Abstract
Regulatory factor X 7 (Rfx7) is an uncharacterized transcription factor belonging to a family involved in ciliogenesis and immunity. Here we found that deletion of Rfx7 leads to a decrease in natural killer (NK) cell maintenance and immunity in vivo. Genomic approaches showed that Rfx7 coordinated a transcriptional network controlling cell metabolism. Rfx7−/− NK lymphocytes presented increased size, granularity, proliferation and energetic state, whereas genetic reduction of mTOR activity mitigated those defects. Notably, Rfx7-deficient NK lymphocytes were rescued by interleukin 15 through engagement of the Janus kinase (Jak) pathway, revealing the importance of this signaling for maintenance of such spontaneously activated NK cells. Rfx7 emerges therefore as a novel transcriptional regulator of NK cell homeostasis and metabolic quiescence.
Regulatory factor X 7 (RFX7, also known as RFX domain containing 2, RFXDC2) belongs to the RFX family, which comprises seven transcription factors1,2. These share a highly conserved winged helix DNA-binding domain (DBD) recognizing X-box motifs3,4. RFX1-4 and RFX6 bear additional domains involved in transactivation or homo/hetero-dimer formation, whereas RFX5 and RFX7 exhibit no identifiable domains besides the DBD2. Based on phylogenetic analyses, RFX family members are subdivided into three clades: RFX1, 2, and 3; RFX4 and 6; and RFX5 and 7.
RFX family members play important roles in several biological processes. For instance, RFX2, 3, and 4 regulate genes essential for the formation of cilia, which are required for normal development of several organs and cell types. RFX2 is necessary for spermatogenesis, as shown by the sterility of knockout mice5,6. Rfx4 deletion leads to incomplete neural tube closure7,8, while Rfx3−/− animals show defects in left-right asymmetry and brain development9,10. Moreover, Rfx3−/− mice also exhibit abnormalities of the pancreatic endocrine compartments11. Along this line, Rfx6 deficiency leads to severe defects in pancreatic islet development and inactivating mutations cause a monogenic form of diabetes in human neonates12. Finally, Rfx5 regulates transcription of major histocompatibility complex (MHC) genes, which are fundamental for proper T cell homeostasis and function. Transcription of MHC genes is coordinated by a multiprotein complex including RFX513,14 and inactivating mutations of RFX5 cause a severe immunodeficiency condition known as bare lymphocyte syndrome.
To date, the only study on the function of Rfx7 reported a role in neural tube closure during Xenopus laevis development15. Rfx7 regulated ciliogenesis through controlling Rfx4 expression. Genome-wide association studies in humans have linked RFX7 to body fat distribution and tumor development16–21. In particular, alterations of RFX7 have been associated to human lymphoid cancers16–19,21 and to the development of lymphoma in murine models22,23. Those findings suggest RFX7 as a potentially interesting gene in the context of metabolic disorders and hematologic malignancies. Yet, its function remains completely uncharacterized in mammalian cells.
Here, we investigated the role of Rfx7. We observed prominent Rfx7 expression in immune system organs and cells. We therefore generated mice with specific Rfx7 deletion in the hematopoietic compartment. These mice exhibited a decrease in the number of NK cells along with defective NK cell-mediated immunity. Through genomic and molecular analyses, we defined Rfx7 target genes, which are largely linked to cellular metabolism. In agreement, Rfx7-deficient NK cells presented features of spontaneous activation. Heterozygosity for Rptor, encoding a cofactor of the metabolic checkpoint kinase mTORC1 complex, partially rescued those defects. Further, interleukin 15 (IL-15) rescued Rfx7-deficient NK lymphocytes by engaging the Jak pathway, which was thus essential for survival of these pseudo-activated NK cells. Rfx7 emerges therefore as an important gatekeeper of NK cell resting state and homeostasis, a prerequisite to warrant optimal responses.
Results
Rfx7 is prominently expressed in lymphoid organs
Rfx7 is conserved among higher vertebrates, exhibiting 93% amino acid identity and an indistinguishable DBD between human and mouse (Supplementary Fig. 1a-c). Thus, analysis of murine Rfx7 is highly relevant to the understanding of its human counterpart. We generated Rfx7 conditional-deficient mice, enabling deletion of the DBD and generating a frame shift (Supplementary Fig. 1d). The deleted exons are common to all annotated isoforms24. To study the role of Rfx7 in the immune system, these mice were crossed to the Vav1-iCre deleter strain, (expressing an improved variant of Cre recombinase (iCre) under the control of the Vav1 promoter; hereafter called Vav Rfx7fl/fl), leading to deletion in hematopoietic cells and their progenitors. In addition, germline deletion of Rfx7 was obtained. Rfx7−/−mice were born from heterozygous parents at a frequency of 7% (n=161), instead of the expected 25%, revealing a central developmental role for Rfx7.
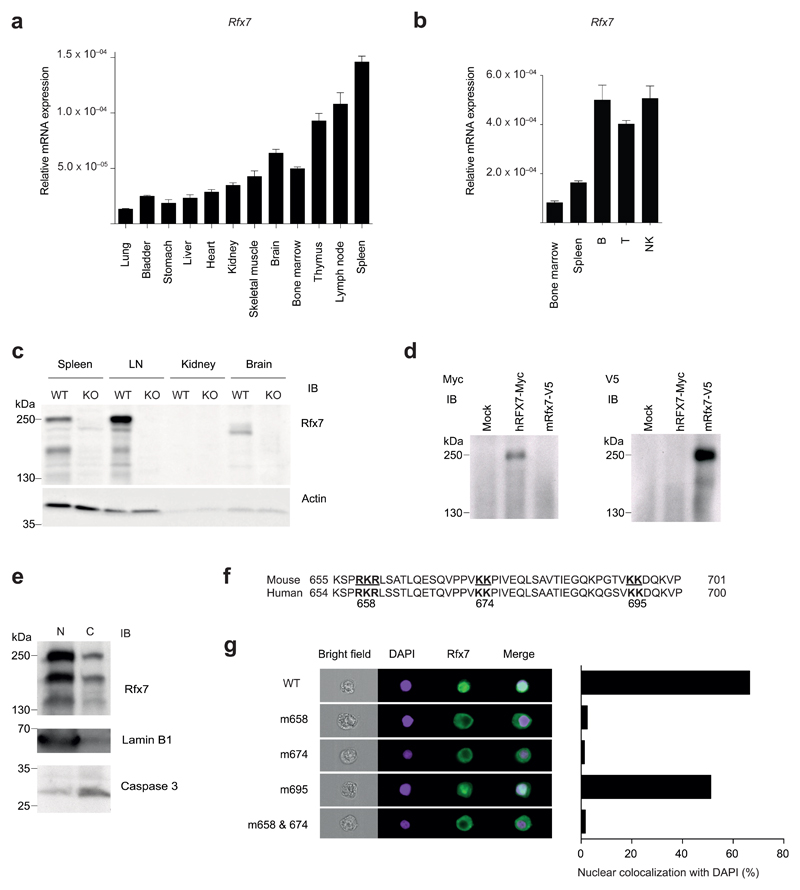
First, we detailed the expression of Rfx7 in different murine tissues. Rfx7 transcript was abundant in lymphoid organs and lymphoid cell subsets (Fig. 1a, b), in agreement with publicly available datasets25. We next tested Rfx7 protein expression in selected organs, using cell lysates derived from Rfx7−/− mice to ensure immunoblot specificity (Fig. 1c). Rfx7 was clearly detected in spleen and lymph node (LN) and – to a lower extent – in the brain. While its predicted molecular weight corresponds to 157 kDa, immunoblot analysis revealed that Rfx7 presented a major isoform close to 250 kDa (Fig. 1c). Overexpressed mouse and human RFX7 migrated similarly (Fig. 1d), suggesting that this factor is heavily modified at the posttranslational level.
Figure 1. Rfx7 cellular and subcellular profiling.
(a,b) The abundance of Rfx7 mRNA was measured by quantitative RT-PCR (qRT-PCR) in the indicated tissues (relative to 18S) (a) and FACS-sorted immune subsets (relative to 18S;Polr2a) (b) derived from C57BL/6 mice. (c) Rfx7 expression in spleen, lymph node (LN), kidney, and brain isolated from wild-type (WT) and Rfx7-deficient (KO) mice was determined by immunoblot analysis. Actin was used as loading control. (d) Migration of human and murine Rfx7 overexpressed in HEK293T cells was determined by immunoblot analysis against the indicated tag. (e) Rfx7 presence in nuclear (N) and cytoplasmic (C) fractions of C57BL/6 splenocytes was analyzed. Lamin B1 and caspase 3 were used as nuclear and cytoplasmic markers, respectively. Predicted bipartite nuclear localization sequences (NLS) in mouse and human proteins (bold). (g) HEK293T cells were co-transfected with expression vectors encoding wild-type or the indicated mutant mouse Rfx7 and mCherry. Rfx7 nuclear localization (colocalization with DAPI) was quantified on mCherry-positive cells by Image Stream analysis; a representative example is shown. Results represent the mean ± SD of three technical replicates (a,b) and are representative of at least two independent experiments (a-e,g).
We next investigated the subcellular localization of this transcription factor. Rfx7 was detected in the nuclear fraction obtained from total splenocytes (Fig. 1e). Based on bioinformatic predictions, three stretches of basic residues, conserved also in human RFX7, were identified as part of potential bipartite nuclear localization signals (NLS) (Fig. 1f). Mutagenesis showed that nuclear localization was lost when Rfx7 was mutated at amino acid positions 658 and/or 674 (Fig. 1g), whereas mutation of position 695 had no effect. These results show that Rfx7 is highly expressed in immune cells and corroborate the notion that it acts as a bona fide transcription factor.
Rfx7 plays a cell-intrinsic role in NK cell homeostasis
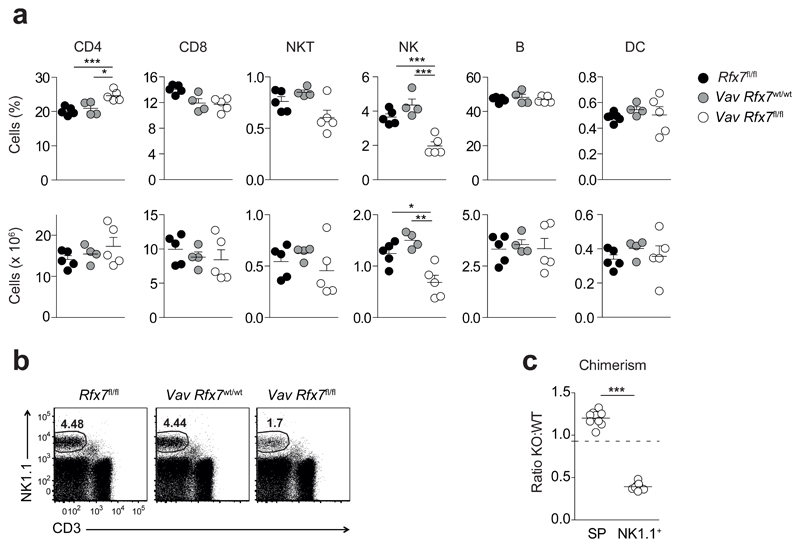
Firstly, we made sure that Rfx7 deletion was effective in multiple hematopoietic subsets from Vav Rfx7fl/fl as compared to Vav Rfx7wt/wt control mice. Deletion was virtually complete in T, B and NK cells (Supplementary Fig. 2a,b). Given the high expression of Rfx7 in hematopoietic cells, we next assessed by flow cytometry the abundance of the main lymphoid populations and of conventional dendritic cells. While most subsets were unaltered, we observed a marked decrease in NK cell percentages and numbers in Vav Rfx7fl/fl mice as compared to Rfx7fl/fl and Vav Rfx7wt/wt control mice (Fig. 2a,b).
Figure 2. NK cells are strongly reduced in Vav Rfx7fl/fl mice.
(a) Percentages and absolute numbers of splenic CD4+ T cells (gated as CD3+CD4+), CD8+ T cells (gated as CD3+CD8+), NKT cells (gated as NK1.1+CD3+), NK cells (gated as NK1.1+CD3−), B cells (gated as CD19+), and conventional dendritic cells (DC; gated as CD11chiCD11bint-hi) from the indicated mice are shown. (b) A representative flow cytometric plot of NK cells in the spleen of Rfx7fl/fl, Vav Rfx7wt/wt, and Vav Rfx7fl/fl mice is shown (gated on lymphocytes). (c) The graph depicts the ratio of Vav Rfx7fl/fl (KO) over Vav Rfx7wt/wt (WT) for total splenocytes (SP) and NK cells (NK1.1+) in the spleen of Vav Rfx7wt/wt:Vav Rfx7fl/fl mixed BM chimeras. The dotted line represents the ratio of the injected mix. Results represent mean ± SEM of n=5 (Rfx7fl/fl), n=4 (Vav Rfx7wt/wt), and n=5 (Vav Rfx7fl/fl) mice (a) and n=9 mice per group (c), and are representative of at least three (a-c) independent experiments. Statistical comparison between the experimental condition lacking Rfx7 and controls were performed (a,c); *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001; Student’s t-test.
To further examine if this defect was intrinsic to NK cells, we generated congenically marked Vav Rfx7wt/wt:Vav Rfx7fl/fl mixed bone marrow (BM) chimeras. Despite hematopoietic cells of Vav Rfx7fl/fl origin efficiently repopulated the spleen of reconstituted mice, NK cells were severely underrepresented (Fig. 2c). These data indicate that Rfx7 is required for NK cell homeostasis in a cell-intrinsic manner.
Rfx7-deficient NK cells present a maintenance problem
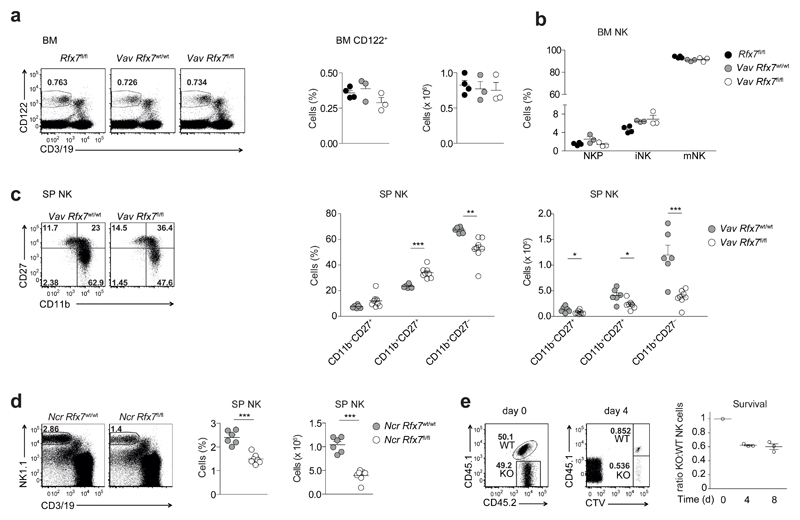
As Rfx7 transcript is abundantly expressed throughout NK cell maturation (Supplementary Fig. 3a), we took a closer look at individual stages of NK cell development. NK cells precursors in the BM are characterized by CD122 expression; acquisition of NK1.1 followed by DX5 denotes progression to the stages of immature and mature NK cells, respectively. We observed that percentages and numbers of total CD122+ cells as well as proportions of CD122+ precursor, immature, and mature NK cells were not altered in the BM of Vav Rfx7fl/fl mice as compared to Rfx7fl/fl and Vav Rfx7wt/wt control mice (Fig. 3a,b). These results suggested that the loss of Rfx7 affected the peripheral population of mature NK cells. Along these lines, when splenic NK cell maturation was measured by the expression of CD11b and CD27, NK cells exhibiting the most mature phenotype (CD11b+CD27−) were more severely reduced among Rfx7-deficient NK cells as compared to control NK cells (Fig. 3c).
Figure 3. Rfx7 deficiency affects mature NK cell survival.
(a) A representative cytometry plot (on lymphocytes), percentages (on total cells), and numbers of NK cells (gated as CD122+CD3/19−) in the bone-marrow (BM) from Rfx7fl/fl, Vav Rfx7wt/wt, and Vav Rfx7fl/fl mice are shown. (b) The graph illustrates precursor (NKp, NK1.1−DX5−), immature (iNK, NK1.1+DX5−), and mature NK cell (mNK, NK1.1+DX5+) percentages in the BM. (c) Representative cytometric profiles of spleen NK cells (gated as NK1.1+CD3−CD19−) stained with CD11b and CD27. Percentages and numbers of CD27 single-positive (CD11b−CD27+), double-positive (DP, CD11b+CD27+), and CD11b single-positive (CD11b+CD27−) populations are shown for Vav Rfx7wt/wt and Vav Rfx7fl/fl mice. (d) A representative flow cytometric image, percentages, and numbers of splenic NK cells are depicted for Ncr Rfx7wt/wt and Ncr Rfx7fl/fl mice. (e) NK cells isolated from Vav Rfx7fl/fl (KO) or Vav Rfx7wt/wt (WT) mice (congenically marked) were CTV-labeled and adoptively co-transferred into C57BL/6 hosts. Flow cytometry plots and graphs illustrate the percentages and the ratios, respectively, of KO and WT NK cells measured in the transferred mix (day 0) and in the spleen four or eight days post-transfer. Results represent mean ± SEM of n=4 (Rfx7fl/fl), n=3 (Vav Rfx7wt/wt), and n=3 (Vav Rfx7fl/fl) (a,b), or n=6 (Vav Rfx7wt/wt) and n=8 (Vav Rfx7fl/fl) (c), or n=6 (Ncr Rfx7wt/wt), and n=7 (Ncr Rfx7fl/fl) mice (d), and n=3 (day 4/8) recipient mice (e), and are representative of at least three independent experiments. Statistical comparisons between the experimental condition lacking Rfx7 and controls were performed (a-d); *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001; Student’s t-test.
To test whether Rfx7 was important after NK cell commitment had occurred, we generated Ncr1-iCre Rfx7fl/fl mice (expressing iCre under the control of the promoter of Ncr1, which encodes an NK cell receptor expressed at the immature-mature stage; hereafter referred to as Ncr Rfx7fl/fl) leading to Rfx7 deletion at the immature to mature transition. These mice exhibited diminished NK cells in the spleen (Fig. 3d) and in other peripheral tissues, such as blood and liver (Supplementary Fig. 3b), as compared to Vav Rfx7wt/wt mice. We further asked whether Rfx7 contributed to maintenance of peripheral NK cells. To this end, congenically marked NK cells from Vav Rfx7fl/fl mice and Vav Rfx7wt/wt were labeled with Cell Trace Violet (CTV), mixed in a 1:1 ratio and co-transferred into wild-type recipient mice. A selective loss of Vav Rfx7fl/fl NK cells was observed already four days after transfer (Fig. 3e). Furthermore, transferred NK cells did not divide under these conditions, as demonstrated by the CTV labeling. These data underline the importance of Rfx7 for peripheral NK cell survival.
Activating cytokines rescue Rfx7-deficient NK cells
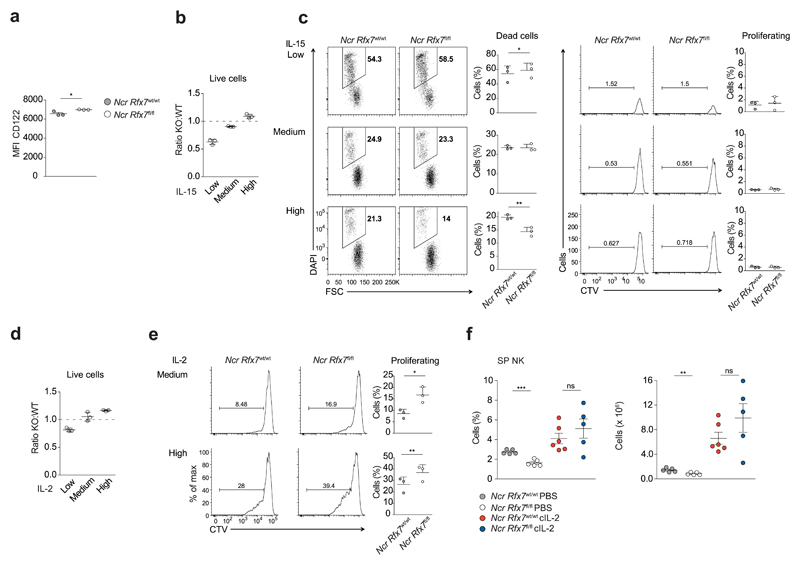
Given the importance of IL-15 for NK cell maintenance, we wondered whether NK cells from Ncr Rfx7fl/fl mice presented a defective response to this cytokine as compared to NK cells from Ncr Rfx7wt/wt mice. Expression of the IL-15 receptor subunit CD122 was not decreased in the absence of Rfx7 (Fig. 4a). We thus set up in vitro co-cultures of congenically marked Rfx7-deficient and control NK cells and supplied with different amounts of IL-15. In the presence of low amount of IL-15 (1 ng/ml), Rfx7-deficient NK cells were preferentially lost after three days of culture (Fig. 4b). This was due to reduced survival, as indicated by the absence of proliferation and the slightly increased percentage of dying cells (as shown by CTV and DAPI staining, respectively; Fig. 4c). Notably, increasing amounts of IL-15 (3 ng/ml or 10 ng/ml) reverted the defect of Rfx7−/− NK cells, which were able to survive equally or even better than their wild-type counterparts (Fig. 4b, c). A similar dose-response curve was observed for IL-2 (Fig. 4d); moreover, after four days of culture at intermediate and high doses (30 ng/ml and 100 ng/ml, respectively) of IL-2, a greater proportion of Rfx7-deficient NK cells underwent cell division (Fig. 4e).
Figure 4. Rfx7-deficient NK cells efficiently respond to high IL-15 doses.
(a) Geometric mean fluorescence intensity (MFI) of CD122 on splenic NK cells (gated as NK1.1+CD3−) from Ncr Rfx7wt/wt and Ncr Rfx7fl/fl mice as measured by flow cytometry. (b-e) Enriched Rfx7-deficient (KO; from Ncr Rfx7fl/fl) and congenically marked control NK cells (WT; from Ncr Rfx7wt/wt) were co-cultured in the presence of IL-15 or IL-2 for three (b-d) or four days (e). (b,d) Graphs depict ratios of Rfx7-deficient to control living NK cells normalized to the mix at the beginning of the culture with IL-15 (b; high 10 ng/ml, medium 3 ng/ml, or low 1 ng/ml) or IL-2 (d; high 100 ng/ml, medium 30 ng/ml, or low 10 ng/ml). (c) Representative flow cytometry plots illustrating dead cells (DAPI+) among Rfx7-deficient and control NK cells or cell division (CTV dilution) among living cells and quantifications thereof. (e) Representative flow cytometry plots illustrate percentages of dividing cells (CTV dilution) among living Rfx7-deficient and control NK cells and quantifications thereof after four days of culture with IL-2. (f) Ncr Rfx7fl/fl and Ncr Rfx7wt/wt mice were treated with PBS or IL-2/S4B6 antibody complexes (cIL-2) and analyzed on day 5. Percentages and numbers of NK cells in the spleen are depicted. Results represent mean ± SEM of n=3 (a), or of n=5 (PBS-treated Ncr Rfx7wt/wt), n=5 (PBS-treated Ncr Rfx7fl/fl), n=6 (cIL-2-treated Ncr Rfx7wt/wt), and n=5 (cIL-2-treated Ncr Rfx7fl/fl) mice (f) and mean ± SD of n=3 technical replicates per condition (b-e). Results are representative of at least three independent experiments (a-f). Statistical comparisons between the experimental condition lacking Rfx7 and control were performed (a,c,e,f); *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001, not significant (ns). Unpaired Student’s t-test (a,f) and paired Student’s t-test (c,e).
We next examined the cytokine-mediated survival response of Rfx7-deficient NK cells in vivo. Exogenous IL-2 was complexed with anti-IL-2 (S4B6), which potentiates the biologic activity of this cytokine26, and the complex was administered to Ncr Rfx7fl/fl mice for four days. This treatment completely restored the defect in the Rfx7-deficient NK cell population, showing that even when starting from lower proportions these lymphocytes expanded to similar percentages and numbers as their control counterparts (Fig. 4f). Therefore, Rfx7−/− NK cells are not inherently defective and even exhibit superior responses to intermediate or high concentrations of IL-15 or IL-2. However, these cells are not adapted to survive at limiting or physiological concentrations of these cytokines.
Rfx7 regulates expression of metabolism-related genes
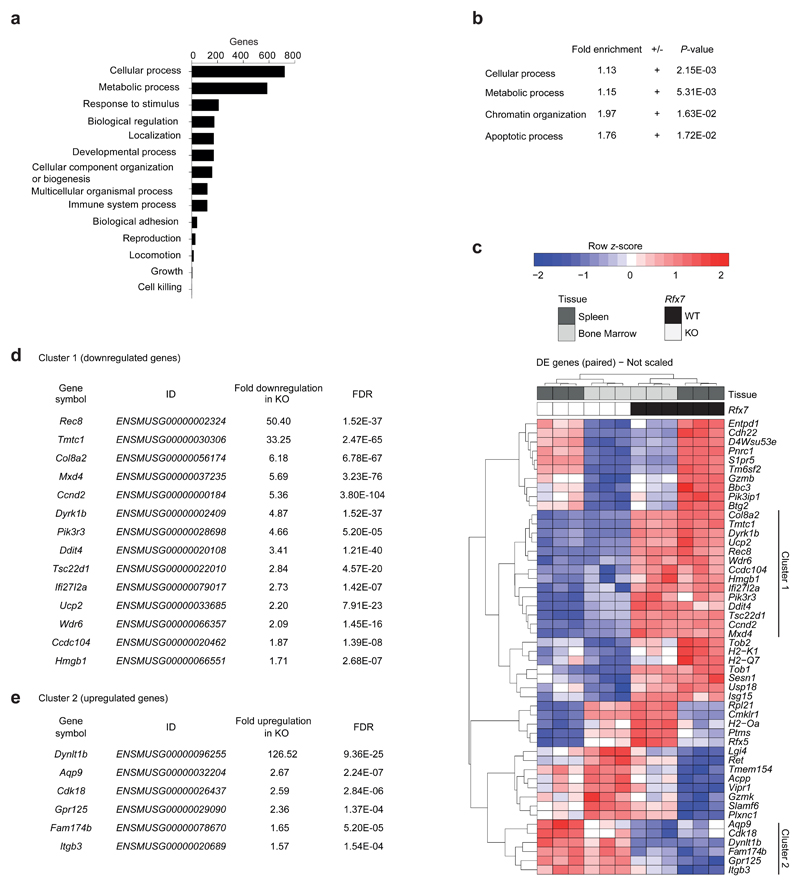
To uncover the underlying molecular mechanisms, we performed global RNA-sequencing analyses on Vav Rfx7wt/wt and Vav Rfx7fl/fl NK cells isolated from BM and spleen. A paired differential expression analysis was performed. Likelihood ratio tests identified 1636 genes significantly altered in Vav Rfx7fl/fl as compared to Vav Rfx7wt/wt NK cells (855 up- and 781 down-regulated, P ≤ 0.05). Supporting the finding that Rfx7 is important in NK cell maintenance, transcription factors and cytokine receptors involved in differentiation were not substantially affected in the absence of Rfx7 (Supplementary Table 1a, b). Functional classification on biological processes and gene ontology (GO) analysis showed that genes related to cellular and metabolic processes constituted the majority of differentially expressed genes and were significantly overrepresented (Fig. 5a,b).
Figure 5. Rfx7-deficient NK cells exhibit deregulated expression of metabolism-related genes.
(a-e) RNA-sequencing was performed on NK cells isolated from BM (CD122+ NK1.1+CD3/19−) and spleen (NK1.1+ Ncr1+CD3/19−) of Vav Rfx7fl/fl (KO) and Vav Rfx7wt/wt (WT) mice. Functional classification (PANTHER GO-Slim, biological processes) (a) and Gene ontology (GO) enrichment analysis results obtained from the online PANTHER overrepresentation test, with Bonferroni correction (b), are illustrated. Analyses were performed on the 1636 differentially expressed genes by likelihood ratio tests, p ≤ 0.05 (a,b). (c-e) Unsupervised hierarchical clustering of the 50 top-ranked differentially expressed genes (False Discovery Rate (FDR) ≤ 0.00015) and two gene clusters consistently altered in BM and spleen. (d,e) Fold differences and FDR values are indicated for each gene of these two clusters.
To validate the RNA-sequencing results, we focused on the 50 top-ranked genes (False Discovery Rate (FDR) ≤ 0.00015) and among these on genes robustly and consistently altered by Rfx7 deficiency across BM- and spleen-derived NK cells. Two clusters clearly separating Rfx7-deficient from -sufficient NK cells in both organs were delineated by the unsupervised hierarchical clustering (Fig. 5c). The first cluster corresponded to 14 downregulated genes (Fig. 5c, d), while the second cluster represented six upregulated genes (Fig. 5c,e). We tested by quantitative RT-PCR (qRT-PCR) selected genes for validation (eleven from cluster 1 and one from cluster 2, Supplementary Fig. 4a). We performed this analysis on NK cells derived from Vav Rfx7wt/wt: Vav Rfx7fl/fl mixed BM chimeras to exclude non-intrinsic alterations. We confirmed the expected differences in transcript abundance for all genes, except for Ifi27l2a – an interferon responsive gene – and Dynlt1b – the most significantly upregulated gene (Supplementary Fig. 4a). We also assessed protein expression of the top targets Rec8, Dyrk1b, and Ddit4. Due to the low number of NK cells, we focused on ex-vivo total T cells, which also presented decreased mRNA abundance for these genes in the absence of Rfx7 (Supplementary Fig. 4b). The expression of these proteins was reduced, albeit DNA damage-inducible transcript 4 protein (Ddit4) was only mildly affected (Supplementary Fig. 4c). Of note, a clear decrease in these proteins was observed in the splenic T cell-depleted fraction from Vav Rfx7fl/fl mice, demonstrating the broad relevance of Rfx7 for the expression of these targets.
Rfx7-deficient NK lymphocytes also revealed slightly reduced mRNA coding for Rfx5, the only Rfx factor found among differentially expressed genes, and for MHC class I genes (Fig. 5c and Supplementary Fig. 4d, e). However, these cells exhibited normal MHC class I surface expression (Supplementary Fig. 4f), consistent with previous findings that mechanisms regulating its display compensate for moderate differences in transcript abundance27. Altogether, data presented in this section indicate that Rfx7 deficiency alters a subset of genes linked to cell metabolism.
Top differentially expressed genes are targets of Rfx7
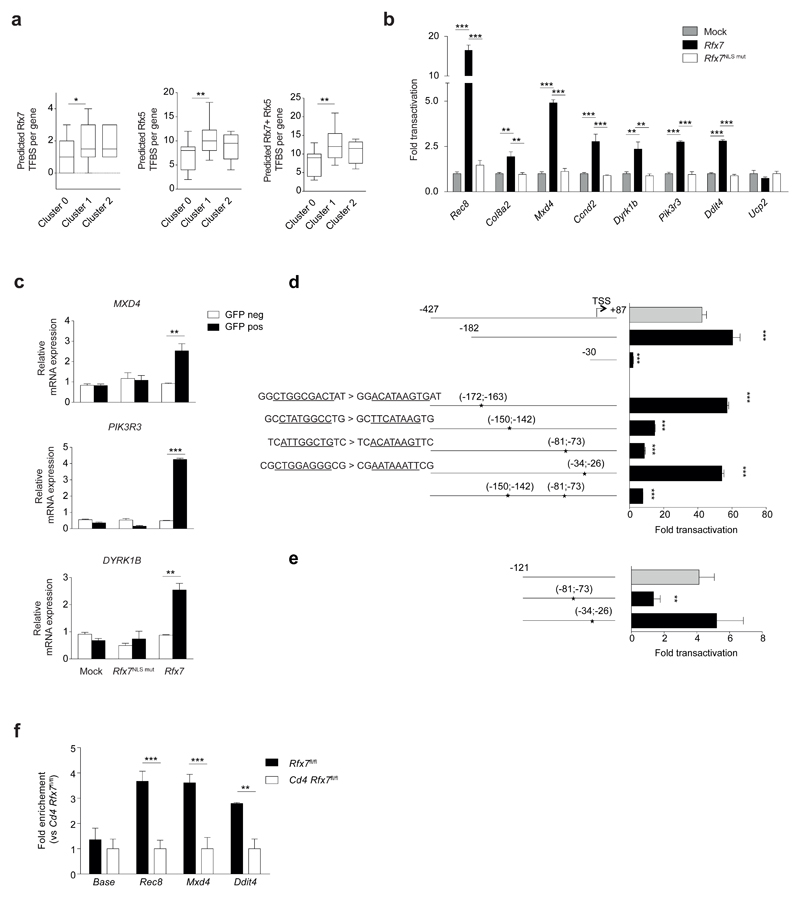
To weigh the likelihood that differentially expressed genes are directly transactivated by Rfx7, we took advantage of in silico binding sites predictions. Promoters of genes in cluster 1 (downregulated in the Rfx7-deficient NK cells), cluster 2 (upregulated), and of a group of non-modulated genes (hereafter called cluster 0) were analyzed for the presence of putative binding sites for Rfx7 and Rfx5, whose DBD has the closest homology to the one of Rfx7. Interestingly, a significant enrichment for both motifs was observed in the promoters of downregulated genes, while a non-significant trend was found for upregulated genes (Fig. 6a). Moreover, analysis for binding sites of two unrelated transcription factors did not show significant enrichment (Supplementary Fig. 5a).
Figure 6. Validation of Rfx7 target genes identified in NK cells.
(a) In Silico analysis of promoter sequences (-700; +300) of differentially expressed genes from cluster 1 (downregulated genes), cluster 2 (upregulated genes), and cluster 0 (non-modulated genes, serving as control group) was performed with JASPAR tool to test for the presence of putative transcription factor binding sites (TFBS) for Rfx7 and Rfx5. The box and whisker plots depict the number of putative TFBS identified with a relative profile score threshold of 0.8. Significance was determined using one-tail Mann-Whitney tests. (b) Luciferase reporter gene assays were performed in HEK293T cells co-transfected with WT, NLS-mutant (m658 & 674) Rfx7-encoding, or empty (mock) vector for the indicated promoters. Results are expressed as fold transactivation over mock-transfected cells. (c) HEK293T cells were co-transfected with empty vector (mock), WT, or NLS-mutant Rfx7 and GFP-encoding vectors. After 48 hours, cells were sorted as GFP-positive or -negative and MXD4, PIK3R3, and DYRK1B transcript abundance was analyzed by qRT-PCR (relative to POLR2A). (d,e) Luciferase assays performed on truncated and mutated versions of full (d) or minimal (e) Rec8 promoter. Positions in bp relative to the transcription start site (TSS, +1), and site-directed mutated sequences (underlined) are indicated for the various constructs. Assays were performed in HEK293T cells co-transfected with Rfx7-encoding or empty (mock) vector for the indicated promoters. Results are expressed as fold transactivation over mock-transfected cells. (f) Binding of Rfx7 to Rec8, Mxd4 and Ddit4 promoters was assessed by quantitative ChIP experiments performed with chromatin extracted from T cells from Cd4-Cre Rfx7fl/fl (Cd4 Rfx7fl/fl) and control mice. Results are expressed as fold enrichment in Rfx7fl/fl over Cd4 Rfx7fl/fl. Results are depicted as the mean ± SD of n=4 (b) or n=3 (c-f) technical replicates and are representative of at least three (b,d,f) or two (c,e) independent experiments. Statistical comparisons between experimental and control condition(s) were performed (b-f); *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001; Student’s t-test; For panel (b) Bonferroni correction was applied.
We therefore cloned the promoters of nine genes from cluster 1 into a luciferase reporter vector, which was co-transfected with plasmids encoding either Rfx7 or its NLS-mutated version, used as a control. All promoters but Ucp2 were transactivated, with the highest induction obtained for Rec8 and Mxd4 (Fig. 6b). We also checked whether enforced Rfx7 expression in HEK293T cells induced transcription of endogenous target genes. For this purpose, Rfx7 encoding plasmid or controls (empty or NLS-mutated version) were co-transfected with a GFP-expressing vector. Notably, GFP+ cells showed increased mRNA abundance of MXD4, PIK3R3, and DYRK1B (Fig. 6c), indicating that even in human kidney-derived cells Rfx7 regulates these genes.
In an attempt to define the sequence recognized by Rfx7, we focused on Rec8 promoter and generated truncated versions. We identified that the region between -182 and -30 base pairs upstream of the transcriptional start site (TSS) was essential for transactivation (Fig. 6d). Four putative binding sites in this region were selected for site-directed mutagenesis, demonstrating the presence of regulatory elements at position -150;-142 and -81;-73 (Fig. 6d). Interestingly, concomitant mutation of these two sequences did not further diminish promoter transactivation, suggesting that their regulatory activity is interdependent. In agreement, a promoter version truncated at position -121 was weakly transactivated by Rfx7 (Fig. 6e). Mutation of the remaining element at position -81;-73 abrogated transactivation in this system, corroborating the functional impact of this putative binding site.
Finally, we set-up chromatin immunoprecipitation (ChIP) assays using anti-Rfx7 to determine whether promoters of selected target genes were occupied by Rfx7. Due to the paucity of NK cells, we performed these experiments in ex-vivo total T cells and confirmed that promoter regions of Rec8, Mxd4, and Ddit4 were enriched by Rfx7 immunoprecipitation (Fig. 6f). Taken together, these results support that the differential gene expression identified through RNA-sequencing is driven by Rfx7.
Loss of Rfx7 leads to a heightened metabolic state
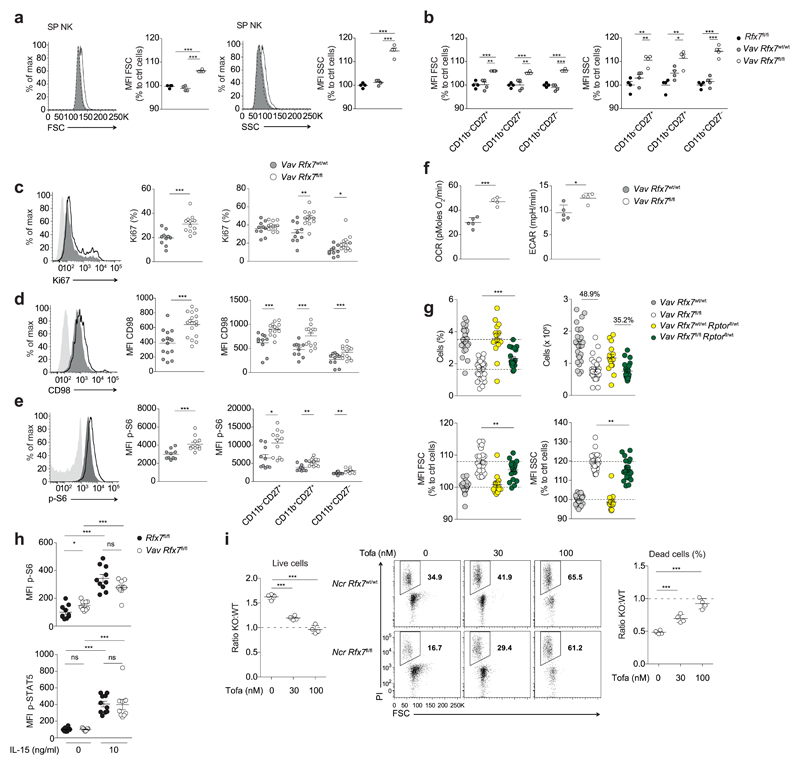
The proteins encoded by the Rfx7 target genes, such as Mxd4 and Ddit4, act as inhibitors of metabolic pathways, suggesting an increased activity in Rfx7-deficient NK cells. We thus assessed cell size and granularity, which are correlated with metabolism, and found both significantly increased in Rfx7-deficient NK cells (Fig. 7a and Supplementary Fig. 6a and b). This phenotype was independent of the maturation stage (Fig. 7b) and present already in the BM (Supplementary Fig. 6c,d). Moreover, Rfx7 deficiency broadly affected size and granularity in immune subsets (Supplementary Fig. 6e), while its overexpression in HEK293T cells led to a reduction in cell size (Supplementary Fig. 6f). Of note, deletion of Rfx5 did not exert similar effects (Supplementary Fig. 6g)14, uncovering a unique function for Rfx7.
Figure 7. Rfx7-deficient NK cells exhibit deregulated metabolism and require Jak1/3 signaling for survival.
(a,b) Representative histograms and/or geometric MFI of forward scatter (FSC) and side scatter (SSC) are shown for total (a), CD11b–CD27+, CD11b+CD27+, and CD11b+CD27– NK cells (b) from Rfx7fl/fl (set as 100%), Vav Rfx7wt/wt, and Vav Rfx7fl/fl mice. (c-e) Percentages of Ki67+ (c), geometric MFI of CD98 (d), p-S6 (e), and representative histograms are illustrated for NK cells, or subsets thereof. (f) NK cell basal oxygen consumption rate (OCR) and extracellular acidification rate (ECAR). (g) Percentages, numbers, FSC, and SSC of NK cells from Vav Rfx7wt/wt, Vav Rfx7fl/fl, Vav Rfx7wt/wt Rptorfl/wt, and Vav Rfx7fl/fl Rptorfl/wt mice. (h) Geometric MFI of p-S6 and p-STAT5 are illustrated for NK cells stimulated with high (10 ng/ml) IL-15 for 40’ or left untreated. (i) The graph on the left depicts ratios of Rfx7-deficient (KO) to control living NK cells (WT; congenically marked) co-cultured in the presence of high IL-15 and Tofacitinib for three days (normalized to initial mix). Flow cytometry plots illustrate dead cell percentage (PI+) and the graph on the right ratios thereof. Results represent mean ± SEM of n=4 mice (a,b), n=10 (Vav Rfx7wt/wt) and n=12 (Vav Rfx7fl/fl) (c,e), n=15 (Vav Rfx7wt/wt) and n=17 (Vav Rfx7fl/fl) (d), n=27 (Vav Rfx7wt/wt), n=29 (Vav Rfx7fl/fl), n=15 (Vav Rfx7wt/wt Rptorfl/wt), and n=21 (Vav Rfx7fl/fl Rptorfl/wt) (g), or n=10 (Rfx7fl/fl) and n=9 (Vav Rfx7fl/fl) mice (h), are a pool of two (c-e,h) or five (g) experiments. Data represent mean ± SD of n=5 (Vav Rfx7wt/wt) and n=4 (Vav Rfx7fl/fl) (f) and n=4 (i) technical replicates. Results are representative of at least two experiments (a-i). Statistical comparisons between control and experimental condition (a-f,h,i), between Vav Rfx7fl/fl, and Vav Rfx7fl/fl Rptorfl/wt mice (g) were performed; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001; Student’s t-test.
Next, we measured the expression of the proliferation marker Ki67, the amino acid transporter CD98, and the activity of mTORC1 by assessing the S6 phosphorylation. We observed that these parameters were augmented in splenic NK cells lacking Rfx7, including at individual maturation stages (Fig. 7c-e). We also assessed the metabolic rate of these NK cells by Seahorse analysis. Oxygen consumption and extracellular acidification rate were both increased at basal level (Fig. 7f). Taken together, we show here that Rfx7-deficient NK cells presented a heightened energetic state.
To evaluate the role of mTORC1 in this phenotype, we bred Vav Rfx7fl/fl mice onto Rptorfl/wt, which is expected to slightly impair mTORC1 signaling28–31. Notably, Rptor heterozygosity moderately restored splenic Rfx7-deficient NK cell percentages and numbers, with respect to Rptorfl/wt controls (Fig. 7g). Mice on a Rptorfl/wt background exhibited spleens of reduced size (Fig. 7g). Rptor heterozygosity also partially normalized Rfx7-deficient NK cell size and granularity (Fig. 7g), indicating that one of the functions of Rfx7 is to limit mTORC1 signaling28.
Yet, Rfx7-deficient NK cells were rescued in vitro by supraphysiological doses of IL-15 (10 ng/ml), which further engage mTORC1. Importantly, IL-15 also activates the Jak 1/3-STAT5 axis, whose basal activity was unaltered in the absence of Rfx7 (Fig. 7h). Notably, “high” IL-15 stimulation (10 ng/ml) engaged both mTORC1 and Jak-STAT5 pathways in control and Rfx7-deficient NK cells and Tofacitinib, a Jak1/3 inhibitor, selectively hampered survival of the latter over three days of culture (Fig 7h,i). This effect was partly due to Myeloid cell leukemia sequence 1, which is a known STAT5 target32 (Supplementary Fig. 6h). Therefore, our results suggest that the increased activation state of Rfx7-deficient NK cells is not detrimental per se, but in the context of insufficient Jak-STAT5 engagement.
Rfx7 is essential for optimal NK cell-mediated immunity
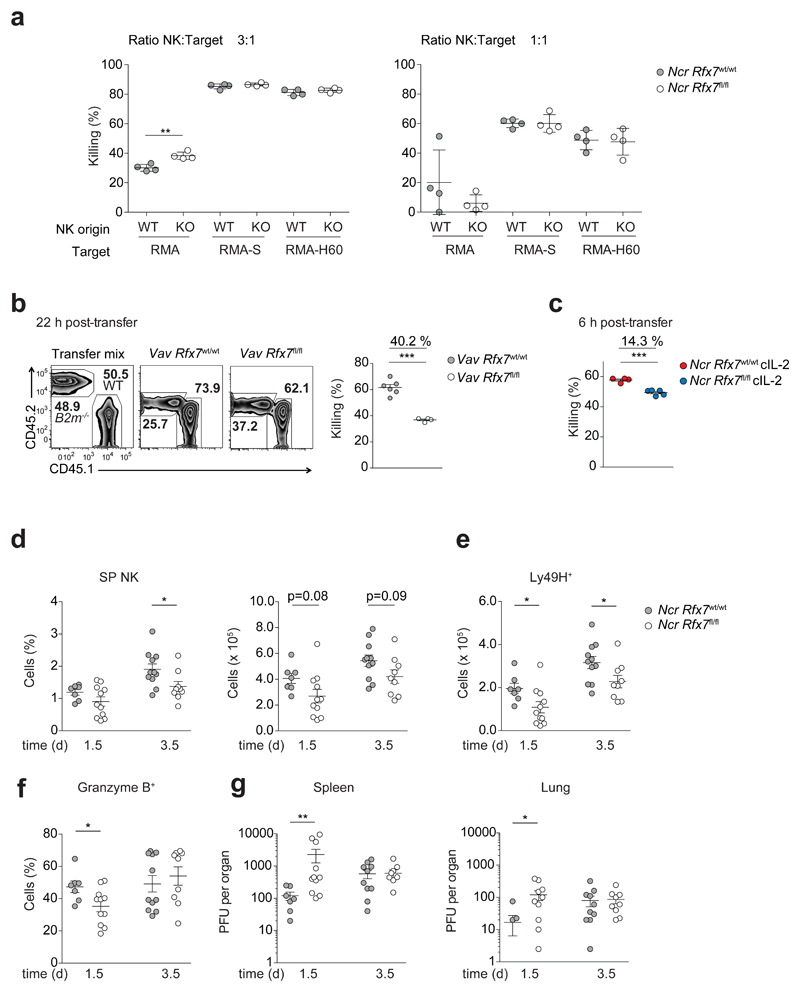
Finally, we sought to understand the functional impact of Rfx7 deficiency on NK cell-mediated immunity. First, we examined whether Rfx7-deficient NK cells presented an altered receptor repertoire. Although expression of several receptors was slightly modulated, the overall expression pattern was maintained (Supplementary Fig. 7a). Next, we assessed whether the killing capacity per cell basis was altered by Rfx7 deficiency. To this end, NK cells were isolated from Ncr Rfx7fl/fl and Ncr Rfx7wt/wt mice and cultured together with RMA-S and RMA-H60 target cells, which lack MHC class I expression and express the activating H60 ligand33, respectively, or with control RMA cells. The killing ability of Rfx7-deficient and control NK cells were comparable (Fig. 8a).
Figure 8. NK cell-mediated immunity is compromised in Vav Rfx7fl/fl mice.
(a) NK cells isolated from polyinosinic:polycytidylic acid-treated Ncr Rfx7wt/wt (WT) or Ncr Rfx7fl/fl mice (KO) were plated with RMA, RMA-S, or RMA-H60 cells at the indicated ratios. The graph depicts percentage killing of target cells, as measured by quantifying PI− living target cells. (b) A representative flow cytometry plot and the quantification of B2m−/− splenocyte rejection analyzed in the spleen one day after transfer are illustrated. (c) Quantification of B2m−/− splenocyte rejection analyzed in the spleen six hours after transfer into Ncr Rfx7fl/fl and Ncr Rfx7wt/wt mice treated with cIL-2 for the preceding four days is illustrated. As control, wild-type splenocytes (CD45.1) were co-injected with B2m−/− splenocytes (CD45.2) (b,c). (d-g) Ncr Rfx7fl/fl and Ncr Rfx7wt/wt mice were infected with MCMV and analyzed 1.5 or 3.5 days post infection. (d,e) Graphs depict percentage and number of total (d) and numbers of Ly49H+ (e) NK cells in the spleen. (f) Graph illustrates percentage of granzyme B+ NK cells. (g) Viral titers in the indicated organs. Results represent mean ± SD of n=4 technical replicates (a), and mean ± SEM of n=6 (Vav Rfx7wt/wt) and n=4 (Vav Rfx7fl/fl) mice (b), n=4 (Ncr Rfx7wt/wt) and n=5 (Ncr Rfx7fl/fl) (c), and n=7 (Ncr Rfx7wt/wt, day 1.5), n=11 (Ncr Rfx7fl/fl, day 1.5), n=11 (Ncr Rfx7wt/wt, day 3.5), and n=9 (Ncr Rfx7fl/fl, day 3.5) (d-g) mice and are representative of (a-c) or a pool of (d-g) two independent experiments. Statistical comparisons between the experimental condition lacking Rfx7 and control were performed; *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001; Student’s t-test.
We therefore investigated whether the altered numbers of Rfx7-deficient NK cells found in vivo had an impact on NK cell-mediated immunity. To this end, β2 microglobulin (B2m)-deficient splenocytes, which also lack expression of MHC class I molecules, were transferred into Vav Rfx7fl/fl and Vav Rfx7wt/wt mice, and their elimination quantified. Notably, the ability of Vav Rfx7fl/fl mice to reject β2m-deficient targets was significantly hampered as compared to Vav Rfx7wt/wt mice (Fig. 8b). To corroborate the hypothesis that this was largely linked to reduced NK cell numbers, we administered IL-2–anti-IL-2 complexes to Rfx7-deficient mice to rescue NK cells (Supplementary Fig. 7b) and then assessed β2m-deficient cell rejection. Under these conditions, recognition of MHC class I-deficient cells in Ncr Rfx7fl/fl mice was substantially closer to that observed in Vav Rfx7wt/wt mice (Fig. 8c).
We thus postulated that Rfx7 might strongly impact antiviral responses at early time points. To investigate this hypothesis, we infected Ncr Rfx7fl/fl and Ncr Rfx7wt/wt mice with murine cytomegalovirus (MCMV). Numbers of total and Ly49H+ Rfx7-deficient NK cells were lower 1.5 days after infection, while the defect was milder at day 3.5 (Fig. 8d, e). In addition, granzyme B production was lower early after infection (Fig. 8f). This finding is in agreement with RNA-sequencing results (Supplementary Table 2), which are further supported by the observation that, in uninfected mice, granzyme A production by Rfx7-deficient NK cells was slightly decreased while interferon-γ level (IFN-γ) was normal as compared to control NK cells (Supplementary Fig. 7c).
We finally assessed resistance of Ncr Rfx7fl/fl mice to MCMV infection. Importantly, spleen and lung of Ncr Rfx7fl/fl mice exhibited significantly higher viral titers early after infection, while the defect was corrected few days after the onset of the inflammatory reaction (Fig. 8g). These data demonstrate the importance of Rfx7 for normal NK cell homeostasis and antiviral immunity in the initial phase of the response.
Discussion
Rfx7 is an understudied protein, which has been ascribed a transcriptional function based on phylogenetic analyses. We found that this factor localized to the nucleus through a bipartite NLS and directly regulated selected genes. Moreover, we identified regulatory sequences required for Rfx7-mediated transactivation. These data further suggested that Rfx7 activity required multiple binding sites, possibly implying DNA loop formation. In agreement with available datasets, our results also indicated that Rfx7 is subject to multiple posttranslational modifications34. A complex regulation of its activity is not surprising, given its functions in immunity and beyond. Indeed, full-body deleted mice are born at submendelian ratios, revealing a developmental role – possibly related to ciliogenesis – that deserves further investigation.
The generation of a conditional knockout mouse model allowed studying Rfx7 in immune cells, in which it is prominently expressed. This highlighted a significant defect in NK lymphocyte percentages and numbers. These cells exhibited an intrinsic survival problem, mainly affecting the most mature, peripheral subsets. While Rfx7-ablated NK lymphocytes displayed only moderately altered receptor repertoire and production of cytotoxic mediators, their decreased numbers affected NK cell-mediated immunity in vivo. This was observed in the context of both “missing-self” and antiviral responses, uncovering the essential role of Rfx7 to contain MCMV replication in the early phases of infection.
Interestingly, GO enrichment analysis revealed that differentially expressed genes were related to metabolic processes. A closer look into the phenotype of Rfx7-deficient NK cells highlighted their heightened metabolic features. The finding that immune populations such as T cells, which were not decreased in numbers, presented morphological and transcriptional alterations similar to peripheral NK cells and the validation of differentially expressed genes as direct Rfx7 targets corroborated the notion that this factor directly limits cell activation and metabolism.
Although counterintuitive, we showed that an increased metabolic rate in Rfx7-deficient NK cells negatively impacted on their survival, highlighting the importance of maintaining quiescence. While this concept is often neglected in immunity, similar examples have been shown. For instance, deletion of Tsc1¸ encoding an inhibitor of mTORC1, in T and NK cells led to improper cellular activation and apoptosis28,35. In agreement, our results showed that Rptor heterozygosity mitigated the defects caused by Rfx7 deficiency.
Indications as to how Rfx7 limits the activation state of NK cells arose from RNA-sequencing analyses. The Rfx7 target Ddit4 inhibits mTORC1 through positive regulation of the TSC complex36. In addition, Mxd4 and Pik3ip1 encode proteins antagonizing Myc and phosphatidylinositol-3-OH kinase, respectively37–40, while the target gene Dyrk1b, which is associated with a form of metabolic syndrome, codes for a protein interfering with sonic hedgehog (Shh) signaling and promoting gluconeogenesis41,42. Rfx7 therefore controls genes, whose products counter proliferative, activating, and anabolic pathways via mTORC1-dependent and independent cascades.
The lower survival of Rfx7-deficient NK cells was corrected by supra-physiological concentrations of IL-15. As features of metabolic deregulation, but not the survival defect, were detected in developing Rfx7-deficient NK cells in the BM, it is tempting to speculate that IL-15 bioavailibility in this primary lymphoid organ are higher than in the periphery, a hypothesis compatible with experimental data43. While IL-15 drives a coordinated engagement of STAT5 and mTORC1, Rfx7 deficiency favors the latter pathway. IL-15 activation rescued Rfx7-deleted splenic NK cells, showing that these lymphocytes were not inherently defective and even presented an advantage if provided with the appropriate Jak-mediated signaling, an effect partly dependent on the STAT5 target Mcl-132,44–46. This underlines the importance of a harmonized activation of these cascades in NK cells.
While regulation of metabolism by Rfx factors has been previously reported – for instance Rfx3 and Rfx6 regulate genes such as those encoding glucokinase in pancreatic cells11,47 – the understanding of the transcriptional mechanisms maintaining cellular quiescence remains fragmentary. Foxo and Foxp1 transcription factors have been involved in such mechanisms39,40,48–50. As compared to Rfx7, reported functions and target genes suggest distinct actions for Foxo transcription factors. Interestingly, Foxp1 maintains quiescence in T cells in part through regulation of Pik3ip1 expression. As this gene is also controlled by Rfx7, future experiments are required to understand their coordinate regulation of gene expression.
RFX7 has been associated with altered body fat distribution; a role as suppressor of metabolic pathways well fits this observation20, predicting Rfx7 to be relevant beyond the immune system. Interestingly, transformed cells are characterized by increased metabolic rates and alterations of RFX7 have been associated with cancer, lymphoid cancers in particular16–19,21–23. While spontaneous activation caused by Rfx7 deficiency leads to cell death in NK cells, we show that this is not the case for other lymphocytes. Our work suggests a role for Rfx7 in limiting cell activation and as a tumor suppressor. Altogether, our data position this transcription factor as a regulator of immunometabolism and its manipulation might be attractive in a range of immune, malignant, and metabolic diseases.
Methods
Mice
Rfx7-floxed mice were generated (IngeniousTargeting) on a C57BL/6 background by targeted gene disruption as described in Supplementary Fig. 1d. For genotyping, the Rfx7-floxed alleles were detected by PCR amplification using a set of primers that amplifies a 315 bp WT band and a 438 bp floxed band (forward primer 5′-GTCACTCCCCAAACAGGAAGTCTATGATG-3′ and reverse primer 5′-CCTCTAGTCCCTCCCATGTTTCTTGTC-3′). Rfx7fl/fl mice were crossed to Vav-iCre and Cd4-Cre (The Jackson Laboratory), or Ncr1-iCre deleter strain51. Rfx7−/− mice were generated by crossing Rfx7fl/fl to the CMV-Cre deleter strain (The Jackson Laboratory), leading to deletion of the loxP-flanked region also in germ cells, thus allowing selection of Rfx7-deleted offspring and elimination of the CMV-Cre transgene in the following generation. Rptorfl/fl mice were previously described52. Conditionally deleted Rfx7fl/fl and control mice, CD45.1+ congenic, B2m−/− (The Jackson Laboratory), and C57BL/6 mice, all on a C57BL/6 (H-2b) background, were bred at the animal facility of the University of Lausanne. Rfx5−/− and littermate controls on a mixed Sv129/C57Bl/6 (H-2b) background were bred at the animal facility of the University of Geneva Medical School53. Sex- and age-matched 6 to 12 week-old mice were used. All animal experimental protocols were approved by the Veterinary Office regulations of the State of Vaud, Switzerland, and all methods were performed in accordance with the Swiss guidelines and regulations.
Mixed bone marrow chimeras
Mixed BM chimeras were generated as previously described54, with the difference that a total of 5 × 106 BM cells were injected intravenously into recipient mice.
NK cell purification and in vitro culture or in vivo transfer
Congenically (CD45.1) marked Rfx7-deficient and control NK cells were purified (STEMCELL technologies Cat. No. 19855).
NK cells were then co-cultured in vitro with recombinant mouse IL-15 or IL-2 (Peprotech) for 3 or 4 days, as indicated. IL-15 was used at 10 ng/ml (“high”), 3 ng/ml (“medium”), or 1 ng/ml (low) dose and IL-2 at 100 ng/ml (“high”), 30 ng/ml (“medium”), or 10 ng/ml (low). The medium used for in vitro experiments was previously described55.
For in vivo transfer, congenically marked NK cells from Vav Rfx7fl/fl or Vav Rfx7wt/wt mice were CTV-labeled and approximately 1.8 x 106 NK cells were adoptively co-transferred into C57BL/6 hosts.
Intracellular staining and PMA/ionomycin stimulation
Intracellular stainings for Ki67 (SolA15), phospho-STAT5 Y694 (SRBCZX) (all from eBioscience) and phospho-S6 ribosomal protein (S235/236; D57.2.2E; from Cell signaling) were performed with the intracellular staining kit from Invitrogen (Ref. 00-5523-00) or using a standard paraformaldehyde/methanol protocol. Briefly, after surface staining, cells were fixed with fixation/permeabilization buffer (Invitrogen (cat. 00-5123)) or with 2% paraformaldehyde, and permeabilized with permeabilization buffer (Invitrogen (cat. 00-8333)) or with 90 °C methanol. Intracellular phospho-stainings were performed for 1 h at 22-28 °C in the dark. A secondary antibody coupled to an APC-fluorochrome (Donkey F(ab’)2, anti-rabbit, IgG, multi species, Southern Biotech) was used in addition to undirectly conjugated phosphorylated antibodies. For resting conditions cells were left in medium at 37 °C for 40 min-1 h, and for IL-15 stimulated conditions cells were stimulated with 10 ng/ml of IL-15 for 40 min at 37 °C before proceeding with the stainings.
For intracellular cytokine detection, freshly isolated splenocytes were cultured in medium at 37 °C and stimulated with 0.1 μM PMA and 1 mg/ml ionomycin for 1 h, followed by Brefeldin A at a final concentration of 10 μg/ml for 3.5 h. Following surface staining, cells were fixed with intracellular fixation buffer (from eBioscience Ref. 00-8222-49) for 15 min, and permeabilized with permeabilization buffer (from eBioscience Ref. 00-8333-56) for 15 min. Intracellular staining for IFN-γ (F3 IGH 48) and granzyme A (3G8.5) was performed with antibodies diluted into the permeabilization buffer. Granzyme B (GB12, Life Technologies) staining was performed after fixation (25 min at 22-28 °C) and permeabilization with the FoxP3 transcription factor staining buffer set of eBioscience (00-5523-00). Intracellular staining was then performed for 30 min at 22-28 °C (with the antibody diluted in permeabilization buffer).
Murine cytomegalovirus infection
MCMV (expressing m157) was derived from the bacterial artificial chromosome pSM3fr 3.356. MCMV was propagated on M2-10B4 cells and viral stocks were purified as described57,58. Mice were infected intravenously with 2 × 105 PFU MCMV. Organs from infected mice were snap-frozen in liquid nitrogen and stored at −80 °C until further use. For determination of the viral load, serial dilutions of organ homogenates were propagated on monolayers of M2-10B4 cells as described58.
Rfx7 constructs and transfection
For expression of the murine Rfx7, the vector encoding a pEF-Dest51-mRfx7-V5/6XHis (C-ter) construct was purchased in Source BioScience LifeSciences. Nucleotides encoding the putative NLS sequences (predicted by: http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi) were mutated by site-directed mutagenesis. For expression of the human protein, the Lenti ORF clone of RFX7, Myc-DDK-tagged (C-ter) was purchased from OriGene. HEK293T cells were transfected using PEI reagent at 1:1.5 (DNA:PEI) ratio.
Preparation and in vivo administration of IL-2/S4B6 antibody complexes
Mice received intraperitoneal injections of IL-2 complexes for 4 consecutive days. Murine IL-2–anti-IL-2 complexes were prepared by mixing in PBS 15-30 min before injection 3 μg murine IL-2 (Peprotech, Cat. No. 212-12) with 15 μg anti-IL-2 (S4B6) per injection (eBioscience, Cat. No. 16-7020-85).
In vivo NK cell-mediated rejection
For testing the ability of NK cells to kill in vivo, 10 × 106 splenocytes from B2m−/− and congenically marked control mice were co-injected intravenously into recipient mice. To distinguish transferred cells, labeling with Cell Trace Violet (CTV, Life Technologies) was performed. Loss of target cells in the spleen was analyzed 22 h (for resting mice) or 6 h (for mice treated 4 days with IL-2–anti-IL-2 complexes) after cell transfer. Data are reported as percentage of rejection normalized to the co-injected wild-type cells and to the initial mix.
NK cell-mediated killing in vitro
Mice were pre-treated with 100 μg of Poly (I:C) (InvivoGen) by intraperitoneal injection 1 day before the experiment. NK cells were purified from spleens using a kit from STEMCELL technologies (Cat. No. 19855). Then NK cells were co-cultured (at the indicated ratios) with 1 × 104 CTV-labeled target cells (RMA, RMA-S, RMA-H60) in round 96-well plates for 4 h at 37°C. Quantitative analysis of the cells was then performed by flow cytometry.
Flow cytometry and cell sorting
For flow cytometry analysis, cells were pre-incubated with anti-CD16/32 (2.4G2) to block Fc receptors and then surface stained using antibodies against B220 (RA3-6B2), CD3e (145-2C11), CD4 (L3T4), CD8a (Ly-2), CD19 (1D3 or 6D5), CD27 (LG.7F9 or LG.3A10), CD45.1 (A20), CD45.2 (104), CD11b (M1/70), CD11c (N418), CD94 (18D3), CD98 (RL388), CD122 (TM-b1), CD226/DNAM (10E5), CD244/2B4 (eBio244F4), DX5/CD49b (DX5), Ncr1 (29A1.4), NK1.1 (PK136), NKG2A/C/E (20d5), H2-Db (28-14-8), H2-Kb (AF6-88.5.5.3), KLRG1 (2F1), Ly49A (A1), Ly49D (4E5), Ly49G2 (4D11), Ly49I (YLI-90), Ly49H (3D10), and Qa2 (69H1-9-9) (purchased from BioLegend or eBioscience). Streptavidin conjugated to different fluorophores were from eBioscience. Stainings were performed with appropriate combinations of fluorophores. Data were acquired with a Becton Dickinson flow cytometer and analyzed using FlowJo software (Tree Star).
For cell sorting, BM and splenic NK cells for RNA sequencing were enriched using a kit from STEMCELL technologies (Cat. No 19855). BM NK cells were sorted by flow cytometry as NK1.1+CD122+CD3–CD19– cells, and splenic NK cells were sorted as NK1.1+ Ncr1+CD3–CD19– cells. Splenic B cells were sorted as CD19+CD3–NK1.1–, and T cells as CD3+CD19–NK1.1–. Sorts were performed using FACSAria, BD Biosciences.
For HEK293T, cells were co-transfected with an empty vector (mock) or vectors encoding wild-type Rfx7- or NLS mutated-Rfx7 and a GFP-encoding vector. Cells were harvested 48 h post-transfection and flow-sorted based on GFP intensity (negative or positive) to distinguish transfected from untransfected cells.
CTV-cell labeling and proliferation analysis
Labeling of cells with Cell Trace Violet (CTV from Life Technologies) was performed in PBS 1% FCS at 37 °C at 5 μM for 20 min. Cell proliferation was measured as dye dilution by flow cytometry.
Seahorse analysis
For extracellular flux assays, 2.5 × 105 flow-sorted NK cells were seeded in a Seahorse Bioscience culture plate coated overnight with Cell-Tak solution (Discovery Labware) in Dulbecco’s modified Eagle’s medium (Sigma) with 10 mM glucose (Sigma), 2 mM glutamine (Life Technology) in a non-CO2 incubator for at least 30 min. Basal OCR and ECAR were then measured by an XF96 Seahorse Extracellular Flux Analyzer following the manufacturer's instruction.
Rfx7 subcellular localization and NK cell size analysis with Image stream
Cells were acquired with Inspire software (Amnis) on a 4-laser 12-channel imaging flow cytometer (Image StreamX MarkII) using 40× magnification. For analysis, cells in focus (using the ‘gradient RMS’ feature for the brightfield image) and single cells (in a plot using ‘area’ versus ‘aspect ratio’) were gated.
For studying the subcellular localization of Rfx7, at least 1 × 106 HEK293T cells for each condition were analyzed. Cells were transfected with mCherry-encoding construct plus wild-type- or mutant Rfx7-encoding vectors. 24 h post transfection, cells were fixed with 4% paraformaldehyde and permeabilized with 0.5% saponin in PBS/1% FCS; the cells were then stained with mouse anti-V5 Ab (mouse IgG1; Thermo Fisher Scientific cat. MA5-15253) followed by rat anti-mouse IgG1 FITC (Southern Biotech cat. 1144-02). In addition, nuclear dye (DAPI; Thermo Fischer Scientific) was added. Rfx7- and mCherry-positive cells were selected and as a measure of nuclear localization the similarity of DAPI and Rfx7 staining in the ‘dilate(object(DAPI, tight)1)’ mask was calculated. Values between 1 and 2.5 are considered as partial nuclear localization.
For NK cell size analysis, at least 3 × 104 cells per mouse and genotype were analyzed. Staining was performed as usual, DAPI was added to discriminate dead cells. The area of NK cells (gated as NK1.1+CD3–19–) was calculated using a morphology mask for NK1.1 and an object mask for the bright field image.
Tissue panel, enrichment of lymphocyte populations and quantitative RT-PCR
For organ collection, mice were perfused with heparin in PBS, and isolated organs were put and processed in TRIzol® reagent (Ambion, Life Technologies) using the TissueLyser (Qiagen) according to the manufacturer’s instructions. For expression analysis of Rfx7, T and B cells were MACS-sorted with anti-CD8a and anti -CD4, or with anti-CD19 magnetic beads (Miltenyi Biotech), or flow-sorted where indicated. NK cells were flow-sorted following enrichment (STEMCELL technologies Cat. No. 19855). Total RNA was extracted according to manufacturer's instructions. Retrotranscription to cDNA and expression analysis were done as previously described54. Expression was determined relative to the indicated housekeeping gene. Primers (forward; reverse) used for qRT-PCR are listed hereafter: 18S (5′-GTAACCCGTTGAACCCCATT-3′; 5′-CCATCCAATCGGTAGTAGCG-3′), Ccnd2 (5′-GACAACTCTGTGAAGCCCCA-3′; 5′-GAAGTCGTGAGGGGTGACTG-3′), Col8a2 (5′-TATGTGCAGCCCATGCAGAA-3′; 5′-CAAGTCCATTGGCAGCATC-3′), Ddit4 (5′-ACCAGTTCGCTCACCCTTC-3′; 5′-GCTGCTAAGGTCCGAATGAA-3′), Dynlt1b (5′-GGAAGACTTCCAGGCCTCA-3′; 5′-GGCGCTTTCTATAGCCTCCT-3′), Dyrk1b (5′-GACACCTGCCCCTCCTCTA-3′; 5′-CGGTAGGCTCTGTTGTCGTT-3′), Ifi27l2a (5′-GCTACCAGGAGGACTCAACA-3′; 5′-CAATGCCTGTCCCAGTGAA-3′), Mxd4 (5′-TCACACAACGAACTAGAAAAGCA-3′; 5′-TAGTGTGACGTGTGCTGTCA-3′), Pik3r3 (5′-GATGTTACGCTTGTTCTGTGG-3′; 5′-GGTTCTGCAAAGCCATATCCT-3′), Polr2a (5′-CCGGATGAATTGAAGCGGATGT-3′; 5′-CCTGCCGTGGATCCATTAGTCC-3′), Ptms (5′-GTGCCAAGGACCTGAAAGA-3′; 5′-TCAGCTCCATTCTCCTCCTC-3′), Rec8 (5′-CCAACAAGGAGCTGGACTTC-3′; 5′-GGACAGCACCAAGAGCAGAT-3′), Tsc22d1 (5′-TCTGGTGCAAGTGTGGTAGC-3′; 5′-TGGCTTTTCACCAGATCCAT-3′), Ucp2 (5′-CCTTCTGCACTCCTGTGTTCT-3′; 5′-GCTGGGAGACGAAACACTTAAA-3′), DYRK1B (5′-GACACCTGCCCCTCTTCC-3′; 5′-ATCGGTTGCTGTAGCGGTAG-3′), MXD4 (5′-AAGCACAGACGAGCCAAACT-3′; 5′-CCTGCTCCTCCAGTTTCTTG-3′), PIK3R3 (5′-ACTGGAGGGAGGTGATGAT-3′; 5′-TTCCATTTGGAACTGCTGAA-3′), POLR2A (5′-CGCACCATCAAGAGAGTCCAGTTC-3′; 5′-GTATTTGATGCCACCCTCCGTCA-3′).
Chromatin immunoprecipitation (ChIP)
For ChIP analysis, T cells were MACS-sorted (see “enrichment of lymphocyte populations”) from spleens of Rfx7fl/fl and Cd4 Rfx7fl/fl mice. Chromatin was prepared as previously described59. Immunoprecipitation was performed using the anti-Rfx7 (NBP1-71819, Novus Biomedicals). Analysis of specific DNA regions was performed by quantitative PCR. The amount of immunoprecipitated DNA was calculated from the standard curves generated with the input chromatin and fold enrichment was determined relative to Cd4 Rfx7fl/fl. Primers (forward; reverse) are listed hereafter: Baseline (5′-CTCGCTTGTCGGACTCTCAG-3′; 5′-CAGCCAAGGAGCTCGTATCC-3′), Ddit4 (5′-GTCACCAGGCAGGAGAGAAC-3′; 5′-GCCCACCTCAGCCAATAG-3′), Mxd4 (5′-GGAGCGCCCACACCTATT-3′; 5′-CTGCCTCAGGCCCTCAAAT-3′), Rec8 (5′-CTTTGCACCTTGGCGAAGTC-3′; 5′-CAATGGCCTCTCGTTCTCG-3′).
Immunoblot analysis
Organs were collected and placed in RIPA buffer containing 50 mM Tris-HCl pH 7.4, 50 mM NaCl, 1 mM EDTA, 1% Nonidet P-40, and 0.1% SDS supplemented with protease inhibitors (cOmplete™ ULTRA Tablets, EDTA-free, Roche) for lysis and protein extraction. For kidney and brain, lysates were further processed in the TissueLyser according to the manufacturer’s instructions (Qiagen) prior to extraction. For splenocyte analysis, T cells were MACS-sorted (see “enrichment of lymphocyte populations”) from Rfx7fl/fl and Vav Rfx7fl/fl mice and the negative fraction (“flow through”, FT) was also collected. Protein concentration was determined for each sample using the Bradford method and equal amounts of protein were loaded. Antibodies used were anti-Rfx7 (NBP1-71819, AA 1313-1363, Novus Biologicals) and anti-Ddit4 (10638-I-AP, Proteintech), anti-Rec8 (EPR16189, Ab192241, Abcam), and anti-Dyrk1b (H-6, sc-390417, Santa Cruz). Control antibodies were polyclonal anti-caspase-3 (from Cell Signaling), anti-lamin B1, and anti-β-actin (from Abcam). Anti-tag used were monoclonal anti-V5 (Thermo Scientific, E10/V4RR) and polyclonal anti-Myc (Millipore, clone 4A6). Chemiluminescence signals were captured by The FUSION Solo S imaging system (Vilber).
Cellular fractionation
Splenocytes were centrifuged at 450g at 4 °C for 5 min and resuspended in 2× volume cytoplasmic lysis buffer: 10 mM HEPES pH 7.9, 1.5 mM MgCl2, 300 mM sucrose, 0.5% NP-40, 10 mM KCl, 0.5 mM DTT, and protease inhibitors (complete ULTRA Tablets, EDTA-free from Roche). The samples were left on ice for 5 min and then centrifuged at 7600g for 45 seconds. The harvested supernatants corresponded to the cytoplasmic fraction. The pellets were washed twice with cytoplasmic buffer and centrifuged 5 min at 7600 xg, and resuspended in 2×volume nuclear lysis buffer: 20 mM HEPES pH 7.9, 100 mM NaCl, 0.2 mM EDTA, 20% glycerol, 100 mM KCl, 0.5 mM DTT, and protease inhibitors. After 5 min, samples were frozen and thawed 3 times and sonicated for 10 seconds. Finally, the extracts were centrifuged at 16000g for 10 min at 4 °C. The supernatant corresponded to the nuclear fraction. Protein concentration was determined for each sample using the Bradford method and equal amounts of protein were loaded.
Luciferase reporter gene assays
DNA fragments corresponding to the indicated promoters, extended with MluI and XhoI restriction sites for cloning, were PCR amplified using the KAPA Hifi PCR kit (KAPA Biosystems) and inserted into the pGl3basic plasmid (Promega). Primers (forward; reverse) used were as follows: Col8a2 (5′-TTTACGCGTTGGTACCACCACCACTGGTTT-3′; 5′-AAACTCGAGCTGGACACCTGGCTGCAT-3′), Mxd4 (5′-TTTACGCGTTATTCCTGCCAGAGACAGGTG-3′; 5′-AAACTCGAGAGGCCCTCAAATTGACACAT-3′), Ccnd2 (5′-TTTACGCGTTAGCTTGAGGGGCATAACCTT-3′; 5′-AAACTCGAGAATGGGAGATTTGGGAGAGG-3′), Dyrk1b (5′-TTTACGCGTTTGTTTCCAATGTGAGCCAAA-3′; 5′-AAACTCGAGCCAGTAATGCAACCAGCAAA-3′), Pik3r3 (5′-TTTACGCGTTTTGCGCCTTCTTTCTTTCTC-3′; 5′-AAACTCGAGGGAAAGCAAAGAGCAGTGGT-3′), Ddit4 (5′-TTTACGCGTTGGTGCCTATTGGCTGAGGT-3′; 5′-AAACTCGAGCGAAGATTGCGGACAAGAG), Ucp2 (5′-TTTACGCGTTGAAACCGACGGAGAGTACCA-3′; 5′-AAACTCGAGACAGGAGTGCAGAAGGCTGT-3′), Rec8(-427;+87) (5′-TTTACGCGTTAGGTCCTATGCTCAGGCAAG-3′; 5′-AAACTCGAGAGGCCTTCTTCGACTCTGCT-3′), Rec8(-182;+87) (5′-TTTACGCGTTGTGATTGGCTGGCGACTATT-3′; 5′-AAACTCGAGAGGCCTTCTTCGACTCTGCT-3′), Rec8(-30;+87) 5′-TTTACGCGTTGAGGGCGACAGGACCTAGAG-3′; 5′-AAACTCGAGAGGCCTTCTTCGACTCTGCT-3′). Rec8 promoter mutants were generated by fusion PCR using overlapping PCR primers (forward; reverse) as follows: Rec8-m(-172;-163) (5′-TTCTCCAAGAGCCTATGGCCTGGAACTGAGTTCTGCTGCC-3′; 5′-TCTTGGAGAATCACTTATGTCCAATCACGAGAATTCTGAGC-3′), Rec8-m(-150;-142) (5′-TTCTCCAAGAGCTTCATAAGTGGAACTGAGTTCTGCTGCC-3′; 5′-TCTTGGAGAATAGTCGCCAGCCAATCACGAGAATTCTGAGC-3′), Rec8-m(-81;-73) (5′-GGTGGTACAAAGCCTTGGAGAGTGTGCACCTGATCCCG-3′; 5′-TTGTACCACCGAACTTATGTGAGAAGCCGCGCCA-3′), Rec8-m(-34;-26) (5′-CGAATAAATTCGACAGGACCTAGAGCAAGG-3′; 5′-CGAATTTATTCGGGATCAGGTGCACAC-3′). All constructs were verified by sequencing.
To assess promoter transactivation, HEK293T cells were co-transfected with the indicated reporter and a vector encoding murine Rfx7, or Rfx7 NLS double mutant, or empty (mock). pRLTK (Renilla) vector was systematically included for normalization. Bioluminescence was measured 20 to 24 h post-transfection using the Dual-Luciferase® Reporter Assay System (Promega) following the manufacturer’s instructions on the EnSpireTM Alpha 2390 Multilabel Reader (PerkinElmer).
Phylogenetic tree of RFX7 molecules and homology analysis
Clustal2.1 software was used to generate the percentage identity matrix and the phylogenetic tree (neighbor-joining method without distance correction) based on full protein sequences. Alignment of RFX7 DNA Binding Domain was performed with JalView program.
RNA-seq library preparation and sequencing
Total RNA was extracted from sorted cells using the RNeasy plus mini kit (Qiagen) according to the manufacturer's instructions. Clontech RNA-seq libraries were prepared for the splenic NK cell preparations. Double stranded cDNA for RNA-seq library preparation was generated using SMART-Seq v4 Ultra Low Input RNA reagents (Catalog Number 634888, Clontech) according to the protocol provided with the reagents beginning with 10 ng of total RNA and using 9 cycles of PCR. 150 pg of the resulting cDNA were used for library preparation with the Illumina Nextera XT DNA Library reagents (Catalog Number 15032354, Illumina) using the single cell RNA-seq library preparation protocol developed for the Fluidigm C1 (Fluidigm).
TruSeq stranded mRNA-seq libraries were prepared for the bone marrow-derived NK cells. RNA-seq libraries were prepared using 400 ng of total RNA and the Illumina TruSeq Stranded mRNA reagents (Illumina) according to the protocol supplied with the reagents and starting with 100ng of total RNA on a Sciclone liquid handling robot (PerkinElmer) using a PerkinElmer-developed automated script.
Library sequencing cluster generation was performed with the resulting libraries using the Illumina TruSeq SR Cluster Kit v4 reagents and sequenced on the Illumina HiSeq 2500 using TruSeq SBS Kit v4 reagents. Sequencing data were processed using the Illumina Pipeline Software version 1.82.
RNA-sequencing data processing
RNA-sequencing reads were first trimmed to remove polyA and Illumina TruSeq adapter sequences using cutadapt60, then aligned to the mouse reference GRCm38 genome using the STAR aligner61. The number of counts were summarized at the gene level using featureCounts62. The R software (version 3.1.0) was then used for subsequent statistical analyses. Gene expression values were computed from the RPKM (reads per kilobase per million) values produced by the RPKM function of the edgeR R package by adding a pseudocount of 1 and log2-transforming the results. These RPKM values were used for drawing the heatmap with the pheatmap R package. Paired differential gene expression analyses were performed with the generalized linear modeling functions glmFit and glmLRT of the edgeR package using integer read counts as input63. Differentially expressed genes were ranked according to the P-values and the top 50 genes (corresponding to a FDR ≤ 0.00015) were selected for plotting.
JASPAR analysis
For transcription factor binding sites analysis, the JASPAR tool (http://jaspar.genereg.net/) was used. Promoter sequences were collected in NCBI (https://www.ncbi.nlm.nih.gov/gene) in order to span -700; +300 bp over the described transcription start site (TSS) of each gene. A relative profile score threshold of 80% was used to screen for similarities with the matrix models described in the JASPAR CORE database64. To generate the cluster = 0, which correspond to a control group of Rfx7 non-modulated genes, the list of genes obtained from RNA sequencing analysis was ranked based on the P-value and among the non-significant genes (average fold difference of one) alphabetically ordered, 16 consecutive genes in the list were selected.
Statistical analysis
Unless otherwise specified in the Figure legends, statistical differences were calculated using Prism software (GraphPad version 5.0) with an unpaired, two-tailed Student’s t test and were considered significant when P ≤ 0.05 (*), very significant when P ≤ 0.01 (**) and extremely significant when P ≤ 0.001 (***). Unless otherwise indicated, only differences statistically significant in comparison to the controls are shown; when two controls are used, only differences statistically significant in comparison to both control conditions are shown.
Supplementary Material
Acknowledgements
We thank M. Thome-Miazza and P. Schneider, UNIL, Lausanne, W. Reith, UNIGE Medical School, Geneva, M. Rüegg and M. Hall, Biozentrum, Basel, and M. Mazzone, VIB, Leuven, for sharing reagents important for this study. We thank N. Fonta, W. Held, R. Bedel, S. Siegert, S. Calderon and K. Harshman, Lausanne, and R. Hertzano, Baltimore, for technical help. Studies in the group of G.G. are funded by the European Research Council (ERC-2012-StG310890) and the Swiss National Science Foundation (PP00P3_139094 and PP00P3_165833). S.P.M.W. is supported by the ETH post-doctoral Fellowship program (FEL29 15-2) and the Horten Foundation. E.V. lab is supported by the ERC under the European Union's Horizon 2020 research and innovation programme (grant agreement 694502), Agence Nationale de la Recherche, Innate Pharma, MSDAvenir, Ligue Nationale contre le Cancer (Equipe labelisée "La Ligue") and Marseille-Immunopole. O.B. is supported by the Swiss National Science Foundation 310030-172978 and the Swiss Cancer Research KFS-4136-02-2017 and M.E.R. is supported by the KFS-MDPhD-3557-06-2015. M.D. was supported by the Fondation Medic, Lausanne. P.-C.H. is supported by Swiss National Science Foundation (31003A_163204), Swiss Cancer League (KFS-3949-08-2016) and Melanoma Research Alliance Young Investigator award.
Footnotes
Data availability.
The data that support the findings of this study are available from the corresponding author upon reasonable request. Source data from RNA-sequencing shown in Fig. 5 are deposited in the GEO repository (GSE113267).
Contributions
W.C., S.T.C., C.N., C.R.-B., S.P.M.W., K.O., H.W., G.R., and L.M., performed the experiments; D.B., S.T.C., and M.D. performed bioinformatics analyses; E.V., M.E.R., O.B., M.D., P.-C.H., and A.O. shared protocols, reagents, help, and advice; W.C., S.T.C., and G.G.; designed the research, analyzed the data, and wrote the manuscript.
Competing Interests
E.V. is cofounder, shareholder, and employee of Innate Pharma. Unrelated projects in G.G. laboratory are supported by OM-Pharma, Galenica, and Novartis Foundation. Three unrelated projects in P.-C.H. laboratory are supported by Roche and Idorsia. O.B. is a shareholder of Anaveon AG. An unrelated project in the M.D. group is supported by Merck KGaA and M.D. owns stocks from the companies Novartis, Roche, and Idorsia. The other authors have no conflicting financial interest to declare.
References
- 1.Emery P, et al. A consensus motif in the RFX DNA binding domain and binding domain mutants with altered specificity. Molecular and cellular biology. 1996;16:4486–4494. doi: 10.1128/mcb.16.8.4486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aftab S, Semenec L, Chu JS, Chen N. Identification and characterization of novel human tissue-specific RFX transcription factors. BMC evolutionary biology. 2008;8:226. doi: 10.1186/1471-2148-8-226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorn A, et al. Conserved major histocompatibility complex class II boxes--X and Y--are transcriptional control elements and specifically bind nuclear proteins. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:6249–6253. doi: 10.1073/pnas.84.17.6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Badis G, et al. Diversity and complexity in DNA recognition by transcription factors. Science. 2009;324:1720–1723. doi: 10.1126/science.1162327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung MI, et al. Coordinated genomic control of ciliogenesis and cell movement by RFX2. eLife. 2014;3:e01439. doi: 10.7554/eLife.01439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kistler WS, et al. RFX2 Is a Major Transcriptional Regulator of Spermiogenesis. PLoS genetics. 2015;11:e1005368. doi: 10.1371/journal.pgen.1005368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blackshear PJ, et al. Graded phenotypic response to partial and complete deficiency of a brain-specific transcript variant of the winged helix transcription factor RFX4. Development. 2003;130:4539–4552. doi: 10.1242/dev.00661. [DOI] [PubMed] [Google Scholar]
- 8.Ashique AM, et al. The Rfx4 transcription factor modulates Shh signaling by regional control of ciliogenesis. Science signaling. 2009;2:ra70. doi: 10.1126/scisignal.2000602. [DOI] [PubMed] [Google Scholar]
- 9.Bonnafe E, et al. The transcription factor RFX3 directs nodal cilium development and left-right asymmetry specification. Molecular and cellular biology. 2004;24:4417–4427. doi: 10.1128/MCB.24.10.4417-4427.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baas D, et al. A deficiency in RFX3 causes hydrocephalus associated with abnormal differentiation of ependymal cells. The European journal of neuroscience. 2006;24:1020–1030. doi: 10.1111/j.1460-9568.2006.05002.x. [DOI] [PubMed] [Google Scholar]
- 11.Ait-Lounis A, et al. Novel function of the ciliogenic transcription factor RFX3 in development of the endocrine pancreas. Diabetes. 2007;56:950–959. doi: 10.2337/db06-1187. [DOI] [PubMed] [Google Scholar]
- 12.Smith SB, et al. Rfx6 directs islet formation and insulin production in mice and humans. Nature. 2010;463:775–780. doi: 10.1038/nature08748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Reith W, Mach B. The bare lymphocyte syndrome and the regulation of MHC expression. Annu Rev Immunol. 2001;19:331–373. doi: 10.1146/annurev.immunol.19.1.331. [DOI] [PubMed] [Google Scholar]
- 14.Ludigs K, et al. NLRC5 Exclusively Transactivates MHC Class I and Related Genes through a Distinctive SXY Module. PLoS genetics. 2015;11:e1005088. doi: 10.1371/journal.pgen.1005088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manojlovic Z, Earwood R, Kato A, Stefanovic B, Kato Y. RFX7 is required for the formation of cilia in the neural tube. Mechanisms of development. 2014;132:28–37. doi: 10.1016/j.mod.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bullinger L, et al. Identification of acquired copy number alterations and uniparental disomies in cytogenetically normal acute myeloid leukemia using high-resolution single-nucleotide polymorphism analysis. Leukemia. 2010;24:438–449. doi: 10.1038/leu.2009.263. [DOI] [PubMed] [Google Scholar]
- 17.Crowther-Swanepoel D, et al. Common variants at 2q37.3, 8q24.21, 15q21.3 and 16q24.1 influence chronic lymphocytic leukemia risk. Nature genetics. 2010;42:132–136. doi: 10.1038/ng.510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Slager SL, et al. Common variation at 6p21.31 (BAK1) influences the risk of chronic lymphocytic leukemia. Blood. 2012;120:843–846. doi: 10.1182/blood-2012-03-413591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berndt SI, et al. Genome-wide association study identifies multiple risk loci for chronic lymphocytic leukemia. Nature genetics. 2013;45:868–876. doi: 10.1038/ng.2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shungin D, et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yau C, et al. A multigene predictor of metastatic outcome in early stage hormone receptor-negative and triple-negative breast cancer. Breast cancer research : BCR. 2010;12:R85. doi: 10.1186/bcr2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogers LM, Olivier AK, Meyerholz DK, Dupuy AJ. Adaptive immunity does not strongly suppress spontaneous tumors in a Sleeping Beauty model of cancer. Journal of immunology. 2013;190:4393–4399. doi: 10.4049/jimmunol.1203227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rusiniak ME, Kunnev D, Freeland A, Cady GK, Pruitt SC. Mcm2 deficiency results in short deletions allowing high resolution identification of genes contributing to lymphoblastic lymphoma. Oncogene. 2012;31:4034–4044. doi: 10.1038/onc.2011.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zerbino DR, et al. Ensembl 2018. Nucleic acids research. 2018;46:D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wu C, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome biology. 2009;10:R130. doi: 10.1186/gb-2009-10-11-r130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyman O, Kovar M, Rubinstein MP, Surh CD, Sprent J. Selective stimulation of T cell subsets with antibody-cytokine immune complexes. Science. 2006;311:1924–1927. doi: 10.1126/science.1122927. [DOI] [PubMed] [Google Scholar]
- 27.Rota G, et al. T Cell Priming by Activated Nlrc5-Deficient Dendritic Cells Is Unaffected despite Partially Reduced MHC Class I Levels. Journal of immunology. 2016 doi: 10.4049/jimmunol.1502084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang M, et al. NK cell development requires Tsc1-dependent negative regulation of IL-15-triggered mTORC1 activation. Nature communications. 2016;7 doi: 10.1038/ncomms12730. 12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai Q, et al. mTOR/Raptor signaling is critical for skeletogenesis in mice through the regulation of Runx2 expression. Cell death and differentiation. 2017;24:1886–1899. doi: 10.1038/cdd.2017.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Polak P, et al. Adipose-specific knockout of raptor results in lean mice with enhanced mitochondrial respiration. Cell metabolism. 2008;8:399–410. doi: 10.1016/j.cmet.2008.09.003. [DOI] [PubMed] [Google Scholar]
- 31.Inoki K, et al. mTORC1 activation in podocytes is a critical step in the development of diabetic nephropathy in mice. The Journal of clinical investigation. 2011;121:2181–2196. doi: 10.1172/JCI44771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sathe P, et al. Innate immunodeficiency following genetic ablation of Mcl1 in natural killer cells. Nature communications. 2014;5:4539. doi: 10.1038/ncomms5539. [DOI] [PubMed] [Google Scholar]
- 33.Doucey MA, et al. Cis association of Ly49A with MHC class I restricts natural killer cell inhibition. Nature immunology. 2004;5:328–336. doi: 10.1038/ni1043. [DOI] [PubMed] [Google Scholar]
- 34.The UniProt, C. UniProt: the universal protein knowledgebase. Nucleic acids research. 2017;45:D158–D169. doi: 10.1093/nar/gkw1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang K, Neale G, Green DR, He W, Chi H. The tumor suppressor Tsc1 enforces quiescence of naive T cells to promote immune homeostasis and function. Nature immunology. 2011;12:888–897. doi: 10.1038/ni.2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brugarolas J, et al. Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes & development. 2004;18:2893–2904. doi: 10.1101/gad.1256804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Boros K, Lacaud G, Kouskoff V. The transcription factor Mxd4 controls the proliferation of the first blood precursors at the onset of hematopoietic development in vitro. Experimental hematology. 2011;39:1090–1100. doi: 10.1016/j.exphem.2011.07.007. [DOI] [PubMed] [Google Scholar]
- 38.Hurlin PJ, et al. Mad3 and Mad4: novel Max-interacting transcriptional repressors that suppress c-myc dependent transformation and are expressed during neural and epidermal differentiation. The EMBO journal. 1995;14:5646–5659. doi: 10.1002/j.1460-2075.1995.tb00252.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wei H, et al. Cutting Edge: Foxp1 Controls Naive CD8+ T Cell Quiescence by Simultaneously Repressing Key Pathways in Cellular Metabolism and Cell Cycle Progression. Journal of immunology. 2016;196:3537–3541. doi: 10.4049/jimmunol.1501896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu Z, et al. PI3K is negatively regulated by PIK3IP1, a novel p110 interacting protein. Biochemical and biophysical research communications. 2007;358:66–72. doi: 10.1016/j.bbrc.2007.04.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lauth M, et al. DYRK1B-dependent autocrine-to-paracrine shift of Hedgehog signaling by mutant RAS. Nat Struct Mol Biol. 2010;17:718–725. doi: 10.1038/nsmb.1833. [DOI] [PubMed] [Google Scholar]
- 42.Keramati AR, et al. A form of the metabolic syndrome associated with mutations in DYRK1B. The New England journal of medicine. 2014;370:1909–1919. doi: 10.1056/NEJMoa1301824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Colpitts SL, et al. Transcriptional regulation of IL-15 expression during hematopoiesis. Journal of immunology. 2013;191:3017–3024. doi: 10.4049/jimmunol.1301389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Eckelhart E, et al. A novel Ncr1-Cre mouse reveals the essential role of STAT5 for NK-cell survival and development. Blood. 2011;117:1565–1573. doi: 10.1182/blood-2010-06-291633. [DOI] [PubMed] [Google Scholar]
- 45.Marcais A, et al. The metabolic checkpoint kinase mTOR is essential for IL-15 signaling during the development and activation of NK cells. Nature immunology. 2014;15:749–757. doi: 10.1038/ni.2936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Huntington ND, et al. Interleukin 15-mediated survival of natural killer cells is determined by interactions among Bim, Noxa and Mcl-1. Nature immunology. 2007;8:856–863. doi: 10.1038/ni1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Piccand J, et al. Rfx6 maintains the functional identity of adult pancreatic beta cells. Cell reports. 2014;9:2219–2232. doi: 10.1016/j.celrep.2014.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Deng Y, et al. Transcription factor Foxo1 is a negative regulator of natural killer cell maturation and function. Immunity. 2015;42:457–470. doi: 10.1016/j.immuni.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wang S, et al. FoxO1-mediated autophagy is required for NK cell development and innate immunity. Nature communications. 2016;7 doi: 10.1038/ncomms11023. 11023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng X, et al. Transcription factor Foxp1 exerts essential cell-intrinsic regulation of the quiescence of naive T cells. Nature immunology. 2011;12:544–550. doi: 10.1038/ni.2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Narni-Mancinelli E, et al. Fate mapping analysis of lymphoid cells expressing the NKp46 cell surface receptor. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:18324–18329. doi: 10.1073/pnas.1112064108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bentzinger CF, et al. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell metabolism. 2008;8:411–424. doi: 10.1016/j.cmet.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 53.Clausen BE, et al. Residual MHC class II expression on mature dendritic cells and activated B cells in RFX5-deficient mice. Immunity. 1998;8:143–155. doi: 10.1016/s1074-7613(00)80467-7. [DOI] [PubMed] [Google Scholar]
- 54.Ludigs K, et al. NLRC5 shields T lymphocytes from NK-cell-mediated elimination under inflammatory conditions. Nature communications. 2016;7 doi: 10.1038/ncomms10554. 10554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Held W, Lowin-Kropf B, Raulet DH. Generation of short-term murine natural killer cell clones to analyze Ly49 gene expression. Methods Mol Biol. 2000;121:5–12. doi: 10.1385/1-59259-044-6:5. [DOI] [PubMed] [Google Scholar]
- 56.Jordan S, et al. Virus progeny of murine cytomegalovirus bacterial artificial chromosome pSM3fr show reduced growth in salivary Glands due to a fixed mutation of MCK-2. Journal of virology. 2011;85:10346–10353. doi: 10.1128/JVI.00545-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Brune W, Hengel H, Koszinowski UH. A mouse model for cytomegalovirus infection. Curr Protoc Immunol. 2001;Chapter 19:Unit 19 17. doi: 10.1002/0471142735.im1907s43. [DOI] [PubMed] [Google Scholar]
- 58.Zurbach KA, Moghbeli T, Snyder CM. Resolving the titer of murine cytomegalovirus by plaque assay using the M2-10B4 cell line and a low viscosity overlay. Virol J. 2014;11:71. doi: 10.1186/1743-422X-11-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Masternak K, Peyraud N, Krawczyk M, Barras E, Reith W. Chromatin remodeling and extragenic transcription at the MHC class II locus control region. Nature immunology. 2003;4:132–137. doi: 10.1038/ni883. [DOI] [PubMed] [Google Scholar]
- 60.Martin M. Cutadapt Removes Adapter Sequences From High-Throughput Sequencing Reads. EMBnet.journal. 2011;17:10–12. [Google Scholar]
- 61.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 63.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wasserman WW, Sandelin A. Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet. 2004;5:276–287. doi: 10.1038/nrg1315. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.