Coxiella burnetii is an intracellular pathogenic bacterium that replicates within a lysosomal vacuole. Biogenesis of the Coxiella-containing vacuole (CCV) requires effector proteins delivered into the host cell cytosol by the type 4B secretion system (T4BSS). Modifications to lysosomal physiology required for pathogen replication within the CCV are poorly understood. Mammalian (or mechanistic) target of rapamycin complex 1 (mTORC1) is a master kinase that regulates lysosome structure and function. Nutrient deprivation inhibits mTORC1, which promotes cell catabolism in the form of accelerated autophagy and increased lysosome biosynthesis. Here, we report that C. burnetii growth is enhanced by T4BSS-dependent inhibition of mTORC1 that does not activate autophagy. Canonical inhibition of mTORC1 by starvation or inhibitor treatment that induces autophagic flux does not benefit C. burnetii growth. Furthermore, hyperactivation of mTORC1 impairs bacterial replication. These findings indicate that C. burnetii inhibition of mTORC1 without accelerated autophagy promotes bacterial growth.
KEYWORDS: Coxiella, Q fever, autophagy, coxiella-containing vacuole, endolysosomal membranes, lysosome, mTor, type IV secretion, vacuole
ABSTRACT
The Q fever agent Coxiella burnetii is a Gram-negative bacterium that invades macrophages and replicates inside a specialized lysosomal vacuole. The pathogen employs a type 4B secretion system (T4BSS) to deliver effector proteins into the host cell that modify the Coxiella-containing vacuole (CCV) into a replication-permissive niche. Mature CCVs are massive degradative organelles that acquire lysosomal proteins. Inhibition of mammalian (or mechanistic) target of rapamycin complex 1 (mTORC1) kinase by nutrient deprivation promotes autophagy and lysosome fusion, as well as activation of the transcription factors TFE3 and TFEB (TFE3/B), which upregulates expression of lysosomal genes. Here, we report that C. burnetii inhibits mTORC1 as evidenced by impaired localization of mTORC1 to endolysosomal membranes and decreased phosphorylation of elF4E-binding protein 1 (4E-BP1) and S6 kinase 1 in infected cells. Infected cells exhibit increased amounts of autophagy-related proteins protein 1A/1B-light chain 3 (LC3) and p62 as well as of activated TFE3. However, C. burnetii did not accelerate autophagy or block autophagic flux triggered by cell starvation. Activation of autophagy or transcription by TFE3/B increased CCV expansion without enhancing bacterial replication. By contrast, knockdown of tuberous sclerosis complex 1 (TSC1) or TSC2, which hyperactivates mTORC1, impaired CCV expansion and bacterial replication. Together, these data demonstrate that specific inhibition of mTORC1 by C. burnetii, but not amplified cell catabolism via autophagy, is required for optimal pathogen replication. These data reveal a complex interplay between lysosomal function and host cell metabolism that regulates C. burnetii intracellular growth.
INTRODUCTION
Coxiella burnetii is a Gram-negative intracellular pathogen that causes human Q fever, a zoonotic disease that is commonly transmitted to humans through inhalation of by-products generated by infected domestic livestock (1). C. burnetii colonizes alveolar macrophages, where it occupies a vacuole that matures canonically within the endosomal pathway, culminating in lysosome fusion (2). By mechanisms not fully understood, C. burnetii resists degradation by acid hydrolases delivered by ongoing fusion of lysosomes with the Coxiella-containing vacuole (CCV). The vacuole contains lysosomal components that include lysosome-associated membrane proteins (LAMPs), cathepsins, and Rab7. The lengthy C. burnetii intracellular replication cycle lasts approximately 6 days, at which point the mature CCV phenotypically resembles an overgrown phagolysosome that can contain hundreds of bacteria (3 – 5). Effector proteins delivered into the host cell by the C. burnetii type 4B secretion system (T4BSS) direct CCV modifications required for pathogen growth (6 – 10). The T4BSS apparatus is encoded by defective in organelle trafficking/intracellular multiplication (dot/icm) genes. Defects in CCV biogenesis and intracellular replication occur in C. burnetii strains containing mutations in critical dot/icm or effector-encoding genes (8, 10 – 13). Several features, including promiscuous fusogenicity, prominent size, temporal stability, and the ability to foster C. burnetii growth, distinguish the CCV from native lysosomal organelles. These characteristics provide clues for identifying host pathways that participate in formation of this specialized replication niche.
Lysosome-vacuole fusion occurs continuously within resting macrophages and generates hybrid organelles, such as phagolysosomes, autolysosomes, and CCVs that degrade vacuole-bound cargo (4, 14 – 17). Lysosomal organelles modulate their degradative capacity to accommodate the metabolic requirements of the cell under a variety of environmental growth conditions. Nutrient sensing and regulatory functions of lysosomal organelles are primarily mediated by the master regulatory kinase mammalian (or mechanistic) target of rapamycin (mTOR) complex 1 (mTORC1), a heteromeric complex composed of the Ser/Thr kinase mTOR, regulatory-associated protein of mTOR (Raptor), mammalian lethal with SEC13 protein 8 (MLST8), proline-rich Akt substrate of 40 kDa (PRAS40), and DEP domain-containing mTOR interacting protein (DEPTOR) (18, 19). mTORC1 sensing of intracellular amino acid levels is mediated by Rag GTPases, whereas sensing of other growth determinants, including glucose, insulin, growth factors, and hypoxia, is mediated by the tuberous sclerosis complex (TSC) (18 – 20). RagA or RagB forms heterodimers with RagC or RagD that exhibit GTPase activity on the lysosomal surface when the levels of amino acids are sufficient (21, 22). TSC is a GTPase-activating protein (GAP) that inhibits Rheb GTPase activity (19). Active GTPases of both Rheb and Rag are required for mTORC1 activation and recruitment to the lysosome surface (18 – 20).
In fully fed cells, active mTORC1 is recruited to the lysosome surface, where it promotes biosynthesis of macromolecules via phosphorylation of numerous downstream effectors, including elF4E-binding protein 1 (4E-BP1) and p70 S6 kinase 1 (S6K1). Phosphorylation by mTORC1 also negatively regulates several factors important for the induction of macroautophagy (here termed autophagy) (18, 19). These include the lysosomal gene transcription factors TFE3 and TFEB (TFE3/B). When cell nutrients are depleted, mTORC1 inactivation triggers a cascade of events that block cell biosynthesis and growth and result in increased cellular catabolism mediated by autophagy. Heightened lysosome fusogenicity in response to mTORC1 inhibition, along with TFE3/B-dependent synthesis of lysosomal components, results in formation of large, fusogenic lysosomal organelles that accommodate the surge of incoming cargo targeted for degradation via autophagy (19, 23). Microtubule-associated protein 1A/1B-light chain 3 (LC3) and sequestome-1 (p62/SQSTM1) are autophagy-related proteins involved in autophagosome biogenesis, cargo selection, and lysosome fusion. Cellular levels of lipidated LC3 (LC3-II) and p62 spike at the onset of accelerated autophagy until degradation increases and LC3-II and p62 levels diminish. Consequently, measurement of LC3 and p62 levels can be used to assess autophagic flux, a measure of degradation activity within cells (24).
The CCV fuses with autophagic vesicles and labels with autophagy-related proteins, including p62, LC3, Beclin-1, and Rab24 GTPase (3, 9, 25 – 29). Inhibition of C. burnetii protein synthesis, or disruption of the T4BSS, reduces GFP-LC3 recruitment to the CCV (27, 28). These observations are consistent with the finding that the T4BSS effector CvpB (Cig2/CBU0021) is required for LC3 recruitment to the CCV (9, 26). Mutation of cvpB, or depletion of autophagy-related proteins ATG5, ATG12, or STX17, produces defects in CCV fusion but not in bacterial replication (9, 30). Data showing recruitment of autophagy-related proteins to the CCV, along with greater CCV expansion when autophagy is activated, led to speculation that cell catabolism by autophagy supplies nutrients for C. burnetii replication (3, 25, 31, 32). Infected cells contain increased levels of LC3 and p62 (9, 28, 32). Levels of p62 remain elevated in infected macrophages after autophagic flux is induced by starvation, suggesting that C. burnetii selectively blocks p62 degradation (32, 33). Autophagy is controlled (in part) by Vps34, a class III phosphoinositide 3-kinase (PI3K) that catalyzes production of phosphatidylinositol 3-phosphate (PI3P) on target membranes, which leads to nucleation of autophagic vesicles (34, 35). Observations that inhibitors of PI3K impair C. burnetii infection (3, 25) and CCV fusion (32) have led to the idea that autophagic trafficking contributes to C. burnetii parasitism. However, the absence of C. burnetii replication defects in cells depleted of essential autophagy components, such as ATG5 and ATG12, suggests that cell catabolism via autophagy is dispensable for pathogen replication (9).
Here, the activity of mTORC1 during C. burnetii infection was characterized to enable better understanding of the importance of autophagy and lysosome biosynthesis in CCV production and pathogen replication. Under conditions of culture in axenic medium, C. burnetii preferentially consumes amino acids and peptides for growth (36). Thus, this study also examined whether C. burnetii subverts amino acid-dependent regulation of mTORC1 to promote growth within the CCV. We report that C. burnetii inhibits mTORC1 by a mechanism that is independent of amino acid-dependent regulatory pathways. Interestingly, inhibition of mTORC1 by C. burnetii promotes replication by a process that does not activate autophagy. In contrast, canonical inhibition of mTORC1 and activation of autophagy do not benefit C. burnetii growth.
RESULTS
C. burnetii-infected cells exhibit decreased endolysosomal localization of mTOR.
The activity of mTORC1 was initially assessed in cells infected with C. burnetii by examining its subcellular localization. HeLa cells infected with C. burnetii for 72 h in RPMI medium with 10% fetal bovine serum (FBS) (complete medium) were incubated in (i) fresh complete medium for 4.5 h, (ii) RPMI medium lacking amino acids and serum (AA− medium) for 4.5 h, or (iii) AA− medium for 4 h followed by complete medium for 30 min. Samples were fixed and immunostained for C. burnetii, mTORC1, and the endolysosomal marker CD63 (Fig. 1A). As predicted, localization of mTORC1 to CD63+ vesicles decreased in cells incubated in AA− medium compared to cells incubated in complete medium. Moreover, incubation of amino-acid-starved cells in complete medium increased mTORC1 CD63 colocalization (Fig. 1B). However, in infected cells incubated in AA− medium or complete medium following starvation, the amount of localization of mTORC1 to CD63+ endolysosomal membranes was significantly less than that seen with uninfected cells treated in parallel. In complete medium, cells infected 72 or 120 h with wild-type C. burnetii showed reduced localization of mTORC1 to CD63+ vesicles, but cells infected with the ΔdotA mutant did not exhibit reduced localization (see Fig. S1 in the supplemental material). In this experiment, mutant ΔdotA replication defects (11) were accounted for by infection with 10-fold more mutant bacteria (10× ΔdotA bacteria) than wild-type C. burnetii (Fig. S2A). These data indicate that C. burnetii inhibits mTORC1 recruitment to the CCV and other endolysosomal membranes.
FIG 1.
C. burnetii-infected cells exhibit decreased endolysosomal localization of mTOR. HeLa cells infected with C. burnetii for 72 h in complete medium were incubated in (i) fresh complete medium for 4.5 h (Comp); (ii) AA− medium for 4.5 h (AA−); or (iii) AA− medium for 4 h followed by complete medium for 30 min (AA+). (A) Representative fluorescence micrographs of HeLa cells stained at 72 hpi for DNA (blue), CD63 (red), mTOR (green), and C. burnetii (gray) (scale bar, 10 µm). (B) Subcellular localization of mTOR in HeLa cells left uninfected (UI) or infected with wild-type C. burnetii (WT). The plot depicts means ± standard deviations for the ratio of lysosomal (CD63+) mTOR signal to cytosolic (lys/cyto) mTOR signal (n = >50 cells). Data are representative of results from three independent experiments. Asterisks indicate statistical significance (***, P < 0.001). Scale bar, 10 µm.
C. burnetii inhibition of mTOR endolysosomal localization is T4BSS dependent. Data represent subcellular localization of mTOR in HeLa cells left uninfected (UI) or infected with wild-type C. burnetii (WT) or the ΔdotA mutant for 72 h (A) or 120 h (B) in complete medium. The plot depicts means ± standard deviations of data representing the ratio of lysosomal (CD63+) mTOR signal to cytosolic (lys/cyto) mTOR signal (n = >100 cells). Data are representative of results from three independent experiments. Asterisks indicate statistical significance (*, P < 0.05). Download FIG S1, PDF file, 0.5 MB (557.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The viability of THP-1 macrophages infected with C. burnetii does not decrease in mTORC1-inhibiting growth media. (A) Representative fluorescence micrographs of THP-1 macrophages infected with wild-type C. burnetii (WT) or the ΔdotA mutant at a multiplicity of infection of 100 or 1,000, respectively, for 72 h in complete medium. DNA, C. burnetii, and host cells were stained blue, green, and magenta, respectively. (B) Percent cytoxicity displayed by THP-1 macrophages infected for 72 h with C. burnetii in complete (Comp), AA−, Torin-1, or Gluc− medium. Percent cytotoxicity was determined using the CytoTox 96 Non-Radioactive Cytotoxicity assay (Promega, Madison, WI) according to the manufacturer’s instructions. The plot depicts means ± standard deviations of the percentage of cytotoxic cells detected within each of the indicated samples for three independent experiments. Scale bar, 20 µm. Download FIG S2, PDF file, 0.8 MB (825.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
C. burnetii inhibits mTORC1 kinase activity in a T4BSS-dependent manner.
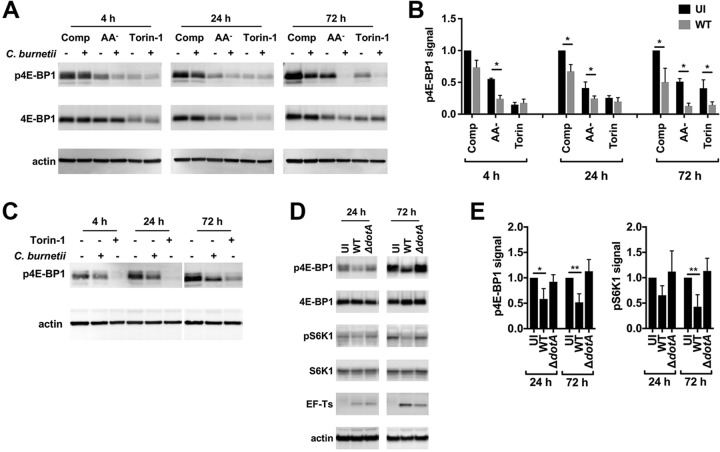
Phosphorylation of 4E-BP1 Thr37/46 and S6K1 Thr389 by mTORC1 is inhibited in cells cultured without amino acids or in the presence of the mTOR catalytic inhibitor Torin-1 (37). The effect of C. burnetii infection on 4E-BP1 phosphorylation was examined in THP-1 macrophages maintained in complete medium, AA− medium, or complete medium plus Torin-1 (Torin-1 medium). AA− medium and Torin-1 medium were included to assess mTORC1 activity during amino acid starvation and as a positive control for mTORC1 inhibition, respectively. Uninfected or infected macrophages cultured in the respective media for 4, 24, or 72 h were lysed and immunoblotted with phosphospecific 4E-BP1 antibody (Fig. 2A). Elevated cell cytotoxicity was not observed here and elsewhere in cells cultured in AA− medium or Torin-1 medium (Fig. S2B). Overall reductions in the levels of phosphorylated 4E-BP1 (p4E-BP1) in cells incubated in AA− medium or Torin-1 medium compared to complete medium indicated that culture in AA− medium or Torin-1 medium inhibited mTORC1 (Fig. 2B). Notably, cells infected with C. burnetii exhibited reduced levels of p4E-BP1 relative to uninfected cells, with significant reductions associated with infected cells cultured in AA− medium at 4 and 24 h postinfection (hpi), in complete medium at 24 hpi, and under all culture conditions at 72 hpi (Fig. 2B). Reduced phosphorylation of 4E-BP1 in infected human peripheral blood mononuclear cell (PBMC)-derived macrophages (Fig. 2C) and infected HeLa cells (Fig. S3) demonstrated that the mTORC1 inhibition was not cell type specific. To determine if mTORC1 inhibition requires an intact dot/icm T4BSS, infections were conducted with 10× ΔdotA mutant bacteria or wild-type bacteria. Phosphorylation of 4E-BP1 and S6K1 was reduced in cells infected for 24 or 72 h with wild-type C. burnetii but not in cells infected with the ΔdotA mutant (Fig. 2D). Collectively, these data demonstrate that C. burnetii inhibits mTORC1 in a T4BSS-dependent fashion.
FIG 2.
C. burnetii inhibits mTORC1 kinase activity in a T4BSS-dependent manner. (A) Immunoblot of lysates from infected or uninfected THP-1 macrophages incubated for 4, 24, or 72 h in complete (Comp), AA−, or Torin-1 medium probed with antibodies against phosphorylated 4E-BP1 Thr37/46 (p4E-BP1), 4E-BP1, or actin. (B) Quantitation of p4E-BP1 signal detected by immunoblot of lysates from THP-1 macrophages left uninfected (UI) or infected with wild-type C. burnetii (WT). The plot depicts means ± standard deviations of p4E-BP1 signal normalized to the actin loading control relative to uninfected cells in complete medium for three independent experiments. (C) Immunoblot of lysates from infected or uninfected human peripheral blood mononuclear cell (PBMC)-derived macrophages incubated for 4, 24, or 72 h in complete medium or complete medium with Torin-1 probed with antibodies against p4E-BP1 or actin. Data are representative of results from three independent experiments. (D) Immunoblot of lysates from THP-1 macrophages left uninfected or infected with wild-type C. burnetii or the ΔdotA mutant for 24 or 72 h in complete medium probed with antibodies against p4E-BP1, 4E-BP1, phosphorylated S6 kinase 1 Thr389 (pS6K1), S6K1, EF-Ts, or actin. (E) Quantitation of p4E-BP1 signal in panel D. Plots depict means ± standard deviations of p4E-BP1 or pS6K1 signal normalized to the actin loading control compared to uninfected cells for three independent experiments. Asterisks indicate statistical significance (*, P < 0.05; **, P < 0.01).
Infected HeLa cells exhibit reduced phosphorylation of 4E-BP1. Immunoblot of lysates from uninfected or C. burnetii-infected HeLa cells incubated for 4, 24, or 72 h in complete (Comp), AA−, or Torin-1 medium. Blots were probed with antibodies against phosphorylated 4E-BP1 Thr37/46 (p4E-BP1) or actin was performed. Download FIG S3, PDF file, 0.2 MB (215.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Nutrient replenishment does not rescue C. burnetii inhibition of mTORC1.
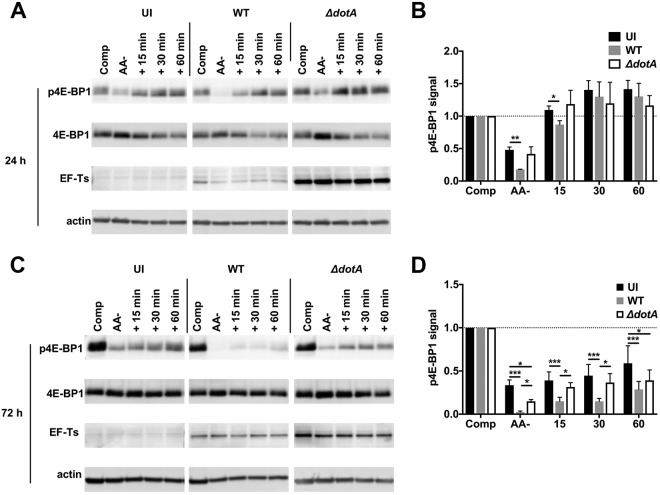
The observation that complete medium does not rescue mTORC1 endolysosomal localization within infected cells (Fig. 1) suggests that C. burnetii might interfere with the activation of mTORC1 by amino acids. To investigate this possibility, uninfected THP-1 macrophages, or macrophages infected with wild-type C. burnetii or 50× ΔdotA mutant, were cultured in AA− medium for 24 or 72 h and then incubated in complete medium for an additional 15, 30, or 60 min before lysis. As controls, lysates of uninfected and infected cells maintained for the entire course of the experiment in complete or AA− medium were also collected. Cell lysates were immunoblotted for p4E-BP1 to asses mTORC1 activity (Fig. 3A and C). After starvation for 24 or 72 h, uninfected cells and cells infected with the ΔdotA mutant showed comparable increases in p4E-BP1 when subsequently incubated in complete medium (Fig. 3B and D). Incubation in complete medium also stimulated phosphorylation of 4E-BP1 in starved cells infected with wild-type C. burnetii; however, p4E-BP1 levels were reduced relative to uninfected cells and cells infected with the ΔdotA mutant. At 24 h, p4E-BP1 levels were reduced in wild-type infected cells starved of amino acids and after incubation for 15 min in complete medium whereas incubation for 30 min restored p4E-BP1 to the control levels (Fig. 3B). At 72 h, p4E-BP1 levels were reduced in starved cells infected with wild-type C. burnetii after incubation for up to 60 min in complete medium (Fig. 3D). Incubation of starved cells infected with wild-type C. burnetii with higher concentrations of amino acids did not rescue p4E-BP1 reductions (Fig. S4). Although wild-type infection reduced 4E-BP1 phosphorylation, the overall increase in the levels of p4E-BP1 stimulated by nutrient replenishment in wild-type-infected cells was comparable to the increase associated with uninfected cells and cells infected with the ΔdotA mutant (Fig. S5). Together, these data indicate that C. burnetii maintains mTORC1 inhibition when amino acids are replete and that mTORC1 is responsive to amino acid activation during infection. Throughout these experiments, we observed an additive inhibitory effect on phosphorylation of 4E-BP1 and recruitment of mTORC1 to the lysosomal surface (Fig. 1) when cells were infected with wild-type bacteria and starved (Fig. 2 and 3; see also Fig. S4). These effects were not rescued by nutrient repletion. Collectively, these results indicate that C. burnetii and amino acids regulate mTORC1 by separate mechanisms.
FIG 3.
Nutrient replenishment does not rescue inhibition of mTORC1. (A and C) Immunoblots of lysates from THP-1 macrophages left uninfected (UI) or infected with wild-type C. burnetii (WT) or the ΔdotA mutant. Cells were incubated for 24 h (A) or 72 h (C) in complete (Comp) or AA− medium or in AA− medium followed by incubation with fresh complete medium for 15, 30, or 60 min. Immunoblots were probed with antibodies against phosphorylated 4E-BP1 Thr37/46 (p4E-BP1), 4E-BP1, EF-Ts, or actin. (B and D) Quantitation of p4E-BP1 signal in panels A and C, respectively. Plots depict means ± standard deviations of p4E-BP1 signal normalized to the actin loading control compared to the respective control cells in complete medium for three independent experiments. Asterisks indicate statistical significance (*, P < 0.05; ***, P < 0.001).
Addition of extra amino acids does not rescue reduced phosphorylation of 4E-BP1 in C. burnetii-infected THP-1 cells. (A) Immunoblot of lysates from infected or uninfected THP-1 macrophages incubated for 72 h in complete (Comp), AA−, or Torin-1 medium with or without additional incubation for 30 min in Comp medium plus additional essential and nonessential amino acids (+AA). Blots were probed with antibodies against phosphorylated 4E-BP1 Thr37/46 (p4E-BP1) or actin. (B) Quantitation of p4E-BP1 signal in panel A. The plot depicts means ± standard deviations of p4E-BP1 signal normalized to the actin loading control relative to respective control cells in complete medium for three independent experiments. Asterisks indicate statistical significance (*, P < 0.05; ***, P < 0.001). Download FIG S4, PDF file, 0.3 MB (258.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Increases in p4EB-BP1 levels are comparable in uninfected and infected cells when they are starved and then incubated with nutrient-rich medium. THP-1 macrophages left uninfected (UI) or infected with wild-type C. burnetii (WT) or the ΔdotA mutant were incubated for 24 h (top) or 72 h (bottom) in AA− medium followed by incubation with fresh complete medium for 15, 30, or 60 min. Immunoblots (Fig. 3A and B) were probed with antibodies against phosphorylated 4E-BP1 Thr37/46 (p4E-BP1) or actin. Plots depict means ± standard deviations with trendlines fitted by linear regression of p4E-BP1 signal normalized to the actin loading control for three independent experiments. Download FIG S5, PDF file, 0.5 MB (546.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Levels of LC3 and p62 are elevated in infected cells, and the proteins are efficiently degraded when autophagy is induced by starvation.
Amplified cell catabolism promoted by inhibition of mTORC1 during cell starvation is mediated (in part) by increased autophagic flux (23). C. burnetii inhibits mTORC1; however, previous reports showed that macrophages infected with C. burnetii displayed elevated amounts of LC3 and p62 without autophagic flux (28). Thus, C. burnetii appears to inhibit mTORC1 without inducing cell catabolism via autophagy. To examine this possibility, LC3 and p62 levels were examined at 72 hpi, a time point corresponding to robust inhibition of mTORC1 by C. burnetii. Immunoblots revealed that infected THP-1 macrophages incubated in complete medium contained significantly more LC3 and p62 than uninfected cells at 72 hpi (Fig. 4A) but not at 4 or 24 hpi (Fig. S6A). These data are consistent with a previous report showing that LC3 and p62 accumulate at later time points postinfection and support the conclusion that C. burnetii does not induce autophagic flux (28). At 72 hpi, macrophages infected with 10× ΔdotA mutant contained less LC3 than cells infected with wild-type C. burnetii, whereas p62 levels were similar (Fig. 4A). This suggested that the increase in LC3 levels during infection is effector driven, which is consistent with previous reports (9, 26). Infected macrophages cultured in AA− or Torin-1 medium for 72 h contained less LC3 and p62 than infected cells in complete medium, but contained more LC3 and p62 than uninfected cells cultured under the same conditions (Fig. S6A and B). To determine whether accumulation of LC3 and p62 by infected cells correlates with defects in autophagic degradation, autophagy was activated by starving cells in Hanks’ balanced salt solution (HBSS). HeLa cells were chosen for these experiments because they exhibit prototypical processing of LC3 and p62 in response to starvation (24). HeLa cells infected for 72 h in complete medium contained more LC3 and p62 than uninfected cells (Fig. 4B). Incubation in HBSS induced autophagic flux in both infected and uninfected cells as evidenced by time-dependent decreases in LC3 and p62 levels (Fig. 4B). Infected cells contained more LC3 and p62 than uninfected cells throughout the starvation time course (Fig. 4C and D). However, a comparison of the time-dependent decreases revealed that the infected cells degraded more LC3 and p62 than the uninfected cells (Fig. S6C). Immunoblots of lysates from THP-1 macrophages for LC3 and p62 confirmed that C. burnetii does not inhibit LC3 or p62 degradation (Fig. S6D). Collectively, these data indicate that C. burnetii does not activate or inhibit autophagic flux. Starvation also results in decreased cell size and cessation of cell proliferation (38). However, the mean cell area occupied by infected HeLa cells was larger than that occupied by uninfected cells (Fig. S7). This result is consistent with a previous report showing that C. burnetii does not inhibit host cell replication (39). Thus, cumulative evidence indicates that inhibition of mTORC1 by C. burnetii does not promote cell catabolism.
FIG 4.
Levels of LC3 and p62 are elevated in infected cells and the proteins are efficiently degraded when autophagy is induced by starvation. (A) Immunoblot of lysates from THP-1 macrophages left uninfected (UI) or infected with wild-type C. burnetii (WT) or the ΔdotA mutant for 72 h in complete medium. Immunoblots were probed with antibodies against LC3, p62, EF-Ts, or actin. Plots depict means ± standard deviations of LC3 or p62 signal normalized to the actin loading control for three independent experiments. (B) Immunoblot of lysates from HeLa cells left uninfected (UI) or infected with wild-type C. burnetii (WT) for 72 h in complete medium (Comp) or subjected to continued incubation in HBSS for the indicated times. Immunoblots were probed with antibodies against LC3, p62, or actin. (C and D) Quantitation of LC3 (C) and p62 (D) signals in panel B. Plots depict means of signal data ± standard deviations of results from three independent experiments. Asterisks indicate statistical significance (*, P < 0.05; **, P < 0.01).
Infected cells contain more LC3 and p62 than uninfected cells and exhibit robust autophagic flux when starved. (A) Immunoblot of lysates from infected or uninfected THP-1 macrophages incubated for 4, 24, or 72 h in complete, AA−, or Torin-1 medium probed with antibodies against LC3, p62, or actin. (B) Quantitation of LC3 (left) or p62 (right) signal in panel A. The plot depicts means ± standard deviations of signal normalized to the actin loading control relative to cells in complete medium at 72 h for three independent experiments. (C) LC3 (left) or p62 (right) degradation rates in HeLa cells left uninfected (UI) or infected with wild-type C. burnetii (WT) for 72 h in complete medium and then incubated for the indicated times with HBSS. Plots depict mean signal data ± standard deviations with trendlines fitted by linear regression for three independent experiments. (D) Immunoblot of lysates from HeLa cells left uninfected (UI) or infected with wild-type C. burnetii (WT) for 72 h in complete medium, then incubated for the indicated times with HBSS and probed with antibodies against LC3, p62, or actin. Asterisks indicate statistical significance (*, P < 0.05). Download FIG S6, PDF file, 0.5 MB (495.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Infected HeLa cells are larger than uninfected cells. The plot depicts means ± standard deviations of the cell area occupied by HeLa cells left uninfected (UI) or infected with wild-type (WT) C. burnetii measured in three independent experiments (n = >10,000 cells measured). Cell area was quantitated using CellProfiler. Each of the three independent data sets was normalized by dividing by the mean area of respective uninfected cells. Asterisks indicate statistical significance (****, P < 0.0001). Download FIG S7, PDF file, 0.2 MB (181KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
TFE3/B activity promotes CCV biogenesis without benefiting C. burnetii replication.
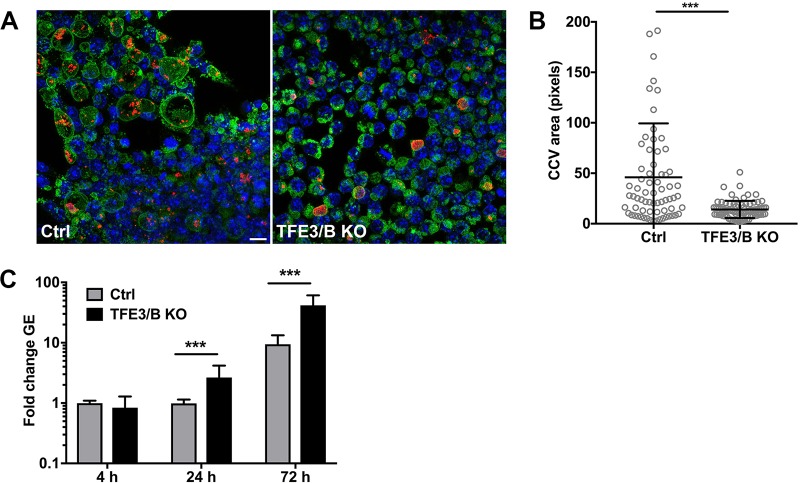
TFE3/B are functionally redundant transcription factors that direct expression of lysosomal genes. Inhibition of mTORC1 allows TFE3/B translocation to the nucleus and transcription of lysosomal genes, leading to enlargement of the lysosomal compartment and enhanced lysosomal degradation (23, 40). Nuclear translocation of active TFE3 was examined in infected cells by immunofluorescence microscopy (Fig. 5A). Infected HeLa cells exhibited greater levels of nuclear TFE3 signal than uninfected cells, with significant increases occurring at 48 and 72 hpi (Fig. 5B). In HeLa cells or THP-1 macrophages infected with 10× ΔdotA mutant, TFE3 nuclear translocation was comparable to that seen with cells infected with wild-type C. burnetii (Fig. S8). This indicates that C. burnetii activates TFE3 independently of T4BSS activity. To investigate the contribution of TFE3/B to C. burnetii infection, pathogen growth was examined in TFE3/B double-knockout RAW mouse macrophages (Fig. 6A) (23). At 72 hpi, knockout macrophages exhibited smaller CCVs than wild-type macrophages (Fig. 6B), indicating that expression of lysosomal genes by TFE3/B promotes CCV expansion. These data correlate with a previous report showing that infected cells have a larger endolysosomal compartment than uninfected cells (41). However, TFE3/B knockout cells supported higher levels of C. burnetii replication than wild-type macrophages (Fig. 6C). Thus, CCV size does not directly correlate with pathogen growth (42). Moreover, the increase in cell catabolism coincident with transcription by TFE3/B does not promote C. burnetii growth in macrophages.
FIG 5.
C. burnetii infection increases nuclear translocation of TFE3. (A) Representative fluorescence micrographs of HeLa cells left uninfected (UI) or infected with wild-type C. burnetii (WT) incubated in complete medium for the indicated times. Cells were fixed and stained for DNA (red), TFE3 (green), CD63 (gray), and C. burnetii (magenta). (B) Quantitation of TFE3 subcellular localization in panel A. The plot depicts means ± standard deviations of the ratio of nuclear TFE3 signal to cytoplasmic TFE3 signal at the indicated time points (n = >50 cells). Data are representative of results from three independent experiments. Asterisks indicate statistical significance (***, P < 0.001). Scale bar, 20 µm.
FIG 6.
Wild-type macrophages contain larger CCVs but are less permissive for C. burnetii replication than TFE3/B knockout macrophages. (A) Representative fluorescence micrographs of control (Ctrl) or TFE3/B double-knockout (KO) RAW macrophages infected with C. burnetii. Cells were stained for DNA (blue), C. burnetii (red), and LAMP1 (green). (B) Quantitation of CCV size in control or TFE3/B double-knockout RAW macrophages 72 hpi (n = >100 cells). Data are representative of results from three independent experiments. (C) Replication of C. burnetii in control or TFE3/B double-knockout RAW macrophages at the indicated time points. The plot depicts means of fold increase ± standard deviations in C. burnetii genome equivalents (GE) relative to the mean at 4 hpi for three independent experiments. Asterisks indicate statistical significance (***, P < 0.001). Scale bar, 10 µm.
C. burnetii infection triggers TFE3 translocation independently of T4BSS activity. Data represent results of quantitation of TFE3 subcellular localization in HeLa cells (A) or THP-1 macrophages (B) left uninfected (UI) or infected with wild-type C. burnetii (WT) or the ΔdotA mutant for 72 h in complete medium. The plots depict means ± standard deviations of the ratio of nuclear TFE3 signal to cytoplasmic TFE3 signal detected in cells (n = >25). Data are representative of results from three independent experiments. Asterisks indicate statistical significance (***, P < 0.001). Download FIG S8, PDF file, 0.3 MB (265.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Autophagy promotes CCV expansion and fusogenicity without benefiting C. burnetii replication.
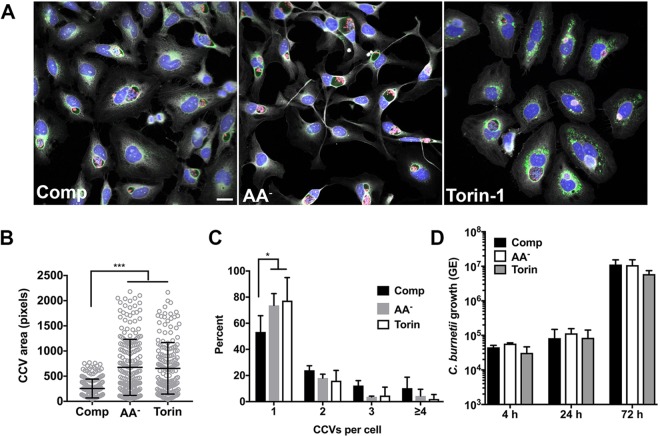
The observation that autophagy increases CCV size and fusogenicity has been used to support the idea that autophagic flux generates nutrients for bacterial growth and membrane for CCV biogenesis (3, 25, 27, 31, 32). However, evidence presented here (Fig. 4C and D; see also Fig. S6B) and elsewhere (28) demonstrates that C. burnetii does not stimulate or impair autophagic flux. To clarify the role of autophagy, we examined C. burnetii growth in cells cultured in AA− or Torin-1 media, conditions that potentiate C. burnetii inhibition of mTORC1 (Fig. 2A and B; see also Fig. 3) and promote autophagic flux (23). HeLa cells cultured in AA− or Torin-1 medium contained larger CCVs (Fig. 7A and B), with fewer cells containing multiple CCVs (Fig. 7C) than were seen with cells cultured in complete medium. These data agree with reports that activation of autophagy promotes CCV biogenesis (3, 27, 32). By contrast, the levels of bacterial replication were comparable in cells incubated in complete, AA−, or Torin-1 medium (Fig. 7D), indicating that induction of autophagy by amino acid starvation or pharmacological inhibition of mTORC1 does not affect C. burnetii intracellular growth.
FIG 7.
Autophagy promotes CCV expansion and fusogenicity without benefitting C. burnetii replication. (A) Representative fluorescence micrographs of infected HeLa cells incubated for 72 h in complete (Comp), AA−, or Torin-1 medium and then stained for DNA (blue), C. burnetii (red), LAMP1 (green), and cells (gray). (B) Quantitation of CCV size in infected HeLa cells incubated in Comp, AA−, or Torin-1 medium. The plot depicts means CCV size ± standard deviations (n = >100 cells) at 72 hpi. Data are representative of results from three independent experiments. (C) Quantitation of the number of CCVs per cell in cells incubated in Comp, AA−, or Torin-1 medium. The plot depicts mean percentages ± standard deviations from cells (n = >100) containing the indicated number of CCVs for three independent experiments. (D) Replication of C. burnetii in THP-1 macrophages incubated in complete, AA−, or Torin-1 medium. The plots depict means ± standard deviations of the fold change in bacterial genome equivalents (GE) relative to the mean at 4 hpi for three independent experiments. Asterisks indicate statistical significance (*, P < 0.05; ***, P < 0.001). Scale bar, 20 µm.
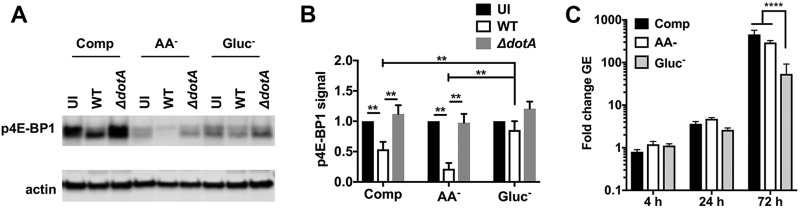
Amino acids are required for growth of C. burnetii in broth culture (36). Thus, it was surprising that growth defects were not observed in cells cultured in medium without amino acids. The AA− medium used in this study contains a limited number of nutrients present in RPMI base medium, such as vitamins, inorganic salts, and glucose. Interestingly, although glucose is not required for C. burnetii growth in broth culture (36), recent transcriptional evidence indicates that C. burnetii preferentially utilizes glucose, and not amino acids, as its primary carbon source inside cells (43). We examined whether culture of THP-1 macrophages in medium replete with amino acids and lacking glucose (Gluc−) would affect mTORC1 activity and intracellular growth of C. burnetii. Infected cells remained viable during glucose deprivation (Fig. S2B) and exhibited larger CCVs than cells maintained in complete medium at 72 hpi (Fig. S9). As expected, cells cultured in Gluc− medium showed overall reductions in p4E-BP1 that were indicative of mTORC1 inhibition in response to starvation (Fig. 8A) (18, 44). Notably, cells infected with wild-type C. burnetii exhibited significantly less p4E-BP1 in complete and AA− medium, but not in Gluc− medium, than uninfected cells and cells infected with the ΔdotA mutant (Fig. 8B). These data indicate that glucose deprivation impairs the inhibition of mTORC1 by C. burnetii. Additionally, C. burnetii replication was reduced ∼10-fold at 72 hpi in cells cultured in Gluc− medium compared to cells cultured in complete or AA− medium (Fig. 8C). These data reinforce our previous finding (Fig. 7D) that cell culture conditions that inhibit mTORC1 and activate autophagy do not promote intracellular replication of C. burnetii. Collectively, our data show that potentiated cell catabolism in response to canonical inhibition of mTORC1 does not benefit C. burnetii intracellular growth.
FIG 8.
Culture of THP-1 macrophages without glucose attenuates C. burnetii-mediated inhibition of mTORC1 and impairs bacterial replication. (A) Immunoblot of lysates from THP-1 macrophages left uninfected (UI) or infected with wild-type C. burnetii (WT) or the ΔdotA mutant for 72 h in Comp, AA−, or Gluc− medium and probed with antibodies against p4E-BP1 or actin. (B) Quantitation of p4E-BP1 signal in panel A. The plot depicts means ± standard deviations of p4E-BP1 signal normalized to the actin loading control relative to uninfected cells for three independent experiments. (C) Replication of C. burnetii in THP-1 macrophages incubated in Comp, AA−, or Gluc− medium. The plots depict means ± standard deviations of the fold change in bacterial genome equivalents (GE) relative to the mean at 4 hpi for three independent experiments. Asterisks indicate statistical significance (***, P < 0.001; ****, P < 0.0001).
CCV size is increased in cells cultured without glucose. Data represent results of quantitation of CCV size in infected HeLa cells incubated in Comp, AA−, or Gluc− medium. The plot depicts means CCV sizes ± standard deviations (n = >100 cells) at 72 hpi. Asterisks indicate statistical significance (***, P < 0.001). Download FIG S9, PDF file, 0.3 MB (349.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Hyperactivation of mTORC1 by disruption of TSC inhibits C. burnetii growth.
TSC integrates signals from a variety of environmental stimuli, including glucose, nutrients, growth factors, hypoxia, and cell stress. Conditions restrictive for cell growth activate TSC, which leads to the inhibition of mTORC1 (18). Knockdown of TSC1 or TSC2 by RNA interference (RNAi) hyperactivates mTORC1, making it resistant to inhibition by glucose or growth factor withdrawal (44, 45). Previous experiments demonstrated that starvation-induced inhibition of mTORC1 that leads to autophagy does not improve C. burnetii growth. However, inhibition of mTORC1 by C. burnetii is distinct from the canonical catabolic response to cell starvation. To determine if the noncanonical response to C. burnetii-mediated inhibition of mTORC1 is important for intracellular growth, we used TSC knockdown to hyperactivate mTORC1 and override C. burnetii inhibition during infection. HeLa cells were treated with TSC1, TSC2, or nontargeting small interfering RNA (siRNA) for 48 h and then infected with C. burnetii. TSC1 or TSC2 levels were depleted throughout the course of infection (Fig. 9A). At 4 and 24 hpi, comparable numbers of bacteria were detected in the siRNA-treated cells (Fig. 9C), indicating that TSC knockdown did not inhibit bacterial internalization. However, at 72 hpi, C. burnetii replication was reduced ∼30% in cells treated with TSC1 or TSC2 siRNA compared to control cells. The defect in bacterial replication correlated with decreased CCV size (Fig. 9D) and increased phosphorylation of 4E-BP1 at 72 hpi (Fig. 9B). mTORC1 is also regulated by amino acids via the Rag GTPases. However, expression of active or inactive forms of RagB (22) did not affect C. burnetii replication (data not shown), further reinforcing the idea that C. burnetii modulates mTORC1 activity independently of amino acid signaling. An overview of host cell responses linked to mTORC1 inhibition by C. burnetii is shown in Fig. S10.
FIG 9.
Hyperactivation of mTORC1 by disruption of TSC inhibits C. burnetii growth. (A) Immunoblots of lysates from infected HeLa cells treated with TSC1, TSC2, or nontargeting (NT) siRNA probed with antibodies against TSC1, TSC2, p4E-BP1, or actin. (B) Quantitation of p4E-BP1 signal in panel A. The plot depicts means ± standard deviations of p4E-BP1 signal normalized to the actin loading control relative to NT-treated cells at 72 hpi for three independent experiments. (C) Replication of C. burnetii in HeLa cells treated with siRNA in panel A. The plots depict means ± standard deviations of fold change in C. burnetii genome equivalents (GE) relative to the mean at 4 hpi for three independent experiments. (D) Quantitation of CCV size in HeLa cells at 72 hpi treated with siRNA as described for panel A (n = >100 cells). Data are representative of results from three independent experiments. Asterisks indicate statistical significance (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
C. burnetii inhibition of mTORC1 triggers a noncanonical response by host cells. The table summarizes host cell responses linked to mTORC1 activation (green) or inhibition (red) under conditions of culture in nutrient-replete or nutrient-deficient medium or infection with C. burnetii. Specific inhibition of mTORC1 by C. burnetii is predicted to promote pathogen replication within the lysosomal CCV. Download FIG S10, PDF file, 0.4 MB (403.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
DISCUSSION
This study was conducted to investigate if C. burnetii modulates lysosomal physiology to generate a vacuole that supports pathogen replication. mTORC1 is a master kinase that regulates lysosome biosynthesis and fusion, as well as trafficking of cargo destined for degradation. When nutrients are replete, mTORC1 is active on the lysosomal surface, where it promotes protein synthesis by phosphorylating 4E-BP1 and S6K1. When nutrients are deficient, mTORC1 is inhibited and localizes to the cell cytoplasm (14, 18, 19). Infection of cells with C. burnetii in complete medium decreases localization of mTORC1 to the surface of lysosomal organelles and reduces the phosphorylation of 4E-BP1 and S6K1. Importantly, infection with a ΔdotA mutant does not inhibit phosphorylation. Simultaneous infection and starvation of amino acids synergistically inhibit localization of mTORC1 to lysosomes and the phosphorylation of 4E-BP1, and inhibition by C. burnetii is maintained when nutrients are replenished. Together, these data demonstrate that C. burnetii inhibits the activity of mTORC1 in a T4BSS-dependent manner.
Starvation-induced inhibition of mTORC1 disrupts cell biosynthesis, which, along with increased breakdown of cellular components, results in decreased cell size and cell cycle arrest (38). A noncanonical response to suppression of mTORC1 during C. burnetii infection is indicated by deficient activation of autophagy and by the observation that infection does not decrease host cell size or impair host cell replication (39). CCV size and fusogenicity are increased when cells infected with wild-type bacteria are incubated in AA− or Gluc− medium, or with the inhibitor Torin-1, all of which increase cell catabolism via autophagy (19, 23, 24). Infection with either wild-type C. burnetii or the ΔdotA mutant stimulated nuclear translocation of TFE3, indicating that activation of TFE3 is not a direct result of T4BSS-dependent inhibition of mTORC1 by C. burnetii. Instead, activation may result from immune detection of the bacterium via Toll-like receptors (46). Macrophages that express TFE3/B contain larger CCVs than macrophages lacking TFE3/B. The increased CCV size is likely due to greater trafficking of membrane and cargo to the compartment along with increased synthesis of lysosomal components. Primarily on the basis of increased CCV size and fusogenicity, previous studies have concluded that autophagy delivers membrane and nutrients to the CCV, which promotes C. burnetii intracellular growth (25, 31, 32). However, our direct measurement of bacterial genomes shows that C. burnetii replication does not increase when autophagy is activated by amino acid or glucose starvation or by treatment with Torin-1. Furthermore, macrophages lacking TFE3/B are more permissive for C. burnetii replication. Thus, although autophagy and TFE3/B benefit CCV expansion, the resultant increase in cell catabolism does not enhance C. burnetii growth.
Activation of autophagy is evidenced by lipidation of LC3, autophagosome biogenesis, and flux of cargo that is degraded in lysosomes. Autophagic flux is gauged by measuring changes in LC3 and p62 levels over time (24). We found that infected cells contain more LC3 and p62 than uninfected cells when cultured in complete medium. When autophagy is activated by starvation, infected cells readily degrade LC3 and p62 but maintain greater amounts of LC3 and p62 than uninfected cells treated in parallel. On the basis of the elevated amounts of p62 in infected cells seen both before and after starvation, previous studies concluded that C. burnetii inhibits degradation of p62 (28, 32, 33). By quantitating changes in LC3 and p62 at multiple time points after autophagy was induced by starvation, we show that infected cells degrade greater amounts of LC3 and p62 over time than uninfected cells. We therefore conclude that infected cells maintain elevated amounts of LC3 and p62 without inhibiting degradation by autophagy. It has been proposed that C. burnetii initiates autophagy without increased autophagic flux based on the respective observations that LC3 is lipidated and p62 accumulates in infected cells (32). However, there is no direct mechanistic evidence linking LC3 lipidation with the activation of autophagy (24). Moreover, autophagic flux can occur in the absence of LC3 lipidation (47). The non-autophagy-related functions of LC3 include stabilization of large phagosomal vacuoles and lysosomal tubules (48, 49). The involvement of LC3 in the formation of large vacuoles, along with the observation that C. burnetii does not activate autophagic flux, highlights the possibility that non-autophagy-related processes may account for the accumulation of LC3 and p62 during C. burnetii infection. Indeed, a recent report suggested that p62 has a cytoprotective role during C. burnetii infection (33).
Inhibition of the Rag GTPases or activation of TSC is sufficient to inhibit mTORC1 activity (22, 44, 45). C. burnetii is auxotrophic for several amino acids, including arginine and leucine (36), which are known to stimulate activation of mTORC1 via the Rag GTPases (18). Sequestration of host cell amino acids by C. burnetii represents a possible mechanism of mTORC1 inhibition. However, accelerated autophagic flux triggered by amino acid starvation does not occur in response to C. burnetii infection. Infection and amino acid starvation exert additive suppressive effects on mTORC1 that are partially rescued by the addition of medium replete with amino acids. This suggests that amino acids and C. burnetii regulate mTORC1 by separate mechanisms, a conclusion that is supported by the observation that C. burnetii intracellular growth is unaffected by expression of constitutively active RagB GTPase (22).
TSC knockdown blocks C. burnetii-mediated inhibition of mTORC1 and impairs pathogen growth. These findings demonstrate that TSC is required for inhibition of mTORC1 by C. burnetii and for optimal bacterial replication. TSC activity is regulated by Akt kinase, extracellular signal-regulated kinase 1/2 (ERK1/2), and AMP kinase (AMPK) signal transduction pathways (18 – 20). When nutrients and growth factors are replete, Akt and ERK1/2 directly phosphorylate and inactivate TSC, thereby promoting mTORC1 activation (18 – 20). Cells infected with C. burnetii exhibit sustained activation of Akt and Erk1/2 (50), indicating that Akt or Erk1/2 signaling via TSC does not promote mTORC1 inhibition during C. burnetii infection. Glucose deprivation activates TSC via AMPK, leading to mTORC1 inhibition (18, 44). However, in pilot experiments, we detected no changes in the phosphorylation of TSC or AMPK during C. burnetii infection (data not shown). Further investigation of signaling pathways that regulate the activity of TSC and mTORC1 during infection will benefit from the identification of the T4BSS effector(s) that targets mTORC1 regulatory mechanisms.
Inhibition of mTORC1 promotes lysosome clustering and fusion, leading to the formation of large lysosomal organelles, such as phagolysosomes and autophagolysosomes. After prolonged starvation, amino acids released by autophagy lead to the reactivation of mTORC1, which regulates recycling of membrane and other components away from lysosomal organelles and the reformation of smaller, discrete primary lysosomes (15 – 17). Reactivation of mTORC1 during prolonged starvation also results in a cessation of autophagy that is predicted to prevent cell death from excessive catabolism (15, 17, 19, 51). Cumulative evidence indicates that inhibition of mTORC1 by C. burnetii results in increased lysosomal fusion that supports CCV biogenesis without amplified cell catabolism or suppression of cell proliferation. Thus, we propose that noncanonical inhibition of mTORC1 by C. burnetii promotes CCV expansion along with metabolic homeostasis that ensures host cell survival during C. burnetii’s lengthy infectious cycle.
MATERIALS AND METHODS
Bacterial and mammalian cell culture.
C. burnetii Nine Mile RSA439 (phase II, clone 4) was cultivated in ACCM-D as previously described (36). HeLa (CCL-2; American Type Culture Collection [ATCC]) human cervical epithelial cells and RAW 264.7 mouse macrophages were cultured in Dulbecco modified Eagle medium (DMEM) (ThermoFisher) containing 10% fetal bovine serum (FBS) (ThermoFisher) at 37°C and 5% CO2. Clustered regularly interspaced short palindromic repeat (CRISPR)-generated TFE3/B double-knockout and control RAW 264.7 macrophages were maintained in DMEM plus 10% FBS and 5 μg/ml puromycin and were a kind gift from the laboratory of Rosa Puertollano (23). RAW 264.7 macrophages were cultured for 48 h without selection prior to infection. THP-1 (TIB-202; ATCC) human monocytic cells were maintained in RPMI 1640 medium (ThermoFisher) containing 10% FBS at 37°C and 5% CO2 and differentiated into macrophage-like cells by overnight treatment with 200 nM phorbol-12-myristate-13-acetate (PMA). PBMCs isolated as previously described (52) from blood obtained from normal healthy donors (institutional review board [IRB]-approved protocol 99-CC-0168) (ClinicalTrials registration no. NCT00001846, National Institutes of Health, Bethesda, MD, USA) were cultured in RPMI 1640 plus 100 ng/ml recombinant human macrophage colony-stimulating factor (M-CSF; PeproTech, Rocky Hill, NJ), 5% human serum, 1 mM sodium pyruvate, nonessential amino acids, 10 mM HEPES buffer, and 2 mM glutamate (ThermoFisher).
Cell infection.
THP-1 cells (5 × 105 per well), HeLa cells (2.5 × 104 per well), PBMCs (2.5 × 105 per well), or RAW 264.7 macrophages (4 × 104 per well) in a 24-well plate were infected with C. burnetii in RPMI medium by centrifugation of plates at 500 × g for 30 min using a multiplicity of infection of approximately 0.1, 25, or 100 for experimental analysis by quantitative PCR (qPCR), immunofluorescence, or immunoblotting, respectively. In all cell types used in this study, the time points of 4 and 24 hpi correspond to the early and late lag phases of growth, respectively. The time points of 48 and 72 hpi correspond to the early and mid-log phases of growth, respectively. Cells were infected with 10-fold or 50-fold more of the ΔdotA mutant than wild-type C. burnetii (11). Cells were washed once with RPMI medium and incubated at 37°C in 5% CO2. Cells were incubated with 500 μl of growth media as follows: complete medium, RPMI 1640 plus 10% FBS (HyClone, GE Healthcare, Pittsburgh, PA); AA− medium, RPMI 1640 without amino acids (US Biologicals, Salem, MA) plus 2 mg/ml glucose; Torin-1 medium, RPMI 1640 plus 10% FBS and 250 nM Torin-1 (Cell Signaling, Danvers, MA); Gluc− medium, RPMI 1640 without glucose (US Biologicals) plus 10% dialyzed serum (ThermoFisher) and 1× nonessential and essential amino acids (Sigma-Aldrich).
Immunoblot and qPCR.
Cells lysed in 75 μl Laemmli sample buffer and boiled for 5 min were separated by SDS-PAGE using 4% to 12% Criterion XT Bis-Tris precast gels (Bio-Rad, Hercules, CA) and were transferred to an Immobilon-P polyvinylidene difluoride membrane (MilliporeSigma, Burlington, MA). Membranes were blocked in Odyssey blocking buffer (Li-Cor Biotechnology, Lincoln, NE) and incubated with the indicated antibodies in Tris-buffered saline containing 0.1% Tween 20 (TBST). Immunoblots were washed extensively in TBST, incubated with SuperSignal West Femto maximum sensitivity substrate (ThermoFisher), and analyzed on an Azure c600 imaging system (Azure Biosystems, Dublin, CA). Immunoreactive bands were quantitated using Image Studio Lite (Li-Cor Biotechnology). C. burnetii genomic equivalents were determined by qPCR analysis of cells lysed and boiled in 0.5 ml of buffer containing 0.05% trypsin, 0.5 mM EDTA, and 20 mM Tris-HCl (pH 8) as previously described (11).
Immunofluorescence microscopy and image quantitation.
Cells seeded on glass coverslips, glass-bottom plates (Greiner Bio-One, Monroe, NC), or plastic-bottom plates (Ibidi, Fitchburg, WI) were fixed for 30 min in phosphate-buffered saline (PBS) containing 4% paraformaldehyde, washed in PBS, and then permeabilized with PBS containing 0.05% saponin and 5% FBS or with PBS containing 0.1% Triton X-100 and 5% FBS. Cells were stained as indicated. Images were collected using a Nikon Eclipse Ti2 inverted epifluorescence microscope fitted with a Nikon DS-Qi2 camera (Nikon Instruments Inc.) and a Lumencor Sola Pad excitation source (Lumencor, Beaverton, OR) or using a LSM710 confocal laser scanning microscope (Carl Zeiss Micro Imaging, Thornwood, NY). The ImageJ selection tool was used to measure CCV size and quantity. Automated image segmentation and analysis with CellProfiler were used to quantitate signal intensity and cell dimensions as previously described (41).
Antibodies and cell stains.
The suppliers and antibodies used were as follows: from Abcam, ab24170 and ab25630 (LAMP1) and ab8224 (actin); from Santa Cruz, 19992 (LAMP1); from Cell Signaling, 9644 (4E-BP1), 2855 (4E-BP1 Thr37/46), 9234 (p70 S6 kinase 1 Thr389), 2775 (LC3B), 6935 (TSC1), 4308 (TSC2), and 2983 (mTOR); from BD Biosciences, 610833 (p62) and 556019 (CD63); from Sigma-Aldrich, HPA023881 (TFE3); from ThermoFisher, Alexa Fluor 568 donkey anti-mouse (A10037) and donkey anti-rabbit (A10042), Alexa Fluor 488 goat anti-mouse (A11029) and goat anti-rabbit (A11034), Hoescht 33342 (62249), HCS CellMask Deep Red stain (H32721), guinea pig anti-C. burnetii serum (11), and rabbit anti-C. burnetii EF-Ts (elongation factor thermostable) serum (53).
Cell transfection.
ON-TARGETplus SMARTpool siRNA duplexes against TSC1 and TSC2, as well as nontargeting siRNA, were obtained from Dharmacon. DharmaFECT1 (Dharmacon) was used to transfect HeLa cells (2 × 104 cells per well) in a 24-well plate with siRNA duplexes according to the manufacturer’s instructions. Knockdown of proteins by targeting siRNAs was confirmed at 48 h posttransfection by immunoblotting, and infections were performed with C. burnetii as previously described (6). Infected HeLa cells were transfected with Flag pLJM1 RagB 99L and Flag pLJM1 RagB 54L (Addgene; plasmids 19314 and 19315), which were a kind gift from David Sabatini (22), as previously described (7).
Statistical analysis.
Statistical analyses were conducted using Prism software (GraphPad Software, Inc.) to perform the unpaired Student's t test or one-way analysis of variance (ANOVA) using Tukey’s posttest.
ACKNOWLEDGMENTS
We thank Carrie M. Long for assistance with the isolation of human macrophages.
This work was supported by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Disease.
Footnotes
Citation Larson CL, Sandoz KM, Cockrell DC, Heinzen RA. 2019. Noncanonical inhibition of mTORC1 by Coxiella burnetii promotes replication within a phagolysosome-like vacuole. mBio 10:e02816-18. https://doi.org/10.1128/mBio.02816-18.
Contributor Information
Jimmy D. Ballard, University of Oklahoma Health Sciences Center.
Yasuko Rikihisa, Ohio State University.
Jason Carlyon, Virginia Commonwealth University School of Medicine.
REFERENCES
- 1.Raoult D, Marrie T, Mege J. 2005. Natural history and pathophysiology of Q fever. Lancet Infect Dis 5:219–226. doi: 10.1016/S1473-3099(05)70052-9. [DOI] [PubMed] [Google Scholar]
- 2.Graham JG, MacDonald LJ, Hussain SK, Sharma UM, Kurten RC, Voth DE. 2013. Virulent Coxiella burnetii pathotypes productively infect primary human alveolar macrophages. Cell Microbiol 15:1012–1025. doi: 10.1111/cmi.12096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beron W, Gutierrez MG, Rabinovitch M, Colombo MI. 2002. Coxiella burnetii localizes in a Rab7-labeled compartment with autophagic characteristics. Infect Immun 70:5816–5821. doi: 10.1128/IAI.70.10.5816-5821.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Howe D, Shannon JG, Winfree S, Dorward DW, Heinzen RA. 2010. Coxiella burnetii phase I and II variants replicate with similar kinetics in degradative phagolysosome-like compartments of human macrophages. Infect Immun 78:3465–3474. doi: 10.1128/IAI.00406-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larson CL, Martinez E, Beare PA, Jeffrey B, Heinzen RA, Bonazzi M. 2016. Right on Q: genetics begin to unravel Coxiella burnetii host cell interactions. Future Microbiol 11:919–939. doi: 10.2217/fmb-2016-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Larson CL, Beare PA, Howe D, Heinzen RA. 2013. Coxiella burnetii effector protein subverts clathrin-mediated vesicular trafficking for pathogen vacuole biogenesis. Proc Natl Acad Sci U S A 110:E4770–E4779. doi: 10.1073/pnas.1309195110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Larson CL, Beare PA, Voth DE, Howe D, Cockrell DC, Bastidas RJ, Valdivia RH, Heinzen RA. 2015. Coxiella burnetii effector proteins that localize to the parasitophorous vacuole membrane promote intracellular replication. Infect Immun 83:661–670. doi: 10.1128/IAI.02763-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber MM, Chen C, Rowin K, Mertens K, Galvan G, Zhi H, Dealing CM, Roman VA, Banga S, Tan Y, Luo ZQ, Samuel JE. 2013. Identification of Coxiella burnetii type IV secretion substrates required for intracellular replication and Coxiella-containing vacuole formation. J Bacteriol 195:3914–3924. doi: 10.1128/JB.00071-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Newton HJ, Kohler LJ, McDonough JA, Temoche-Diaz M, Crabill E, Hartland EL, Roy CR. 2014. A screen of Coxiella burnetii mutants reveals important roles for Dot/Icm effectors and host autophagy in vacuole biogenesis. PLoS Pathog 10:e1004286. doi: 10.1371/journal.ppat.1004286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martinez E, Cantet F, Fava L, Norville I, Bonazzi M. 2014. Identification of OmpA, a Coxiella burnetii protein involved in host cell invasion, by multi-phenotypic high-content screening. PLoS Pathog 10:e1004013. doi: 10.1371/journal.ppat.1004013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beare PA, Gilk SD, Larson CL, Hill J, Stead CM, Omsland A, Cockrell DC, Howe D, Voth DE, Heinzen RA. 2011. Dot/Icm type IVB secretion system requirements for Coxiella burnetii growth in human macrophages. mBio 2:e00175. doi: 10.1128/mBio.00175-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carey KL, Newton HJ, Lührmann A, Roy CR. 2011. The Coxiella burnetii Dot/Icm system delivers a unique repertoire of type IV effectors into host cells and is required for intracellular replication. PLoS Pathog 7:e1002056. doi: 10.1371/journal.ppat.1002056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Newton HJ, McDonough JA, Roy CR. 2013. Effector protein translocation by the Coxiella burnetii Dot/Icm type IV secretion system requires endocytic maturation of the pathogen-occupied vacuole. PLoS One 8:e54566. doi: 10.1371/journal.pone.0054566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luzio JP, Pryor PR, Bright NA. 2007. Lysosomes: fusion and function. Nat Rev Mol Cell Biol 8:622–632. doi: 10.1038/nrm2217. [DOI] [PubMed] [Google Scholar]
- 15.Yu L, McPhee CK, Zheng L, Mardones GA, Rong Y, Peng J, Mi N, Zhao Y, Liu Z, Wan F, Hailey DW, Oorschot V, Klumperman J, Baehrecke EH, Lenardo MJ. 2010. Termination of autophagy and reformation of lysosomes regulated by mTOR. Nature 465:942–946. doi: 10.1038/nature09076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rong Y, Liu M, Ma L, Du W, Zhang H, Tian Y, Cao Z, Li Y, Ren H, Zhang C, Li L, Chen S, Xi J, Yu L. 2012. Clathrin and phosphatidylinositol-4,5-bisphosphate regulate autophagic lysosome reformation. Nat Cell Biol 14:924–934. doi: 10.1038/ncb2557. [DOI] [PubMed] [Google Scholar]
- 17.Chen Y, Yu L. 2013. Autophagic lysosome reformation. Exp Cell Res 319:142–146. doi: 10.1016/j.yexcr.2012.09.004. [DOI] [PubMed] [Google Scholar]
- 18.Saxton RA, Sabatini DM. 2017. mTOR signaling in growth, metabolism, and disease. Cell 169:361–371. doi: 10.1016/j.cell.2017.03.035. [DOI] [PubMed] [Google Scholar]
- 19.Puertollano R. 2014. mTOR and lysosome regulation. F1000Prime Rep 6:52. doi: 10.12703/P6-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laplante M, Sabatini DM. 2012. mTOR signaling in growth control and disease. Cell 149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bar-Peled L, Schweitzer LD, Zoncu R, Sabatini DM. 2012. Ragulator is a GEF for the rag GTPases that signal amino acid levels to mTORC1. Cell 150:1196–1208. doi: 10.1016/j.cell.2012.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sancak Y, Peterson TR, Shaul YD, Lindquist RA, Thoreen CC, Bar-Peled L, Sabatini DM. 2008. The Rag GTPases bind raptor and mediate amino acid signaling to mTORC1. Science 320:1496–1501. doi: 10.1126/science.1157535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Martina JA, Diab HI, Lishu L, Jeong AL, Patange S, Raben N, Puertollano R. 2014. The nutrient-responsive transcription factor TFE3 promotes autophagy, lysosomal biogenesis, and clearance of cellular debris. Sci Signal 7:ra9. doi: 10.1126/scisignal.2004754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, Adhihetty PJ, Adler SG, Agam G, Agarwal R, Aghi MK, Agnello M, Agostinis P, Aguilar PV, Aguirre-Ghiso J, Airoldi EM, Ait-Si-Ali S, Akematsu T, Akporiaye ET, Al-Rubeai M, Albaiceta GM, Albanese C, Albani D, Albert ML, Aldudo J, Algül H, Alirezaei M, Alloza I, Almasan A, Almonte-Beceril M, Alnemri ES, Alonso C, Altan-Bonnet N, Altieri DC, Alvarez S, Alvarez-Erviti L, Alves S, Amadoro G, Amano A, Amantini C, Ambrosio S, Amelio I, Amer AO, Amessou M, Amon A, An Z, et al. 2016. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12:1–222. doi: 10.1080/15548627.2015.1100356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gutierrez MG, Vazquez CL, Munafo DB, Zoppino FC, Beron W, Rabinovitch M, Colombo MI. 2005. Autophagy induction favours the generation and maturation of the Coxiella-replicative vacuoles. Cell Microbiol 7:981–993. doi: 10.1111/j.1462-5822.2005.00527.x. [DOI] [PubMed] [Google Scholar]
- 26.Kohler LJ, Reed SR, Sarraf SA, Arteaga DD, Newton HJ, Roy CR. 2016. Effector protein Cig2 decreases host tolerance of infection by directing constitutive fusion of autophagosomes with the Coxiella-containing vacuole. mBio 7:e011127-16. doi: 10.1128/mBio.01127-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Romano PS, Gutierrez MG, Beron W, Rabinovitch M, Colombo MI. 2007. The autophagic pathway is actively modulated by phase II Coxiella burnetii to efficiently replicate in the host cell. Cell Microbiol 9:891–909. doi: 10.1111/j.1462-5822.2006.00838.x. [DOI] [PubMed] [Google Scholar]
- 28.Winchell CG, Graham JG, Kurten RC, Voth DE. 2014. Coxiella burnetii type IV secretion-dependent recruitment of macrophage autophagosomes. Infect Immun 82:2229–2238. doi: 10.1128/IAI.01236-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vazquez CL, Colombo MI. 2010. Coxiella burnetii modulates Beclin 1 and Bcl-2, preventing host cell apoptosis to generate a persistent bacterial infection. Cell Death Differ 17:421–438. doi: 10.1038/cdd.2009.129. [DOI] [PubMed] [Google Scholar]
- 30.Martinez E, Allombert J, Cantet F, Lakhani A, Yandrapalli N, Neyret A, Norville IH, Favard C, Muriaux D, Bonazzi M. 2016. Coxiella burnetii effector CvpB modulates phosphoinositide metabolism for optimal vacuole development. Proc Natl Acad Sci U S A 113:E3260–E3269. doi: 10.1073/pnas.1522811113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gutierrez MG, Colombo MI. 2005. Autophagosomes: a fast-food joint for unexpected guests. Autophagy 1:179–181. [DOI] [PubMed] [Google Scholar]
- 32.Latomanski EA, Newton HJ. 2018. Interaction between autophagic vesicles and the Coxiella-containing vacuole requires CLTC (clathrin heavy chain). Autophagy 14:1710–1725. doi: 10.1080/15548627.2018.1483806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Winchell CG, Dragan AL, Brann KR, Onyilagha FI, Kurten RC, Voth DE. 2018. Coxiella burnetii subverts p62/sequestosome 1 and activates Nrf2 signaling in human macrophages. Infect Immun 86:e00608-17. doi: 10.1128/IAI.00608-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dall'Armi C, Devereaux KA, Di Paolo G. 2013. The role of lipids in the control of autophagy. Curr Biol 23:R33–R45. doi: 10.1016/j.cub.2012.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jean S, Kiger AA. 2014. Classes of phosphoinositide 3-kinases at a glance. J Cell Sci 127:923–928. doi: 10.1242/jcs.093773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sandoz KM, Beare PA, Cockrell DC, Heinzen RA. 2016. Complementation of arginine auxotrophy for genetic transformation of Coxiella burnetii by use of a defined axenic medium. Appl Environ Microbiol 82:3042–3051. doi: 10.1128/AEM.00261-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang Y, Nicholatos J, Dreier JR, Ricoult SJ, Widenmaier SB, Hotamisligil GS, Kwiatkowski DJ, Manning BD. 2014. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature 513:440–443. doi: 10.1038/nature13492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang RC, Levine B. 2010. Autophagy in cellular growth control. FEBS Lett 584:1417–1426. doi: 10.1016/j.febslet.2010.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baca OG, Crissman HA. 1987. Correlation of DNA, RNA, and protein content by flow cytometry in normal and Coxiella burnetii-infected L929 cells. Infect Immun 55:1731–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou J, Tan SH, Nicolas V, Bauvy C, Yang ND, Zhang J, Xue Y, Codogno P, Shen HM. 2013. Activation of lysosomal function in the course of autophagy via mTORC1 suppression and autophagosome-lysosome fusion. Cell Res 23:508–523. doi: 10.1038/cr.2013.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Larson CL, Heinzen RA. 2017. High-content imaging reveals expansion of the endosomal compartment during Coxiella burnetii parasitophorous vacuole maturation. Front Cell Infect Microbiol 7:48. doi: 10.3389/fcimb.2017.00048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schulze-Luehrmann J, Eckart RA, Ölke M, Saftig P, Liebler-Tenorio E, Lührmann A. 2016. LAMP proteins account for the maturation delay during the establishment of the Coxiella burnetii-containing vacuole. Cell Microbiol 18:181–194. doi: 10.1111/cmi.12494. [DOI] [PubMed] [Google Scholar]
- 43.Kuley R, Bossers-deVries R, Smith HE, Smits MA, Roest HI, Bossers A. 2015. Major differential gene regulation in Coxiella burnetii between in vivo and in vitro cultivation models. BMC Genomics 16:953. doi: 10.1186/s12864-015-2143-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Inoki K, Zhu T, Guan KL. 2003. TSC2 mediates cellular energy response to control cell growth and survival. Cell 115:577–590. [DOI] [PubMed] [Google Scholar]
- 45.Lee CH, Inoki K, Karbowniczek M, Petroulakis E, Sonenberg N, Henske EP, Guan KL. 2007. Constitutive mTOR activation in TSC mutants sensitizes cells to energy starvation and genomic damage via p53. EMBO J 26:4812–4823. doi: 10.1038/sj.emboj.7601900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pastore N, Brady OA, Diab HI, Martina JA, Sun L, Huynh T, Lim JA, Zare H, Raben N, Ballabio A, Puertollano R. 2016. TFEB and TFE3 cooperate in the regulation of the innate immune response in activated macrophages. Autophagy 12:1240–1258. doi: 10.1080/15548627.2016.1179405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nishida Y, Arakawa S, Fujitani K, Yamaguchi H, Mizuta T, Kanaseki T, Komatsu M, Otsu K, Tsujimoto Y, Shimizu S. 2009. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 461:654–658. doi: 10.1038/nature08455. [DOI] [PubMed] [Google Scholar]
- 48.Florey O, Kim SE, Sandoval CP, Haynes CM, Overholtzer M. 2011. Autophagy machinery mediates macroendocytic processing and entotic cell death by targeting single membranes. Nat Cell Biol 13:1335–1343. doi: 10.1038/ncb2363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanjuan MA, Dillon CP, Tait SW, Moshiach S, Dorsey F, Connell S, Komatsu M, Tanaka K, Cleveland JL, Withoff S, Green DR. 2007. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature 450:1253–1257. doi: 10.1038/nature06421. [DOI] [PubMed] [Google Scholar]
- 50.Voth DE, Heinzen RA. 2009. Sustained activation of Akt and Erk1/2 is required for Coxiella burnetii antiapoptotic activity. Infect Immun 77:205–213. doi: 10.1128/IAI.01124-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Munson MJ, Allen GF, Toth R, Campbell DG, Lucocq JM, Ganley IG. 2015. mTOR activates the VPS34-UVRAG complex to regulate autolysosomal tubulation and cell survival. EMBO J 34:2272–2290. doi: 10.15252/embj.201590992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fuss IJ, Kanof ME, Smith PD, Zola H. 2009. Isolation of whole mononuclear cells from peripheral blood and cord blood. Curr Protoc Immunol Chapter 7:Unit 7.1. doi: 10.1002/0471142735.im0701s85. [DOI] [PubMed] [Google Scholar]
- 53.Stead CM, Omsland A, Beare PA, Sandoz KM, Heinzen RA. 2013. Sec-mediated secretion by Coxiella burnetii. BMC Microbiol 13:222. doi: 10.1186/1471-2180-13-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
C. burnetii inhibition of mTOR endolysosomal localization is T4BSS dependent. Data represent subcellular localization of mTOR in HeLa cells left uninfected (UI) or infected with wild-type C. burnetii (WT) or the ΔdotA mutant for 72 h (A) or 120 h (B) in complete medium. The plot depicts means ± standard deviations of data representing the ratio of lysosomal (CD63+) mTOR signal to cytosolic (lys/cyto) mTOR signal (n = >100 cells). Data are representative of results from three independent experiments. Asterisks indicate statistical significance (*, P < 0.05). Download FIG S1, PDF file, 0.5 MB (557.5KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
The viability of THP-1 macrophages infected with C. burnetii does not decrease in mTORC1-inhibiting growth media. (A) Representative fluorescence micrographs of THP-1 macrophages infected with wild-type C. burnetii (WT) or the ΔdotA mutant at a multiplicity of infection of 100 or 1,000, respectively, for 72 h in complete medium. DNA, C. burnetii, and host cells were stained blue, green, and magenta, respectively. (B) Percent cytoxicity displayed by THP-1 macrophages infected for 72 h with C. burnetii in complete (Comp), AA−, Torin-1, or Gluc− medium. Percent cytotoxicity was determined using the CytoTox 96 Non-Radioactive Cytotoxicity assay (Promega, Madison, WI) according to the manufacturer’s instructions. The plot depicts means ± standard deviations of the percentage of cytotoxic cells detected within each of the indicated samples for three independent experiments. Scale bar, 20 µm. Download FIG S2, PDF file, 0.8 MB (825.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Infected HeLa cells exhibit reduced phosphorylation of 4E-BP1. Immunoblot of lysates from uninfected or C. burnetii-infected HeLa cells incubated for 4, 24, or 72 h in complete (Comp), AA−, or Torin-1 medium. Blots were probed with antibodies against phosphorylated 4E-BP1 Thr37/46 (p4E-BP1) or actin was performed. Download FIG S3, PDF file, 0.2 MB (215.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Addition of extra amino acids does not rescue reduced phosphorylation of 4E-BP1 in C. burnetii-infected THP-1 cells. (A) Immunoblot of lysates from infected or uninfected THP-1 macrophages incubated for 72 h in complete (Comp), AA−, or Torin-1 medium with or without additional incubation for 30 min in Comp medium plus additional essential and nonessential amino acids (+AA). Blots were probed with antibodies against phosphorylated 4E-BP1 Thr37/46 (p4E-BP1) or actin. (B) Quantitation of p4E-BP1 signal in panel A. The plot depicts means ± standard deviations of p4E-BP1 signal normalized to the actin loading control relative to respective control cells in complete medium for three independent experiments. Asterisks indicate statistical significance (*, P < 0.05; ***, P < 0.001). Download FIG S4, PDF file, 0.3 MB (258.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Increases in p4EB-BP1 levels are comparable in uninfected and infected cells when they are starved and then incubated with nutrient-rich medium. THP-1 macrophages left uninfected (UI) or infected with wild-type C. burnetii (WT) or the ΔdotA mutant were incubated for 24 h (top) or 72 h (bottom) in AA− medium followed by incubation with fresh complete medium for 15, 30, or 60 min. Immunoblots (Fig. 3A and B) were probed with antibodies against phosphorylated 4E-BP1 Thr37/46 (p4E-BP1) or actin. Plots depict means ± standard deviations with trendlines fitted by linear regression of p4E-BP1 signal normalized to the actin loading control for three independent experiments. Download FIG S5, PDF file, 0.5 MB (546.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Infected cells contain more LC3 and p62 than uninfected cells and exhibit robust autophagic flux when starved. (A) Immunoblot of lysates from infected or uninfected THP-1 macrophages incubated for 4, 24, or 72 h in complete, AA−, or Torin-1 medium probed with antibodies against LC3, p62, or actin. (B) Quantitation of LC3 (left) or p62 (right) signal in panel A. The plot depicts means ± standard deviations of signal normalized to the actin loading control relative to cells in complete medium at 72 h for three independent experiments. (C) LC3 (left) or p62 (right) degradation rates in HeLa cells left uninfected (UI) or infected with wild-type C. burnetii (WT) for 72 h in complete medium and then incubated for the indicated times with HBSS. Plots depict mean signal data ± standard deviations with trendlines fitted by linear regression for three independent experiments. (D) Immunoblot of lysates from HeLa cells left uninfected (UI) or infected with wild-type C. burnetii (WT) for 72 h in complete medium, then incubated for the indicated times with HBSS and probed with antibodies against LC3, p62, or actin. Asterisks indicate statistical significance (*, P < 0.05). Download FIG S6, PDF file, 0.5 MB (495.2KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
Infected HeLa cells are larger than uninfected cells. The plot depicts means ± standard deviations of the cell area occupied by HeLa cells left uninfected (UI) or infected with wild-type (WT) C. burnetii measured in three independent experiments (n = >10,000 cells measured). Cell area was quantitated using CellProfiler. Each of the three independent data sets was normalized by dividing by the mean area of respective uninfected cells. Asterisks indicate statistical significance (****, P < 0.0001). Download FIG S7, PDF file, 0.2 MB (181KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
C. burnetii infection triggers TFE3 translocation independently of T4BSS activity. Data represent results of quantitation of TFE3 subcellular localization in HeLa cells (A) or THP-1 macrophages (B) left uninfected (UI) or infected with wild-type C. burnetii (WT) or the ΔdotA mutant for 72 h in complete medium. The plots depict means ± standard deviations of the ratio of nuclear TFE3 signal to cytoplasmic TFE3 signal detected in cells (n = >25). Data are representative of results from three independent experiments. Asterisks indicate statistical significance (***, P < 0.001). Download FIG S8, PDF file, 0.3 MB (265.8KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
CCV size is increased in cells cultured without glucose. Data represent results of quantitation of CCV size in infected HeLa cells incubated in Comp, AA−, or Gluc− medium. The plot depicts means CCV sizes ± standard deviations (n = >100 cells) at 72 hpi. Asterisks indicate statistical significance (***, P < 0.001). Download FIG S9, PDF file, 0.3 MB (349.6KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.
C. burnetii inhibition of mTORC1 triggers a noncanonical response by host cells. The table summarizes host cell responses linked to mTORC1 activation (green) or inhibition (red) under conditions of culture in nutrient-replete or nutrient-deficient medium or infection with C. burnetii. Specific inhibition of mTORC1 by C. burnetii is predicted to promote pathogen replication within the lysosomal CCV. Download FIG S10, PDF file, 0.4 MB (403.4KB, pdf) .
This is a work of the U.S. Government and is not subject to copyright protection in the United States. Foreign copyrights may apply.