Summary
In Rspondin-based three-dimensional cultures, Lgr5 stem cells from multiple organs form ever-expanding epithelial organoids that retain their tissue identity. Here we report the establishment of tumor organoid cultures from 20 consecutive colorectal carcinoma (CRC) patients. For most, organoids were also generated from adjacent normal tissue. Organoids closely resemble the original tumor. The spectrum of genetic changes within the 'living biobank' agrees well with previous large-scale mutational analyses of CRC. Gene expression analysis indicates that the major CRC molecular subtypes are represented. Tumor organoids are amenable to high-throughput drug screens allowing detection of gene-drug associations. As an example, a single organoid culture was exquisitely sensitive to Wnt secretion (porcupine) inhibitors and carried a mutation in the negative Wnt feedback regulator RNF43, rather than in APC. Organoid technology may fill the gap between cancer genetics and patient trials, complement cell line- and xenograft-based drug studies and allow personalized therapy design.
Introduction
Colorectal carcinoma (CRC) represents one of the major forms of cancer. Seminal studies have revealed a series of molecular pathways that are critical to the pathogenesis of CRC, including WNT, RAS-MAPK, PI3K, P53, TGF-β, and DNA mismatch repair (Fearon, 2011; Fearon and Vogelstein, 1990). Large-scale sequencing analyses have dramatically extended the list of recurrently mutated genes and chromosomal translocations (Garraway and Lander, 2013; Vogelstein et al., 2013). CRC cases are characterized by either microsatellite instability (MSI; associated with a hyper-mutator phenotype), or as microsatellite-stable (MSS) but chromosomally unstable (CIN) (Lengauer et al., 1997). The absolute number and combination of genetic alterations in CRC confounds our ability to unravel the functional contribution of each of these potential cancer genes. Thus, while genome changes in tumors of individual patients can be assessed in great detail and at low cost, these data are difficult to interpret in terms of prognosis, drug response or patient outcome, necessitating model systems for analysis of genotype-to-phenotype correlations.
Self-renewal of the intestinal epithelium is driven by Lgr5 stem cells located in crypts (Barker et al., 2007). We have recently developed a long-term culture system that maintains basic crypt physiology (Sato et al., 2009). Wnt signals are required for the maintenance of active crypt stem cells (Korinek et al., 1998; Kuhnert et al., 2004; Pinto et al., 2003). Indeed, the Wnt agonist R-spondin1 induces dramatic crypt hyperplasia in vivo (Kim et al., 2005). R-spondin-1 is the ligand for Lgr5 (Carmon et al., 2011; de Lau et al., 2011). Epidermal growth factor (EGF) signaling is associated with intestinal proliferation (Wong et al., 2012), while transgenic expression of Noggin induces a dramatic increase in crypt numbers (Haramis et al., 2004). The combination of R-spondin-1, EGF, and Noggin in Basement Membrane Extracts (BME) sustains ever-expanding small intestinal organoids, which display all hallmarks of the original tissue in terms of architecture, cell type composition, and self-renewal dynamics. We adapted the culture condition for long-term expansion of human colonic epithelium and primary colonic adenocarcinoma, by adding nicotinamide, A83-01 (Alk inhibitor), Prostaglandin E2 and the p38 inhibitor SB202190 (Sato et al., 2011). Of note, a two-dimensional culture method for cells from normal and malignant primary tissue has been described by Schlegel and colleagues (Liu et al., 2012).
Here, we explore organoid technology to routinely establish and phenotypically annotate ‘paired organoids’ derived from adjacent tumor and healthy epithelium from CRC patients.
Results
Establishment of a living CRC biobank
Surgically resected tissue was obtained from previously untreated CRC patients. Tissue from rectal cancer patients was excluded because they routinely undergo irradiation before surgery. For multiple tissues, we observe that normal tissue derived organoids outcompete tumor organoids under the optimized culture conditions, presumably due to genomic instability and resulting apoptosis in the latter. Combination of Wnt3A and the Wnt amplifier R-spondin1 is essential to grow organoids from normal epithelium. Over 90% of CRC cases harbor mutations that aberrantly activate the Wnt signaling pathway, so we exploited the Wnt-dependency of normal colonic stem cells to selectively expand tumor organoids (Cancer Genome Atlas Network, 2012). A total of 22 tumor organoid cultures and 19 normal-adjacent organoid cultures were derived from 20 patients (P19 and P24 each carried two primary tumors separated by >10 cm; Figure 1A). We successfully generated organoid cultures from 22 of 27 tumor samples. For one, we never observed growth. Four were lost due to bacterial/yeast infection. Since then we have added next-generation antibiotics (see methods), and currently observe a ~90% success rate.
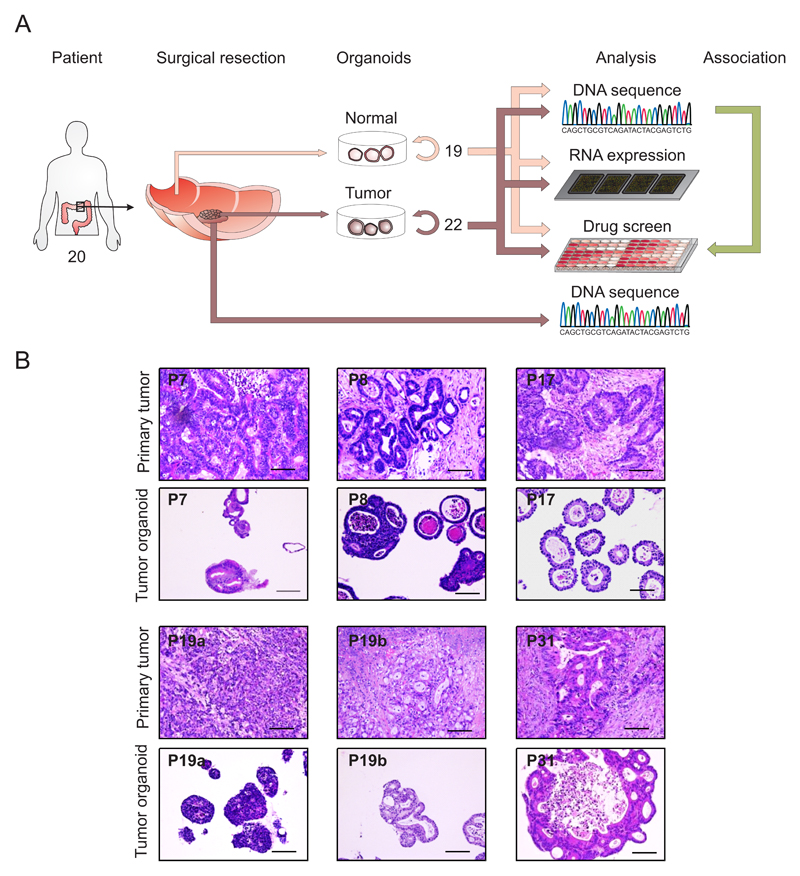
Figure 1. Derivation of organoids from primary tissue.
A) Overview of the procedure. A total of 22 tumor organoids and 19 normal control organoids were derived and analyzed by exome-sequencing, RNA expression analysis and high-throughput drug screening. To determine the concordance between tumor organoids and primary tumor, DNA from the primary tumor was also isolated.
B) Organoids architecture resembles primary tumor epithelium. H&E staining of primary tumor and the tumor organoids derived of these. A feature of most organoids is the presence of one or more lumens, resembling the tubular structures of the primary tumor (e.g. P8 and P19b). Tumors devoid of lumen give rise to compact organoids without lumen (P19a). Scale bar = 100 μM. See also Supplemental Data File S1
The number of primary tumor organoids varied between patient samples, with some tumors rendering thousands of primary organoids whereas others yielded only 10 – 20 primary organoids. This difference in derivation likely reflects the heterogeneous composition of tumors, with proliferative areas intermingled with regions of differentiated cells, stromal cells or necrosis. The growth rate of the organoids from patients 5 and 27 decreased over time, which prohibited their inclusion in the drug screen. All other organoids could be readily expanded and frozen to create a master cell bank. Upon thawing, cell survival was typically > 80%. Unlike healthy tissue derived organoids, tumor-derived organoids presented with a range of patient-specific morphologies, ranging from thin-walled cystic structures to compact organoids devoid of a lumen. H&E staining on primary tumors and the corresponding organoids revealed that the ‘cystic vs. solid’-organization of the epithelium was generally preserved. Yet marker expression analysis (KI67, OLFM4, KRT 20, Alcian blue) revealed heterogeneity both between patients and individual organoids within each culture (Figure 1B and Supplemental data file S1).
Genomic characterization of tumor-derived organoids
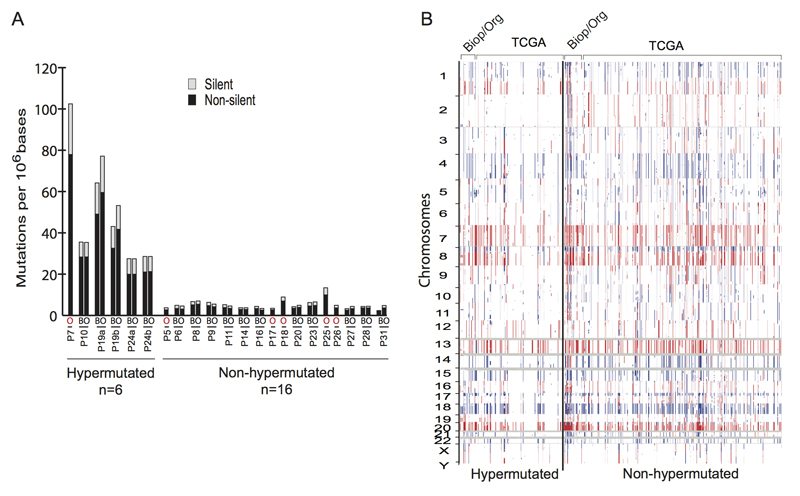
Genomic DNA was isolated from tumor and matched normal organoid cultures for whole-exome sequencing in order to identify tumor-specific somatic mutations (Cancer Genome Atlas Network, 2012). Genomic DNA from the corresponding biopsy specimens were available for comparative analysis for 16 of these cases (Table S1A). The mutation rates per Mb varied widely for different tumor organoids (range 2.0 to 77.9), with a median value of 3.7 in the tumor organoids, similar to the median rate of 3.6 in the biopsy samples (Figure 2A and Table S1B). Mutations were predominantly CpG to T transitions, consistent with results from large-scale CRC sequencing (Figure S1A-B and Table S1C). Of the 22 tumor organoids, 6 displayed hypermutation (>10 mutations/Mb): P7, P10 and the organoids from the two patients with two tumors each (P19a and –b, P24a and –b). Interestingly, the P19a and P19b tumors share TP53 R273C and BRAF V600E alterations, suggesting they arose from the same somatically altered progenitor cell but then diverged to acquire independent secondary alterations (Figure S1C and Figure S1D). In contrast, the P24a and P24b tumors share 80% (469/590) of somatic alterations but then have discordant driving alterations in APC and TP53, indicating that the hypermutator phenotype may have been present prior to the acquisition of growth promoting mutations (Figure S1E and Figure S1F). The frequency of hypermutated organoid cultures in our patient panel (20%; 4 of 20), agreed with the reported frequency in a much larger cohort of clinical samples and display comparable somatic copy number alterations (SCNAs) (Figure 2B and Table S1D) (Bass et al., 2011; Cancer Genome Atlas Network, 2012). The successful derivation of both hypermutated and non-hypermutated organoids implies an absence of culture-based bias.
Figure 2. CRC subtypes are present in organoid cultures.
A) Whole exome sequencing of the tumor and corresponding biopsy, when available, revealed the presence of hypermutated (>10mutations/Mb) and non-hypermutated subtypes within the organoids. Comparable rates of mutations were observed in the tumor organoid (O) and tumor biopsy (B). Organoids without corresponding biopsy are indicated in with red (O).
B) Comparison of somatic copy number alterations found in the biopsies and corresponding organoids (Biop/Org) and TCGA CRC in hypermutated and non-hypermutated samples.
Somatic variants within the coding regions in organoid cultures were highly concordant with the corresponding biopsy specimen for both hypermutated and non-hypermutated patients (median = 0.88 frequency of concordance, range 0.62 to1.00) (Figure 3A and Table S1E). Indeed, combined analysis of SCNAs and SNVs to infer Cancer Cell Fractions (CCF) (Carter et al., 2012; Landau et al., 2013) in the biopsy and tumor organoids, revealed that the common CRC driver mutations were maintained in culture. In 13 out of 14 organoid-biopsy pairs tested, tumor subclones sharing common CRC drivers were detected in the biopsy. In 50% of the organoids, a dominant subclone from the biopsy was present, likely representing sampling during derivation but it could also indicate loss in culture (Figure S2A-B, Table S1F and Table S1G). Transcriptome analysis of single organoids showed subtle differences in gene expression within an organoid culture, confirming their heterogeneous composition. The differences in overall gene expression were more pronounced in the organoids derived from the hypermutant tumors. (Figure S2C).
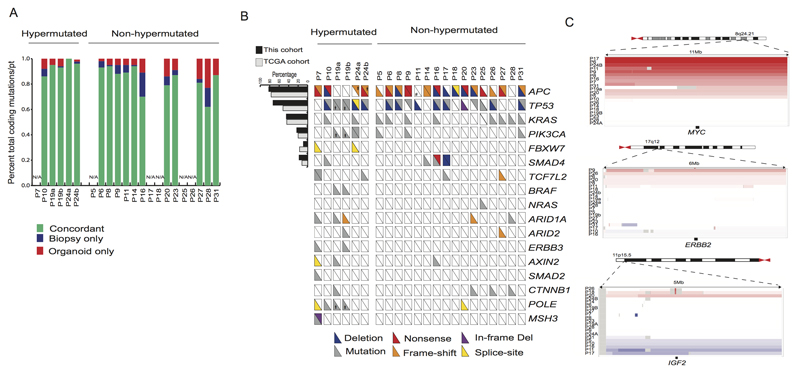
Figure 3. Genomic alterations found in CRC are represented in organoid cultures.
A) Concordance of somatic mutations detected in organoid and corresponding biopsies. Bar graph represents the proportion of coding alterations that are concordant between the biopsy and the corresponding organoid culture, and those that are found only in organoid or biopsy specimen. N/A indicates cases in which exome-sequencing was not performed on the corresponding biopsy.
B) Overview of the mutations found in the tumor organoids. The hash-mark in each box represents each allele and whether it was subject to deletion, mutation, frame-shift alteration, nonsense mutation or splice site mutation. Those alterations present in greater than 10% of cases are compared to the percentage of cases reported by the TCGA CRC. * indicates discordant mutations targeting the same gene between the two sites in P19 and P24. See also Tables S1I-J.
C) Somatic copy number alterations in organoids amongst commonly amplified genes identified in TCGA CRC.
Discordant mutations were assessed for their likely biological significance in cancer, based on Cancer Gene Census and data reported from the PanCancer analysis of 5000 whole exomes (Futreal et al., 2004; Lawrence et al., 2014). Only 4% (27/679) of discordant mutations found in organoids affected cancer-related genes, including a third hit to APC, which was already biallelically inactivated in P14, SMAD4 mutation in P16, and POLE mutation in P19b (Table S1H). Cancer-significant genes that were discordant in the biopsy represented 4.4% (12/271) (Table S1H). The discordant mutations had a mean allelic frequency of 10.3% and 34.1% for the biopsy and organoids, respectively. This could represent the enrichment or depletion of a sub-clonal population in the organoid culture present within the original tumor, as well as acquisition of additional mutations during derivation or propagation.
The most commonly altered genes in CRC (Bass et al., 2011; Cancer Genome Atlas Network, 2012; Lawrence et al., 2014) were well represented in the organoid cultures (Figure 3B and Tables S1I-J). Inactivating alterations to the tumor suppressors APC, TP53, FBXW7 and SMAD4, as well as activating mutations in KRAS (codon 12 and 146) and PIK3CA (codon 545 and 1047) were observed. Activating mutations in BRAF and TGFBR1/2 mutations were observed in the hypermutated organoids, consistent with previous reports for primary CRC (Cancer Genome Atlas Network, 2012).
Mutations of genes in DNA mismatch repair (MMR)-associated pathways are associated with a hypermutated phenotype (Boland and Goel, 2010). Consistent with their classification as hypermutated CRC cases (Cancer Genome Atlas Network, 2012), missense mutations were present in MSH3 in P7, and POLE mutations were detected in P10 and P19a/b. We did not observe mutations in MMR-associated genes in P24a and P24b and expression analysis showed normal levels of the pertinent genes. The culprit for hyper mutability thus remains to be identified for P24. The limited cohort size did not allow a statistical analysis for somatic copy number alterations to identify significant regions of amplification and deletions. However, manual inspection of the top regions identified by TCGA did reveal the presence of ERBB2-, MYC- and IGF2-amplified organoids, as well as a reported gain of 13q in the non-hypermutated group (Figure 3C) In aggregate, these analyses demonstrate that organoid cultures faithfully capture the genomic features of the primary tumor from which they derive and much of the genomic diversity of CRC.
Most CRC cases carry activating mutations in the WNT pathway: inactivation mutations in APC, FBXW7, AXIN2, and FAM123B, or activating mutations in CTNNB1 (Cancer Genome Atlas Network, 2012). Gene fusions involving the Wnt-agonistic RSPO2 and RSPO3 genes have been observed in 5-10% of CRC (Seshagiri et al., 2012). RNF43 encodes a negative regulator of the Wnt pathway, which serves to remove the Wnt receptor FZ in a negative feedback loop (Hao et al., 2012; Koo et al., 2012, de Lau et al., 2014). Recent sequencing efforts of gastric, ovary and pancreatic neoplasias identified RNF43 mutations (Jiao et al., 2014; Ryland et al., 2013; Wang et al., 2014), and RNF43 mutations have been observed in CRC (Giannakis et al., 2014; Ivanov et al., 2006; Koo et al., 2012)
We found APC alterations in all but four of the organoids (P11, P19a/b, P28). Western blotting revealed P11 to express a truncated APC protein, pointing to a mutational event not covered by our exome-sequencing (Figure S3). The wtAPC organoid P28 carries an activating mutation in CTNNB1 (T41A). In both P19a and P19b, we detected RNF43 mutations: frameshifts at aa positions 659 and 355 respectively. Only the latter is predicted to affect protein function.
RNA Analysis of normal and tumor-derived organoids
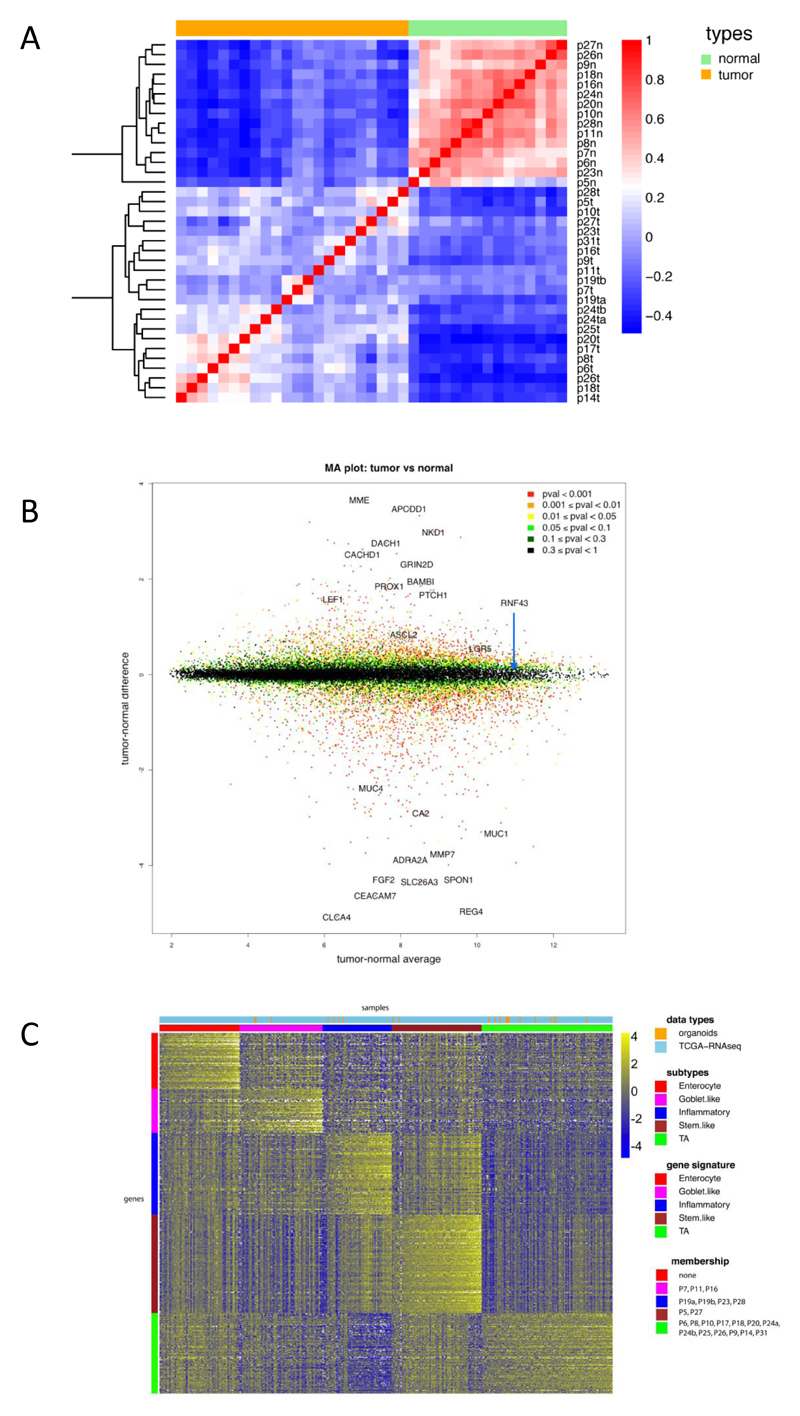
Organoid cultures consist purely of epithelial cells. Therefore, the system allows for direct gene expression analysis without a contamination from mesenchyme, blood vessels, immune cells etc. Normal colon derived and tumor-derived organoids were plated under identical conditions in complete medium (+Wnt). After three days, RNA was analysed using Affymetrix single transcript arrays. Figure 4A shows the correlation heat-map of the organoid samples. Normal colon derived organoids clustered tightly together, while the tumor-derived organoids exhibited much more heterogeneity. Next, we searched for genes differentially expressed between normal and tumor organoids. Normal colon derived organoids (Figure 4B) expressed genes of differentiated cells (e.g. the goblet cell markers MUC1 and MUC4 and the colonocyte marker CA2). Genes enriched in tumor organoids included cancer-associated genes such as PROX1, BAMBI and PTCH1, and the Wnt target gene APCDD1 (Takahashi et al., 2002).
Figure 4. RNA expression analysis.
A) Correlation heat map of normal organoids versus tumor organoids based on 2186 genes (the top 10% of genes in terms of standard deviation). The normal organoids are very highly correlated with each other, whereas the tumor samples exhibit more heterogeneity. The colors represent pairwise Pearson correlations after the expression values have been logged and mean-centered for every gene. The hierarchical clustering is based on one minus correlation distance. The affix N = normal, T = tumor.
B) MA plot of logged normal versus tumor gene expression. P-values are computed with the R package limma, by comparing normal versus tumor gene expression. Cancer associated genes, e.g. APCDD1, PROX1 and PTCH1 are shown in the top half.
C) CRC molecular subtypes are represented by the organoid panel. Genes by samples heat map of normalized gene expression of 22 organoid samples and 431 TCGA RNA-seq tumor tissue samples, organized by subtype. Within each subtype, samples are sorted by their mean gene expression for the signature genes associated with that specific subtype.
Several CRC classifications have been proposed based on RNA expression. We combined expression data from organoid samples and TCGA tissue samples and classified these in subtypes using the gene signatures by Sadanandam (Sadanandam et al., 2013). Figure 4C displays the subtyping of the 22 organoid samples and 431 TCGA RNA-seq tumor tissue samples. The heat map shows the normalized scores of genes by samples, both sorted by subtype (see Methods). Organoid samples were spread across the subtypes, with the transit-amplifying (TA) subtype being most frequently represented. The enterocyte subtype was not represented. In addition, the RNA expression data allowed expression analysis of individual genes in organoids. MLH1 expression was absent from two tumor organoids from patient 19 as well as from patient 7 (that is also mutant in MSH3) (Figure S4). In the two tumor organoids from P24, we did not detect expression changes in MLH1 or any other MSI-associated gene.
Effect of Porcupine inhibitor on RNF43 mutant organoids
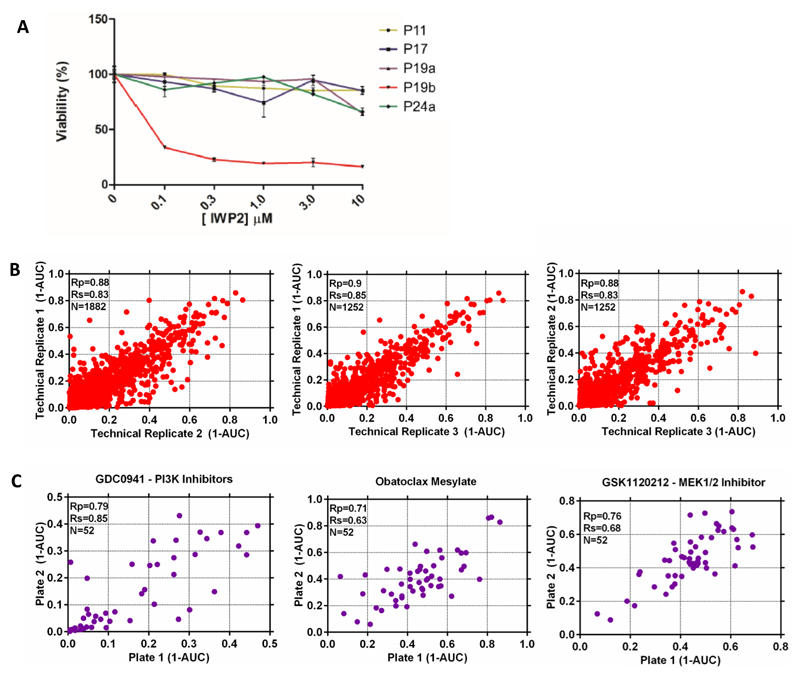
Unlike most other WNT pathway mutations, RNF43 mutations yield a cell that is hypersensitive to -yet still dependent on- secreted WNT. Array data confirmed the expression of several WNTs by the organoids (Figure S5A). The O-acyltransferase Porcupine is required for the secretion of WNTs and its inhibition prevents autocrine/paracrine activation of the pathway (Kadowaki et al., 1996). The small molecule porcupine inhibitor IWP2 (Chen et al., 2009) was tested on a small panel of the tumor organoids and strongly affected the RNF43-mutant P19b organoid (Figure 5A). This observation implied that porcupine inhibition may be evaluated for treatment of the small subset of cancer patients mutant in RNF43.
Figure 5. Development of a high-throughput drug screening assay utilizing organoid models.
A) Autocrine/paracrine WNT signaling in P19b. A small panel of tumor organoids was incubated with increasing amounts of the Porcupine inhibitor IWP2. Growth of the RNF43 mutant P19b was inhibited, indicative of dependency on autocrine/paracrine WNT signaling. See also Figure S5
B) Scatterplot of (1-AUC) values for all technical replicates of drug screening data. Plots show the correlation between the 3 different technical replicates and each data point represents the (1-AUC) value for an individual organoid.
C) Scatterplots of the correlation in (1-AUC) values for 3 compounds (GDC0941, obatoclax mesylate and trametinib) screened twice during every screening run. Values are the mean of 3 technical replicates.
Organoid proof-of-concept drug screen
Prompted by this, we developed a robotized drug sensitivity screen in 3D-organoid culture and correlated drug sensitivity with genomic features to identify molecular signatures associated with altered drug response. Organoid cultures were gently disrupted and plated on BME-coated 384-well plates in a 2% BME solution. Organoids were left overnight before being drugged and left for 6 days before measuring cell number using CellTitre-Glo® reagent. Drug sensitivity was represented by the half-maximal inhibitory concentration (IC50), the slope of the dose response curve, and area under the dose response curve (AUC).
A bespoke 83 compound library was assembled for screening, including drugs in clinical use (n = 25), chemotherapeutics (n = 10), drugs previously investigated in or currently undergoing studies in clinical trials (n = 29), and experimental compounds to a diverse range of cancer targets (n = 29) (Table S2A). The library included the anti-EGFR antibody cetuximab, used clinically for KRAS/NRAS/BRAF wild-type CRC, as well as oxaliplatin and 5-FU, first line chemotherapeutics for CRC treatment. In total, 19 of 20 tumor organoids (from 18 different patients) were successfully screened in experimental triplicate, generating >5000 measurements of organoid-drug interactions (Table S2B).
We incorporated a number of controls into the assay design. The median Z-factor score, a measure assay plate quality, across all screening plates was 0.62 (n = 119; upper and lower quartile = 0.85 and 0.3 respectively), consistent with an experimentally robust assay. We did observe some unexplained organoid-specific variation in assay plate quality. Dose response measurements were performed in experimental triplicate or duplicate (on separate plates) and replicate AUC values were highly correlated (Pearson correlation (Rp) > 0.87) (Figure 5B). Furthermore, the compounds trametinib, GDC0941 and obatoclax mesylate were screened twice independently on separate assay plates and a good correlation was observed between the experimentally determined AUC values (Rp = 0.79, 0.71 and 0.76, respectively) (Figure 5C).
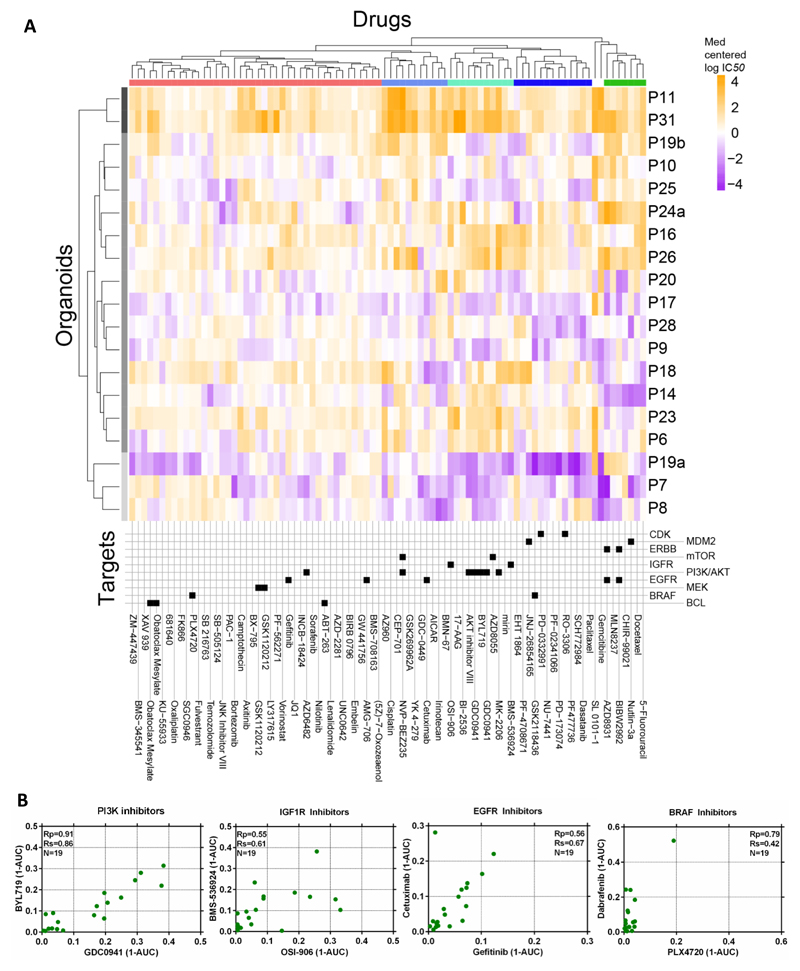
As a first validation, the only tumor organoid in the panel that was sensitive to the Porcupine inhibitor LGK974 was P19b (Figure S5B), confirming the observations made with IWP2 (Figure 5A). The clustering of compounds based on their IC50 values demonstrated a diverse range of sensitivities across the organoids and identified 3 major sub-groups (Figure 6A). One group was associated with sensitivity to a majority of the compounds (organoids P8, P7 and P19a), in contrast to the cluster (P31, P11) exhibiting insensitivity. The remaining organoids had intermediate sensitivity. Interestingly, the multifocal tumors P19a and P19b, derived from the same patient and both carrying the BRAF V600E mutation, differed in their overall drug response profile. We observed clustering of drugs that inhibit the IGF1R and PI3K-AKT signaling pathways (Figure 6A), and compounds with similar nominal targets had comparable activity across the organoid collection. For example, a similar sensitivity pattern was observed for the PI3K inhibitors GDC0941 and BYL719 (α-selective), the IGF1R inhibitors OSI-906 and BMS-536924, EGFR inhibitors cetuximab and gefitinib, and the BRAF inhibitors dabrafenib and PLX4720 (Figure 6B). All but one of the organoids displayed a lack of sensitivity to BRAF inhibition. P19a, a BRAF V600E mutant organoid, displayed partial sensitivity to dabrafenib with an IC50 of 0.5 μM, comparable to IC50 values of BRAF V600E colorectal cancer cell lines (range 0.004-2.55 μM; average 0.96 μM).
Figure 6. Heatmap of IC50’s of all 85 compounds against 19 colorectal cancer organoids.
A) Organoids have been clustered based on their IC50 values across the drug panel. The drug names and their nominal target(s) are provided in the bottom panel.
B) Drugs with the same nominal targets have similar activity profiles across the organoid panel. (1-AUC) values are plotted for inhibitor of PI3K (GDC0941 and BYL719), IGF1R (OSI-906 and BMS-536924), EGFR (cetuximab and gefitinib) and BRAF (PLX4720 and dabrafenib).
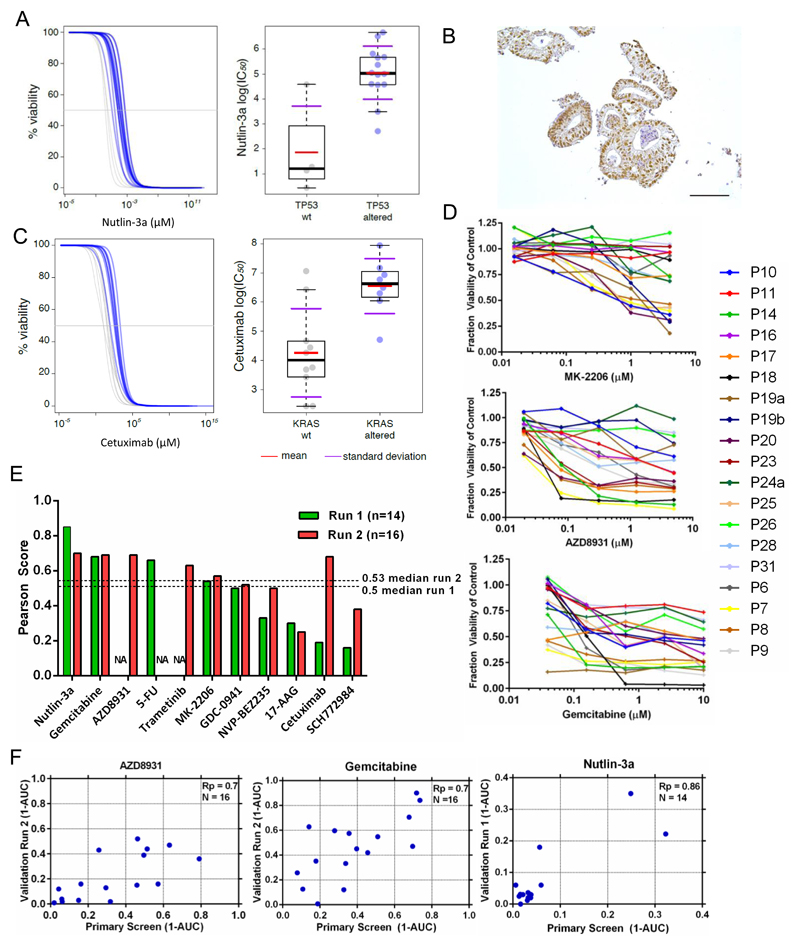
To identify genetic correlates between individual oncogenic mutations and drug response, we performed a multivariate analysis of variance (MANOVA) incorporating IC50 values and slopes of the corresponding dose response curves, with MSI-status as a covariate. Complete drug sensitivity and genomic datasets were available for 18 organoids and used for this analysis. The analysis included 16 genes identified as mutated, amplified or deleted in CRC (referred to as mutant genes) as described by Lawrence et al., 2014 (Table S3). The MANOVA identified a subset (12 of 864, ~1%) of gene-drug associations as statistically significant (p<0.005, incorporating a 30% false discovery rate (FDR)) (Table S4). These results were further filtered based on the magnitude of the effect size on the IC50 values of wild-type versus mutant cell line populations (effect size > 2; Cohen’s D), and correlations identified due to a singlet outlier organoids were removed. This resulted in the identification of one high confidence gene-drug association already reported in the literature (Vassilev et al., 2004). Loss of function mutations of the tumor suppressor TP53 were associated with resistance to nutlin-3a (p = 0.0018), an inhibitor of MDM2 (Figure 7A). Of the 4 organoids that were wild-type for TP53 by DNA sequencing, only P18 was (unexpectedly) insensitive to nutlin-3a. However, immunohistochemistry of p53 in P18 revealed the protein to be stabilized, indicative of functional inactivation of the p53 pathway (Figure 7B).
Figure 7. Gene-drug associations and differential drug sensitivity profiles of interest.
(A) Association of TP53 mutational status with nutlin-3a response. Viability response curves of the altered (blue) and wild-type organoids (grey) as well as scatter plots of cell line IC50 (μM) values. IC50 values are on a log scale comparing TP53 mutant and wild type (WT) cell lines. Each circle represents the IC50 of one cell line and the red bar is the geometric mean.
(B) Immunohistochemical staining showing stabilization of TP53 in organoid P18 (Scale bar = 100 μM).
(C) Association of KRAS status and cetuximab response.
(D) Dose-response curves after 6 days treatment with MK2206, AZD8931 and gemcitabine. Error bars are the standard deviation of triplicate measurements.
(E) Reproducibility of drug response profiles for 11 drugs. The Pearson correlation score of (1-AUC) values from the primary screen compared to (1-AUC) values from validation screens are used for comparison. The validation screen was performed twice (run 1 and 2) with >1 month elapsed between each screen. NA - data unavailable for this drug.
(F) The correlation of 1-AUC values from the primary and validation screens for AZD8931, gemcitabine and nutlin-3a.
We could also readily detect resistance to the anti-EGFR inhibitors cetuximab and BIBW2992 (afatinib) in the setting of KRAS mutant organoids (p = 0.008/FDR 37% and p=0.029/FDR 54%, respectively), although these associations were below statistical significance when considering an FDR < 30% (Figure 7C and Figure S6). Of the KRAS wild-type organoids, a subset (2/11) were insensitive to cetuximab, including P19b which has a BRAF mutation, a known mediator of cetuximab resistance (Di Nicolantonio et al., 2008). For the remaining organoids, further mechanisms beyond mutated KRAS/NRAS/BRAF are likely to be involved in cetuximab resistance (De Roock et al., 2010; Vecchione, 2014).
We also identified a number of compounds with differential activity in the absence of an apparent genetic biomarker (Figure 7D). For example, a subset of organoids were exquisitely sensitive to the AKT1/2 inhibitor MK2206. Similarly, we observed distinct subsets of organoids which are exquisitely sensitive to the pan-ERBB inhibitor AZD8931 and the chemotherapeutic gemcitabine. We also performed a validation screen with 11 of the original 83 compounds across the organoid panel, and compared the measured responses (Table S5 and Figure S7). We observed positive correlation for all compounds and 9 exhibited good to fair reproducibility as indicated by an Rp of 0.5 or greater (Figure 7E and Figure 7F). Variation within the assay was likely due to inherent technical noise, biological variation, and sensitivity to outlier data points due to the small number of organoids.
In summary, the successful application of organoids in a systematic and unbiased high-throughput drug screen to identify clinically relevant biomarkers demonstrates the feasibility and utility of organoid technology for investigating the molecular basis of drug response. Furthermore, the identification of putative novel molecular markers has opened new avenues for further investigation of drug sensitivity in CRC. The current analysis is still constrained by the relatively small number of patients. The derivation of a significantly larger organoid collection would increase the representation of rare genotypes and the statistical power to detect molecular markers of drug response.
Discussion
Cancer cell lines have served for many years as the workhorse model in cancer research. Recent studies have exploited high-throughput screening of large panels of cancer cell lines to identify drug-sensitivity patterns and to correlate drug sensitivity to genomic alterations (Barretina et al., 2012; Garnett et al., 2012). From these high-throughput cell line-based studies, a picture emerges of a complex network of biological factors that affect sensitivity to the majority of cancer drugs. For instance, no direct relationship may exist between sensitivity to a certain drug and a single genomic alteration. Instead, difficult-to-find, complex interactions between multiple genomic alterations may determine drug sensitivity outcome. Thus, with currently available insights, it remains a challenge to develop algorithms that accurately predict the drug sensitivity of a patient's tumor based on the spectrum of genomic alterations present, in the context of the unique genetic background.
Two approaches to determine, in a patient-derived sample, directly the drug sensitivity have been quite widely exploited, namely the short-term culture of tumor sections (Centenera et al., 2013), and xeno-transplantation of the tumor into immunodeficient mice (Jin et al., 2010; Tentler et al., 2012). Short-term culture allows for in vitro screening at a reasonably large scale, but is constrained by the limited proliferative capacity of the cultures. Xenotransplantation allows for in vivo screening but is resource-intensive due to the need for large mouse colonies. It thus appears of interest to develop additional technologies that allow the combination of sequencing and high-throughput drug screening in patient-derived samples. Here, we demonstrate that the organoid culture platform can be exploited for genomic and functional studies at the level of the individual patient at a scale that cannot be achieved by existing approaches. Our organoid drug screening assay generates reproducible high quality drug sensitivity data, positive correlation of biological replicates and reproducible activity of compounds inhibiting the same target. By connecting genetic and drug sensitivity data, we were able to confirm the activity of cetuximab in a subset of KRAS wild-type organoids reflecting observations made in the clinic (De Roock et al., 2010) as well as Nutlin-3a effectiveness in TP53 wild-type organoids. Furthermore, we describe the differential activity of a handful of clinical and preclinical compounds (gemcitabine, MK2206 and AZD8941).
Tumors are composed of a mixture of sub-clones that coevolve through a Darwinian selection process. This cellular heterogeneity and phenotypic variation allows the emergence of a complex clonal architecture, which underpins important features such as drug resistance and metastatic potential (Burrell et al., 2013). Our CCF analysis of clonal structure determined that almost all of the biopsies were polyclonal at the time of resection, and this is reflected to varying extent in the corresponding organoid culture. The ability to capture sub-clonal populations in in vitro organoid culture should enable more predictive modeling of patient responses to therapy. In many respects, the clonal selection and heterogeneity observed in organoids is similar to PDX models of cancers (Eirew et al., 2014). For both models, understanding the factors that affect tumor heterogeneity and evolution, and how heterogeneity impacts on drug response, will be important to fully exploit their potential for predicting patient responses.
We perceive patient-derived organoids to be used to directly test drug sensitivity of the tumor in a personalized treatment approach. For this, we envision organoids to be tested against a limited number of clinically approved drugs within weeks after derivation. While building this pilot biobank, we observed that normal epithelial tissue always yield good numbers of organoids within weeks, while significant differences in 'take rates' were observed between patients' tumor organoids. Crucial for this approach to be effective, is to decrease the time needed to derive and expand the organoids. In conclusion, tumor organoids may fill the gap between cancer genetics and patient trials, complement cell line- and xenograft-based drug studies and allow personalized therapy design.
Experimental procedures
Human Tissues
Colonic tissues were obtained from The Diakonessen Hospital Utrecht with informed consent and the study was approved by the ethical committee. All patients were diagnosed with colorectal cancer. From the resected colon segment, normal as well as tumor tissue was isolated. The isolation of healthy crypts and tumor epithelium was performed essentially as described by Sato et al. (Sato et al., 2011).
Organoid culture
Healthy tissue derived organoids were cultured in Human Intestinal Stem Cell medium (HISC). The composition of HISC is: Basal culture medium with 50% Wnt conditioned medium, 20% R-Spondin conditioned medium, 10% Noggin conditioned medium, 1 x B27, 1,25 mM n-Acetyl Cysteine, 10 mM Nicotinamide, 50 ng/ml human EGF, 10 nM Gastrin, 500 nM A83-01, 3 uM SB202190, 10 nM Prostaglandine E2 and 100 μg/ml Primocin (Vivogen). Tumor organoids were cultured in HICS minus Wnt. See the extended experimental procedures section for a detailed description.
Whole-exome sequencing and copy number analysis
For each sample, approximately 250 ng of DNA was sheared and subject to whole-exome sequencing using the Agilent v2 capture probe set and sequenced by HiSeq2500 using 76 base pair reads, as previously described (Fisher et al., 2011; Imielinski et al., 2012). A median 9.6 Gb of unique sequence was generated for each sample (Table S1A).
Sequence data was locally realigned to improve sensitivity and reduce alignment artifacts prior to identification of mutations, insertions and deletions as previously described (Cibulskis et al., 2013; DePristo et al., 2011; Ojesina et al., 2014).
Somatic copy number analysis was performed using segmented copy number profiles generated from whole-exome sequencing using the SegSeq algorithm (Table S1D) (Chiang et al., 2009). The procedure is described in detail in the extended experimental procedures section.
Organoid data processing
RNA from 22 organoid tumor samples and 15 paired normal samples was hybridized on Affymetrix Human Gene 2.0 ST arrays. The raw CEL files were processed with Affymetrix Power Tools using the Hg19 genome build and NetAffx annotation dating from 09-30-2012. Between-array normalisation was performed using rma-sketch, within APT. This resulted in an intensity matrix of 21681 genes by 37 samples. For analysis of individual genes, data were analyzed using the R2 web application, which is freely available at http://r2.amc.nl.
To subtype the samples, we used the gene signature published by Sadanandam et al. (Sadanandam et al., 2013). The procedure is described in detail in the extended experimental procedures section.
Organoid viability assays
8μl of ~7mg/ml BME was dispensed in to 384-well microplates and allowed to polymerize. Organoids were mechanically dissociated by pipetting before being resuspended in 2% BME/growth media (15-20,000 organoids/ml) and dispensed into drug wells. The following day a 5-point 4-fold dilution series of each compound was dispensed using liquid handling robotics and cell viability assayed using CellTiter-Glo® (Promega) following 6 days of drug incubation. All screening plates were subjected to stringent quality control measures and a Z-factor score comparing negative and positive control wells calculated. Dose–response curves were fitted to the luminescent signal intensities utilizing a method previously described (Garnett et al., 2012). Further information of the compounds used, data-fitting algorithm and validation screen can be found in the Supplementary Methods
Systematic Multivariate Analysis of Variance (MANOVA)
We excluded from the analysis drugs with no IC50 values falling within the range of tested concentrations. For each of the remaining drugs, we assembled an 18 × 2 matrix Y composed by two vectors of length n=18, containing IC50 values and dose-response curve slopes β respectively, obtained by treating 18 organoids with the drug under consideration. A multivariate analysis of variance (MANOVA) model was then fitted to this drug response data matrix with factors including the microsatellite stability status of the organoids and the status (altered or wild-type) of 16 genomic features (Supplementary Methods). Significance and effect size scores were obtained for each of the genomic-feature/drug pairs. Q-values were subsequently obtained by correcting the MANOVA p-values for multiple hypotheses testing, and a threshold of 30% of positive false discovery rate, IC50 and effect size > 2 (as quantified by the Cohen’s D) was used to identify significant associations.
Supplementary Material
Acknowledgements
Thanks to the Broad Institute Genomics Platform for processing and generating the sequence data, the cell line screening and analysis team at the Sanger Institute, Liliane Wijnaendts and Joost Oudejans for help with tumor dissections, Harry Begthel and Jeroen Korving for histology, Sepideh Darakhshan for help with organoids and the members of the contributing labs for support. Marc van de Wetering is supported by Stichting Virtutis Opus and Stichting Vrienden van het Hubrecht. Robert G.J. Vries, Sridevi Jaksani and Joyce Blokker are supported by a Grant from Alpe dHuzes/KWF. This work was supported with a grant from the Dutch Cancer Society to Michael Stratton (H1/2014-6919). The organoids are available for academic research upon evaluation of a research proposal by the Medical Ethical Comity. M. Meyerson reports receiving a commercial research grant from Bayer and has ownership interest (including patents) in and is a consultant/advisory board member for Foundation Medicine.
Footnotes
Accession numbers
The healthy and tumor organoid array data are available under GEO number GSE64392. The GEO number for the single organoid RNA-seq data is GSE65253.
Author Contributions
M.v.d.W derived, maintained and analyzed organoid cultures. H.E.F developed, performed and analyzed the organoid drug screen. J.M.F analyzed sequencing data. G.B analyzed RNA expression data. R.G.J.V organized ethical approval. F.I. performed analyses and statistical inferences on the drug screening data supervised by J.SR.. A.P and W.v.H. performed surgery. J.v.G. isolated tumor and normal tissue from resected material. A.T.M. performed cancer cell fraction analysis, L.K. performed single organoid transcriptomics supervised by A.v.O., A.M assisted in drug screening. J.B. performed immunostainings and assisted in culturing organoids together with S.J. and S.B.. P.v.d.S. and R.V. processed and analyzed RNA, supervised by R.V.. V.S.W.L. performed APC western analysis. S.S processed DNA samples for exome sequencing. C.S.P., K.C., S.L.C., A.M., M.S.L., L.L. and C.S. helped in processing and analyzing sequencing data. G.G and M.M. supervised the sequencing and analysis. L.W. supervised the RNA analysis. M.R.S., U.M., M.G. and H.C. participated in the development of the project concept. M.G. aided in drug data analysis. M.v.d.W. and H.E.F. participated in data analysis and project design. M.v.d.W., H.E.F., J.M.F., M.M., M.J.G. and H.C. wrote the manuscript.
References
- Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, Haegebarth A, Korving J, Begthel H, Peters PJ, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass AJ, Lawrence MS, Brace LE, Ramos AH, Drier Y, Cibulskis K, Sougnez C, Voet D, Saksena G, Sivachenko A, et al. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet. 2011;43:964–968. doi: 10.1038/ng.936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boland CR, Goel A. Microsatellite Instability in Colorectal Cancer. Gastroenterology. 2010;138:2073–2087.e3. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burrell RA, McGranahan N, Bartek J, Swanton C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature. 2013;501:338–345. doi: 10.1038/nature12625. [DOI] [PubMed] [Google Scholar]
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487:330–337. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmon KS, Gong X, Lin Q, Thomas A, Liu Q. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/beta-catenin signaling. Proc Natl Acad Sci U S A. 2011;108:11452–11457. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter SL, Cibulskis K, Helman E, McKenna A, Shen H, Zack T, Laird PW, Onofrio RC, Winckler W, Weir BA, et al. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30:413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Centenera MM, Raj GV, Knudsen KE, Tilley WD, Butler LM. Ex vivo culture of human prostate tissue and drug development. Nat Rev Urol. 2013;10:483–487. doi: 10.1038/nrurol.2013.126. [DOI] [PubMed] [Google Scholar]
- Chen B, Dodge ME, Tang W, Lu J, Ma Z, Fan C-W, Wei S, Hao W, Kilgore J, Williams NS, et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5:100–107. doi: 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang DY, Getz G, Jaffe DB, O’Kelly MJT, Zhao X, Carter SL, Russ C, Nusbaum C, Meyerson M, Lander ES. High-resolution mapping of copy-number alterations with massively parallel sequencing. Nat Methods. 2009;6:99–103. doi: 10.1038/nmeth.1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanfna M, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eirew P, Steif A, Khattra J, Ha G, Yap D, Farahani H, Gelmon K, Chia S, Mar C, Wan A, et al. Dynamics of genomic clones in breast cancer patient xenografts at single-cell resolution. Nature. 2014 doi: 10.1038/nature13952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- Fisher S, Barry A, Abreu J, Minie B, Nolan J, Delorey TM, Young G, Fennell TJ, Allen A, Ambrogio L, et al. A scalable, fully automated process for construction of sequence-ready human exome targeted capture libraries. Genome Biol. 2011;12:R1. doi: 10.1186/gb-2011-12-1-r1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Futreal PA, Coin L, Marshall M, Down T, Hubbard T, Wooster R, Rahman N, Stratton MR. A census of human cancer genes. Nat Rev Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett MJ, Edelman EJ, Heidorn SJ, Greenman CD, Dastur A, Lau KW, Greninger P, Thompson IR, Luo X, Soares J, et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature. 2012;483:570–575. doi: 10.1038/nature11005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garraway LA, Lander ES. Lessons from the cancer genome. Cell. 2013;153:17–37. doi: 10.1016/j.cell.2013.03.002. [DOI] [PubMed] [Google Scholar]
- Giannakis M, Hodis E, Jasmine Mu X, Yamauchi M, Rosenbluh J, Cibulskis K, Saksena G, Lawrence MS, Qian ZR, Nishihara R, et al. RNF43 is frequently mutated in colorectal and endometrial cancers. Nat Genet. 2014 doi: 10.1038/ng.3127. advance online publication. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao H-X, Xie Y, Zhang Y, Charlat O, Oster E, Avello M, Lei H, Mickanin C, Liu D, Ruffner H, et al. ZNRF3 promotes Wnt receptor turnover in an R-spondin-sensitive manner. Nature. 2012;485:195–200. doi: 10.1038/nature11019. [DOI] [PubMed] [Google Scholar]
- Haramis A-PG, Begthel H, van den Born M, van Es J, Jonkheer S, Offerhaus GJA, Clevers H. De novo crypt formation and juvenile polyposis on BMP inhibition in mouse intestine. Science. 2004;303:1684–1686. doi: 10.1126/science.1093587. [DOI] [PubMed] [Google Scholar]
- Imielinski M, Berger AH, Hammerman PS, Hernandez B, Pugh TJ, Hodis E, Cho J, Suh J, Capelletti M, Sivachenko A, et al. Mapping the hallmarks of lung adenocarcinoma with massively parallel sequencing. Cell. 2012;150:1107–1120. doi: 10.1016/j.cell.2012.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanov I, Lo KC, Hawthorn L, Cowell JK, Ionov Y. Identifying candidate colon cancer tumor suppressor genes using inhibition of nonsense-mediated mRNA decay in colon cancer cells. Oncogene. 2006;26:2873–2884. doi: 10.1038/sj.onc.1210098. [DOI] [PubMed] [Google Scholar]
- Jiao Y, Yonescu R, Offerhaus GJA, Klimstra DS, Maitra A, Eshleman JR, Herman JG, Poh W, Pelosof L, Wolfgang CL, et al. Whole-exome sequencing of pancreatic neoplasms with acinar differentiation. J Pathol. 2014;232:428–435. doi: 10.1002/path.4310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin K, Teng L, Shen Y, He K, Xu Z, Li G. Patient-derived human tumour tissue xenografts in immunodeficient mice: a systematic review. Clin Transl Oncol Off Publ Fed Span Oncol Soc Natl Cancer Inst Mex. 2010;12:473–480. doi: 10.1007/s12094-010-0540-6. [DOI] [PubMed] [Google Scholar]
- Kadowaki T, Wilder E, Klingensmith J, Zachary K, Perrimon N. The segment polarity gene porcupine encodes a putative multitransmembrane protein involved in Wingless processing. Genes Dev. 1996;10:3116–3128. doi: 10.1101/gad.10.24.3116. [DOI] [PubMed] [Google Scholar]
- Kim K-A, Kakitani M, Zhao J, Oshima T, Tang T, Binnerts M, Liu Y, Boyle B, Park E, Emtage P, et al. Mitogenic influence of human R-spondin1 on the intestinal epithelium. Science. 2005;309:1256–1259. doi: 10.1126/science.1112521. [DOI] [PubMed] [Google Scholar]
- Koo B-K, Spit M, Jordens I, Low TY, Stange DE, van de Wetering M, van Es JH, Mohammed S, Heck AJR, Maurice MM, et al. Tumour suppressor RNF43 is a stem-cell E3 ligase that induces endocytosis of Wnt receptors. Nature. 2012;488:665–669. doi: 10.1038/nature11308. [DOI] [PubMed] [Google Scholar]
- Korinek V, Barker N, Moerer P, van Donselaar E, Huls G, Peters PJ, Clevers H. Depletion of epithelial stem-cell compartments in the small intestine of mice lacking Tcf-4. Nat Genet. 1998;19:379–383. doi: 10.1038/1270. [DOI] [PubMed] [Google Scholar]
- Kuhnert F, Davis CR, Wang H-T, Chu P, Lee M, Yuan J, Nusse R, Kuo CJ. Essential requirement for Wnt signaling in proliferation of adult small intestine and colon revealed by adenoviral expression of Dickkopf-1. Proc Natl Acad Sci U S A. 2004;101:266–271. doi: 10.1073/pnas.2536800100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landau DA, Carter SL, Stojanov P, McKenna A, Stevenson K, Lawrence MS, Sougnez C, Stewart C, Sivachenko A, Wang L, et al. Evolution and impact of subclonal mutations in chronic lymphocytic leukemia. Cell. 2013;152:714–726. doi: 10.1016/j.cell.2013.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lau W, Barker N, Low TY, Koo B-K, Li VSW, Teunissen H, Kujala P, Haegebarth A, Peters PJ, van de Wetering M, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- De Lau W, Peng WC, Gros P, Clevers H. The R-spondin/Lgr5/Rnf43 module: regulator of Wnt signal strength. Genes Dev. 2014;28:305–316. doi: 10.1101/gad.235473.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Mermel CH, Robinson JT, Garraway LA, Golub TR, Meyerson M, Gabriel SB, Lander ES, Getz G. Discovery and saturation analysis of cancer genes across 21 tumour types. Nature. 2014;505:495–501. doi: 10.1038/nature12912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]
- Liu X, Ory V, Chapman S, Yuan H, Albanese C, Kallakury B, Timofeeva OA, Nealon C, Dakic A, Simic V, et al. ROCK inhibitor and feeder cells induce the conditional reprogramming of epithelial cells. Am J Pathol. 2012;180:599–607. doi: 10.1016/j.ajpath.2011.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Nicolantonio F, Martini M, Molinari F, Sartore-Bianchi A, Arena S, Saletti P, De Dosso S, Mazzucchelli L, Frattini M, Siena S, et al. Wild-type BRAF is required for response to panitumumab or cetuximab in metastatic colorectal cancer. J Clin Oncol Off J Am Soc Clin Oncol. 2008;26:5705–5712. doi: 10.1200/JCO.2008.18.0786. [DOI] [PubMed] [Google Scholar]
- Ojesina AI, Lichtenstein L, Freeman SS, Pedamallu CS, Imaz-Rosshandler I, Pugh TJ, Cherniack AD, Ambrogio L, Cibulskis K, Bertelsen B, et al. Landscape of genomic alterations in cervical carcinomas. Nature. 2014;506:371–375. doi: 10.1038/nature12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinto D, Gregorieff A, Begthel H, Clevers H. Canonical Wnt signals are essential for homeostasis of the intestinal epithelium. Genes Dev. 2003;17:1709–1713. doi: 10.1101/gad.267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, Kalogeras KT, Kotoula V, Papamichael D, Laurent-Puig P, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11:753–762. doi: 10.1016/S1470-2045(10)70130-3. [DOI] [PubMed] [Google Scholar]
- Ryland GL, Hunter SM, Doyle MA, Rowley SM, Christie M, Allan PE, Bowtell DDL, Australian Ovarian Cancer Study Group. Gorringe KL, Campbell IG. RNF43 is a tumour suppressor gene mutated in mucinous tumours of the ovary. J Pathol. 2013;229:469–476. doi: 10.1002/path.4134. [DOI] [PubMed] [Google Scholar]
- Sadanandam A, Lyssiotis CA, Homicsko K, Collisson EA, Gibb WJ, Wullschleger S, Ostos LCG, Lannon WA, Grotzinger C, Del Rio M, et al. A colorectal cancer classification system that associates cellular phenotype and responses to therapy. Nat Med. 2013;19:619–625. doi: 10.1038/nm.3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato T, Vries RG, Snippert HJ, van de Wetering M, Barker N, Stange DE, van Es JH, Abo A, Kujala P, Peters PJ, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459:262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- Sato T, Stange DE, Ferrante M, Vries RGJ, Van Es JH, Van den Brink S, Van Houdt WJ, Pronk A, Van Gorp J, Siersema PD, et al. Long-term expansion of epithelial organoids from human colon, adenoma, adenocarcinoma, and Barrett’s epithelium. Gastroenterology. 2011;141:1762–1772. doi: 10.1053/j.gastro.2011.07.050. [DOI] [PubMed] [Google Scholar]
- Seshagiri S, Stawiski EW, Durinck S, Modrusan Z, Storm EE, Conboy CB, Chaudhuri S, Guan Y, Janakiraman V, Jaiswal BS, et al. Recurrent R-spondin fusions in colon cancer. Nature. 2012;488:660–664. doi: 10.1038/nature11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi M, Fujita M, Furukawa Y, Hamamoto R, Shimokawa T, Miwa N, Ogawa M, Nakamura Y. Isolation of a novel human gene, APCDD1, as a direct target of the beta-Catenin/T-cell factor 4 complex with probable involvement in colorectal carcinogenesis. Cancer Res. 2002;62:5651–5656. [PubMed] [Google Scholar]
- Tentler JJ, Tan AC, Weekes CD, Jimeno A, Leong S, Pitts TM, Arcaroli JJ, Messersmith WA, Eckhardt SG. Patient-derived tumour xenografts as models for oncology drug development. Nat Rev Clin Oncol. 2012;9:338–350. doi: 10.1038/nrclinonc.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- Vecchione L. Optimization of Anti-EGFR Treatment of Advanced Colorectal Cancer. Curr Colorectal Cancer Rep. 2014;10:263–271. [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Jr, Kinzler KW. Cancer genome landscapes. Science. 2013;339:1546–1558. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Yuen ST, Xu J, Lee SP, Yan HHN, Shi ST, Siu HC, Deng S, Chu KM, Law S, et al. Whole-genome sequencing and comprehensive molecular profiling identify new driver mutations in gastric cancer. Nat Genet. 2014;46:573–582. doi: 10.1038/ng.2983. [DOI] [PubMed] [Google Scholar]
- Wong VWY, Stange DE, Page ME, Buczacki S, Wabik A, Itami S, van de Wetering M, Poulsom R, Wright NA, Trotter MWB, et al. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat Cell Biol. 2012;14:401–408. doi: 10.1038/ncb2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.