Abstract
Transcriptional regulators include a superfamily of nuclear proteins referred to as co-activators and co-repressors, both of which are involved in controlling the functions of several nuclear receptors (NRs). The Nuclear Receptor Signaling Atlas (NURSA) has cataloged the composition of NRs, co-regulators, and ligands present in the human cell and their effort has been identified in more than 600 potential molecules. Given the importance of co-regulators in steroid, retinoid, and thyroid hormone signaling networks, hypothesizing that NRs/co-regulators are implicated in a wide range of pathologies are tempting. The co-activators known as peroxisome proliferator-activated receptor gamma co-activator 1 (PGC-1) and their key nuclear partner, the estrogen-related receptor (ERR), are emerging as pivotal transcriptional signatures that regulate an extremely broad repertoire of mitochondrial and metabolic genes, making them very attractive drug targets for cancer. Several studies have provided an increased understanding of the functional and structural biology of nuclear complexes. However, more comprehensive work is needed to create different avenues to explore the therapeutic potential of NRs/co-activators in precision oncology. Here, we discuss the emerging data associated with the structure, function, and molecular biology of the PGC-1/ERR network and address how the concepts evolving from these studies have deepened our understanding of how to develop more effective treatment strategies. We present an overview that underscores new biological insights into PGC-1/ERR to improve cancer outcomes against therapeutic resistance. Finally, we discuss the importance of exploiting new technologies such as single-particle cryo-electron microscopy (cryo-EM) to develop a high-resolution biological structure of PGC-1/ERR, focusing on novel drug discovery for precision oncology.
Introduction
Transcriptional regulators comprise of nuclear proteins known as co-activators and co-repressors, which bind and control the functions of nuclear receptors (NRs) and transcription factors (TFs).1–4 The essential role of NRs and their co-factors in many aspects of mammalian development and physiology raises the possibility that dysfunctions in biological signaling networks controlled by receptors or co-activators, which could be associated with metabolic diseases.5,6
Under normal physiological conditions, the proliferator-activated receptor gamma co-activator 1/estrogen-related receptor (PGC-1/ERR) transcriptional axis is involved in the control of mitochondrial biogenesis.7,8 Mitochondria are considered to be key regulatory organelles that control cellular survival and death mechanisms, including biomass and energy production for rapid cell growth and apoptosis, respectively.9 Hence, many reports support the concept that the PGC-1/ERR pathway plays a dual role in cancer, depending on the specific cellular or tissue context and the environmental stimuli.10–15 Notably, the PGC-1/ERR axis has been shown to be essential for functional cancer cell motility and metastasis, leading to malignant transformation in breast and melanoma cancer progression.14,16,17 In contrast, this pathway has also been shown to suppress prostate cancer progression and metastasis.13,18,19 Although substantial progress has been made in increasing the understanding of the function and molecular biology of NRs and their co-activators, a considerable gap still exists in comprehending how the PGC-1/ERR axis integrates mitochondrial activity through oxidative phosphorylation (OxPhos) leading to cell survival or cell death and how this regulatory function is connected to its dual role in cancer progression.
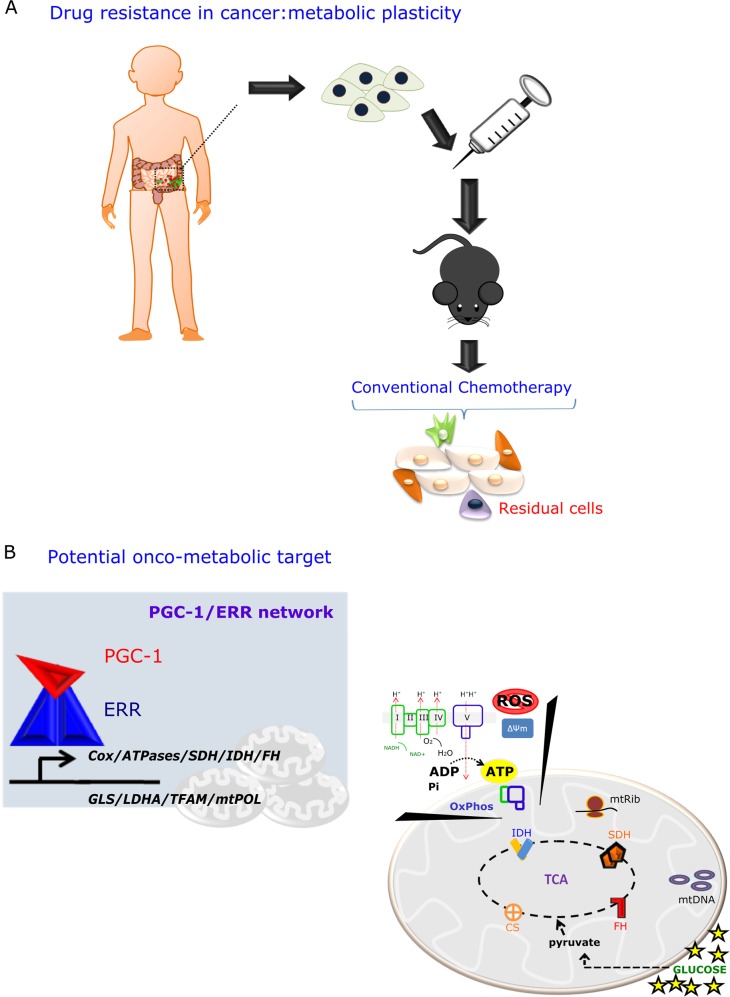
Additional studies have been conducted that have led to a more comprehensive knowledge of the structure, function, and molecular biology of PGC-1/ERR signaling in cancer biology.20–24 Notably, accumulating evidence supports the importance of the PGC-1/ERR transcriptional axis in the context of metabolic-addicted cancer cells.12,25–29 This increases the significance of exploiting the therapeutic potential of these targets in an effort to predict the efficacy of therapeutic resistance, the mechanism of which relies on mitochondrial metabolic plasticity (Fig. 1).12,20,30–32 This therapeutic potential depends on the function of PGC-1/ERR in coordinating the activity of a broad repertoire of target gene expression associated with mitochondrial biogenesis, OxPhos, energy homeostasis, and glucose, glutamine and lipid metabolism (Table 1).22,33 Notably, targeting the PGC-1/ERR network could be accessed by exploring the potential of mitochondrial-linked weak spots, where selective inhibitors of the PGC-1/ERR axis and mitochondrial metabolism might have to be used in combination to target the metabolic addiction of specific cancer cells (Table 1 and Fig. 2).34
Fig. 1.
The PGC-1/ERR network as a potential onco-metabolic target in cancer treatment. a Cancer translational research methodology based on the patient-derived xenograft (PDX) model highlighting drug resistance in cancer. The survival mechanism of residual cells after conventional chemotherapy relies on metabolic plasticity. b The concept of metabolic vulnerability associated with cancer progression can be exploited by targeting a combination of the PGC-1/ERR network and several other mitochondrial weak spots, such as respiratory chain defects, TCA cycle enzymes, including citrate synthase (CS), isocitrate dehydrogenase (IDH), succinate dehydrogenase (SDH) and fumarate hydratase (FH). These enzymes might be exploited as potential onco-metabolic targets, depending on the specific type of cancer
Table 1.
Examples of mitochondrial-targeted enzymes regulated by PGC-1/ERR
| Metabolic process | Major enzymes | Drug | References |
|---|---|---|---|
| Glycolysis | Hexokinase 2 (HK2) | 2-DG, XCT790 | 140– 142 |
| Lactate dehydrogenases (LDHA, LDHB) | AT-101, FX11, Cpd29 | 29, 143 | |
| Pyruvate kinase (PKM2) | TLN-232/CAP-232 | 144, 145 | |
| TCA cycle | Succinate dehydrogenase (SDHB) | 3-BrPA, XCT790 | 25, 146, 147 |
| Isocitrate dehydrogenases−1 and −2 (IDH1, IDH2, IDH3A) | Enasidenib, ivosidenib, Cpd29 | 17, 25, 148 | |
| Fumarate hydratase (FH) | Cpd29 | 17, 25 | |
| OxPhos | Mitochondrial complex I | Metformin, phenformin, Cpd29 | 11, 17, 149, 150 |
| Amino acid metabolism | Glutamic-oxaloacetic transaminase−1 and −2 (GOT1, GOT2) | Aminooxyacetate | 25, 151 |
| Lipid metabolism | Carnitine palmitoyltransferase 1 (CPT1) | Etomoxir | 140, 152 |
| Fatty acid synthase (FASN) | Orlistat, cerulenin, TVB-2640 | 14, 110 |
Fig. 2.
Cryo-EM technology as an important tool for precision oncology. a The new cyro-EM technology could provide a better understanding of the native/physiological state of protein complexes for the development of promising therapeutic targets and drug discovery for precision oncology. Reprinted from publication: Merk, A. et al., Breaking Cryo-EM Resolution Barriers to Facilitate Drug Discovery, 1698-707, 2016, with permission from Elsevier.126 b Mitochondrial inhibitors, like metformin, enasidenib or etomoxir, have been used to treat cancer patients. An ideal scenario to exploit the resistance mechanism in cancer cells that seems to rely on OxPhos activity could be achieved using the concept of metabolic vulnerability, when a combination of targetable genes leads to a lethal phenotype. The PGC-1/ERR axis has great potential to be included as a potential metabolic target for precision oncology for the treatment of non-responder patients. Thus, the combination of mitochondrial inhibitors and inverse agonists of the PGC-1/ERR axis, like compound 29, might provide new hope for treating non-responder patients whose cancer cell survival mechanism relies on mitochondrial metabolism
In an effort to provide new insights into understanding the role of the PGC-1/ERR network, we suggest that new technologies, such as cryo-electron microscopy (cryo-EM), could provide mechanistic comprehension of the biological processes of these protein complexes. The use of cryo-EM could explain the conflicting observations of biochemistry and crystallography that focused only on individual domains or peptides of some of the molecules involved. Moreover, using cryo-EM will allow the investigation of the native state of the PGC-1/ERR complex, which might lead to the identification of novel mechanistic insights into the PGC-1/ERR biology, as well as the discovery of new binding-partners, which could open new therapeutic windows for targeting the PGC-1/ERR complex and its metabolism in precision oncology. For example, Yi et al., used cryo-EM and observed a novel biological insight in the transcriptional activity of the estrogen-receptor (ER) co-activator complex on DNA.35 The authors suggested that ER recruits two steroid receptor co-activator 3 proteins and one p300 protein from a DNA-bound complex. The structure of the ER-co-activator complex provided an initial step toward the understanding of the assembly of a full transcriptionally active NR-co-activator complex. Thus, studying the PGC-1/ERR complex in its native state that could shed new light on mechanisms of cancer resistance, which could be better exploited as a therapeutic strategy.
Also, applications of novel technologies that can shed new light on high-resolution biological structures are urgently needed as potential tools to fully elucidate the function and molecular biology of the PGC-1/ERR network in order to be able to further develop promising therapeutic targets and drug discovery for precision oncology (Fig. 2).36 Advances in cryo-EM are enabling structure determination of smaller protein complexes without additional modifications such as those required for crystallization that restricts the ability to fully access the mechanistic basis of how cancer metabolism can be orchestrated by these NRs and co-factors.
The main purpose of this review is to provide a critical understanding of the structural biology and function of the PGC-1/ERR network derived from work over the past decade. Moreover, focusing on promising therapeutic targets for precision oncology, this review will explore the underlying potential mechanisms of mitochondrial metabolic targets that could be exploited in combination with the PGC-1/ERR network to improve patient care against therapeutic resistance (Table 1).30,37 Understanding this signaling axis could yield crucial insights for the development of novel drugs and therapeutic strategies. This knowledge could lead to a better understanding of the specific type of cancer and patients who are responders and who would benefit from the pharmacological targeting of the PGC-1/ERR network. Thus, the PGC-1/ERR transcriptional axis fits into a novel category of targets that could be useful for exploitation in future research in personalized cancer medicine, so called precision oncology (Fig. 2).
Structure, function, and molecular biology of PGC-1 and ERR
The PGC-1 co-activator family comprises three different members, PGC-1α, PGC-1β, and PGC-1 related co-activator (PRC). Peroxisome proliferator-activated receptor gamma co-activator 1 alpha (PGC-1α) was first reported to regulate thermogenesis, interacting with the nuclear receptor peroxisome proliferator-activated receptor gamma (PPARγ) in brown adipose tissue (BAT).38 The other two members of the PGC-1 family, PGC-1β (PERC) and PRC were described by using sequence homology against PGC-1α.39,40
All members of the PGC-1 co-activator family share extensive protein sequence similarity and distinct domains, which could explain their similar physiological role and protein binding-patterns that have already been described.22 The human PPARGC1A gene on chromosome 4 encodes a 798-amino acid protein also known as PGC-1α. Its homologues, PGC-1β and PRC, comprise 1023 amino acids and 1664 amino acids, respectively. They are encoded by the human PPARGC1B gene on chromosome 5 and the human PPRC1 gene on chromosome 10. Notably, several truncated variants of PGC-1α with distinct transcripts and protein structures have been described. Basically, these variants are generated by alternative splicing and/or differential promoter usage.41,42 The presence of different variants of PGC-1α suggests that different protein variants might have distinct transcripts and protein structures with diverse functions, expression levels, and protein–protein interactions, depending on tissue-type or specific disease-context.41 More structural and functional studies are needed to address the mechanisms of the regulation of PGC-1α variants and to determine the presence of different variants associated with PGC-1β and PRC.
Structurally, all three members of the PGC-1 co-activator family and all nine variants of PGC-1α have one activation domain localized at their N-terminal region, which contains at least two LXXLL nuclear receptor box motifs.38,43 These specific leucine-rich repeats can bind with several NRs, especially the estrogen-related receptor family (ERRα, ERRβ, and ERRγ).44 The C-terminal region of all full-length PGC-1s contains a well-conserved RNA-binding domain, including short serine/arginine-rich stretches (RS), a nuclear localization signal (NLS), and the RNA recognition motif (RRM). The RS domain is present only in PGC-1α and PRC, suggesting that both members regulate RNA splicing and processing of mRNAs. However, further investigation is needed for determining whether the RS domain could be involved in processing PGC-1α variants by alternative splicing.45 The NLS domain plays a role in the maintenance of PGC-1s inside the nucleus. This domain is missing in alternative PGC-1α variants, which might be found in different cellular compartments, such as the cytosol or mitochondria.46,47 The RRM motif also regulates RNA splicing processing of mRNAs, whose function still needs to be substantiated. In addition, other conserved domains have been described in the co-activator family. For example, the aspartic acid (D), histidine (H), aspartic acid (D), and tyrosine (Y) tetrapeptide has been reported to be a binding-partner of the host cell factor (HCF) protein, a transcription factor that regulates gene expression during cell cycle progression. Another motif, including three threonine and four proline amino acids (TPPTTPP), is present in all full-length protein members but the function still needs to be determined.48
Functionally, PGC-1α was described as a docking platform for the assembly of transcriptional machinery, forming a macromolecular complex at specific DNA sequences to drive target gene expression.38 The same group that first described its activity reported that PGC-1α, even without any histone acetyl transferase activity, promotes gene transcription activity through the formation of a multi-protein complex encompassed by histone acetyltransferase proteins, such as cAMP response element-binding protein-binding protein/p300 and steroid receptor co-activator 1 (SRC-1).49 Later, PGC-1α was reported to bind with protein acetyl transferase p300 and the TRAP/mediator complex, mediator of RNA polymerase II transcription subunit 1, leading to the coordination of an important mechanism of chromatin remodeling.50 The idea that the interaction between a co-activator and NRs could recruit proteins responsible for chromatin remodeling, histone acetyl transferase activity, and transcriptional activity has emphasized the complex biological network involved in the PGC-1 axis. In vivo studies using double knockout (PGC-1α and PGC-1β) mice suggested that both members of the family share a similar role in the maintenance of mitochondrial function and energetic metabolic demand in many tissues.51,52 Conversely, the attempt to generate PRC-knockout mice failed because deleting this gene resulted in embryonic lethality.53 Nonetheless, in vitro data have shown that PRC plays an important role in mitochondrial biogenesis, but responds to proliferative signals leading to increased cell growth.54,55
The molecular biology of PGC-1 has been extensively exploited in different fields of health-related research, including cancer, diabetes, cardiovascular disease, and obesity.56 The expression of PGC-1α is characterized by high expression levels in tissues, including kidney, skeletal muscle, liver, heart, neural tissue, and blood mononuclear cells, which exhibit greater energy demand caused by increased mitochondrial activity.57–59 The vast number of different tissues or physiological contexts in which PGC-1α is expressed reflects the large number of different NRs and TFs that are regulated by PGC-1α, possibly including all three estrogen-related receptors (ERRα, ERRβ, and ERRγ), SRC-1, and glucocorticoid receptors (GR), as well as the tumor suppressor p53, PPARγ, forkhead box protein O1 (FoxO1), hepatocyte nuclear factor 4α (HNF-4α), nuclear respiratory factor 1 (NRF-1), the cAMP response element binding protein (CREB), and the signal transducer and activator of transcription 6 (STAT-6).44,60–62
Most intriguing, the activity of PGC-1α can also be regulated by post-translational modification (PTM) mechanisms.63 In fact, most of these regulatory mechanisms are dictated by PTMs, including phosphorylation, acetylation, methylation, ubiquitylation, and O-glycosylation. Clearly, this emphasizes the complex molecular biology and function of the PGC-1 family of proteins. For example, PGC-1α acts as a master regulator of mitochondrial metabolism mediating the entire demand of acetyl-groups and methyl-groups through the tri-carboxylic acid (TCA) cycle and amino acid metabolism, leading to feedback responses of PTM mechanisms, such as acetylation and methylation.64,65 Moreover, PGC-1α contributes to ATP production through the OxPhos process to supply the demand of ATP necessary for phosphorylation processes.66 Thus, PGC-1α is emerging as a fascinating transcriptional metabolic co-regulator playing a role in the maintenance of a tight equilibrium between metabolic precursors and energy production for sustaining PTM mechanisms.67 Besides the complex molecular biology of PGC-1α, more studies need to be conducted to characterize the function of other members of the family.
The molecular biology of PGC-1α is also associated with oncogenes and tumor suppressors. Several studies have demonstrated the interplay between PGC-1α and several oncogenes, including hypoxia-inducible factor 1-alpha (HIF-1α), oncogene carried by the Avian virus, Myelocytomatosis (c-Myc), vascular endothelial growth factor, protein kinase B, and B-Raf proto-oncogene, serine/threonine kinase/microphthalmia-associated transcription factor (MITF), as well as tumor suppressor p53 and 5’ AMP-activated protein kinase (AMPK).16,62,68–70 For example, higher PGC-1α expression predicts poor outcome in human melanoma, when the expression of PGC-1α is regulated by the MITF increasing mitochondrial function and resistance to oxidative stress.16 Moreover, in wild-type p53 lung adenocarcinoma, PGC-1α binds with p53, promoting cell survival in the presence of metabolic stress.62,71 However, the mechanism by which PGC-1α directly modulates oncogenic and tumor suppressor signaling in cancer cells is still unclear.
Different members of the PGC-1 family can act through similar or different molecular mechanisms depending on the cancer type and the stage of disease.22 Both PGC-1α and PGC-1β exhibit a similar tissue-specific expression pattern.44 As indicated above for PGC-1α, the molecular biology of PGC-1β in cancer is also associated with oncogenes, such as HIF-1α and c-Myc, and the tumor suppressor gene known as Von Hippel-Lindau (VHL).72 c-Myc is known to control the transcription of the gene encoding PGC-1β73 and the VHL/HIF-1 pathway can act as a repressor of c-Myc.72 For instance, the loss of PGC-1β and PGC-1α expression is a major factor contributing to impaired mitochondrial respiration in VHL-deficient renal carcinoma cells.72,74 Interestingly, the molecular biology of PGC-1β and PGC-1α seems to be modulated toward a metabolic rewiring through the VHL/HIF-1 pathway, leading to “the Warburg effect”, instead of mitochondrial OxPhos metabolism, through the inhibition of the transcriptional c-Myc/PGC-1β axis. Therefore, targeting PGC-1β and PGC-1α and rescuing the mitochondrial metabolic phenotype could be exploited as a therapeutic approach for the treatment of VHL-deficient renal carcinoma cells.74 Furthermore, a genomic study designed to assess the activity of ERRα in eight hundred breast tumor samples suggested that the molecular biology of PGC-1β in breast cancer progression relies on the c-Myc pathway. The authors had shown that the insulin-like growth factor 1 receptor pathway controls the stabilization of the c-Myc protein, leading to the up-regulation of PGC-1β.75
PGC-1α and PGC-1β were also reported to play a role in the resistance of ER-positive/tamoxifen-sensitive breast tumors.76 The interaction between PGC-1β and ERRα mediates a positive transcriptional regulation of receptor tyrosine-protein kinase (ERBB2) expression and co-amplifies genes associated with the ERBB2 amplicon. This biological mechanism was reported as a major factor contributing to tamoxifen resistance in a breast cancer model.76 Moreover, PGC-1β mediates adaptive resistance to genotoxic stress in lung cancer associated with mitochondrial DNA mutations.77
Taken together, these findings highlight a similar oncogenic network and resistance-related mechanisms in the cancer biology of both members of the PGC-1 family. On the other hand, the third member of the PGC-1 family, called PRC, seems to be restricted to the regulation of the expression of the mitochondrial biogenesis genes in proliferating cells. In contrast to both PGC-1α and PGC-1β, more studies are still needed to determine whether PRC plays a role in cancer progression by associating with oncogenic pathways.44 However, one study has recently suggested that the molecular biology of PRC could be associated with the c-Myc pathway in response to mitochondrial stress.78
Progress has been made in understanding the signaling network between PGC-1α and ERRα. This network has emerged as an important nuclear transcriptional axis and metabolic signaling pathway in regulating metabolic adaptation in specific cancer types, including breast cancer, prostate cancer, and melanoma.12 A majority of the literature has described the function of the founding members of the co-activator family, PGC-1α and ERRα, as a pivotal axis that might be linked to metabolic addiction of specific cancer cells that rely on mitochondrial metabolism for survival.
ERR
The ERR family encompasses three different members, ERRα (NR3B1), ERRβ (NR3B2), and ERRγ (NR3B3).79,80 All three members belong to a subfamily of orphan NRs, sharing sequence homology with the estrogen receptor (ER), but do not require endogenous ligands for activation. In fact, ERRs were first discovered by cDNA library screening using the ERα homology sequence to identify novel steroid receptors.80 The DNA-binding domains (DBD) and the ligand-binding domains (LBD) of ERRα and ERα are present in both classical estrogen receptor and orphan receptor families of NRs, but display different molecular biology and function, such as hormone-independent transcriptional activation and ERRα binding with co-activators.81 For instance, the ERR-LBD is only 36% similar with the ERs, which could explain why some ERα ligands, such as 17β-estradiol, estrone, and estriol do not activate ERRα.
Structurally, the N-terminal region of ERRs contains a DNA-binding domain and a ligand-independent transcriptional activation function (AF-1) that is poorly conserved within the ERR family members. The N-terminal AF-1 domain can weakly bind several TFs. The recent advances in structural biology have shown that this domain is subjected to various post-translational modifications (e.g., sumoylation and phosphorylation). Only a few groups have described the high-resolution structure of AF-1, which is at least partially due to the very flexible structure and low-affinity nuclear co-activator-binding.82,83 The DBD domain can also be acetylated, which controls the affinity by which ERR can bind with its own element responsive sequence (ERRE).84 Interestingly, the DBD domain is well-conserved across the members of this subfamily of NRs and contains two highly conserved zinc finger motifs with a specific-DNA binding sequence, TCAAGGTCA. The ERRE is exclusive for the DNA-binding sequence of all three ERR members. However, ERR members have been shown to bind with the responsive element sequence of ER (ERE), suggesting that ERRs play an important role in regulating similar downstream genes controlled by the ER pathway.82 The C-terminal region of ERRs comprises an LBD containing a conserved AF-2 helix motif. The LBD of ERR is required for the physical protein interaction with co-activator and co-repressor proteins, such as PGC-1α and PGC-1β or receptor-interacting protein 140 and nuclear receptor co-repressor 1, respectively. The conformation of the LBD of ERR, even in the absence of ligand, is responsible for constitutive transcriptional activation of ERR, due to its unique conformation that facilitates the recruitment of nuclear co-activators.79 The LBD domain of ERR binds with the nuclear receptor box motif LXXLL of PGC-1α, forming a binary complex, the stoichiometry of which is defined as two molecules of ERR (dimer) and one molecule of PGC-1α.21 Despite the importance of co-repressors in the context of ERR function, we focus herein on the structure, function, and molecular biology of ERR and nuclear co-activators, concentrating on how the PGC-1/ERR network can be exploited as a promising therapeutic target to improve patient outcome. The role of co-repressors and ERRs has been extensively reviewed.82
Previously, the observation of a largely occluded ligand-binding pocket in the transcriptionally active conformation of ERRα that has led some to the conclusion that ERRα does not lend itself to direct activation by small molecule agents. However, the crystal structure of the ERRα–LBD has presented an opportunity for generating selective inverse agonists.85,86 Currently, a series of diaryl ether-based thioazolidinediones has been screened resulting in the identification of specific inverse agonists of ERRα.87 Interestingly, compound 29 was obtained for the ERRα–LBD by solving the X-ray crystal structure.85 Compound 29 acts as a ligand of ERRα through a covalent interaction leading to conformational changes (in the amino acid Phe328) that disrupts the interaction between ERRα and PGC-1α.87 The biological consequence of this binding has shown growth-inhibitory therapeutic effects in certain cancers such as breast cancer and melanoma.17,29,88 Furthermore, several other compounds have been developed as inverse agonists of ERRα, such as compound 1a, compound 3, N-arylindole, XCT790, and GSK0903. However, more biological study is needed to determine the specificity of these compounds in targeting only the PGC-1/ERR network. For instance, XCT790 was previously developed as a specific inverse agonist of ERRα with the capacity to disrupt the interaction between ERRα and PGC-1α,89 leading to growth-inhibitory therapeutic effects in breast cancer.29 However, XCT790 does not seem to be a very specific inverse agonist of ERRα because at nanomolar concentrations (10-fold lower than the concentration required to inhibit ERRα), XCT790 is a potent mitochondrial uncoupler, leading to a rapid depletion of ATP and activation of AMPK. The authors suggested that XCT790 is a potent, fast-acting, mitochondrial uncoupler that acts independent of its inhibition of ERRα.90
Notably, Kallen and co-workers showed for the first time the X-ray crystal structure of the ERRα-LBD with one co-activator peptide derived from PGC-1α, leading to a ligand-independent transcriptional activation by ERR.91 In 2007, the same group used an inverse agonist called compound 1a to show that the binding interface of this compound with the ERRα-LBD comprised the helix H12 together with helices H3 and H4. Interestingly, the compound imposed dramatic conformational changes in the amino acid Phe328 located at H3, moving away the amino acid Phe510 of H12, which contains the co-activator groove of the ERRα-LBD. Based on this evidence, they proposed a novel molecular mechanism supporting the idea that the helix H12 binds with the co-activator peptide or compound 1a filling the co-activator groove of the AF-2 domain of ERRα. Hence, no co-activators and co-repressors are allowed to interact with ERRα.85 The X-ray crystal structure of the ERRα-LBD with compound 29 displayed similar dramatic conformational changes and side chain rotation of the amino acid Phe328 (H3). The significant change in structure was observed in the loop between helices H11 and H12. Hence, the C-terminal AF-2 domain that is responsible for transducing the constitutive activity for ERRα is no longer functional.87
As indicated earlier, the function and molecular biology of ERRs under normal physiological conditions are associated with the regulation of metabolic genes that are involved in glycolysis, the TCA cycle, and mitochondrial metabolism. These receptors also influence enzymes participating in OxPhos (e.g., several components of mitochondrial respiratory complexes), amino acid metabolism, and lipid synthesis (Table 1).
However, in the context of cancer, the different members of the ERR family seem to exhibit distinct functions in cancer progression. ERRα expression is associated with poor prognosis in breast tumors because it appears to drive lapatinib-resistance and tamoxifen-resistance in those patients.27,92 Several studies have shown that ERRα expression is associated with an increased risk of recurrence and worse prognosis, as well as drug resistance in patients with breast cancer.92–94 ERRα has been shown to increase the expression of ERBB2, mediating endocrine-resistant ERα-positive cells.76,95 Interestingly, ERRα has been shown to mediate pro-survival functions and represents a novel therapeutic target in a particularly aggressive melanoma phenotype, known as PGC-1α-positive melanomas.17
In contrast, the role of the ERRγ isoform in cancer biology seems to be paradoxical, whether this NR functions as an oncogene or as a tumor suppressor. Recently, genomic analysis in gastric cancer revealed that ERRγ acts as a tumor suppressor by directly targeting the Wnt signaling pathway. In fact, activating ERRγ expression by a specific agonist, DY131, inhibits gastric cancer cell growth and improved patient prognosis.96 Furthermore, ERRγ was described as an anti-proliferative target in androgen-sensitive and androgen-insensitive prostate cancer cells.97 Conversely, ERRγ is up-regulated in liver cancer and its inhibition suppresses cancer cell survival through the p21 and p27 proteins.98
Most intriguing is the paradoxical function of ERRγ in breast cancer progression. Recently, several studies reported that the hyper-activation of ERRγ induces a pro-survival transcriptional program in tamoxifen-treated breast cancer, as reported to the isoform ERRα.99,100 Conversely, in breast tumors co-expressing ER and PR, ERRγ induces E-cadherin expression and promotes the mesenchymal-to-epithelial transition (MET), resulting in the inhibition of tumor growth.101,102 In spite of the inconsistencies, the expression of ERRα and ERRγ in breast cancer and prostate cancer seems to be inversely correlated, where the ERRα is associated with a more aggressive disease and the expression of ERRγ is associated with a favorable prognosis of patients with breast and skin cancer.81,103 For instance, in androgen-dependent and castration-resistant prostate cancer, the progression of the disease is associated with a loss of ERRγ expression, whereas strategies to reactivate ERRγ expression could be exploited as a generalized therapeutic approach to manage prostate cancer.19
Overall, ERRα and ERRγ are considered key regulators of metabolic reprogramming in breast and prostate cancer. However, how this nuclear receptors network influences the metabolic state of cancer cells seems to be very complex and diverse, depending on the cancer type. Regarding the third isoform of this family, in prostate cancer the expression of ERRβ transactivates a promoter upstream of the cyclin-dependent kinase inhibitor, p21 gene, resulting in the inhibition of cell cycle progression, whereas the potential role of ERRβ in breast cancer remains unclear.104
Clearly a close relationship exists between PGC-1/ERR activity and cancer therapeutic resistance. New insights into the PGC-1α/ERRα network in cancer will be discussed next.
New biological insights into the PGC-1α co-activator and ERRα
Reprogramming of energy metabolism and evading immune destruction have been recently included in the select list of biological capabilities or hallmarks acquired during the development of cancer.105 Notably, the plasticity of cancer cells toward metabolic reprogramming has gained attention in the mechanisms of drug resistance.30,106–108 The entire molecular network that orchestrates the inherent ability of tumor cells to switch between different metabolic profiles, depending on the microenvironment stimuli, still needs to be fully elucidated.
The most important metabolic plasticity mechanisms in cancer rely on glycolysis-dependent or mitochondrial OxPhos-addiction activities.109,110 Both are considered metabolic hallmarks of cancer cells because they are involved in the direct activation of many oncogenic pathways.107,111–113 However, not all reprogrammed metabolic activities contribute equally to cancer progression. Yet, deregulation of mitochondrial metabolism could be considered a potential therapeutic target in tumor resistance.114 Because the PGC-1α/ERRα network is a master regulator of mitochondrial biogenesis, it could be considered a nodal regulatory step capable of controlling the entire cellular metabolism and, at least in part, in modulating this resistance-related mechanism in cancer, leading to cancer recurrence (Fig. 1).115,116
Recent studies have provided a rationale for therapeutically targeting mitochondria in certain types of cancer.11,16,117,118 Different modulators of mitochondrial activity that have been approved by The Food and Drug Administration (FDA) to treat chronic diseases, such as type 2 diabetes, cardiovascular disease, obesity, and acute myelogenous leukemia (AML), are currently being explored in clinical trials to determine potential efficacy to treat cancer. For example, anti-diabetic drugs like metformin and phenformin, the anti-obesity drug, etomoxir, or the anti-AML drug, enasidenib (IDHIFA), might provide new hope for treating responder patients whose cancer cell survival mechanism relies on mitochondrial metabolism.119 Although the clinical use of these drugs in combination with conventional chemotherapeutic drugs has led to clinical improvement outcomes, the drug-resistance mechanisms of cancer cells still remain unclear.
From metabolism to precision oncology
To focus on precision oncology and the development of effective drug design to eradicate drug-resistant cancer cells, the signaling network associated with PGC-1α/ERRα must be considered a novel targetable vulnerability in cancer cells. In examining the concept of metabolic rewiring, targeting this particular transcriptional/mitochondrial metabolic network might expose other vulnerabilities to oxidative stress in tumors (Table 1).34 Thus, targeting PGC-1α/ERRα in combination with additional metabolic vulnerabilities such as the respiratory chain defects, antioxidant programs, and TCA cycle enzymes might lead to the disruption of the nutrient sensing pathways responsible for survival of residual cells (Fig. 2b).120
A thorough analysis of the literature shows that many of the human NRs and their co-activators have been extensively studied using traditional structural analysis. However, only studies with partial protein structure and limited protein–protein interaction have been purposed, which has led to an incomplete understanding of the entire functional mechanism of the PGC-1/ERR network.21,85,91,121,122 Therefore, new insights into the overall structure of the PGC-1/ERR complex could provide insights for effectively targeting cancer resistance mechanisms and answering important questions associated with the function of this nodal transcriptional signaling network. Essential questions that could be answered include whether the full-length protein structure of both components would impose particular changes in the protein–protein interaction model that could affect the discovery and development of new therapies targeting this complex. In addition, structural insights could assist in determining whether the PGC-1α and ERRα proteins interact with specific oncogenes or tumor suppressors and thus play different roles in metabolic plasticity favoring drug resistance or decreasing cancer progression.
In this context, recent advances have been made in successfully determining high-resolution biological structures. Solving the PGC-1/ERR complex structure in its physiological state might lead to the discovery of novel mechanistic insights into the biology of the PGC-1/ERR axis, as well as the identification of novel binding partners that might have clinical relevance to treat cancers that rely on mitochondrial activity.17,123 Therefore, more studies must be pursued to predict how this complex might be exploited in basic and clinical research, leading to the elucidation of dynamic biological processes in their native states and drug discovery for personalized medicine.
Cryo-EM as a potential tool for the visualization of protein complexes
Cryo-EM is an outstanding new technology that is based on transmission electron microscopy in which a protein sample is examined in its native state at cryogenic temperatures, which can lead to successful resolution of the protein’s structure at the subatomic or atomic level. This technology has several advantages over X-ray crystallography because the protein is frozen in its native state, which can overcome technical problems with proteins that are refractory to crystallization or are just difficult to crystallize. Cryo-EM has been used successfully to resolve proteins of greater than 300 kDa to produce images with resolution as good as 2.2 Å. For example, the large β-galactosidase protein (465 kDa) has been successfully resolved by cryo-EM.124 Notably, high resolution cryo-EM images could reveal protein–protein interactions, conformational changes, and interactions between proteins and drug targets at an atomic level of precision.125 Despite the lower size limitation of protein structures that can be resolved by cryo-EM, one study showed that cryo-EM is suitable to solve the structure of small metabolic enzymes at near-atomic resolution.126 The authors presented the structure of a known therapeutic cancer target, isocitrate dehydrogenase (IDH, 93 kDa), with a resolution of 3.8 Å, which could facilitate research toward therapeutic targets and drug discovery. This is possible because crossing the 3 Å resolution level and obtaining protein structures with sizes <100 kDa might allow scientists to investigate drug-target interactions and dynamic conformational states of protein complexes (Fig. 2a).126,127 This could even benefit patients already treated with conventional chemotherapies, such as lapatinib and tamoxifen in metastatic breast cancer, epidermal growth factor inhibitors (EGFRi) in EGFR-driven lung adenocarcinoma, mitogen-activated protein kinase inhibitors (MAPKi) in melanoma, and 5-fluorouracil in Myc/PGC-1α-driven pancreatic cancer.30
The PGC-1/ERR network as a promising therapeutic target for precision oncology
Panomics data, including genomics, transcriptomics, proteomics and metabolomics, in combination with patient-matched data, are currently being used for designing treatments for personalized medicine.128 New technologies, including the clustered regularly interspaced short palindromic repeats-associated 9 system for targeted genome editing and cryo-EM, can provide mechanistic understanding of complex biological processes and are the potential tools for identifying promising therapeutic targets for precision oncology.36
The relevance of deciphering the role of the PGC-1/ERR signaling network and the therapeutic implication for precision oncology relies on several factors. First, the PGC-1/ERR transcriptional network is responsible for metabolic plasticity, which corresponds well with therapeutic resistance.42 For example, like all cancers, breast cancer is considered a heterogeneous disease and currently the approach to treat breast cancer is still based on histopathological markers that rely on tumor subtypes to evaluate and treat each patient.129–131 In this era of precision oncology, seeking clinically relevant biomarkers that might be exploited for therapeutic purposes is highly pertinent and could be combined with conventional therapies to generate synthetic lethality in breast cancer.132,133 Synthetic lethality is defined as any genetic mutation, chemical or drug perturbation, and environmental conditions that have a unique effect on cell viability but when exploited in combination results in cell death.134 For instance, the concept of synthetic lethality is used to treat breast cancer based on the treatment of BRCA1-deficient patients with PARP inhibitors.135 However, the means by which the PGC-1/ERR axis can be targeted to interfere with the metabolic synthetic lethality of mitochondrial enzymes still remains largely unknown.34 Second, the PGC-1/ERR complex is an important nuclear transcriptional axis that orchestrates the mitochondrial bioenergetic requirements of tumors and thereby it could be therapeutically exploited in metabolic-addictive cancers as a new metabolic vulnerability.136 Third, preventing or bypassing drug resistance is arguably the most important medical need in cancer research.114 Cleary, the identification of biomarker-defined patient populations that will most likely respond to specific drugs is critical.114 Thus, silencing the PGC-1/ERR axis in drug-resistant cancers with a high level of OxPhos might culminate in specific elimination of these cells. Finally, the attempt to access the native and physiological state of PGC-1/ERR axis will allow us to a greater understanding of the PGC-1/ERR axis in cancer biology. Moreover, the potential discovery of important oncogenes or tumor suppressors that interact with this complex will lead to the ability to fully access the mechanistic basis of how these NRs and co-activators orchestrate metabolic plasticity toward drug-resistance in cancer treatment.
Concluding remarks
Despite the progress that has been made in using structural biology for potential drug discovery to increase patient outcomes underlying precision oncology, new therapies that effectively eradicate drug-resistant cancer cells are an immediate clinical necessity. The PGC-1/ERR network holds promise as a therapeutic target for precision medicine, because this transcriptional axis orchestrates the expression of several genes involved in mitochondrial biogenesis and cell metabolism. Bosc et al. have suggested that the resistance mechanism in cancer might be associated with a shift toward an increased OxPhos status that should be considered a distinctive characteristic of drug resistance.30 The master PGC-1/ERR axis that controls mitochondrial OxPhos activity should be considered as a new pathway that drives resistance in tumor progression. The metabolic vulnerability concept that explores potential mitochondrial targets to treat cancer, rather than conventional chemotherapy, extends the concept that the up-stream mitochondrial biogenesis PGC-1/ERR network must be included as a novel targetable metabolic vulnerability (Fig. 2b). Furthermore, a native-state high-resolution structure of this nuclear complex is urgently needed.89,137–139 Overall, further studies are needed to determine the role of PGC-1/ERR network as a key metabolic vulnerability associated with cancer cell progression by using cryo-EM as a promising tool for drug discovery in precision oncology. Such refinements could provide opportunities to be exploited in therapeutic resistance.
Reporting summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This work was supported by The Hormel Foundation and National Institutes of Health grants CA166011, CA187027, and CA196639, and CAPES 9281-13-4. We thank Dr. Ferdinando Chiaradonna for important discussion regarding this manuscript.
Author contributions
A.M.B. and Z.D. supervised the conception of the work and participated in discussion and revision of the manuscript. H.D.V. wrote the manuscript and produced figures. H.D.V., A.M.B., and Z.D. revised the manuscript. All authors approved the final version.
Data availability
Data sharing not applicable to this article as no datasets were generated or analyzed for the current study.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies the paper on the npj Precision Oncology website (10.1038/s41698-019-0081-6).
References
- 1.Dasgupta S, Lonard DM, O’Malley BW. Nuclear receptor coactivators: master regulators of human health and disease. Annu Rev. Med. 2014;65:279–292. doi: 10.1146/annurev-med-051812-145316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lonard DM, O’Malley BW. Molecular pathways: targeting steroid receptor coactivators in cancer. Clin. Cancer Res. 2016;22:5403–5407. doi: 10.1158/1078-0432.CCR-15-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans RM, Mangelsdorf DJ. Nuclear receptors, RXR, and the big bang. Cell. 2014;157:255–266. doi: 10.1016/j.cell.2014.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baek SH, Kim KI. Emerging roles of orphan nuclear receptors in cancer. Annu Rev. Physiol. 2014;76:177–195. doi: 10.1146/annurev-physiol-030212-183758. [DOI] [PubMed] [Google Scholar]
- 5.Sonoda J, Pei L, Evans RM. Nuclear receptors: decoding metabolic disease. FEBS Lett. 2008;582:2–9. doi: 10.1016/j.febslet.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sever R, Glass CK. Signaling by nuclear receptors. Cold Spring Harb. Perspect. Biol. 2013;5:a016709. doi: 10.1101/cshperspect.a016709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schreiber SN, Knutti D, Brogli K, Uhlmann T, Kralli A. The transcriptional coactivator PGC-1 regulates the expression and activity of the orphan nuclear receptor estrogen-related receptor alpha (ERRalpha) J. Biol. Chem. 2003;278:9013–9018. doi: 10.1074/jbc.M212923200. [DOI] [PubMed] [Google Scholar]
- 8.Fan W, et al. ERRγ promotes angiogenesis, mitochondrial biogenesis, and oxidative remodeling in PGC1α/β-deficient muscle. Cell Rep. 2018;22:2521–2529. doi: 10.1016/j.celrep.2018.02.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duchen MR. Mitochondria in health and disease: perspectives on a new mitochondrial biology. Mol. Asp. Med. 2004;25:365–451. doi: 10.1016/j.mam.2004.03.001. [DOI] [PubMed] [Google Scholar]
- 10.Gravel SP. Deciphering the dichotomous effects of PGC-1α on tumorigenesis and metastasis. Front. Oncol. 2018;8:75. doi: 10.3389/fonc.2018.00075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LeBleu VS, et al. PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat. Cell Biol. 2014;16:1001–1015. doi: 10.1038/ncb3039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tan Z, et al. The role of PGC1α in cancer metabolism and its therapeutic implications. Mol. Cancer Ther. 2016;15:774–782. doi: 10.1158/1535-7163.MCT-15-0621. [DOI] [PubMed] [Google Scholar]
- 13.Torrano V, et al. The metabolic co-regulator PGC1α suppresses prostate cancer metastasis. Nat. Cell Biol. 2016;18:645–656. doi: 10.1038/ncb3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deblois G, St-Pierre J, Giguere V. The PGC-1/ERR signaling axis in cancer. Oncogene. 2013;32:3483–3490. doi: 10.1038/onc.2012.529. [DOI] [PubMed] [Google Scholar]
- 15.Yun SH, Han SH, Park JI. Peroxisome proliferator-activated receptor. PPAR Res. 2018;2018:6727421. doi: 10.1155/2018/6727421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vazquez F, et al. PGC1alpha expression defines a subset of human melanoma tumors with increased mitochondrial capacity and resistance to oxidative stress. Cancer Cell. 2013;23:287–301. doi: 10.1016/j.ccr.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Luo C, et al. ERRα maintains mitochondrial oxidative metabolism and constitutes an actionable target in PGC1α-elevated melanomas. Mol. Cancer Res. 2017;15:1366–1375. doi: 10.1158/1541-7786.MCR-17-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wallace M, Metallo CM. PGC1α drives a metabolic block on prostate cancer progression. Nat. Cell Biol. 2016;18:589–590. doi: 10.1038/ncb3365. [DOI] [PubMed] [Google Scholar]
- 19.Audet-Walsh Eacute. Androgen-dependent repression of ERRγ reprograms metabolism in prostate cancer. Cancer Res. 2017;77:378–389. doi: 10.1158/0008-5472.CAN-16-1204. [DOI] [PubMed] [Google Scholar]
- 20.Deblois G, Giguere V. Oestrogen-related receptors in breast cancer: control of cellular metabolism and beyond. Nat. Rev. Cancer. 2013;13:27–36. doi: 10.1038/nrc3396. [DOI] [PubMed] [Google Scholar]
- 21.Devarakonda S, et al. Disorder-to-order transition underlies the structural basis for the assembly of a transcriptionally active PGC-1alpha/ERRgamma complex. Proc. Natl Acad. Sci. USA. 2011;108:18678–18683. doi: 10.1073/pnas.1113813108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo C, Widlund HR, Puigserver P. PGC-1 coactivators: shepherding the mitochondrial biogenesis of tumors. Trends Cancer. 2016;2:619–631. doi: 10.1016/j.trecan.2016.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tam IS, Giguère V. There and back again: the journey of the estrogen-related receptors in the cancer realm. J. Steroid Biochem Mol. Biol. 2016;157:13–19. doi: 10.1016/j.jsbmb.2015.06.009. [DOI] [PubMed] [Google Scholar]
- 24.Mastropasqua F, Girolimetti G, Shoshan M. PGC1α: friend or foe in cancer? Genes. 2018;9:48. doi: 10.3390/genes9010048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chang CY, McDonnell DP. Molecular pathways: the metabolic regulator estrogen-related receptor α as a therapeutic target in cancer. Clin. Cancer Res. 2012;18:6089–6095. doi: 10.1158/1078-0432.CCR-11-3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martin-Martin N, Carracedo A, Torrano V. Metabolism and transcription in cancer: merging two classic tales. Front. Cell Dev. Biol. 2017;5:119. doi: 10.3389/fcell.2017.00119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deblois G, et al. ERRα mediates metabolic adaptations driving lapatinib resistance in breast cancer. Nat. Commun. 2016;7:12156. doi: 10.1038/ncomms12156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McGuirk S, et al. PGC-1α supports glutamine metabolism in breast cancer. Cancer Metab. 2013;1:22. doi: 10.1186/2049-3002-1-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park S, et al. ERRα-regulated lactate metabolism contributes to resistance to targeted therapies in breast cancer. Cell Rep. 2016;15:323–335. doi: 10.1016/j.celrep.2016.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bosc C, Selak MA, Sarry JE. Resistance is futile: targeting mitochondrial energetics and metabolism to overcome drug resistance in cancer treatment. Cell Metab. 2017;26:705–707. doi: 10.1016/j.cmet.2017.10.013. [DOI] [PubMed] [Google Scholar]
- 31.Audet-Walsh Eacute, et al. The PGC-1α/ERRα axis represses one-carbon metabolism and promotes sensitivity to anti-folate therapy in breast cancer. Cell Rep. 2016;14:920–931. doi: 10.1016/j.celrep.2015.12.086. [DOI] [PubMed] [Google Scholar]
- 32.Gentric G, Mieulet V, Mechta-Grigoriou F. Heterogeneity in cancer metabolism: new concepts in an old field. Antioxid. Redox Signal. 2017;26:462–485. doi: 10.1089/ars.2016.6750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Valcarcel-Jimenez L, Gaude E, Torrano V, Frezza C, Carracedo A. Mitochondrial metabolism: yin and yang for tumor progression. Trends Endocrinol. Metab. 2017;28:748–757. doi: 10.1016/j.tem.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zecchini V, Frezza C. Metabolic synthetic lethality in cancer therapy. Biochim Biophys. Acta. 2017;1858:723–731. doi: 10.1016/j.bbabio.2016.12.003. [DOI] [PubMed] [Google Scholar]
- 35.Yi P, et al. Structure of a biologically active estrogen receptor-coactivator complex on DNA. Mol. Cell. 2015;57:1047–1058. doi: 10.1016/j.molcel.2015.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shin SH, Bode AM, Dong ZG. Addressing the challenges of applying precision oncology. Npj Precision. NPJ Precision Oncol. 2017;1:10. doi: 10.1038/s41698-017-0018-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zong WX, Rabinowitz JD, White E. Mitochondria and cancer. Mol. Cell. 2016;61:667–676. doi: 10.1016/j.molcel.2016.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Puigserver P, et al. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998;92:829–839. doi: 10.1016/S0092-8674(00)81410-5. [DOI] [PubMed] [Google Scholar]
- 39.Andersson U, Scarpulla RC. Pgc-1-related coactivator, a novel, serum-inducible coactivator of nuclear respiratory factor 1-dependent transcription in mammalian cells. Mol. Cell Biol. 2001;21:3738–3749. doi: 10.1128/MCB.21.11.3738-3749.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kressler D, Schreiber SN, Knutti D, Kralli A. The PGC-1-related protein PERC is a selective coactivator of estrogen receptor alpha. J. Biol. Chem. 2002;277:13918–13925. doi: 10.1074/jbc.M201134200. [DOI] [PubMed] [Google Scholar]
- 41.Martínez-Redondo V, et al. Peroxisome proliferator-activated receptor γ coactivator-1 α isoforms selectively regulate multiple splicing events on target genes. J. Biol. Chem. 2016;291:15169–15184. doi: 10.1074/jbc.M115.705822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang G, et al. Targeting mitochondrial biogenesis to overcome drug resistance to MAPK inhibitors. J. Clin. Invest. 2016;126:1834–1856. doi: 10.1172/JCI82661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knutti D, Kaul A, Kralli A. A tissue-specific coactivator of steroid receptors, identified in a functional genetic screen. Mol. Cell Biol. 2000;20:2411–2422. doi: 10.1128/MCB.20.7.2411-2422.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Villena JA. New insights into PGC-1 coactivators: redefining their role in the regulation of mitochondrial function and beyond. FEBS J. 2015;282:647–672. doi: 10.1111/febs.13175. [DOI] [PubMed] [Google Scholar]
- 45.Monsalve M, et al. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell. 2000;6:307–316. doi: 10.1016/S1097-2765(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 46.Aquilano K, et al. Peroxisome proliferator-activated receptor gamma co-activator 1alpha (PGC-1alpha) and sirtuin 1 (SIRT1) reside in mitochondria: possible direct function in mitochondrial biogenesis. J. Biol. Chem. 2010;285:21590–21599. doi: 10.1074/jbc.M109.070169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chang JS, et al. Regulation of NT-PGC-1alpha subcellular localization and function by protein kinase A-dependent modulation of nuclear export by CRM1. J. Biol. Chem. 2010;285:18039–18050. doi: 10.1074/jbc.M109.083121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin J, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- 49.Puigserver P, et al. Activation of PPARgamma coactivator-1 through transcription factor docking. Science. 1999;286:1368–1371. doi: 10.1126/science.286.5443.1368. [DOI] [PubMed] [Google Scholar]
- 50.Wallberg AE, Yamamura S, Malik S, Spiegelman BM, Roeder RG. Coordination of p300-mediated chromatin remodeling and TRAP/mediator function through coactivator PGC-1alpha. Mol. Cell. 2003;12:1137–1149. doi: 10.1016/S1097-2765(03)00391-5. [DOI] [PubMed] [Google Scholar]
- 51.Lai L, et al. Transcriptional coactivators PGC-1alpha and PGC-lbeta control overlapping programs required for perinatal maturation of the heart. Genes Dev. 2008;22:1948–1961. doi: 10.1101/gad.1661708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zechner C, et al. Total skeletal muscle PGC-1 deficiency uncouples mitochondrial derangements from fiber type determination and insulin sensitivity. Cell Metab. 2010;12:633–642. doi: 10.1016/j.cmet.2010.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.He X, et al. Peri-implantation lethality in mice lacking the PGC-1-related coactivator protein. Dev. Dyn. 2012;241:975–983. doi: 10.1002/dvdy.23769. [DOI] [PubMed] [Google Scholar]
- 54.Gleyzer N, Scarpulla RC. PGC-1-related coactivator (PRC), a sensor of metabolic stress, orchestrates a redox-sensitive program of inflammatory gene expression. J. Biol. Chem. 2011;286:39715–39725. doi: 10.1074/jbc.M111.291575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gleyzer N, Scarpulla RC. Activation of a PGC-1-related coactivator (PRC)-dependent inflammatory stress program linked to apoptosis and premature senescence. J. Biol. Chem. 2013;288:8004–8015. doi: 10.1074/jbc.M112.426841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Handschin C, Spiegelman BM. Peroxisome proliferator-activated receptor gamma coactivator 1 coactivators, energy homeostasis, and metabolism. Endocr. Rev. 2006;27:728–735. doi: 10.1210/er.2006-0037. [DOI] [PubMed] [Google Scholar]
- 57.Lin J, Handschin C, Spiegelman BM. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005;1:361–370. doi: 10.1016/j.cmet.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 58.Puigserver P. Tissue-specific regulation of metabolic pathways through the transcriptional coactivator PGC1-alpha. Int J. Obes. 2005;29:S5–S9. doi: 10.1038/sj.ijo.0802905. [DOI] [PubMed] [Google Scholar]
- 59.Fabregat-Andrés Oacute. Induction of PGC-1α expression can be detected in blood samples of patients with ST-segment elevation acute myocardial infarction. PLoS ONE. 2011;6:e26913. doi: 10.1371/journal.pone.0026913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huss JM, Kopp RP, Kelly DP. Peroxisome proliferator-activated receptor coactivator-1alpha (PGC-1alpha) coactivates the cardiac-enriched nuclear receptors estrogen-related receptor-alpha and -gamma. Identification of novel leucine-rich interaction motif within PGC-1alpha. J. Biol. Chem. 2002;277:40265–40274. doi: 10.1074/jbc.M206324200. [DOI] [PubMed] [Google Scholar]
- 61.Kamei Y, et al. PPARgamma coactivator 1beta/ERR ligand 1 is an ERR protein ligand, whose expression induces a high-energy expenditure and antagonizes obesity. Proc. Natl Acad. Sci. USA. 2003;100:12378–12383. doi: 10.1073/pnas.2135217100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sen N, Satija YK, Das S. PGC-1α, a key modulator of p53, promotes cell survival upon metabolic stress. Mol. Cell. 2011;44:621–634. doi: 10.1016/j.molcel.2011.08.044. [DOI] [PubMed] [Google Scholar]
- 63.Fernandez-Marcos PJ, Auwerx J. Regulation of PGC-1α, a nodal regulator of mitochondrial biogenesis. Am. J. Clin. Nutr. 2011;93:884S–890S. doi: 10.3945/ajcn.110.001917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Martinez-Pastor B, Cosentino C, Mostoslavsky R. A tale of metabolites: the cross-talk between chromatin and energy metabolism. Cancer Discov. 2013;3:497–501. doi: 10.1158/2159-8290.CD-13-0059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wong CC, Qian Y, Yu J. Interplay between epigenetics and metabolism in oncogenesis: mechanisms and therapeutic approaches. Oncogene. 2017;36:3359–3374. doi: 10.1038/onc.2016.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Ardito F, Giuliani M, Perrone D, Troiano G, Lo Muzio L. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review) Int J. Mol. Med. 2017;40:271–280. doi: 10.3892/ijmm.2017.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fan J, Krautkramer KA, Feldman JL, Denu JM. Metabolic regulation of histone post-translational modifications. ACS Chem. Biol. 2015;10:95–108. doi: 10.1021/cb500846u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Arany Z, et al. HIF-independent regulation of VEGF and angiogenesis by the transcriptional coactivator PGC-1alpha. Nature. 2008;451:1008–1012. doi: 10.1038/nature06613. [DOI] [PubMed] [Google Scholar]
- 69.Li X, Monks B, Ge Q, Birnbaum MJ. Akt/PKB regulates hepatic metabolism by directly inhibiting PGC-1alpha transcription coactivator. Nature. 2007;447:1012–1016. doi: 10.1038/nature05861. [DOI] [PubMed] [Google Scholar]
- 70.Sancho P, et al. MYC/PGC-1α balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell Metab. 2015;22:590–605. doi: 10.1016/j.cmet.2015.08.015. [DOI] [PubMed] [Google Scholar]
- 71.Cioce M, Blandino G. PGC1α confers specificity-metabolic stress and p53-dependent transcription. Mol. Cell. 2011;44:515–516. doi: 10.1016/j.molcel.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 72.Zhang H, et al. HIF-1 inhibits mitochondrial biogenesis and cellular respiration in VHL-deficient renal cell carcinoma by repression of C-MYC activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 73.Zeller KI, et al. Global mapping of c-Myc binding sites and target gene networks in human B cells. Proc. Natl Acad. Sci. USA. 2006;103:17834–17839. doi: 10.1073/pnas.0604129103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.LaGory EL, et al. Suppression of PGC-1α is critical for reprogramming oxidative metabolism in renal cell carcinoma. Cell Rep. 2015;12:116–127. doi: 10.1016/j.celrep.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chang CY, et al. The metabolic regulator ERRα, a downstream target of HER2/IGF-1R, as a therapeutic target in breast cancer. Cancer Cell. 2011;20:500–510. doi: 10.1016/j.ccr.2011.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Deblois G, et al. Transcriptional control of the ERBB2 amplicon by ERRalpha and PGC-1beta promotes mammary gland tumorigenesis. Cancer Res. 2010;70:10277–10287. doi: 10.1158/0008-5472.CAN-10-2840. [DOI] [PubMed] [Google Scholar]
- 77.Yao Z, et al. PGC-1β mediates adaptive chemoresistance associated with mitochondrial DNA mutations. Oncogene. 2013;32:2592–2600. doi: 10.1038/onc.2012.259. [DOI] [PubMed] [Google Scholar]
- 78.Gleyzer N, Scarpulla RC. Concerted Action of PGC-1-related Coactivator (PRC) and c-MYC in the stress response to mitochondrial dysfunction. J. Biol. Chem. 2016;291:25529–25541. doi: 10.1074/jbc.M116.719682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Giguere V. To ERR in the estrogen pathway. Trends Endocrinol. Metab. 2002;13:220–225. doi: 10.1016/S1043-2760(02)00592-1. [DOI] [PubMed] [Google Scholar]
- 80.Giguere V, Tremblay A, Tremblay GB. Estrogen receptor beta: re-evaluation of estrogen and antiestrogen signaling. Steroids. 1998;63:335–339. doi: 10.1016/S0039-128X(98)00024-5. [DOI] [PubMed] [Google Scholar]
- 81.Misawa A, Inoue S. Estrogen-related receptors in breast cancer and prostate cancer. Front Endocrinol. 2015;6:83. doi: 10.3389/fendo.2015.00083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Huss JM, Garbacz WG, Xie W. Constitutive activities of estrogen-related receptors: transcriptional regulation of metabolism by the ERR pathways in health and disease. Biochim Biophys. Acta. 2015;1852:1912–1927. doi: 10.1016/j.bbadis.2015.06.016. [DOI] [PubMed] [Google Scholar]
- 83.Misra J, Kim DK, Choi HS. ERRγ: a junior orphan with a senior role in metabolism. Trends Endocrinol. Metab. 2017;28:261–272. doi: 10.1016/j.tem.2016.12.005. [DOI] [PubMed] [Google Scholar]
- 84.Wilson BJ, Tremblay AM, Deblois G, Sylvain-Drolet G, Giguere V. An acetylation switch modulates the transcriptional activity of estrogen-related receptor alpha. Mol. Endocrinol. 2010;24:1349–1358. doi: 10.1210/me.2009-0441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kallen J, et al. Crystal structure of human estrogen-related receptor alpha in complex with a synthetic inverse agonist reveals its novel molecular mechanism. J. Biol. Chem. 2007;282:23231–23239. doi: 10.1074/jbc.M703337200. [DOI] [PubMed] [Google Scholar]
- 86.Abad MC, et al. Structural determination of estrogen-related receptor gamma in the presence of phenol derivative compounds. J. Steroid Biochem Mol. Biol. 2008;108:44–54. doi: 10.1016/j.jsbmb.2007.06.006. [DOI] [PubMed] [Google Scholar]
- 87.Patch RJ, et al. Identification of diaryl ether-based ligands for estrogen-related receptor α as potential antidiabetic agents. J. Med. Chem. 2011;54:788–808. doi: 10.1021/jm101063h. [DOI] [PubMed] [Google Scholar]
- 88.Riggins RB, Mazzotta MM, Maniya OZ, Clarke R. Orphan nuclear receptors in breast cancer pathogenesis and therapeutic response. Endocr. Relat. Cancer. 2010;17:R213–R231. doi: 10.1677/ERC-10-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Willy PJ, et al. Regulation of PPARgamma coactivator 1alpha (PGC-1alpha) signaling by an estrogen-related receptor alpha (ERRalpha) ligand. Proc. Natl Acad. Sci. USA. 2004;101:8912–8917. doi: 10.1073/pnas.0401420101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Eskiocak B, Ali A, White MA. The estrogen-related receptor α inverse agonist XCT 790 is a nanomolar mitochondrial uncoupler. Biochemistry. 2014;53:4839–4846. doi: 10.1021/bi500737n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kallen J, et al. Evidence for ligand-independent transcriptional activation of the human estrogen-related receptor alpha (ERRalpha): crystal structure of ERRalpha ligand binding domain in complex with peroxisome proliferator-activated receptor coactivator-1alpha. J. Biol. Chem. 2004;279:49330–49337. doi: 10.1074/jbc.M407999200. [DOI] [PubMed] [Google Scholar]
- 92.Manna S, et al. ERRα is a marker of tamoxifen response and survival in triple-negative breast cancer. Clin. Cancer Res. 2016;22:1421–1431. doi: 10.1158/1078-0432.CCR-15-0857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Suzuki T, et al. Estrogen-related receptor alpha in human breast carcinoma as a potent prognostic factor. Cancer Res. 2004;64:4670–4676. doi: 10.1158/0008-5472.CAN-04-0250. [DOI] [PubMed] [Google Scholar]
- 94.Berman AY, et al. ERRα regulates the growth of triple-negative breast cancer cells via S6K1-dependent mechanism. Signal. Transduct. Target Ther. 2017;2:e17035. doi: 10.1038/sigtrans.2017.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Thewes V, et al. Reprogramming of the ERRα and ERα target gene landscape triggers tamoxifen resistance in breast cancer. Cancer Res. 2015;75:720–731. doi: 10.1158/0008-5472.CAN-14-0652. [DOI] [PubMed] [Google Scholar]
- 96.Kang MH, et al. Estrogen-related receptor gamma functions as a tumor suppressor in gastric cancer. Nat. Commun. 2018;9:1920. doi: 10.1038/s41467-018-04244-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yu S, Wang X, Ng CF, Chen S, Chan FL. ERRgamma suppresses cell proliferation and tumor growth of androgen-sensitive and androgen-insensitive prostate cancer cells and its implication as a therapeutic target for prostate cancer. Cancer Res. 2007;67:4904–4914. doi: 10.1158/0008-5472.CAN-06-3855. [DOI] [PubMed] [Google Scholar]
- 98.Kim JH, et al. Estrogen-related receptor γ is upregulated in liver cancer and its inhibition suppresses liver cancer cell proliferation via induction of p21 and p27. Exp. Mol. Med. 2016;48:e213. doi: 10.1038/emm.2015.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Heckler MM, Thakor H, Schafer CC, Riggins RB. ERK/MAPK regulates ERRγ expression, transcriptional activity and receptor-mediated tamoxifen resistance in ER + breast cancer. FEBS J. 2014;281:2431–2442. doi: 10.1111/febs.12797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Madhavan S, Gusev Y, Singh S, Riggins RB. ERRγ target genes are poor prognostic factors in Tamoxifen-treated breast cancer. J. Exp. Clin. Cancer Res. 2015;34:45. doi: 10.1186/s13046-015-0150-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ariazi EA, Clark GM, Mertz JE. Estrogen-related receptor alpha and estrogen-related receptor gamma associate with unfavorable and favorable biomarkers, respectively, in human breast cancer. Cancer Res. 2002;62:6510–6518. [PubMed] [Google Scholar]
- 102.Tiraby C, Hazen BC, Gantner ML, Kralli A. Estrogen-related receptor gamma promotes mesenchymal-to-epithelial transition and suppresses breast tumor growth. Cancer Res. 2011;71:2518–2528. doi: 10.1158/0008-5472.CAN-10-1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Audet-Walsh Eacute, Giguére V. The multiple universes of estrogen-related receptor α and γ in metabolic control and related diseases. Acta Pharmacol. Sin. 2015;36:51–61. doi: 10.1038/aps.2014.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yu S, et al. Orphan nuclear receptor estrogen-related receptor-beta suppresses in vitro and in vivo growth of prostate cancer cells via p21(WAF1/CIP1) induction and as a potential therapeutic target in prostate cancer. Oncogene. 2008;27:3313–3328. doi: 10.1038/sj.onc.1210986. [DOI] [PubMed] [Google Scholar]
- 105.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 106.Sancho P, Barneda D, Heeschen C. Hallmarks of cancer stem cell metabolism. Br. J. Cancer. 2016;114:1305–1312. doi: 10.1038/bjc.2016.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhao Y, Butler EB, Tan M. Targeting cellular metabolism to improve cancer therapeutics. Cell Death Dis. 2013;4:e532. doi: 10.1038/cddis.2013.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.DeBerardinis RJ, Chandel NS. Fundamentals of cancer metabolism. Sci. Adv. 2016;2:e1600200. doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Vander Heiden MG, DeBerardinis RJ. Understanding the intersections between metabolism and cancer biology. Cell. 2017;168:657–669. doi: 10.1016/j.cell.2016.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Denko NC. Hypoxia, HIF1 and glucose metabolism in the solid tumour. Nat. Rev. Cancer. 2008;8:705–713. doi: 10.1038/nrc2468. [DOI] [PubMed] [Google Scholar]
- 112.Nagarajan A, Malvi P, Wajapeyee N. Oncogene-directed alterations in cancer cell metabolism. Trends Cancer. 2016;2:365–377. doi: 10.1016/j.trecan.2016.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Viale A, et al. Oncogene ablation-resistant pancreatic cancer cells depend on mitochondrial function. Nature. 2014;514:628–632. doi: 10.1038/nature13611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Morandi A, Indraccolo S. Linking metabolic reprogramming to therapy resistance in cancer. Biochim Biophys. Acta. 2017;1868:1–6. doi: 10.1016/j.bbcan.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 115.Kuntz EM, et al. Targeting mitochondrial oxidative phosphorylation eradicates therapy-resistant chronic myeloid leukemia stem cells. Nat. Med. 2017;23:1234–1240. doi: 10.1038/nm.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Lee KM, et al. MYC and MCL1 cooperatively promote chemotherapy-resistant breast cancer stem cells via regulation of mitochondrial oxidative phosphorylation. Cell Metab. 2017;26:633–647 e637. doi: 10.1016/j.cmet.2017.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Hensley CT, et al. Metabolic heterogeneity in human lung tumors. Cell. 2016;164:681–694. doi: 10.1016/j.cell.2015.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Porporato PE, Filigheddu N, Pedro JMB, Kroemer G, Galluzzi L. Mitochondrial metabolism and cancer. Cell Res. 2018;28:265–280. doi: 10.1038/cr.2017.155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Trousil S, et al. Phenformin enhances the efficacy of ERK inhibition in NF1-mutant melanoma. J. Invest. Dermatol. 2017;137:1135–1143. doi: 10.1016/j.jid.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Efeyan A, Comb WC, Sabatini DM. Nutrient-sensing mechanisms and pathways. Nature. 2015;517:302–310. doi: 10.1038/nature14190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Takacs M, et al. The asymmetric binding of PGC-1α to the ERRα and ERRγ nuclear receptor homodimers involves a similar recognition mechanism. PLoS ONE. 2013;8:e67810. doi: 10.1371/journal.pone.0067810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang L, et al. X-ray crystal structures of the estrogen-related receptor-gamma ligand binding domain in three functional states reveal the molecular basis of small molecule regulation. J. Biol. Chem. 2006;281:37773–37781. doi: 10.1074/jbc.M608410200. [DOI] [PubMed] [Google Scholar]
- 123.Weinberg SE, Chandel NS. Targeting mitochondria metabolism for cancer therapy. Nat. Chem. Biol. 2015;11:9–15. doi: 10.1038/nchembio.1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Bartesaghi A, et al. 2.2A resolution cryo-EM structure of beta-galactosidase in complex with a cell-permeant inhibitor. Science. 2015;348:1147–1151. doi: 10.1126/science.aab1576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kuhlbrandt W. Cryo-EM enters a new era. Elife. 2014;3:e03678. doi: 10.7554/eLife.03678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Merk A, et al. Breaking Cryo-EM resolution barriers to facilitate drug discovery. Cell. 2016;165:1698–1707. doi: 10.1016/j.cell.2016.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Banerjee S, et al. 2.3 Å resolution cryo-EM structure of human p97 and mechanism of allosteric inhibition. Science. 2016;351:871–875. doi: 10.1126/science.aad7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Sandhu C, Qureshi A, Emili A. Panomics for precision medicine. Trends Mol. Med. 2018;24:85–101. doi: 10.1016/j.molmed.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Polyak K. Heterogeneity in breast cancer. J. Clin. Invest. 2011;121:3786–3788. doi: 10.1172/JCI60534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Yersal O, Barutca S. Biological subtypes of breast cancer: prognostic and therapeutic implications. World J. Clin. Oncol. 2014;5:412–424. doi: 10.5306/wjco.v5.i3.412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Koren S, Bentires-Alj M. Breast tumor heterogeneity: source of fitness, hurdle for therapy. Mol. Cell. 2015;60:537–546. doi: 10.1016/j.molcel.2015.10.031. [DOI] [PubMed] [Google Scholar]
- 132.Rehman FL, Lord CJ, Ashworth A. Synthetic lethal approaches to breast cancer therapy. Nat. Rev. Clin. Oncol. 2010;7:718–724. doi: 10.1038/nrclinonc.2010.172. [DOI] [PubMed] [Google Scholar]
- 133.La Thangue NB, Kerr DJ. Predictive biomarkers: a paradigm shift towards personalized cancer medicine. Nat. Rev. Clin. Oncol. 2011;8:587–596. doi: 10.1038/nrclinonc.2011.121. [DOI] [PubMed] [Google Scholar]
- 134.Lord CJ, Ashworth A. PARP inhibitors: synthetic lethality in the clinic. Science. 2017;355:1152–1158. doi: 10.1126/science.aam7344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Farmer H, et al. Targeting the DNA repair defect in BRCA mutant cells as a therapeutic strategy. Nature. 2005;434:917–921. doi: 10.1038/nature03445. [DOI] [PubMed] [Google Scholar]
- 136.Wolpaw AJ, Dang CV. Exploiting metabolic vulnerabilities of cancer with precision and accuracy. Trends Cell Biol. 2018;28:201–212. doi: 10.1016/j.tcb.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Busch BB, et al. Identification of a selective inverse agonist for the orphan nuclear receptor estrogen-related receptor alpha. J. Med. Chem. 2004;47:5593–5596. doi: 10.1021/jm049334f. [DOI] [PubMed] [Google Scholar]
- 138.Lanvin O, Bianco S, Kersual N, Chalbos D, Vanacker JM. Potentiation of ICI182,780 (Fulvestrant)-induced estrogen receptor-alpha degradation by the estrogen receptor-related receptor-alpha inverse agonist XCT790. J. Biol. Chem. 2007;282:28328–28334. doi: 10.1074/jbc.M704295200. [DOI] [PubMed] [Google Scholar]
- 139.Sharabi K, et al. Selective chemical inhibition of PGC-1α gluconeogenic activity ameliorates type 2 diabetes. Cell. 2017;169:148–160.e115. doi: 10.1016/j.cell.2017.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Giguère V. Transcriptional control of energy homeostasis by the estrogen-related receptors. Endocr. Rev. 2008;29:677–696. doi: 10.1210/er.2008-0017. [DOI] [PubMed] [Google Scholar]
- 141.Simons AL, Ahmad IM, Mattson DM, Dornfeld KJ, Spitz DR. 2-Deoxy-D-glucose combined with cisplatin enhances cytotoxicity via metabolic oxidative stress in human head and neck cancer cells. Cancer Res. 2007;67:3364–3370. doi: 10.1158/0008-5472.CAN-06-3717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Michalek RD, et al. Estrogen-related receptor-α is a metabolic regulator of effector T-cell activation and differentiation. Proc. Natl Acad. Sci. USA. 2011;108:18348–18353. doi: 10.1073/pnas.1108856108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Yu Y, et al. Selective active site inhibitors of human lactate dehydrogenases A4, B4, and C4. Biochem. Pharmacol. 2001;62:81–89. doi: 10.1016/S0006-2952(01)00636-0. [DOI] [PubMed] [Google Scholar]
- 144.Cai Q, Lin T, Kamarajugadda S, Lu J. Regulation of glycolysis and the Warburg effect by estrogen-related receptors. Oncogene. 2013;32:2079–2086. doi: 10.1038/onc.2012.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Porporato PE, Dhup S, Dadhich RK, Copetti T, Sonveaux P. Anticancer targets in the glycolytic metabolism of tumors: a comprehensive review. Front. Pharmacol. 2011;2:49. doi: 10.3389/fphar.2011.00049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Pereira da Silva AP, et al. Inhibition of energy-producing pathways of HepG2 cells by 3-bromopyruvate. Biochem. J. 2009;417:717–726. doi: 10.1042/BJ20080805. [DOI] [PubMed] [Google Scholar]
- 147.Casaburi I, et al. Estrogen related receptor α (ERRα) a promising target for the therapy of adrenocortical carcinoma (ACC) Oncotarget. 2015;6:25135–25148. doi: 10.18632/oncotarget.4722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Mullard A. FDA approves first-in-class cancer metabolism drug. Nat. Rev. Drug Discov. 2017;16:593. doi: 10.1038/nrd.2017.174. [DOI] [PubMed] [Google Scholar]
- 149.Brown EL, et al. Estrogen-related receptors mediate the adaptive response of brown adipose tissue to adrenergic stimulation. iScience. 2018;2:221–237. doi: 10.1016/j.isci.2018.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Janzer A, et al. Metformin and phenformin deplete tricarboxylic acid cycle and glycolytic intermediates during cell transformation and NTPs in cancer stem cells. Proc. Natl Acad. Sci. USA. 2014;111:10574–10579. doi: 10.1073/pnas.1409844111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hao Y, et al. Oncogenic PIK3CA mutations reprogram glutamine metabolism in colorectal cancer. Nat. Commun. 2016;7:11971. doi: 10.1038/ncomms11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Reaven GM, Chang H, Hoffman BB. Additive hypoglycemic effects of drugs that modify free-fatty acid metabolism by different mechanisms in rats with streptozocin-induced diabetes. Diabetes. 1988;37:28–32. doi: 10.2337/diab.37.1.28. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data sharing not applicable to this article as no datasets were generated or analyzed for the current study.