Abstract
Background/Aims
Inflammatory bowel disease (IBD) is regarded as a kind of chronic and unspecific intestinal inflammatory disorder. Its exact pathogenesis has not been elucidated. Curcumin, as an herbal drug, has been used in the treatment of IBD due to its immunoregulation. Autophagy has been reported to play an important role in the mechanism of IBD. In the present study, we focused on the autophagic regulation role of curcumin in the murine model of dextran sulfate sodium (DSS)-induced colitis.
Materials and Methods
We investigated the effects of curcumin on the progress of DSS-induced acute colitis in mice by evaluating the disease activity index (DAI) and histopathological score. Meanwhile, the mRNA and protein expression of autophagy-related key genes from colon tissues comprising autophagy-related 5 (ATG5), LC3-phosphatidylethanolamine conjugate (LC-3II), beclin-1, and B cell lymphoma 2 (bcl-2) was examined by quantitative reverse transcription polymerase chain reaction and Western blot. Furthermore, the mRNA and protein expression of cytokines, including tumor necrosis factor (TNF)-α, interleukin (IL) 6, IL-10, and IL-17A, was examined. Autophagosome was also examined under transmission electron microscopy.
Results
Both DAI and histopathological score increased in mice with DSS-induced colitis and obviously decreased after curcumin intervention. The expression levels of TNF-α, IL-6, IL-17, ATG5, LC-3II, and beclin-1 were significantly higher in mice with colitis than in normal ones, whereas those of IL-10 and bcl-2 decreased accordingly. However, curcumin intervention adjusted the expression level of those factors toward normal level. The number of autophagosome in the colon epithelia increased after DSS stimulation and decreased after curcumin administration.
Conclusion
Curcumin could prevent the development of DSS-induced colitis through the inhibition of excessive autophagy and regulation of following cytokine networks.
Keywords: Inflammatory bowel disease, curcumin, autophagy, cytokine
INTRODUCTION
Inflammatory bowel disease (IBD), comprising ulcerative colitis and Crohn’s disease, is characterized by chronic and relapsing intestinal inflammation of unidentified origins. Accumulating evidence indicates that IBD occurs due to several complex factors, including specific external infection, imbalanced microbe-host interaction, dysfunction of intestinal immunity, and genetic susceptibility (1). Along with its increasing incidence, currently, available treatment for IBD has not reached a most satisfying result and is still under intense discussion. Thus, exploration of the exact pathogenesis of IBD is of great importance to discover novel therapies and to improve prognosis for IBD.
Autophagy, characterized by a highly evolutionarily conserved progress, is a process that degrades excessive, damaged, or aged proteins and organelles to maintain cellular homeostasis (2). It contributes to a variety of organic biochemical reaction, consisting of cellular homeostasis, development and differentiation, survival, and innate and adaptive immunity. Dysregulated autophagy is linked to a multitude of diseases, such as diabetes, tumors, and rheumatic and neurodegenerative diseases (3). During the past decades, genome-wide association studies (GWASs) and further analyses have identified that aberrant autophagy is closely associated with the pathogenesis of IBD, and that several certain single-nucleotide polymorphisms are involved in the autophagy pathway (4). Autophagy plays multiple roles in IBD pathogenesis by altering the processes that include antimicrobial peptide secretion by Paneth cells, goblet cell function, absorption by epithelial cells, proinflammatory cytokine production and bacterial clearance by macrophages, antigen presentation by dendritic cells, and adaptive immune reaction by T and B cells (5,6).
Curcumin, a compound extracted from Curcuma longa, was used for the treatment of biliary disorders and rheumatism in Indian traditional medicine (7). It has anti-inflammatory, antioxidant, antitumor, and antiproliferative properties (8). Studies have indicated that curcumin application can ameliorate intestinal inflammation condition in the murine colitis model and in patients with IBD along with regular therapy (9,10). It has been noted that the mechanism is associated with immune regulation, and that curcumin may take effect on a couple of signaling pathways, such as mitogen-activated protein kinase (MAPK)/extracellular signal-regulated kinase, nuclear factor (NF)-κB, and inflammatory cytokine secretion, including TNF-α, interferon gamma, cyclooxygenase 1/2, and inducible nitric oxide synthase (11). However, how curcumin relieves the IBD state through acting on autophagy has not been described yet until now, although curcumin was reported to promote autophagy by inhibiting the growth of several common tumors (12–14).
In the present study, we investigated the in vivo effect of curcumin on the overall situation and intestinal inflammation status in dextran sulfate sodium (DSS)-induced murine colitis model. Then, we evaluated how curcumin performed its role via influencing autophagy pathway and proinflammatory cytokine expression. Our study revealed a novel mechanism of curcumin reducing IBD inflammation through acting on the autophagy pathway.
MATERIALS AND METHODS
Mice and reagent
Forty-eight-week-old male BALB/c mice were purchased from the Chinese Academy of Science (Shanghai). They weighed approximately 25 g and were housed in a pathogen-free facility. The experimental procedures were in accordance with the Animal Care and Use Committee at Fudan University. The following reagents were used: DSS (MW50000; MP Biomedicals, USA) diluted with distilled water into 5% solution, curcumin (LUYE Pharma, Shanghai, China), anti-TNF-α (sc-52746; Santa Cruz, USA), anti-IL-6 (sc-57315; Santa Cruz), anti-IL-10 (sc-365858; Santa Cruz), anti-IL-17 (sc-52567; Santa Cruz), anti-autophagy related 5 (ATG5, ab106551; Abcam, USA), anti-beclin 1 (CST3738; Cell Signaling Technology, USA), anti-LC3-phosphatidylethanolamine conjugate (LC3, sc-376404; Santa Cruz), anti-B cell lymphoma 2 (bcl-2, sc-492; Santa Cruz), and anti-β-actin (sc-47778; Santa Cruz).
Ulcerative colitis model
Forty mice were randomly assigned into five groups: normal control group (group A), colitis model group (group B), low-dose intervention group (group C), moderate-dose intervention group (group D), and large-dose intervention group (group E). According to the classical model method described by Cooper (15), group B to group E were provided with 5.0% DSS solution as drinking water for 1–7 days to induce acute intestinal inflammation, whereas group A drank distilled water ad libitum. Simultaneously, groups C, D, and E received curcumin injection intraperitoneally once on each day, with a dose of 15 mg/kg each time for group C, 30 mg/kg for group D, and 60 mg/kg for group E, respectively. Group A and group B received equivalent sodium injection intraperitoneally as control. All mice were sacrificed on day 8, and the whole colon from the cecum to the anus of each mouse was dissected completely. Then, the entire colon was divided into four segments and processed for further detection.
Evaluation of clinical parameters
Each mouse of every group was under close observation per day for weight loss, feces property, and hematochezia, and records were summarized to evaluate the disease activity index (DAI) scores according to Cooper description (15). Mental status, movement capacity, and hair condition were also observed.
Histological assessment
The murine colon tissues were embedded in paraffin and stained with hematoxylin and eosin (H&E). Histological scoring was calculated in terms of the classic scoring system by Cooper (15) and performed blindly. The scoring system included the following parameters: inflammation, lesion depth, crypt destruction, and lesion extent.
Transmission electron microscopy
The cleaned colon tissues were fixed with 2.5% glutaraldehyde, followed by 0.1 M phosphate-buffered saline (pH 7.4) washed three times. Then, the tissues were post-fixed with 1% osmium tetroxide for 2 h at room temperature, processed in a standard manner of dehydration and permeation, and embedded in Embed 812 (Electron Microscopy Sciences) for 48 h at 60 °C. Ultrathin sections were cut at 70 nm, mounted on 200-mesh copper grids, and stained with uranyl acetate and lead citrate. Stained grids were examined under FEI (Tecnai G2 20 TWIN) transmission electron microscopy (TEM) and photographed using a digital camera.
Reverse transcription polymerase chain reaction
Total RNA was extracted from the colon tissues using TRIzol (Invitrogen, USA), and cDNA was generated using PrimeScript RT Master Mix (TaKaRa Biotechnology, China). For quantitative reverse transcription polymerase chain reaction, Power SYBR Green Master kit (ABI) and Applied Biosystems 7500 were used according to the manufacturer’s instructions. The relative expression of target genes, including TNF-α, IL-6, IL-10, IL-17A, ATG5, LC-3II, beclin-1, and bcl-2, was calculated using the 2−ΔC(t) method. Table 1 shows the gene primer sequences.
Table 1.
Primer sequences used in the study
| Gene name | Primers (5′-3′) |
|---|---|
| TNF-α | Forward: CCCTCACACTCAGATCATCTTCT |
| Reverse: GCTACGACGTGGGCTACAG | |
| IL-6 | Forward: TAGTCCTTCCTACCCCAATTTCC |
| Reverse: TTGGTCCTTAGCCACTCCTTC | |
| IL-10 | Forward: GCTCTTACTGACTGGCATGAG |
| Reverse: CGCAGCTCTAGGAGCATGTG | |
| IL-17 | Forward: TTTAACTCCCTTGGCGCAAAA |
| Reverse: CTTTCCCTCCGCATTGACAC | |
| ATG5 | Forward: AGCCAGGTGATGATTCACGG |
| Reverse: GGCTGGGGGACAATGCTAA | |
| LC-3II | Forward: TTATAGAGCGATACAAGGGGGAG |
| Reverse: CGCCGTCTGATTATCTTGATGAG | |
| Beclin-1 | Forward: ATGGAGGGGTCTAAGGCGTC |
| Reverse: TCCTCTCCTGAGTTAGCCTCT | |
| Bcl-2 | Forward: TACCGTCGTGACTTCGCAGAG |
| Reverse: GGCAGGCTGAGCAGGGTCTT | |
| β-Actin | Forward: GGCTGTATTCCCCTCCATCG |
| Reverse: CCAGTTGGTAACAATGCCATGT |
Western blot
Protein extracts were prepared by lysing colon tissues. Total protein concentrations were determined by BCA assay (Sangon Biotech, China). Aliquots of extracts were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to a polyvinylidene fluoride membrane (Amersham Pharmacia Biotech, UK) by an electroblotter (Bio-Rad, USA). The membranes were blocked and incubated overnight with primary antibodies against TNF-α, IL-6, IL-10, IL-17A, ATG5, LC-3II, beclin-1, and bcl-2 (1:1000 dilution) and β-actin (1:2000 dilution) at 4 °C. Then, the membranes were incubated with horseradish peroxidase-conjugated anti-rabbit or anti-mouse IgG (1:5000 dilution) for 1 h at room temperature. The membranes were subsequently incubated using ECL kits (Pierce/Thermo Fisher Scientific, USA) and developed by the chemiluminescence imaging system (ImageQuant™ LAS 4000). The result of each experimental group was repeated three times.
Statistical analysis
For analyzing inflammatory cytokines, autophagy-related gene expression, and physical indexes, differences between two groups and among five groups were assessed by unpaired Student’s t-test and one-way ANOVA analysis, respectively. For analyzing DAI and histological scoring, differences between two groups and among five groups were assessed by Mann-Whitney U test and Kruskal-Wallis H analysis, respectively. All procedures were performed using the Statistical Package for Social Sciences 17.0 software (SPSS Inc.; Chicago, IL, USA). A p value <0.05 was considered statistically significant.
RESULTS
Curcumin improved DAI and other clinical parameters of colitis
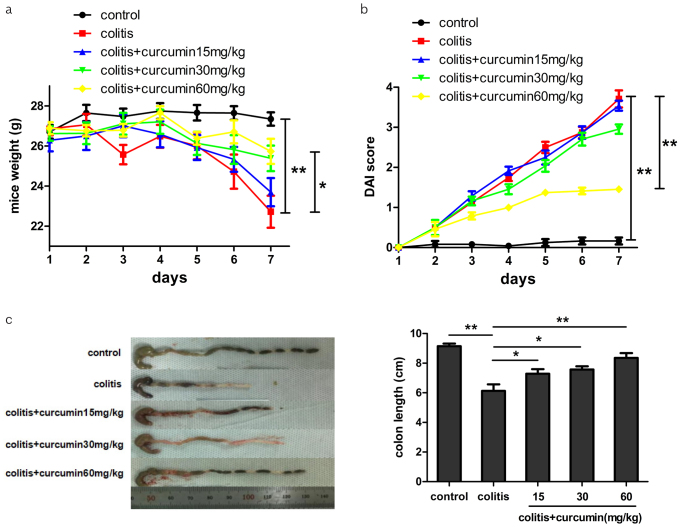
As shown in Figure 1a, mice in the control group maintained a stable weight during the experimental process, whereas mice in the colitis group significantly suffered weight loss (p<0.01). However, curcumin intervention could reverse this effect in a dose-dependent manner (p<0.05), implying that malnutrition or exhaustion condition induced by DSS could be relieved by curcumin administration. Furthermore, DAI was an integrated index reflecting the overall severity of colitis. Mice in the control group had low DAI score and were in good mental and activity state with normal feces during the process, whereas mice in the colitis group had drooped spirit, reduced activity, hematochezia, and a rather high DAI score (p<0.01). However, curcumin treatment in the other three colitis groups could help reduce DAI score in accordance with dose supply (p<0.01), indicating that curcumin could relieve symptoms of colitis to some extent (Figure 1b). Finally, measurement of colon length after dissection uncovered that DSS-induced colitis could lead to shortened colon length, which was consistent with colitis severity, whereas curcumin administration might perform a reverse role dose-dependently (p<0.05, Figure 1c).
Figure 1. a–c.
Curcumin ameliorated the manifestations of DSS-induced colitis in mice. (a, b) Mice weight and DAI scores were measured at day 7. (c) Representative colons of mice from each group dissected at day 8. Colon lengths were measured.
Data are presented as mean±SEM **p<0.01, *p<0.05. n=8 for each group
Curcumin relieved histological features of colitis
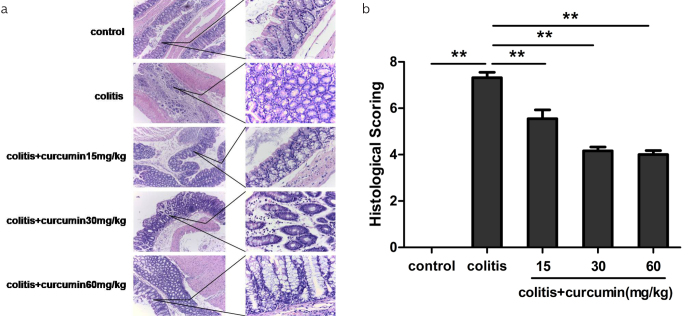
Since we found that curcumin could improve the systematical features of DSS-induced colitis, we thus further investigated how curcumin influenced colon inflammation histologically. As shown in Figure 2, it was discovered from H&E staining of colon slices that the colon mucosa of the control group was intact without any ulcer or inflammatory cell infiltration, whereas that of the colitis group was characterized by varied ulcers, epithelial defect, inflammatory cell infiltration, and glandular destruction. In addition, the curcumin intervention groups showed less severity of the above manifestations, indicating that curcumin might reduce inflammation injury caused by DSS. Therefore, histological score was lowest in the control group and highest in the colitis group (p<0.01), and the score gradually decreased according to dose of curcumin supply (p<0.01).
Figure 2. a, b.
Curcumin improved the histological features of DSS-induced colitis. (a) Representative photomicrographs of H&E staining of the colons of mice from each group (100× magnification for the left and 400× magnification for the right). (b) Histological scores were measured.
Data are presented as mean±SEM **p<0.01. n=8 for each group
Curcumin regulated cytokine expression in colon tissues of colitis
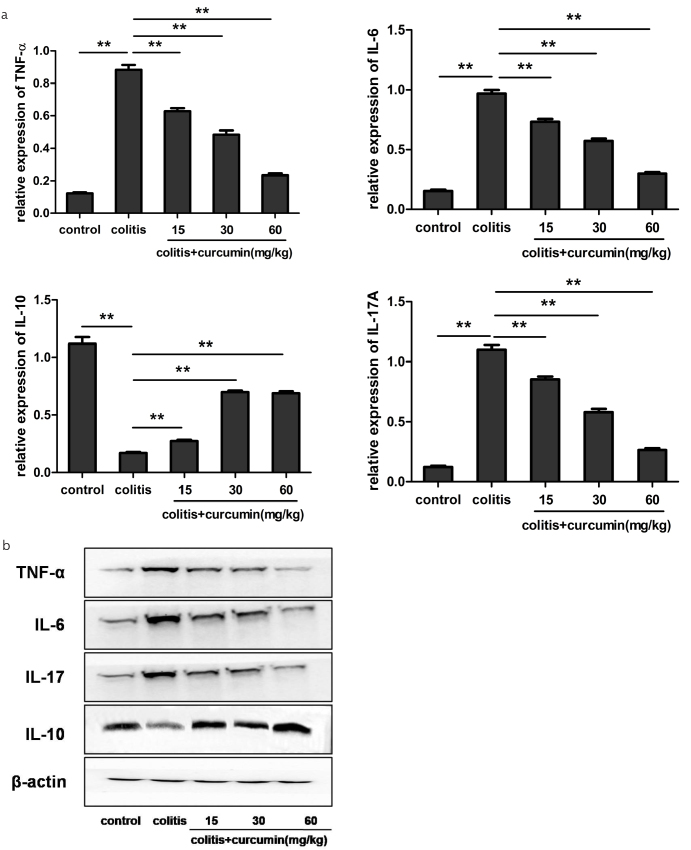
Dysregulated intestinal immune environment, including aberrant cytokine expression, has been reported to play a pivotal role in IBD pathogenesis (16). Then, we explored the expression profile of typical cytokines altered by curcumin administration. We found that DSS drinking led to an obviously increased expression of proinflammatory cytokines of TNF-α, IL-6, and IL-17 and, however, a decreased expression of anti-inflammatory cytokine of IL-10 compared with the control group (p<0.01). Curcumin intervention could re-store the expression level of the above cytokines toward normal state in line with its dose (p<0.01), indicating that curcumin could exert anti-inflammatory effects on colitis, thus relieving inflammation degree (Figure 3a, b).
Figure 3. a, b.
Curcumin regulated the cytokine expression in the colon mucosa of DSS-induced colitis. (a) Quantitative reverse transcription polymerase chain reaction analysis of the mRNA expression of TNF-α, IL-6, IL-10, and IL-17A in each group. Data are presented as mean±SEM. **p<0.01. n=8 for each group. (b) Western blot analysis of the protein levels of TNF-α, IL-6, IL-10, and IL-17A in each group. β-Actin was used as the control
Curcumin adjusted autophagy process and expression profile of autophagy-associated key proteins in colon tissues of colitis
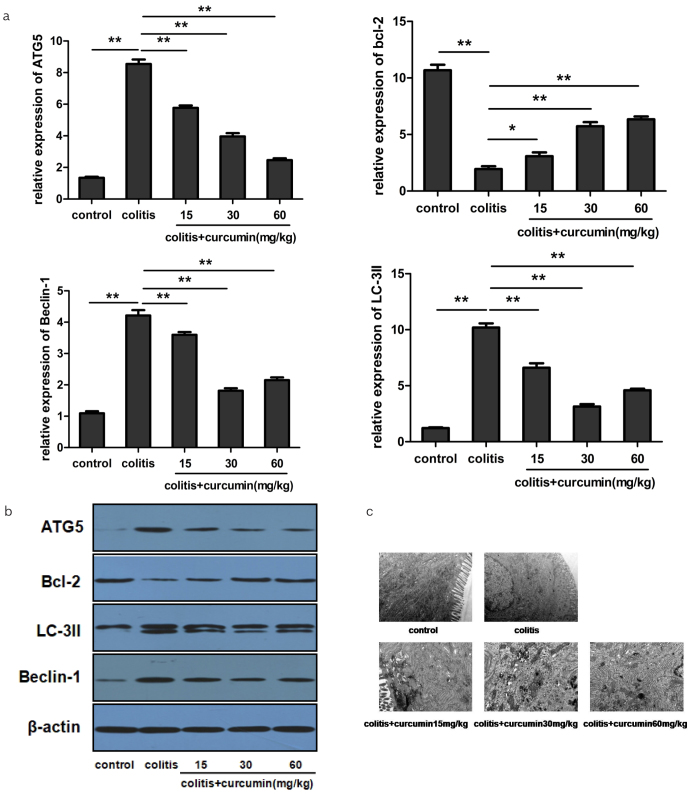
Since autophagy plays an important role in regulating cytokine expression, we next investigated the effect of curcumin on key proteins involved in the autophagy-related signaling pathway. We discovered that DSS drinking gave rise to an increased expression of autophagic factors of ATG5, LC-3II, and beclin-1 and, however, a decreased expression of anti-autophagic factor of bcl-2 on mRNA and protein level compared with the control group (p<0.01), implying the occurrence of enhanced autophagy in colitis. However, curcumin administration could re-adjust the expression level of the above factors toward normal condition, consistent with its dose supply (p<0.01), indicating that curcumin could inhibit the over-activated autophagy state, thus contributing to inflammation manipulation (Figure 4a, b). Furthermore, autophagosome formation, which fuses with lysosome for further degradation, is the major step for the autophagy process. We found under TEM that rare autophagosome was detected in the intestinal epithelial cells of the control group with normal endoplasmic reticulum and tight conjunction. However, an increased number of autophagosome was seen in the epithelial cells of the colitis group with loose conjunction and swollen endoplasmic reticulum. Additionally, curcumin administration could reduce the number of autophagosome and improve the condition of the endoplasmic reticulum, implying the inhibitory effect of curcumin on the autophagy progress (Figure 4c).
Figure 4. a–c.
Curcumin inhibited the autophagy process by regulating autophagy-related gene expression. (a) Quantitative reverse transcription polymerase chain reaction analysis of the mRNA expression of ATG5, LC-3II, beclin-1, and bcl-2 in each group. Data are presented as mean±SEM. **p<0.01, *p<0.05. n=8 for each group. (b) Western blot analysis of the protein levels of ATG5, LC-3II, beclin-1, and bcl-2 in each group. β-Actin was used as the control. (c) Autophagosome detection under transmission electron microscopy
DISCUSSION
In the present study, we demonstrated that curcumin could inhibit the development of DSS-induced colitis in mice, and this action was performed through the regulation of autophagy-related signaling pathway and the following several cytokine expression.
Curcumin, as a traditional herbal medicine, has been investigated in the clinic and in the laboratory to treat IBD due to its special anti-inflammation and antioxidant effects (17). In the present study, we discovered that after curcumin administration to mice suffering from colitis, the DAI score that reflected overall common situation and the histological score that reflected inflammation severity were both significantly improved along with increased curcumin dose, indicating that curcumin performed an inflammation-inhibitory role in DSS-induced colitis.
With regard to detailed mechanisms, previous studies demonstrated that curcumin may target several classical signaling pathways, such as MAPK and NF-κB, and regulate immunocyte activity and a variety of cytokine expression. For example, curcumin could inhibit the activity of TNF-α, which was also the target of infliximab (18), IL-1β, and up-regulated IL-10 level (19). Furthermore, curcumin could inhibit the activity of neutrophils, promote the proliferation of Treg cells, and act on a series of proteases and adhesion molecules (20,21). It was worthwhile to note that curcumin could also inhibit Toll-like receptor 4 (TLR4) activation and subsequent Myd88-NF-κB pathway, thus ameliorating 2,4,6-trinitrobenzene sulfonic acid-induced colitis in mice (22), which was in accordance with our previous finding that TLR4 blockade suppressed DSS-induced colitis (23). In our study, we discovered that curcumin administration could reduce the expression level of TNF-α, IL-6, and IL-17A and increase that of IL-10. In addition, these functions possessed a dose-effect relationship. As is widely acknowledged that TNF-α, IL-6, and IL-17A performed proinflammatory activity, and IL-10 had potential of anti-inflammation (24), therefore, our results implied that curcumin might inhibit DSS-induced colitis through the regulation of cytokine networks, which was similar with previous study.
Autophagy, a pivotal process for cell survival, refers to an intracellular lysosomal degradation process by recycling long-lived and damaged proteins in the maintenance of normal cellular homeostasis. Numerous GWAS reports have confirmed that several autophagy-related genetic variations, including ATG16L1 and IRGM, were closely associated with enhanced risk of IBD (25,26). Moreover, medicines for IBD treatment might take effect through autophagy regulation (27). However, how curcumin improves IBD by adjusting autophagy has not been reported yet. With regard to the autophagy progress, beclin-1 accompanied with phosphoinositide 3-kinase complex contributes to ATG proteins recruiting in the initiation phase (28). However, bcl-2, which combines with beclin-1, can prevent the process of autophagy. In the following elongation phase, ATG5 belonging to the ATG12-ATG5-ATG16L1 conjunction system and LC-3II belonging to the LC3 conjunction system play important roles for autophagosome formation (29).
In our study, we detected that in the control group, the expression levels of beclin-1, ATG5, and LC-3II in mice colon were rather low, whereas those of bcl-2 was rather high, implying autophagy kept at a basal level without any stimuli. However, DSS drinking caused obviously elevated levels of beclin-1, ATG5, and LC-3II and decreased level of bcl-2, indicating that colitis as a potent stress gave rise to enhanced autophagy. Furthermore, curcumin administration down-regulated the levels of beclin-1, ATG5, and LC-3II, whereas up-regulated those of bcl-2, resulting in ameliorated colitis, indicating the autophagy-inhibitory role of curcumin. Additionally, the number of autophagosome varying among different groups validated the change of autophagy level. From the above findings, we speculated that the basal level of autophagy could prevent the normal intestine from inflammation or injury (30). When intestinal inflammation occurred due to external invading factors, such as DSS, excessive autophagy was initiated, followed by cascade-like reactions, including enhanced proinflammatory cytokine secretion, leading to uncontrolled inflammation, cell death, and tissue injury. Curcumin could adjust autophagy to a reasonable level, and proper autophagy could further dominate the cytokine framework, thus relieving colon inflammation. Therefore, we believed that autophagy was a double-edged sword, and curcumin could control autophagy within the appropriate range encountering inflammation.
Based on the above findings, we could conclude that curcumin ameliorated DSS-induced colitis through immune regulation and autophagy adjustment. Although the immune regulation role of curcumin in colitis has been reported previously, and autophagy was believed to be an upstream regulator for immune reaction, to our knowledge, our study, for the first time, discovered the role of curcumin via manipulating autophagy and subsequent cytokine expression in colitis. Our study had several limitations. First, the exact dose of curcumin, which could just maximally control autophagy and inflammation without side effects, needs to be further demonstrated. Second, whether curcumin could influence autophagy in peripheral circulation even in patients with IBD required further investigation. Finally, monitoring of change of autophagy during the disease process was required since we only selected the terminal as the observation point, and however, autophagy was a complex and dynamic process, the level of which might vary at the early or late stage of a specific disease.
In summary, our study revealed a novel autophagy regulation function of curcumin in DSS-induced colitis. Since targeting autophagy-associated pathway might lead to new strategies for the management of IBD, curcumin provided a rather promising application as adjuvant therapy for patients with IBD.
Footnotes
Ethics Committee Approval: The experimental procedures were approved by the Animal Care and Use Committee at Fudan University.
Peer-review: Externally peer-reviewed.
Author Contributions: Concept - J.L.; Design - J.L., Y.L.; Supervision - Y.L., W.Y.; Resource - J.L.; Materials - W.Y.; Data Collection and/or Processing - X.L.; Analysis and/or Interpretation - L.L.; Literature Search - J.H.; Writing Manuscript - W.Y.; Critical Reviews - Y.L.
Conflict of Interest: The authors have no conflict of interest to declare.
Financial Disclosure: This study was supported by grants from the National Natural Science Foundation of China (No: 81630016).
REFERENCES
- 1.Mehta M, Ahmed S, Dryden G. Immunopathophysiology of inflammatory bowel disease: how genetics link barrier dysfunction and innate immunity to inflammation. Innate Immun. 2017;23:497–505. doi: 10.1177/1753425917722206. [DOI] [PubMed] [Google Scholar]
- 2.Shibutani ST, Saitoh T, Nowag H, et al. Autophagy and autophagy-related proteins in the immune system. Nature Immun. 2015;16:1014–24. doi: 10.1038/ni.3273. [DOI] [PubMed] [Google Scholar]
- 3.Yang Z, Klionsky DJ. Eaten alive: A history of macroautophagy. Nat Cell Biol. 2010;12:814–22. doi: 10.1038/ncb0910-814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pugazhendhi S, Baskaran K, Santhanam S, et al. Association of ATG16L1 gene haplotype with inflammatory bowel disease in Indians. PLoS One. 2017;12:e0178291. doi: 10.1371/journal.pone.0178291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salzman NH, Hung K, Haribhai D, et al. Enteric defensins are essential regulators of intestinal microbial ecology. Nat Immunol. 2010;11:76–83. doi: 10.1038/ni.1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Conway K, Kuballa P, Song J. Atg16L1 is Required for autophagy in intestinal epithelial cells and protection of mice from Salmonella infection. Gastroenterology. 2013;145:1–20. doi: 10.1053/j.gastro.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gupta SC, Kismali G, Aggarwal BB. Curcumin, a component of turmeric: From farm to pharmacy. Biofactors. 2013;39:2–13. doi: 10.1002/biof.1079. [DOI] [PubMed] [Google Scholar]
- 8.Kumar A, Ahuja A, Ali J, et al. Conundrum and therapeutic potential of curcumin in drug delivery. Crit Rev Ther Drug Carrier Syst. 2010;27:279–312. doi: 10.1615/CritRevTherDrugCarrierSyst.v27.i4.10. [DOI] [PubMed] [Google Scholar]
- 9.Lang A, Salomon N, Wu JC, et al. Curcumin in combination with mesalamine induces remission in patients with mild-to-moderate ulcerative colitis in a randomized controlled trial. Clin Gastroenterol Hepatol. 2015;13:1444–9. doi: 10.1016/j.cgh.2015.02.019. [DOI] [PubMed] [Google Scholar]
- 10.Deguchi Y, Andoh A, Inatomi O, et al. Curcumin prevents the development of dextran sulfate Sodium (DSS)-inducedexperimental colitis. Dig Dis Sci. 2007;52:2993–8. doi: 10.1007/s10620-006-9138-9. [DOI] [PubMed] [Google Scholar]
- 11.Baliga MS, Joseph N, Venkataranganna MV, et al. Curcumin, an active component of turmeric in the prevention and treatment of ulcerative colitis: preclinical and clinical observations. Food Funct. 2012;3:1109–17. doi: 10.1039/c2fo30097d. [DOI] [PubMed] [Google Scholar]
- 12.Liu F, Gao S, Yang Y, et al. Curcumin induced autophagy anticancer effects on human lung adenocarcinoma cell line A549. Oncol Lett. 2017;14:2775–82. doi: 10.3892/ol.2017.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu Y, Bu S. Curcumin induces autophagy, apoptosis, and cell cycle arrest in human pancreatic cancer cells. Evid Based Complement Alternat Med. 2017;2017 doi: 10.1155/2017/5787218. 5787218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun Q, Zhang W, Guo Y, et al. Curcumin inhibits cell growth and induces cell apoptosis through upregulation of miR-33b in gastric cancer. Tumor Biology. 2016;37:13177–84. doi: 10.1007/s13277-016-5221-9. [DOI] [PubMed] [Google Scholar]
- 15.Cooper HS, Murthy SN, Shah RS, et al. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab Invest. 1993;69:238–49. [PubMed] [Google Scholar]
- 16.Katsanos KH, Papadakis KA. Inflammatory Bowel Disease: updates on molecular targets for biologics. Gut Liver. 2017;11:455–63. doi: 10.5009/gnl16308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ammon HP, Wahl MA. Pharmacology of Curcuma longa. Planta Med. 1991;57:1–7. doi: 10.1055/s-2006-960004. [DOI] [PubMed] [Google Scholar]
- 18.Beloqui A, Memvanga PB, Coco R, et al. A comparative study of curcumin-loaded lipid-based nanocarriers in the treatment of inflammatory bowel disease. Colloids Surf B Biointerfaces. 2016;143:327–35. doi: 10.1016/j.colsurfb.2016.03.038. [DOI] [PubMed] [Google Scholar]
- 19.Epstein J, Docena G, MacDonald TT, et al. Curcumin suppresses p38 mitogen-activated protein kinase activation, reduces IL-1beta and matrix metalloproteinase-3 and enhances IL-10 in the mucosa of children and adults with inflammatory bowel disease. Br J Nutr. 2010;103:824–32. doi: 10.1017/S0007114509992510. [DOI] [PubMed] [Google Scholar]
- 20.Larmonier CB, Midura-Kiela MT, Ramalingam R, et al. Modulation of neutrophil motility by curcumin: implications for inflammatory bowel disease. Inflamm Bowel Dis. 2011;17:503–15. doi: 10.1002/ibd.21391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohno M, Nishida A, Sugitani Y, et al. Nanoparticle curcumin ameliorates experimental colitis via modulation of gut microbiota and induction of regulatory T cells. PLoS One. 2017;12:e0185999. doi: 10.1371/journal.pone.0185999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lubbad A, Oriowo MA, Khan I. Curcumin attenuates inflammation through inhibition of TLR-4 receptor in experimental colitis. Mol Cell Biochem. 2009;322:127–35. doi: 10.1007/s11010-008-9949-4. [DOI] [PubMed] [Google Scholar]
- 23.Liu Y, Zhang Z, Wang L, et al. TLR4 monoclonal antibody blockade suppresses dextran-sulfate-sodium-induced colitis in mice. J Gastroenterol Hepatol. 2010;25:209–14. doi: 10.1111/j.1440-1746.2009.06046.x. [DOI] [PubMed] [Google Scholar]
- 24.Soufli I, Toumi R, Rafa H, et al. Overview of cytokines and nitric oxide involvement in immuno-pathogenesis of inflammatory bowel diseases. World J Gastrointest Pharmacol Ther. 2016;7:353–60. doi: 10.4292/wjgpt.v7.i3.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salem M, Ammitzboell M, Nys K, et al. ATG16L1: A multifunctional susceptibility factor in Crohn disease. Autophagy. 2015;11:585–94. doi: 10.1080/15548627.2015.1017187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rufini S, Ciccacci C, Di Fusco D, et al. Autophagy and inflammatory bowel disease: Association between variants of the autophagy-related IRGM gene and susceptibility to Crohn’s disease. Dig Liver Dis. 2015;47:744–50. doi: 10.1016/j.dld.2015.05.012. [DOI] [PubMed] [Google Scholar]
- 27.Hooper KM, Barlow PG, Stevens C, et al. Inflammatory Bowel Disease drugs: A focus on autophagy. J Crohns Colitis. 2017;11:118–27. doi: 10.1093/ecco-jcc/jjx002.208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wirth M, Joachim J, Tooze SA. Autophagosome formation-the role of ULK1 and Beclin1-PI3KC3 complexes in setting the stage. Semin Cancer Biol. 2013;23:301–9. doi: 10.1016/j.semcancer.2013.05.007. [DOI] [PubMed] [Google Scholar]
- 29.Tanida I, Waguri S. Measurement of autophagy in cells and tissues. Methods Mol Biol. 2010;648:193–214. doi: 10.1007/978-1-60761-756-3_13. [DOI] [PubMed] [Google Scholar]
- 30.Kroemer G, Marino G, Levine B. Autophagy and the integrated stress response. Mol Cell. 2010;40:280–93. doi: 10.1016/j.molcel.2010.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]