Abstract
Protein dynamics in crowded environments is important for understanding protein functions in vivo and is especially relevant for membrane proteins because of the roles of protein-protein interactions in membrane protein functions and their regulation. Here, using solid-state NMR spectroscopy in combination with coarse-grained molecular dynamics simulations, we report that the rotational correlation time for the transmembrane domain of the influenza A M2 proton channel in lipid bilayers increases dramatically at an elevated protein/lipid ratio. This increase is attributable to persistent protein-protein interactions, thus revealing for the first time, to the best of our knowledge, extensive cluster formation of the M2 tetrameric channel. Such clustering appears to have direct biological relevance during budding of the nascent influenza virus, which does not use the endosomal sorting complexes required for transport machinery. Indeed, initial coarse-grained molecular dynamics simulations of the longer M2 construct known as the conductance domain suggest clustering-induced membrane curvature formation.
Introduction
M2 from influenza A is a tetrameric transmembrane (TM) α-helical protein with multiple functions (1), including the facilitation of viral budding and the conductance of protons (2). The structure of the TM and juxtamembrane helices (together constituting the conductance domain, M2CD), determined in liquid-crystalline lipid bilayers, has a fourfold symmetric pyramidal shape (3) (Protein Data Bank (PDB): 2L0J) that is proposed to be important for inducing membrane curvature essential for the viral budding process (1, 2). Unlike many viruses, influenza A does not sequester the endosomal sorting complexes required for transport (ESCRT) of the host cell for viral budding; instead, this task falls on M2 (2, 4). Although much has been learned about how membrane and membrane mimetic environments influence the structures of membrane proteins and specifically of M2 (5, 6, 7, 8), little is known about how the lipid/protein tetramer (L/T) ratio affects the interactions between protein tetramers or how the latter may contribute to the pinching off of viral particles. This process likely requires the induction of dramatic membrane curvature that may involve the clustering of the pyramidal-shaped M2CD tetramers in the “neck” of the viral particle.
M2 is expressed at nearly the same level as neurominidase and hemagglutinin but is found at much lower concentrations in the cholesterol- and sphingomyelin-rich mature viral membrane because M2 is localized primarily in liquid-crystalline regions of the cellular membrane during viral budding (4, 9). On the other hand, M2 has been shown to bind cholesterol (10, 11) and thereby stabilize the juxtamembrane helix in the membrane-water interface (10), and so a high concentration of M2 can be anticipated in the vicinity of the cholesterol-rich, raft-like domains where budding occurs. This points to a scenario in which M2 concentrates in the relatively cholesterol-poor liquid-crystalline phase surrounding the lipid rafts, where neurominidase and hemagglutinin are localized.
Protein oligomerization in crowded cell membranes has been predicted on theoretical grounds (12). Many studies have shown that membrane proteins at high concentrations (i.e., low lipid/protein (L/P) ratios) experience slowed diffusion attributable to protein clustering, as demonstrated by electron paramagnetic resonance (EPR) for the Tordepo marmorata acetylcholine receptor (13) and more recently in a coarse-grained (CG) simulation study of diverse types of membrane proteins embedded in lipid bilayers (14). Likewise, EPR and fluorescence experiments have indicated a 50–100% increase in rotational correlation time (RCT) for rhodopsin at L/P = 43:1 relative to samples at 240:1 (15). The increase was explained as due to the formation of aggregates, fitting for a protein that naturally occurs in semicrystalline arrays. For the M13 coat protein, a reduction in the L/P to less than a single annulus of lipids around the single TM α-helix resulted in a nearly fivefold increase in the RCT (15, 16, 17, 18). Similarly, an EPR study of the sarco/endoplasmic reticulum Ca2+-ATPase, which has been observed in clusters in both native and reconstituted membranes (19), showed a strong dependence of the protein RCT on the L/P (20), again suggesting the involvement of protein-protein interactions.
The functional importance of protein oligomerization or clustering within lipid bilayers has been explored in a number of studies. The trimerization of human immunodeficiency virus envelope protein was shown to be correlated with human immunodeficiency virus maturation and infectivity (21). The dynamical dimerization of G protein-coupled receptors (GPCRs) and its implications for GPCR function was studied by fluorescence and molecular dynamics simulations (22, 23). The association of β-barrel outer membrane proteins in small patches of the Escherichia coli membrane has been linked with their turnover process (24). Very recently, the functional clustering of proteins has been observed in the plasma membrane via fluorescence microscopy (25, 26). In quiescent T cells, T-cell receptors formed nanometer- to micrometer-sized clusters; the function of the SNARE machinery, responsible for docking and fusion of synaptic vesicles, was shown to be dependent on stoichiometric specific clustering of proteins.
So far, no evidence has been presented that the M2 tetrameric channel forms clusters in cell membranes, but intertetramer contacts between the transmembrane helices of two antiparallel tetramers of S31N M2CD have been observed by solid-state NMR (ssNMR) in proteoliposome samples at L/T = 24:1 (27). Such clustering of S31N M2CD was not observed at L/T = 60:1 (28). Although the M2CD includes both the TM helix (residues 22–46) and a juxtamembrane amphipathic helix (residues 47–62), the TM helix (M2TM) alone forms a proton-selective channel, albeit with a somewhat lower conductance rate than the full-length or conductance-domain constructs (29, 30). Here, we report the measurement of M2 RCTs at different L/T ratios by ssNMR and the quantitative explanation of the NMR data by CG molecular dynamics simulations. The measurement by ssNMR required a site-specific isotopically labeled M2 sample, in which a fully deuterated Ala29 (d4Ala29) residue was incorporated into M2TM by chemical synthesis (31). We found a 25-fold increase in RCT upon lowering L/T from 80:1 to 30:1. To explain this dramatic change, we carried out CG molecular dynamics simulations of M2TM at different L/T ratios. The simulations suggest that the slowed rotational motion at the higher protein mole fraction is due to oligomerization of M2 tetramers. The results, together with initial simulations of the larger M2CD, implicate an important role for M2 in viral budding.
Materials and Methods
Peptide synthesis
The M2TM peptide with fully deuterated Ala29 (d4Ala29) and 15N Ile42 (sequence SSDPLVVd4A29AS IIGILHLILW15NI42LDRL; herein referred to as M2TM d4Ala29) was synthesized using an Applied Biosystems (Foster City, CA) 433A peptide synthesizer and fluorenylmethyloxycarbonyl (Fmoc) solid phase chemistry (31). After synthesis, the 2-chlorotrityl resin beads containing the elongated peptide chain were dried overnight under vacuum and stored at −20°C. The peptide chain was cleaved from the resin, and the Fmoc group and other protection groups were removed from side chains by exposing the resin-peptide conjugate to a solution containing 95% (v/v) trifluoroacetic acid, 4% (v/v) 1,2 ethanedithiol, and 1% (w/v) indole for 2 h. The synthesized peptide was recovered by precipitation with cold ether. The precipitated peptide pellet was washed with cold ether four times to remove all trifluoroacetic acid. The peptide was dried under vacuum for 2 h. To remove small fragments of truncated peptide, the pellet was resuspended in a minimal amount of methanol, and ice-cold deionized water was added dropwise to the methanol solution until precipitation was observed. The precipitate containing the peptide was recovered by centrifugation and the pellet was lyophilized. Peptide purity was assessed by electron spray ionization mass spectrometry that indicated a molecular weight of 2732 and estimated purity of 90%.
Proteoliposome preparation
M2TM d4Ala29 was incorporated into dioleoylphosphatidylcholine (DOPC) and dioleoylphosphadylethanolamine (DOPE) in a 4:1 molar ratio, the same lipid composition as for M2CD structure determination (3). Lipids were purchased from Avanti Polar Lipids (Alabaster, AL) and used as is. Between 10 to 20 mg of the peptide was dissolved in methanol, to which DOPC and DOPE lipids in chloroform stock solution were added to a final L/T ratio of 30:1 or 80:1. The organic solvent mixture was removed by exposure to a stream of N2 gas to form a lipid-protein film on the walls of a round-bottom flask. The film was vacuum dried overnight to insure maximal removal of the organic solvent. Between 10 to 20 mL of 10 mM Tris (pH 7.5) was added to the dry film and placed in a 30°C incubator with mild rotation. This suspension was bath sonicated for 5 to 10 min to clarify the cloudy solution. The proteoliposome suspension was then transferred to a 3.5 kDa cutoff Spectra/Por dialysis membrane (Spectrum Labs, Rancho Dominguez, CA) and allowed to equilibrate against 2 L of 10 mM Tris (pH 7.5) for 24 h. The proteoliposome suspension was centrifuged at 3000 × g for 10 min to remove any precipitate or large aggregates. The resulting supernatant was centrifuged at 400,000 × g for 4 h. For 2H spectroscopy, proteoliposomes were washed four times with buffer prepared with deuterium-depleted water to remove natural abundance deuterium from the water used during the sample preparation. The clear proteoliposome pellets were placed in a sample cell immediately after centrifugation and sealed to avoid dehydration.
NMR spectroscopy
All 2H spectra were acquired with a quadrupolar echo pulse sequence using a 2.5 μs π/2 pulse and a recycle delay of 1 s with various echo times of 10 to 60 μs at three field strengths: 16.9, 19.6, and 21.1 T. For experiments at 19.6 T, a single-channel 4 mm diameter coil probe was used with room temperature air circulating over the sample; the temperature for the sample was calibrated using the powder pattern of lead nitrate and determined to be 20 ± 2°C. At 16.9 and 21.1 T, a probe coil of 5 mm diameter was used, and temperatures were as reported by a probe thermocouple. An average of 100,000 scans were acquired for each sample at each echo time with an interscan recycle delay of 1 s. Either 800 or 1000 Hz exponential decay line broadening was applied to the spectra.
Line shape calculations
The 2H line shape calculations were performed using Express (32), a MATLAB (The MathWorks, Natick, MA)-based software. In short, the 2H quadrupole echo spectra are calculated by solving the stochastic Louiville equation for the spin density matrix; time evolution is treated as Markovian jumps between discrete orientational sites. The modes of motion that affect the 2H powder pattern of Ala are described in the Supporting Materials and Methods, Session 1. The simulation algorithm takes into account the magnetic field, the pulse length, and the echo time, τ. The methyl rotation, libration, and the channel rotational motion are explicitly accounted for by building individual frames where the orientation of each motional axis with respect to the previous frame is given in terms of α, β and γ, respectively, where α is kept at 0, β is the angle between the quadrupolar interaction and the motional axis (as given in Figs. S1–S3), and γ represents the Markovian jumps between discrete orientational states. The order of the frames matters: the faster motions must be evaluated before the slower motions.
The line shape simulations yield a rotational diffusion constant (DR, in s−1) and an amplitude angle. The axisymmetric rotational diffusion in this study is reported as an RCT; the conversion between DR and RCT is RCT = 1/(2DR) (33). The best fit between experimental spectra and simulated line shapes was evaluated by visual inspection of their overlay over the full range of echo times for each sample. The parameters that gave the overall best fit might not produce line shapes that match perfectly with spectra for one or more echo times. Such mismatch could be due to spectral artifacts. At small echo times especially, artifacts coming from the receiver and probe ring down can perturb the line shape. Also, the sample could be slightly heterogeneous, and slightly different motional frequencies that are either more or less sensitive to the T2 relaxation can influence the analysis of the echo experiment, making a global evaluation of the spectra at different echo times even more important.
The central line at 0 kHz present in all spectra came from natural abundance deuterium in water. Even though the samples were washed with deuterium-depleted water several times, water from the air exchanged rapidly with the samples.
Setup of CG models
The CG model of M2TM (residue 25–46) was built in the Martini force field (34) using a crystal structure (PDB: 3LBW) (35). The backbone structures of 3LBW and 2L0J are very similar (36). Martini on average maps four heavy atoms (and the bonded hydrogen atoms) into a single bead and restrains secondary structures throughout a simulation. Initial M2TM-lipid models were set up by using the program insane (37). Systems with two L/T ratios, 80:1 and 28:1, and two scenarios of M2TM tetrameric channel orientations, mixed and parallel, were prepared. All the systems contained 16 copies of M2TM tetramer, and the two L/T ratios were obtained by changing the number of lipids. Specifically, the lower L/T ratio was obtained with 360 DOPC, 90 DOPE, and 5439 water beads (four atomistic water molecules were mapped onto a CG water bead), along with 59 Na+ and 59 Cl− ions (representing a salt concentration of 0.15 M); the higher L/T ratio was obtained with 1025 DOPC, 256 DOPE, and 8921 water beads, along with 98 Na+ and 98 Cl− ions (salt concentration again at 0.15 M). In the mixed orientation case, half of the M2TM tetramers had one orientation, whereas the other half had the opposite orientation. For the parallel orientation case, all the M2TM tetramers had the same orientation.
Similarly, the CG model of M2CD (S22SDPLVVAASIIGILHLILWILDRLF FKSIYRFFEHGLKRG62) was built using the ssNMR structure 2L0J at an L/T ratio of 85:1. This system contained 16 copies of M2CD tetramer in parallel orientation, 1084 DOPC, 271 DOPE, and 10,710 water beads, along with 54 Na+ and 182 Cl− ions (with salt concentration at 0.15 M and the entire system charge neutral).
CG simulation parameters and protocols
All CG simulations were run using GROMACS-4.6.3 (www.gromacs.org) (38). For the proteins (M2TM or M2CD tetramer), we used the Martini 2.2 force field, whereas for lipids, water, and ions, we used the Martini 2.0 force field. Electrostatic interactions were smoothly shifted to zero over interbead distance of 0–12 Å. Lennard-Jones interactions were smoothly shifted to zero over 9–12 Å. All simulations were performed at constant temperature, pressure, and number of particles. The temperatures of the protein plus lipid region and the water plus ion region were separately controlled at 293 K using velocity rescaling with a stochastic term (39) having a coupling constant of 1 ps. The pressure was semiisotropically coupled using the Parrinello-Rahman algorithm at 1 bar with a coupling constant τP = 12 ps and a compressibility of 3 × 10−4 bar−1. The time step for integration was 20 fs, and coordinates were saved every 100 ps for subsequent analysis. Simulations were run for 200 μs in each case of M2TM. For the L/T = 28:1 cases, two repeat simulations were run to improve statistics. The same protocol was followed for the M2CD simulation, which totaled 122 μs.
Analysis and visualization
Clusters of M2TM tetramers were calculated and visualized using the open visualization tool OVITO (www.ovito.org) (40). Contact frequency, defined as the fraction of snapshots in which a residue was involved in intertetramer contact, was calculated using VMD (41). The results, after normalization by the highest contact frequency, are reported as relative contact frequencies.
Results
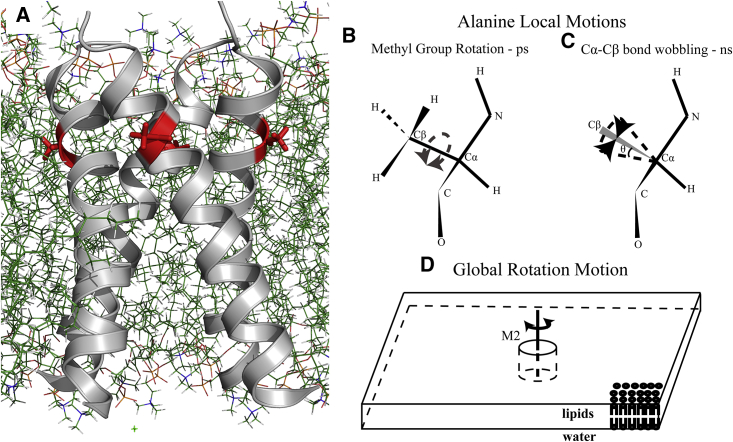
2H ssNMR spectra are exceptionally sensitive to molecular motions with rates on the order of 10−4–10−6 Hz, a range often found for the global motions of small membrane proteins in lipid bilayers (42). The 2H nuclear quadrupolar coupling constant (QCC) is anisotropic, depending on the orientation of the C-2H bond with respect to the principal motional axis of the protein. The main modes of motion that affect the 2H ssNMR spectra of a deuterated amino acid residue in a membrane protein are the picosecond (ps) local motion of side chains; the nanosecond (ns) small amplitude librations of the peptide plane that manifest as a wobbling motion of the Cα-Cβ bond; and the microsecond to millisecond (μs–ms) rotational motion of the protein around an axis parallel to the bilayer normal (43). The interpretation of the resulting 2H NMR spectra relies on knowledge of the orientations of the molecular motional axes with respect to the deuterium quadrupolar coupling tensor defined within the molecule structure (molecular frame). A detailed description of these modes of motions and their effects on the 2H ssNMR spectra is given in Supporting Materials and Methods. In proteoliposome preparations with no preferred orientation of the bilayer normal (and thus the protein rotational axis) with respect to the external magnetic field, a broad 2H resonance, known as a “powder pattern,” reflects molecular motions that are not sufficiently fast to average the quadrupolar interaction to an isotropic value but lead to a reduction of the “intensity” of the QCC as well as characteristic changes to the powder-pattern line shape (Figs. S1–S3). Such powder-pattern line shapes are particularly informative for accurate measurements of protein dynamics (44, 45, 46, 47, 48, 49). The powder patterns, however, cannot be resolved in the case of multiple labeled sites; hence the need for a site-specific labeled peptide. The 2H ssNMR spectra for the deuterated methyl group in the d4Ala29-labeled M2TM peptide incorporated in lipid bilayers allowed for a characterization of the rotational motion of the tetrameric channel as well as the methyl side-chain and peptide-plane local motions. Ala29 in M2 projects into the fatty acyl chains of the lipid environment, away from both the aqueous pore of the channel and from the helix-helix interface of the tetrameric structure (Fig. 1 A). Hence, this residue’s methyl deuterons are sensitive to the fast methyl rotation, Cα-Cβ bond wobbling, and rotational motion of the tetramer within the bilayer environment (Fig. 1, B–D) but relatively immune from influences of water in the pore and intersubunit relative motion.
Figure 1.
Different modes of motions of a labeled side chain in the M2TM domain. (A) TM helices of the M2CD structure (PDB: 2L0J) in DOPC/DOPE lipids are shown. In red, Ala29, is shown facing the lipid acyl chains. The 2H NMR spectrum of deuterated Ala29 methyl is affected by three types of motion: (B) methyl rotation about the Cα-Cβ bond on the ps timescale, which can only be damped at temperatures far below 100 K; (C) the wobbling motion of the Cα-Cβ bond, illustrated as the rotation about a cone with amplitude defined by a half angle θ, resulting from small-amplitude motions (libration) of the peptide plane on the ns timescale; and (D) the overall rotational correlation of M2TM, modeled as a cylinder in a two-dimensional slab of lipid bilayer (43).
To extract motional parameters, local and global modes of motion need to be resolved. The effects of multiple modes of motion on 2H NMR spectra have been described in many references (45, 50, 51, 52, 53). The deconvolution of the local and global motions from the 2H methyl powder pattern can be performed by numerical calculations (line shape simulations) in which the modes of motion are separated into different frames and modeled as jumps having a given amplitude and rate (32), as described in the Materials and Methods.
Local motions of Ala29 in M2TM
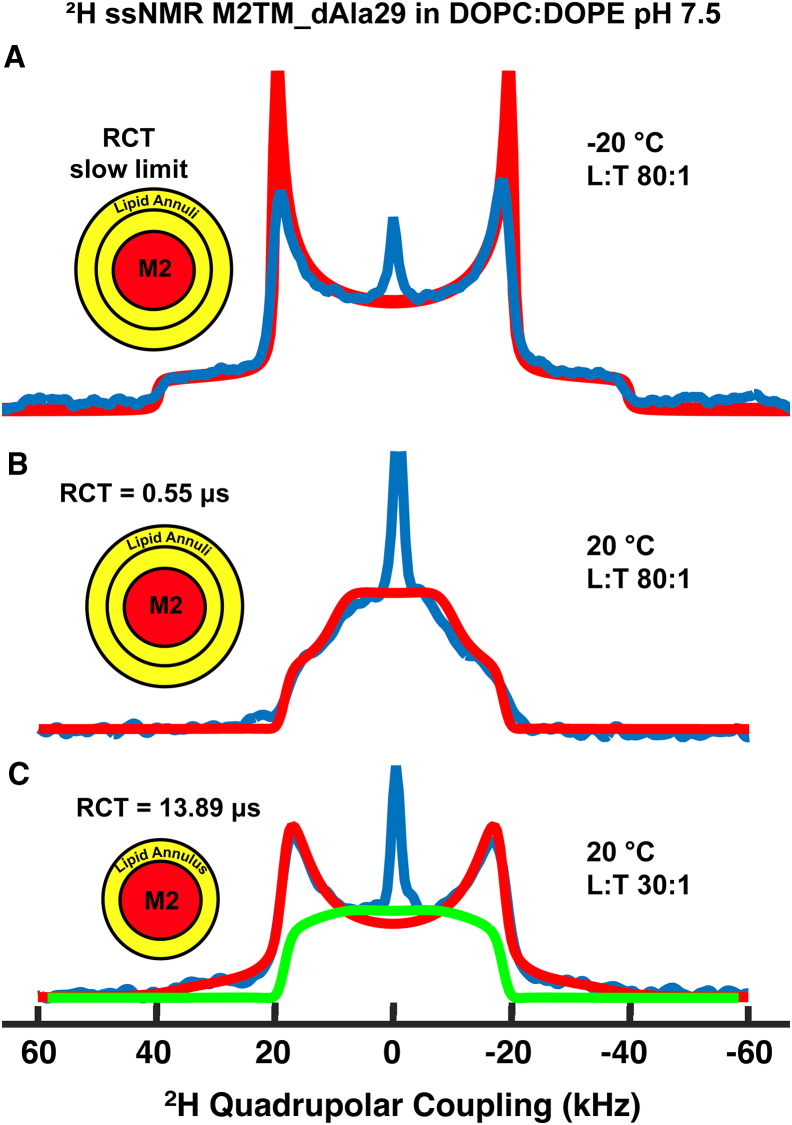
The process of deconvoluting local and global modes of motion in membrane proteins can be facilitated by means of slowing the rotational correlation motion of the protein below the NMR timescale detection limit by reducing the temperature below the gel to liquid-crystalline phase transition of the lipids. The 2H ssNMR spectrum of M2TM d4Ala29 incorporated in DOPC/DOPE (4:1 molar ratio) at an L/T of 80:1 was obtained at −20 and −45°C; the latter is well below the phase transition temperature. At these two temperatures, the spectral patterns were similar (Fig. S4), implying very slow rotational motion compared to the width of the quadrupolar interaction. Only fast rotation of the methyl and wobbling of the Cα-Cβ axis influenced these spectra. Fig. 2 A shows an overlay of the spectrum of M2TM d4Ala29 at −20°C with a line shape simulation that assumed ps methyl rotation and a ±8° amplitude wobbling motion for the Ala29 Cα-Cβ bond on the ns timescale. The ps rotational motion of the methyl deuterons was insensitive to the temperatures sampled in this study.
Figure 2.
2H ssNMR spectra of M2TM d4Ala29 in DOPC/DOPE bilayers at pH 7.5. (A) Blue shows the spectrum for L/T = 80:1 acquired at −20°C in a 16.9 T magnet with 10 μs echo time. Red shows a simulated line shape for a methyl deuteron assuming only ps methyl rotation and ns Cα-Cβ wobbling (with amplitude = 8°). (B) Blue shows the spectrum for L/T = 80:1 at 20°C in a 19.6 T magnet with 30 μs echo time. Red shows a simulated line shape with RCT = 0.55 μs. (C) Blue shows the spectrum for L/T = 30:1 acquisition parameters, the same as in (B). Red shows a simulated line shape with RCT = 13.9 μs. Green shows a simulated line shape with RCT = 1.11 μs. Inset: an L/T of 80:1 generates ∼2 lipid annuli around the M2TM tetramer, whereas an L/T of 30:1 generates a single lipid annulus. All line shape simulations were performed using Express (32).
Slowdown of rotational diffusion at low L/T
From the 2H spectral analysis at temperatures below the phase transition of the lipids, the lower limit of the local modes of motion was determined. 2H spectra of M2TM d4Ala29 were then acquired at 20°C to probe the channel’s rotational motion. The spectrum for an L/T ratio of 80:1 (Fig. 2 B) shows a collapsed powder pattern lacking sharp discontinuities, indicative of motions on the intermediate timescale in reference to the 2H QCC. Line shape simulation of this spectrum (red line Fig. 2 B) resulted in a higher wobbling amplitude for the M2TM tetrameric channel Ala29 Cα-Cβ bond of ±15° occurring on the ns timescale and an RCT of 0.55 ± 0.1 μs. This RCT is consistent with the theoretical value for a single tetrameric M2 channel in a liquid-crystalline lipid slab (43), as described in the Supporting Materials and Methods. At L/T = 80:1, each tetrameric M2TM is surrounded by two lipid annuli (Fig. 2 B inset), and neighboring tetramers are separated by ∼4 lipid annuli. A reduction of L/T to 30:1 increases the protein concentration within the proteoliposome and results in approximately a single lipid annulus around each tetrameric M2TM. The 2H ssNMR spectrum (Fig. 2 C) has clear discontinuities, indicating motions on a much slower timescale. An RCT of 13.9 ± 2 μs was obtained from the line shape simulation for this sample (red line in Fig. 2 C), corresponding to a 25-fold slowdown in rotational diffusion compared to the 80:1 sample. For comparison, a twofold slowdown (RCT increasing from 0.55 to 1.11 μs; Fig. 2 C green) results in a spectrum indicative of intermediate timescale motion (Fig. S5) and cannot explain the observed spectrum at L/T = 30:1. The observed dramatic slowdown in rotational diffusion suggests extensive protein-protein interactions. The methyl rotational motion and the Cα-Cβ wobbling amplitude for Ala29 remained unchanged for the two L/T ratios. The fitting of this motional model to the spectra at the two L/T ratios was also performed as a function of echo times (Fig. S6) to ensure robustness of the line shape calculations as described in Materials and Methods.
Clustering of M2TM at low L/T
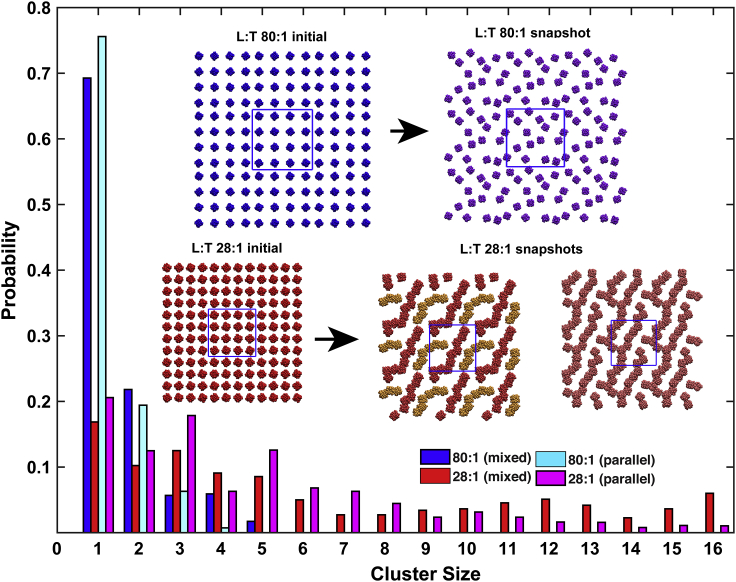
To gain insights into the slowdown in rotational diffusion, CG molecular dynamics simulations of M2TM in DOPC/DOPE (4:1) lipids at L/T ratios of 80:1 (two lipid annuli) and 28:1 (one lipid annulus) were conducted. Two scenarios for the orientations of the M2TM tetramers with respect to the bilayer were studied: 1) mixed, in which half of the tetramers had one orientation and the other half had the opposite orientation, to mimic the random orientation in the experimental samples; and 2) parallel, in which all the molecules had the same orientation, as in cellular membranes. In each case, the RCT of M2TM was obtained by an exponential fitting of the time-correlation function for the axial orientation (Figs. S7 and S8), as described in Materials and Methods. The RCTs in the mixed orientation were 0.73 μs for L/T = 80:1 and 17.7 μs for L/T = 28:1. These values correspond to a ∼24-fold slowdown in rotational diffusion, in excellent agreement with the experimental results.
In the simulations, the M2TM channels largely remained monomeric at L/T = 80:1, as indicated by a high green bar at cluster size = 1 in the distribution and illustrated by a snapshot in Fig. 3. In contrast, at L/T = 28:1, cluster sizes are much more evenly distributed from 1 to 16 (red bars in the cluster-size distribution in Fig. 3). Note that cluster size = 16 corresponds to a single cluster within the unit cell of the simulations (blue squares). Two snapshots shown in Fig. 3 provide further illustration of the strong tendency to cluster. In the parallel orientation, the RCTs (Fig. S8) and cluster-size distributions (Fig. 3, blue and magenta bars) again showed dramatic slowdown in rotational diffusion and extensive M2 tetramer clustering, suggesting that the conclusions drawn here are relevant for the M2 channel in cell membranes.
Figure 3.
Cluster size distributions from CG simulations of M2TM. Results are shown for two L/T ratios (80:1 and 28:1) and two scenarios of M2TM orientations (mixed and parallel), as indicated by bars of different colors. Inset: initial and subsequent snapshots of the mixed orientation CG simulations at L/T = 80:1 and 28:1 (the distinct colors in 28:1 inset correspond to different clusters).
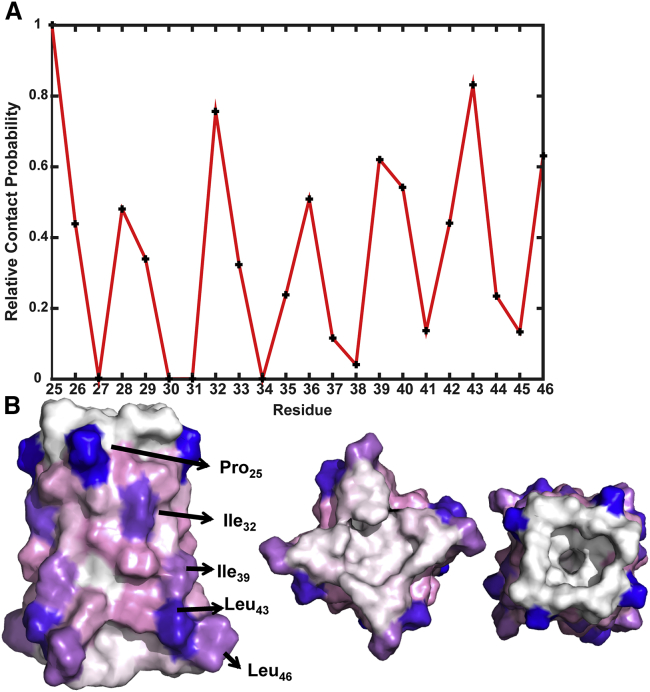
In the CG simulations with mixed orientation at L/T = 28:1, clusters were formed without selection for parallel or antiparallel orientations between neighboring tetramers, indicating that the clustering is not site specific. Indeed, both parallel and antiparallel interactions of the tetramers were observed, and most of the lipid-facing surface of the tetramer had comparable intertetramer contact frequencies (Fig. 4 A). The contacts were mostly formed by bulky lipid-facing residues, Pro25, Ile32, Ile39, Leu43, and Leu46, as illustrated in Fig. 4 B. Although there is no obvious site specificity for the tetramer-tetramer interactions, most of the tetramers interact with two other tetramers and only rarely with one or three tetramers. Overall, this leads to string-like clustering (Fig. 3 inset).
Figure 4.
Relative contact frequencies of individual residues in the mixed orientation CG simulations at L/T = 28:1. (A) Contact frequency was calculated as the fraction of snapshots in which a residue was involved in intertetramer contact and then normalized by the highest value of all residues. (B) The M2TM surface colored according to residue contact frequency, from the maximal 1 (blue) to 0.5 (pink) to 0 (white). In the side view on the left, five residues with the highest contact frequencies are labeled. In the top view on the right, an example of antiparallel interactions of two tetramers is shown in a corner-to-face pose, illustrating shape complementarity.
Cluster formation in the CG simulations with a parallel orientation at L/T = 28:1 exhibited very similar features. The most prominent contact-forming residues for these clusters are Ile32, Leu36, Leu40, and Leu43 (Fig. S9), showing some difference with the antiparallel interactions, but again, the interactions appear to be driven by the exposed bulky side chains. In addition, the string-like clustering pattern is similar to the mixed-orientation CG simulations.
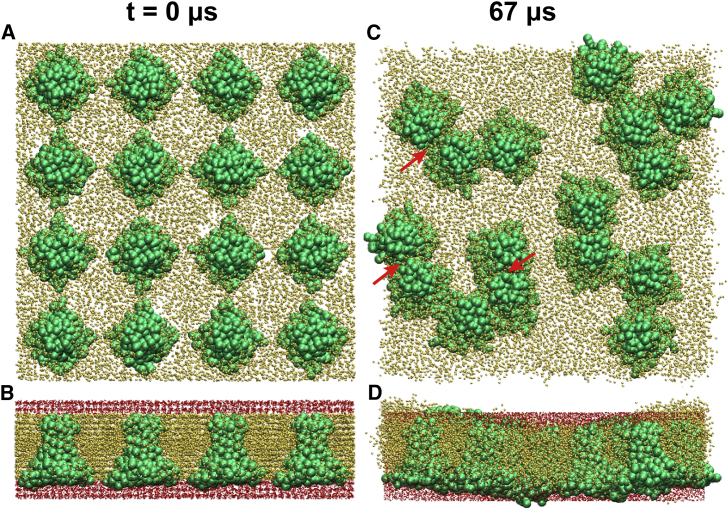
Initial CG simulations were also performed for M2CD, corresponding to residues 22–62, in the all-parallel configuration at an L/T ratio of 85:1. This construct contains the juxtamembrane amphipathic helix essential for the budding process. At the L/T ratio of 85:1, each channel is surrounded by two lipid annuli around the TM region and at least one lipid annulus around the amphipathic helix. The CG simulations also show string-like clustering by M2CD (Fig. 5 C). However, although the clustering of the TM channel was driven by exposed hydrophobic residues in the TM region, the clustering of M2CD is dominated by contacts between the amphipathic helices of neighboring channels. The snapshot at 67 μs shows the presence of isolated lipid molecules between the TM regions of the tetramers. Interestingly, the snapshot also shows contacts between the TM region of channels (indicated by red arrows in Fig. 5). The contact frequency between oligomers is shown in Fig. 6. The larger contact frequencies are concentrated at the C-terminus of the TM domain (residues 44–46) and to the amphipathic helix (residues 47–62). Although the contact between the cores of the TM regions is less frequent, contacts were observed for residues Pro25, Leu36, Leu40, Trp41, and Leu43. Much like with the M2TM CG simulation, these correspond to large hydrophobic residues exposed to the lipid surface, except for Trp41, which projects into the channel pore but is accessible from a lateral fenestration. Importantly, the snapshot in Fig. 5 D shows formation of negative curvature in the bilayers. An analysis of the position of the lipid headgroup nitrogen atom along the simulation box Z axis (Fig. S10) shows that the lipid headgroups developed a curved surface, with the C-terminus of M2CD having the amphipathic helix on the convex surface and the N-termini of M2CD on the concave surface.
Figure 5.
M2CD CG simulation at L/T 85:1. M2CD (res 22–62 in green), lipids represented by small gold beads and water in red. (A and B) Top and side views of CG starting point (t = 0) are shown. (C and D) Top and side views of snapshot at 67 μs are shown. The oligomerization contacts happen predominately via the amphipathic helix region, whereas the TM regions are observed to make less frequent contact. Red arrows in (C) indicate the contacts between TM regions. (D) shows a side view from the snapshot and indicates membrane curvature.
Figure 6.
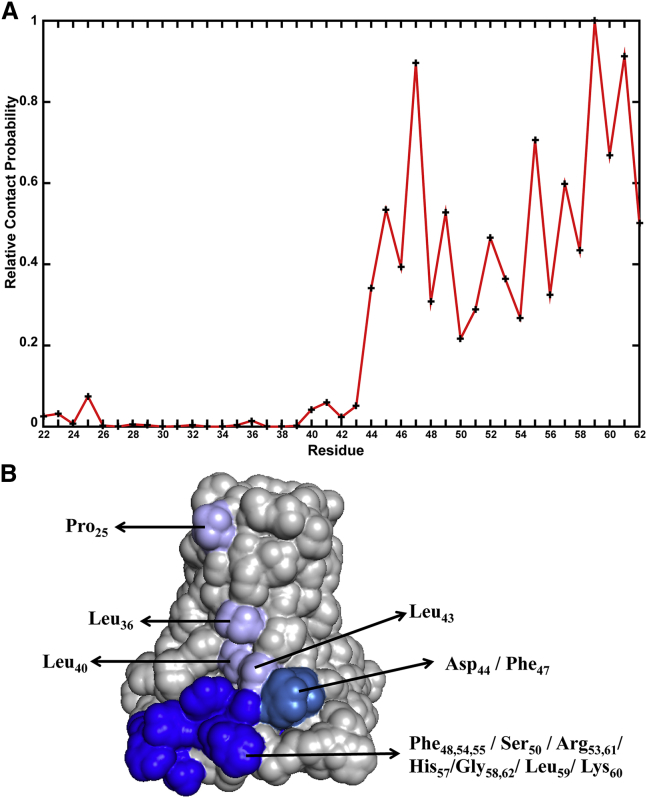
Relative contact frequencies of individual residues in the all-parallel orientation CG simulations of M2CD at L/T = 85:1. (A) Contact frequency was calculated as the fraction of snapshots in which a residue was involved in intertetramer contact, and then normalized by the highest value of all residues. (B) The M2CD surface was colored according to residue contact frequency: light blue, low frequency contacts in the TM; dark cyan, midrange contact frequency of the C-terminal portion of the TM segment; dark blue, high-frequency contacts of the amphipathic helix.
Discussion
The influenza A virion buds through a raft-like domain that forms a favored environment for neuraminidase and hemagglutinin proteins of the virus. Although M2 is expressed in a similar quantity to neuraminidase and hemagglutinin (2, 9) the copy number of M2 is relatively low in the viral coat. This is consistent with the affinity that neuraminidase and hemagglutinin have for raft-like domains and the low affinity that M2 has for such raft-like domains and yet some M2 is found in the viral capsid. Now that it is known that M2 replaces the ESCRT proteins that facilitate viral budding in other viruses, we can discuss how that can be achieved with M2. M2 is highly soluble in liquid-crystalline lipid bilayers and model membranes such as those used in this study. From previous studies, it is known that cholesterol binds M2 (10, 11), consistent with a high concentration of cholesterol surrounding raft-like domains. Furthermore, cholesterol in binding to the amphipathic helix stabilizes the amphipathic helix in the bilayer and hence stabilizes M2’s pyramidal shape (10, 11, 54).
The 2H spectra of M2TM d4Ala29 in DOPC/DOPE lipids reflect motions of the protein in the μs range, and these motions were highly sensitive to the L/T ratio. The spectral analysis demonstrates that the RCT of the M2 tetramer was increased 25-fold by a reduction from two annuli of lipids to just one, suggesting dramatic clustering of the tetramers. These results from a transmembrane helical site, the Ala29 methyl deuterons, were corroborated by CG simulations of the M2TM tetramer in the same L/T ratios, suggesting interactions between the tetramers primarily in the form of linear arrays. The 25-fold increase in RCT observed here differs dramatically from the 1.5- to 2-fold increases for other proteins in proteoliposome preparations reported by EPR and fluorescence (15, 17, 18). As shown in Figs. 4 B and S9 B, the M2 intertetramer interactions are formed by bulky hydrophobic residues and enforced by shape complementarity. It is not clear to what extent these physical characteristics are generalizable to other transmembrane proteins.
Although the contact surface for the M2TM tetramers from the CG simulations does not follow the common intercalated pattern of large and small hydrophobic side chains observed for helix-helix interactions in proteins or in protein complexes (55), what we described here has some resemblance to leucine zippers (56). The string-like clusters of M2TM are, however, substantially different from the clusters observed in other membrane proteins, including the purple membrane containing bacteriorhodopsin and, in mammalian cells, the oligomerization of aquaporin. The first cryo-fracture electron microscopy of the purple membrane showed highly ordered protein aggregates in which the trimeric bacteriorhodopisin makes contact with up to six other trimers (57). In the case of the oligomeric aquaporin 4, the aggregate organizes as a regular square array, first observed by cryo-fracture electron microscopy and now observed in cells by fluorescence microscopy, forming a supramolecular structure in which one channel contacts three or four other channels (58).
As support for the role of M2 in viral budding, we observed the lipid headgroups in CG simulations of M2CD at an L/T ratio of 85:1 in which all channels were oriented in a parallel fashion as in the native environment. Clear evidence for membrane curvature was obtained. Moreover, contacts between the TM regions of M2CD tetramers were also observed. Although intertetramer contacts between transmembrane helices of two antiparallel tetramers of the M2CD have been observed by ssNMR (27), and although we have also observed antiparallel interactions between tetramers here in CG simulations, we are focusing on the native-like interactions between parallel copies of M2TM where we have also observed extensive linear clustering, and in short M2CD CG simulations, lipid bilayer curvature has been observed. Importantly, this is the role that ESCRT proteins play for many viruses during their budding process (59). The extensive string-like clustering is likely to be involved in influenza A’s mechanism for viral budding, for which M2 is essential. The M2 tetramer can be thought of as a frustrum with the top and base parallel to the bilayer and four sides spanning the membrane. A linear organization of M2 tetramers or frustrums could form a narrow protein-lipid bilayer neck for the budding virus.
M2 is found in a liquid-crystalline lipid bilayer environment surrounding raft-like domains that solubilize viral hemagglutinin and neurominidase. It binds cholesterol at considerable concentration at the edge of the raft-like domains. The frustrum shape has the potential, demonstrated here through its linear organization, to dramatically induce curvature in the lipid bilayer at the edge of the raft-like domain and thus facilitate the budding process that leads to a small number of M2 proton channels being incorporated into the viral capsid for one of its other essential roles in the life cycle of influenza A. Future studies on the clustering of the M2CD tetramer in the presence cholesterol could further demonstrate the importance of cholesterol binding and further confirm the linear clustering of the tetramers as the mechanism by which the negative curvature neck of the budding virion is formed, leading to virion excision.
Author Contributions
J.P.: conceptualization; formal analysis; investigation; writing—original draft; sample preparation NMR data acquisition. X.P.: visualization; writing—review and editing; CG molecular dynamics simulations. I.H.: writing—review and editing; initial instruction on NMR data acquisition and data analysis. H.-X.Z.: conceptualization; resources; supervision; methodology; writing—review and editing; CG molecular dynamics simulations. T.A.C.: conceptualization; resources; supervision; writing—original draft; writing—review and editing.
Acknowledgments
This work was supported by National Institutes of Health Grants AI023007, AI119178, and GM122698. The NMR experiments were performed at the National High Magnetic Field Laboratory, funded by the National Science Foundation Division of Materials Research (DMR-1644779) and the State of Florida.
Editor: Michael Brown.
Footnotes
Supporting Materials and Methods and 10 figures are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(19)30114-6.
Supporting Citations
References (60, 61) can be found in the Supporting Material.
Supporting Material
References
- 1.Rossman J.S., Lamb R.A. Influenza virus assembly and budding. Virology. 2011;411:229–236. doi: 10.1016/j.virol.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rossman J.S., Jing X., Lamb R.A. Influenza virus m2 ion channel protein is necessary for filamentous virion formation. J. Virol. 2010;84:5078–5088. doi: 10.1128/JVI.00119-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sharma M., Yi M., Cross T.A. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science. 2010;330:509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holsinger L.J., Nichani D., Lamb R.A. Influenza A virus M2 ion channel protein: a structure-function analysis. J. Virol. 1994;68:1551–1563. doi: 10.1128/jvi.68.3.1551-1563.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou H.X., Cross T.A. Influences of membrane mimetic environments on membrane protein structures. Annu. Rev. Biophys. 2013;42:361–392. doi: 10.1146/annurev-biophys-083012-130326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cross T.A., Murray D.T., Watts A. Helical membrane protein conformations and their environment. Eur. Biophys. J. 2013;42:731–755. doi: 10.1007/s00249-013-0925-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cross T.A., Ekanayake V., Wright A. Solid state NMR: The essential technology for helical membrane protein structural characterization. J. Magn. Reson. 2014;239:100–109. doi: 10.1016/j.jmr.2013.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Murray D.T., Li C., Cross T.A. Membrane protein structural validation by oriented sample solid-state NMR: diacylglycerol kinase. Biophys. J. 2014;106:1559–1569. doi: 10.1016/j.bpj.2014.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rossman J.S., Jing X., Lamb R.A. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 2010;142:902–913. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ekanayake E.V., Fu R., Cross T.A. Structural influences: cholesterol, drug, and proton binding to full-length influenza A M2 protein. Biophys. J. 2016;110:1391–1399. doi: 10.1016/j.bpj.2015.11.3529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Elkins M.R., Williams J.K., Hong M. Cholesterol-binding site of the influenza M2 protein in lipid bilayers from solid-state NMR. Proc. Natl. Acad. Sci. USA. 2017;114:12946–12951. doi: 10.1073/pnas.1715127114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou H.X. Crowding effects of membrane proteins. J. Phys. Chem. B. 2009;113:7995–8005. doi: 10.1021/jp8107446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rousselet A., Cartaud J., Changeux J.P. The rotational diffusion of the acetylcholine receptor in Torpeda marmorata membrane fragments studied with a spin-labelled alpha-toxin: importance of the 43 000 protein(s) EMBO J. 1982;1:439–445. doi: 10.1002/j.1460-2075.1982.tb01188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Javanainen M., Martinez-Seara H., Vattulainen I. Diffusion of integral membrane proteins in protein-rich membranes. J. Phys. Chem. Lett. 2017;8:4308–4313. doi: 10.1021/acs.jpclett.7b01758. [DOI] [PubMed] [Google Scholar]
- 15.Kusumi A., Sakaki T., Ohnishi S. Protein-lipid interaction in rhodopsin recombinant membranes as studied by protein rotational mobility and lipid alkyl chain flexibility measurements. J. Biochem. 1980;88:1103–1111. doi: 10.1093/oxfordjournals.jbchem.a133063. [DOI] [PubMed] [Google Scholar]
- 16.Kusumi A., Hyde J.S. Spin-label saturation-transfer electron spin resonance detection of transient association of rhodopsin in reconstituted membranes. Biochemistry. 1982;21:5978–5983. doi: 10.1021/bi00266a039. [DOI] [PubMed] [Google Scholar]
- 17.Sanders J.C., Ottaviani M.F., Hemminga M.A. A small protein in model membranes: a time-resolved fluorescence and ESR study on the interaction of M13 coat protein with lipid bilayers. Eur. Biophys. J. 1992;21:305–311. doi: 10.1007/BF00188342. [DOI] [PubMed] [Google Scholar]
- 18.Wolkers W.F., Spruijt R.B., Hemminga M.A. Conventional and saturation-transfer EPR of spin-labeled mutant bacteriophage M13 coat protein in phospholipid bilayers. Biochim. Biophys. Acta. 1997;1327:5–16. doi: 10.1016/s0005-2736(97)00038-2. [DOI] [PubMed] [Google Scholar]
- 19.Maguire P.B., Ohlendieck K. Oligomerization of sarcoplasmic reticulum Ca2+-ATPase from rabbit skeletal muscle. FEBS Lett. 1996;396:115–118. doi: 10.1016/0014-5793(96)01106-4. [DOI] [PubMed] [Google Scholar]
- 20.James Z.M., McCaffrey J.E., Thomas D.D. Protein-protein interactions in calcium transport regulation probed by saturation transfer electron paramagnetic resonance. Biophys. J. 2012;103:1370–1378. doi: 10.1016/j.bpj.2012.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chojnacki J., Staudt T., Kräusslich H.G. Maturation-dependent HIV-1 surface protein redistribution revealed by fluorescence nanoscopy. Science. 2012;338:524–528. doi: 10.1126/science.1226359. [DOI] [PubMed] [Google Scholar]
- 22.Gahbauer S., Böckmann R.A. Membrane-mediated oligomerization of G protein coupled receptors and its implications for GPCR function. Front. Physiol. 2016;7:494. doi: 10.3389/fphys.2016.00494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kasai R.S., Kusumi A. Single-molecule imaging revealed dynamic GPCR dimerization. Curr. Opin. Cell Biol. 2014;27:78–86. doi: 10.1016/j.ceb.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 24.Rassam P., Copeland N.A., Kleanthous C. Supramolecular assemblies underpin turnover of outer membrane proteins in bacteria. Nature. 2015;523:333–336. doi: 10.1038/nature14461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hu Y.S., Cang H., Lillemeier B.F. Superresolution imaging reveals nanometer- and micrometer-scale spatial distributions of T-cell receptors in lymph nodes. Proc. Natl. Acad. Sci. USA. 2016;113:7201–7206. doi: 10.1073/pnas.1512331113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sajman J., Trus M., Sherman E. The L-type voltage-gated calcium channel co-localizes with syntaxin 1A in nano-clusters at the plasma membrane. Sci. Rep. 2017;7:11350. doi: 10.1038/s41598-017-10588-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreas L.B., Reese M., Griffin R.G. Structure and mechanism of the influenza A M218-60 dimer of dimers. J. Am. Chem. Soc. 2015;137:14877–14886. doi: 10.1021/jacs.5b04802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright, A. K. 2015. Application of solid state nuclear magnetic resonance in drug discovery. PhD thesis (Florida State University).
- 29.Duff K.C., Ashley R.H. The transmembrane domain of influenza A M2 protein forms amantadine-sensitive proton channels in planar lipid bilayers. Virology. 1992;190:485–489. doi: 10.1016/0042-6822(92)91239-q. [DOI] [PubMed] [Google Scholar]
- 30.Ma C., Polishchuk A.L., Pinto L.H. Identification of the functional core of the influenza A virus A/M2 proton-selective ion channel. Proc. Natl. Acad. Sci. USA. 2009;106:12283–12288. doi: 10.1073/pnas.0905726106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kovacs F.A., Cross T.A. Transmembrane four-helix bundle of influenza A M2 protein channel: structural implications from helix tilt and orientation. Biophys. J. 1997;73:2511–2517. doi: 10.1016/S0006-3495(97)78279-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vold R.L., Hoatson G.L. Effects of jump dynamics on solid state nuclear magnetic resonance line shapes and spin relaxation times. J. Magn. Reson. 2009;198:57–72. doi: 10.1016/j.jmr.2009.01.008. [DOI] [PubMed] [Google Scholar]
- 33.McNaughton B.H., Shlomi M., Kopelman R. Magnetic confinement of Brownian rotation to a single axis and application to Janus and cluster microparticles. Appl. Phys. Lett. 2010;97:144103. [Google Scholar]
- 34.Marrink S.J., Risselada H.J., de Vries A.H. The MARTINI force field: coarse grained model for biomolecular simulations. J. Phys. Chem. B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- 35.Acharya R., Carnevale V., Klein M.L. Structure and mechanism of proton transport through the transmembrane tetrameric M2 protein bundle of the influenza A virus. Proc. Natl. Acad. Sci. USA. 2010;107:15075–15080. doi: 10.1073/pnas.1007071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dong H., Yi M., Zhou H.X. Ab initio calculations and validation of the pH-dependent structures of the His37-Trp41 quartet, the heart of acid activation and proton conductance in the M2 protein of Influenza A virus. Chem. Sci. 2013;4:2776–2787. doi: 10.1039/C3SC50293G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wassenaar T.A., Ingólfsson H.I., Marrink S.J. Computational lipidomics with insane: a versatile tool for generating custom membranes for molecular simulations. J. Chem. Theory Comput. 2015;11:2144–2155. doi: 10.1021/acs.jctc.5b00209. [DOI] [PubMed] [Google Scholar]
- 38.Berendsen H.J.C., van der Spoel D., van Drunen R. GROMACS: a message-passing parallel molecular dynamics implementation. Comput. Phys. Commun. 1995;91:43–56. [Google Scholar]
- 39.Bussi G., Donadio D., Parrinello M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007;126:014101. doi: 10.1063/1.2408420. [DOI] [PubMed] [Google Scholar]
- 40.Alexander S. Visualization and analysis of atomistic simulation data with OVITO–the Open Visualization Tool. Model Simul. Mater. Sci. 2010;18:015012. [Google Scholar]
- 41.Humphrey W., Dalke A., Schulten K. VMD: visual molecular dynamics. J. Mol. Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27–28. [DOI] [PubMed] [Google Scholar]
- 42.Duer M. Blackwell Science Ltd.; London, England: 2002. Solid State NMR Spectroscopy: Principles and Applications; pp. 267–268. [Google Scholar]
- 43.Saffman P.G., Delbrück M. Brownian motion in biological membranes. Proc. Natl. Acad. Sci. USA. 1975;72:3111–3113. doi: 10.1073/pnas.72.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davis J.H. The description of membrane lipid conformation, order and dynamics by 2H-NMR. Biochim. Biophys. Acta. 1983;737:117–171. doi: 10.1016/0304-4157(83)90015-1. [DOI] [PubMed] [Google Scholar]
- 45.Gall C.M., Cross T.A., Opella S.J. Protein dynamics by solid-state NMR: aromatic rings of the coat protein in fd bacteriophage. Proc. Natl. Acad. Sci. USA. 1982;79:101–105. doi: 10.1073/pnas.79.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Griffin R.G. Solid state nuclear magnetic resonance of lipid bilayers. Methods Enzymol. 1981;72:108–174. doi: 10.1016/s0076-6879(81)72010-x. [DOI] [PubMed] [Google Scholar]
- 47.Nicholson L.K., Teng Q., Cross T.A. Solid-state nuclear magnetic resonance derived model for dynamics in the polypeptide backbone of the gramicidin A channel. J. Mol. Biol. 1991;218:621–637. doi: 10.1016/0022-2836(91)90706-c. [DOI] [PubMed] [Google Scholar]
- 48.Opella S.J. Protein dynamics by solid state nuclear magnetic resonance. Methods Enzymol. 1986;131:327–361. doi: 10.1016/0076-6879(86)31048-6. [DOI] [PubMed] [Google Scholar]
- 49.Seelig J. Deuterium magnetic resonance: theory and application to lipid membranes. Q. Rev. Biophys. 1977;10:353–418. doi: 10.1017/s0033583500002948. [DOI] [PubMed] [Google Scholar]
- 50.Beshah K., Griffin R.G. Deuterium quadrupole echo NMR study of methyl group dynamics in N-Acetyl-dl-(γ-d6)-valine. J. Magn. Reson. 1989;84:268–274. [Google Scholar]
- 51.Vugmeyster L., Ostrovsky D., Vold R.L. Glassy dynamics of protein methyl groups revealed by deuteron NMR. J. Phys. Chem. B. 2013;117:1051–1061. doi: 10.1021/jp311112j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lee K.C., Hu W., Cross T.A. 2H NMR determination of the global correlation time of the gramicidin channel in a lipid bilayer. Biophys. J. 1993;65:1162–1167. doi: 10.1016/S0006-3495(93)81150-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lee K.C., Cross T.A. Side-chain structure and dynamics at the lipid-protein interface: Val1 of the gramicidin A channel. Biophys. J. 1994;66:1380–1387. doi: 10.1016/S0006-3495(94)80928-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Elkins M.R., Sergeyev I.V., Hong M. Determining cholesterol binding to membrane proteins by cholesterol 13C labeling in yeast and dynamic nuclear polarization NMR. J. Am. Chem. Soc. 2018;140:15437–15449. doi: 10.1021/jacs.8b09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dong H., Sharma M., Cross T.A. Glycines: role in α-helical membrane protein structures and a potential indicator of native conformation. Biochemistry. 2012;51:4779–4789. doi: 10.1021/bi300090x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Landschulz W.H., Johnson P.F., McKnight S.L. The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science. 1988;240:1759–1764. doi: 10.1126/science.3289117. [DOI] [PubMed] [Google Scholar]
- 57.Baldwin J.M., Henderson R., Zemlin F. Images of purple membrane at 2.8 A resolution obtained by cryo-electron microscopy. J. Mol. Biol. 1988;202:585–591. doi: 10.1016/0022-2836(88)90288-4. [DOI] [PubMed] [Google Scholar]
- 58.Rossi A., Moritz T.J., Verkman A.S. Super-resolution imaging of aquaporin-4 orthogonal arrays of particles in cell membranes. J. Cell Sci. 2012;125:4405–4412. doi: 10.1242/jcs.109603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen B.J., Lamb R.A. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology. 2008;372:221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ottiger M., Bax A. How tetrahedral are methyl groups in proteins? A liquid crystal NMR study. J. Am. Chem. Soc. 1999;121:4690–4695. [Google Scholar]
- 61.Wu Y., Stefl M., Kuimova M.K. Molecular rheometry: direct determination of viscosity in Lo and Ld lipid phases via fluorescence lifetime imaging. Phys. Chem. Chem. Phys. 2013;15:14986–14993. doi: 10.1039/c3cp51953h. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.