Abstract
Background:
High-risk human papillomavirus (HPV) types are the main etiological factors for cervical cancer. HPV16 and HPV18 are generally the most common forms associated with development of high-grade cervical lesions. This study was undertaken to identify intratypic variants of HPV16 and HPV18 among women with cervical lesions in Tunisia.
Materials and Methods:
DNA was extracted from cervical samples collected from 49 women. using a PureLinkTM Genomic DNA mini Kit (Invitrogen). E6 and L1 open reading frames (ORF) were amplified by PCR and viral DNA amplicons were subjected to automated sequencing using Big Dye Terminators technology (Applied Biosystems). The obtained sequences were analyzed using an appropriate software program to allow phylogenetic trees to be generated.
Results:
HPV16 and HPV18 were detected in 15 and 5 cases, respectively. HPV16 E6 sequences clustered with the European German lineage (A2) whereas one isolate diverged differently in the L1 region and clustered with the African sub-lineage (B1). HPV 18 E6 sequences clustered with the European sub-lineage (A1) but L1 sequences clustered as a new clade which diverged from A1-A5.
Conclusions:
Our results suggest that the distribution of HPV16 and HPV18 sequences in women with cervical lesions in Tunisia is mainly related to European epidemiological conditions and point to the presence of recombinant HPV forms.
Keywords: HPV16, HPV18, phylogeny, Tunisia
Introduction
Human papillomaviruses (HPV), belonging to the Papillomaviridae family, are small, non-enveloped virus with a circular double-stranded DNA genome (Zur Hausen and de Villiers, 1994; Walboomers et al., 1999). Over 150 HPV genotypes are identified and grouped into five genera: Alpha, Beta, Gamma, Mu, and Nu papillomaviruses (de Villiers et al., 2004; Bernard et al., 2010). About forty of these genotypes infect the genital tract and are referred to as genital HPVs (Zur Haussen, 2002; Zur Haussen, 2009). Genital HPVs are closely associated with cervical cancer and are grouped into high-risk and low-risk HPVs according to their degree of oncogenicity. Among the high risk HPV genotypes, HPV 16 and 18 alone account for about 80% of total cervical cancer cases (Bosch et al., 1995; Bello et al., 2009; Li et al., 2011).
The prevalence of genital HPV infection in women differs considerably between countries and regions (Clifford et al., 2005; Seoud, 2012) as well as between different risk groups. In Tunisia, a 13.2% prevalence in the Grand Tunis region has been reported (Ardhaoui et al., 2016). In studies conducted on prostitutes, HPV-DNA prevalence was 39%-45% (Hassen et al., 2003; Demarco et al., 2006). HPV typing in these high-risk groups as well as in Tunisian women with precancerous and cancerous lesions showed the predominance of HPV16 as expected from the data of other countries (Missaoui et al., 2010).
It was reported that HPV16 and HPV18 variants are reflective of the migration patterns of Homo sapiens and suggested that HPV16 and HPV18 variant lineages might have diverged through genetic isolation at approximately the same time as Homo sapiens began establishing residence in different continental regions (Fu Xi et al., 2006; Chen et al., 2005; Chen et al., 2009; Pimenoff et al., 2016). Indeed, HPV16 and HPV18 isolates worldwide can be classified into 4 and 3 lineages, respectively, according to nucleotide variations in their genomes (Chen et al., 2009). To our knowledge, only two reports on intratypic HPV variants in Tunisia have been published to date and HPV16 and HPV18 variants of European sublineage were found to be the most prevalent (KrennHrubec et al., 2011; Ghedira et al., 2016). It has been reported that HPV intratypic variants differ in oncogenic potential (Villa et al., 200; Hang et al., 2016). In the present study in order to identify the HPV16 and HPV18 intratypic variants in women with cervical disease, we have sequenced and analyzed the E6 and L1 open reading frames (ORF) of both viruses identified among a sample of Tunisian women with precancerous or cancerous cervical lesions.
Materials and Methods
Patients and cervical samples
The study population consisted of 49 Tunisian women with cervical lesions consulting either at the Department of Gynecology and Obstetrics, La Rabta Maternity and Neonatology Center, Tunis, or in a private gynecologic office. Their mean age was 45 year (range, 21-64 years). A questionnaire about personal information was filled for each woman; clinical characteristics of patients are summarized in Table 1. Cervical samples were collected in SurePath™ vials (BD Diagnostics, France) for Pap smear assay and HPV typing. The study was conducted in compliance with the Helsinki Declaration and approved by the Habib Thameur Hospital local Ethical Committee, Tunis. Tunisia.
Table 1.
Study Population Characteristics
| characteristic | Total n=49 (%) |
|---|---|
| Age (years) | |
| <25 | 12 (24.5) |
| 26-40 | 15 (30.6) |
| >40 | 22 (44.9) |
| Marital status | |
| Never maried | 1 (2.0) |
| Maried/cohabiting | 40 (81.6) |
| Divorced/separated | 2(4.0) |
| Widowed | 3(6.0) |
| Not known | 3(6.0) |
| Sexual behavior | |
| Age at first sex | |
| <20 | 8 (16.3) |
| 20-25 | 13 (26.5) |
| >26 | 28(57.2) |
| Lifetime sex partners | |
| 0 | 1 (2.0) |
| 1 | 44 (89.8) |
| 2-5 | 4 (8.2) |
| Number of pregnancies | |
| 0 | 9 (18.0) |
| 1-3 | 26 (53.0) |
| ≥ 4 | 14 (29.0) |
| Contraceptive use | |
| Hormonal | 15 (30.6) |
| Non Hormonal | 25 (51.0) |
| No contraceptive | 9 (18.4) |
| Cervical cytology | |
| Normal | 9 (18.4) |
| ASCUS | 5 (10.2) |
| LSIL | 12 (24.5) |
| HSIL | 13 (26.5) |
| Cancer | 10 (20.4) |
DNA Extraction and HPV amplification
DNA was extracted from cervical samples using PureLinkTM Genomic DNA mini Kit (Invitrogen, Carlsbad, CA, USA) according to manufacturer’s instructions. DNA extracts were stored at -20°C until use.
HPV-DNA amplification was performed in the presence of consensus primers for genital HPVs and in the presence of type-specific primers for HPV 16 and HPV 18. The primer pair MY09/MY11 (Manos et al., 1989) was used as external primers followed by GP5+/GP6+ (Snijders et al., 1990) as internal primers for nested PCR to amplify a region of about 150 bp in ORF L1. Type-specific PCR was performed in the presence of either primer pair E6-HPV16U/E6-HPV16D for HPV 16 or E6-HPV18U/E6-HPV18D for HPV 18. These primers specify a region of 420 bp for HPV16 or 456 bp for HPV18. Primer sequences are shown in Table 2.
Table 2.
Primer Sequences
| Primer designation | Orientation | Sequence | Product size (bp) |
|---|---|---|---|
| E6-HPV16U | Sense | 5’-GCAAGCAACAGTTACTGCGA-3’ | 420 |
| E6-HPV16D | Antisense | 5’-GTTGTCTCTGGTTGCAAATC-3’ | |
| E6-HPV18U | Sense | 5’-ATGGCGCGCTTTGAGGATCC-3’ | 456 |
| E6-HPV18D | Antisense | 5’-TCGTTGGAGTCGTTCCTGTC-3’ |
Target DNA was amplified in a 50 µl reaction mixture containing: 1x Taq polymerase buffer, 1.5 mM MgCl2, 200µM each dNTP, 1.25 units HotStart Taq DNA polymerase (Invitrogen, Courtaboeuf, France), 0.5 µM each primer, 10µl DNA extract for simple PCR or 2 µl of first round PCR product for nested PCR. Thermocycler programs were as follows. For simple PCR with MY09/MY11 and for type-specific PCR: 40 cycles of denaturation at 94°C for 1 minute, annealing for 1 minute at 55°C, extension at 72°C for 1 minute and a final extension step at 72°C for 7 minutes. For the second round of nested PCR the temperature program was as above except for annealing which was performed at 50°C. PCR reactions were carried out in a GeneAmp PCR system 9700 Thermal Cycler (Applied Biosystems). Each PCR run included a blank control containing water instead of target DNA, a negative control DNA from HEp-2 cells and a positive control containing either HPV 16 DNA from SiHa cells or HPV18 DNA from HeLa cells (Adey et al., 2013). The quality of DNA extracts was assessed by amplification of a 536 bp sequence of the β globin gene using the primer pair KM29/RS42. All PCR products were visualized on 2% agarose gels in the presence of ethidium bromide.
HPV Sequencing and Phylogenetic Analysis
Sequencing of the purified PCR products was carried out in the forward and reverse directions using the corresponding primer pairs and Big Dye Terminators Cycle Sequencing Ready Reaction kit (Applied Biosystems, Foster City, CA, USA). Products of the sequencing PCRs were analyzed in an automatic ABI Prism 3100 Avant-Genetic Analyser (Applied Biosystems).
The obtained sequences were analyzed, using BioEdit (Hall, 1999) and Mega 6 (Tamura et al., 2013) softwares. Multiple alignments were generated by ClustalW integrated into BioEdit. Phylogenetic trees were generated using TreeView and DNA Maximum Likelihood program with molecular clock (DNAMLK) in Mega 6. The obtained isolates were aligned against references sequences shown in Table 3.
Table 3.
HPV Lineages Included in the Study
| Accession number | Geographic localization | |
|---|---|---|
| Genotype 16 | ||
| Lineage A | ||
| A1 (P) | K02718.1 | European Prototype |
| A2 | AF536179.1 | European German |
| A3 | HQ644236.1 | Europe |
| A4 | AF534061.1 | Europe/Asia |
| Lineage B | ||
| B1 | AF536180 | African1a |
| B2 | HQ644298 | African 1b |
| Lineage C | ||
| C1 | AF472509 | African 2a |
| Lineage D | ||
| D1 | HQ644257 | North American 1 |
| D2 | AY686579 | Asian American 2 |
| D3 | AF402678 | Asian American 1 |
| Genotype 18 | ||
| Lineage A | ||
| A1 | AY262282.1 | Asian Amerindian |
| A2 | EF202146.1 | Asian Amerindian |
| A3 | EF202147.1 | European |
| A4 | EF202151.1 | European |
| A5 | GQ180787.1 | European |
| Lineage B | ||
| B1 | EF202155.1 | African, African 1 |
| B2 | KC470225.1 | African |
| B3 | EF202152.1 | African 2 |
| Lineage C | ||
| C1 | KC470229 | African |
Results
Among the 49 woman included in the study, 15 (30.6%) and 5 (10.2%) were positive for HPV16 and HPV18, respectively and the remaining 29 samples were positive for other HPV types. As shown in Table 4, 65% (13/20) of the women positive for HPV16 or HPV18 had precancerous (HSIL) or cancerous lesions.
Table 4.
Distribution of HPV16 and HPV18 According to the Cytological Results
| Strain | Genotype | Type of lesion |
|---|---|---|
| MJR1 | HSIL | |
| MJR2 | HSIL | |
| MJR3 | HSIL | |
| MJR4 | LSIL | |
| MJR5 | LSIL | |
| MJR6 | Carcinoma | |
| MJR7 | LSIL | |
| MJR8 | HPV16 | LSIL |
| MJR9 | LSIL | |
| MJR10 | HSIL | |
| MJR11 | Adenocarcinoma | |
| MJR12 | LSIL | |
| MJR20 | Carcinoma | |
| MJR21 | HSIL | |
| MJR22 | Carcinoma | |
| MJR13 | HSIL | |
| MJR14 | HSIL | |
| MJR15 | HPV18 | Adenocarcinoma |
| MJR16 | LSIL | |
| MJR17 | Carcinoma |
All PCR products were sequenced and sequences from each region were submitted to GenBank under accession numbers KT931993 to KT932004, KU175625 to KU175627 and KX75949to KX759668.
Variant distribution was determined through L1 and E6 ORF sequences. HPV16 isolates were initially examined for nucleotide variation within a 114-nucleotide (nt) sequence in ORF L1 (nt 6654 to 6767). Sequence polymorphism was identified at 1 nt position through the ORF L1. All HPV16 strains appeared A2-like in E6 but one strain (MJR9) diverged differently within the L1 region. Indeed, within the 392-nt sequence in ORF E6 (nt 228 to 620), HPV16 strains showed a T350G nt change which is specific to the European German lineage A2 [27] and lead to a L83V substitution, whereas in ORF L1, one isolate showed the (G6719A) substitution, only found in the African sub-lineage B1.
All HPV18 strains were similar to the European sub-lineage (A1-A5) through the E6 region, but interestingly, all the HPV18 isolates showed a G to A silent transition through ORF L1 at position 6731.
Phylogenetic analysis
Four phylogenetic trees based on L1 and E6 nt sequences for HPV16 and HPV18 were constructed using Maximum Likelihood Tree integrated into MEGA6 program (Figure 1).
Figure 1.
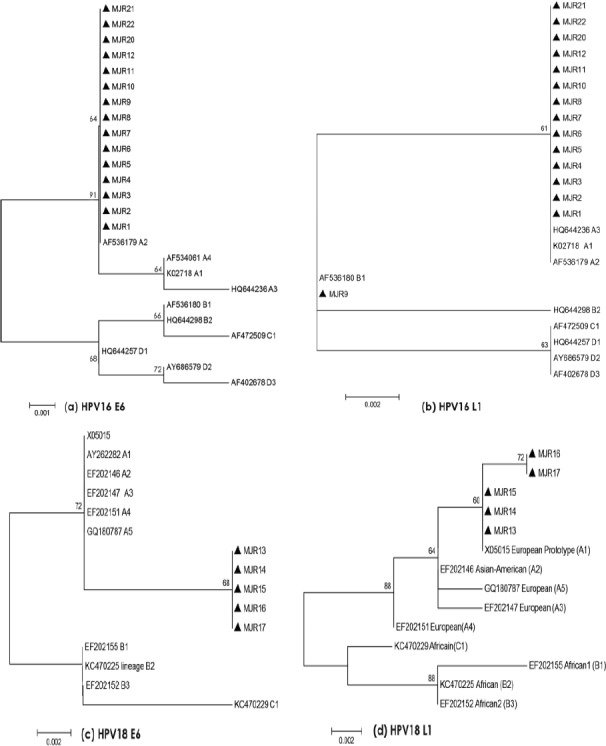
Phylogenetic Trees of HPV16 and HPV18 Based on L1 and E6 Nucleotide were Constructed Using Maximum Likelihood Tree Integrated into MEGA6 Program.
For HPV16, the phylogenetic tree corresponding to E6 (Figure 1a) showed that all isolates clustered with European German sub-lineage (A2), whereas the phylogenetic tree corresponding to L1 HPV16 (Figure 1b) showed that fourteen HPV16 isolates clustered closely with European variants (A1-A3), while one isolate (MJR9) clustered with African sub-lineage (B1). This suggests that MJR9 could be a recombinant between European and African variants.
For HPV18, the phylogenetic tree corresponding to E6 (Figure 1c) showed that all isolates clustered with the European variant (A1), while for L1, phylogenetic tree (Figure 1d) showed that these five isolates constitute a new clade due to G to A silent transition at position 6731, suggesting the possibility of a hitherto undescribed HPV18 variant.
Discussion
Intratypic variants of HPV16 and HPV18 show a particular geographical and ethnical distribution worldwide (Ong et al.,1993; Cornet et al., 2012; Chen et al., 2015).
Contrary to the previous studies from Tunisia (Hassen et al., 2003; Ghedira et al., 2016) women enrolled in the present study were selected on the basis of cervical lesions. Therefore, the high frequency of precancerous or cancerous cervical lesions in these women positive for HPV16 or HPV18 is not surprising and, as expected, HPV16 was the most frequent type. Ennaifer et al., (2011) have reported predominance of HPV16 and HPV18 (92.1%) in cervical cancer cases from Tunisian women (Ennaifer et al., 2011). A similar result has been reported from a neighbor country of Algeria with a prevalence of 61.4% for HPV16 and 15.8% for HPV18 (Hammouda et al., 2011).
All HPV16 and HPV18 isolates identified in this study were closely related to European lineages. This finding tallies with worldwide reported data (Li et al., 2011; Bravo and Felez-Sanchez., 2015; Kabekkodu et al., 2015; Pimenoff et al., 2016, KrennHrubec et al., 2011). The dominance of HPV16 European variants among this group of Tunisian women is in agreement with data obtained in Morocco showing the predominance (58.3%) of European variant among Moroccan woman (Qmichou et al., 2013). However the proportion of African variants (31.1%) was higher in Morocco than in the present and a previous Tunisian study (Ghedira et al., 2016). However, the observation of a probable European/African HPV16 recombinant might reflect the contribution of populations from sub-Saharan African origin in the epidemiology of HPV in Tunisia. On the other hand, the fact that all HPV18 isolates constituted a new clade suggests a particular virus host co-evolution in this North African country.
It has been reported that intratypic HPV variants present a different oncogenic potential (Villa et al., 2000; Hang et al., 2016). In the present study, the homogeneity in HPV strains precluded to perform this analysis. Further studies will be necessary to determine whether the HPV16 recombinant or the HPV18 variants identified in this study have a particular oncogenic potential.
This study is the first to date to propose an evolutionary scenario for both HPV16 and HPV18 in Tunisia. The current study provides a phylogenetic classification among a group of Tunisian woman and confirms that HPV16 and HPV18 infections in Tunisia are much more due to European variants than to African variants. This suggests that the distribution of HPV16 and HPV18 are closely related to the European pattern and could reflect historical links and migrations between Tunisia and the European continent.
Further studies are required to analyze the relationship between HPV16 and HPV18 Tunisian variants and viral pathogenicity and the possible impact of the variability observed in ORF L1 on the HPV vaccine efficacy.
Acknowledgements
This study was supported by the Ministry of higher education and Research of Tunisia and was conducted in Viral and Molecular Tumor Diagnostics Unit, Habib Thameur Hospital, Tunis, Tunisia. We thank Prof. Aldo Venuti from National Cancer Institute Regina Elena, Rome, Italy for having designed and provided the primers pairs E6-HPV16 and E6 HPV18. We thank Dr Ignacio Gonzalez Bravo for his thoughtful advice on phylogenetic analysis
References
- 1.Adey A, Burton JN, Kitzman JO, et al. The haplotype-resolved genome and epigenome of the aneuploid HeLa cancer cell line. Nature. 2013;500:207–11. doi: 10.1038/nature12064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ardhaoui M, Ennaifer E, Letaief H, et al. Prevalence, genotype distribution and risk factors for cervical Human Papillomavirus infection in the Grand Tunis Region, Tunisia. PLoS One. 2016;11:e0157432. doi: 10.1371/journal.pone.0157432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bello BD, Spinillo A, Alberizz P, et al. Cervical infections by multiple human papillomavirus (HPV) genotypes:Prevalence and impact on the risk of precancerous epithelial lesions. J Med Virol. 2009;81:703–12. doi: 10.1002/jmv.21429. [DOI] [PubMed] [Google Scholar]
- 4.Bernard HU, Burk RD, Chen Z, et al. Classification of papillomaviruses (PVs) based on 189 PV types and proposal of taxonomic amendments. Virology. 2010;401:70–9. doi: 10.1016/j.virol.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bosch FX, Manos MM, Munoz N, et al. Prevalence of human papillomavirus in cervical cancer:a worldwide perspective, International biological study on cervical cancer (IBSCC) Study Group. J Natl Cancer Inst. 1995;87:796–802. doi: 10.1093/jnci/87.11.796. [DOI] [PubMed] [Google Scholar]
- 6.Bravo IG, Felez Sanchez M. Papillomaviruses Viral evolution, cancer and evolutionary medicine. Evol Med Public Health. 2015;1:32–51. doi: 10.1093/emph/eov003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen AA, Gheit T, Franceschi S, et al. Human papillomavirus 18 genetic variations and cervical cancer risk worldwide. J Virol. 2015;89:10680–87. doi: 10.1128/JVI.01747-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen Z, Terai M, Fu L, et al. Diversifying selection in human papillomavirus type 16 lineages based on complete genome analyses. J Virol. 2005;79:7014–23. doi: 10.1128/JVI.79.11.7014-7023.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Z, DeSalle R, Schiffman M, et al. Evolutionary dynamics of variant genomes of human papillomavirus types 18, 45, and 97. J Virol. 2009;83:1443–55. doi: 10.1128/JVI.02068-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clifford GM, Gallus S, Herrero R, et al. Worldwide distribution of human papillomavirus types in cytologically normal women in the International Agency for Research on Cancer HPV prevalence surveys:a pooled analysis. Lancet. 2005;366:991–98. doi: 10.1016/S0140-6736(05)67069-9. [DOI] [PubMed] [Google Scholar]
- 11.Cornet I, Gheit T, Iannacone MR, et al. HPV16 genetic variation and the development of cervical cancer worldwide. Br J Cancer. 2013;108:240–44. doi: 10.1038/bjc.2012.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Marco F, Houissa-Kchouk F, Khelifa R, Marcante ML. High-risk HPV types in Tunisia. A pilot study reveals an unexpectedly high prevalence of types 58 and 82 and lack of HPV 18 among female prostitutes. J Med Virol. 2006;78:950–53. doi: 10.1002/jmv.20646. [DOI] [PubMed] [Google Scholar]
- 13.de Villiers EM, Fauquet C, Broker TR, Bernard HU, Zur Hausen H. Classification of papillomaviruses. Virology. 2004;324:17–27. doi: 10.1016/j.virol.2004.03.033. [DOI] [PubMed] [Google Scholar]
- 14.Ennaifer E, Salhi F, Laassili T, et al. Type-specific human papillomavirus distribution in invasive squamous cervical carcinomas in Tunisia and vaccine impact. Asian Pac J Cancer Prev. 2015;16:6769–72. doi: 10.7314/apjcp.2015.16.15.6769. [DOI] [PubMed] [Google Scholar]
- 15.Fu Xi L, Kiviat NB, Hildesheim A, et al. Human papillomavirus type 16 and 18 variants:Race-related distribution and persistence. J Nat Cancer Instit. 2006;98:1045–52. doi: 10.1093/jnci/djj297. [DOI] [PubMed] [Google Scholar]
- 16.Ghedira R, Mahfoudh W, Hadhri S, et al. Human papillomavirus genotypes and HPV-16 variants distribution among Tunisian women with normal cytology and squamous intraepithelial lesions. Infect Agent Cancer. 2016;11:61. doi: 10.1186/s13027-016-0109-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall TA. BioEdit:A user-friendly biological sequence alignment editor and analysis program for windows 98/98/NT. Nucleic Acids Symp Ser (Oxf) 1999;41:95–8. [Google Scholar]
- 18.Hammouda D, Clifford GM, Pallardy S, et al. Human papillomavirus infection in a population-based sample of women in Algiers, Algeria. Int J Cancer. 2011;128:2224–9. doi: 10.1002/ijc.25539. [DOI] [PubMed] [Google Scholar]
- 19.Hang D, Yin Y, Han J, et al. Analysis of human papillomavirus 16 variants and risk for cervical cancer in Chinese population. Virology. 2016;488:156–61. doi: 10.1016/j.virol.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 20.Hassen E, Chaieb A, Letaief M, et al. Cervical human papillomavirus infection in Tunisian women. Infection. 2003;31:143–8. doi: 10.1007/s15010-003-3112-7. [DOI] [PubMed] [Google Scholar]
- 21.Kabekkodu SP, Bhat S, Pandey D, et al. Prevalence of human papillomavirus types and phylogenetic analysis of HPV-16 L1 variants from Southern India. Asian Pac J Cancer Prev. 2015;16:2073–80. doi: 10.7314/apjcp.2015.16.5.2073. [DOI] [PubMed] [Google Scholar]
- 22.KrennHrubec K, Mrad K, Sriha B, et al. HPV types and variants among cervical cancer tumors in three regions of Tunisia. J Med Virol. 2011;83:651–7. doi: 10.1002/jmv.22011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li N, Franceschi S, Howell-Jones R, et al. Human papillomavirus type distribution in 30,848 invasive cervical cancers worldwide:Variation by geographical region, histological type and year of publication. Int J Cancer. 2011;128:927–35. doi: 10.1002/ijc.25396. [DOI] [PubMed] [Google Scholar]
- 24.Manos MM, Ting Y, Wright DK, et al. The use of polymerase chain reaction amplification for the detection of genital human papillomaviruses. Cancer Cell. 1989;7:209–14. [Google Scholar]
- 25.Missaoui N, Hmissa S, Trabelsi A, et al. Prevalence of HPV infection in precancerous and cancerous lesions of the uterine cervix in Tunisia. Ann Biol Clin. 2010;68:297–303. doi: 10.1684/abc.2010.0431. [DOI] [PubMed] [Google Scholar]
- 26.Ong CK, Chan SY, Campo MS, et al. Evolution of human papillomavirus type 18:ancient phylogenetic roots in Africa and intratype diversity reflect coevolution with human ethnic groups. J Virol. 1993;67:6424–31. doi: 10.1128/jvi.67.11.6424-6431.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.PaVE Website:Papilloma virus genome database. https://pave.niaid.nih.gov/
- 28.Pimenoff VN, Mendes de Oliveira C, Bravo IG. Transmission between archaic and modern human ancestors during the evolution of the oncogenic human papillomavirus 16. Mol Biol Evol. 2016;34:4–19. doi: 10.1093/molbev/msw214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qmichou Z, Khyatti M, Berraho M, et al. Analysis of mutations in the E6 oncogene of human papillomavirus 16 in cervical cancer isolates from Moroccan women. BMC Infect Dis. 2013;13:378. doi: 10.1186/1471-2334-13-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seoud M. Burden of human papillomavirus-related cervical disease in the extended middle East and north Africa-a comprehensive literature review. J Low Genit Tract Di. 2012;16:106–20. doi: 10.1097/LGT.0b013e31823a0108. [DOI] [PubMed] [Google Scholar]
- 31.Snijders PJ, van den Brule AJ, Schrijnemakers HF, et al. The use of general primers in the polymerase chain reaction permits the detection of a broad spectrum of human papillomavirus genotypes. J Gen Virol. 1990;71:173–81. doi: 10.1099/0022-1317-71-1-173. [DOI] [PubMed] [Google Scholar]
- 32.Tamura K, Stecher G, Peterson D, et al. MEGA6:Molecularevolutionary genetics analysis Version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Villa LL, Sichero L, Rahal B, et al. Molecular variants of human papillomavirus type 16 and 18 preferentially associated with cervical neoplasia. J Gen Virol. 2000;81:2959–68. doi: 10.1099/0022-1317-81-12-2959. [DOI] [PubMed] [Google Scholar]
- 34.Walboomers JM, Jacobs MV, Manos MM, et al. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol. 1999;189:12–9. doi: 10.1002/(SICI)1096-9896(199909)189:1<12::AID-PATH431>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 35.Zur Hausen H. Papillomaviruses in the causation of human cancers –a brief historical account. Virology. 2009;384:260–5. doi: 10.1016/j.virol.2008.11.046. [DOI] [PubMed] [Google Scholar]
- 36.Zur Hausen H, de Villiers EM. Human papillomaviruses. Annu Rev Microbiol. 1994;48:427–47. doi: 10.1146/annurev.mi.48.100194.002235. [DOI] [PubMed] [Google Scholar]
- 37.Zur Hausen H. Papillomaviruses and cancer:from basic studies to clinical application. Nat Rev Cancer. 2002;2:342–50. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]


