Summary
Evolution of gene-regulatory sequences is considered the primary driver of morphological variation [1, 2, 3]. In animals, the diversity of body plans between distantly related phyla is due to the differential expression patterns of conserved “toolkit” genes [4]. In plants, variation in expression domains similarly underlie most of the reported diversity of organ shape both in natural evolution and in the domestication of crops [5, 6, 7, 8, 9]. The heart-shaped fruit from members of the Capsella genus is a morphological novelty that has evolved after Capsella diverged from Arabidopsis ∼8 mya [10]. Comparative studies of fruit growth in Capsella and Arabidopsis revealed that the difference in shape is caused by local control of anisotropic growth [11]. Here, we show that sequence variation in regulatory domains of the fruit-tissue identity gene, INDEHISCENT (IND), is responsible for expansion of its expression domain in the heart-shaped fruits from Capsella rubella. We demonstrate that expression of this CrIND gene in the apical part of the valves in Capsella contributes to the heart-shaped appearance. While studies on morphological diversity have revealed the importance of cis-regulatory sequence evolution, few examples exist where the downstream effects of such variation have been characterized in detail. We describe here how CrIND exerts its function on Capsella fruit shape by binding sequence elements of auxin biosynthesis genes to activate their expression and ensure auxin accumulation into highly localized maxima in the fruit valves. Thus, our data provide a direct link between changes in expression pattern and altered hormone homeostasis in the evolution of morphological novelty.
Keywords: morphological diversity, Brassicaceae, Capsella rubella, gene regulation, INDEHISCENT, localized auxin biosynthesis, fruit shape
Highlights
-
•
Fruit-shape defect observed in crind mutant is rescued by exogenous auxin application
-
•
Auxin dynamics are perturbed in crind mutant
-
•
CrIND directly controls expression of auxin-biosynthesis genes in fruit valves
-
•
CrIND regulatory sequences contribute to the morphological novelty of Capsella fruits
Dong et al. demonstrate that diversification of INDEHISCENT expression in the Capsella genus has contributed to morphological changes of the heart-shaped fruit compared to the cylindrical Arabidopsis fruit and that INDEHISCENT mediates its effect via localized activation of auxin-biosynthesis genes.
Results and Discussion
The INDEHISCENT Gene Controls Fruit Shape in Capsella rubella
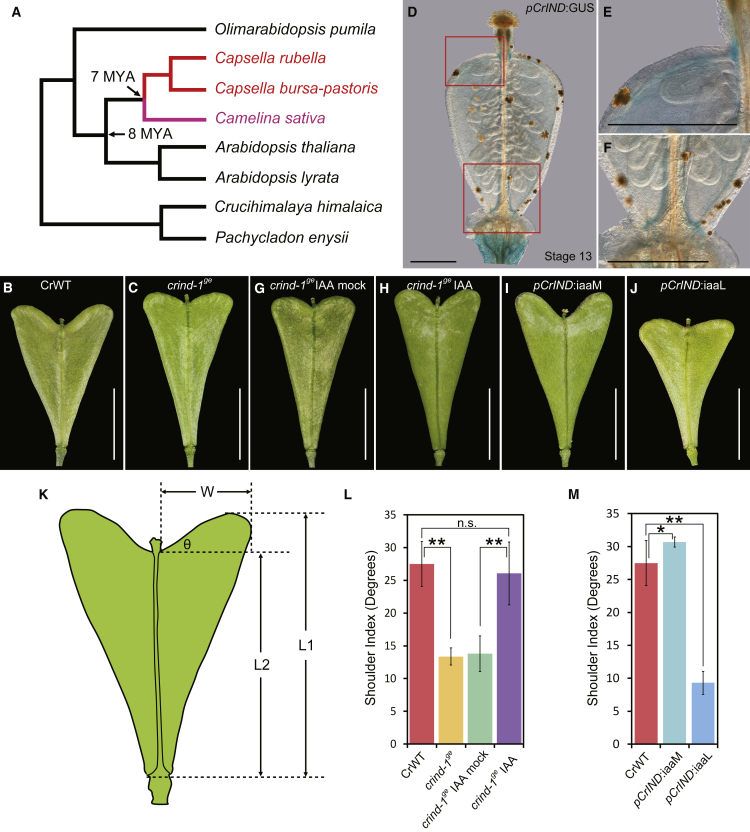
The Brassicaceae family is characterized by a remarkable diversity in fruit shape between different genera [12]. Even so, the overall tissue composition of Brassicaceae fruits is highly conserved with valves enclosing the seeds, a replum in the center of the fruit, and valve margins that form the border between the valves and the replum [13]. The overall fruit-shape diversity is primarily due to variation in valve morphology. For example, fruits from the Capsella genus develop valves that are extended at the apical end, giving them a heart-shaped appearance [11, 12] (Figure 1A and 1B). This shape is unique to fruits from Capsella species; fruits from the closest relative, Camelina, are spherical, while Arabidopsis produce cylindrically shaped fruits [10, 14] (Figure 1A). Comparative studies between the development of fruits from Capsella and other Brassicaceae therefore provides an excellent system to study the molecular mechanisms underlying morphological changes [15].
Figure 1.
Effect of CrIND and Manipulation of Auxin Levels on Capsella Fruit Shape
(A) A simplified phylogeny of Capsella and its close relatives according to [10]. The heart-shaped fruit from Capsella (red) shares a common ancestor of Camelina (magenta), which develops spherical siliques. The black branches in the phylogeny represent the species with cylindrical fruit.
(B and C) Fruit morphology of CrWT (B) and crind-1ge (C) at developmental stage 17.
(D–F) Expression pattern of CrIND during fruit development with pCrIND:GUS line. (E) and (F) show enlarged pictures of the regions outlined with red box in (D) with valve expression (E) and valve margin expression (F), respectively.
(G and H) Fruit morphology of crind-1ge after mock (G) or IAA (H) treatment at stage 17.
(I and J) Fruit morphology of pCrIND:iaaM (I) and pCrIND:iaaL (J) at stage 17.
(K) Schematic drawing to illustrate the shoulder index calculation.
(L) Shoulder index measurements of fruits from CrWT, crind-1ge, and crind-1ge± IAA treatment. Error bars represent SD of 30 individual fruits.
(M) Shoulder index measurements of fruits from WT, pCrIND:iaaM, and pCrIND:iaaM plants. Error bars represent SD of 30 individual fruits.
Scale bars represent 5 mm for (B), (C), and (G)–(J) and 100 μm for (D)–(F). ∗∗p < 0.01 (Student’s t test) in (L) and (M).
See also Figure S1.
In a previous study, we demonstrated that the master regulator of valve development, FRUITFULL (FUL), has conserved functions in both Arabidopsis and Capsella based on highly similar ful loss-of-function phenotypes [11]. In a continued effort to test for diversity of function between known key regulators of fruit development in the Brassicaceae family, we created a knockout line of the Capsella rubella IND gene (CrIND) using CRISPR/Cas9, leading to a 107-bp deletion within the coding region (Figures 1B and S1A). This mutant allele was named crind-1ge, where “ge” stands for “genome editing” according to guidelines recently published for Marchantia gene nomenclature [16]. In agreement with the function of IND in both Arabidopsis and Brassica valve-margin formation [17, 18], the crind-1ge mutant fruits do not form valve margins and are as a consequence completely indehiscent (Figures S1B and S1C). Additionally, mature crind-1ge fruits exhibit a reduction in the development of the shoulders (measured as a shoulder index, Figure 1K) compared to wild-type, indicating that CrIND has a role in Capsella fruit-shape formation (Figure 1A, 1B, and 1L). In Arabidopsis, the IND gene (AtIND) is specifically expressed in the valve margins after fertilization of the ovary [17, 19]. We tested whether the role of CrIND in valve-shape formation could be due to a change in expression pattern compared to AtIND in Arabidopsis or could be a result of differential growth caused by loss of valve-margin tissue. In support of the former, we detected expression of CrIND in Capsella valves by quantitative RT-PCR (qRT-PCR) (Figure S1D). To examine more specifically the CrIND expression pattern in the Capsella valves, we constructed a pCrIND:GUS reporter and found GUS signal in the apical parts in addition to the signal in the valve margin (Figures 1D–1F and S1E–S1H). These data suggest that CrIND affects fruit-shape formation cell autonomously due to an expansion of its expression domain in the developing shoulders.
The function of IND in both valve-margin specification and earlier during gynoecium development has been closely associated with auxin dynamics [19, 20]. Therefore, we investigated whether a link to auxin could also be established for CrIND in fruit-shape formation. We found that application of exogenous auxin (indole-3-acetic acid or IAA) to the apex of crind-1ge mutant fruit rescued the growth defect observed in the valves (Figure 1G, 1H, and 1L). Moreover, expression of a bacterial auxin biosynthesis gene, iaaM [21], under the CrIND promoter in a wild-type background led to shoulders that were extended further than in wild-type (Figure 1I and 1M). In contrast, depleting free IAA in the same domain by expressing the iaaL gene [22] under control of the CrIND promoter significantly reduces the shoulder index of the heart-shaped fruits (Figures 1J and 1M).
CrIND Is Required to Maintain Auxin Homeostasis in Capsella Fruit Valves
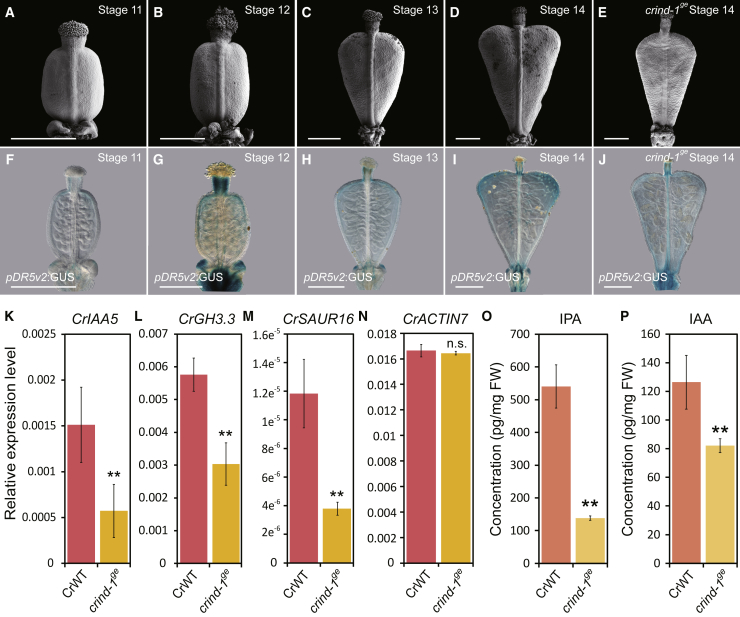
The auxin-signaling reporter pDR5:revGFP has previously been used to map the dynamics of auxin during Arabidopsis gynoecium development [20, 23]. It has been demonstrated that auxin is required to mediate a symmetry transition in the apical style through its tightly controlled accumulation in specific auxin maxima [20]. Here, we used two modified DR5 reporters, pDR5v2:GFP and pDR5v2:GUS [24] transformed into C. rubella. The pDR5v2:GFP reporter mimicked the pattern in the style observed in Arabidopsis during the early developmental stages (Figure S2A-H) [20]. In contrast to Arabidopsis, DR5 signal was also observed in the vascular tissue of the Capsella valves at developmental stage 10 (stages defined in [13, 25]) (Figure S2H). At later stages, when the Capsella gynoecium develops from an oblate spheroid (flat disc) into an emerging heart shape [11] (Figures 2A–2D), the DR5v2 reporter is clearly observed in the apical part of the valves with very specific maxima in the shoulders (Figures 2F–2I). Interestingly, this expression pattern is reduced in stage-14 fruits of crind-1ge, when the defect in shoulder development has emerged (Figures 2E and 2J). The reduction of auxin signaling in the crind-1ge fruit shoulders correlated with qRT-PCR data showing that expression of three different auxin-responsive genes is significantly reduced in fruit shoulders from crind-1ge compared to wild-type (Figures 2K–2M). In contrast, expression of the house-keeping gene, CrACTIN7, was not significantly different (Figure 2N). In line with this observation, direct measurements of both the predominant natural auxin, indole-3-acetic acid (IAA), and its precursor, indole-3-pyruvate (IPA), show a significant reduction in the shoulders of crind-1ge mutant (Figures 2O and 2P). Together, these results show that CrIND mediates its function on Capsella fruit shape by local control of auxin dynamics in the shoulders of the valves.
Figure 2.
Auxin Dynamics during Capsella Fruit Development in Wild-Type and the crind Mutant
(A–E) SEM images of fruits from CrWT at developmental stages 11 (A), 12 (B), 13 (C), and 14 (D) and from crind-1ge at stage 14 (E).
(F–J) Auxin signaling visualized by pDR5v2:GUS in CrWT fruit of developmental stages 11 (F), 12 (G), 13 (H), and 14 (I) and in the crind-1ge mutant at stage 14 (J).
(K–N) Expression analysis by qRT-PCR of CrIAA5 (K), CrGH3.3 (L), CrSAUR16 (M), and CrACTIN7 (N) in the fruit shoulders of WT and crind-1ge at stage 14. Error bars represent SD of three biological replicates.
(O and P) Measurements of IPA (O) and IAA (P) in fruit shoulders of WT and crind-1ge stage-14 fruits. Error bars represent SD of three biological replicates.
Scale bars represent 150 μm (A–J). ∗∗p < 0.01 (Student’s t test) in (K–P).
See also Figure S2.
CrIND Directly Regulates Auxin Biosynthesis Genes to Control Capsella Fruit Shape
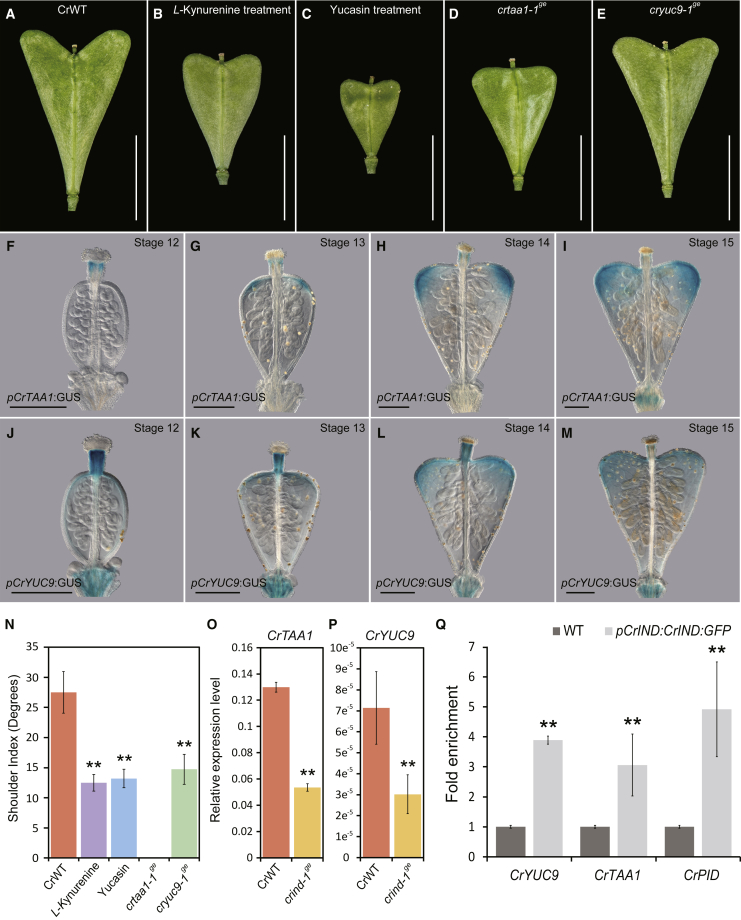
The rescue of the crind-1ge valve-shape phenotype by application of auxin (Figures 1G, 1H, and 1L) combined with the reduced auxin levels measured in the crind-1ge mutant (Figures 2O and 2P) suggest that CrIND is involved in controlling auxin biosynthesis. We first tested if inhibition of auxin biosynthesis affects the development of wild-type fruits. Therefore, we applied inhibitors of the two steps leading to the synthesis of IAA from tryptophan. The first step leading to IPA is catalyzed by enzymes belonging to the TAA1/TAR family [26] and can be inhibited by L-Kynurenine [27], whereas the second step from IPA to IAA is mediated by members of the YUCCA family [28], which are inhibited by Yucasin [29]. Treatment with L-Kynurenine and Yucasin led to fruits with strongly reduced shoulder indices (Figures 3A–3C and 3N). We next carried out a comparative expression analysis of members of the TAA1/TAR and YUCCA gene families in Capsella to screen for genes that are more highly expressed in the shoulders relative to the base (Figure S3A). For the three members of the TAA1/TAR family, only TAA1 showed this pattern (Figures S3B–S3D), and a pCrTAA1:GUS reporter line exhibited very specific expression in the developing shoulders (Figures 3F–3I), suggesting highly localized auxin biosynthesis. The YUCCA family consists of 11 genes that can be divided into five clades based on their sequence identity (Figure S3E). We carried out qRT-PCR for representatives of each clade—namely, CrYUC2, CrYUC4, CrYUC7, CrYUC9, and CrYUC10. Of these, expression of CrYUC2 and CrYUC9 was significantly higher in the shoulders compared to the base of the fruit (Figures S3F–S3J). GUS-reporter lines were developed for CrYUC2 and CrYUC9; however, a signal could only be detected for the pCrYUC9:GUS reporter. Similar to CrTAA1, pCrYUC9:GUS exhibited specific expression in the shoulders, compatible with a role in mediating auxin biosynthesis at these sites (Figures 3J–3M). Therefore, expression of both CrTAA1 and CrYUC9 overlap with expression of the DR5v2 reporter line (Figures 2G–2I). In agreement with the recognized importance for local auxin biosynthesis throughout plant development [30], these data suggest a specific role for the TAA/YUC auxin-biosynthesis pathway in generating a highly specific auxin maximum at the valve apices. To test if CrTAA1 and CrYUC9 are required for fruit-shape formation in Capsella, we generated knockout lines using CRISPR/Cas9, leading to a 104-bp deletion in Exon II of CrTAA1 (crtaa1-1ge) and a 1-bp deletion in Exon I of CrYUC9 (cryuc9-1ge) (Figure S3K). Both mutations resulted in reduced valve growth (Figures 3D, 3E, and 3N) similar to the treatments with the auxin-biosynthesis inhibitors. However, the cryuc9-1ge mutant fruits were less severely affected in shoulder growth compared to crtaa1-1ge, which may be due to residual activity of CrYUC2 in the cryuc9-1ge background.
Figure 3.
CrIND-Induced Expression of Auxin Biosynthesis Gene
(A–C) Whole-mount images showing morphologies of CrWT fruits 8 DPA of mock-treated (A), treated with L-Kynurenine (B) and Yucasin (C).
(D and E) Whole-mount images of crtaa1-1ge and cryuc9-1ge.
(F–M) Expression pattern of CrTAA1 and CrYUC9 shown by GUS staining of the pCrTAA1:GUS (F–I) and pCrYUC9:GUS (J–M) reporter lines at developmental stages 12 (F and J), 13 (G and K), 14 (H and L) and 15 (I and M).
(N) Shoulder indices of fruits from CrWT, L-Kynurenine treatment, Yucasin-treatment, crtaa1-1ge, and cryuc9-1ge fruits. N.D. indicate not determinable. Error bars represent SD of 30 individual fruits.
(O and P) Expression analysis of CrTAA1 (N) and CrYUC9 (O) in the fruit shoulders of CrWT and crind-1ge stage-14 fruits. Error bars represent SD of three biological replicates.
(Q) Chromatin Immuno-Precipitation (ChIP) analysis of CrIND associated with the CrTAA1 and CrYUC9 promoter. The CrPINOID (CrPID) was used as a positive control, the potential E boxes bound by CrIND are shown below each gene. Error bars represent SD of three biological replicates.
Scale bars represent 5 mm (A–E) and 150 μm (F–M). ∗∗p < 0.01 (Student’s t test) in (N–R).
See also Figure S3.
To test whether CrIND regulates CrTAA1 and CrYUC9, we performed qRT-PCR using RNA extracted from wild-type and crind-1ge mutant fruits and found significantly reduced levels of both CrTAA1 and CrYUC9 mRNA in the mutant (Figures 3O and 3P). FUL in Arabidopsis (AtFUL) is a repressor of AtIND, excluding AtIND expression from the valves and restricting it to the valve margins. As a consequence, the AtIND expression level is elevated in atful mutant fruits [17, 31]. Similarly, in fruits from the Capsella crful-1 mutant [11], CrIND expression was upregulated (Figure S3L). In agreement with CrIND positively regulating the expression of CrTAA1 and CrYUC9, we found that these genes were upregulated in crful-1, while this effect was abolished in the crful-1 crind-1ge double mutant (Figures S3M and S3N).
The crful-1 mutant fruits have a severe growth defect similar to that reported for heegeri, which is a natural variant of the tetraploid C. bursa-pastoris [11, 32]. This clearly shows that other factors than CrIND are involved in determining the heart shape. As in Arabidopsis, loss of IND leads to a significant rescue of the growth defects of the crful-1 mutant. However, this is not accompanied by the development of shoulders, which supports that CrIND is required for the local induction of CrTAA1 and CrYUC9 expression (Figure S3O).
In previous studies, we have found that IND directly regulates genes that affect auxin dynamics such as the protein kinase genes PINOID (PID) and WAG2 by binding to a variant “E-box” (CACGCG) in the regulatory regions [19, 33]. An analysis of the promoter regions of CrTAA1 and CrYUC9 identified potential CrIND recognition sites (CACGAG for CrTAA1 and CGCGTC for CrYUC9). Using crind-1ge plants complemented with pCrIND:CrIND:GFP (Figure 4G), we performed chromatin immunoprecipitation (ChIP) on fruit tissue and showed that CrIND directly interacts with promoter regions of both CrTAA1 and CrYUC9 (Figure 3Q). The specific binding of CrIND protein to the identified elements was further tested by yeast one-hybrid (Figure S3P). Taken together, these results suggest that rather than initiating shoulder formation per se, CrIND promotes growth after shoulder initiation by inducing localized expression of auxin biosynthesis genes. We hypothesize that establishment of localized auxin maxima at the shoulder tips provides polarity and thus stimulates anisotropic growth in their direction. This is similar to the effects of auxin maxima observed in other developmental contexts such as lateral root growth and gynoecium development [20, 23].
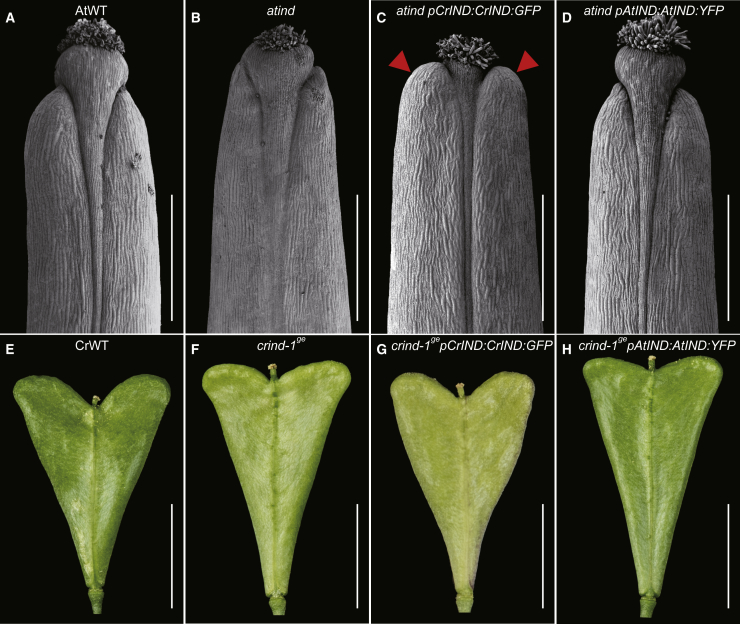
Figure 4.
Morphological Effects of AtIND and CrIND Expression
(A–D) SEM images of the apex of stage-17 fruits of AtWT (A), atind-2 (B), atind-2 pCrIND:CrIND:GFP (C), and atind-2 pAtIND:AtIND:YFP (D). The red triangles in (D) indicate the expanded apical growth of the valve tips.
(E–H) Fruit morphology at stage 17 of CrWT (E), crind-1ge (F), crind-1gepCrIND:CrIND:GFP (G), and crind-1gepAtIND:AtIND:YFP (H).
Scale bars represent 5 mm (E–H) and 400 μm (A–D).
See also Figure S4.
Regulatory Divergence in the IND Genes of Capsella and Arabidopsis Contributes to the Morphological Differences in Fruit Shape
The heart-shaped fruit is unique to the Capsella genus in the Brassicaceae family and evolved after Camelina and Capsella diverged ∼7 mya [10] (Figure 1A). In both animals and plants, morphological novelties most often arise from mutations in regulatory sequences that alter gene expression patterns rather than in protein-encoding regions [1, 2, 3, 4, 34, 35]. The expanded expression of CrIND in the fruit valves of Capsella strongly suggests that the CrIND promoter has diverged from other Brassicaceae IND sequences. Previously, we found that the valve margin-specific expression of AtIND was governed by sequence contained in a 406-bp promoter element [18]. We extracted ∼2.1 kb regulatory sequences of nine Brassicaceae IND genes, including four from the Capsella genus. A phylogenetic shadowing analysis was carried out using the mVISTA software [36] to assess regional conservation across species. In this analysis, pairwise comparison of the CrIND promoter sequence against IND sequences from the other eight species revealed a highly conserved region, which includes the region required for valve-margin expression (Figure S4A). Interestingly, this analysis also revealed large regions of the promoter where the Capsella sequences are highly conserved but diverge from other species. Conceivably, these Capsella-specific regions contain elements that have allowed for the expanded IND expression in Capsella (Figure S4A).
While the expanded expression of CrIND in the valves compared to IND in Arabidopsis could be due to changes in the regulatory sequence of the CrIND gene itself, it is also possible that CrIND expression in the valve apices is caused by differential expression of an upstream regulator. To distinguish between these two possibilities, we first transformed the atind-2 mutant from Arabidopsis with a pCrIND:CrIND:GFP construct. Resulting transgenic lines were fully dehiscent, demonstrating that the pCrIND:CrIND:GFP gene complemented the indehiscence phenotype of the atind-2 mutant similarly to the effect of the pAtIND:AtIND:YFP construct (Figures 4A–4D and S4B–S4E). However, while the atind-2 pAtIND:AtIND:YFP fruits had a wild-type shape, fruits from atind-2 pCrIND:CrIND:GFP plants have abnormal apices where valve growth expands above the style (Figures 4C and 4D). Conversely, the pAtIND:AtIND:YFP construct only restored the dehiscence defect of the crind-1ge mutant, but not the shape change (Figures 4E–4H and S4F–S4I). This is supported by the expression pattern of the pAtIND:AtIND:YFP, which was undetectable in the valves but observed in valve margins (Figure S4J). In contrast, the pCrIND:CrIND:GFP construct complemented both defects (Figures 4G and S4H). These results show that expansion of CrIND expression into the valves in Capsella is due to changes in cis-regulatory sequences in the CrIND gene itself and that this has contributed to the change in fruit shape between these two genera. It is interesting to speculate that the modified expression pattern of CrIND may have led to novel genetic interactions such as described for KNOX genes in leaf development [9], thereby facilitating the recognition of the auxin biosynthesis target genes in the valves.
Concluding Remarks
In animals, changes in cis-regulatory elements of otherwise conserved “toolkit” genes is the primary driver of morphological evolution [2, 3, 37]. A similar pattern is emerging in plants, where modifications of regulatory sequences have been revealed as the major determinant of developmental variation both during domestication and natural evolution [5, 6, 7]. Even so, examples have also been reported where changes in protein-coding sequence are either fully or partly responsible for the evolution of morphological diversity [38, 39]. The work described here directly links changes in expression domain of a fruit-tissue-identity gene to effects on hormone homeostasis resulting in a morphological novelty. Given the stunning variation in fruit shape among members of the Brassicaceae family, it is possible that direct effects of gene-expression diversity on hormone dynamics is a common driver in the evolution of fruit-shape diversity.
STAR★Methods
Key Resources Table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-GFP monoclonal antibody | Roche | 11814460001 |
| Bacterial Strains | ||
| DH5-alpha competent E. coli | New England Biolabs | C29871 |
| Agrobacterium tumefaciens strain LBA4404 | N/A | N/A |
| Biological Samples | ||
| Capsella rubella (22.5) | This paper | N/A |
| Arabidopsis thaliana (Col-0) | This paper | N/A |
| atind-2 | [17] | N/A |
| atind-2 pAtIND:AtIND:YFP | [40] | N/A |
| crind-1ge | This paper | N/A |
| crful-1 | [11] | N/A |
| crful-1 crind-1ge | This paper | N/A |
| crtaa1-1ge | This paper | N/A |
| cryuc9-1ge | This paper | N/A |
| Chemicals Peptides, and Recombinant Proteins | ||
| Phusion High-Fidelity DNA polymerase | New England Biolabs | M0530L |
| DnaseI | QIAGEN | 79254 |
| In-Fusion Cloning Recombinase | Clontech | 638909 |
| Proteinase K | Invitrogen | 59895 |
| L-Kynurenine | Sigma-Aldrich | K8625 |
| Yucasin | Carbosynth | FC1222381801 |
| Indole-3-acetic acid (IAA) | Sigma-Aldrich | I5148 |
| Chlorohydrate | Sigma-Aldrich | 15307 |
| DMSO | Sigma-Aldrich | D8418 |
| Formaldehyde | Sigma-Aldrich | F8775 |
| K3Fe(CN)6 | Sigma-Aldrich | P8131 |
| K4Fe(CN)6 | Sigma-Aldrich | P9387 |
| Triton X-100 | Sigma-Aldrich | T8787 |
| Cysteamine | Sigma-Aldrich | M9768 |
| X-gluc | MELFORD | MB1121 |
| Oligonucleotides | ||
| A list of oligonucleotides is given in Methods S1 | N/A | |
| Other | ||
| QIAprep Spin MiniPrep Kit | QIAGEN | 27104 |
| DNeasy Plant Mini Kit | QIAGEN | 69104 |
| QIAquick PCR Purification Kit | QIAGEN | 28104 |
| RNeasy Plant Mini Kit | QIAGEN | 74104 |
| Pierce Protein G Magnetic Beads | ThermoFisher | 19958500 |
| SuperScript™ IV First-Strand Synthesis System | ThermoFisher | 18091050 |
| SYBR Green JumpStart Taq ReadyMix | Sigma-Aldrich | S4438 |
| Oasis HLB 1 cc Vac Cartridge | Waters | WAT094225 |
| Recombinant DNA | ||
| pDR5v2:GUS | This Paper | N/A |
| pDR5v2:GFP | This Paper | N/A |
| pCrIND:GUS | This Paper | N/A |
| pCrTAA1:GUS | This Paper | N/A |
| pCrYUC9:GUS | This Paper | N/A |
| pCrIND:iaaM | This Paper | N/A |
| pCrIND:iaaL | This Paper | N/A |
| pCrIND:CrIND:GFP | This Paper | N/A |
| pAtIND:AtIND:YFP | [40] | N/A |
| Software and Algorithms | ||
| ImageJ | [41] | https://imagej.nih.gov/ij/ |
| VISTA | [36] | http://genome.lbl.gov/vista |
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Lars Østergaard (lars.ostergaard@jic.ac.uk).
Experimental Model and Subject Details
Capsella rubella Cr22.5 and Arabidopsis thaliana (Col-0) were used in all experiments of this study. For Capsella rubella, the seeds were germinated on MS medium containing 10 μM Gibberellin at 22°C. 10-day-old seedlings were then transplanted into a controlled environment room at 22°C, 16 hr light/8 hr dark conditions. For Arabidopsis thaliana, the seeds were germinated on MS medium and 7 days old seedlings were then transplanted to soil and grown in the glasshouse at 22°C, 16 hr light/8 hr dark conditions.
Method Details
Plasmids construction and plant transformation
For the construction of the promoter:GUS reporter plasmids of CrIND, CrTAA1 and CrYUC9, ∼2.0kb promoter was isolated by PCR on genomic DNA and inserted upstream of the GUS gene of pCambia1301 vectors. For construction of the pDR5v2:GFP/GUS plasmid, a 207-bp promoter fragment containing eight repeats of the auxin response element and 47-bp CaMV 35S minimal promoter [24] was inserted upstream of the GFP and GUS reporter genes of pCambia1301 and pCambia1302 vectors, respectively. For construction of the pCrIND:CrIND:GFP plasmid, a ∼2.6kb genomic fragment of CrIND containing the ∼2.0kb promoter and full length coding sequence of CrIND was isolated and fused in-frame with the GFP coding sequence of pCambia1302 vector. For construction of the pCrIND:iaaL/iaaM plasmids, ∼2.0kb CrIND promoter was isolated and fused with the full length of the iaaL/iaaM coding sequence and then inserted into the pCambia1301 vector. For construction of the RNA-guided genome editing plasmids, DNA sequences encoding gRNAs adjacent to the PAM sequences (NGG) were designed to target two specific sites in the exons of CrIND, CrTAA1 and CrYUC9. The gRNAs were synthesized as oligonucleotides with Golden-gate cloning adapters and were then inserted downstream of U6 promoters. The resulting gRNA plasmids were then recombined with pRPS5a:Cas9z:E9t and Fast-Red selection marker using golden-gate cloning methods to produce the binary vectors. All vectors were verified by sequencing and introduced into Agrobacterium tumefaciens strain LBA4404 by electroporation. Primers used in the construction of the vectors are listed in Methods S1.
Transformation of Arabidopsis and Capsella followed the floral dipping method with minor modifications in Capsella. Specifically, the Agrobacterium was cultured to 2.0 (OD600) and resuspended with 5% sucrose solution plus 0.02% Silwet. Capsella seedlings with 10cm long inflorescences were subjected to the first round of dipping, after which, the plants were kept in the dark for 36 hours at 22°C. The floral dipping process was repeated twice in five-day intervals. For each construct, at least 10 independent transformants were obtained for further analysis.
Phenotyping and Microscopy
The mature fruits of each genotype were collected and recorded photographically with Nikon D610 camera with a 105mm prime lens.
To quantify the shoulder phenotype, three parameters were measured: W (denotes the half width of the fruits), L1 (denotes the length of the fruit from the fruit shoulder tips to the fruit base) and L2 (denotes the length of the fruit from the style base to the fruit base). The angle of the shoulders was calculated with the anti-trigonometric function θ = Arctan((L1-L2)/W).
For Scanning Electron Microscopy (SEM), the young inflorescences and mature fruits of each genotype were fixed in FAA and infiltrated under vacuum. Gynoecia from distinct developmental stages were dissected with a needle in 70% ethanol under a light microscope. The materials were critically-point dried in CO2 and spotter-coated with gold. The samples were subsequently examined using a Zeiss Supra 55VP field Scanning Electron Microscope with an acceleration voltage of 3.0 kV.
Confocal microscopy was performed on a Leica SP5 laser scanning microscope equipped with an Argon Krypton laser (Leica Microsystems). The 488-nm excitation line of an argon ion laser was used to excite GFP, the 514-nm excitation line of an argon ion laser was used to excite YFP. GFP/YFP emission spectra were collected between 497 and 551 nm. For the top views of the gynoecium, the samples were dissected and placed vertically on a slide, we used the X25/0.95 water dipping objective lens to visualize the GFP signal of the specimens. Images were processed in ImageJ software.
Chemical Treatment and Auxin Metabolite Quantification
The auxin biosynthesis inhibitors L-Kynurenine and Yucasin were dissolved in DMSO, the Indole-3-acetic acid (IAA) was dissolved in ethanol. For L-Kynurenine and Yucasin treatment, 100 μM working solutions were prepared with water and silwet (0.02%) and dipped onto the 10-cm inflorescences. For IAA application, 100 μM working solutions were prepared with water and silwet (0.02%) and applied specifically to the apical part of fruits from WT or crind-1ge Capsella plants using a needle. The control plants were mock-treated with the same concentration of either Dimethyl sulfoxide (DMSO) or ethanol used to dissolve the chemicals.
To quantify auxin metabolite levels in the fruit shoulders, WT and crind-1ge fruits were dissected under a light microscope. Extraction, purification and the LC-MS/MS analysis of endogenous IAA and specific IAA metabolites was carried out according to the method described previously [42]. Briefly, around 20 mg of frozen material per sample was homogenized and extracted in 1 mL of 50 mM sodium phosphate buffer containing 1% sodium diethyldithiocarbamate and a mixture of 13C6 or deuterium labeled internal standards. After centrifugation (14,000 RPM, 15 min, 4°C), the supernatant was divided in two aliquots, the first was derivatised by cysteamine (0.25 M, pH 8, 1h, room temperature, Sigma-Aldrich), the second one was immediately further processed as following. The pH of sample was adjusted to 2.5 by 1 M HCl and the sample was applied on a preconditioned solid-phase extraction column (Oasis HLB, 30 mg, 1 cc, Waters Inc., USA). After sample application, the column was rinsed with 2 mL 5% methanol. Compounds of interest were then eluted with 2 mL 80% methanol. The derivatised fraction was purified alike. Mass spectrometry quantification was performed by LC-MS/MS, using a 1290 Infinity Binary LC System coupled to a 6490 Triple Quad LC/MS System with Jet Stream and Dual Ion Funnel technologies (Agilent Technologies, USA).
RNA extraction and expression analysis
The fruit shoulders and basal fruit were sampled from stage-13 fruits of CrWT and crind-1ge, respectively. For the crful-1, crful-1 crind-1ge mutants, the whole stage-13 fruits were collected. Total RNA was isolated from the samples using the RNeasy Plant Mini Kit (QIAGEN). Next, 1 ug of total RNA was reverse transcribed into cDNA with the SuperScript IV First-Strand Synthesis System (ThermoFisher) according to the manufacturer’s instructions.
For real-time qPCR, gene specific primers were designed, and verified by PCR and sequencing. The efficiency of the primers (95% to 105%) was determined by creating a standard curve. The SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich) was used to perform real-time qPCR with ROX as a reference dye on a BioRad CFX96 Q-PCR System (BioRad). The CT value of each gene was determined by normalizing the fluorescence threshold. The relative expression level of the target gene was determined using the ratio = 2-ΔCT method, and CrUBQ10 was used as an internal control. Statistical analysis was done in Microsoft Excel.
For GUS histochemical assay, fruit samples were fixed in acetone for 20 min at −80°C, washed twice for 5 min in 100 mM sodium phosphate buffer, and processed in 100 mM sodium phosphate buffer containing 1mM K3Fe(CN)6, 1 mM K4Fe(CN)6 at room temperature for 30 min. The staining was incubated at 37°C in the X-Gluc solution for 6-8h. The X-Gluc solution contains 100 mM sodium phosphate buffer, 10 mM EDTA, 0.5 mM K3Fe(CN)6, 3 mM K4Fe(CN)6, 0.1% Triton X-100 and 1 mg/mL of β-glucoronidase substrate X-gluc (5-bromo-4-chloro-3-indolyl glucuronide, MELFORD) dissolved in DMSO. After staining, the reaction buffer was replaced with 70% ethanol until chlorophyll was completely washed out from the samples. Fruits were dissected, mounted in Chlorohydrate (Sigma) solution and analyzed using a Zeiss Axio Imager light microscope.
Chromatin immunoprecipitation and Yeast one-hybrid analysis
Stage-16 fruits from pCrIND:CrIND:GFP and WT plants were collected and fixed with 1% formaldehyde and immediately frozen in liquid nitrogen. Approximately 3.0 g of tissue was ground in liquid nitrogen and chromatin fragments were prepared after sonication. After sonication, a 1/20 sample was taken out as DNA Input. The remaining samples underwent immunoprecipitation. GFP tagged protein together with the associated DNAs were immunoprecipitated by using Pierce Protein G Magnetic Beads (ThermoFisher) coated with monoclonal anti-GFP antibody (Roche) according to the manufacturer’s instructions. Beads were washed two times with the immunoprecipitation buffer followed by two washes with TE buffer. Reverse crosslinking was done by boiling the beads at 65°C for 12 hours in presence of 10% SDS followed by Proteinase K treatment at 50°C for 1 hour. DNA was ethanol precipitated following phenol/chloroform extraction. qPCR was performed using SYBR Green JumpStart Taq ReadyMix (Sigma-Aldrich) on a BioRad CFX96 Q-PCR System (BioRad).
To perform yeast-one-hybrid (Y1H) analysis, the full length CrIND coding sequence was amplified and inserted into the pDEST22 vector (used as the effector plasmid). Synthetic fragments were produced (Sigma) containing wild-type and mutant versions of the putative CrIND binding sites from CrTAA1, CrYUC9 and CrPID promoter repeated four times separated by 8-bp spacers. The sequences were then amplified by PCR and inserted into the pHISLEU vector (used as the reporter plasmid). The constructs were co-transformed into the yeast strain, AH109, by using the LiAc method following the instructions for the yeast transformation. The yeasts transformants were selected on synthetic defined SD/–Trp/–Leu (–WL) agar medium plates and cultured at 28°C. Twelve individual transformants were randomly selected and mixed by three in four Eppendorf tubes, dropped on SD/–Trp/–Leu/–His (–WLH) agar plates and grow at 28°C for 2-3 days to test the interactions. Different concentrations of 3-aminotriazole (3-AT) was applied on the plate to prevent the unspecific interactions.
Quantification and Statistical Analysis
All statistics were calculated in Microsoft Excel. All measured data are presented as means ± SD specified along with sample sizes (n) in the methods and in figure legends. Comparisons between groups for the analysis of qRT-PCR was performed with Microsoft Excel Student’s t test, and significance levels are marked as: ∗ p < 0.05, ∗∗ p < 0.01.
Acknowledgments
We are grateful to Heather Bland, Yuli Ding, Lauren Grubb, André Kuhn, Mikhaela Neequaye, Pauline Stephenson, Tongbing Su, and Billy Tasker-Brown for critically reading the manuscript and providing comments prior to submission. We thank Andrew Davies and Phil Robinson for photographic assistance. This study was supported by a grant from the Biotechnological and Biological Research Council (BBSRC) to L.Ø. (BB/P020747/1) and an Institute Strategic Programme Grant from the BBSRC to the John Innes Centre (BB/P013511/1). K.L. and J.Š. acknowledge the Knut and Alice Wallenberg Foundation (KAW), the Swedish Governmental Agency for Innovation Systems (VINNOVA), the Swedish Research Council (VR), and the Swedish Metabolomics Centre (https://www.swedishmetabolomicscentre.se/) for access to instrumentation.
Author Contributions
Y.D. and L.Ø. designed the research. Y.D. performed most of the experiments with assistance from N.S. and scientific input from L.M. F.J. produced the crind-1ge CRISPR allele, Ł.Ł. developed the DR5v2 Capsella lines, J.Š. and K.L. carried out IAA and IPA measurements, and L.Ø. did the phylogenetic shadowing analysis. Y.D. and L.Ø. outlined and wrote the manuscript, and L.Ø. supervised the project. All authors participated in the discussion of the data and in the production of the final version of the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: February 28, 2019
Footnotes
Supplemental Information can be found with this article online at https://doi.org/10.1016/j.cub.2019.01.057.
Supplemental Information
References
- 1.Carroll S.B. Evolution at two levels: on genes and form. PLoS Biol. 2005;3:e245. doi: 10.1371/journal.pbio.0030245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carroll S.B. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- 3.Wray G.A. The evolutionary significance of cis-regulatory mutations. Nat. Rev. Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 4.Carroll S.B. Endless forms: the evolution of gene regulation and morphological diversity. Cell. 2000;101:577–580. doi: 10.1016/s0092-8674(00)80868-5. [DOI] [PubMed] [Google Scholar]
- 5.Wang R.-L., Stec A., Hey J., Lukens L., Doebley J. The limits of selection during maize domestication. Nature. 1999;398:236–239. doi: 10.1038/18435. [DOI] [PubMed] [Google Scholar]
- 6.Konishi S., Izawa T., Lin S.Y., Ebana K., Fukuta Y., Sasaki T., Yano M. An SNP caused loss of seed shattering during rice domestication. Science. 2006;312:1392–1396. doi: 10.1126/science.1126410. [DOI] [PubMed] [Google Scholar]
- 7.Arnaud N., Lawrenson T., Østergaard L., Sablowski R. The same regulatory point mutation changed seed-dispersal structures in evolution and domestication. Curr. Biol. 2011;21:1215–1219. doi: 10.1016/j.cub.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 8.Lemmon Z.H., Bukowski R., Sun Q., Doebley J.F. The role of cis regulatory evolution in maize domestication. PLoS Genet. 2014;10:e1004745. doi: 10.1371/journal.pgen.1004745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rast-Somssich M.I., Broholm S., Jenkins H., Canales C., Vlad D., Kwantes M., Bilsborough G., Dello Ioio R., Ewing R.M., Laufs P. Alternate wiring of a KNOXI genetic network underlies differences in leaf development of A. thaliana and C. hirsuta. Genes Dev. 2015;29:2391–2404. doi: 10.1101/gad.269050.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hohmann N., Wolf E.M., Lysak M.A., Koch M.A. A time-calibrated road map of Brassicaceae species radiation and evolutionary history. Plant Cell. 2015;27:2770–2784. doi: 10.1105/tpc.15.00482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Eldridge T., Łangowski Ł., Stacey N., Jantzen F., Moubayidin L., Sicard A., Southam P., Kennaway R., Lenhard M., Coen E.S., Østergaard L. Fruit shape diversity in the Brassicaceae is generated by varying patterns of anisotropy. Development. 2016;143:3394–3406. doi: 10.1242/dev.135327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Łangowski Ł., Stacey N., Østergaard L. Diversification of fruit shape in the Brassicaceae family. Plant Reprod. 2016;29:149–163. doi: 10.1007/s00497-016-0278-6. [DOI] [PubMed] [Google Scholar]
- 13.Roeder A.H.K., Yanofsky M.F. Fruit development in Arabidopsis. Arabidopsis Book. 2006;4:e0075. doi: 10.1199/tab.0075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bailey C.D., Koch M.A., Mayer M., Mummenhoff K., O’Kane S.L., Jr., Warwick S.I., Windham M.D., Al-Shehbaz I.A. Toward a global phylogeny of the Brassicaceae. Mol. Biol. Evol. 2006;23:2142–2160. doi: 10.1093/molbev/msl087. [DOI] [PubMed] [Google Scholar]
- 15.Sicard A., Lenhard M. Capsella. Curr. Biol. 2018;28:R920–R921. doi: 10.1016/j.cub.2018.06.033. [DOI] [PubMed] [Google Scholar]
- 16.Bowman J.L., Araki T., Arteaga-Vazquez M.A., Berger F., Dolan L., Haseloff J., Ishizaki K., Kyozuka J., Lin S.-S., Nagasaki H. The naming of names: Guidelines for gene nomenclature in Marchantia. Plant Cell Physiol. 2016;57:257–261. doi: 10.1093/pcp/pcv193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liljegren S.J., Roeder A.H.K., Kempin S.A., Gremski K., Østergaard L., Guimil S., Reyes D.K., Yanofsky M.F. Control of fruit patterning in Arabidopsis by INDEHISCENT. Cell. 2004;116:843–853. doi: 10.1016/s0092-8674(04)00217-x. [DOI] [PubMed] [Google Scholar]
- 18.Girin T., Stephenson P., Goldsack C.M.P., Kempin S.A., Perez A., Pires N., Sparrow P.A., Wood T.A., Yanofsky M.F., Østergaard L. Brassicaceae INDEHISCENT genes specify valve margin cell fate and repress replum formation. Plant J. 2010;63:329–338. doi: 10.1111/j.1365-313X.2010.04244.x. [DOI] [PubMed] [Google Scholar]
- 19.Sorefan K., Girin T., Liljegren S.J., Ljung K., Robles P., Galván-Ampudia C.S., Offringa R., Friml J., Yanofsky M.F., Østergaard L. A regulated auxin minimum is required for seed dispersal in Arabidopsis. Nature. 2009;459:583–586. doi: 10.1038/nature07875. [DOI] [PubMed] [Google Scholar]
- 20.Moubayidin L., Østergaard L. Dynamic control of auxin distribution imposes a bilateral-to-radial symmetry switch during gynoecium development. Curr. Biol. 2014;24:2743–2748. doi: 10.1016/j.cub.2014.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Romano C.P., Robson P.R., Smith H., Estelle M., Klee H. Transgene-mediated auxin overproduction in Arabidopsis: hypocotyl elongation phenotype and interactions with the hy6-1 hypocotyl elongation and axr1 auxin-resistant mutants. Plant Mol. Biol. 1995;27:1071–1083. doi: 10.1007/BF00020881. [DOI] [PubMed] [Google Scholar]
- 22.Romano C.P., Hein M.B., Klee H.J. Inactivation of auxin in tobacco transformed with the indoleacetic acid-lysine synthetase gene of Pseudomonas savastanoi. Genes Dev. 1991;5:438–446. doi: 10.1101/gad.5.3.438. [DOI] [PubMed] [Google Scholar]
- 23.Benková E., Michniewicz M., Sauer M., Teichmann T., Seifertová D., Jürgens G., Friml J. Local, efflux-dependent auxin gradients as a common module for plant organ formation. Cell. 2003;115:591–602. doi: 10.1016/s0092-8674(03)00924-3. [DOI] [PubMed] [Google Scholar]
- 24.Liao C.Y., Smet W., Brunoud G., Yoshida S., Vernoux T., Weijers D. Reporters for sensitive and quantitative measurement of auxin response. Nat. Methods. 2015;12:207–210. doi: 10.1038/nmeth.3279. 2, 210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Smyth D.R., Bowman J.L., Meyerowitz E.M. Early flower development in Arabidopsis. Plant Cell. 1990;2:755–767. doi: 10.1105/tpc.2.8.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stepanova A.N., Robertson-Hoyt J., Yun J., Benavente L.M., Xie D.Y., Dolezal K., Schlereth A., Jürgens G., Alonso J.M. TAA1-mediated auxin biosynthesis is essential for hormone crosstalk and plant development. Cell. 2008;133:177–191. doi: 10.1016/j.cell.2008.01.047. [DOI] [PubMed] [Google Scholar]
- 27.He W., Brumos J., Li H., Ji Y., Ke M., Gong X., Zeng Q., Li W., Zhang X., An F. A small-molecule screen identifies L-kynurenine as a competitive inhibitor of TAA1/TAR activity in ethylene-directed auxin biosynthesis and root growth in Arabidopsis. Plant Cell. 2011;23:3944–3960. doi: 10.1105/tpc.111.089029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao Y., Christensen S.K., Fankhauser C., Cashman J.R., Cohen J.D., Weigel D., Chory J. A role for flavin monooxygenase-like enzymes in auxin biosynthesis. Science. 2001;291:306–309. doi: 10.1126/science.291.5502.306. [DOI] [PubMed] [Google Scholar]
- 29.Nishimura T., Hayashi K., Suzuki H., Gyohda A., Takaoka C., Sakaguchi Y., Matsumoto S., Kasahara H., Sakai T., Kato J. Yucasin is a potent inhibitor of YUCCA, a key enzyme in auxin biosynthesis. Plant J. 2014;77:352–366. doi: 10.1111/tpj.12399. [DOI] [PubMed] [Google Scholar]
- 30.Zhao Y. Essential roles of local auxin biosynthesis in plant development and in adaptation to environmental changes. Annu. Rev. Plant Biol. 2018;69:417–435. doi: 10.1146/annurev-arplant-042817-040226. [DOI] [PubMed] [Google Scholar]
- 31.Ferrándiz C., Liljegren S.J., Yanofsky M.F. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science. 2000;289:436–438. doi: 10.1126/science.289.5478.436. [DOI] [PubMed] [Google Scholar]
- 32.Shull G.H. Duplicate genes for capsule-form in Capsella bursa-pastoris. Z. Abst. u. Vererbl. 1914;12:97–149. [Google Scholar]
- 33.Girin T., Paicu T., Stephenson P., Fuentes S., Körner E., O’Brien M., Sorefan K., Wood T.A., Balanzá V., Ferrándiz C. INDEHISCENT and SPATULA interact to specify carpel and valve margin tissue and thus promote seed dispersal in Arabidopsis. Plant Cell. 2011;23:3641–3653. doi: 10.1105/tpc.111.090944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rebeiz M., Tsiantis M. Enhancer evolution and the origins of morphological novelty. Curr. Opin. Genet. Dev. 2017;45:115–123. doi: 10.1016/j.gde.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jacob F. Evolution and tinkering. Science. 1977;196:1161–1166. doi: 10.1126/science.860134. [DOI] [PubMed] [Google Scholar]
- 36.Mayor C., Brudno M., Schwartz J.R., Poliakov A., Rubin E.M., Frazer K.A., Pachter L.S., Dubchak I. VISTA : visualizing global DNA sequence alignments of arbitrary length. Bioinformatics. 2000;16:1046–1047. doi: 10.1093/bioinformatics/16.11.1046. [DOI] [PubMed] [Google Scholar]
- 37.Koshikawa S., Giorgianni M.W., Vaccaro K., Kassner V.A., Yoder J.H., Werner T., Carroll S.B. Gain of cis-regulatory activities underlies novel domains of wingless gene expression in Drosophila. Proc. Natl. Acad. Sci. USA. 2015;112:7524–7529. doi: 10.1073/pnas.1509022112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H., Studer A.J., Zhao Q., Meeley R., Doebley J.F. Evidence that the origin of naked kernels during maize domestication was caused by a single amino acid substitution in tga1. Genetics. 2015;200:965–974. doi: 10.1534/genetics.115.175752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vuolo F., Mentink R.A., Hajheidari M., Bailey C.D., Filatov D.A., Tsiantis M. Coupled enhancer and coding sequence evolution of a homeobox gene shaped leaf diversity. Genes Dev. 2016;30:2370–2375. doi: 10.1101/gad.290684.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Simonini S., Deb J., Moubayidin L., Stephenson P., Valluru M., Freire-Rios A., Sorefan K., Weijers D., Friml J., Østergaard L. A noncanonical auxin-sensing mechanism is required for organ morphogenesis in Arabidopsis. Genes Dev. 2016;30:2286–2296. doi: 10.1101/gad.285361.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abràmoff M.D., Magalhães P.J., Ram S.J. Image processing with ImageJ. Biophoton. Int. 2004;11:36–42. [Google Scholar]
- 42.Novák O., Hényková E., Sairanen I., Kowalczyk M., Pospíšil T., Ljung K. Tissue-specific profiling of the Arabidopsis thaliana auxin metabolome. Plant J. 2012;72:523–536. doi: 10.1111/j.1365-313X.2012.05085.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.