Abstract
Heroin use disorder (HUD) is a complex disease resulting from interactions among genetic and other factors (e.g., environmental factors). The mechanism of HUD development remains unknown. Newly developed network medicine tools provide a platform for exploring complex diseases at the system level. This study proposes that protein–protein interactions (PPIs), particularly those among proteins encoded by casual or susceptibility genes, are extremely crucial for HUD development. The giant component of our constructed PPI network comprised 111 nodes with 553 edges, including 16 proteins with large degree (k) or high betweenness centrality (BC), which were further identified as the backbone of the network. JUN with the largest degree was suggested to be central to the PPI network associated with HUD. Moreover, PCK1 with the highest BC and MAPK14 with the secondary largest degree and 9th highest BC might be involved in the development HUD and other substance diseases.
Introduction
Heroin was originally synthesized in the late nineteenth century. Abstaining from heroin use is difficult, and it leads to a high relapse rate among past users1,2. New heroin users easily become addicted to the drug, tend to have serious withdrawal symptoms, and finally develop heroin use disorder (HUD). It has recently become a serious problem in South and East Asia3–5, and heroin users have the highest mortality rate among all the users of addictive substances in Taiwan6.
Similar to the other addictive disorders, HUD is a complex disorder resulting from the interplay between the environmental factors and the genetic predisposing factors7–10. Studies have suggested that HUD is a polygenic disorder and identified various susceptibility genes contributing to HUD through different mechanisms at different stages of HUD development8,11,12. However, the pathogenesis remains unclear. Previous studies evaluated the genetic influence on HUD rather than gene expression at the protein level. In addition to the genetic influence engendered by DNA, all biochemical processes are controlled by proteins. We propose that protein–protein interactions (PPIs), particularly those among proteins encoded by the aforementioned casual or susceptibility genes, are essential for HUD development.
In this study, we used the relevant gene biosignatures as the seeds to construct the PPI network associated with the development of HUD. The regulatory pathways were explored through topological analysis of the PPI network for the further understanding of the HUD mechanism. In a PPI network, nodes represent proteins, and edges represent interactions13. According to graph theory, the topological structure of the PPI network provides basic and direct information regarding the network and is associated with biological functions14. The combination of the topological structure of the PPI network with the relevant biological knowledge provides a promising tool for understanding the biological mechanisms of species.
Recently, topological analyses have been applied to molecular networks including protein interaction networks13–19, gene regulatory networks20–22, and metabolic networks23–26. Connectivity degree (k), betweenness centrality (BC), closeness centrality (CC), eigenvector centrality (EC), and eccentricity are the fundamental measures of nodes in network theory27,28. In a PPI network, nodes with large degree, defined as hub proteins, are crucial proteins, because they might be corresponding to the disease-causing genes15,29, whereas nodes with high BC, defined as bottlenecks, tend to indicate essential genes since they can be analogized to heavily used intersection in major highways or bridges14,30,31. In this study, we mainly focused on the hubs or bottlenecks that were central to the PPI network, identified the proteins with large degree or high BC as the key proteins, and considered the sub-network of these key proteins as the backbone worth further investigating the signaling pathways involved in HUD development.
Drug addiction is a psychiatric disorder resulting in maladaptive neuroplasticity that underlies the development of compulsive drug seeking and vulnerability to relapse during periods of attempted abstinence32. MiRNAs are small non-coding RNAs (18–25 nucleotides) that post- transcriptionally modulated gene expression by either repressing translation or inducing degradation of mRNA33,34. Recent studies indicate that miRNAs are considered to be ‘master regulators’ of gene expression, and they control the translation of target mRNAs, thereby regulating critical aspects of neurogenesis, synaptic plasticity and neurological disorders35–38. Consequently, discovering miRNA-disease association makes an important contribution to understanding the pathogenesis of diseases as well as designing diagnostic and therapeutic approaches for diseases39–42.
Materials and Methods
The identification of the susceptibility genes associated with HUD
We have identified the susceptibility genes associated with HUD in our previous case-control studies43,44 that included 124 Han Chinese men from Taiwan as the cases fulfilling the diagnostic criteria of HUD according to the Diagnostic and Statistical Manual of Mental Disorders, 5th edition (DSM-5), and 124 demographically similar patients as the controls getting regular medical checkups at a local medical center. The details of the 248 participants and the investigating methods are in the previous studies43,44. Briefly, we recruited no controls with substance-related disorders or substance use disorders except nicotine, and no participants with the other psychiatric diagnoses such as neurodevelopmental disorders, schizophrenia spectrum disorders, bipolar-related disorders, depressive disorders, neurocognitive disorders, etc.
From each participant, a total of 8 mL of venous blood was collected to establish the lymphoblastoid cell lines (LCLs) for RNA extraction and real-time quantitative PCR (qPCR) analyses. The study protocol was approved by the Ethical Committee of Bali Psychiatric Center (approval number: IRB970609-03), the written informed consents were obtained from the participants after full explanation of the protocol, and we performed all methods in accordance with the relevant guidelines and regulations. The identified results of the susceptibility genes associated with HUD were AUTS2, CD74, CEBPB, CEBPG, ENO2, HAT1, IMPDH2, JUN, MBD1, PDK1, PRKCB, RASA1, RGS3 (listed in Table S1), and we considered them as the seed proteins to construct the PPI network associated with HUD.
The construction of the PPI network associated with HUD
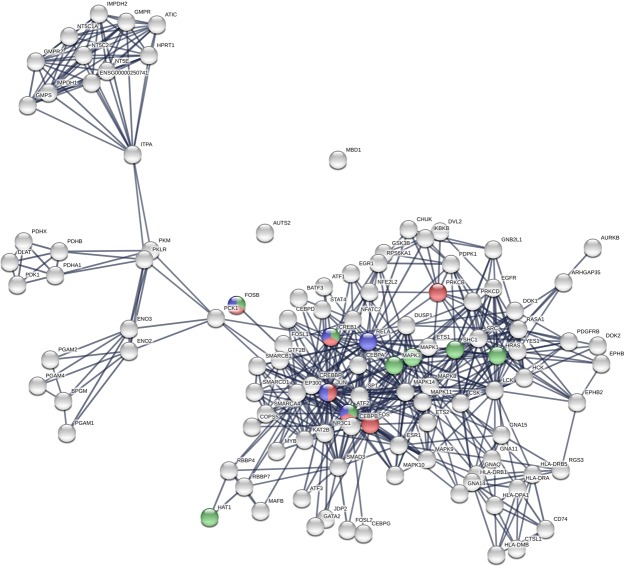
We used the Search Tool for the Retrieval of Interacting Genes/Proteins database (STRING v10.5)45 to construct the PPI network associated with HUD. Given a list of the proteins as input, STRING can search for their neighbor interactors, the proteins that have direct interactions with the inputted proteins; then STRING can generate the PPI network consisting of all these proteins and all the interactions between them. Based on the seed proteins as input, we first constructed the PPI network (Fig. 1) associated with HUD containing the seed proteins and their neighbors. All the interactions between them were derived from high-throughput lab experiments and previous knowledge in curated databases at high level of confidence (sources: experiments, databases; score ≥ 0.90).
Figure 1.
The PPI network.
We further searched for the interactions derived from three sources, lab experiments, curated databases, and gene expression data, with the same confidence to construct the PPI network with the co-expression interactions (Fig. S1) for comparison. In addition, Gephi46, a program for large network analysis, was used to analyze the structure of the PPI networks.
The topological analysis of the PPI network
To evaluate the nodes in the PPI networks, we adopted several topological measures including degree (k), between centrality (BC), eccentricity, closeness centrality (CC), eigenvector centrality (EC), and clustering coefficient47,48. The first two measures, degree (k) and BC, are often used for detecting the hub or bottleneck in a network. Degree (k) of a node is defined as the number of edges linked to it. A node with high degree (k) denotes a hub having many neighbors. BC of a node is the proportion of the number of shortest paths passing through it to the number of all the shortest paths in the network, quantifying how often a node acts as a bridge along the shortest paths between two other nodes. A node with high BC has great influence on what flows in the network and has more control over the network. It can represent the bottleneck in the network.
Eccentricity and CC of a node are the measures of centrality in the network, defined as the maximum distance from the node to all other nodes and the inverse of the average length of the shortest paths between the node and all other nodes, respectively. A node with lower eccentricity or higher CC is closer to the other nodes and more central in the network. Moreover, the maximum eccentricity is the diameter of a network; the minimum eccentricity is the radius of a network. The center of a network is the set of nodes of eccentricity equal to the radius.
EC assigns relative score to all nodes in the network based on the concept that connections to high-scoring nodes contribute more to the score of the node in question than equal connections to low-scoring nodes. Clustering coefficient of a node is the proportion of the edges to all the possible edges within its neighbors, quantifying the closeness among its neighbors, and evaluating how small its neighbors’ world is. A node with higher clustering coefficient has its neighbors closer to one another, and the world of its neighbors is smaller.
Global topological measurements of networks include average degree (<k>), mean shortest path length (mspl), diameter (D), and average clustering coefficient (acc). The clustering coefficient is a measure of the local interconnectedness of the graph, whereas the shortest path length is an indicator of its overall connectedness. A graph is considered small-world if it has a low mspl and a high acc49–51. According to Watts and Strogatz, small-world networks are a class of networks that are “highly clustered, like regular lattices, yet have small characteristic path lengths, like random graphs”.
The retrieval of the backbone from the PPI network
We considered the protein nodes with high degree (k) or BC as the hubs or bottlenecks. They were key to the PPI network and constituted the backbone of the network27. Given the PPI network, we retrieved the protein nodes with top 10% highest degree (k) or BC, and defined the graph of these proteins as the backbone. With these proteins as input, we used STRING45 again to construct the 2nd extended PPI network for further comprehensive analysis.
Results
The giant component of the PPI network
The giant component (Fig. 1) of the PPI network generated by STRING45 consisted of 111 nodes (Table S2) and 553 edges. The results of the topological analyses of each node were list in Table S3, including degree, BC, eccentricity, CC, EC, clustering coefficient, etc. The number of edges is larger than the expected for random network of the same size significantly (p-value ≤ 10−16); the nodes were more connected than randomly. It suggested that the PPI network could be considered as a relatively small world in comparison with random graph, and the proteins might be biologically relevant. The similar findings were also revealed in the results of the global topological measurements including average degree, mspl, diameter D, and average clustering coefficient listed in Table 1. In addition, the giant component of the PPI network with co-expression interactions (Fig. S1; Tables S4, S5) demonstrated the similar results, too.
Table 1.
Global topological measurements of four PPI networks.
| Symbol | Description | Giant component | Giant component (co-expression) | 2nd extended network | 2nd extended network (co-expression) |
|---|---|---|---|---|---|
| N | number of nodes | 111 | 111 | 116 | 116 |
| <k> | average degree | 9.96 ± 7.32 | 9.24 ± 5.94 | 10.86 ± 9.14 | |
| D | diameter | 7 | 8 | 6 | 7 |
| mspl | mean shortest path length | 3.31 | 3.62 | 2.56 | 2.71 |
| acc | average clustering coefficient | 0.672 | 0.707 | 0.647 | 0.7640 |
Note: The average degrees of 16 key proteins in the giant component are 21.31 ± 9.78 and 18.88 ± 7.80.
The backbone in the PPI network
The results of the topological analyses of each node in Table S3 showed that JUN was a hub (with the largest degree k = 43) and bottleneck (with the fourth highest BC = 0.1383) in the PPI network; PCK1 was only a bottleneck (with the highest BC = 0.35) but not a hub (with lower degree than average). We retrieved JUN, PCK1, and the other 14 proteins with top 10% largest degree (k) or highest BC and considered them as the hubs or bottlenecks and constituted the backbone of the giant component network (Fig. 2; Tables 2 and S6–S7). They were extensively connected with their neighbors in the network (the average degree: 21.31 ± 9.78) and had very much control over the network (the average BC: 0.11 ± 0.08). The backbone network consisted of 51 edges and 16 nodes. Among them, six proteins, JUN, MAPK14, CREEP, EP300, FOS, and RELA were both with top 10% largest degree (k) and highest BC, while PCK1 remained the most important bottleneck in the backbone.
Figure 2.
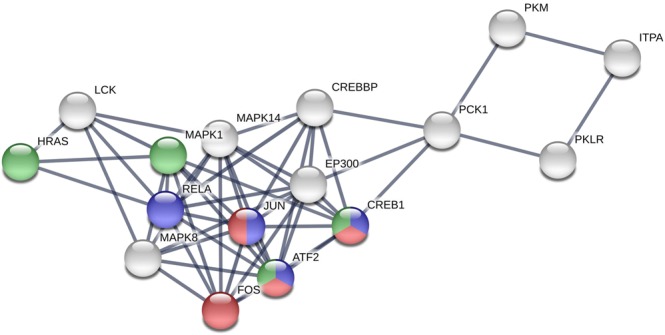
The backbone network.
Table 2.
The proteins in the backbone network.
| Label | Name | Description | Degree | Betweenness centrality |
|---|---|---|---|---|
| 1 | JUN | Jun proto-oncogene | 43 | 0.138335165 |
| 2 | MAPK14 | Mitogen-activated protein kinase 14 | 32 | 0.083166142 |
| 3 | FOS | FBJ murine osteosarcoma viral oncogene homolog | 26 | 0.059760577 |
| 4 | LCK | Lymphocyte-specific protein tyrosine kinase | 26 | 0.058523406 |
| 5 | RELA | V-rel reticuloendotheliosis viral oncogene homolog A (avian) | 25 | 0.069572515 |
| 6 | MAPK1 | Mitogen-activated protein kinase 1 | 25 | 0.046624799 |
| 7 | CREBBP | CREB binding protein | 24 | 0.163843625 |
| 8 | ATF2 | Activating transcription factor 2 | 24 | 0.022118131 |
| 9 | EP300 | E1A binding protein p300 | 23 | 0.129821489 |
| 10 | MAPK8 | Mitogen-activated protein kinase 8 | 22 | 0.044774657 |
| 11 | HRAS | v-Ha-ras Harvey rat sarcoma viral oncogene homolog | 22 | 0.027715087 |
| 12 | CREB1 | cAMP responsive element binding protein 1 | 17 | 0.128192989 |
| 13 | ITPA | Inosine triphosphatase (nucleoside triphosphate pyrophosphatase) | 13 | 0.181820962 |
| 14 | PCK1 | Phosphoenolpyruvate carboxykinase 1 (soluble) | 7 | 0.354489853 |
| 15 | PKLR | Pyruvate kinase, liver and RBC | 6 | 0.135446205 |
| 16 | PKM | Pyruvate kinase, muscle | 6 | 0.135446205 |
| Average | 21.31 ± 9.78 | 0.112282 ± 0.082026 |
Note: The bold ones are both in the backbone networks with and without co-expression setting in the STRING.
We also retrieved the backbone of the PPI network with co-expression interactions (Fig. S2 and Table S8). Comparing the two backbones with and without co-expression interactions, we discovered 13 proteins in common, and the 13-protein-subnetworks in the two backbones had the same structure (Fig. S3). The 13-protein-subnetworks contained most nodes and edges of the backbones. As a result, the main part of the backbones was robust, no matter with or without co-expression interactions derived from gene expression data. It suggested that only the rest part of the backbones could be influenced by the perturbations of gene expression through co-expression interactions.
The 2nd extended PPI network
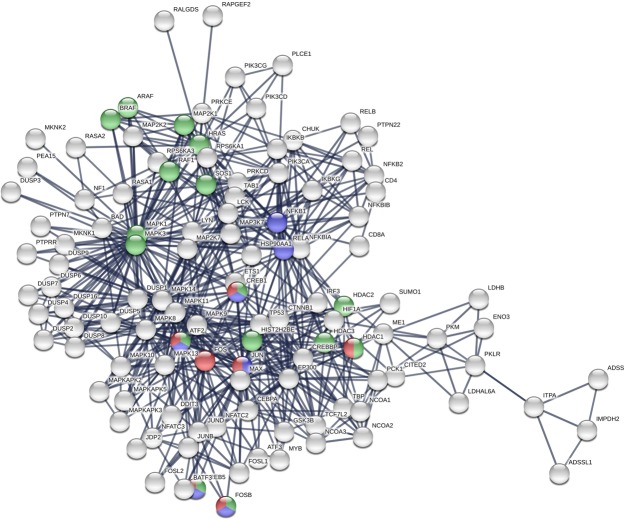
With use of the backbone nodes as input, we implemented STRING again to construct the 2nd extended PPI network associated with HUD. The global topological characteristics of four PPI networks in this study were listed in Table 1. The 2nd extended PPI network without co-expression interactions demonstrated 16 nodes for the KEGG pathways52 of alcoholism (ARAF, ATF2, BRAF, CREB1, CREB5, FOSB, HDAC1, HDAC2, HDAC3, HIST2H2BE, HRAS, MAP2K1, MAPK1, MAPK3, RAF1, and SOS1), 7 nodes of amphetamine addiction (ATF2, CREB1, CREB5, FOS, FOSB, HDAC1, and JUN), and 7 nodes of cocaine addiction (ATF2, CREB1, CREB5, FOSB, JUN, NFKB1, and RELA) in Table 3. The nodes of the 2nd extended PPI network involved in the KEGG pathways of amphetamine addiction, cocaine addiction, and alcoholism were colored in red, blue, and green, respectively, in Fig. 3. Moreover, the 2nd extended PPI network with co-expression interactions was Fig. S4 which demonstrated 10 nodes for the KEGG pathways of alcoholism, 5 nodes of amphetamine addiction, and 6 nodes of cocaine addiction in Table S9. In addition, the three KEGG pathways52 were shown in Figs S5–S7.
Table 3.
The proteins in the 2nd extended PPI network involved in the KEGG pathways of alcoholism, amphetamine addiction, and cocaine addition.
| KEGG pathway | Name | Description |
|---|---|---|
| Alcoholism | ARAF | V-raf murine sarcoma 3611 viral oncogene homolog |
| Alcoholism | ATF2 | Activating transcription factor 2 |
| Alcoholism | BRAF | V-raf murine sarcoma viral oncogene homolog B1 |
| Alcoholism | CREB1 | cAMP responsive element binding protein 1 |
| Alcoholism | CREB5 | cAMP responsive element binding protein 5 |
| Alcoholism | FOSB | FBJ murine osteosarcoma viral oncogene homolog B |
| Alcoholism | HDAC1 | Histone deacetylase 1 |
| Alcoholism | HDAC2 | Histone deacetylase 2 |
| Alcoholism | HDAC3 | Histone deacetylase 3 |
| Alcoholism | HIST2H2BE | Histone cluster 2, H2be |
| Alcoholism | HRAS | v-Ha-ras Harvey rat sarcoma viral oncogene homolog |
| Alcoholism | MAPK1 | Mitogen-activated protein kinase 1 |
| Alcoholism | MAP2K1 | Mitogen-activated protein kinase kinase 1 |
| Alcoholism | MAPK3 | Mitogen-activated protein kinase 3 |
| Alcoholism | RAF1 | V-raf-1 murine leukemia viral oncogene homolog 1 |
| Alcoholism | SOS1 | Son of sevenless homolog 1 (Drosophila) |
| Amphetamine | ATF2 | Activating transcription factor 2 |
| Amphetamine | CREB1 | cAMP responsive element binding protein 1 |
| Amphetamine | CREB5 | cAMP responsive element binding protein 5 |
| Amphetamine | FOS | FBJ murine osteosarcoma viral oncogene homolog |
| Amphetamine | FOSB | FBJ murine osteosarcoma viral oncogene homolog B |
| Amphetamine | JUN | Jun proto-oncogene |
| Amphetamine | HDAC1 | Histone deacetylase 1 |
| Cocaine | ATF2 | Activating transcription factor 2 |
| Cocaine | CREB1 | cAMP responsive element binding protein 1 |
| Cocaine | CREB5 | cAMP responsive element binding protein 5 |
| Cocaine | FOSB | FBJ murine osteosarcoma viral oncogene homolog B |
| Cocaine | JUN | Jun proto-oncogene |
| Cocaine | NFKB1 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| Cocaine | RELA | V-rel reticuloendotheliosis viral oncogene homolog A (avian) |
Figure 3.
The 2nd extended PPI network. *The red ones are nodes in amphetamine addiction pathway. +The blue ones are nodes in cocaine addiction pathway. #The green ones are nodes in alcoholism pathway.
Discussion
Several studies have been conducted on HUD and several related susceptibility genes have been reported; however, the potential mechanism underlying HUD development remains unclear53,54. The proteins encoded by susceptibility genes may determine an individual’s susceptibility to HUD through their encoded PPIs. Here, we studied the potential key proteins through topological analysis18,55,56. We used degree (k) and BC, the 2 measures widely used in network theory, as the main parameters for evaluating the nodes in the PPI network27.
This is the first study to investigate the PPI network of HUD and explore the contributions of the proteins encoded by the susceptibility genes associated with HUD. Initially, a total of 111 proteins were included in our giant component network. By considering the important topological measures (degree and BC) in a large network, we selected 16 proteins to construct the backbone network: 11 with large degree, 11 with high BC, and 4 with both large degree and high BC. In the PPI network, there were 16 proteins involved in the alcoholism pathway, 7 proteins involved in the amphetamine addiction pathway, and 7 proteins involved in the cocaine addiction pathway in the KEGG. Nevertheless, although both morphine and heroin are synthesized from opium, proteins involved in the KEGG pathway of morphine addiction were absent in the network. According to gateway drug theory, alcohol and cannabis are frequently abused before illicit drugs such as cocaine and heroin. Cannabis use is not popular in Taiwan; therefore, the 16 proteins involved in alcoholism might be important for the further development of HUD. In Taiwan, the frequency of cocaine use is low; however, the frequency of amphetamine use, a cocaine-like stimulant, is high in clients with HUD57. It might be due to most individuals with HUD abusing other substance such as amphetamine58,59.
Seven nodes including JUN, FOS, RELA, MAPK1, ATF2, HRAS, and CREB1 in the backbone are the recorded genes of alcoholism, amphetamine addiction, or cocaine addiction in the KEGG pathways. They are the identified key proteins in substance diseases involved in HUD development revealed in this study.
The most important one among them is JUN, one of the biosignatures for detecting HUD in men43 and a seed protein in the initial PPI network. JUN (with the largest degree and the fourth highest BC) is central to the PPI network, the backbone network and the 2nd extended PPI network. It has been implicated in cancer-related studies60 as well as studies on psychological disorders such as Alzheimer disease and schizophrenia61,62. JUN has been reported to be involved in amphetamine and cocaine addiction and their respective KEGG pathways63,64.
FOS, RELA, and HRAS in the backbone are known to be in the KEGG pathway associated with substance diseases. RELA, or v-rel avian reticuloendotheliosis viral oncogene homolog A, is also named as p65 or NF-κB (nuclear factor kappa-light-chain-enhancer of activated B cells) is a protein complex that controls transcription of DNA, cytokine production and cell survival and was found to be correlated with cancers and Alzheimer’s disease65–68. RELA (NF-κB) is known as an induced transcriptional targets of ΔFosB associated with addiction to a stimulus such as cocaine and the effect of reward system69–72. HRAS is a small G protein in the Ras subfamily associated small GTPases, and familial alcohol dependence was associated with hypomethylation of CpG sites in the HRAS promoter region73. FOS is a 380 amino acid protein with a basic leucine zipper region for dimerisation and DNA-binding and a transactivation domain at C-terminus and FOS is unable to make FOS-FOS homodimers. JUN–FOS heterodimers are more stable and have stronger DNA-binding activity than JUN–JUN homodimers. FOS is known to have interaction in an animal study74. Moreover, ATF2, CREB1, CREB5, and FOSB in all three substance diseases (alcoholism, amphetamine, and cocaine addiction pathways). All of them except FOSB are selected in to the backbone network. CREB1 and CREB5 belong to CREB (cAMP response element-binding protein) family and are correlated with substance diseases75–80. FOSB belongs to FOS gene family including FOS. The FOS family play a role in the development and maintenance of drug addictions70,71. ATF2 interacting with JUN, CREB family, and other proteins in the backbone network would be discussed latter81.
Some other nodes in the backbone network are worth to study although they are not recorded in the alcoholism, amphetamine addiction, and cocaine addiction pathways. MAPK14, is not recognized in substance diseases in these KEGG pathways. However, MAPK14, also called p38-α, is an enzyme in humans encoded by the MAPK14 gene and a member of the MAP kinase family. The substrates of this kinase also include transcription regulator ATF2 on stress-activated protein kinases82. ATF2 is in the backbone network and is involved in all three substance diseases (alcoholism, amphetamine addiction, and cocaine addiction) in the KEGG pathways. PCK1, an enzyme in humans encoded by the PCK1 gene, is an important control point for the regulation of gluconeogenesis. PCK1 and MAPK14 were not found to be involved in any substance diseases previously but they were noticed in the study of schizophrenia83. MAPK14 has the 2nd large degree and 9th high BC and PCK1 has the highest BC in our giant component network. The two nodes provide us the new cues for further study in HUD and other substance diseases.
In addition, changes in miRNA expression levels are linked to neurodegeneration84 with mounting of evidence supporting the dysregulation of miRNA expression in psychiatric and neurological disorders35–37,85–87. MiRNAs might play important functions in moderating the central stress response within different brain regions via the regulation of genes35. As a result, miRNAs can not only serve as biomarkers of addition, but also as promising therapies for the prevention or treatment of neurodevelopmental and neuropathological disorders37. Based on proteins in Table 2, we found that some miRNAs target FOS, JUN, MAPK1, MAPK14, and RELA (Table S10) in the study of the brains genomic response to environmental stress35, and some of them involve in addiction such as cocaine, alcohol37,85. We searched for miRNA-substance-use-disorder associations from HMDD v3.0 database42, and there were few information related to substance use disorders (Table S11). In addition, researches of the miRNA-HUD associations are few38, therefore, it still remains much unknown. The recent advances of specific miRNAs have emerged as key regulator leading to addiction, and further studies may be central for developing novel therapeutic approaches85. Implementing predictive computational models might be potential to discovery miRNA biomarkers for HUD in the future39–42,88.
A limitation of this study is the lack of proteins strongly correlated with morphine addiction or HUD in the backbone network. This may be due to early-life exposure to alcohol or amphetamine having a greater impact on persons with HUD than later-life heroin use. Another limitation of this study is that we used peripheral blood as the sample, rather than brain tissue from areas such as nucleus accumbens89, based on a previous human study90.
Conclusion
Our finding suggests that HUD develops through an integrated PPI network with JUN (the largest degree and the 4th highest BC) at its center. JUN is also involved in the development of amphetamine and cocaine addiction. However, the role of JUN in HUD requires further clarification. FOS, RELA, ATF2, and HRAS in the backbone network are also recorded of one of three substance diseases in KEGG pathways. (Alcoholism, amphetamine addiction, cocaine addiction) ATF2, CREB1, CREB5, and FOSB in all three substance disease KEGG pathways are suspected be related with CREB and FOS family, and JUN. Furthermore, MAPK14 (the second largest degree and the 9th high BC) and PCK1 (the highest BC) have a major role in the PPI network. The present study marks the beginning of the formulation of the PPI network of HUD, with JUN and other two key proteins MAPK14 and PCK1.
Supplementary information
Acknowledgements
The authors appreciate the support from Tzu Chi University and Ministry of Science and Technology in Taiwan (MOST 104-2221-E-320-004, MOST 106-2221-E-320-003).
Author Contributions
C.C. and K.C. planned and conceived the experiment design. S.C., T.W. and K.C. were the main contributors of manuscript text, figures and tables, and responsible for analyzing the data/results. S.C. and T.W. were responsible for experiments/methods. D.L. and C.C. contributed reagents and expertise. All authors reviewed the manuscript and provided edits and suggestions.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-41552-z.
References
- 1.Bell J, Zador D. A risk-benefit analysis of methadone maintenance treatment. Drug Safety. 2000;22:179–190. doi: 10.2165/00002018-200022030-00002. [DOI] [PubMed] [Google Scholar]
- 2.Trujols J, et al. Multi-episode survival analysis: an application modelling readmission rates of heroin dependents at an inpatient detoxification unit. Addict Behav. 2007;32:2391–2397. doi: 10.1016/j.addbeh.2007.02.008. [DOI] [PubMed] [Google Scholar]
- 3.Kulsudjarit K. Drug problem in Southeast and Southwest Asia. Annals of the New York Academy of Sciences. 2004;1025:446–457. doi: 10.1196/annals.1316.055. [DOI] [PubMed] [Google Scholar]
- 4.Glatt SJ, et al. Genome-wide linkage analysis of heroin dependence in Han Chinese: Results from Wave Two of a multi-stage study. Drug & Alcohol Dependence. 2008;98:30–34. doi: 10.1016/j.drugalcdep.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li J, Ha TH, Zhang C, Liu H. The Chinese government’s response to drug use and HIV/AIDS: A review of policies and programs. Harm Reduction. Journal. 2010;7:4–4. doi: 10.1186/1477-7517-7-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen CY, Wu PN, Su LW, Chou YJ, Lin KM. Three-year mortality and predictors after release: a longitudinal study of the first-time drug offenders in Taiwan. Addiction. 2010;105:920–927. doi: 10.1111/j.1360-0443.2009.02894.x. [DOI] [PubMed] [Google Scholar]
- 7.Reder LM, Weber K, Shang J, Vanyukov PM. The adaptive character of the attentional system: statistical sensitivity in a target localization task. J Exp Psychol Hum Percept Perform. 2003;29:631–649. doi: 10.1037/0096-1523.29.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nat Rev Genet. 2005;6:521–532. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- 9.Uhl GR, et al. “Higher order” addiction molecular genetics: convergent data from genome-wide association in humans and mice. Biochem Pharmacol. 2008;75:98–111. doi: 10.1016/j.bcp.2007.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Levran O, et al. Stress-related genes and heroin addiction: a role for a functional FKBP5 haplotype. Psychoneuroendocrinology. 2014;45:67–76. doi: 10.1016/j.psyneuen.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bailey A, et al. Immediate withdrawal from chronic “binge” cocaine administration increases μ-opioid receptor mRNA levels in rat frontal cortex. Molecular Brain Research. 2005;137:258–262. doi: 10.1016/j.molbrainres.2005.02.017. [DOI] [PubMed] [Google Scholar]
- 12.Nielsen DA, et al. Genome-wide association study identifies genes that may contribute to risk for developing heroin addiction. Psychiatric Genetics. 2010;20:207–214. doi: 10.1097/YPG.0b013e32833a2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rual J-F, et al. Towards a proteome-scale map of the human protein-protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 14.Raman K. Construction and analysis of protein–protein interaction networks. Automated Experimentation. 2010;2:2. doi: 10.1186/1759-4499-2-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stelzl U, et al. A human protein-protein interaction network: A resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 16.Lim J, et al. A protein-protein interaction network for human inherited ataxias and disorders of Purkinje cell degeneration. Cell. 2006;125:801–814. doi: 10.1016/j.cell.2006.03.032. [DOI] [PubMed] [Google Scholar]
- 17.Oti M, Snel B, Huynen MA, Brunner HG. Predicting disease genes using protein–protein interactions. Journal of Medical Genetics. 2006;43:691–698. doi: 10.1136/jmg.2006.041376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li M, Zhang H, Wang JX, Pan Y. A new essential protein discovery method based on the integration of protein-protein interaction and gene expression data. BMC Syst Biol. 2012;6:9. doi: 10.1186/1752-0509-6-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rakshit H, Rathi N, Roy D. Construction and analysis of the protein-protein interaction networks based on gene expression profiles of Parkinson’s disease. PLoS ONE. 2014;9:e103047. doi: 10.1371/journal.pone.0103047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carninci P, et al. The transcriptional landscape of the Mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 21.Margolin AA, et al. ARACNE: An algorithm for the reconstruction of gene regulatory networks in a Mammalian cellular context. BMC Bioinformatics. 2006;7(S7):s1–s7. doi: 10.1186/1471-2105-7-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chatterjee P, Pal NR. Construction of synergy networks from gene expression data related to disease. Gene. 2016;590:250–262. doi: 10.1016/j.gene.2016.05.029. [DOI] [PubMed] [Google Scholar]
- 23.Jeong H, Tombor B, Albert R, Oltvai ZN, Barabasi AL. The large-scale organization of metabolic networks. Nature. 2000;407:651–654. doi: 10.1038/35036627. [DOI] [PubMed] [Google Scholar]
- 24.Hatzimanikatis V, Li C, Ionita JA, Broadbelt LJ. Metabolic networks: Enzyme function and metabolite structure. Current Opinion in Structural Biology. 2004;14:300–306. doi: 10.1016/j.sbi.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 25.Duarte NC, et al. Global reconstruction of the human metabolic network based on genomic and bibliomic data. Proceedings of the National Academy of Sciences. 2007;104:1777–1782. doi: 10.1073/pnas.0610772104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee D-S, et al. The implications of human metabolic network topology for disease comorbidity. Proceedings of the National Academy of Sciences. 2008;105:9880–9885. doi: 10.1073/pnas.0802208105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ran J, et al. Construction and analysis of the protein-protein interaction network related to essential hypertension. BMC Syst Biol. 2013;7:1–12. doi: 10.1186/1752-0509-7-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen H, et al. Pathway mapping and development of disease-specific biomarkers: protein-based network biomarkers. Journal of Cellular and Molecular Medicine. 2015;19:297–314. doi: 10.1111/jcmm.12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Barabási A-L, Gulbahce N, Loscalzo J. Network medicine: A network-based approach to human disease. Nature Reviews. Genetics. 2011;12:56–68. doi: 10.1038/nrg2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yu H, Kim PM, Sprecher E, Trifonov V, Gerstein M. The importance of bottlenecks in protein networks: Correlation with gene essentiality and expression dynamics. PLoS Comput Biol. 2007;3:e59. doi: 10.1371/journal.pcbi.0030059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ghasemi O, Ma Y, Lindsey ML, Jin Y-F. Using systems biology approaches to understand cardiac inflammation and extracellular matrix remodeling in the setting of myocardial infarction. Wiley Interdisciplinary Reviews. Systems Biology and Medicine. 2014;6:77–91. doi: 10.1002/wsbm.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kalivas PW, Volkow ND. The neural basis of addiction: A pathology of motivation and choice. Am J Psychiatry. 2005;162(8):1403–1413. doi: 10.1176/appi.ajp.162.8.1403. [DOI] [PubMed] [Google Scholar]
- 33.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 34.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 35.Hollins SL, Cairns MJ. MicroRNA: small RNA mediators of the brains genomic response to environmental stress. Progress in Neurobiology. 2016;143:61–81. doi: 10.1016/j.pneurobio.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 36.Oliver RJ, Mandyam CD. Regulation of adult neurogenesis by non-coding RNAs: Implications for substance use disorders. Frontiers in Neuroscience. 2018;12:849. doi: 10.3389/fnins.2018.00849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith ACW, Kenny PJ. MicroRNAs regulate synaptic plasticity underlying drug addiction. Genes, Brain and Behavior. 2018;17:e12424. doi: 10.1111/gbb.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yan B, et al. MiR-218 targets MeCP2 and inhibits heroin seeking behavior. Scientific Reports. 2017;7:40413. doi: 10.1038/srep40413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X, et al. BNPMDA: Bipartite network projection for miRNA–disease association prediction. Bioinformatics. 2018;34:3178–3186. doi: 10.1093/bioinformatics/bty333. [DOI] [PubMed] [Google Scholar]
- 40.Chen X, Wang L, Qu J, Guan N-N, Li J-Q. Predicting miRNA–disease association based on inductive matrix completion. Bioinformatics. 2018;34:4256–4265. doi: 10.1093/bioinformatics/bty503. [DOI] [PubMed] [Google Scholar]
- 41.You ZH, et al. PBMDA: A novel and effective path-based computational model for miRNA-disease association prediction. PLoS Comput Biol. 2017;13(3):e1005455. doi: 10.1371/journal.pcbi.1005455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang Z, et al. HMDDv3.0: a database for experimentally supported human microRNA-disease associations. Nucleic Acids Research. 2019;47:D1013–D1017. doi: 10.1093/nar/gky1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen S-J, et al. Genetic signatures of heroin addiction. Medicine. 2016;95:e4473. doi: 10.1097/MD.0000000000004473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liao DL, Cheng MC, Lai CH, Tsai HJ, Chen CH. Comparative gene expression profiling analysis of lymphoblastoid cells reveals neuron-specific enolase gene (ENO2) as a susceptibility gene of heroin dependence. Addict Biol. 2011;19(1):102–10. doi: 10.1111/j.1369-1600.2011.00390.x. [DOI] [PubMed] [Google Scholar]
- 45.Szklarczyk D, et al. STRINGv10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Research. 2015;43:D447–D452. doi: 10.1093/nar/gku1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bastian M., Heymann S. & Jacomy M. Gephi: an open source software for exploring and manipulating networks. International AAAI Conference on Weblogs and Social Media (ICWSM′09).
- 47.Albert R, Barabasi A-L. Statistical mechanics of complex networks. Rev Modern Physic. 2002;74:47–85. doi: 10.1103/RevModPhys.74.47. [DOI] [Google Scholar]
- 48.Barabasi A-L, Oltvai ZN. Network biology: understanding the cell’s functional organization. Nat Rev Genet. 2004;5:101–113. doi: 10.1038/nrg1272. [DOI] [PubMed] [Google Scholar]
- 49.Humphries MD, Gurney K, Prescott TJ. The brainstem reticular formation is a small-world, not scale-free. network. Proceedings. Biological Sciences. 2006;273:503–511. doi: 10.1098/rspb.2005.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Telesford QK, Joyce KE, Hayasaka S, Burdette JH, Laurienti PJ. The ubiquity of small-world networks. Brain Connectivity. 2011;1:367–375. doi: 10.1089/brain.2011.0038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 52.Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017;45:D353–D361. doi: 10.1093/nar/gkw1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gelernter J, Kranzler HR. Genetics of drug dependence. Dialogues in Clinical Neuroscience. 2010;12:77–84. doi: 10.31887/DCNS.2010.12.1/jgelernter. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang J-C, Kapoor M, Goate AM. The genetics of substance dependence. Annual Review of Genomics and Human Genetics. 2012;13:241–261. doi: 10.1146/annurev-genom-090711-163844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Goñi J, et al. A computational analysis of protein-protein interaction networks in neurodegenerative diseases. BMC Systems Biology. 2008;2:52. doi: 10.1186/1752-0509-2-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hashimoto TB, Nagasaki M, Kojima K, Miyano S. BFL: a node and edge betweenness based fast layout algorithm for large scale networks. BMC Bioinformatics. 2009;10:19. doi: 10.1186/1471-2105-10-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ahn W-Y, Vassileva J. Machine-learning identifies substance-specific behavioral markers for opiate and stimulant dependence. Drug and Alcohol Dependence. 2016;161:247–257. doi: 10.1016/j.drugalcdep.2016.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meacham MC, et al. Prevalence and correlates of heroin–methamphetamine co-injection among persons who inject drugs in San Diego, California, and Tijuana, Baja California, Mexico. Journal of Studies on Alcohol and Drugs. 2016;77:774–781. doi: 10.15288/jsad.2016.77.774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Preston KL, Jobes ML, Phillips KA, Epstein DH. Real-time assessment of alcohol drinking and drug use in opioid-dependent polydrug users. Behavioural Pharmacology. 2016;27:579–584. doi: 10.1097/FBP.0000000000000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Behrens A, Jochum W, Sibilia M, Wagner EF. Oncogenic transformation by ras and fos is mediated by c-Jun N-terminal phosphorylation. Oncogene. 2000;19:2657–2663. doi: 10.1038/sj.onc.1203603. [DOI] [PubMed] [Google Scholar]
- 61.Helbecque N, et al. Islet-brain1//C-Jun N-terminal kinase interacting protein-1 (IB1//JIP-1) promoter variant is associated with Alzheimer’s disease. Mol Psychiatry. 2003;8:413–422. doi: 10.1038/sj.mp.4001344. [DOI] [PubMed] [Google Scholar]
- 62.Ahn YM, et al. Reduction in the protein level of c-Jun and phosphorylation of Ser73–c-Jun in rat frontal cortex after repeated MK-801 treatment. Psychiatry Research. 2009;167:80–87. doi: 10.1016/j.psychres.2007.12.013. [DOI] [PubMed] [Google Scholar]
- 63.Chambers JW, Howard S, LoGrasso PV. Blocking c-Jun N-terminal kinase (JNK) translocation to the mitochondria prevents 6-hydroxydopamine-induced toxicity in vitro and in vivo. Journal of Biological Chemistry. 2013;288:1079–1087. doi: 10.1074/jbc.M112.421354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nagelová V, Pirník Z, Železná B, Maletínská L. CART (cocaine- and amphetamine-regulated transcript) peptide specific binding sites in PC12 cells have characteristics of CART peptide receptors. Brain Research. 2014;1547:16–24. doi: 10.1016/j.brainres.2013.12.024. [DOI] [PubMed] [Google Scholar]
- 65.Meffert MK, Chang JM, Wiltgen BJ, Fanselow MS, Baltimore D. NF-κB functions in synaptic signaling and behavior. Nature Neuroscience. 2003;6:1072. doi: 10.1038/nn1110. [DOI] [PubMed] [Google Scholar]
- 66.Perkins ND. Integrating cell-signalling pathways with NF-κB and IKK function. Nature Reviews Molecular Cell Biology. 2007;8:49. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 67.Wiciński M, et al. Neuroprotective activity of sitagliptin via reduction of neuroinflammation beyond the incretin effect: Focus on Alzheimer’s disease. BioMed Research International. 2018;2018:6091014. doi: 10.1155/2018/6091014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Concetti J, Wilson CL. NFKB1 and cancer: Friend or foe? Cells. 2018;7:133. doi: 10.3390/cells7090133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nestler EJ. Transcriptional mechanisms of addiction: role of ΔFosB. Philosophical Transactions of the Royal Society B: Biological Sciences. 2008;363:3245–3255. doi: 10.1098/rstb.2008.0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Robison AJ, Nestler EJ. Transcriptional and epigenetic mechanisms of addiction. Nat Rev Neurosci. 2011;12:623–637. doi: 10.1038/nrn3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nestler EJ. Cellular basis of memory for addiction. Dialogues in Clinical Neuroscience. 2013;15:431–443. doi: 10.31887/DCNS.2013.15.4/enestler. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ruffle JK. Molecular neurobiology of addiction: what’s all the (Δ)FosB about? The American Journal of Drug and Alcohol Abuse. 2014;40:428–437. doi: 10.3109/00952990.2014.933840. [DOI] [PubMed] [Google Scholar]
- 73.Hill SY, Rompala G, Homanics GE, Zezza N. Cross-generational effects of alcohol dependence in humans on HRAS and TP53 methylation in offspring. Epigenomics. 2017;9:1189–1203. doi: 10.2217/epi-2017-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chavez C, et al. Differential effect of amphetamine on c- < em > fos < /em > expression in female aromatase knockout (ArKO) mice compared to wildtype controls. Psychoneuroendocrinology. 2011;36:761–768. doi: 10.1016/j.psyneuen.2010.10.013. [DOI] [PubMed] [Google Scholar]
- 75.Konradi C, Cole R, Heckers S, Hyman S. Amphetamine regulates gene expression in rat striatum via transcription factor CREB. The Journal of Neuroscience. 1994;14:5623–5634. doi: 10.1523/jneurosci.14-09-05623.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nazarian A, et al. Sex differences in basal and cocaine-induced alterations in PKA and CREB proteins in the nucleus accumbens. Psychopharmacology. 2008;203:641. doi: 10.1007/s00213-008-1411-5. [DOI] [PubMed] [Google Scholar]
- 77.DiRocco DP, Scheiner ZS, Sindreu CB, Chan GCK, Storm DR. A role for calmodulin-stimulated adenylyl cyclases in cocaine sensitization. The Journal of Neuroscience: the Official Journal of the Society for Neuroscience. 2009;29:2393–2403. doi: 10.1523/JNEUROSCI.4356-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang Y, Ghezzi A, Yin JCP, Atkinson NS. CREB regulation of BK channel gene expression underlies rapid drug tolerance. Genes, Brain, and Behavior. 2009;8:369–376. doi: 10.1111/j.1601-183X.2009.00479.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Madsen HB, et al. CREB1 and CREB-binding protein in striatal medium spiny neurons regulate behavioural responses to psychostimulants. Psychopharmacology. 2012;219:699–713. doi: 10.1007/s00213-011-2406-1. [DOI] [PubMed] [Google Scholar]
- 80.Zhang H, et al. Adolescent alcohol exposure epigenetically regulates CREB signaling in the adult amygdala. Scientific Reports. 2018;8:10376. doi: 10.1038/s41598-018-28415-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Gruber T. Toward principles for the design of ontologies used for knowledge sharing. Int J Hum-Comput Stud. 1995;43:907–928. doi: 10.1006/ijhc.1995.1081. [DOI] [Google Scholar]
- 82.Watson G, Ronai ZeA, Lau E. ATF2, a paradigm of the multifaceted regulation of transcription factors in biology and disease. Pharmacological Research. 2017;119:347–357. doi: 10.1016/j.phrs.2017.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Olsen L, et al. The estrogen hypothesis of schizophrenia implicates glucose metabolism: Association study in three independent samples. BMC Medical Genetics. 2008;9:39. doi: 10.1186/1471-2350-9-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Nelson PT, et al. MicroRNAs (miRNAs) in neurodegenerative diseases. Brain Pathol. 2008;18:130–138. doi: 10.1111/j.1750-3639.2007.00120.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dreyer JL. New insights into the roles of microRNAs in drug addiction and neuroplasticity. Genome. Medicine. 2010;2:92. doi: 10.1186/gm213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Geaghan M, Cairns MJ. MicroRNA and posttranscriptional dysregulation in psychiatry. Biol Psychiatry. 2014;78(4):231–239. doi: 10.1016/j.biopsych.2014.12.009. [DOI] [PubMed] [Google Scholar]
- 87.Miller BH, Wahlestedt C. MicroRNA dysregulation in psychiatric disease. Brain Res. 2010;1338:89–99. doi: 10.1016/j.brainres.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yang Z, et al. dbDEMC 2.0: updated database of differentially expressed miRNAs in human cancers. Nucleic acids research. 2017;45:D812–D818. doi: 10.1093/nar/gkw1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Albertson DN, Schmidt CJ, Kapatos G, Bannon MJ. Distinctive profiles of gene expression in the human nucleus accumbens associated with cocaine and heroin abuse. Neuropsychopharmacology. 2006;31:2304–2312. doi: 10.1038/sj.npp.1301089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Mas S, Gasso P, Parellada E, Bernardo M, Lafuente A. Network analysis of gene expression in peripheral blood identifies mTOR and NF-[kappa]B pathways involved in antipsychotic-induced extrapyramidal symptoms. Pharmacogenomics J. 2015;15:452–460. doi: 10.1038/tpj.2014.84. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.