Abstract
Dissections or ruptures of aortic aneurysms remain a leading cause of death in the developed world, with the majority of deaths being preventable if individuals at risk are identified and properly managed. Genetic variants predispose individuals to these aortic diseases. In the case of thoracic aortic aneurysm and dissections (TAD), genetic data can be used to identify some at risk individuals and dictate management of the associated vascular disease. For abdominal aortic aneurysms (AAAs), genetic associations have been identified, which provide insight on the molecular pathogenesis but cannot be used clinically yet to identify individuals at risk for AAAs. This compendium will discuss our current understanding of the genetic basis of TAD and AAA disease. While both diseases share several pathogenic similarities, including proteolytic elastic tissue degeneration and smooth muscle dysfunction, they also have several distinct differences, including population prevalence and modes of inheritance.
Keywords: thoracic aortic aneurysm, abdominal aortic aneurysm, aorta, acute aortic dissection genetics
Subject Terms: Genetics, Aneurysm, Aortic Dissection
Aortic aneurysms are typically asymptomatic and often go undiagnosed until catastrophic complications like aortic rupture or dissection occur. The Centers for Disease Control has ranked the downstream consequences of aortic aneurysms, aortic dissections and ruptures, as high as the 19th leading cause of death in the United States (accounting for 43,000 – 47,000 deaths annually).1 These deaths are preventable if individuals at risk are identified and proper management of the aneurysm initiated to prevent these ruptures and dissections.2 Evidence has accumulated that genetic variants predispose individuals to these aortic diseases and these data can be used to identify individuals at risk to prevent premature deaths due to aortic rupture or dissections.
When considering the genetics of aortic aneurysmal disease, it is important to differentiate between thoracic and abdominal aortic disease. While both diseases share several pathogenic similarities, including proteolytic elastic tissue degeneration and smooth muscle cell (SMC) loss, they also have several distinct differences, including population prevalence, modes of inheritance and predisposing genes. Approximately 20% of individuals with thoracic aortic aneurysms or dissections exhibit an autosomal dominant pattern of inheritance of the condition in the family, i.e., thoracic aortic disease results from a mutation in the single gene, whereas abdominal aortic aneurysm (AAA) does not typically demonstrate such inheritance. Causative genes that confer a high risk for thoracic aortic disease have been identified for this heritable risk for thoracic aortic disease (HTAD). Importantly, these genes typically do not confer an increased risk for AAA and no such high-risk genes have been identified specifically for AAA.
Genetic Risks for Disease
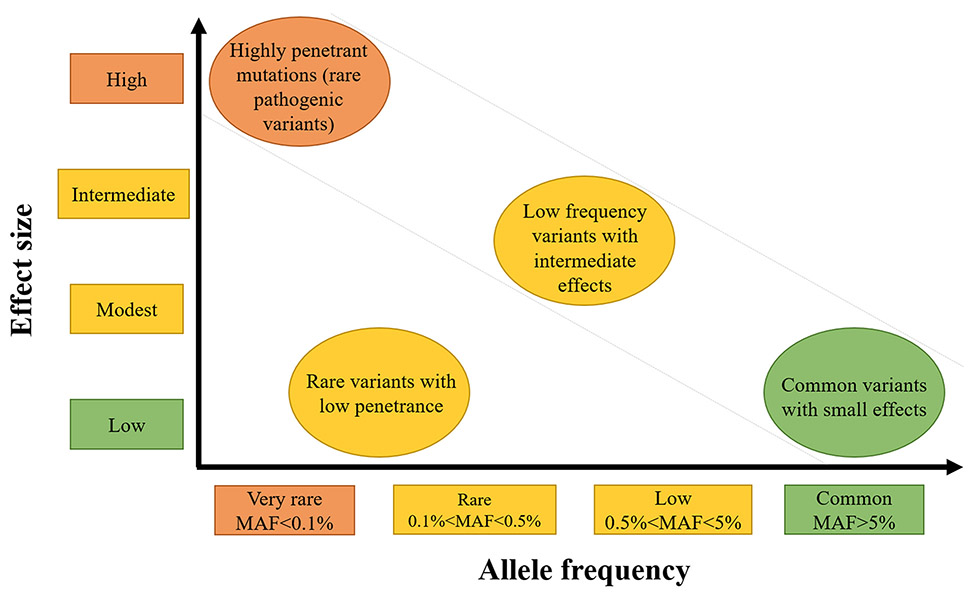
The genetic risk for diseases extends from rare and highly penetrant genetic variants that trigger disease in all individuals who inherit the alteration (e.g., Marfan syndrome due to FBN1 mutations) to common variants found in the general population that confer only a minimal risk for disease. These genetic risks are illustrated in Figure 1 and graphed based on the frequency of the risk allele in the population and the strength of genetic effect (i.e., odds ratio).3 Allele frequency and effect size are generally inversely related, with rare variants having large effects (i.e., mutations or pathogenic variants) and common variants having low effect size. Note that common variants with large effects are rare and subject to strong purifying selection, and rare variants with small effects are difficult to identify.
Figure 1:
Thoracic aortic disease risk associated to variant frequencies (adapted from Manolio et al 2009)3.
Pathogenic variants (rare variants in disease-causing genes) are categorized using the American College of Medical Genetics (ACMG) classification framework, which is based on the variant conferring a high penetrance for the disease, segregation of the variant with disease in families, presence of the variant in unrelated cases, and absence of the variant in population databases (e.g., the Genome Aggregation Database (GnomAD), http://gnomad.broadinstitute.org/).4 The current approach to identify these rare pathogenic variants is exome or whole genome sequencing. These rare but highly penetrant variants for disease are represented in the upper left of the graph (Figure 1). In contrast, low penetrant variants that increase the risk for dissection only in combination with “environmental” insults or with other low risk variants (i.e., two genetic “hits”) are more difficult to validate as disease-predisposing alleles and typically require large cohorts to confirm an association. Genome wide, case control association studies (GWAS) are commonly used to identify these low risk and common variants, which are represented in the lower right of the graph.
Thoracic Aortic Aneurysm and Acute Aortic Dissections
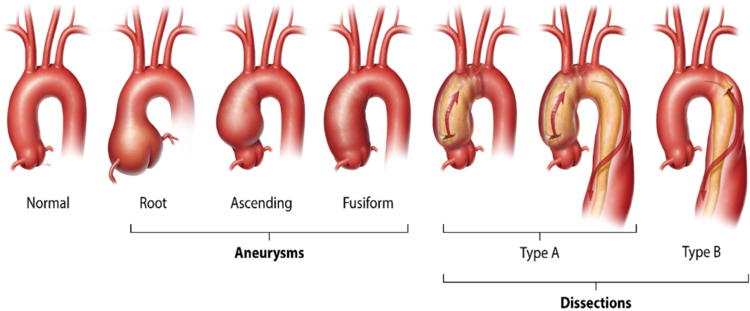
The major diseases affecting the thoracic aorta are aortic aneurysm and acute aortic dissection, termed collectively as thoracic aortic disease (TAD). The natural history of a thoracic aortic aneurysm, involving the aortic root or ascending aorta or both, is an asymptomatic enlargement over time until the aorta becomes unstable and an acute tear in the intimal layer leads to an ascending aortic dissection (classified as Stanford type A dissections, Figure 2). With dissection, blood penetrates the aortic wall and separates the aortic layers, causing aortic rupture and other complications. Type A aortic dissections, which originate in the ascending aorta, may or may not extend into the descending aorta and cause sudden death in up to 50% of individuals.5 Survivors of the acute event continue to have a high mortality rate despite emergency surgery to repair the dissected ascending aorta. The majority of the deaths in patients that die prior to hospital admission are due to blood dissecting retrograde and rupturing into the pericardial sac, causing pericardial tamponade.6 The thoracic aortic disease spectrum also includes aortic dissections originating from the descending thoracic aorta just distal to the branching of the subclavian artery, termed type B dissections (Figure 2). Type B aortic dissections are less likely to result in death and occur with little to no enlargement of the thoracic descending aorta.
Figure 2:
Schematic representation of thoracic aortic aneurysms and aortic dissections.
Although medical treatments (e.g. β-adrenergic and angiotensin receptor type I antagonists) can slow the enlargement of an aneurysm, the mainstay of treatment to prevent premature deaths due to life-threatening aortic dissection is surgical repair of the aneurysm. Prophylactic surgical repair of an aortic aneurysm can completely alter the course and outcome of the disease; timely aortic root aneurysm repair extends the life expectancy in Marfan syndrome (MFS) patients by 25 years or more.7, 8 This repair is typically recommended when the aortic diameter reaches 5.0 – 5.5 cm9; however, studies on patients presenting with acute type A dissections indicate that up to 60% present with aortic diameters smaller than 5.5 cm.10 Therefore, clinical predictors and biomarkers are needed to identify who is at risk for aortic dissection occurrence with the aortic diameter smaller than 5.0 cm and use this information to dictate timely surgical repair to prevent a dissection. One such biomarker is the specific gene altered to cause thoracic aortic disease; the gene alteration can identify other family members who are at risk for thoracic aortic disease but also predict when to pursue prophylactic repair of the aorta, thereby optimizing the timing of aortic surgery to prevent dissections.11–13
The major risk factors for thoracic aortic disease are hypertension and an underlying genetic alteration. Other risk factors include pregnancy, activities that increase forces on the ascending aorta (e.g., cocaine or methamphetamine abuse, bodybuilding weight lifting), and the presence of a bicuspid aortic valve (BAV). Thoracic aortic disease can occur with a genetic syndrome due to a single autosomal gene mutation, as is the case for Marfan and Loeys-Dietz syndromes.14–16 Furthermore, 20% of non-syndromic thoracic aortic disease patients have similarly affected first-degree relatives, indicating many patients with thoracic aortic disease most likely have an underlying gene mutation even in the absence of a genetic syndrome.17, 18 The disease in these families is typically inherited in an autosomal dominant manner with decreased penetrance, particularly in women.19 Collectively, thoracic aortic disease due to a single gene mutation is termed HTAD.
Heritable Thoracic Aortic Disease Due to Highly Penetrant, Pathogenic Variants in Single Genes
Based on curation of genes that predispose to thoracic aortic disease by the ClinGen Aortopathy Working Group, 11 genes have sufficient data to confirm that mutations in these genes confer a highly penetrant risk for thoracic aortic aneurysms and acute aortic dissections, with or without syndromic features.20 These genes encode proteins involved in vascular SMC contraction and adhesion to the extracellular matrix (ECM), transforming growth factor (TGF)-β signaling pathways, or SMC metabolism and include the following genes: FBN1 (encodes fibrillin-1), LOX (lysyl oxidase), MYH11 (smooth muscle myosin heavy chain 11), ACTA2 (smooth muscle α-actin 2), MYLK (myosin light chain kinase), PRKG1 (cGMP-dependent protein kinase type I), COL3A1 (α−1 procollagen, type III), TGFBR2 (TGF-β receptor type II), TGFBR1 (TGF-β receptor type I), TGFB2 (TGF-β2), and SMAD3 (mothers against decapentaplegic homolog 3).15, 16, 21–30 Mutations in these genes can be identified in the majority of HTAD families with systemic features of MFS or Loeys-Dietz syndrome (LDS), but only 30% of HTAD families without these syndromic features have mutations in these genes, indicating there are more genes yet to be discovered. Table 1 is a complete list of the validated and proposed genes predisposing to HTAD in humans, the name of the corresponding protein, and the domains in the corresponding protein in which pathogenic missense mutations occur.
Table 1:
Genes validated and proposed to predispose to heritable thoracic aortic disease (HTAD) in humans
| Risk associated with HTAD | Genes name | Protein | Inheritance | Associated syndrome‡ | OMIM | HI | Domain location for missense, inframe deletion/duplication | Pathways impacted |
|---|---|---|---|---|---|---|---|---|
| Definitive | ACTA2 | smooth muscle actin α2 | AD | Smooth muscle dysfunction syndrome | 613834 | No* | † | SMC contraction |
| COL3A1 | procollagen type III α1 | AD | Vascular Ehlers-Danlos syndrome | 130050 | Yes | Triple helical domain (disruptglycines) | ECM | |
| FBN1 | fibrillin-1 | AD | Marfan syndrome | 154700 | Yes | Mainly EGF-like domains (disruption cysteines and calcium binding) | ECM | |
| MYH11 | smooth muscle myosin heavy chain 11 | AD | 132900 | No* | Missense mutations and inframe deletions in coiled coil domain | SMC contraction | ||
| SMAD3 | mothers against decapentaplegic drosophila homolog 3 | AD | Loeys-Dietz syndrome 3 | 613795 | Yes | Mainly MH2 domain, but also MH1 domain | TGF-β | |
| TGFB2 | transforming growth factor β2 | AD | Loeys-Dietz syndrome 4 | 614816 | Yes | Furin cleavage site and cytokine domain | TGF-β | |
| TGFBR1 | transforming growth factor β receptor type I | AD | Loeys-Dietz syndrome 1 | 609192 | Yes | Intracellular kinase domain | TGF-β | |
| TGFBR2 | transforming growth factor β receptor type II | AD | Loeys-Dietz syndrome 2 | 610168 | Yes | Intracellular kinase domain | TGF-β | |
| MYLK | myosin light chain kinase | AD | 613780 | Yes | Kinase and calmodulin-binding domain | SMC contraction | ||
| Strong | LOX | lysyl oxydase | AD | Musculoskeletal manifestations of Marfan syndrome | 617168 | Yes | Catalytic domain | ECM |
| PRKG1 | protein kinase cGMP-dependent type 1 | AD | 615436 | No* | Onegain of function mutation, p.Arg177Gln | SMC contraction | ||
| Moderate | EFEMP2 | EGF containing fibulin-like extracellular matrix protein 2 (fibulin-4) | AR | Cutis laxa type 1B syndrome | 614437 | ECM | ||
| Limited | ELN | elastin | AD | Cutis Laxa syndrome | 123700 | ECM | ||
| FBN2 | fibrillin 2 | AD | Congenital contractural arachnodactyly | 121050 | EGF-like domain | ECM | ||
| FLNA | filamin A | X-linked Recessive | Cardiac valvular dysplasia | 314400 | SMC contraction | |||
| NOTCH1 | notch 1 | AD | 109730 | EGF-like domain | ||||
| SLC2A10 | solute carrier family 2 member 10 | AR | Arterial tortuosity syndrome | 208050 | ||||
| SMAD4 | mothers against decapentaplegic drosophila homolog 4 | AD | Juvenile polyposis/hereditary hemorrhagic telangiectasia syndrome | 175050 | Yes | Mainly MH2 domain | TGF-β | |
| SKI | SKI proto-oncogene | AD | Shprintzen-Goldberg syndrome | 182212 | SMAD-binding domain | TGF-β | ||
| Uncertain (recent reported genes) | BGN | biglycan | X-linked Recessive | Musculoskeletal manifestations of Marfan syndrome | 300989 | ECM | ||
| FOXE3 | forkhead box E3 | AD | 617349 | Forkhead DNA-binding domain | ||||
| HCN4 | hyperpolarization activated cyclic nucleotide-gated potassium channel 4 | AD | 163800 | |||||
| MAT2A | methionine adenosyltransferase II α | AD | 607086 | Catalytic domain | ||||
| MFAP5 | microfibrillar associated protein 5 | AD | Musculoskeletal manifestations of Marfan syndrome | 616166 | Yes | N-terminal domain | ECM | |
| SMAD2 | mothers against decapentaplegic drosophila homolog 2 | AD | MH2 domain | TGF-β | ||||
| TGFB3 | transforming growth factor β3 | AD | Loeys-Dietz syndrome 5 | 615582 | Furin cleavage site and cytokine domains | TGF-β | ||
| LTBP3 | latent transforming growth factor β binding protein 3 | AR | Dental anomalies and short stature syndrome | 601216 | EGF-like domain | ECM and TGF-β | ||
| ARIH1 | ariadne drosophila homolog 1 | AD | Yes | Glycine-rich domain |
Footnotes: HI: Haploinsufficiency in the gene can cause disease, AD: Autosomal Dominant, AR: Autosomal Recessive.
Based on current data,
pathogenic variants are found throughout the gene,
Individuals can have pathogenic variants in these genes and may have no syndromic features but have thoracic aortic disease or other vascular diseases.
Based on the ClinGen Aortopathy Working Group classification,20 “Definitive” and “Strong” genes are established to predispose to heritable thoracic aortic disease. “Moderate” and “Limited” genes are potentially important in terms of proper diagnosis in an individual with thoracic aortic enlargement but these genes do not carry significant risk for progression to acute aortic dissections. “Uncertain” category indicates recently published genes for which the data are limited and no categorization is possible until additional data are available.
Through clinical characterization of HTAD families with mutations in novel genes, we have determined that the underlying gene predicts not only who in the family is at risk for thoracic aortic disease, but also (a) associated syndromic features, including those typical of MFS and LDS and other additional syndromic complications; (b) aortic disease presentation (age, dissection versus aneurysm), (c) risk for dissection at a given aortic diameter, and (d) risk for additional vascular diseases. As examples of the risk for other vascular disease, TGFBR2 mutations predispose to TAD, as well as intracranial aneurysms and aneurysms of other arteries, whereas ACTA2 mutations lead to TAD and occlusive vascular disease, including early onset stroke and coronary artery disease (CAD).15, 30 Thus, identifying the underlying HTAD gene triggering TAD provides powerful information for vascular disease management to prevent aortic dissections and other vascular disease complications. Based on the strength of these clinical correlates, the ACCF/AHA Treatment Guidelines for Thoracic Aortic Diseases emphasize a gene-based management of aortic disease if the defective gene is identified.9
HTAD Genes Encoding Proteins Involved in SMC Adhesion and Contraction Highlights the Importance of Maintaining the Elastin-Contractile Unit
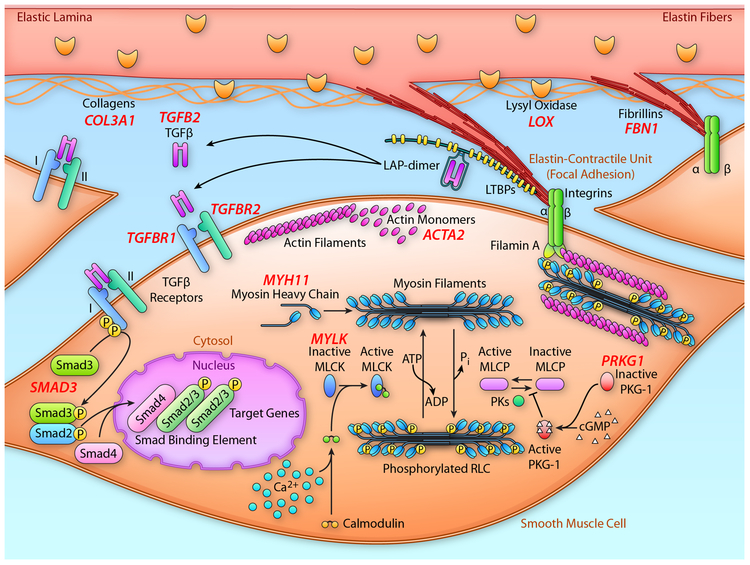
The aorta is continually subjected to biomechanical forces from pulsatile blood pressure and flow from the beating heart. The aorta has a unique role in blood flow because it not only acts as a conduit for blood but the aorta’s elastic properties allow it to serve as a reservoir for blood. The aorta distends when the blood pressure rises with cardiac systole and recoils when the pressures fall during diastole, the so-called Windkessel effect. The thoracic aorta is a tube composed of three layers: an intima comprising endothelial cells on a basement membrane, an elastin- and smooth muscle-rich media, and a collagen- and fibroblast-rich adventitia. It is the medial layer of the aorta that confers elasticity and strength to the aortic wall and it is composed of greater than 50 alternating layers of SMC and elastic lamellae in humans. The SMC elastin-contractile unit is the structural unit that links the elastin lamellae to the SMCs (Figure 3).31 Microfibril extensions from the elastic lamellae are obliquely anchored to the surface of SMCs through focal adhesions (also termed dense plaques), thus providing connection of the SMCs to the elastic fibers and allowing the propagation of mechanical forces between elastin and SMCs via integrin receptors.31 The oblique orientation of the elastin-contractile units reverses direction in successive SMC layers in a herringbone-like pattern, a unique design that minimizes the biomechanical forces on individual aortic SMCs (reviewed in32, 33). This “elastin-contractile unit” is uniquely designed to coordinate SMC contractile and elastic tensions in response to mechanical stresses imposed on the vessel wall from pulsatile blood flow. Consistent with the hypothesis that the architecture of the SMC contractile-elastin unit is a functional and structural element in the aorta, many of the altered genes that predispose to thoracic aortic disease disrupt components of this unit (Figure 3).
Figure 3:
Schematic representation elastin lamellae and SMCs highlighting the proteins that are disrupted by mutations in genes, leading to heritable thoracic aortic disease. Extensions from the elastin lamellae with fibrillin-1-containing microfibrils at the end link to integrin receptors on the cell surface of SMCs. The integrin receptors then link to the contractile filaments inside the cells, thus forming the elastin-contractile unit. Also illustrated is the proteins involved in canonical TGF-β signaling that are disrupted by mutations in the corresponding genes to also lead to heritable thoracic aortic disease. The validated genes predisposing to thoracic aortic disease are shown in red and are adjacent to their corresponding protein. MLCK; myosin light chain kinase; PKG-1, type I cGMP-dependent protein kinase. (Illustration Credit: Ben Smith).
The first gene identified for HTAD was the gene for Marfan syndrome, FBN1 (OMIM 134797), which encodes fibrillin-1, a large cysteine-rich glycoprotein (350 kDa, 2871 amino acids). Fibrillin-1 is made as a proprotein and cleaved by a furin-like protease as it is secreted from the cells, then polymerizes to form microfibrils that are found at the end of the elastin extensions, connecting the SMCs to the elastin lamellae. Fibrillin-1 contains 47 epidermal growth factor (EGF)-like motifs interspersed with 7 TGF-β1 binding protein (TB) motifs and the majority of EGF-like motifs contain a calcium binding sequence (cb-EGF).34 Each EGF-like motif contains 6 cysteine amino acids, which form disulfide bonds, and each TB motif contains 8 cysteine amino acids. These cysteines are the key residues for stabilizing the quaternary structure of the fibrillin-1.
Marfan syndrome (MFS, OMIM 154700) is an autosomal dominant condition characterized by highly penetrant aortic root aneurysms, leading to type A dissections in the absence of surgical repair. In addition, individuals with MFS have a constellation of skeletal features and ocular complications.35 Almost 2000 unique FBN1 mutations have been identified in patients with MFS and the majority of these mutations are missense mutations, but frameshift, nonsense, splicing errors or complete FBN1 deletion have also been identified.36 Disease-causing FBN1 variants and cell biology have provided unique insights into the biological and physiological role of this complex glycoprotein.37, 38 A subset of FBN1 mutations, specifically frameshift and nonsense mutations leading to decay of the message and gene deletions, are predicted to lead to production of half the normal amount of fibrillin. Fibroblasts explanted from MFS patients exhibit decreased fibrillin-1 incorporation into the extracellular matrix, regardless of the underlying mutation.39, 40 Many missense mutations alter a cysteine codon within one of the FBN1 gene’s numerous EGF-like domains, disrupting disulfide pairing and proper folding of the protein41, decreasing fibrillin-1 secretion from the cell and its assembly into microfibrils42, or increasing its susceptibility for proteolysis.43 EGF domains also mediate direct interaction between fibrillin-1 and SMCs via integrin αVβ3 binding to fibrillin-1.44, 45 One of the MFS animal models, the Fbn1mgR/mgR mouse, which makes significantly less normal fibrillin-1 and the connections between SMCs and elastin fibers are completely lost in the aortic media in the mutant mice aortas.46 FBN1 mutations decrease the fibrillin-1-containing microfibrils in the aorta that provide the structural attachment of the elastin fibers to the SMCs in the medial layer of the aorta.
Two additional genes encoding proteins important for ECM integrity also predispose to thoracic aortic disease when mutated. LOX (OMIM 153455) encodes a copper-dependent lysyl oxidase responsible for cross-linking of collagen and elastin by catalyzing the formation of aldehydes from lysine residues in the precursors of these two proteins. This cross-linking is essential for stabilization of collagen fibrils and for the integrity and elasticity of mature elastin in the ECM, and the heterozygous mutations identified in HTAD families lead to decreased lysyl oxidase activity.25, 47 Patients with mutations in LOX (OMIM 617168) present with fusiform enlargement of the root and ascending thoracic aorta, leading to ascending aortic dissections, in addition to variable presence of skeletal manifestations similar to MFS. COL3A1 encodes the type III pro-collagen which is part of the fibrillar collagen family with types I, II, V and XI. Procollagens are the precursor form, which are secreted, and then cleaved by procollagen protease to allow extracellular assembly. Patients with vascular Ehlers-Danlos syndrome (vEDS) (OMIM 130050), an autosomal dominant syndrome, present a thin translucent skin, extensive bruising, characteristic facial appearance, arterial, intestinal, and/or uterine fragility, and premature aging of the skin, resulting from mutation in COL3A1. Patients with vEDS have a greater risk of developing aneurysms, dissection and rupture of large and medium sized arteries, and, less commonly, aortic dissection and aneurysms.48
The importance of elastin and microfibrils in maintaining the structural integrity of the thoracic aorta to prevent disease is further emphasized by recently reported novel mutant genes that predispose to thoracic aortic disease. We recently found that homozygous LTBP3 pathogenic variants predispose to TAD, along with the previously described skeletal alterations, leading to short stature and dental abnormalities.49 Preliminary data also indicate that missense variants in LTBP3 may also predispose to later onset TAD. LTBP3 encodes latent TGF-β binding protein-3 (LTBP-3), which belongs to a family of proteins that regulate TGF-β activity by enabling its secretion, directing it to specific sites in the ECM, and participating in its activation.50 TGF-β is secreted from cells bound to a complex that includes its dimeric pro-peptide (termed latency-associated peptide; LAP) and one of three LTBPs. LTBP-3 binds to all three TGF-β ligands, and the secretion of LTBP-3 from cells requires LTBP-3 to be complexed with TGF-β and LAP.51 Once secreted, LTBP-3, with LAP and inactive TGF-β, associates with fibrillin-1 in the microfibrils. Loss of fibrillin-1 in cells or aortic tissues diminishes LTBP-3 immunofluorescence in the ECM.52 Thus, loss of LTBP-3 would be predicted to decrease LTBP-3 levels in fibrillin-1-containing microfibrils but also decrease TGF-β secretion from cells.
Heterozygous loss-of-function mutations in MFAP5 disrupt one of the microfibril associated proteins, MAGP-2, and predispose to thoracic aortic disease.53 Mutations in another microfibril associated protein, emilin1, have been also been reported to predispose to thoracic aortic aneurysm.54 Emilin and MAGP-2 are both components of microfibrils. For all three genes, additional families need to be identified to validate the genes before they can be used clinically to identify at risk individuals. Mutations in the gene for elastin itself (ELN) cause dominant cutis laxa, and there are reports of these patients developing thoracic aortic aneurysms.55 Finally, recessive mutations in a gene for another protein involved in maintenance of elastin, fibulin 4 (EFEMP2), also cause cutis laxa with aortic aneurysms, along with arterial tortuosity and stenoses.56–58
Genes that cause HTAD when altered also encode proteins that are major components of the SMC contractile unit and kinases that control contraction, thus disrupting the intracellular component of the elastin-contractile unit (Figure 3). For contractile function, SMCs express smooth muscle-specific isoforms of α-actin and myosin (encoded by ACTA2 and MYH11, respectively), which polymerize to form thin and thick filaments, respectively. The thick filaments are composed of a SM-specific isoform of myosin heavy chain dimer (MHC, encoded by MYH11), two regulatory light chains (RLC), and two essential light chains. Force generation by SMCs requires ATP-dependent cyclic interaction between these two types of filaments, which is triggered by phosphorylation of RLCs. This intracellular SMC contractile unit is linked to focal adhesions (also called dense plaques) and this unique configuration is designed to transit forces from the elastin fiber to the SMCs via integrin receptors. MYLK and PRKG1 encode kinases that control phosphorylation and de-phosphorylation of the RLCs, respectively, thus controlling contraction and relaxation of the SMCs.
ACTA2 (OMIM 102620) encodes the SMC-specific isoform of α-actin, which polymerizes to form the thin filaments of the SMC contractile unit. In SMCs, smooth muscle α-actin is the most expressed protein, representing 40% of the total proteins.59 Patients with mutations in ACTA2 are at risk for thoracic aortic disease, along with early onset coronary artery disease (CAD) and ischemic strokes (defined as an age of onset less than 55 years in men and less than 60 years in women) (OMIM 611788). Specific ACTA2 mutations predispose to either early-onset CAD, or moyamoya-like cerebrovascular disease (OMIM 614042).30 ACTA2 missense pathogenic variants that specifically disrupt the arginine 179 residue cause multisystemic smooth muscle dysfunction syndrome (OMIM 613834),59 which is characterized by an extensive vasculopathy (moyamoya disease and thoracic aortic disease), pulmonary hypertension, congenital mydriasis, patent ductus arteriosus, hypoperistalsis and hypotonic bladder. Mutations in ACTA2 are responsible for disease in 14% of the non-syndromic HTAD patients.23
ACTA2 is a highly conserved gene and relatively invariant in the general population. ACTA2 mutations disrupt amino acids located in all 4 subdomains of actin and are predicted to produce structurally altered actin monomers, leading to the disruption of the force generation by SMCs, and thereby resulting in defective contractile function and contributing to the pathogenesis of TAAD. Molecular characterization of a disease-causing ACTA2 variant (p.Arg258Cys) indicates that the mutation makes the actin filaments more unstable and susceptible to severing by cofilin.60 Additionally, profilin binds more tightly to the mutant actin, which is predicted to increase the pool of monomeric actin in SMCs, and myosin moves more slowly across mutant actin filaments. All these observations indicate that the mutant actin will decrease the ability of the SMC to contract in response to pulse pressures.
MYH11 (OMIM 160745) encodes the SMC-specific myosin heavy chain, a major component of the SMC contractile unit. Patients with mutations in MYH11 gene have thoracic aortic disease that is often associated with patent ductus arteriosus (OMIM 132900).29 Most mutations are located in the C-terminal coiled-coil region or rod domain and are predicted to disrupt the ability of the mutant myosin to polymerize into thick filaments and form their quaternary structure. Note that deletions of 16p13.1 occur in the general population and include MYH11, but there is no evidence that these deletions predispose to thoracic aortic disease. Thus, haploinsufficiency for MYH11 does not appear to cause thoracic aortic disease. In contrast, duplications of 16p13.1 also occur in the general population and are predicted to increase expression of MYH11 and do confer an increased risk for acute aortic dissections (see below).61
MYLK (OMIM 600922) encodes the myosin light chain kinase (MLCK), which is a ubiquitously expressed kinase that takes part in phosphorylation of the RLC of the smooth and nonmuscle myosin II. RLC phosphorylation increases actin-dependent myosin II ATPase activity, and thus initiates SMC contraction. Patients with mutations in the MYLK gene, present with aortic dissection with little to no aortic enlargement.62, 63 MYLK mutations are located in the short form of myosin light chain kinase, the only form expressed in human aorta64, and thus rare variants disrupting amino acids 1 to 922 should not cause thoracic aortic disease. Mutations lead to either haploinsufficiency or are missense mutations that disrupt the kinase activity and are predicted to decrease the phosphorylation of the RLC, and thus diminish aortic SMC contraction. More recently, MYLK rare variants have been identified in the kinase domain that segregate with disease in large families, indicating they are disease-causing alleles, but do not disrupt kinase function. 62, 63
PRKG1 (OMIM 615436) encodes PKG-1, the type I cGMP-dependent protein kinase, which is activated on binding of cGMP, and controls SMC relaxation. PKG-1α is the major isoform present in vascular SMC and is activated by nitric oxide, which stimulates soluble guanylyl cyclase and increases cellular cGMP and subsequently regulates many cellular systems that lead to relaxation of SMCs, including activation of the regulatory myosin-binding subunit of the phosphatase that dephosphorylates RLC. Only one rare recurrent gain of function variant p.Arg177Gln in PRKG1 gene has been identified to cause TAD in patients who present with aortic aneurysm and acute aortic dissection at relatively young ages (63% of affected individuals present an aortic dissection at mean age of 31 years) and, in some cases, also have vascular abnormalities including aneurysms of the descending thoracic aorta, abdominal aorta and coronary arteries.24 This mutation is located in the high-affinity cGMP binding site of the protein, and abolishes the binding of cGMP, but paradoxically leads to constitutive activation of PRK-1α. Because it is no longer regulated by cGMP, the mutant protein becomes constitutively active. The increased kinase activity associated with PKG-1α leads to decreased levels of phosphorylated (active) RLC and promotes SMC relaxation.
In SMC, integrins link the microfibrils to the actin filaments through filamin A, an actin-associated protein (Fig. 3). Mutations in the gene for filamin A (FLNA) cause periventricular nodular heterotopia and otopalatodigital spectrum disorders, but also predispose to TAD.65 With loss of function mutations in FLNA, 18% of cases will have thoracic aortic disease, often associated with PDA and valvular abnormalities.66
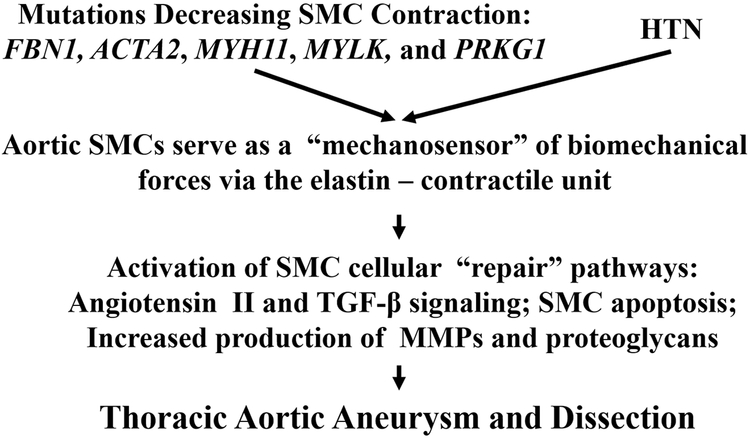
Collectively, mutations in the genes listed above disrupt the ability of aortic SMCs to properly sense and respond to forces through the elastin-contractile units, implying that altering this elastin-contractile structural element may be a primary driver for thoracic aortic aneurysms and dissections (Figure 4). Specifically, mutated genes that predispose to TAD alter SMC force generation by disruption of SMC contractile unit or the matrix to which the SMCs attach for effective contraction. This disruption of the elastin–contractile unit will alter mechanosensing by the SMCs, thus triggering changes in the SMCs signaling that lead to thoracic aortic disease. The hypothesis that altered mechanosensing through the elastin–contractile unit is the major driver of thoracic aortic disease provides an explanation for other trigger of the disease, hypertension. In a hypertensive individual, the elastin–contractile unit may not be altered due to genetic variants but the forces across the unit will be increased because of the hypertension, thus potentially altering similar SMC cellular pathways downstream of the elastin–contractile unit and leading to TAD.
Figure 4: A proposed mechanism by which mechanotransduction or mechanosensing across the elastin-contractile units predispose to thoracic aortic disease.
Thoracic aortic aneurysms and dissections are driven by environmental factors, such as hypertension (HTN), or mutations that disrupt proteins that are components of elastin-contractile units. In the case of gene mutations, the elastin-contractile unit is predicted or has been shown to be disrupted,46, 162 and thus may upregulate cellular pathways in SMCs that lead to disease. In terms of hypertension triggering thoracic aortic disease, the elastin-contractile unit may not be disrupted, or may be only altered in a minor way, but the hypertension increases the forces across the elastin-contractile unit, potentially activating the same downstream pathways leading to thoracic aortic aneurysms and dissections.
HTAD Genes Disrupting TGF-β Signaling
Mutations in the TGF-β receptor type II (TGFBR2) were first identified in individuals with Marfanoid features, and the condition was initially named Marfan syndrome 2.16 Subsequently, mutations in genes encoding other proteins in the canonical TGF-β signaling pathway have been identified that predispose to thoracic aortic disease, including TGF-β receptor type I (TGFBR1), SMAD3 (SMAD3), SMAD4 (SMAD4), and one of the three TGF-β ligands, TGF-β2 (TGFB2). In addition to Marfanoid skeletal features and thoracic aortic disease, mutations in these genes predispose to aneurysms and dissections beyond the aorta, including arterial branches of the aorta and intracranial arteries. Additional systemic features can also be present, including bifid or wide uvula, and thin or translucent skin with delayed wound healing similar to that observed in individuals with vascular Ehlers-Danlos syndrome. TGFBR1 and TGFBR2 mutations also cause Loeys-Dietz syndrome (LDS), characterized by craniosynostosis, cleft palate/bifid uvula, developmental delay, congenital heart disease, and aggressive and early onset of both thoracic aortic disease and aneurysms and dissections involving other arteries67 These genes can also lead to an inherited predisposition to thoracic aortic disease in the absence of syndromic features. Inheritance of a risk for thoracic aortic disease was first described in a family with nine affected members over two generations, who did not have phenotypic features of MFS, vascular Ehlers-Danlos syndrome or systemic hypertension.68 An underlying TGFBR1 mutation was later identified as the cause of disease in this family.69 It has been confirm that many HTAD families without syndromic features have underlying mutations in these genes encoding proteins in the canonical TGF-β signaling pathway. Thus, it is important to emphasize that all the genes that cause HTAD associated with syndromic features, e.g., FBN1, TGFBR1, and TGFBR2, can also lead to autosomal dominant inheritance of TAD in the absence of syndromic features.69, 70
The causative mutations identified in these genes are predicted or have been shown to decrease canonical TGF-β signaling.21, 71, 72 TGF-β signaling is initiated when the cytokine binds to the cell surface TGF-β type II receptor, which then recruits and phosphorylates the TGF-β type I receptor (Figure 3). The type I receptor phosphorylates SMAD2 and SMAD3 (mothers against decapentaplegic homolog 2 (OMIM 601366) and 3 (OMIM 603109), respectively), which complex with SMAD4 and translocate to the nucleus to alter gene transcription. Validated genes mutated to cause HTAD in this pathway include: genes encoding one of the three TGF-β ligand family members, TGFB2 (TGF-β2; OMIM 190220), the TGF-β cellular receptors, TGFBR2 (OMIM 190182) and TGFBR1 (OMIM 190181), and SMAD3. Pathogenic variants in these genes have been shown or are predicted to decrease cellular TGF-β signaling due to haploinsufficiency (e.g., nonsense or frameshift mutations) or missense variants that disrupt the protein function (e.g., decrease the intracellular kinase activity of the TGF-β receptors).21, 71, 73 In addition, mutations in a gene for another TGF-β ligand, TGF-β3 (TGFB3) and SMAD2 were recently reported but additional data is needed to validate these genes for HTAD.74, 75 Finally, mutations in SMAD4 cause a syndrome characterized by juvenile polyposis (JPS) and hereditary hemorrhagic telangiectasia (HHT) and these individuals are at risk for thoracic aortic disease.74, 75
Pathogenic variants in TGFBR1 and TGFBR2 predisposing to HTAD are located in the intracellular kinase domain of the proteins and result in decreased kinase function. TGFB2 encodes TGF-β type 2 in the form of a synthetized precursor of 442 amino acid polypeptide from which a mature 112 amino acid active protein is derived by proteolytic cleavage. Patients with heterozygous mutations in the TGFB2 gene, present with TAD and share some clinical features (skin, skeletal) with MFS and LDS. This disease is called LDS4 (OMIM 614816). One percent of HTAD patients have been reported as having a pathogenic variant in the TGFB2 gene.21 SMAD3 (OMIM 603109) encodes mothers against decapentaplegic drosophila homolog 3, an intracellular effector of TGF-β signaling. Patients with aneurysm-osteoarthritis syndrome (AOS) (OMIM 613795), an autosomal dominant syndrome, present early-onset joint abnormalities and aortic aneurysms and dissections throughout the arterial tree, resulting from mutations in the SMAD3 gene.26, 27 Some clinical features overlap with MFS and LDS, and AOS has also been labeled LDS3. The MH2 domain of the protein mediates the oligomerization of SMAD3 with SMAD4 and SMAD-dependent transcriptional activation and mutations located in this domain have a high impact on these processes. It is important to note that the phenotypes resulting from any one of these TGF-β signaling genes can range from no to minimal features of MFS, LDS or vEDS. Although marked aortic and arterial tortuosity has been associated with mutations of these genes, tortuosity is increase in both vascular beds for the majority of HTAD-related genes.76
Although initial studies in MFS mouse models suggested increased TGF-β signaling was the primary driver for TAD, the mutations disrupting TGF-β pathway are predicted or have been shown to decrease TGF-β signaling.21, 71, 72, 77 Subsequent studies in mice have shown that decreasing TGF-β signaling in MFS mouse models early augments rather decreases aortic disease.78, 79 Exposure to TGF-β drives differentiation of SMCs, and this differentiation is defined by increased levels of the contractile proteins, including smooth muscle α-actin and smooth muscle myosin heavy chain. Thus, TGF-β signaling loss-of-function mutations may disrupt proper differentiation of SMCs, resulting in reduced numbers of contractile units within aortic SMCs and decreased SMC force generation, thus leading to TAD. TGF-β signaling is also critical for the differentiation of neural crest cells into SMCs populating the ascending thoracic aorta during development; thus, lower TGF-β signaling may influence proper organogenesis and the resulting structure of the ascending aortic wall.
HTAD Genes Disrupting Other Pathways
Current data in two additional genes that have not been validated as HTAD genes, MAT2A and FOXE3, indicate that these genes predispose to disease through other pathways.80, 81 FOXE3 encodes a fork head transcription factor and mutations in this gene decrease the ability of SMCs to withstand stress-induced apoptosis and blocking this SMC apoptosis prevented TAD in the Foxe3−/− mice. 80 MAT2A encodes methionine adenosyltransferase II alpha (MAT IIα) that catalyzes the transfer of the adenosyl moiety from ATP to L-methionine to synthesize S-adenosylmethionine (SAM). SAM serves as the methyl group donor for methylation reactions involving DNA, RNA, and protein.82 After donating its methyl group, SAM is converted to S-adenosylhomocysteine (SAH), which is a competitive inhibitor of methyltransferases and is rapidly hydrolysed to homocysteine.83 Further studies are needed to determine if disruption of the production of SAM predisposes to thoracic aortic disease.
ARIH1 encodes a protein that is part of the LINC (linker of nucleoskeleton and cytoskeleton) complex. The LINC complex binds to emerin through nesprins and lamins and these protein influence many different regulatory proteins involved in chromatin modification, transcriptional regulation, and mRNA processing.84 ARIH1 rare variants have been identified in individuals with TAD.85 Aortic SMCs from patients with aortic or cerebrovascular aneurysms and ARIH mutations displayed aberrant nuclear morphology compared to aortic SMCs from controls. Further studies are needed confirm ARIH1 mutations cause TAD before these variants can be used clinically to identify at risk individuals and to further define how disruption of the LINC complex predisposes to TAD.
Genetic Predisposition in Thoracic Aortic Disease Patients who do not have HTAD
Patients with thoracic aortic disease who do not have a family history of the disease or syndromic features of MFS represent 80% of cases and are termed “sporadic” thoracic aortic disease (STAD). Initially, GWAS were used to identify common variants or SNPs in the genome that associated with thoracic aortic disease. Approximately 800 STAD patients who were hospitalized with either an acute aortic dissection or surgical repair of a thoracic aneurysm were used for the initial GWAS. Interestingly, the only GWAS peak for common variants associated with STAD was within FBN1 on chromosome 15.86 Thus, both rare pathogenic variants and common variants in FBN1 predispose to thoracic aortic disease. In contrast to the high risk associated with FBN1 mutations in patients with MFS, common variants in FBN1 only confer a 1.6 – 1.8-fold increased risk. This increased risk was for both presentation with either an aneurysm or dissection, and was also associated with STAD in patients with bicuspid aortic valve.
Another genetic variation in the general population that is associated with thoracic aortic disease involved another validated HTAD gene, MYH11. Copy number variant (CNV) analyses of the same GWAS data on 800 STAD patients identified a greater than12-fold increased risk specifically for aortic dissections that was associated with a recurrent duplication of 16p13.1, a region that involves 9 genes, including MYH11.61 In contrast to the risk associated with FBN1 common variants, the 16p13.1 duplication confers a risk sufficiently high to possibly be clinically actionable in terms of controlling risk factors and screening for aortic enlargement. These initial studies indicate that genetic variants in known HTAD genes, FBN1 and MYH11, contribute to the pathogenesis of sporadic thoracic aortic aneurysms and dissections.
More recently, we sought to identify genetic triggers specifically for sporadic aortic dissections and identified common variants associated with a low risk of dissection in novel genes, ULK4 and LRP1.87 Encouraged by the possibility that genetic variants specific for aortic dissections could be identified, exome sequencing was performed using samples from 355 STAD cases with acute aortic dissections and less than 56 years of age. Individuals who met the clinical diagnostic criteria of a genetic syndrome associated with acute dissections, such as MFS, along with individuals who had a family history of thoracic aortic disease or a known HTAD gene mutation, were excluded from this study. For this study, variants of unknown significance (VUSs) were defined as variants with a minor allele frequency < 0.005 and cannot be classified as either pathogenic or benign using established criteria.4 In contrast, pathogenic variants (i.e., mutations) are defined based on a minor allele frequency < 0.005 and are confirmed to confer a highly penetrant risk for TAD. The frequency of individuals with pathogenic rare variants in HTAD genes in this dissection cohort was 9.3%, 4% of which were FBN1 mutations.88 Perhaps more interesting, the frequency of VUSs identified in HTAD genes within the cohort was significantly higher than in controls, with 30% of the cases having 1 to 3 VUSs. The burden of VUSs was greatest for ACTA2, TGFBR2, TGFBR1, and MYLK. For example, TGFBR2 has been routinely sequenced by genetic diagnostic labs for over 10 years and thus, TGFBR2 mutations are well established. At the same time, there are no common variants in TGFBR2, as defined by a MAF > 0.005. VUSs in TGFBR2 were present in 3.5% of the dissection cases, significantly higher than the controls (0.2%). Furthermore, some of these VUSs decreased the function of the corresponding protein but to a lesser extent than mutations.89 These data emphasize the need for a classification system for variants in disease-causing genes beyond the current system of either pathogenic or benign, which includes low penetrant risk variants that trigger dissections in a stochastic manner or in combination with additional environmental or genetic risks.
In the past, HTAD due to known genes may have been considered to be either syndromic (part of a set of clinical findings) or non-syndromic (occurring as an isolated finding), but the distinction between them has become increasingly blurred. Indeed, current data indicates that variants in HTAD genes can lead to a phenotypic spectrum that ranges from syndromic to non-syndromic risk for TAD. In addition, some variants confer a high penetrant risk for disease, while other variants in the same gene confer a low penetrant risk for disease.89 Functional assays and bioinformatics analyses support the hypothesis that some of these VUS are disease-predisposing variants that trigger disease in a stochastic manner or when other risk factors are present, i.e., hypertension, but are not, in of themselves, highly penetrant pathogenic variants (Figure 1, bottom left). These data indicate that we need to improve the classification of VUSs in HTAD genes so that accurate risk profiling for aortic dissection in the general population can be done, and we have proposed to term variants that increase risk but not to the extent of a pathogenic variant as “risk variants”.89
Clinical Implications of Genes for Thoracic Aortic Disease
As indicated above, disruption of the ability of aortic SMCs to properly mechano-transduce through the elastin-contractile units may be a primary driver for thoracic aortic aneurysms and dissections (Figure 3). These genetic findings further support that hypertension should be aggressively treated and controlled in individuals with HTAD, including at-risk family members even when the specific genetic cause of HTAD has not been identified. In fact, treatment with β-blockers is recommended in individuals with aneurysms with or without hypertension. Individuals who have aortic dilatation or a pathogenic variant in an HTAD-related gene are advised to avoid isometric exercises and contact sports. Other cardiovascular risk factors, including smoking and hyperlipidemia, should also be addressed. Although β-blockers can slow the rate of enlargement of a thoracic aortic aneurysm, the mainstay of prevention of premature deaths from dissection of a type A thoracic aortic aneurysm is surgical repair. Surgery is typically recommended when the diameter of the aorta is approximately twice normal. This recommendation is based on the observation that at aortic diameters greater than 5.5 to 6.0 cm the risk of an adverse event (dissection, rupture, death) exceeds the risk associated with elective repair. However, the timing of aneurysm repair should also be based on the underlying gene altered to trigger the disease.11–13
Genetic testing for rare disease-causing variants in HTAD genes is available worldwide. It is crucial to identify individuals with an increased risk for thoracic aortic disease since dissections and the associated premature deaths are preventable. Identifying the underlying HTAD gene triggering TAD provides powerful information for aortic and other vascular disease management, along with identifying at risk family members. Variants in these genes need to be investigated in a patient with thoracic aortic disease and syndromic features, a family history, or early-onset disease (defined as an aortic dissection less than 56 years of age). It is important to remember that variants in HTAD genes can result in a phenotypic spectrum that ranges from syndromic to a risk for thoracic aortic disease in the absence of any associated syndromic features. In the absence of syndromic features, the only clinical clue for a HTAD gene running in the family is a history of aortic dissections, thoracic aortic aneurysms or unexplained sudden death. Therefore, family history is a powerful tool to identify individuals at risk for aortic dissections. We are striving to establish a precision medicine model with dissection “risk” scores that integrates specific predisposing genetic variant with other risk factors (aortic size, arterial tortuosity, age, sex, hypertension, plans for pregnancy, etc.) to predict an individual’s risk for an acute aortic dissection, and thus can be used to timing of surgical repair of the thoracic aorta. Going forward, a main challenge for identifying all the genes and variants that predispose to TAD is the significant genetic heterogeneity for this disease. In addition, the classification of rare variants in known HTAD genes as to whether they are pathogenic variants, low penetrant risk variants or benign needs to be improved.
The Genetics of Abdominal Aortic Aneurysm
Abdominal aortic aneurysm (AAA) is a common chronic disorder90 that shares many risk factors with occlusive forms of atherosclerosis but also appears to be a distinct pathologic entity. For example, diabetes is negatively associated with both AAA susceptibility and progression (AAA growth rate), which is in contrast to atherosclerosis.91 Nevertheless, it is significant to note the correlation between aneurysmal or pre-aneurysmal infra-renal aortic dimensions and non-AAA related cardiovascular mortality,92 emphasizing a substantive degree of overlap between these disorders.
Population epidemiology evidence suggests a strong genetic component in AAA risk, with twin studies indicating that heritability (the proportion of variance attributable to genetic effects) may be as high as 70%.93 In population studies, approximately 20% of AAA patients report an affected first-degree relative, which is substantially higher than the 2–10% typically reported by unaffected controls.94–98 Correspondingly, a positive family history has been shown to be associated with an approximate doubling of an individual’s risk of developing AAA.99 In the vast majority of AAA patients, the pattern of inheritance appears to be autosomal, although evidence for both recessive and dominant models have been suggested.100 These facts highlight a point of caution when considering the genetic risk of AAA, as it may be important to note the potential distinction between patients presenting with familial versus sporadic disease. Family-based linkage analysis studies may well be investigating a different genetic entity to that identified through GWAS focused on individual patients with AAAs, the majority of which will not have a family history.
Nevertheless, it is clear that AAA does cluster within some families94, 96, 97, 101, 102 and this along with observations of the co-occurrence of thoracic aortic aneurysm (TAA) and AAA,103, 104 have led to investigations into a potential shared genetic risk between dilating arterial phenotypes, such as TAA, AAA and intracranial aneurysm.105 Although some potentially pan-aneurysmal effects, notably in the 9p21 (CDKN2B-AS1)105, 106 and 15q21 (near the FBN1 gene)105 loci, have been reported, the overarching impression appears to be, at best, a very modest genetic overlap between these aneurysmal phenotypes.
The past era of candidate gene association studies
Approximately 100 studies have performed candidate gene based investigations into the genetic basis of AAA. Bradley and colleagues reviewed the state of reported AAA genetic associations in 2014107 and showed that while over 250 genes had been investigated, resulting in 87 positive associations, only six were supported by strong evidence, with a further three deemed to have moderate supporting evidence. A range of genetics study design issues were catalogued, including failure to correct for multiple testing, potential systematic genotyping errors (no assessment of Hardy-Weinberg equilibrium), flexible post-hoc inheritance model testing, small sample size, population stratification, lack of validation in independent cohorts and quality of control samples. Unfortunately, the entire early population genetics era was blighted by such deficiencies, leading to a situation whereby the majority of previously reported candidate gene associations are likely false positives.
Family-based linkage studies
The most extensive family-based linkage analysis study for familial AAA was conducted by Shibamura and colleagues and examined 119 families.108 Two loci, 4q31 and 19q13 were shown to be in linkage with AAA, but only when sex and the number of affected individuals in a family group were included as covariates. The large size of the regions implicated in this analysis makes it very difficult to identify the causative genes, however, these regions do harbour genes of potential interest. These include the endothelin receptor type A within 4q31 and a number of kallikrien genes within 19q13. SNP variants of endothelin receptor type A (EDNRA) are robustly associated with coronary artery disease (CAD)109, while genes such as kallikrien 1 (KLK1) have been shown to have altered expression in AAA tissue.110 Unfortunately, due in part to the late age of onset of AAA disease, the difficulties associated with collecting large numbers of affected families has resulted in most researchers shifting towards case-control genome-wide association studies. As highlighted above, familial AAA, as the focus of linkage studies and the more common sporadic AAA presentation, may have different genetic architectures. Further work is needed to validate the results of previous familial AAA linkage analysis associations. Such opportunities may yet develop as AAA population screening becomes increasingly more available throughout the world.
The GWAS era: Independently validated associations at a genome-wide level of significance
To date the largest analysis of AAA genetics was performed by a multi-national consortium that examined over ten thousand AAA cases. The study consisted of a meta-analysis of six GWAS from five countries (The United States, United Kingdom, Netherlands, Iceland and New Zealand), with validation analysis in a further eight independent cohorts.111 It should be noted that family history was not collected in each of the contributing GWAS and it was therefore not possible to differentiate between familial and sporadic AAA cases. In the New Zealand cohorts, which contributed 20 percent of the discovery meta-GWAS cases, approximately 17 percent of cases had an affected first degree relative. The results of the AAA meta-GWAS are therefore likely to be far more pertinent to sporadic, rather than familial AAA. It is also unclear if treating familial and sporadic AAA as the same affection status had any substantive effect on the resulting association analysis.
Nevertheless, ten genetic loci were associated with AAA, nine of which remained below a genome wide level of significance (p=5×10−8) when validated within independent cohorts.
The locus that did not reach genome-wide significance, when combined with the validation cohorts, was 12q13 (LRP1), however, it should be noted that previous reports have suggested genome wide significance for a SNP in this locus.112 The peak meta-GWAS SNP (rs1385526111, in complete linkage disequilibrium with the original SNP reported for this locus, rs1466535112) is within an intron of the LRP1 gene. In addition, as noted above, LRP1 variants have also been associated with sporadic acute aortic dissections.87 Of note, the peak AAA (rs1385526) and STAAD (rs11172113) SNPs are in modest linkage disequilibrium (r2=0.54, CEU), with the aortic dissection risk SNP being significantly associated with AAA (P<3.8×10−8, I2=0, pHet=0.58) in the meta-GWAS discovery cohort. The balance of evidence appears to suggest that the LRP1 locus should be considered to be associated with AAA. Therefore, at this point in time, a total of ten loci should be considered to be robustly associated with AAA.
The loci, in chronological order of identification and with the nearest gene(s), are 9p21 (CDKN2BAS1/ANRIL)106, 9q33 (DAB2IP)113, 12q13 (LRP1)112, 1q21.3 (IL6R)114, 1p13.3 (PSRC1/CELSR2/SORT1)115 and 19p13.2 (LDLR)116 followed by the four loci identified in the meta-GWAS111, 1q32.3 (SMYD2), 13q12.11 (LINC00540), 20q13.12 (PLTP/PCIF1/MMP9/ZNF335), and 21q22.2 (ERG).
Although the recent AAA meta-GWAS performed an extensive series of bioinformatic analyses and lookups, interpreting the functional effects of these AAA SNPs remains a challenge. Unlike monogenic disorders, such as Marfan syndrome that can be linked to a functional variant influencing a biologically plausible gene, many of the AAA variants are not intragenic nor do they appear to directly alter functional elements of genes, such as promoters or CpG islands. However, by examining potential effects through tools, such as expression quantitative trait loci (eQTLs), phenome-wide association (PheWAS) and chromatin interaction databases (such as GWAS3D), a series of (biologically plausible) gene associations were identified. When placed in a series of different network analysis tools genes such as MMP9, IL6R and LDLR were highlighted as hub features in the pathway networks. This is encouraging given that metalloproteinase activity117, tissue inflammation118–120 and lipoprotein accumulation120 are documented features of AAA pathobiology.
Genetic overlap between CAD and AAA
Consistent with the phenotypic overlap noted above, several of these genetic features appear to overlap with the genetic associations reported for CAD.109, 121 As noted above, AAA is now recognised as a related but distinct entity from occlusive atherosclerotic disease, such as CAD. Nevertheless, given that over 70 robust genetic associations have now been reported for CAD109, 121 it is worth considering the potential overlap between validated CAD and AAA associations.
The AAA loci 9p21 (CDKN2BAS1/ANRIL),106 1q21.3 (IL6R),114 1p13.3 (PSRC1/CELSR2/SORT1),115 19p13.2 (LDLR)116 and 20q13.12 (PLTP/PCIF1/MMP9/ZNF335)111 appear to have the strongest overlap with occlusive atherosclerotic diseases.109, 121, 122 Of note, other potential AAA associations, such as SNPs associated with the LPA gene,123 have been shown to lose their statistical significance when those cases with concurrent CAD were excluded from the analysis, suggesting that the observed unadjusted AAA associations were likely being confounded by the presence of concurrent atherosclerotic disease. The concept of ‘pan-atherosclerotic’ genetic association is well recognised for markers such as 9p21(CDKN2BAS1/ANRIL) and also matches the broad arterial disease phenotype associations of circulating markers such as Lp(a) protein.124 There will undoubtedly be numerous other regions of genetic overlap between AAA and ‘occlusive’ atherosclerotic diseases, however the identification of these common loci is hampered by the limited size of the AAA meta-GWAS. For example, the SNP rs964184 (11q23.1 near ZNF259, APOA5- A4-C3-A) is significantly and reproducibly associated with CAD (P=1×10−17)125, while this SNP in the AAA meta-GWAS summary data also suggests a potential association (P<5×10−5, I2=0, pHet=0.84), but the association falls below a genome-wide multiple testing significance threshold.
9p21 (CDKN2BAS1/ANRIL)
Both the lead SNP in the AAA meta-GWAS (rs1075727)111 and the original SNP reported at this locus (rs10757278)106 are in strong linkage disequilibrium (r2>0.9 CEU). The 9p21 locus is strongly associated with a broad range of arterial pathologies, including coronary artery disease.106 These SNPs appear to influence CDKN2A and CDKN2B expression via alterations in the neighboring non-coding antisense CDKN2BAS1.126, 127 These cyclin dependent kinase inhibitors are strong mediators of vascular tissue proliferation and inflammation.127
1q21.3 (IL6R)
Both the lead SNP in the AAA meta-GWAS (rs4129267)111 and the original SNP reported at this locus (rs7529229)114 are in linkage disequilibrium (r2=1 CEU) and are located within introns of the interleukin 6 receptor (IL6R) gene. Circulating levels of IL-6 have been shown to be associated with the presence of small AAAs114 and both of these AAA SNPs are in high LD (r2>0.95, CEU) with a SNP (rs2228145) which has been shown to be strongly associated with circulating IL-6 levels.128 Nevertheless, this locus does not appear to be AAA specific as it has also been shown to be associated with a wide range of chronic conditions including coronary artery disease109 and asthma.129
1p13.3 (PSRC1/CELSR2/SORT1)
Both the lead SNP in the AAA meta-GWAS (rs602633)111 and the original SNP reported at this locus (rs599839)115 are in linkage disequilibrium (r2=1 CEU) and are located near the promoter of PSRC1. There are however, several other genes in this region, including CELSR2 and SORT1; and eQTL and chromatin interaction data suggest that SORT1 is likely to be the most significantly influenced by these SNPs.111, 130 The sortilin-1 gene (SORT1) is expressed in the liver and regulates plasma LDL-cholesterol via interactions with PCSK9 acidic sphingomyelinase and apolipoprotein B100 in hepatocytes.131 The gene is also expressed in the aortic wall115 and recent work suggests that SORT1 may play an important role as a regulator of vascular smooth muscle calcification.132 SNPs in this locus are also associated with CAD125 and given its association with both dyslipidemia and arterial wall calcification, this locus appears to have a broad cardiovascular risk association.
19p13.2 (LDLR)
The same variant (rs6511720) was reported as the peak SNP for this locus in both the meta-GWAS111 and original report for this AAA association.116 LDLR encodes for a member of the LDL-cholesterol cell surface receptor family and this SNP variant (or others in high LD) have a strong association with both increased plasma LDL-cholesterol133 and CAD.109 Importantly, LDLR mutations are the most common cause of familial hypercholesterolemia. Due to its strong association with dyslipidemia this locus appears to confer pan-atherosclerotic risk and, as with the variants in 9p21, 1q21.3 and 1p13.3, does not appear to have a AAA specific association. This notwithstanding, Mendelian randomization studies have suggested that dyslipidemia, and in particular elevated LDL, are potentially causally associated with AAA.134, 135
20q13.12 (PLTP/PCIF1/MMP9/ZNF335)
The peak SNP (rs3827066) for this locus is located within an intron of the ZNF335 gene, however there are several other genes in the surrounding 100kb region, including PLTP, PCIF1, NEURL2 and MMP9. MMP9 is an obvious gene of interest, given the large body of work implicating this metalloproteinase in end-stage AAA pathobiology.136 However, it should be noted that this SNP most-likely has the strongest association with PLTP, as evidenced by independent eQTL data derived from aortic tissue and chromatin interaction analysis. PLTP plays a role in cholesterol transport, especially HDL.137 Induced overexpression of PLTP in mice results in an elevated atherogenic lipid profile, increased atherosclerotic lesion area, and reduced plaque stability (as assessed by collagen content).138 PLTP expression is significantly higher in AAA tissue than in control aorta.111 Interestingly, there may be an indirect link between this SNP and MMP9, since PLTP inhibition has been shown to alter ERK and NF-κB activation, and in turn, MMP-9 levels within lung tissues.139 There is no literature that would suggest a potential link with the nearest gene (ZNF335) and AAA, with the only reported genetic association for this gene being with celiac disease.140
While this locus was reported as a potential AAA specific association111, a recent meta-analysis identified the rs3827066 SNP as also being associated with CAD.122 The cumulative evidence therefore suggests that this locus is likely to exert its pathogenic influences in a similar fashion to that of the SORT1 locus, primarily through aberrations of lipid metabolism but with potential influences on vessel wall inflammation and extracellular matrix composition.
Potential AAA specific associations
The observation of cross-phenotype genetic associations with other cardiometabolic traits is not particularly surprising given the epidemiology of AAA. Unfortunately, however, conventional cardiovascular risk reduction therapies, such as antihypertensive and anti-dyslipidemic medications, have no significant effects on AAA growth rates.141 This lack of effective medical therapies highlights the need to improve our understanding of the disease specific pathobiological features of AAA. The recently reported AAA meta-GWAS identified several loci with no apparent overlap with either CAD burden or cardiovascular disease-related metabolic traits such as diabetes, hypertension or dyslipidemia.
1q32.3 (SMYD2)
The peak SNP (rs1795061) in this locus is located near the promoter of SMYD2. The SET and MYND domain containing 2 (SMYD2) gene is highly expressed in muscle cells and is associated with myofibril organisation. The SMYD2 protein interacts with three functional groups of molecules, involved with chromatin remodelling and histone modification, DNA replication and repair and molecular chaperones such as heat shock protein 90 (Hsp90).142 The ability of SMYD2 to methylate Hsp90 is potentially significant as methylated Hsp90 and Smyd2 can form a complex with the sarcomere protein Titin’s N2A domain to alter sarcomere stability. SMYD2 also appears to regulate macrophage associated inflammatory cytokine production by methylating the promoters of the interleukin 6 (IL6) and tumor necrosis factor alpha (TNF) genes. This in turn leads to inhibition of NF-κB and ERK signalling143, with both of these pathways having been implicated as regulators of vascular remodelling and AAA formation.144, 145 The potential biological association between SMYD2 and AAA is further supported by observations such as the inhibition of Hsp90 reducing AAA formation in murine models146 and the significant hypomethylation of the SMYD2 promoter in smooth muscle cells extracted from AAAs compared to controls.147 SMYD2 methylation in aortic smooth muscle correlates with gene expression and the protein appears to have lower cytoplasmic abundance in smooth muscle within the residual AAA tunica media compared with that of non-aneurysmal controls.147
12q13 (LRP1)
As discussed above, SNPs in this locus have also been shown to be associated with thoracic aortic dissection. LRP1 encodes for low-density lipoprotein receptor-related protein 1, a gene that is known to be highly expressed in vascular smooth muscle, macrophages, and fibroblasts. Smooth muscle cell specific LRP1 knockout mice have altered smooth muscle function, including enhanced migration, abnormally activated TGF-β and PDGF signalling148, resulting in aortic wall structural alterations leading to aneurysm formation.149 Although some LRP1 SNPs are linked with circulating lipid levels, eQTL data suggests that the AAA associated LRP1 variants influence aortic, but not hepatic, gene expression.112 In addition, the AAA associated LRP1 variants (rs1385526111 and rs1466535112) appear to be independently associated from dyslipidemia, diabetes, blood pressure or history of CAD.112 The LRP1 pathway is therefore of considerable interest as a potentially disease specific driver of both thoracic and abdominal aortic aneurysm.
13q12.11 (LINC00540),
The AAA associated SNPs in this locus are in a rather gene barren region, though it is located 3’ to a long non-coding RNA (LINC00540). While LINC00540 has no currently known function, both chromatin interaction and eQTL analyses independently suggest a distal association between the peak SNP in this locus and the fibroblast growth factor-9 (FGF9) gene. A subsequent lookup of an independent abdominal aortic wall whole genome mRNA expression dataset indicated increased FGF9 expression within aneurysmal aortic tissue. The FGF9 gene has been shown to be a key regulator of respiratory smooth muscle cell differentiation.150, as well as vascular smooth muscle and endothelial cell apoptosis thereby influencing post-infarct angiogenesis in cardiac tissue.151 Extensive angiogenesis is an important pathogenic feature of AAA, being associated with medial atrophy, adventitial inflammation120 and sac rupture sites.152
21q22.2 (ERG)
The peak SNP in this locus (rs2836411) is located within an intron of the ETS (erythroblast transformation-specific) -related gene (ERG) and was shown to be associated with ERG expression in an independent eQTL dataset derived from mammary artery tissue.111 ERG encodes a transcriptional regulator protein that is normally present in hematopoietic and endothelial cells. ERG has a role in vascular endothelial growth factor mediated vascular development, and has also been shown to control endothelial cell activation.153 As such, ERG is proposed as a key mediator of vascular angiogenesis and inflammation, both of which play key roles in the development of AAA.120, 152 ERG also influences embryonic development of the aorta.154 Consistent with the ‘fetal origins of adult disease hypothesis’, such alterations in in utero aortic development may have a role in later-life risk of developing AAA.155 ERG expression correlates with matrix metalloproteinase 9 expression in human prostate cancer cells.156 Taken together this data, though limited, points to several potential roles by which ERG may influence the development of AAA.
Pathway analysis
The AAA SNP loci and their predicted gene associations allow for gene pathway analyses to be performed.111 Using the integrated gene pathway function analysis tool DEPICT the strongest predicted gene set was long bone epiphyseal plate formation. This is intriguing given that previous studies have reported tall stature as a risk factor for AAA157 and conversely short stature as being associated with occlusive CAD.158, 159 Other predicted gene pathway sets were also consistent with our current pathophysiological understanding of AAA. These included plasma lipoprotein clearance, abnormal vascular smooth muscle physiology/ increased vasodilation, TGF-β regulation, inflammation-induced extracellular matrix remodeling, acute phase response including IL-6 secretion, abnormal T and B cell function/ activation, apoptosis, hyperglycemia and the phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha, c-Jun N-terminal kinase, and mitogen-activated kinase-like protein cascades.
Ingenuity Pathway Analysis (IPA) and Consensus PathDB network analysis both demonstrated similar results, and suggested a potential central role for MMP9 in AAA. IPA identified direct interactions between ERG, IL6R and LDLR with MMP9. Concordantly, ConsensusPathDB suggested a direct interaction between ERG and MMP9, with secondary interactions between both SMYD2 and LDLR and MMP9.
Importantly, particularly in the context of a polygenic disorder, these results suggest that the novel loci could act in concert, either synergistically or antagonistically, to affect the AAA phenotype. These potential interactions require further investigation but may provide much needed insights into novel, AAA disease-specific, therapeutic developments.
While the AAA meta-analysis brought together the largest case series to date, it is a long way from reaching the statistical power for the detection of genetic associations that have been reported for related phenotypes such as CAD. While it represents a good starting point, a considerable amount of further work is required to generate a more complete picture of the genetic basis of this disorder. Furthermore, while our understanding of the genetic basis of AAA has substantially evolved over the last decade, the therapeutic implications of these observations remain unclear.
Potential gene-environment interactions
Given that smoking is by far the strongest environmental risk factor for AAA, and smoking consumption rates are significantly influenced by genetic factors, it is intriguing to consider whether such variants may be over-represented amongst AAA patients. If this were the case then it would represent an example of a gene influencing environment effect. Thorgeirsson and colleagues160 identified a series of SNPs in the CHRNB3-CHRNA6 (nicotinic acetylcholine receptor subunits) gene region of chromosome 15, which were associated with daily cigarette consumption rates. By far the strongest association was for rs1051730 (A effect allele). While a lookup of the AAA metaGWAS indicates that the same allele is overrepresented in AAA patients (odds ratio 1.10 (95%CI 1.04–1.15, P=1.2×10−3, I2=0.41) this association falls well below genome-wide significance.
Although outside the scope of this review, another interesting mode of gene-environment interaction can be investigated via epigenetics, including differential DNA methylation and microRNA expression. AAA associated DNA methylation has been previously postulated,161 and global differences in DNA methylation levels have been reported both for the presence and severity of aneurysmal disease.147 In addition, as described above, a targeted analysis of AAA genetic risk loci indicated that the SMYD2 locus is also differentially methylated.147 AAA epigenome wide association analysis is needed to fully ascertain the DNA methylation epigenetic associations in AAA, fortunately such a study, performed by members of the International Aneurysm Consortium, is in the final stages of completion and should be published in the near future.
This compendium summarises the current understanding of the inherited basis for aortic aneurysmal disease, and clearly highlights the need to differentiate between thoracic and abdominal aortic manifestations. Approximately 20% of thoracic aortic aneurysms are associated with an autosomal pattern of inheritance from a mutation in a single gene, whereas abdominal aortic aneurysms do not typically demonstrate such inheritance, but rather appear to present as a polygenic disorder involving variants of weaker effect. This notwithstanding, there are some distinct biological pathway similarities between TAAD and AAA, these include the TGF-β and LRP1 pathways, as well as other aspects of vascular smooth muscle cell function.
Currently, over a dozen causative genes have been validated for heritable thoracic aortic disease. Each confers a high risk for disease and genetic testing for these genes is now offered clinically. The situation is far less advanced for AAA, and while polygenic AAA risk scores are being developed, using genetic variants to predict AAAs has not progressed to the extent that it can be used to identify at risk individuals.
Acknowledgments
Sources of Funding:
This work was supported by the National Heart, Lung and Blood Institute (RO1 HL62594 and P01HL110869–01), the John Ritter and Remembrin’ Benjamin Foundations to Dr. Milewicz and the Health Research Council of New Zealand (08–75 and 14–155) to Dr. Jones.
Non-standard Abbreviations and Acronyms:
- AAA
abdominal aortic aneurysm
- ACMG
American college of medical genetics
- AOS
aneurysm-osteoarthritis syndrome
- BAV
bicuspid aortic valve
- CAD
coronary artery disease
- CNV
copy number variation
- ECM
extracellular matrix
- EGF
epithelial growth factor
- eQTL
expression quantitative trait loci
- GWAS
genome wide association study
- HTAD
heritable risk for thoracic aortic disease
- HHT
hereditary hemorrhagic telangiectasia
- JPS
juvenile polyposis
- LDS
Loeys-Dietz syndrome
- LINC
linker of nucleoskeleton and cytoskeleton
- MAF
minor allele frequency
- MFS
Marfan syndrome
- OMIM
online mendelian inheritance in man
- PDGF
platelet-derived growth factor
- SAM
S-adenosylmethionine
- SMC
smooth muscle cell
- SNP
single nucleotide polymorphism
- STAD
sporadic thoracic aortic disease
- TAD
thoracic aortic disease
- TGF
transforming growth factor
- TNF
tumor necrosis factor
- RLC
regulatory light chains
- vEDS
vascular Ehlers-Danlos syndrome
- VUS
variant of unknown significance
Footnotes
Disclosures section:
None.
References:
- 1.Hoyert DL, Arias E, Smith BL, Murphy SL, Kochanek KD. Deaths: Final data for 1999. Natl Vital Stat Rep. 2001;49:1–113 [PubMed] [Google Scholar]
- 2.Huynh TT, Starr JE. Diseases of the thoracic aorta in women. J Vasc Surg. 2013;57:11S–17S [DOI] [PubMed] [Google Scholar]
- 3.Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461:747–753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, Voelkerding K, Rehm HL, ACMB Laboratory Quality Assurance Committee. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the american college of medical genetics and genomics and the association for molecular pathology. Genet Med. 2015;17:405–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Howard DP, van Lammeren GW, Redgrave JN, Moll FL, de Vries JP, de Kleijn DP, de Borst GJ, Pasterkamp G, Rothwell PM. Histological features of carotid plaque in patients with ocular ischemia versus cerebral events. Stroke. 2013;44:734–739 [DOI] [PubMed] [Google Scholar]
- 6.Prakash SK, Haden-Pinneri K, Milewicz DM. Susceptibility to acute thoracic aortic dissections in patients dying outside the hospital: An autopsy study. Am Heart J. 2011;162:474–479 [DOI] [PubMed] [Google Scholar]
- 7.Finkbohner R, Johnston D, Crawford ES, Coselli J, Milewicz DM. Marfan syndrome. Long-term survival and complications after aortic aneurysm repair. Circulation. 1995;91:728–733 [DOI] [PubMed] [Google Scholar]
- 8.Silverman DI, Burton KJ, Gray J, Bosner MS, Kouchoukos NT, Roman MJ, Boxer M, Devereux RB, Tsipouras P. Life expectancy in the marfan syndrome. Am J Cardiol. 1995;75:157–160 [DOI] [PubMed] [Google Scholar]
- 9.Hiratzka LF, Bakris GL, Beckman JA, et al. 2010 ACCF/AHA/AATS/ACR/ASA/SCA/SCAI/SIR/STS/SVM guidelines for the diagnosis and management of patients with thoracic aortic disease. A report of the american college of cardiology foundation/american heart association task force on practice guidelines, american association for thoracic surgery, american college of radiology, american stroke association, society of cardiovascular anesthesiologists, society for cardiovascular angiography and interventions, society of interventional radiology, society of thoracic surgeons, and society for vascular medicine. J Am Coll Cardiol. 2010;55:e27–e129 [DOI] [PubMed] [Google Scholar]
- 10.Pape LA, Tsai TT, Isselbacher EM, et al. Aortic diameter >or = 5.5 cm is not a good predictor of type a aortic dissection: Observations from the international registry of acute aortic dissection (IRAD). Circulation. 2007;116:1120–1127 [DOI] [PubMed] [Google Scholar]
- 11.Jondeau G, Ropers J, Regalado E, et al. International registry of patients carrying tgfbr1 or tgfbr2 mutations: Results of the MAC (montalcino aortic consortium). Circ Cardiovasc Genet. 2016;9:548–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Regalado ES, Guo DC, Prakash S, et al. Aortic disease presentation and outcome associated with acta2 mutations. Circ Cardiovasc Genet. 2015;8:457–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Milewicz DM, Regalado ES, Guo DC. Treatment guidelines for thoracic aortic aneurysms and dissections based on the underlying causative gene. J Thorac Cardiovasc Surg. 2010;140:S2–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dietz HC, Cutting GR, Pyeritz RE, Maslen CL, Sakai LY, Corson GM, Puffenberger EG, Hamosh A, Nanthakumar EJ, Curristin SM, et al. Marfan syndrome caused by a recurrent de novo missense mutation in the fibrillin gene. Nature. 1991;352:337–339 [DOI] [PubMed] [Google Scholar]
- 15.Loeys BL, Schwarze U, Holm T, et al. Aneurysm syndromes caused by mutations in the tgf-beta receptor. N Engl J Med. 2006;355:788–798 [DOI] [PubMed] [Google Scholar]
- 16.Mizuguchi T, Collod-Beroud G, Akiyama T, et al. Heterozygous tgfbr2 mutations in marfan syndrome. Nat Genet. 2004;36:855–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Albornoz G, Coady MA, Roberts M, Davies RR, Tranquilli M, Rizzo JA, Elefteriades JA. Familial thoracic aortic aneurysms and dissections - incidence, modes of inheritance, and phenotypic patterns. Ann Thorac Surg. 2006;82:1400–1406 [DOI] [PubMed] [Google Scholar]
- 18.Biddinger A, Rocklin M, Coselli J, Milewicz DM. Familial thoracic aortic dilatations and dissections: A case control study. J Vasc Surg. 1997;25:506–511 [DOI] [PubMed] [Google Scholar]
- 19.Milewicz DM, Chen H, Park ES, Petty EM, Zaghi H, Shashidhar G, Willing M, Patel V. Reduced penetrance and variable expressivity of familial thoracic aortic aneurysms/dissections. Am J Cardiol. 1998;82:474–479 [DOI] [PubMed] [Google Scholar]
- 20.Renard M, Francis CA, Ghosh R, et al. Evaluation of the clinical validity of genes for heritable thoracic aortic aneurysm and dissections: Application of the clinical genome resource framework. J Am Coll Cardiol. 2018;72:605–615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boileau C, Guo DC, Hanna N, et al. TGFB2 mutations cause familial thoracic aortic aneurysms and dissections associated with mild systemic features of marfan syndrome. Nat Genet. 2012;44:916–921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Byers PH, Holbrook KA, Barsh GS, Smith LT, Bornstein P. Altered secretion of type iii procollagen in a form of type iv ehlers-danlos syndrome. Biochemical studies in cultured fibroblasts. Lab Invest. 1981;44:336–341 [PubMed] [Google Scholar]
- 23.Guo DC, Pannu H, Tran-Fadulu V, et al. Mutations in smooth muscle alpha-actin (ACTA2) lead to thoracic aortic aneurysms and dissections. Nat Genet. 2007;39:1488–1493 [DOI] [PubMed] [Google Scholar]
- 24.Guo DC, Regalado E, Casteel DE, et al. Recurrent gain-of-function mutation in prkg1 causes thoracic aortic aneurysms and acute aortic dissections. Am J Hum Genet. 2013;93:398–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo DC, Regalado ES, Gong L, et al. Lox mutations predispose to thoracic aortic aneurysms and dissections. Circ Res. 2016;118:928–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Regalado ES, Guo DC, Villamizar C, et al. Exome sequencing identifies smad3 mutations as a cause of familial thoracic aortic aneurysm and dissection with intracranial and other arterial aneurysms. Circ Res. 2011;109:680–U220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van de Laar IMBH, Oldenburg RA, Pals G, et al. Mutations in SMAD3 cause a syndromic form of aortic aneurysms and dissections with early-onset osteoarthritis. Nat Genet. 2011;43:121–U165 [DOI] [PubMed] [Google Scholar]
- 28.Wang L, Guo DC, Cao JM, et al. Mutations in myosin light chain kinase cause familial aortic dissections. Am J Hum Genet. 2010;87:701–707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhu L, Vranckx R, Khau Van Kien P, Lalande A, Boisset N, Mathieu F, Wegman M, Glancy L, Gasc JM, Brunotte F, Bruneval P, Wolf JE, Michel JB, Jeunemaitre X. Mutations in myosin heavy chain 11 cause a syndrome associating thoracic aortic aneurysm/aortic dissection and patent ductus arteriosus. Nat Genet. 2006;38:343–349 [DOI] [PubMed] [Google Scholar]
- 30.Guo DC, Papke CL, Tran-Fadulu V, et al. Mutations in smooth muscle alpha-actin (ACTA2) cause coronary artery disease, stroke, and moyamoya disease, along with thoracic aortic disease. Am J Hum Genet. 2009;84:617–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davis EC. Smooth muscle cell to elastic lamina connections in developing mouse aorta. Role in aortic medial organization. Lab Invest. 1993;68:89–99 [PubMed] [Google Scholar]
- 32.Humphrey JD, Milewicz DM, Tellides G, Schwartz MA. Cell biology. Dysfunctional mechanosensing in aneurysms. Science. 2014;344:477–479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Humphrey JD, Schwartz MA, Tellides G, Milewicz DM. Role of mechanotransduction in vascular biology: Focus on thoracic aortic aneurysms and dissections. Circ Res. 2015;116:1448–1461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Corson GM, Chalberg SC, Dietz HC, Charbonneau NL, Sakai LY. Fibrillin binds calcium and is coded by cdnas that reveal a multidomain structure and alternatively spliced exons at the 5’ end. Genomics. 1993;17:476–484 [DOI] [PubMed] [Google Scholar]
- 35.Pyeritz RE, McKusick VA. The marfan syndrome: Diagnosis and management. N Engl J Med. 1979;300:772–777 [DOI] [PubMed] [Google Scholar]
- 36.Pinard A, Salgado D, Desvignes JP, Rai G, Hanna N, Arnaud P, Guien C, Martinez M, Faivre L, Jondeau G, Boileau C, Zaffran S, Beroud C, Collod-Beroud G. WES/WGS reporting of mutations from cardiovascular “actionable” genes in clinical practice: A key role for umd knowledgebases in the era of big databases. Hum Mutat. 2016;37:1308–1317 [DOI] [PubMed] [Google Scholar]
- 37.Milewicz DM, Guo DC, Tran-Fadulu V, Lafont AL, Papke CL, Inamoto S, Kwartler CS, Pannu H. Genetic basis of thoracic aortic aneurysms and dissections: Focus on smooth muscle cell contractile dysfunction. Annu Rev Genomics Hum Genet. 2008;9:283–302 [DOI] [PubMed] [Google Scholar]
- 38.Milewicz DM. Identification of defects in the fibrillin gene and protein in individuals with the marfan syndrome and related disorders. Tex Heart Inst J. 1994;21:22–29 [PMC free article] [PubMed] [Google Scholar]
- 39.Hollister DW, Godfrey M, Sakai LY, Pyeritz RE. Immunohistologic abnormalities of the microfibrillar-fiber system in the marfan syndrome. N Engl J Med. 1990;323:152–159 [DOI] [PubMed] [Google Scholar]
- 40.Milewicz DM, Pyeritz RE, Crawford ES, Byers PH. Marfan syndrome: Defective synthesis, secretion, and extracellular matrix formation of fibrillin by cultured dermal fibroblasts. J Clin Invest. 1992;89:79–86 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dietz HC, Saraiva JM, Pyeritz RE, Cutting GR, Francomano CA. Clustering of fibrillin (FBN1) missense mutations in marfan syndrome patients at cysteine residues in egf-like domains. Hum Mutat. 1992;1:366–374 [DOI] [PubMed] [Google Scholar]
- 42.Whiteman P, Hutchinson S, Handford PA. Fibrillin-1 misfolding and disease. Antioxid Redox Signal. 2006;8:338–346 [DOI] [PubMed] [Google Scholar]
- 43.Suk JY, Jensen S, McGettrick A, Willis AC, Whiteman P, Redfield C, Handford PA. Structural consequences of cysteine substitutions c1977y and c1977r in calcium-binding epidermal growth factor-like domain 30 of human fibrillin-1. J Biol Chem. 2004;279:51258–51265 [DOI] [PubMed] [Google Scholar]
- 44.Jones JA, Spinale FG, Ikonomidis JS. Transforming growth factor-beta signaling in thoracic aortic aneurysm development: A paradox in pathogenesis. J Vasc Res. 2009;46:119–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ramirez F, Pereira L. The fibrillins. Int J Biochem Cell Biol. 1999;31:255–259 [DOI] [PubMed] [Google Scholar]
- 46.Bunton TE, Biery NJ, Myers L, Gayraud B, Ramirez F, Dietz HC. Phenotypic alteration of vascular smooth muscle cells precedes elastolysis in a mouse model of marfan syndrome. Circ Res. 2001;88:37–43 [DOI] [PubMed] [Google Scholar]
- 47.Lee VS, Halabi CM, Hoffman EP, Carmichael N, Leshchiner I, Lian CG, Bierhals AJ, Vuzman D, Brigham Genomic M, Mecham RP, Frank NY, Stitziel NO. Loss of function mutation in lox causes thoracic aortic aneurysm and dissection in humans. Proc Natl Acad Sci U S A. 2016;113:8759–8764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gdynia HJ, Kuhnlein P, Ludolph AC, Huber R. Connective tissue disorders in dissections of the carotid or vertebral arteries. J Clin Neurosci. 2008;15:489–494 [DOI] [PubMed] [Google Scholar]
- 49.Guo DC, Regalado ES, Pinard A, et al. LTBP3 pathogenic variants predispose individuals to thoracic aortic aneurysms and dissections. Am J Hum Genet. 2018;102:706–712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robertson IB, Horiguchi M, Zilberberg L, Dabovic B, Hadjiolova K, Rifkin DB. Latent tgf-beta-binding proteins. Matrix Biol. 2015;47:44–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Y, Dabovic B, Annes JP, Rifkin DB. Latent tgf-beta binding protein-3 (LTBP-3) requires binding to tgf-beta for secretion. FEBS Lett. 2002;517:277–280 [DOI] [PubMed] [Google Scholar]
- 52.Zilberberg L, Todorovic V, Dabovic B, Horiguchi M, Courousse T, Sakai LY, Rifkin DB. Specificity of latent TGF-beta binding protein (LTBP) incorporation into matrix: Role of fibrillins and fibronectin. J Cell Physiol. 2012;227:3828–3836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barbier M, Gross MS, Aubart M, et al. Mfap5 loss-of-function mutations underscore the involvement of matrix alteration in the pathogenesis of familial thoracic aortic aneurysms and dissections. Am J Hum Genet. 2014;95:736–743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Capuano A, Bucciotti F, Farwell KD, Tippin Davis B, Mroske C, Hulick PJ, Weissman SM, Gao Q, Spessotto P, Colombatti A, Doliana R. Diagnostic exome sequencing identifies a novel gene, emilin1, associated with autosomal-dominant hereditary connective tissue disease. Hum Mutat. 2016;37:84–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Szabo Z, Crepeau MW, Mitchell AL, Stephan MJ, Puntel RA, Yin Loke K, Kirk RC, Urban Z. Aortic aneurysmal disease and cutis laxa caused by defects in the elastin gene. J Med Genet. 2006;43:255–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Al-Hassnan ZN, Almesned AR, Tulbah S, Hakami A, Al-Omrani A, Al Sehly A, Mohammed S, Majid S, Meyer B, Al-Fayyadh M. Recessively inherited severe aortic aneurysm caused by mutated EFEMP2. Am J Cardiol. 2012;109:1677–1680 [DOI] [PubMed] [Google Scholar]
- 57.Sasaki T, Hanisch FG, Deutzmann R, Sakai LY, Sakuma T, Miyamoto T, Yamamoto T, Hannappel E, Chu ML, Lanig H, von der Mark K. Functional consequence of fibulin-4 missense mutations associated with vascular and skeletal abnormalities and cutis laxa. Matrix Biol. 2016;56:132–149 [DOI] [PubMed] [Google Scholar]
- 58.Hibino M, Sakai Y, Kato W, Tanaka K, Tajima K, Yokoyama T, Iwasa M, Morisaki H, Tsuzuki T, Usui A. Ascending aortic aneurysm in a child with fibulin-4 deficiency. Ann Thorac Surg. 2018;105:e59–e61 [DOI] [PubMed] [Google Scholar]
- 59.Milewicz DM, Ostergaard JR, Ala-Kokko LM, et al. De novo ACTA2 mutation causes a novel syndrome of multisystemic smooth muscle dysfunction. Am J Med Genet A. 2010;152A:2437–2443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu H, Fagnant PM, Bookwalter CS, Joel P, Trybus KM. Vascular disease-causing mutation r258c in ACTA2 disrupts actin dynamics and interaction with myosin. Proc Natl Acad Sci U S A. 2015;112:E4168–4177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kuang SQ, Guo DC, Prakash SK, et al. Recurrent chromosome 16p13.1 duplications are a risk factor for aortic dissections. PLoS Genet. 2011;7:e1002118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shalata A, Mahroom M, Milewicz DM, et al. Fatal thoracic aortic aneurysm and dissection in a large family with a novel MYLK gene mutation: Delineation of the clinical phenotype. Orphanet J Rare Dis. 2018;13:41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wallace SE, Regalado ES, Gong L, et al. MYLK pathogenic variants aortic disease presentation, pregnancy risk, and characterization of pathogenic missense variants. Genet Med. 2018;doi: 10.1038/s41436-018-0038-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Herring BP, Dixon S, Gallagher PJ. Smooth muscle myosin light chain kinase expression in cardiac and skeletal muscle. Am J Physiol Cell Physiol. 2000;279:C1656–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sheen VL, Jansen A, Chen MH, et al. Filamin a mutations cause periventricular heterotopia with ehlers-danlos syndrome. Neurology. 2005;64:254–262 [DOI] [PubMed] [Google Scholar]
- 66.Chen MH, Choudhury S, Hirata M, Khalsa S, Chang B, Walsh CA. Thoracic aortic aneurysm in patients with loss of function filamin a mutations: Clinical characterization, genetics, and recommendations. Am J Med Genet A. 2018;176:337–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Loeys BL, Chen J, Neptune ER, et al. A syndrome of altered cardiovascular, craniofacial, neurocognitive and skeletal development caused by mutations in TGFBR1 or TGFBR2. Nat Genet. 2005;37:275–281 [DOI] [PubMed] [Google Scholar]
- 68.Nicod P, Bloor C, Godfrey M, Hollister D, Pyeritz RE, Dittrich H, Polikar R, Peterson KL. Familial aortic dissecting aneurysm. J Am Coll Cardiol. 1989;13:811–819 [DOI] [PubMed] [Google Scholar]
- 69.Tran-Fadulu V, Pannu H, Kim DH, et al. Analysis of multigenerational families with thoracic aortic aneurysms and dissections due to TGFBR 1 or TGFBR 2 mutations. J Med Genet. 2009;46:607–613 [DOI] [PubMed] [Google Scholar]
- 70.Pannu H, Fadulu V, Chang J, Lafont A, Hasham SN, Sparks E, Giampietro PF, Zaleski C, Estrera AL, Safi HJ, Shete S, Willing MC, Raman CS, Milewicz DM. Mutations in transforming growth factor-beta receptor type ii cause familial thoracic aortic aneurysms and dissections. Circulation. 2005;112:513–520 [DOI] [PubMed] [Google Scholar]
- 71.Inamoto S, Kwartler CS, Lafont AL, et al. TGFBR2 mutations alter smooth muscle cell phenotype and predispose to thoracic aortic aneurysms and dissections. Cardiovasc Res. 2010;88:520–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lindsay ME, Schepers D, Bolar NA, et al. Loss-of-function mutations in TGFB2 cause a syndromic presentation of thoracic aortic aneurysm. Nat Genet. 2012;44:922–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Horbelt D, Guo G, Robinson PN, Knaus P. Quantitative analysis of TGFBR2 mutations in marfan-syndrome-related disorders suggests a correlation between phenotypic severity and smad signaling activity. J Cell Sci. 2010;123:4340–4350 [DOI] [PubMed] [Google Scholar]
- 74.Micha D, Guo DC, Hilhorst-Hofstee Y, et al. SMAD2 mutations are associated with arterial aneurysms and dissections. Hum Mutat. 2015;36:1145–1149 [DOI] [PubMed] [Google Scholar]
- 75.Schepers D, Tortora G, Morisaki H, et al. A mutation update on the LDS-associated genes TGFB2/3 and SMAD2/3. Hum Mutat. 2018;39:621–634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Morris SA, Orbach DB, Geva T, Singh MN, Gauvreau K, Lacro RV. Increased vertebral artery tortuosity index is associated with adverse outcomes in children and young adults with connective tissue disorders. Circulation. 2011;124:388–396 [DOI] [PubMed] [Google Scholar]
- 77.Habashi JP, Judge DP, Holm TM, et al. Losartan, an at1 antagonist, prevents aortic aneurysm in a mouse model of marfan syndrome. Science. 2006;312:117–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cook JR, Clayton NP, Carta L, Galatioto J, Chiu E, Smaldone S, Nelson CA, Cheng SH, Wentworth BM, Ramirez F. Dimorphic effects of transforming growth factor-beta signaling during aortic aneurysm progression in mice suggest a combinatorial therapy for marfan syndrome. Arterioscler Thromb Vasc Biol. 2015;35:911–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wei H, Hu JH, Angelov SN, Fox K, Yan J, Enstrom R, Smith A, Dichek DA. Aortopathy in a mouse model of marfan syndrome is not mediated by altered transforming growth factor beta signaling. J Am Heart Assoc. 2017;6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuang SQ, Medina-Martinez O, Guo DC, et al. FOXE3 mutations predispose to thoracic aortic aneurysms and dissections. J Clin Invest. 2016;126:948–961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guo DC, Gong L, Regalado ES, et al. MAT2A mutations predispose individuals to thoracic aortic aneurysms. Am J Hum Genet. 2015;96:170–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Frau M, Feo F, Pascale RM. Pleiotropic effects of methionine adenosyltransferases deregulation as determinants of liver cancer progression and prognosis. J Hepatol. 2013;59:830–841 [DOI] [PubMed] [Google Scholar]
- 83.Ryan BM, Weir DG. Relevance of folate metabolism in the pathogenesis of colorectal cancer. J Lab Clin Med. 2001;138:164–176 [DOI] [PubMed] [Google Scholar]
- 84.Wang N, Tytell JD, Ingber DE. Mechanotransduction at a distance: Mechanically coupling the extracellular matrix with the nucleus. Nat Rev Mol Cell Biol. 2009;10:75–82 [DOI] [PubMed] [Google Scholar]
- 85.Tan KL, Haelterman NA, Kwartler CS, et al. Ari-1 regulates myonuclear organization together with parkin and is associated with aortic aneurysms. Dev Cell. 2018;45:226–244 e228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.LeMaire SA, McDonald MLN, Guo DC, et al. Genome-wide association study identifies a susceptibility locus for thoracic aortic aneurysms and aortic dissections spanning fbn1 at 15q21.1. Nat Genet. 2011;43:996–U109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Guo DC, Grove ML, Prakash SK, et al. Genetic variants in lrp1 and ulk4 are associated with acute aortic dissections. Am J Hum Genet. 2016;99:762–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Guo DC, Hostetler EM, Fan Y, Kulmacz RJ, Zhang D, Gen TACI, Nickerson DA, Leal SM, LeMaire SA, Regalado ES, Milewicz DM. Heritable thoracic aortic disease genes in sporadic aortic dissection. J Am Coll Cardiol. 2017;70:2728–2730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kwartler CS, Gong L, Chen J, Wang S, Kulmacz RJ, Duan X, Janda A, Huang J, Kamm K, Stull JT, Guo DC, Milewicz DM. Variants of unknown significance in genes associated with heritable thoracic aortic disease can be low penetrant “risk variants”. Am J Hum Genet. 2018;103:138–143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Lederle FA. In the clinic. Abdominal aortic aneurysm. Ann Intern Med. 2009;150:ITC5–1–15 [DOI] [PubMed] [Google Scholar]
- 91.De Rango P, Farchioni L, Fiorucci B, Lenti M. Diabetes and abdominal aortic aneurysms. Eur J Vasc Endovasc Surg. 2014;47:243–261 [DOI] [PubMed] [Google Scholar]
- 92.Forsdahl SH, Solberg S, Singh K, Jacobsen BK. Abdominal aortic aneurysms, or a relatively large diameter of non-aneurysmal aortas, increase total and cardiovascular mortality: The tromso study. Int J Epidemiol. 2010;39:225–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wahlgren CM, Larsson E, Magnusson PK, Hultgren R, Swedenborg J. Genetic and environmental contributions to abdominal aortic aneurysm development in a twin population. J Vasc Surg. 2010;51:3–7 [DOI] [PubMed] [Google Scholar]
- 94.Johansen K, Koepsell T. Familial tendency for abdominal aortic aneurysms. JAMA. 1986;256:1934–1936 [PubMed] [Google Scholar]
- 95.Jones GT, Hill BG, Curtis N, Kabir TD, Wong LE, Tilyard MW, Williams MJ, van Rij AM. Comparison of three targeted approaches to screening for abdominal aortic aneurysm based on cardiovascular risk. Br J Surg. 2016;103:1139–1146 [DOI] [PubMed] [Google Scholar]
- 96.Kuivaniemi H, Shibamura H, Arthur C, et al. Familial abdominal aortic aneurysms: Collection of 233 multiplex families. J Vasc Surg. 2003;37:340–345 [DOI] [PubMed] [Google Scholar]
- 97.Ogata T, MacKean GL, Cole CW, Arthur C, Andreou P, Tromp G, Kuivaniemi H. The lifetime prevalence of abdominal aortic aneurysms among siblings of aneurysm patients is eightfold higher than among siblings of spouses: An analysis of 187 aneurysm families in nova scotia, canada. J Vasc Surg. 2005;42:891–897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Rossaak JI, Hill TM, Jones GT, Phillips LV, Harris EL, van Rij AM. Familial abdominal aortic aneurysms in the otago region of new zealand. Cardiovasc Surg. 2001;9:241–248 [DOI] [PubMed] [Google Scholar]
- 99.Larsson E, Granath F, Swedenborg J, Hultgren R. A population-based case-control study of the familial risk of abdominal aortic aneurysm. J Vasc Surg. 2009;49:47–50 [DOI] [PubMed] [Google Scholar]
- 100.Sandford RM, Bown MJ, London NJ, Sayers RD. The genetic basis of abdominal aortic aneurysms: A review. Eur J Vasc Endovasc Surg. 2007;33:381–390 [DOI] [PubMed] [Google Scholar]
- 101.Clifton MA. Familial abdominal aortic aneurysms. Brit J Surg. 1977;64:765–766 [DOI] [PubMed] [Google Scholar]
- 102.Linne A, Lindstrom D, Hultgren R. High prevalence of abdominal aortic aneurysms in brothers and sisters of patients despite a low prevalence in the population. J Vasc Surg. 2012;56:305–310 [DOI] [PubMed] [Google Scholar]
- 103.Larsson E, Vishnevskaya L, Kalin B, Granath F, Swedenborg J, Hultgren R. High frequency of thoracic aneurysms in patients with abdominal aortic aneurysms. Ann Surg. 2011;253:180–184 [DOI] [PubMed] [Google Scholar]
- 104.Svensjo S, Bengtsson H, Bergqvist D. Thoracic and thoracoabdominal aortic aneurysm and dissection: An investigation based on autopsy. Br J Surg. 1996;83:68–71 [DOI] [PubMed] [Google Scholar]
- 105.van ‘t Hof FN, Ruigrok YM, Lee CH, et al. Shared genetic risk factors of intracranial, abdominal, and thoracic aneurysms. J Am Heart Assoc. 2016;5:e002603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Helgadottir A, Thorleifsson G, Magnusson KP, et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008;40:217–224 [DOI] [PubMed] [Google Scholar]
- 107.Bradley DT, Badger SA, McFarland M, Hughes AE. Abdominal aortic aneurysm genetic associations: Mostly false? A systematic review and meta-analysis. Eur J Vasc Endovasc Surg. 2016;51:64–75 [DOI] [PubMed] [Google Scholar]
- 108.Shibamura H, Olson JM, van Vlijmen-Van Keulen C, et al. Genome scan for familial abdominal aortic aneurysm using sex and family history as covariates suggests genetic heterogeneity and identifies linkage to chromosome 19q13. Circulation. 2004;109:2103–2108 [DOI] [PubMed] [Google Scholar]
- 109.Nikpay M, Goel A, Won HH, et al. A comprehensive 1,000 genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015;47:1121–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Biros E, Norman PE, Walker PJ, Nataatmadja M, West M, Golledge J. A single nucleotide polymorphism in exon 3 of the kallikrein 1 gene is associated with large but not small abdominal aortic aneurysm. Atherosclerosis. 2011;217:452–457 [DOI] [PubMed] [Google Scholar]
- 111.Jones GT, Tromp G, Kuivaniemi H, et al. Meta-analysis of genome-wide association studies for abdominal aortic aneurysm identifies four new disease-specific risk loci. Circ Res. 2017;120:341–353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bown MJ, Jones GT, Harrison SC, et al. Abdominal aortic aneurysm is associated with a variant in low-density lipoprotein receptor-related protein 1. Am J Hum Genet. 2011;89:619–627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gretarsdottir S, Baas AF, Thorleifsson G, et al. Genome-wide association study identifies a sequence variant within the DAB2IP gene conferring susceptibility to abdominal aortic aneurysm. Nat Genet. 2010;42:692–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Harrison SC, Smith AJ, Jones GT, et al. Interleukin-6 receptor pathways in abdominal aortic aneurysm. Eur Heart J. 2013;34:3707–3716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Jones GT, Bown MJ, Gretarsdottir S, et al. A sequence variant associated with sortilin-1 (SORT1) on 1p13.3 is independently associated with abdominal aortic aneurysm. Hum Mol Genet. 2013;22:2941–2947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bradley DT, Hughes AE, Badger SA, et al. A variant in LDLR is associated with abdominal aortic aneurysm. Circ Cardiovas Genet. 2013;6:498–504 [DOI] [PubMed] [Google Scholar]
- 117.Baxter BT. Could medical intervention work for aortic aneurysms? Am J Surg. 2004;188:628–632 [DOI] [PubMed] [Google Scholar]
- 118.Freestone T, Turner RJ, Coady A, Higman DJ, Greenhalgh RM, Powell JT. Inflammation and matrix metalloproteinases in the enlarging abdominal aortic aneurysm. Arterioscler Thromb Vasc Biol. 1995;15:1145–1151 [DOI] [PubMed] [Google Scholar]
- 119.Jones GT, Phillips LV, Williams MJ, van Rij AM, Kabir TD. Two c-c family chemokines, eotaxin and rantes, are novel independent plasma biomarkers for abdominal aortic aneurysm. J Am Heart Assoc. 2016;5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Jones GT. The pathohistology of abdominal aortic aneurysm In: Grundmann RT, ed. Diagnosis, screening and treatment of abdominal, thoracoabdominal and thoracic aortic aneurysms. InTech, Rijeka, Croatia; 2011:414. [Google Scholar]
- 121.Howson JMM, Zhao W, Barnes DR, et al. Fifteen new risk loci for coronary artery disease highlight arterial-wall-specific mechanisms. Nat Genet. 2017;49:1113–1119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.van der Harst P, Verweij N. Identification of 64 novel genetic loci provides an expanded view on the genetic architecture of coronary artery disease. Circ Res. 2018;122:433–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Helgadottir A, Gretarsdottir S, Thorleifsson G, et al. Apolipoprotein(a) genetic sequence variants associated with systemic atherosclerosis and coronary atherosclerotic burden but not with venous thromboembolism. J Am Coll Cardiol. 2012;60:722–729 [DOI] [PubMed] [Google Scholar]
- 124.Jones GT, van Rij AM, Cole J, Williams MJ, Bateman EH, Marcovina SM, Deng M, McCormick SP. Plasma lipoprotein(a) indicates risk for 4 distinct forms of vascular disease. Clin Chem. 2007;53:679–685 [DOI] [PubMed] [Google Scholar]
- 125.Schunkert H, Konig IR, Kathiresan S, et al. Large-scale association analysis identifies 13 new susceptibility loci for coronary artery disease. Nat Genet. 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Visel A, Zhu Y, May D, Afzal V, Gong E, Attanasio C, Blow MJ, Cohen JC, Rubin EM, Pennacchio LA. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature. 2010;464:409–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Pilbrow AP, Folkersen L, Pearson JF, et al. The chromosome 9p21.3 coronary heart disease risk allele is associated with altered gene expression in normal heart and vascular tissues. PLoS One. 2012;7:e39574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.van Dongen J, Jansen R, Smit D, Hottenga JJ, Mbarek H, Willemsen G, Kluft C, Collaborators A, Penninx BW, Ferreira MA, Boomsma DI, de Geus EJ. The contribution of the functionalIL6R polymorphism rs2228145, eQTLs and other genome-wide SNPs to the heritability of plasma sIL-6R levels. Behav Genet. 2014;44:368–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ferreira MA, Matheson MC, Duffy DL, et al. Identification of IL6R and chromosome 11q13.5 as risk loci for asthma. Lancet. 2011;378:1006–1014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Musunuru K, Strong A, Frank-Kamenetsky M, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Kjolby M, Andersen OM, Breiderhoff T, Fjorback AW, Pedersen KM, Madsen P, Jansen P, Heeren J, Willnow TE, Nykjaer A. SORT1, encoded by the cardiovascular risk locus 1p13.3, is a regulator of hepatic lipoprotein export. Cell Metab. 2010;12:213–223 [DOI] [PubMed] [Google Scholar]
- 132.Zhang Z, Jiang W, Yang H, Lin Q, Qin X. The mir-182/SORT1 axis regulates vascular smooth muscle cell calcification in vitro and in vivo. Exp Cell Res. 2018;362:324–331 [DOI] [PubMed] [Google Scholar]
- 133.Teslovich TM, Musunuru K, Smith AV, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466:707–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Harrison SC, Holmes MV, Burgess S, et al. Genetic association of lipids and lipid drug targets with abdominal aortic aneurysm: A meta-analysis. JAMA Cardiol. 2018;3:26–33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Weng LC, Roetker NS, Lutsey PL, Alonso A, Guan W, Pankow JS, Folsom AR, Steffen LM, Pankratz N, Tang W. Evaluation of the relationship between plasma lipids and abdominal aortic aneurysm: A mendelian randomization study. PLoS One. 2018;13:e0195719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Thompson RW, Holmes DR, Mertens RA, Liao S, Botney MD, Mecham RP, Welgus HG, Parks WC. Production and localization of 92-kilodalton gelatinase in abdominal aortic aneurysms. An elastolytic metalloproteinase expressed by aneurysm-infiltrating macrophages. J Clin Invest. 1995;96:318–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chowaniec Z, Skoczynska A. Plasma lipid transfer proteins: The role of pltp and cetp in atherogenesis. Adv Clin Exp Med. 2018 [DOI] [PubMed] [Google Scholar]
- 138.Moerland M, Samyn H, van Gent T, van Haperen R, Dallinga-Thie G, Grosveld F, van Tol A, de Crom R. Acute elevation of plasma PLTP activity strongly increases pre-existing atherosclerosis. Arterioscler Thromb Vasc Biol. 2008;28:1277–1282 [DOI] [PubMed] [Google Scholar]
- 139.Brehm A, Geraghty P, Campos M, Garcia-Arcos I, Dabo AJ, Gaffney A, Eden E, Jiang XC, D’Armiento J, Foronjy R. Cathepsin g degradation of phospholipid transfer protein (pltp) augments pulmonary inflammation. FASEB J. 2014;28:2318–2331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Coleman C, Quinn EM, Ryan AW, et al. Common polygenic variation in coeliac disease and confirmation of znf335 and nifa as disease susceptibility loci. Eur J Hum Genet. 2016;24:291–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Sweeting MJ, Thompson SG, Brown LC, Powell JT, collaborators R. Meta-analysis of individual patient data to examine factors affecting growth and rupture of small abdominal aortic aneurysms. Br J Surg. 2012;99:655–665 [DOI] [PubMed] [Google Scholar]
- 142.Du SJ, Tan X, Zhang J. Smyd proteins: Key regulators in skeletal and cardiac muscle development and function. Anat Rec (Hoboken). 2014;297:1650–1662 [DOI] [PubMed] [Google Scholar]
- 143.Xu G, Liu G, Xiong S, Liu H, Chen X, Zheng B. The histone methyltransferase smyd2 is a negative regulator of macrophage activation by suppressing interleukin 6 (IL-6) and tumor necrosis factor alpha (TNF-alpha) production. J Biol Chem. 2015;290:5414–5423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Ghosh A, DiMusto PD, Ehrlichman LK, Sadiq O, McEvoy B, Futchko JS, Henke PK, Eliason JL, Upchurch GR Jr., The role of extracellular signal-related kinase during abdominal aortic aneurysm formation. J Am Coll Surg. 2012;215:668–680 e661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Saito T, Hasegawa Y, Ishigaki Y, Yamada T, Gao J, Imai J, Uno K, Kaneko K, Ogihara T, Shimosawa T, Asano T, Fujita T, Oka Y, Katagiri H. Importance of endothelial nf-kappab signalling in vascular remodelling and aortic aneurysm formation. Cardiovasc Res. 2013;97:106–114 [DOI] [PubMed] [Google Scholar]
- 146.Qi J, Yang P, Yi B, Huo Y, Chen M, Zhang J, Sun J. Heat shock protein 90 inhibition by 17-dmag attenuates abdominal aortic aneurysm formation in mice. Am J Physiol, Heart Circ Physiol. 2015;308:H841–852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Toghill BJ, Saratzis A, Freeman PJ, Sylvius N, collaborators U, Bown MJ. SMYD2 promoter DNA methylation is associated with abdominal aortic aneurysm (AAA) and SMYD2 expression in vascular smooth muscle cells. Clin Epigenetics. 2018;10:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Zhou L, Takayama Y, Boucher P, Tallquist MD, Herz J. Lrp1 regulates architecture of the vascular wall by controlling PDGFRbeta-dependent phosphatidylinositol 3-kinase activation. PLoS One. 2009;4:e6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Boucher P, Gotthardt M, Li WP, Anderson RG, Herz J. Lrp: Role in vascular wall integrity and protection from atherosclerosis. Science. 2003;300:329–332 [DOI] [PubMed] [Google Scholar]
- 150.Yi L, Domyan ET, Lewandoski M, Sun X. Fibroblast growth factor 9 signaling inhibits airway smooth muscle differentiation in mouse lung. Dev Dyn. 2009;238:123–137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Singla D, Wang J. Fibroblast growth factor-9 activates c-kit progenitor cells and enhances angiogenesis in the infarcted diabetic heart. Oxid Med Cell Longev. 2016;2016:5810908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Choke E, Thompson MM, Dawson J, Wilson WR, Sayed S, Loftus IM, Cockerill GW. Abdominal aortic aneurysm rupture is associated with increased medial neovascularization and overexpression of proangiogenic cytokines. Arterioscler Thromb Vasc Biol. 2006;26:2077–2082 [DOI] [PubMed] [Google Scholar]
- 153.Dryden NH, Sperone A, Martin-Almedina S, Hannah RL, Birdsey GM, Khan ST, Layhadi JA, Mason JC, Haskard DO, Gottgens B, Randi AM. The transcription factor erg controls endothelial cell quiescence by repressing activity of nuclear factor (NF)-kappaB p65. J Biol Chem. 2012;287:12331–12342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wythe JD, Dang LT, Devine WP, Boudreau E, Artap ST, He D, Schachterle W, Stainier DY, Oettgen P, Black BL, Bruneau BG, Fish JE. Ets factors regulate vegf-dependent arterial specification. Dev Cell. 2013;26:45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Norman PE, Powell JT. Site specificity of aneurysmal disease. Circulation. 2010;121:560–568 [DOI] [PubMed] [Google Scholar]
- 156.Tian TV, Tomavo N, Huot L, Flourens A, Bonnelye E, Flajollet S, Hot D, Leroy X, de Launoit Y, Duterque-Coquillaud M. Identification of novel tmprss2:Erg mechanisms in prostate cancer metastasis: Involvement of MMP9 and PLXNA2. Oncogene. 2014;33:2204–2214 [DOI] [PubMed] [Google Scholar]
- 157.Reed D, Reed C, Stemmermann G, Hayashi T. Are aortic aneurysms caused by atherosclerosis? Circulation. 1992;85:205–211 [DOI] [PubMed] [Google Scholar]
- 158.Nelson CP, Hamby SE, Saleheen D, et al. Genetically determined height and coronary artery disease. N Engl J Med. 2015;372:1608–1618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Paajanen TA, Oksala NK, Kuukasjarvi P, Karhunen PJ. Short stature is associated with coronary heart disease: A systematic review of the literature and a meta-analysis. Eur Heart J. 2010;31:1802–1809 [DOI] [PubMed] [Google Scholar]
- 160.Thorgeirsson TE, Gudbjartsson DF, Surakka I, et al. Sequence variants at chrnb3-chrna6 and cyp2a6 affect smoking behavior. Nat Genet. 2010;42:448–453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Toghill BJ, Saratzis A, Harrison SC, Verissimo AR, Mallon EB, Bown MJ. The potential role of DNA methylation in the pathogenesis of abdominal aortic aneurysm. Atherosclerosis. 2015;241:121–129 [DOI] [PubMed] [Google Scholar]
- 162.Chen J, Peters A, Papke CL, et al. Loss of smooth muscle alpha-actin leads to nf-kappab-dependent increased sensitivity to angiotensin ii in smooth muscle cells and aortic enlargement. Circ Res. 2017;120:1903–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]