Abstract
Objectives:
Gastrointestinal disorders, such as inflammatory bowel diseases (IBD) and functional gastrointestinal disorders (FGID), involve disrupted homeostatic interactions between the microbiota and the host. Both disorders are worsened during stress, and in laboratory mice, stress exposure has been shown to change the composition of the gut microbiome. Stress-induced changes to the microbiome exacerbate intestinal inflammation and alter intestinal motility in mice. However, it is not yet known whether microbiota-derived short chain fatty acids (butyrate, propionate, and acetate) and their receptors contribute to this effect.
Methods:
Mice were exposed to a social disruption (SDR) stress, or left undisturbed as a control. After the first stress exposure, mice were orally challenged with Citrobacter rodentium or with vehicle. The levels of SCFAs were measured using gas chromatography-mass spectrometry. SCFA receptors were measured via real time PCR. Microbial community composition was assessed using 16S rRNA gene sequencing.
Results:
Stress exposure reduced colonic SCFA levels. However, stress exposure and C. rodentium significantly increased SCFA levels and changed the expression of SCFA receptors. The levels of SCFAs did not correlate with the severity of colonic inflammation, but the colonic expression of the SCFA receptor GPR41 was positively associated with inflammatory cytokines and colonic histopathology scores. The relative abundances of several taxa of colonic bacteria were significantly changed by stress exposure, including SCFA producers.
Conclusions:
Social stress can have a significant effect on infection-induced colonic inflammation, and stress-induced changes in microbial-produced metabolites and their receptors may be involved.
Keywords: Microbiome, stress, colitis, GPR41, Citrobacter rodenium, social disruption stress, Parabacteroides
Introduction:
Inflammatory bowel diseases (IBD) and functional gastrointestinal disorders (FGID) involve disrupted homeostatic interactions between the gut microbiota and the host leading to heightened inflammatory responses (in IBD)1 or altered pain perception and motility (in FGIDs)2–4. The severity of IBD and self-reported health related quality of life (HRQoL) is often worsened during stressful periods5–8. Likewise, associations between stressful life events and HRQoL have been found in patients with FGIDs9,10.
The microbiota-gut-brain axis involves bidirectional interactions between the gastrointestinal mucosal immune system, commensal microbiota, and the brain1,11. These host-microbe interactions can be significantly affected during stress exposure12,13, but how stress leads to homeostatic imbalance in the intestines is not clear. Social stress, such as social disruption (SDR), significantly increases plasma levels of serum corticosterone, epinephrine, norepinephrine, and affects gut microbiome diversity12,14–16. These changes in the gut microbiome can significantly impact immune system activity17,18.
Gut microbes ferment carbohydrates to produce short chain fatty acids (SCFAs), including acetic, propionic, and butyric acids, that affect mucosal inflammation through direct anti-inflammatory effects on CD4+ T cells and increases in the number of regulatory T cells19,20. SCFAs can activate G-protein coupled receptors on gut epithelial cells, lamina propria leukocytes, and cells of the enteric nervous system21,22. We hypothesized that stress would affect the production of SCFAs, the commensal gut microbiome, and the expression of SCFA receptors.
METHODS:
Experimental design:
Male C57BL/6 mice (Charles River Laboratories), 6–8 weeks of age were housed 3 per cage on a 12 hr light:dark schedule (lights on 0600). Standard diet and water were provided ad libitum. All experimental procedures were approved by and performed in accordance with The Ohio State University Animal Care and Use Committee.
Mice were exposed to the SDR stress over a 2 hr period (1630–1830) for 6 consecutive days23, which involves placing an aggressive male into the experimental animal’s home cage. Immediately after the first exposure to SDR, ½ of the stress-exposed and ½ of non-stress-exposed mice were challenged with Citrobacter rodentium by oral gavage. Half of the mice were euthanized the morning after the 6th night of SDR (6 days post-infection) and remaining mice were euthanized 12 days post-infection. The non-stress-exposed mice were euthanized at identical times (Fig. 1a and b). The experiments were conducted as discovery and validation experiments with the discovery experiment consisting of 1 cage (3 mice) per group and the validation experiment consisting of 2 cages (6 mice) per group. A total of 8 different groups with data combined from discovery and validation experiments (for a total of 9 mice per group) are presented in the manuscript, with data from individual discovery and validation experiments shown in the supplement.
Figure 1: Experimental design.
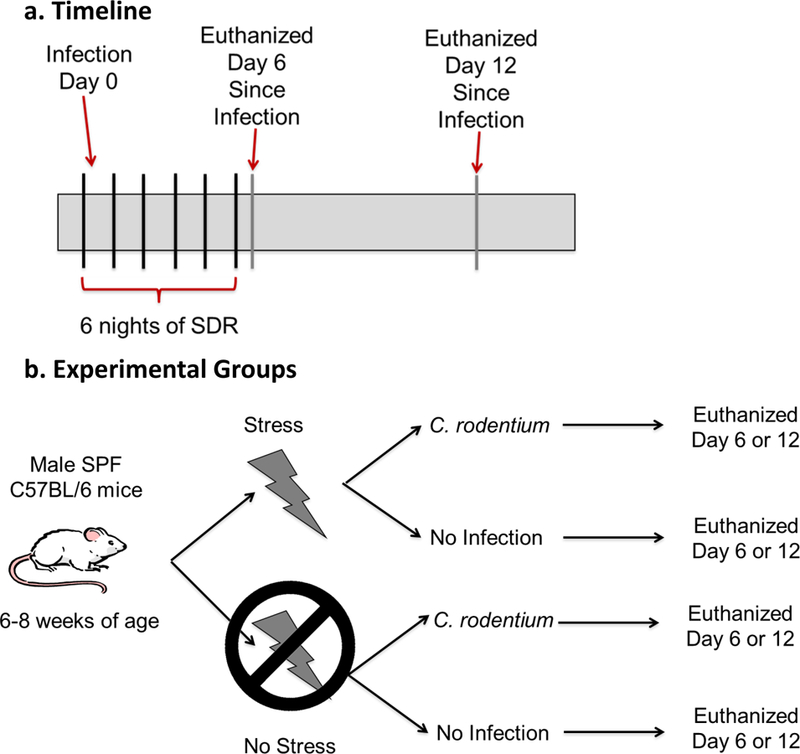
a and b. Mice were exposed to the stress for 6 nights in a row. After the first exposure to SDR, half of the stress exposed and half of non-stress exposed mice were challenged with Citrobacter rodentium by oral gavage. Half of the mice exposed to SDR were euthanized the morning after the 6th night of SDR. The other mice exposed to SDR were euthanized on 12 days post-challenge. The non-stress exposed mice were euthanized at the identical time points. N=9 mice per group.
Citrobacter rodentium:
Citrobacter rodentium is a Gram-negative bacterium that causes an effacing lesion to the brush border in mice similar to that in humans with enteropathogenic and enterohemorrhagic Escherichia coli24. It induces mild/moderate inflammation in the descending colon24 that is dependent upon the amount of C. rodentium administered. Citrobacter rodentium strain DBS120 (pCRP1::Tn5) was grown in Difco Lennox broth overnight. Mice in the infection group were challenged via oral gavage with 100 μl of C. rodentium containing 3 × 106 CFU in PBS. Fecal shedding of C. rodentium was determined on days 6 and 12 post-infection by plating stool on MacConkey agar with kanamycin (40µg/ml).
Histopathology:
Histopathology was scored in the distal colon in a blinded fashion using a validated scoring system; a score of 0 represented no inflammation and a score of 4 represented severe inflammation with lamina propria infiltration, architectural distortion, crypt abscesses, and ulcers25.
Semiquantitative real-time PCR:
RNA was isolated from proximal colons using TriZOL26. Real-time PCR was performed on the ABI Prism 7000 system14. 18S was used as a housekeeping gene. SYBR green was used for GPR41, GPR43, GPR109A, IL-1β, IFNγ, IL-22, and REG3γ (Supplemental Table 1). Data are expressed as a fold change from uninfected-non-stress-exposed group, day 6.
Short chain fatty acid quantification:
Fecal samples were lyophilized for 22 hrs. Extraction and gas chromatography-mass spectrometry (GC-MS) was performed as previously published27. The GC-MS consisted of a Trace Ultra GC with an AS3000 automatic liquid sampler coupled to a DSQ II mass spectrometer (Thermo Scientific, Waltham, MA). A stabilwax-DA (Restek, cat# 11023) highly polar column with polyethylene glycol stationary phase was installed. Split injection mode with split flow and split ratio set at 10. Ethyl acetate was run between every sample, and each sample was injected twice. Calculated amounts are the average from two injections. A standard curve was generated for acetic, butyric, and propionic acid at the beginning and standards were used throughout each run. SCFA identification was based on retention times (RT) and EI-MS spectra compared to NIST/EPA/NIH mass spectral libraries (NIST Mass Spectral Search Program v2.0, 2005)27.
The processing setup module in Xcalibur (Thermo Scientific, Waltham, MA) was used for rapid quantification of the absolute amount of SCFAs in each sample using data obtained from the standards. Target ions selected to generate extracted ion chromatograms (EICs) for quantification of acetic, propionic, and butyric acids were m/z 60, 74, and 60. The ICIS peak detection algorithm was used to detect the highest peak in the expected RT range for acetic (RT= 4.08, 20 second window), propionic (RT = 4.75, 20 second window), and butyric acid (RT = 5.53, 30 second window). The quality of the peaks was validated by enabling the resolution, symmetry and peak classification parameters.
Mucosa-associated microbiome:
The QIAamp DNA minikit protocol (Qiagen, Valencia, CA) was used for DNA recovery of the colonic mid-section. DNA was amplified targeting the V3–4 hypervariable region of the 16S rRNA gene (F:5’TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG; R:5’GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC). Libraries were prepared (NexteraXT kit; Illumina) and equimolar samples pooled. Sequencing was performed via the Illumina MiSeq platform resulting in 34 million paired-end reads (MiSeq Reagent Kit v3 600cycle; Illumina). Forward and reverse reads were merged using Quantitative Insights into Microbial Ecology (QIIME) version 1.9.1 with an overlap length of 40 and 95% similarity in the overlap region. Trimming and filtering at Q20 resulted in ~8 million reads. Closed reference OTU picking and green genes version 13.8 were used to produce OTUs incorporating 77% of the input reads. De novo OTU picking was performed using AbundantOTU+ version 0.92b which incorporated 98% of the input reads after removing chimeric and contaminant OTUs. Taxa were retained if they had ≥ 0.005% of the total count28. Linear mixed effect model followed by ANOVA was conducted with Group, Infection, and Day as fixed effects and cage as a random effect29. All P-values were FDR corrected. Alpha diversity was assessed using Chao1 and Shannon diversity indexes (SDI) using rarefied counts. Beta diversity was assessed using PCoAs designed from Bray-Curtis dissimilarity using Log 10 normalized counts29,30.
Statistical analysis:
Three-way ANOVAs were used with stress exposure (i.e., stress vs. no stress), infection exposure (i.e., C. rodentium vs. no C. rodentium), and sac day (i.e., day 6 vs. day 12) as the between-subjects variables. Modified Bonferroni method was used for multiplicity of significance tests between groups. Pearson correlation analysis was performed with multiple regression analysis to determine significant associations. An alpha level of p<.05 was set as the rejection criteria for the null hypothesis. All data were analyzed using SPSS statistical software version 21 (IBM Corp, Armonk, NY). Combined data and analyses are shown in the paper’s main text. Discovery and validation experimental data and analyses are shown in supplemental figures.
Results:
Stress exposure and infection affects colonic inflammation:
Infection-challenged mice that were not exposed to the stress had low levels of C. rodentium (approximately 1×104 CFU/g). However, exposure to stress during infection significantly increased C. rodentium levels by approximately 100 fold on both day 6 and 12 post-infection (Fig. 2a; p<.05). Mice exposed to the infection and the stress did not exhibit any differences in stool consistency, weight differences, or observed overt behavioral changes.
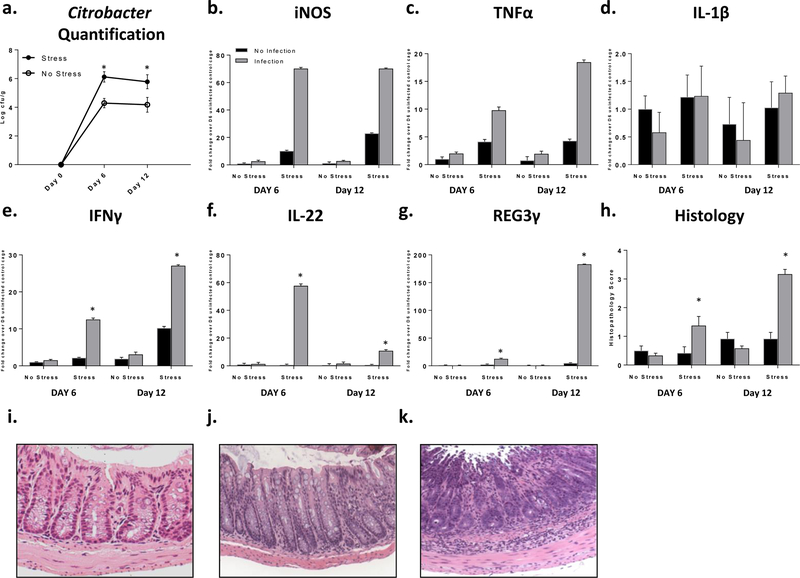
Figure 2: Stress exposure increased C. rodentium levels and inflammation.
a. Citrobacter rodentium levels increased with stress (p<.05). b and c. iNOS and TNFα main effect of stress and infection exposure (p<.05). d. IL-1β main effect of stress exposure (p<.05). e. IFNγ *p<.05 stress x infection interaction. f. IL-22 *p<.05 stress x infection interaction. g. REG3γ *p<.05 stress x infection interaction. h. Histopathology *p<.05 vs no infection same day. i-k. Representative images i. mild inflammation, infection exposed euthanized day 6. j. moderate inflammation, infection and stress exposed euthanized day 6. k. severe inflammation, infection and stress exposed euthanized day 12. 20X magnification.
Stress exposure regardless of infection increased iNOS, TNFα, and IL-1β mRNA levels in the colon (Fig. 2b-d; p<.05). C. rodentium also significantly increased iNOS and TNFα mRNA levels regardless of stress exposure (Fig. 2b-c; p<.05). In contrast, IFNγ, IL-22, and REG3γ mRNA expression were only increased in mice exposed to stress during infection (Fig. 2e-g; p<.05).
Exposure to the stress in the absence of C. rodentium did not impact histopathology scores (Fig. 2h). However, histopathology scores were significantly increased in stress-exposed mice on day 6 and day 12 post-infection when compared to non-stress exposed mice (p<.05), even though both groups of mice were challenged with the same dose of C. rodentium. Mice exposed to stress and C. rodentium had an average colonic pathology score of 3.17 on day 12, which was significantly different than all other groups (Fig. 2h; p<.05). On day 6 post-infection, histopathology scores were significantly higher in mice exposed to SDR during infection compared to mice not exposed to the stress (p<.05). Representative histologic sections are provided in Fig. 2i-k. The effects of stress on colonic gene expression and histopathology were similar in both discovery and validation experiments (Supplemental Fig. 1).
SCFA levels and receptors are influenced by stress exposure:
SCFA levels were not significantly changed by C. rodentium alone, but there were significant interactions between stress exposure and C. rodentium on acetic and butyric acid (Fig. 3a-b; p<.05). In the absence of infection, acetic and butyric acid levels were reduced on day 6 in stress-exposed mice compared with non-stressed mice (p<.05). However, in mice exposed to stress during infection, acetic and butyric acid levels were significantly increased in comparison to uninfected mice (p<.05). This increase persisted through day 12 for acetic acid (p<.05). Propionic acid levels were not affected by C. rodentium, but were significantly increased in mice exposed to SDR regardless C. rodentium challenge (Fig. 3c; p<.05).
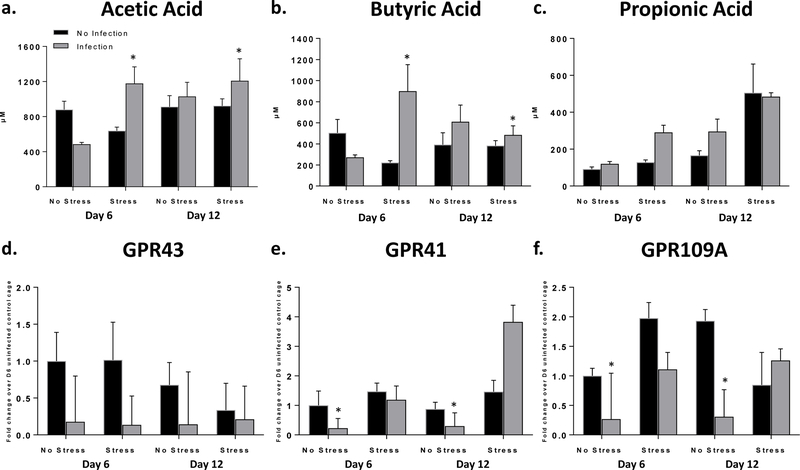
Figure 3: C. rodentium and stress exposure affects SCFA levels and SCFA receptor expression.
a. Acetic acid analysis *p<.05 stress x infection interaction. b. Butyric acid analysis *p<.05 stress x infection interaction. c. Propionic acid analysis main effect of stress (p<.05). d. GPR43 receptor expression qPCR analysis main effect of infection exposure (p<.05). e. GPR41 receptor expression qPCR analysis *p<.05 stress x infection interaction. f. GPR109A receptor expression qPCR analysis *p<.05 stress x infection interaction.
Challenge with C. rodentium reduced mRNA expression for the SCFA receptor GPR43 regardless of stress exposure (Fig. 3d; p<.05). However, the effects on GPR41 and GPR109A were dependent on whether the mice were stress-exposed during infection (Fig. 3e-f). GPR41 mRNA was significantly reduced on days 6 and 12 in infection-exposed mice not exposed to the stress vs infected-stress-exposed mice (p<.05). However, infected-stress-exposed mice exposed GPR41 mRNA expression on day 12 was significantly increased in comparison to all groups not exposed to stress (p<.05). GPR109A mRNA exposure was significantly reduced in mice challenged with C. rodentium in the absence of stress exposure compared to infected-stress-exposed mice (p<.05). Although there was considerable variability when samples were analyzed separately based on whether they were collected as part of the discovery or validation experiments, the patterns of SCFA levels and receptor gene expression were similar in the replicate experiments (Supplemental Fig. 2).
Stress exposure and C. rodentium altered the composition of the mucosa-associated microbiome:
Alpha diversity, which measures species diversity within a sample, was assessed using the SDI and Chao1 index. It was only significantly affected by infection and it was lower in mice infected with C. rodentium compared to mice that were not infected (Fig. 4a-b; p<.05). Alpha diversity was not significantly affected by stress exposure.
Figure 4: Stress exposure and infection effected alpha and beta diversity.
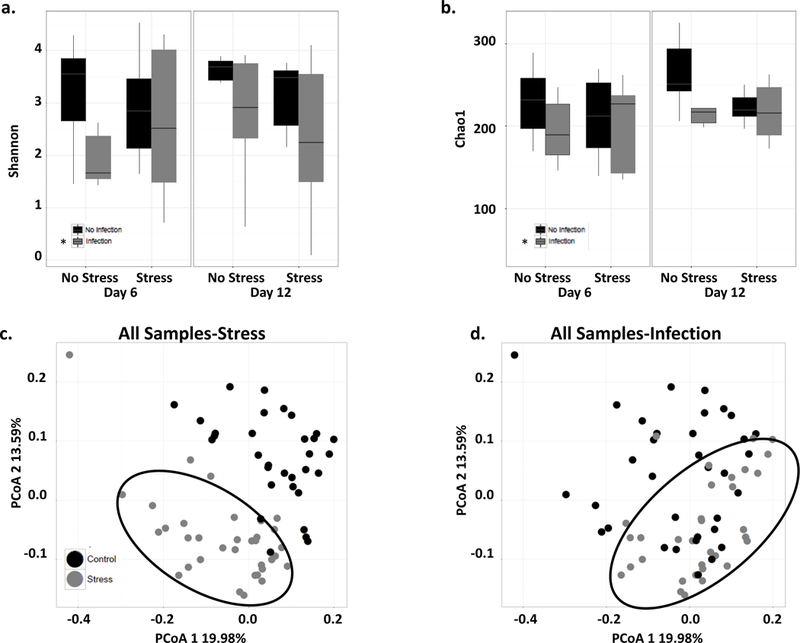
a. SDI decreased when exposed to the infection (*p<.05). b. Chao1 diversity decreased when exposed to the infection (*p<.05). c. All samples depicted on a PCoA separated by stress exposure. Mice exposed to the stress (black circle around) clustered separately from not exposed to the stress regardless of infection, significant PCoA axis 1 and 2 (p<.05). d. All samples depicted on a PCoA separated by infection. Mice challenged with the infection clustered separately from mice not exposed to the infection (black circle around) regardless of stress exposure had a significant PCoA axis 1 and 2 (p<.05).
Beta diversity was significantly different in stress-exposed mice. The samples clustered separately from mice not exposed to the stress regardless of C. rodentium (Fig. 4c) with significant differences on PCoA axis 1 and 2 (p<.05). When samples were classified only based upon infection, it was evident that infected mice clustered separately from uninfected mice on PCoA axis 1 and 2 (Fig. 4d; p<.05).
Taxonomic analyses at the genus level showed main effects of stress-exposure on the relative abundances of Akkermansia, Anaerostipes, Butyricicoccus, Coprococcus, Parabacteroides, and SMB53 that were decreased in stress-exposed mice (Supplemental Fig. 3a-f; p<.05). Bacteroides and Butyricimonas relative abundances were decreased with stress exposure and infection-exposure (Supplemental Fig. 3g and h; p<.05). There were also main effects of stress-exposure on the relative abundances of Odoribacter, Sutterela, AF12, Helicobacter, and Prevotella that were increased in stress-exposed mice (Supplemental Fig. 3i and j and Supplemental Fig. 4a-c; p<.05). AF12, Helicobacter, and Prevotella relative abundances were also decreased with a main effect of infection-exposure (p<.05). Anaerococcus, Anaeroplasma, Bradyrhizobium, Enhydrobacter, Mucispirillum, Oscillospira, Peptoniphilus, and Roseburia relative abundances decreased with infection-exposure (Supplemental Fig. 4d-k; p<.05). Enterobacter, Flavobacterium, Flexispira, and Trabulsiella relative abundances were increased in infected mice (Supplemental Fig. 4l-o; p<.05). Similar patterns were evident in both discovery and validation experiments (Supplemental fig. 5–8).
Multiple regression analysis indicated that the expression of GPR41 was positively associated with inflammatory cytokines TNFα and iNOS (p<.001) and colonic histopathology scores (p<.001). Propionic acid was also positively associated with inflammatory cytokines TNFα and iNOS (p<.001) and colonic histopathology scores (p<.01), but not associated with GPR41. Analyses were performed to determine whether specific genera were associated with SCFA levels and histopathology scores. Sequences classified at the genus level were normalized by finding the square root of the proportion of total sequences, followed by the arcsine of the square root. Propionic acid was positively correlated with Sutterella (p<.001), whereas butyric acid correlated with Anaerofustis, Anaerotruncus, Butyricicoccus, Clostridium, Coprococcus, Dehalobacterium, Dorea, Oscillospira, and Ruminococcus (p<.006). Increases in the percentages of Dehalobacterium and Dorea were associated with higher concentrations of acetic acid (p<.006). In addition, there was a negative association between Parabacteroides and histopathology scores (p<.01).
Discussion:
Stress exposure has a profound effect on microbiome diversity, multiple genera, and mucosal inflammation, which are significantly affected when mice are challenged with C. rodenium5–7,13,31,32,. Stress prior to pathogen challenge increased the susceptibility to, and severity of, colonic inflammation14. Although C. rodentium levels were increased, stress led to dysregulation of the colonic inflammatory response, which was not directly related to pathogen load. In our previous study, we showed that administering the probiotic L. reuteri prevented the exacerbating effects of stress on colonic inflammation, even though pathogen levels remained high in stress-exposed mice18. This suggests that an increase in pathogen load is not the only factor contributing to excessive inflammatory responses. In fact, using experimental fecal transplants, we have found that the gut microbiota contribute to dysregulation of mucosal inflammatory responses during stress17. In the current study, stress during infection increased inflammation and this was negatively associated with the abundance of Parabacteroides. Interestingly, similar inverse associations have been seen in patients with IBD where the abundance of Parabacteroides was decreased in patients with inflammation vs healthy control groups33. The negative association between Parabacteroides and degree of inflammation suggests this genus has protective effects in the colon, but the exact effects are not known.
Changes in the microbiome in stress exposed mice could also be due to contamination/coprophagy of aggressor stool. Helicobacter was increased in stress-exposed mice, and some species of Helicobacter can exacerbate colonic inflammation34. We cannot rule out that this increase in Helicobacter is not due to direct contamination from the aggressor because the microbiome of the aggressor was not assessed. It is known that Helicobacter growth can be increased by stress-induced hormones such as epinephrine and norepinephrine35. Other stress paradigms, such as prolonged restraint, which do not involve contamination from other mice, also affect microbiome composition36. Thus, we do not believe that all of the effects of stress exposure on microbial community composition are due to contamination/coprophagy of aggressor stool and follow up research is necessary to understand the factors that lead to the increase in Helicobacter.
Stress led to a decrease in known SCFA producing genera Anaerostipes, Butyricicoccus, Coprococcus, Parabacteroides, and Butyricimonas, but an increase in Odoribacter37–41, whereas infection led to a significant decrease in known SCFA producers Butyricimonas, Anaerococcus, and Roseburia39,41,42 and increase in Enterobacter. Several studies have suggested that SCFAs play a protective role in the intestines. For example, intestinal inflammation improved in mice treated with butyric and acetic acid43, and clinically, SCFA enemas have been successfully used to treat diversion colitis and proctosigmoiditis44,45. The benefits of SCFAs are likely due to an increase in regulatory T cells, since SCFAs given to germ-free mice cause an increase in regulatory T cells20,46.
We showed that exposure to stress leads to a decrease in butyric and acetic acid levels, as well as an increase in propionic acid levels after 6 days of stress exposure, but only in the absence of the infection. In contrast, mice exposed to stress during infection had an increase in SCFA levels, particularly on day 6. This was somewhat surprising given the importance of SCFAs in other models, and clinical cases, of colonic inflammation. We have also seen similar results when mice are exposed to a restraint stress, which led to decreases in acetic, butyric, and propionic acids in the absence of infection, but increases during infection36. These unexpected results may be due to the nature of the C. rodentium challenge. Inflammation starts in the cecum and spreads distally to cause mild/moderate colonic hyperplasia in the descending colon24. In mice, the cecum is a key location for microbial fermentation, thus, murine models of colitis that cause pancolitis may affect SCFA production to a greater degree than C. rodentium-induced colitis, a hypothesis worth testing in future studies.
In our study, receptor expression, rather than SCFA levels, were more directly related to the severity of colonic inflammation. SCFAs exert their mechanism of action via G-protein coupled receptors and through inhibition of histone deacetylases43,47. Stress exposure during infection significantly changed SCFA receptor expression. Infection led to a down regulation of GPR41, GPR43, and GPR109A receptor expression. These mice had little evidence of inflammation. However, mice exposed to stress during infection had significant increases in SCFA receptors and intestinal inflammation, suggesting that SCFA receptors play a role in stress-induced exacerbation of colonic inflammation. In particular, GPR41 had a positive association with TNFα and iNOS mRNA, as well as histopathology. Our results suggest that stress-induced upregulation of GPR41 contributes to enhanced disease pathology in C. rodentium-challenged mice. A pro-inflammatory role of GPR41 is consistent with studies in GPR41−/− mice showing significant improvements in colonic inflammation22.
It is not yet clear why SCFA levels did not significantly decrease in the colon when the relative abundance of SCFA-producing taxa was significantly reduced. However, this may reflect a strong limitation of studies that use 16S rRNA gene sequencing to access the mucosa-associated microbiome. While 16S rRNA gene sequencing can identify differences in community diversity and the relative abundance of specific bacterial genera, it is often not predictive of bacterial function/activity48. Future studies will use additional methods to identify microbial gene functions and metabolic activities.
The samples for this study were collected during two experiments, a discovery experiment and a follow-up validation experiment, and data from the experiments were combined for our primary analysis. Although the majority of findings showed the same patters when discovery, validation, and combined data were assessed, the results from the discovery samples (which contained an n=3) did not match the validated samples when comparing Citrobacter quantification, butyric acid, propionic acid, and GPR109A. When considered together, the findings support previous findings indicating that stress-induced changes of the mucosa-associated microbiome influence colonic immune responses. Because the exploratory nature of this study, further in depth investigations is warranted to confirm the important implications for GI illnesses and conditions that are associated with differences in the gut microbiome (such as IBD or FGIDs) and are often worsened during stressful periods. It was predicted that bacterial-produced SCFAs would be directly correlated with disease severity. However, only changes in the SCFA receptor GPR41 were directly related to colonic inflammation. Our results, along with findings that GPR41−/− mice have reduced colonic inflammation22 have led us to surmise GPR41 plays an important role in linking the gut microbiome to stress-induced exacerbation of colonic inflammation. Ongoing studies are utilizing additional models of murine inflammation that are similar to IBD, and are prospectively assessing IBD and FGID patients, to better understand the effects of stress on the microbiome and metabolome.
Supplementary Material
Whats is Known:
Inflammatory bowel diseases (IBD) and functional gastrointestinal disorders (FGID) are worsened with stress exposure and both involve a dysbiotic microbiome.
Stress exposure has been shown to change the composition of the gut microbiome and worsen intestinal inflammation.
What is New:
Social stress induced an upregulation of GPR41 and was associated with colonic inflammation.
The relative abundances of short chain fatty acid producing bacteria and short chain fatty acid levels were significantly affected by exposure to social stress.
Negative associations between histopathology scores and the abundance of Parabacteroides in this study are similar to associations seen in IBD patients.
Acknowledgments
Conflicts of Interest and Source of Funding:
This works was supported by the National Center for Complementary & Integrative Health of the National Institutes of Health under Award Number R01AT006552 (to M.T.B.). The author(s) report no conflict of interests or competing financial interests.
References
- 1.Sartor RB Microbial influences in inflammatory bowel diseases. Gastroenterology 134, 577–594, doi: 10.1053/j.gastro.2007.11.059 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Crouzet L et al. The hypersensitivity to colonic distension of IBS patients can be transferred to rats through their fecal microbiota. Neurogastroenterology and motility : the official journal of the European Gastrointestinal Motility Society 25, e272–282, doi: 10.1111/nmo.12103 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Murakami T et al. Changes in Intestinal Motility and Gut Microbiota Composition in a Rat Stress Model. Digestion 95, 55–60, doi: 10.1159/000452364 (2017). [DOI] [PubMed] [Google Scholar]
- 4.O’Mahony SM et al. Disturbance of the gut microbiota in early-life selectively affects visceral pain in adulthood without impacting cognitive or anxiety-related behaviors in male rats. Neuroscience 277, 885–901, doi: 10.1016/j.neuroscience.2014.07.054 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Bernstein CN et al. A prospective population-based study of triggers of symptomatic flares in IBD. The American journal of gastroenterology 105, 1994–2002, doi: 10.1038/ajg.2010.140 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Traue HC & Kosarz P Everyday stress and Crohn’s disease activity: a time series analysis of 20 single cases. International journal of behavioral medicine 6, 101–119, doi: 10.1207/s15327558ijbm0602_1 (1999). [DOI] [PubMed] [Google Scholar]
- 7.Mardini HE, Kip KE & Wilson JW Crohn’s disease: a two-year prospective study of the association between psychological distress and disease activity. Digestive diseases and sciences 49, 492–497 (2004). [DOI] [PubMed] [Google Scholar]
- 8.Herzer M, Denson LA, Baldassano RN & Hommel KA Patient and parent psychosocial factors associated with health-related quality of life in pediatric inflammatory bowel disease. Journal of pediatric gastroenterology and nutrition 52, 295–299, doi: 10.1097/MPG.0b013e3181f5714e (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devanarayana NM et al. Abdominal pain-predominant functional gastrointestinal diseases in children and adolescents: prevalence, symptomatology, and association with emotional stress. Journal of pediatric gastroenterology and nutrition 53, 659–665, doi: 10.1097/MPG.0b013e3182296033 (2011). [DOI] [PubMed] [Google Scholar]
- 10.Devanarayana NM, Rajindrajith S & Benninga MA Op-20 the Association between Adverse Life Events and Abdominal Pain-Predominant Functional Gastrointestinal Disorders. Journal of pediatric gastroenterology and nutrition 61, 517–518, doi: 10.1097/01.mpg.0000472224.86421.3d (2015). [DOI] [PubMed] [Google Scholar]
- 11.Kostic AD, Xavier RJ & Gevers D The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 146, 1489–1499, doi: 10.1053/j.gastro.2014.02.009 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bailey MT et al. Exposure to a social stressor alters the structure of the intestinal microbiota: implications for stressor-induced immunomodulation. Brain, behavior, and immunity 25, 397–407, doi: 10.1016/j.bbi.2010.10.023 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galley JD et al. Exposure to a social stressor disrupts the community structure of the colonic mucosa-associated microbiota. BMC microbiology 14, 189, doi: 10.1186/1471-2180-14-189 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bailey MT et al. Stressor exposure disrupts commensal microbial populations in the intestines and leads to increased colonization by Citrobacter rodentium. Infection and immunity 78, 1509–1519, doi: 10.1128/IAI.00862-09 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bailey MT, Lubach GR & Coe CL Prenatal stress alters bacterial colonization of the gut in infant monkeys. Journal of pediatric gastroenterology and nutrition 38, 414–421 (2004). [DOI] [PubMed] [Google Scholar]
- 16.Hanke ML, Powell ND, Stiner LM, Bailey MT & Sheridan JF Beta adrenergic blockade decreases the immunomodulatory effects of social disruption stress. Brain, behavior, and immunity 26, 1150–1159, doi: 10.1016/j.bbi.2012.07.011 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Galley JD, Parry NM, Ahmer BM, Fox JG & Bailey MT The commensal microbiota exacerbate infectious colitis in stressor-exposed mice. Brain, behavior, and immunity, doi: 10.1016/j.bbi.2016.09.010 (2016). [DOI] [PMC free article] [PubMed]
- 18.Mackos AR et al. Social stress-enhanced severity of Citrobacter rodentium-induced colitis is CCL2-dependent and attenuated by probiotic Lactobacillus reuteri. Mucosal immunology, doi: 10.1038/mi.2015.81 (2015). [DOI] [PMC free article] [PubMed]
- 19.Cummings JH, Pomare EW, Branch WJ, Naylor CP & Macfarlane GT Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28, 1221–1227 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith PM et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 341, 569–573, doi: 10.1126/science.1241165 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thangaraju M et al. GPR109A is a G-protein-coupled receptor for the bacterial fermentation product butyrate and functions as a tumor suppressor in colon. Cancer research 69, 2826–2832, doi: 10.1158/0008-5472.CAN-08-4466 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim MH, Kang SG, Park JH, Yanagisawa M & Kim CH Short-chain fatty acids activate GPR41 and GPR43 on intestinal epithelial cells to promote inflammatory responses in mice. Gastroenterology 145, 396–406 e391–310, doi: 10.1053/j.gastro.2013.04.056 (2013). [DOI] [PubMed] [Google Scholar]
- 23.Avitsur R, Stark JL & Sheridan JF Social stress induces glucocorticoid resistance in subordinate animals. Hormones and behavior 39, 247–257, doi: 10.1006/hbeh.2001.1653 (2001). [DOI] [PubMed] [Google Scholar]
- 24.Borenshtein D, Nambiar PR, Groff EB, Fox JG & Schauer DB Development of fatal colitis in FVB mice infected with Citrobacter rodentium. Infection and immunity 75, 3271–3281, doi: 10.1128/IAI.01810-06 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rath HC et al. Normal luminal bacteria, especially Bacteroides species, mediate chronic colitis, gastritis, and arthritis in HLA-B27/human beta2 microglobulin transgenic rats. The Journal of clinical investigation 98, 945–953, doi: 10.1172/JCI118878 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mackos AR, Eubank TD, Parry NM & Bailey MT Probiotic Lactobacillus reuteri attenuates the stressor-enhanced severity of Citrobacter rodentium infection. Infection and immunity 81, 3253–3263, doi: 10.1128/IAI.00278-13 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garcia-Villalba R et al. Alternative method for gas chromatography-mass spectrometry analysis of short-chain fatty acids in faecal samples. Journal of separation science 35, 1906–1913, doi: 10.1002/jssc.201101121 (2012). [DOI] [PubMed] [Google Scholar]
- 28.Bokulich NA et al. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nature methods 10, 57–59, doi: 10.1038/nmeth.2276 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCafferty J et al. Stochastic changes over time and not founder effects drive cage effects in microbial community assembly in a mouse model. The ISME journal 7, 2116–2125, doi: 10.1038/ismej.2013.106 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arthur JC et al. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nature communications 5, 4724, doi: 10.1038/ncomms5724 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Galley JD, Mackos AR, Varaljay VA & Bailey MT Stressor exposure has prolonged effects on colonic microbial community structure in Citrobacter rodentium-challenged mice. Scientific reports 7, 45012, doi: 10.1038/srep45012 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sartor RB Genetics and environmental interactions shape the intestinal microbiome to promote inflammatory bowel disease versus mucosal homeostasis. Gastroenterology 139, 1816–1819, doi: 10.1053/j.gastro.2010.10.036 (2010). [DOI] [PubMed] [Google Scholar]
- 33.Zitomersky NL et al. Characterization of adherent bacteroidales from intestinal biopsies of children and young adults with inflammatory bowel disease. PloS one 8, e63686, doi: 10.1371/journal.pone.0063686 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X et al. SCID/NCr mice naturally infected with Helicobacter hepaticus develop progressive hepatitis, proliferative typhlitis, and colitis. Infection and immunity 66, 5477–5484 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Doherty NC, Tobias A, Watson S & Atherton JC The effect of the human gut-signalling hormone, norepinephrine, on the growth of the gastric pathogen Helicobacter pylori. Helicobacter 14, 223–230, doi: 10.1111/j.1523-5378.2009.00682.x (2009). [DOI] [PubMed] [Google Scholar]
- 36.Maltz RM et al. Prolonged restraint stressor exposure in outbred CD-1 mice impacts microbiota, colonic inflammation, and short chain fatty acids. PloS one 13, e0196961, doi: 10.1371/journal.pone.0196961 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Q et al. Accelerated dysbiosis of gut microbiota during aggravation of DSS-induced colitis by a butyrate-producing bacterium. Scientific reports 6, 27572, doi: 10.1038/srep27572 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eeckhaut V et al. Butyricicoccus pullicaecorum in inflammatory bowel disease. Gut 62, 1745–1752, doi: 10.1136/gutjnl-2012-303611 (2013). [DOI] [PubMed] [Google Scholar]
- 39.Sakamoto M et al. Butyricimonas synergistica gen. nov., sp. nov. and Butyricimonas virosa sp. nov., butyric acid-producing bacteria in the family ‘Porphyromonadaceae’ isolated from rat faeces. International journal of systematic and evolutionary microbiology 59, 1748–1753, doi: 10.1099/ijs.0.007674-0 (2009). [DOI] [PubMed] [Google Scholar]
- 40.Clarke JM et al. Butyrate esterified to starch is released in the human gastrointestinal tract. The American journal of clinical nutrition 94, 1276–1283, doi: 10.3945/ajcn.111.017228 (2011). [DOI] [PubMed] [Google Scholar]
- 41.Zhong Y, Nyman M & Fak F Modulation of gut microbiota in rats fed high-fat diets by processing whole-grain barley to barley malt. Molecular nutrition & food research 59, 2066–2076, doi: 10.1002/mnfr.201500187 (2015). [DOI] [PubMed] [Google Scholar]
- 42.Imai K, Yamada K, Tamura M, Ochiai K & Okamoto T Reactivation of latent HIV-1 by a wide variety of butyric acid-producing bacteria. Cellular and molecular life sciences : CMLS 69, 2583–2592, doi: 10.1007/s00018-012-0936-2 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maslowski KM et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461, 1282–1286, doi: 10.1038/nature08530 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Senagore AJ, MacKeigan JM, Scheider M & Ebrom JS Short-chain fatty acid enemas: a cost-effective alternative in the treatment of nonspecific proctosigmoiditis. Diseases of the colon and rectum 35, 923–927 (1992). [DOI] [PubMed] [Google Scholar]
- 45.Kiely EM, Ajayi NA, Wheeler RA & Malone M Diversion procto-colitis: response to treatment with short-chain fatty acids. Journal of pediatric surgery 36, 1514–1517, doi: 10.1053/jpsu.2001.27034 (2001). [DOI] [PubMed] [Google Scholar]
- 46.Mishiro T et al. Butyric acid attenuates intestinal inflammation in murine DSS-induced colitis model via milk fat globule-EGF factor 8. Laboratory investigation; a journal of technical methods and pathology 93, 834–843, doi: 10.1038/labinvest.2013.70 (2013). [DOI] [PubMed] [Google Scholar]
- 47.Macia L et al. Metabolite-sensing receptors GPR43 and GPR109A facilitate dietary fibre-induced gut homeostasis through regulation of the inflammasome. Nature communications 6, 6734, doi: 10.1038/ncomms7734 (2015). [DOI] [PubMed] [Google Scholar]
- 48.Su Y, Chen X, Liu M & Guo X Effect of three lactobacilli with strain-specific activities on the growth performance, faecal microbiota and ileum mucosa proteomics of piglets. Journal of animal science and biotechnology 8, 52, doi: 10.1186/s40104-017-0183-3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.