Abstract
Klebsiella michiganensis is a newly emerging human pathogen. We describe a case of bloodstream infection in an immunocompromised patient. The pathogen was repeatedly isolated from blood and one rectal swab, and was identified using routine standard procedures. Further investigations revealed that the K. michiganensis was multidrug resistant, carrying a plasmid harbouring a Klebsiella pneumoniae carbapenemase (KPC)-3 carbapenemase gene. This plasmid has been frequently encountered in K. pneumoniae isolates in Europe but has never been described in K. michiganensis.
Keywords: Antibiotic resistance, carbapenemase, ceftazidim/avibactam, Enterobacterales, KPC
Introduction
The fast and reliable detection and characterization of multidrug-resistant pathogens is of utmost importance in order to support therapy and infection control. Laboratories are challenged by a notoriously growing number of variants of extended-spectrum β-lactamase and carbapenemase (CP) enzymes. With the newly approved antibiotic ceftazidime/avibactam, showing activity against class A and C β-lactamases but not against B and D [1], the reporting of CP positivity alone is no longer sufficient; the determination of the underlying CP mechanism is important to establish an efficient antibiotic therapy. Immediate tracing of resistant pathogens is crucial for infection control purposes, especially in low-prevalence countries, in order to contain resistant strains and prevent their spread.
Case description and results
We report the first case of an invasive infection with Klebsiella pneumoniae CP (KPC)-3–producing Klebsiella michiganensis from St Gall, Switzerland. K. michiganensis was originally been discovered on a toothbrush holder during a study on microbial hot spots in residential US households in 2012 [2]. Since then, this potential emerging pathogen has been reported in clinically relevant contexts all over the world [3], [4], [5]. Our K. michiganensis was retrieved from a blood culture (Becton Dickinson, Franklin Lakes, NJ, USA) while performing routine diagnostics in a febrile neutropenic patient with acute myeloid leukaemia. Several follow-up blood cultures (n = 16) and one rectal swab from the same patient were also culture positive for K. michiganensis.
Identification and susceptibility testing were done following routine procedures using MALDI-TOF MS (Bruker Daltonics, Billerica, MA, USA), the commercial NMIC-417 susceptibility Phoenix panel (Becton Dickinson) and the UMIC Colistin Kit (Biocentric, Bandol, France). The susceptibility test, interpreted according to the European Committee on Antimicrobial Susceptibility Testing guidelines (version 7.1), revealed resistance against first- to fourth-generation cephalosporins and the carbapenem ertapenem, intermediate resistance against imipenem and meropenem (both MIC = 8 mg/L) and susceptibility towards ceftazidime/avibactam (MIC = 1 mg/L). In addition, the isolate was resistant to almost all other standard antibiotics except for amikacin, colistin, tigecycline and fosfomycin (Supplementary Table S1).
The patient was initially treated with high-dose meropenem (prolonged infusion) in combination with colistin. Because of persistent fever, treatment was changed to ceftazidime/avibactam instead of meropenem, tigecycline and amikacin were added to the regimen and the central venous catheter was removed. Blood cultures remained negative thereafter; however, the patient died several weeks after the last positive blood culture of his underlying illness.
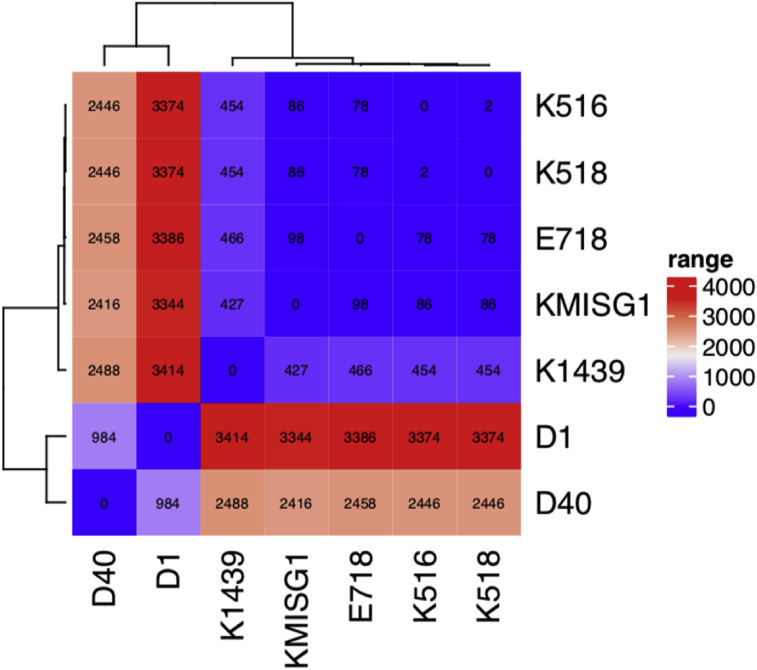
Initial MALDI-TOF MS–based identification indicated an infection with Klebsiella oxytoca. However, whole-genome sequencing (WGS) identified the pathogen as K. michiganensis (95.24% query coverage, 96.7% reference coverage). WGS was performed using Illumina MiSeq with the Nextera XT library preparation kit (Illumina, San Diego, CA, USA) according to the manufacturer's instructions. Assembly of the sequences was accomplished with Spades (http://cab.spbu.ru/software/spades/), and the genetic content, including the identification of the species, was examined using GoSeqIt (https://www.goseqit.com/). Analysing the WGS data with an overall coverage of 81× indicated that the isolate carried the β-lactamase genes blaKPC-3, blaTEM-1B and blaOXY-1-1 along with resistance genes against aminoglycoside (aac(3)-IId, aph(3′)-Ia, aadA5, strA, strB), macrolide (mph(A)), sulphonamide (su11, su12), tetracycline (tet(B)) and trimethoprim (dfrA17). The core-genome calculations, performed by Roary (version 3.11.2), indicated that our isolate was genetically related with 98 to 427 single nucleotide polymorphisms (SNP)/core genome to clinical isolates K1439 (human blood, unpublished data), K518/K516 from China [3] and E718 from Taiwan [4]. With 2416 to 3344 SNPs/core genome, our isolate was also distantly related to isolates D1 and D40 (unpublished data) that originated from cows with mastitis (Fig. 1).
Fig. 1.
Single nucleotide polymorphism core-genome heat map of KMISG1 and closely related Klebsiella michiganensis isolates. Calculations were performed using Roary, a software tool for rapid large-scale prokaryote pan-genome analysis (https://academic.oup.com/bioinformatics/article/31/22/3691/240757, version 3.11.2). National Center for Biotechnology Information BioProject numbers of isolates used for calculation (year, location and source of isolation) are as follows: K516 (GCA_002216835, 2016, China, human), K518 (GCA_002290285, 2016, China, human), E718 (GCA_000276705, 2010, Taiwan, human), K1439 (GCA_002265195, 2014, China, human), D1 (GCA_002857365, 2016, USA, cow), D40 (GCA_002856965, 2016, USA, cow) and KMISG1 (PRJNA485881, 2017, Switzerland, human).
Plasmid multilocus sequence typing algorithms, which are part of the GoSeqIt analysis, revealed that the isolate was harbouring at least two plasmids, IncQ and IncFIIK2-FIB [6]. The IncFIIK2-FIB plasmid carrying the blaKPC-3 gene, encoded on a Tn4401a isoform, has been subsequently closed using specific primers and Sanger sequencing (BioProject PRJNA485881) (Supplementary Table S1). Comparing the plasmid sequence with GenBank data using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi) revealed a remarkable homology to pIT-11C07 (100% query coverage, >99.9% maximum nucleotide identity) that originated from a clinical K. pneumoniae isolated from Italy [7]. The plasmids, both having blaKPC-3 as the only resistance gene, shared a high similarity to the blaKPC-3-carrying plasmid pKpQIL, which originated from a clonal outbreak in Israel and has since been isolated all over the world [7], [8]. High homology was also seen with plasmids originating from Switzerland [9] (99% query coverage, 99% maximum nucleotide identity to CP015824), Greece [7] (97% query coverage, 99% maximum nucleotide identity to GR-1870) and the United States [10] (91% query coverage, 99% maximum nucleotide identity to p500_1420).
Conclusion
To our knowledge, this is the first clinical case of a patient with an invasive multidrug-resistant K. michiganensis infection in Europe. Moreover, the isolate harboured a blaKPC-3-carrying IncFIIK2-FIB plasmid, a combination that has not been published before. The lack of epidemiologic risk factors as the exposure to other patients with CP-producing pathogens, recent travel history or hospital admission abroad raises concerns about potential CP circulation within Switzerland. Our case highlights the necessity of fast and precise diagnostics. Moreover, conclusive diagnostics increasingly require molecular data such as WGS in order to provide clinicians and epidemiologists with reliable information.
Acknowledgements
We thank our laboratory staff members for their excellent work, and we thank the members of the infectious diseases and hospital epidemiology division for their close collaboration.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.nmni.2019.100516.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Kazmierczak K.M., de Jonge B.L.M., Stone G.G., Sahm D.F. In vitro activity of ceftazidime/avibactam against isolates of Enterobacteriaceae collected in European countries: INFORM global surveillance, 2012–15. J Antimicrob Chemother. 2018;73:2782–2788. doi: 10.1093/jac/dky266. [DOI] [PubMed] [Google Scholar]
- 2.Saha R., Farrance C.E., Verghese B., Hong S., Donofrio R.S. Klebsiella michiganensis sp. nov., a new bacterium isolated from a tooth brush holder. Curr Microbiol. 2013;66:72–78. doi: 10.1007/s00284-012-0245-x. [DOI] [PubMed] [Google Scholar]
- 3.Zheng B., Xu H., Yu X., Lv T., Jiang X., Cheng H. Identification and genomic characterization of a KPC-2–, NDM-1– and NDM-5–producing Klebsiella michiganensis isolate. J Antimicrob Chemother. 2018;73:536–538. doi: 10.1093/jac/dkx415. [DOI] [PubMed] [Google Scholar]
- 4.Liao T.L., Lin A.C., Chen E., Huang T.W., Liu Y.M., Chang Y.H. Complete genome sequence of Klebsiella oxytoca E718, a New Delhi metallo-β-lactamase-1–producing nosocomial strain. J Bacteriol. 2012;194:5454. doi: 10.1128/JB.01216-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pedersen T., Sekyere J.O., Govinden U., Moodley K., Sivertsen A., Samuelsen Ø. Spread of plasmid-encoded NDM-1 and GES-5 carbapenemases among extensively drug-resistant and pandrug-resistant clinical Enterobacteriaceae in Durban, South Africa. Antimicrob Agents Chemother. 2018;62 doi: 10.1128/AAC.02178-17. e02178-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.García-Fernández A., Villa L., Carta C., Venditti C., Giordano A., Venditti M. Klebsiella pneumoniae ST258 producing KPC-3 identified in Italy carries novel plasmids and OmpK36/OmpK35 porin variants. Antimicrob Agents Chemother. 2012;56:2143–2145. doi: 10.1128/AAC.05308-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Papagiannitsis C.C., Di Pilato V., Giani T., Giakkoupi P., Riccobono E., Landini G. Characterization of KPC-encoding plasmids from two endemic settings, Greece and Italy. J Antimicrob Chemother. 2016;71:2824–2830. doi: 10.1093/jac/dkw227. [DOI] [PubMed] [Google Scholar]
- 8.Leavitt A., Chmelnitsky I., Carmeli Y., Navon-Venezia S. Complete nucleotide sequence of KPC-3-encoding plasmid pKpQIL in the epidemic Klebsiella pneumoniae sequence type 258. Antimicrob Agents Chemother. 2010;54:4493–4496. doi: 10.1128/AAC.00175-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nicolet S., Goldenberger D., Schwede T., Page M., Creus M. Plasmid-mediated colistin resistance in a patient infected with Klebsiella pneumoniae. Lancet Infect Dis. 2016;16:998–999. doi: 10.1016/S1473-3099(16)30197-9. [DOI] [PubMed] [Google Scholar]
- 10.Wright M.S., Perez F., Brinkac L., Jacobs M.R., Kaye K., Cober E. Population structure of KPC-producing Klebsiella pneumoniae isolates from midwestern US hospitals. Antimicrob Agents Chemother. 2014;58:4961–4965. doi: 10.1128/AAC.00125-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.