with the size of the reproductive organ as an important trait. Most studies suggest that the balance and amount of nutrition (Nutrition/Growth model) drive the size polyphenism between queens and workers. Our genetic and rearing results now suggest that the response to nutrition relies on a genetic program that is switched on by the fem gene. The genetic females with a mutant fem gene show large size reproductive organ (large polyphenism), while WT genetic females (Fig 5A) reared on the same worker nutrition have only small reproductive organs (small polyphenism). Genetic females that have a mutated dsx gene (operating downstream of fem) do show small reproductive organs (small size polyphenism; Fig 5A). dsx mutants produce intersex reproductive organs and male-like gonads that are all of small size, demonstrating that small size does not rely on female development of the tissue. The small size polyphenism also did not result from dsx malfunction because (i) small phenotypes were consistently observed irrespective of the different degrees of dsx malfunctions we introduced by missense and nonsense mutations (Fig 4) and (ii) dsx mutations in other insects did not influence the size of the reproductive organs [42, 52, 53]. Thus, the results together suggest that the fem gene is required for the small size polyphenism. We conclude that the fem gene must be switched “ON” so that size polyphenism can be executed (Fig 5B). The essential role of the fem gene in small size polyphenism assigns a further key function to the fem gene. Previous studies demonstrated that the fem gene is also required to (i) induce entire female development in response to the primary signal csd [19, 38] and to (ii) maintain the female signal during development via a positive regulatory feedback loop [19]. Whether fem also instructs the large size polyphenism of queens needs further functional testing once a queen-only rearing protocol has been developed for the laboratory [7].
Fig 5. The role of the sex-determining genes fem and dsx in size polyphenism.
(a) Schematic presentation of the mutant effects of fem and dsx gene on size polyphenism. Genetic female bees reared on worker nutrition produce only small reproductive organs. Genetic females with a mutant fem gene show no small size polyphenism of reproductive organs. Genetic females that have a mutated dsx (operating downstream of fem) do show size polyphenism of the intersex reproductive organ and male-like gonads. Thus, we conclude that the fem gene is required for the small size polyphenism. Crosses mark the genes that we compromised using CRISPR/Cas9-induced mutations. (b) The role of the fem gene for caste development. The gene products of the sex determination pathway (Fem, DsxF, DsxM) are shown in red (female) and blue (male) boxes. The nutrition-mediated process is shown in pink. Arrows indicate regulatory relationships. CRISPR/Cas9, clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9.
The genetic instruction via the fem gene provides an entry point to dissect nutrition-mediated control. Our results suggest that the fem gene switches “ON” the machinery that is required for sensing the worker nutrition and for implementing the size polyphenism. Because the fem gene encodes a serine arginine rich (SR)-type protein, the direct targets of the fem gene involved in size polyphenism may also be activated by sexual splicing. The fem-controlled candidate genes can be functionally tested by determining whether they affect the size polyphenism. The function will be directly tested in mutated genetic females as demonstrated in this study.
Our mutant analysis further demonstrate that dsx controls female differentiation of the reproductive organs. The mutant honeybee phenotypes of the reproductive organs in honeybees yielded similar phenotypes as in female D. melanogaster. Female dsx-mutant fruit flies have reproductive organs of varying intersex phenotypes. The organs are often underdeveloped with occasionally developed ovaries, but are frequently of the “male type” [52, 54, 55]. The internal duct system can develop into a mixture of female/male or single poorly differentiated ducts [52]. RNAi-mediated knockdown studies on the beetle Tribolium molitor, housefly Musca domestica, and sawfly Athalia rosae, as well as conditional expression and CRISPR/Cas9 experiments on the silkworm Bombyx mori, have revealed sex-related effects on internal reproductive organ development [42–46, 53]. Our results support a conserved role for dsx in the sexual development of the reproductive organ. However, in honeybees there is a nutrition-driven size control of reproductive organ development that operates upstream of or in parallel with dsx-regulated sexual development.
The first CRISPR/Cas9-induced morphological mutants in honeybees introduced a new genetic screening method for worker bees. We efficiently induced mutations in injected embryos using the CRISPR/Cas9 method [34, 35] and directly screened for somatic mutations in the reared honeybees (somatic mutation approach). Up to 100% of the embryos were mutated, and mosaicism among the mutated embryos was rare (<10%). The previous studies in honeybees using CRISPR/Cas9-induced mutations report on 1 out of 2 queens with only 12% and 2 out of 4 queens with only 5% and 10% mutant drone offspring, suggesting that the previously published method has a substantial lower rate and produced strong mosaicism in the queens [36, 56]. These previous studies generated no worker bees that would require further crossing experiments. With very early embryonic injections [37] and a selection step to identify the most efficient sgRNAs and Cas9 concentrations, we generated mutation rates of up to 100% and no mosaicism in worker bees directly. The rearing of the mutated embryos to worker bees was performed under controlled conditions in the laboratory [16, 33]. This required no rearing of queens and drones and crossing experiments. The procedure was demonstrated for mutations at two target sites for two genes and their morphological changes (Figs 3 and 4). The absence of mosaicism and completeness of mutagenesis of this procedure were shown by the results that most mutated bees lost the WT allele (they were double mutants; Figs 2, 3D and 4) and that double fem nonsense mutations produced an entire female to male switch, including dsx splice products (Fig 3C). This somatic mutation approach does not require further crossing experiments and laborious maintenance of hundreds of colonies and therefore offers the prospect of larger genetic screens in honeybees. In other insects in which somatic mutation approaches have been applied [57, 58], the adults were genetic mosaics in which parts of the butterfly wing were WT while other parts were mutated. Enhancing the efficiency of mutagenesis can thus provide an opportunity for somatically testing gene functions in insects that are not yet genetically trackable.
Methods
sgRNA and mRNA syntheses
Cas9 mRNA was synthesized from the Cas9 gene [59] (Vector MLM3613, ID #42251, Addgene, Cambridge, MA) using a linearized plasmid via the T7 promoter and the mMESSAGE mMACHINE Kit (Ambion, Darmstadt, Germany). mRNAs were polyadenylated using the Poly(A) Tailing Kit (Ambion). Target sites for the sgRNAs were identified via Optimal Target Finder software (http://tools.flycrispr.molbio.wisc.edu/targetFinder/). sgRNAs were 20 nt long with a G nucleotide at the 5´ end. sgRNAs with no off-target effects or with at least three nucleotide mismatches to alternative target sites were selected. sgRNAs were generated via PCR without a template using two overlapping oligonucleotide sequences containing the sequence of the T7 RNA polymerase transcription start site, the gene-specific target site and the Cas9 protein-binding site. sgRNAs were synthesized using a RiboMax Kit (Promega, Madison, WI) according to the manufacturer’s instructions. RNAs were purified using the MEGAclear Kit (Ambion).
Microinjection and rearing
Embryos were microinjected 0 to 1.5 hours after egg deposition [19, 37, 60] using 53-mm injection pipettes (Hilgenberg, Malsfeld, Germany). Cas9 mRNA or protein (New England Biolabs, Ipswich, MA) was applied at 400 to 2,000 ng/μl and mixed with sgRNAs using a molar ratio of 1:2 to 1:0.75. The number of injected embryos that hatch can vary greatly between experiments and sgRNAs (5% to 40%). Rearing was performed using a mass rearing technique for the worker bees [16, 33]. Freshly hatched larvae were provisioned only once with the worker larval diet (50%–53% RJ, 4% glucose, 8% fructose, 1% yeast extract, and 30%–34% water), approximately 120 to 170 mg of which was consumed [16, 33]. The larvae were incubated at 34°C and 90% humidity until the larval stage 5 or to adults. For pupal rearing we also used a slightly different diet for larvae at stage 5 (50 mg diet 2 [50% RJ, 12% fructose, 6% glucose, 2% yeast extract, and 30% water]).
DNA preparation, RNA isolation, and cDNA synthesis
For genotyping, genomic DNA was isolated from freshly hatched L1 or L5 larvae [61] using the peqGOLD Tissue DNA Mini Kit (VWR, Darmstadt, Germany). RNA was isolated using the TRIZOL method (Thermo Scientific, Braunschweig, Germany), and cDNA was synthesized using the RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific). Second-strand cDNA synthesis was performed by adding 10 μl of 10× DNA Polymerase Buffer, 40 U DNA Polymerase I, 0.8 U Ribonuclease H, and 65.68 μl of dH2O to 20 μl of the cDNA first-strand synthesis product. Double-stranded cDNA was purified using the EZNA Cycle Pure kit (Omega Bio-Tek Inc., Norcross, GA).
PCR, sequencing, and FL analysis
All mutant bees were genotyped by sequencing the amplicons of the targeted site. PCR amplifications were performed using standard conditions [62] and GoTaq polymerase (Promega). Oligonucleotide sequences were synthesized at Eurofins (Ebersberg, Germany). Amplicons were either cloned and sequenced (Sanger sequencing [Eurofins]) or sequenced via NGS. NGS index PCR was performed using the Nextera XT Index Kit (Illumina, San Diego, CA), and purification of the Index PCR products was performed using Agencourt AMPure XP beads (Beckman Coulter, Brea, CA). NGS was performed on an Illumina MiSeq system using the MiSeq Reagent Kit version 2 (500 cycles; Illumina), generating 800,000 paired-end reads with a read length of 2 × 250 bp, resulting in approximately 15,000 paired-end reads per sample. We removed contamination by removing sequences that were less frequent than 5%. The FLs of hexachlorofluorescein (HEX)-labeled amplicons were determined using an ABI 3130XL Genetic Analyzer (Applied Biosystems, Darmstadt, Germany) and Peak Scanner software (Thermo Scientific). For the fem mutants, we conducted fragment and sequence analysis on the amplicons of the cDNAs to ensure that the many fem-related sequences observed at the genomic fem locus (derived from duplication events) [63] were not amplified.
Supporting information
Genomic organization of the genes fru (a), loc552773 (b), dsx (c), and fem (d) with the designated sgRNA target sites (black arrows). Boxes indicate exons. If genes transcribe sexual splice variants, they are presented. Green boxes indicate common, red the female-specific, and blue the male specific ORF of the sexual transcripts. dsx, doublesex; fem, feminizer; fru, fruitless; ORF, open reading frame; sgRNA, single guide RNA.
(PDF)
(a) Diagrams of the FL analysis for each of the 4 individuals and WT worker bee examples. (b) The nucleotide sequences. We conducted fragment and sequence analysis on amplicons of cDNA to ensure that the many fem-related sequences observed at the fem locus (derived from duplication events) [63] were not amplified. The designated binding sites of the sgRNAs are underlined. Sequence b in larvae #4 resulted from fusion of exon 3 with exon 5. The sequences in larvae #4 resulted from fusion between exon 3 and other fem-related sequences [63]. The WT sequences were obtained from a sample of 5 WT worker larvae (5 clones each). cDNA, complementary DNA; FL, fragment length; WT, wild type.
(PDF)
The dsx WT nucleotide sequences are represented as a reference sequence. NGS, next-generation sequencing; WT, wild type.
(PDF)
The deleted conserved histidine at position 68 of the honeybee sequence (Am) is highlighted with an arrow.
(PDF)
Scale bar, 1 mm. The genetic females were double mutant for dsx and reared on worker nutrition. For further details, see legend of Fig 4 in the main text.
(PDF)
These females have the typical reduced reproductive organ of workers and the fully developed reproductive organs of males. Head and (a) and (c) and reproductive organ (b) and (d). Gonads were stained with aceto-orcein (reddish coloring) to facilitate the dissection process. Scale bar = 1 mm.
(PDF)
(PDF)
(PDF)
Sequences complementary to the designated genomic target site are shown in bold letters. sgRNA, single guide RNA.
(PDF)
WT, wild type.
(PDF)
At least 10 clones for each larvae were sequenced. These nucleotide changes were consistently not observed in 7 nontreated (WT) larvae. The sequence complementary to the sgRNAs are underlined. sgRNA, single guide RNA; WT, wild type.
(PDF)
CRISPR/Cas9, clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Acknowledgments
We thank Eva Theilenberg, Marion Müller-Borg, and Cahit Ozturk for their assistance with bee handling and Max Ewinger for the support provided in the sequence and fragment length analyses.
Abbreviations
- CRISPR/Cas9
clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9
- csd
complementary sex determiner
- dsx
doublesex
- fem
feminizer
- FL
fragment length
- fru
fruitless
- HEX
hexachlorofluorescein
- IIS
insulin/IGF signaling
- NGS
next-generation sequencing
- ORF
open reading frame
- PAM
Protospacer adjacent motif
- RJ
royal jelly
- RNAi
interference RNA
- RT-PCR
reverse transcription PCR
- sgRNA
single guide RNA
- SR
serine arginine rich
- TOR
target of rapamycin
- WT
wild type
- WJ
worker jelly
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by grants from the Deutsche Forschungsgemeinschaft (http://www.dfg.de/) to MB (BE 2194). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Evans JD, Wheeler DE. Gene expression and the evolution of insect polyphenisms. Bioessays. 2001;23(1):62–8. . [DOI] [PubMed] [Google Scholar]
- 2.Simpson SJ, Sword GA, Lo N. Polyphenism in insects. Curr Biol. 2011;21(18):R738–49. 10.1016/j.cub.2011.06.006 . [DOI] [PubMed] [Google Scholar]
- 3.Trible W, Kronauer DJ. Caste development and evolution in ants: it's all about size. The Journal of experimental biology. 2017;220(Pt 1):53–62. 10.1242/jeb.145292 . [DOI] [PubMed] [Google Scholar]
- 4.Maleszka R. Beyond Royalactin and a master inducer explanation of phenotypic plasticity in honey bees. Commun Biol. 2018;1(1):8 10.1038/s42003-017-0004-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.West-Eberhard MJ. Developmental plasticity and evolution. New York: Oxford University Press; 2003. [Google Scholar]
- 6.Corona M, Libbrecht R, Wheeler DE. Molecular mechanisms of phenotypic plasticity in social insects. Curr Opin Insect Sci. 2016;13:55–60. 10.1016/j.cois.2015.12.003 . [DOI] [PubMed] [Google Scholar]
- 7.Buttstedt A, Ihling CH, Pietzsch M, Moritz RF. Royalactin is not a royal making of a queen. Nature. 2016;537(7621):E10–2. 10.1038/nature19349 . [DOI] [PubMed] [Google Scholar]
- 8.Haydak MH. Honey Bee Nutrition. Annual Review of Entomology. 1970;15:143-. 10.1146/annurev.en.15.010170.001043 PubMed PMID: WOS:A1970F046400007. [DOI] [Google Scholar]
- 9.Asencot M, Lensky Y. The effect of soluble sugars in stored royal jelly on the differentiation of female honeybee (Apis mellifera L.) larvae to queens. Insect Biochem. 1988;18(2):127-. 10.1016/0020-1790(88)90016-9 PubMed PMID: WOS:A1988N288000001. [DOI] [PubMed] [Google Scholar]
- 10.Asencot M, Lensky Y. The effect of sugars and juvenile hormone on the differentiation of the female honeybee larvae (Apis mellifera L.) to queens. Life Sci. 1976;18(7):693–9. . [DOI] [PubMed] [Google Scholar]
- 11.Leimar O, Hartfelder K, Laubichler MD, Page RE Jr. Development and evolution of caste dimorphism in honeybees—a modeling approach. Ecol Evol. 2012;2(12):3098–109. 10.1002/ece3.414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rembold H, Lackner B. Rearing of honeybee larvae in vitro: Effect of yeast extract on queen differentiation. Journal of Apicultural Research. 2015;20(3):165–71. 10.1080/00218839.1981.11100492 [DOI] [Google Scholar]
- 13.Von Rhein W. Über die Entstehung des weiblichen Dimorphismus im Bienenstaate. Wilhelm Roux' Archiv fur Entwicklungsmechanik der Organismen. 1933;129(4):601–65. Epub 1933/12/01. 10.1007/BF00656581 . [DOI] [PubMed] [Google Scholar]
- 14.Rembold H, Lackner B, Geistbeck I. The chemical basis of honeybee, Apis mellifera, caste formation. Partial purification of queen bee determinator from royal jelly. Journal of Insect Physiology. 1974;20(2):307–14. 10.1016/0022-1910(74)90063-8 [DOI] [Google Scholar]
- 15.Kamakura M. Royalactin induces queen differentiation in honeybees. Nature. 2011;473(7348):478–83. 10.1038/nature10093 . [DOI] [PubMed] [Google Scholar]
- 16.Kaftanoglu O, Linksvayer TA, Page RE. Rearing honey bees, Apis mellifera, in vitro 1: effects of sugar concentrations on survival and development. J Insect Sci. 2011;11:96 10.1673/031.011.9601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nijhout HF. Development and evolution of adaptive polyphenisms. Evol Dev. 2003;5(1):9–18. . [DOI] [PubMed] [Google Scholar]
- 18.Dedej S, Hartfelder K, Aumeier P, Rosenkranz P, Engels W. Caste determination is a sequential process: effect of larval age at grafting on ovariole number, hind leg size and cephalic volatiles in the honey bee (Apis mellifera carnica). Journal of Apicultural Research. 1998;37(3):183–90. 10.1080/00218839.1998.11100970 PubMed PMID: WOS:000078695300006. [DOI] [Google Scholar]
- 19.Gempe T, Hasselmann M, Schiott M, Hause G, Otte M, Beye M. Sex determination in honeybees: two separate mechanisms induce and maintain the female pathway. PLoS Biol. 2009;7(10):e1000222 10.1371/journal.pbio.1000222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Patel A, Fondrk MK, Kaftanoglu O, Emore C, Hunt G, Frederick K, et al. The making of a queen: TOR pathway is a key player in diphenic caste development. PLoS ONE. 2007;2(6):e509 10.1371/journal.pone.0000509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolschin F, Mutti NS, Amdam GV. Insulin receptor substrate influences female caste development in honeybees. Biol Lett. 2011;7(1):112–5. 10.1098/rsbl.2010.0463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wheeler DE, Buck N, Evans JD. Expression of insulin pathway genes during the period of caste determination in the honey bee, Apis mellifera. Insect Mol Biol. 2006;15(5):597–602. 10.1111/j.1365-2583.2006.00681.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Azevedo SV, Hartfelder K, Amdam GV. Insulin-like peptides (AmILP1 and AmILP2) differentially affect female caste development in the honey bee (Apis mellifera L.). The Journal of Experimental Biology. 2013;216(Pt 23):4347–57. 10.1242/jeb.085779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ikeya T, Galic M, Belawat P, Nairz K, Hafen E. Nutrient-dependent expression of insulin-like peptides from neuroendocrine cells in the CNS contributes to growth regulation in Drosophila. Curr Biol. 2002;12(15):1293–300. . [DOI] [PubMed] [Google Scholar]
- 25.Colombani J, Raisin S, Pantalacci S, Radimerski T, Montagne J, Leopold P. A nutrient sensor mechanism controls Drosophila growth. Cell. 2003;114(6):739–49. . [DOI] [PubMed] [Google Scholar]
- 26.Slaidina M, Delanoue R, Gronke S, Partridge L, Leopold P. A Drosophila insulin-like peptide promotes growth during nonfeeding states. Dev Cell. 2009;17(6):874–84. 10.1016/j.devcel.2009.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Barchuk AR, Cristino AS, Kucharski R, Costa LF, Simoes ZL, Maleszka R. Molecular determinants of caste differentiation in the highly eusocial honeybee Apis mellifera. BMC Developmental Biology. 2007;7(1):70 10.1186/1471-213X-7-70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cameron RC, Duncan EJ, Dearden PK. Biased gene expression in early honeybee larval development. BMC Genomics. 2013;14:903 10.1186/1471-2164-14-903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wojciechowski M, Lowe R, Maleszka J, Conn D, Maleszka R, Hurd PJ. Phenotypically distinct female castes in honey bees are defined by alternative chromatin states during larval development. Genome Research. 2018;28(10):1532–42. 10.1101/gr.236497.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Foret S, Kucharski R, Pellegrini M, Feng S, Jacobsen SE, Robinson GE, et al. DNA methylation dynamics, metabolic fluxes, gene splicing, and alternative phenotypes in honey bees. Proc Natl Acad Sci U S A. 2012;109(13):4968–73. 10.1073/pnas.1202392109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319(5871):1827–30. 10.1126/science.1153069 . [DOI] [PubMed] [Google Scholar]
- 32.Mandla R, Kumar NR. Comparison of carbohydrates in the worker, drone and queen brood food of Apis mellifera during spring. Journal of Global Biosciences. 2016;5(3):3. [Google Scholar]
- 33.Kaftanoglu O, Linksvayer TA, Page RE. Rearing honey bees (Apis mellifera L.) in vitro: effects of feeding intervals on survival and development. Journal of Apicultural Research. 2010;49(4):311–7. 10.3896/Ibra.1.49.4.03 PubMed PMID: WOS:000283013700003. [DOI] [Google Scholar]
- 34.Wiedenheft B, Sternberg SH, Doudna JA. RNA-guided genetic silencing systems in bacteria and archaea. Nature. 2012;482(7385):331–8. 10.1038/nature10886 . [DOI] [PubMed] [Google Scholar]
- 35.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337(6096):816–21. 10.1126/science.1225829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kohno H, Suenami S, Takeuchi H, Sasaki T, Kubo T. Production of knockout mutants by CRISPR/Cas9 in the european honeybee, Apis mellifera L. Zoolog Sci. 2016;33(5):505–12. 10.2108/zs160043 . [DOI] [PubMed] [Google Scholar]
- 37.Schulte C, Theilenberg E, Muller-Borg M, Gempe T, Beye M. Highly efficient integration and expression of piggyBac-derived cassettes in the honeybee (Apis mellifera). Proc Natl Acad Sci U S A. 2014;111(24):9003–8. Epub 2014/05/14. 10.1073/pnas.1402341111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hasselmann M, Gempe T, Schiott M, Nunes-Silva CG, Otte M, Beye M. Evidence for the evolutionary nascence of a novel sex determination pathway in honeybees. Nature. 2008;454(7203):519–22. 10.1038/nature07052 . [DOI] [PubMed] [Google Scholar]
- 39.Beye M, Hasselmann M, Fondrk MK, Page RE, Omholt SW. The gene csd is the primary signal for sexual development in the honeybee and encodes an SR-type protein. Cell. 2003;114(4):419–29. 10.1016/S0092-8674(03)00606-8 PubMed PMID: WOS:000184928800006. [DOI] [PubMed] [Google Scholar]
- 40.Cline TW, Meyer BJ. Vive la difference: males vs females in flies vs worms. Annu Rev Genet. 1996;30:637–702. 10.1146/annurev.genet.30.1.637 . [DOI] [PubMed] [Google Scholar]
- 41.Williams TM, Carroll SB. Genetic and molecular insights into the development and evolution of sexual dimorphism. Nature Reviews Genetics. 2009;10(11):797–804. 10.1038/nrg2687 . [DOI] [PubMed] [Google Scholar]
- 42.Hediger M, Burghardt G, Siegenthaler C, Buser N, Hilfiker-Kleiner D, Dubendorfer A, et al. Sex determination in Drosophila melanogaster and Musca domestica converges at the level of the terminal regulator doublesex. Dev Genes Evol. 2004;214(1):29–42. 10.1007/s00427-003-0372-2 . [DOI] [PubMed] [Google Scholar]
- 43.Suzuki MG, Funaguma S, Kanda T, Tamura T, Shimada T. Role of the male BmDSX protein in the sexual differentiation of Bombyx mori. Evol Dev. 2005;7(1):58–68. 10.1111/j.1525-142X.2005.05007.x . [DOI] [PubMed] [Google Scholar]
- 44.Shukla JN, Palli SR. Doublesex target genes in the red flour beetle, Tribolium castaneum. Scientific reports. 2012;2:948 Epub 2012/12/12. 10.1038/srep00948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mine S, Sumitani M, Aoki F, Hatakeyama M, Suzuki MG. Identification and functional characterization of the sex-determining gene doublesex in the sawfly, Athalia rosae (Hymenoptera: Tenthredinidae). Appl Entomol Zool. 2017;52(3):497–509. 10.1007/s13355-017-0502-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matson CK, Zarkower D. Sex and the singular DM domain: insights into sexual regulation, evolution and plasticity. Nature Reviews Genetics. 2012;13(3):163–74. Epub 2012/02/09. 10.1038/nrg3161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhu L, Wilken J, Phillips NB, Narendra U, Chan G, Stratton SM, et al. Sexual dimorphism in diverse metazoans is regulated by a novel class of intertwined zinc fingers. Gene Dev. 2000;14(14):1750–64. [PMC free article] [PubMed] [Google Scholar]
- 48.Cristino AS, do Nascimento AM, Costa LD, Simoes ZLP. A comparative analysis of highly conserved sex-determining genes between Apis mellifera and Drosophila melanogaster. Genet Mol Res. 2006;5(1):154–68. PubMed PMID: WOS:000203011700021. [PubMed] [Google Scholar]
- 49.Dearden PK, Wilson MJ, Sablan L, Osborne PW, Havler M, McNaughton E, et al. Patterns of conservation and change in honey bee developmental genes. Genome Research. 2006;16(11):1376–84. 10.1101/gr.5108606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cho S, Huang ZY, Zhang JZ. Sex-specific splicing of the honeybee doublesex gene reveals 300 million years of evolution at the bottom of the insect sex-determination pathway. Genetics. 2007;177(3):1733–41. 10.1534/genetics.107.078980 PubMed PMID: WOS:000251368800037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Murphy MW, Lee JK, Rojo S, Gearhart MD, Kurahashi K, Banerjee S, et al. An ancient protein-DNA interaction underlying metazoan sex determination. Nature Structural & Molecular Biology. 2015;22(6):442–51. Epub 2015/05/26. 10.1038/nsmb.3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hildreth PE. Doublesex, recessive gene that transforms both males and females of Drosophila into intersexes. Genetics. 1965;51:659–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Xu J, Zhan S, Chen S, Zeng B, Li Z, James AA, et al. Sexually dimorphic traits in the silkworm, Bombyx mori, are regulated by doublesex. Insect Biochem Mol Biol. 2017;80:42–51. 10.1016/j.ibmb.2016.11.005 . [DOI] [PubMed] [Google Scholar]
- 54.Schupbach T. Autosomal mutations that interfere with sex determination in somatic cells of Drosophila have no direct effect on the germline. Dev Biol. 1982;89(1):117–27. . [DOI] [PubMed] [Google Scholar]
- 55.Bownes M, Dempster M, Blair M. Expression of the yolk-protein genes in the mutant doublesex dominant (dsxD) of Drosophila melanogaster. J Embryol Exp Morphol. 1983;75:241–57. . [PubMed] [Google Scholar]
- 56.Kohno H, Kubo T. mKast is dispensable for normal development and sexual maturation of the male European honeybee. Scientific Reports. 2018;8(1):11877 10.1038/s41598-018-30380-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang L, Reed RD. Genome editing in butterflies reveals that spalt promotes and Distal-less represses eyespot colour patterns. Nature Communications. 2016;7:11769 10.1038/ncomms11769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mazo-Vargas A, Concha C, Livraghi L, Massardo D, Wallbank RWR, Zhang L, et al. Macroevolutionary shifts of WntA function potentiate butterfly wing-pattern diversity. Proc Natl Acad Sci U S A. 2017;114(40):10701–6. 10.1073/pnas.1708149114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hwang WY, Fu Y, Reyon D, Maeder ML, Tsai SQ, Sander JD, et al. Efficient genome editing in zebrafish using a CRISPR-Cas system. Nat Biotechnol. 2013;31(3):227–9. 10.1038/nbt.2501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Beye M, Hartel S, Hagen A, Hasselmann M, Omholt SW. Specific developmental gene silencing in the honey bee using a homeobox motif. Insect Molecular Biology. 2002;11(6):527–32. 10.1046/j.1365-2583.2002.00361.x PubMed PMID: WOS:000179068300002. [DOI] [PubMed] [Google Scholar]
- 61.Hunt GJ, Page RE Jr. Linkage map of the honey bee, Apis mellifera, based on RAPD markers. Genetics. 1995;139(3):1371–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hasselmann M, Beye M. Signatures of selection among sex-determining alleles of the honey bee. Proc Natl Acad Sci USA. 2004;101(14):4888–93. 10.1073/pnas.0307147101 PubMed PMID: WOS:000220761200032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koch V, Nissen I, Schmitt BD, Beye M. Independent evolutionary origin of fem paralogous genes and complementary sex determination in hymenopteran insects. PLoS ONE. 2014;9(4):e91883 Epub 2014/04/20. 10.1371/journal.pone.0091883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nissen I, Muller M, Beye M. The Am-tra2 gene is an essential regulator of female splice regulation at two levels of the sex determination hierarchy of the honeybee. Genetics. 2012;192(3):1015–26. Epub 2012/09/04. 10.1534/genetics.112.143925 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Genomic organization of the genes fru (a), loc552773 (b), dsx (c), and fem (d) with the designated sgRNA target sites (black arrows). Boxes indicate exons. If genes transcribe sexual splice variants, they are presented. Green boxes indicate common, red the female-specific, and blue the male specific ORF of the sexual transcripts. dsx, doublesex; fem, feminizer; fru, fruitless; ORF, open reading frame; sgRNA, single guide RNA.
(PDF)
(a) Diagrams of the FL analysis for each of the 4 individuals and WT worker bee examples. (b) The nucleotide sequences. We conducted fragment and sequence analysis on amplicons of cDNA to ensure that the many fem-related sequences observed at the fem locus (derived from duplication events) [63] were not amplified. The designated binding sites of the sgRNAs are underlined. Sequence b in larvae #4 resulted from fusion of exon 3 with exon 5. The sequences in larvae #4 resulted from fusion between exon 3 and other fem-related sequences [63]. The WT sequences were obtained from a sample of 5 WT worker larvae (5 clones each). cDNA, complementary DNA; FL, fragment length; WT, wild type.
(PDF)
The dsx WT nucleotide sequences are represented as a reference sequence. NGS, next-generation sequencing; WT, wild type.
(PDF)
The deleted conserved histidine at position 68 of the honeybee sequence (Am) is highlighted with an arrow.
(PDF)
Scale bar, 1 mm. The genetic females were double mutant for dsx and reared on worker nutrition. For further details, see legend of Fig 4 in the main text.
(PDF)
These females have the typical reduced reproductive organ of workers and the fully developed reproductive organs of males. Head and (a) and (c) and reproductive organ (b) and (d). Gonads were stained with aceto-orcein (reddish coloring) to facilitate the dissection process. Scale bar = 1 mm.
(PDF)
(PDF)
(PDF)
Sequences complementary to the designated genomic target site are shown in bold letters. sgRNA, single guide RNA.
(PDF)
WT, wild type.
(PDF)
At least 10 clones for each larvae were sequenced. These nucleotide changes were consistently not observed in 7 nontreated (WT) larvae. The sequence complementary to the sgRNAs are underlined. sgRNA, single guide RNA; WT, wild type.
(PDF)
CRISPR/Cas9, clustered regularly interspaced short palindromic repeats/CRISPR-associated protein 9.
(PDF)
(PDF)
(PDF)
(PDF)
(PDF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.