Abstract
Single-molecule and super-resolution imaging relies on successful, sensitive, and accurate detection of the emission from fluorescent molecules. Yet, despite the widespread adoption of super-resolution microscopies, single-molecule data processing algorithms can fail to provide accurate measurements of the brightness and position of molecules in the presence of backgrounds that fluctuate significantly over time and space. Thus, samples or experiments that include obscuring backgrounds can severely, or even completely, hinder this process. To date, no general data analysis approach to this problem has been introduced that is capable of removing obscuring backgrounds for a wide variety of experimental modalities. To address this need, we present the Single-Molecule Accurate LocaLization by LocAl Background Subtraction (SMALL-LABS) algorithm, which can be incorporated into existing single-molecule and super-resolution analysis packages to accurately locate and measure the intensity of single molecules, regardless of the shape or brightness of the background. Accurate background subtraction is enabled by separating the foreground from the background based on differences in the temporal variations of the foreground and the background (i.e., fluorophore blinking, bleaching, or moving). We detail the function of SMALL-LABS here, and we validate the SMALL-LABS algorithm on simulated data as well as real data from single-molecule imaging in living cells.
Main Text
Single-molecule super-resolution imaging has revolutionized microscopy (1, 2, 3, 4, 5) through a variety of experimental modalities, such as stochastic optical reconstruction microscopy (STORM) (6) photoactivated localization microscopy (PALM) (7, 8), and points accumulation for imaging in nanoscale topography (PAINT) (9). Yet, these experimental techniques all rely on accurately and precisely localizing single emitters with successful data processing algorithms (10, 11, 12, 13, 14). Realistic backgrounds vary in time and space and decrease the signal-to-noise ratio (SNR); these backgrounds can severely obscure super-resolution imaging by reducing the localization precision, introducing systematic biases, and even preventing successful detection through both false-positive and false-negative errors. In addition to improving single-molecule localization, a particular challenge in the field is to attain unbiased measurements of single-molecule intensities for single-molecule counting experiments, single-molecule fluorescence resonance energy transfer (FRET) experiments, and as a single-molecule probe of the local environment.
Experimental measures can certainly decrease backgrounds. However, popular methods for this purpose, such as confined illumination via light sheets (15, 16, 17) and total internal reflection (TIRF) (18), reduce out-of-focus fluorescence but do not address in-plane backgrounds. Longer wavelength excitation decreases cellular autofluorescence (19) but sacrifices the resolution improvement of imaging at a shorter wavelength. Additional fluorescent objects, such as plasmonic antennas for fluorescence enhancement or fiducial markers for drift compensation, can be incorporated into the sample for added functionality or to improve imaging, but themselves produce a punctate spot in the background that can be misidentified as a fluorescent molecule (fluorophore) or that can obfuscate nearby fluorophores (20, 21, 22). Moreover, these adaptations tend to complicate or restrict experiments. As a broadly applicable alternative to modifying experimental designs to reduce backgrounds, we report here a general algorithm: SMALL-LABS (Single-Molecule Accurate LocaLization by LocAl Background Subtraction), which accurately locates and measures the intensity of single molecules, regardless of the shape or brightness of the background. Accurate background subtraction is enabled by separating the foreground from the background based on differences in the temporal variations of the foreground and the background due to fluorophore blinking, bleaching, or moving.
To our knowledge, no other background removal algorithm to date can eliminate the systematic bias in intensity measurements and position determination (localization) for a wide range of experimental systems (Supporting Materials). For instance, although several new approaches can accurately localize single molecules within a dense ensemble (10, 11), these algorithms assume a background shaped like the image of an overlapping neighboring molecule and fail for arbitrary backgrounds. Additionally, such high-density approaches indiscriminately identify as molecules all signals that look like the system point spread function (PSF) regardless of the temporal dynamics. In general, approaches that attempt to subtract the background without first identifying the foreground (23) will inevitably introduce distortions by subtracting some of the image of a fluorophore from itself (Supporting Materials). SMALL-LABS provides the true background-subtracted image for single-molecule data by specifically distinguishing the foreground from the background; the only requirement is that the local background changes more slowly than the characteristic on/off timescale of the fluorophores. In this Computational Tools article, we present SMALL-LABS and detail its function, validate its performance on simulated single-molecule fluorescence data, and demonstrate its capability on measured live-cell single-molecule data. We also provide open-source MATLAB (The MathWorks, Natick, MA) code that implements the SMALL-LABS algorithm.
SMALL-LABS operating principles
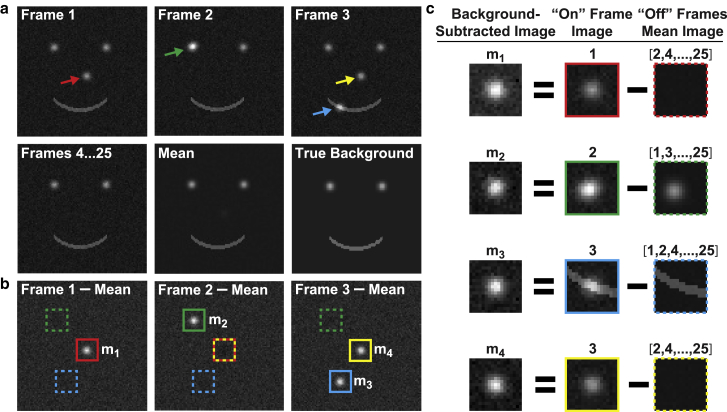
The SMALL-LABS algorithm comprises a workflow described in detail below and summarized here. First, an “approximate background” calculated from the running average is subtracted from the raw movie, making single fluorescent molecules detectable with standard image analysis techniques (Fig. 1, a and b, step 1). This approximate background correction (23) removes the obscuring background but will also subtract part of the true image from itself, reducing the apparent intensity and possibly introducing systematic biases (Supporting Materials). Therefore SMALL-LABS uses the approximate background subtraction only for this initial molecule detection step (Fig. 1 b, step 2). Next, for each detection, SMALL-LABS identifies which frames contained detections at or near the position of the current detection. Fluorophores can turn on and off due to blinking, bleaching, or moving, so this check produces a list of “off” imaging frames in which no other molecule is detected in the local vicinity of each detected molecule (step 3). The “true local background” is defined in SMALL-LABS as the average of these “off”-frame images at the molecule position. Finally, this true background is removed locally for each detected (“on” frame) molecule (Fig. 1 c, step 4). Importantly, this algorithm does not subtract the image of a molecule from itself, ensuring that further analysis of the background-subtracted image provides accurate super-resolution information and avoids the systematic biases in the determination of each molecule’s brightness that would arise from incorrect background subtraction (step 5).
Figure 1.
Schematic illustration of the SMALL-LABS algorithm. (a) Simulated raw data (imaging frames), the mean of the entire movie, and the true background (all on the same grayscale) are shown. Frames 1–3 have fluorescent molecules, indicated with colored arrows; frames 4–25 are identical except for detection noise and only contain the background; the mean includes a faint image of the real molecules over the true background. (b) Molecule detection in the approximate background-subtracted movie is shown. Solid colored boxes indicate a detected molecule, and dashed colored boxes indicate the local background for that molecule in “off” frames. Box colors correspond to the arrows in (a). (c) The SMALL-LABS background subtraction process is shown. The true image of each molecule is obtained by locally subtracting the mean of the “off” frames from the raw image. To see this figure in color, go online.
The SMALL-LABS algorithm is described in this article. We also provide open-source code that implements this algorithm. The code we provide is customizable for diverse data sets (for details, see the user guide), although it is optimized here for the case of low-density single-molecule data. Additionally, the SMALL-LABS algorithm is modular, and we encourage users to incorporate it into their own code (for instance, to use different detection, analysis, or filtering tools than the ones we provide). For example, researchers imaging high-density fluorophores will need to modify several parts of our code or use their own code to implement the SMALL-LABS algorithm for background removal. Specifically, in the high-density case in which there are many in-frame overlapping molecule PSFs, our method here of determining “off” frames (step 3) will need to be modified. We show here that for a range of experimental conditions and desired measurements, SMALL-LABS successfully detects single molecules and accurately estimates the background to reduce biases due to background subtraction and thereby increases the accuracy of single-molecule position and brightness estimations.
The workflow for the SMALL-LABS algorithm is as follows:
-
1)
Approximate background subtraction from the raw_movie to produce the avgsub_movie
-
2)
Molecule detection in the avgsub_movie
-
3)
“Off”-frame identification for each detected molecule
-
4)
Accurate background subtraction for each molecule
-
5)
Further analysis of the true background-subtracted image of each molecule (intensity measurement, position determination, tracking, etc.)
Step 1: Approximate background subtraction
To enable the initial single-molecule detection step (2), an approximate background is subtracted from the original movie (the raw_movie). This background is only an approximation because the foreground has not been distinguished from the background. The simplest method to calculate the approximate background, which we employ in the provided code, is to calculate a moving temporal mean (or median or similar statistical measure; Figs. S1–S3; Tables S1–S4) for the raw_movie. This mean image is shown in Fig. 1 a. For simplicity, the example shown in Fig. 1 calculates an approximate background from the mean of the entire movie. In general, the characteristic on/off frequency of the molecules (from blinking, photobleaching, photoswitching, or motion) should be considered: the choice of the window length over which to calculate an average (or median) should be the longest window possible that does not include slow background changes at lower frequencies than this characteristic frequency. Although the window time is a fairly weak parameter, if this length is too short, fluorophores that do not blink, bleach, or move over the window length will be erroneously removed with the more static background. Having a long window time relative to the characteristic on/off time increases the accuracy of the approximate calculation of the background by minimizing contributions from molecules to the mean. The mean raw_movie image is then subtracted from each frame of the raw_movie to produce the avgsub_movie, the approximate background-subtracted movie (20, 23, 24), as shown in Fig. 1 b.
Step 2: Molecule detection
An obscuring background in the raw_movie could produce a large number of false-positive or false-negative errors in single-molecule detection. The approximate background removal in step 1 allows molecules to be identified in the avgsub_movie (Fig. 1 b) with standard image analysis techniques. Any suitable detection algorithm can be used for step 2 in the SMALL-LABS algorithm. For example, the detection algorithm we provide in our code applies a bandpass to the image in spatial frequency, then identifies spots brighter than a user-supplied threshold percentile and that have an equivalent diameter (calculated with regionprops in MATLAB) near the PSF size.
Although the accuracy and precision of these detections may be hindered by the approximate background (Supporting Materials), detecting molecules in the avgsub_movie rather than in the raw_movie greatly reduces the likelihood of false-positive and false-negative detection errors. For example, molecular detection in the raw_movie would likely have missed molecule 3 in Fig. 1 (a false-negative error). Similarly, single-molecule detection in the raw_movie would have incorrectly identified the eyes in the background smiley face in Fig. 1 as molecules, giving several false-positive errors. Doing molecule detection in the avgsub_movie avoids such errors. Furthermore, as long as the false-negative rate is low, a substantial false-positive rate is permissible, and the SMALL-LABS algorithm is largely insensitive to any accuracy or precision loss in this detection step because step 5 below repeats the characterization of each single molecule to provide high accuracy and precision measurements and to allow for further false-positive screening.
Step 3: “Off”-frame identification
To accurately calculate the true background, it is essential to exclude the foreground (images of single molecules). For each molecule detected in step 2, a local “off”-frames list is constructed; this list enumerates all frames in which no molecule was detected in the local region. Because we expect a diffraction-limited single-molecule image with a shape given by the microscope PSF, this local region is a box about the molecule position with a side length approximately double the PSF width, although the local region can be changed for different imaging conditions like defocus. SMALL-LABS is agnostic to whether the same molecule is on in multiple frames. Rather, the “off”-frames list depends only on if any molecule is detected in the same local region in other imaging frames, regardless of whether this molecule is the same molecule fluorescing for sequential frames, a molecule that blinks on and off, or distinct molecules that appear at the same location in different frames. The “off”-frames list can be calculated over the entire movie, as in Fig. 1, or for a smaller number of frames based on the window length considerations discussed in step 1.
For example, in Fig. 1, molecule 2 (green arrow) appears in frame 2 and is the only molecule ever detected in that local region (green box); the “off”-frames list for molecule 2 therefore consists of all the other frames in the movie (i.e., frames [1,3,…,25]). Similarly, molecule 3 (Fig. 1, blue arrow) is only fluorescent in frame 3; its “off”-frames list is frames [1,2,4,…,25]. On the other hand, molecules 1 and 4 (Fig. 1) appear in the same local region (yellow and red boxes) in different frames, and thus the “off”-frames list is the same for both molecules: this list excludes both the frame in which molecule 1 appears and the frame in which molecule 4 appears (Fig. 1 c).
Step 4: Accurate background subtraction
In this key step of SMALL-LABS, the true background is calculated by taking the temporal mean (or median or similar statistical measure) over only frames in the “off”-frames list of the raw_movie in the local region around a molecule detection (Fig. 1 c, dashed boxes). Whereas an approximate background is removed in step 1 and nonspecific spatial bandpassing can be used to suppress some backgrounds in step 2, the true background is subtracted here in step 4. This accurate background does not contain partial images of the molecule itself or of any other molecule (the foreground). This accurate background is subtracted from the original raw_movie image of the molecule (Fig. 1 c, solid boxes) to produce to a local background-free image of the molecule (Fig. 1 c, boxes m1 through m4). For example, for molecule 3 (Fig. 1 c), the local region around the molecule is averaged over frames [1,2,4,…,25] to produce the true background, which is subtracted from the image of molecule 3 in the raw_movie frame 3, thereby completely removing the background from the smiley face mouth.
Step 5: Further single-molecule analysis
Once the background has been accurately removed, any further single-molecule analysis can be performed. For instance, PSF fitting the background-free single-molecule image provides super-resolution localization (1, 2, 3, 25) while avoiding any biases that could be introduced by the background or by an inaccurate background removal. We discuss these biases in depth in the Supporting Materials. Importantly, although an approximate background removal like that in step 1 typically preserves the localization precision for very sparse samples, SMALL-LABS is essential for providing unbiased measurements of single-molecule intensities. Thus, in addition to enabling precise position determination, the emission intensity of each fluorescent molecule can be accurately measured based on PSF fitting or by summing pixel intensities after accurate background subtraction.
Validating SMALL-LABS with simulated data
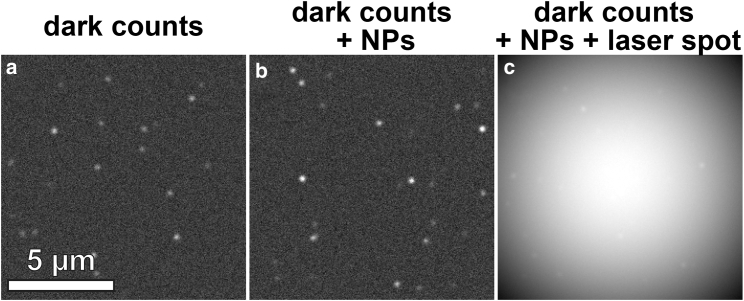
To test the scope and performance of SMALL-LABS, we simulated realistic single-molecule data with increasingly difficult realistic backgrounds and compared the measured results from the algorithm to the ground truth input to the simulations. Three different simulated movies were analyzed. The first movie (Fig. 2 a) has only the simple intensity offset background (nonzero dark counts) common to most electron-multiplying charge-coupled device and scientific complementary metal-oxide-semiconductor cameras. In addition to the constant intensity offset of Fig. 2 a, the second movie (Fig. 2 b) has several static bright background spots identical to fluorophore images in brightness and size but invariant over time. This background condition is common when fiduciary markers or photoluminescent nanoparticles (NPs) are incorporated into a sample (22, 26, 27). The third movie (Fig. 2 c) contains the same background as in Fig. 2 b and additionally has a wide, bright Gaussian image overlaid on the entire movie to mimic the spatially varying background that can result from spatial variations in the excitation laser beam.
Figure 2.
Representative frames from the simulated movies with similar foregrounds and different backgrounds. (a) The background consists of only dark counts. (b) The background contains dark counts and static fluorescent nanoparticles (NPs). (c) The movie has dark counts, static fluorescent NPs, and a spatially varying background.
The simulated movies were created with signal intensity distributions and noise parameters that realistically occur in single-molecule experiments with fluorescent probes detected on an electron-multiplying charge-coupled device detector (20). The purpose of this data set is to test the background removal ability of SMALL-LABS and not to push the algorithm to find extremely low SNR molecules or to try to use the algorithm to achieve high-density localization; to analyze such data sets, users will need to modify the code that implements the SMALL-LABS algorithm. Thus, the simulated movies contained reasonable SNRs (here defined as the ratio of the single-molecule fluorescence amplitude to the standard deviation (SD) of the movie noise) ranging from 1.25 to 10 (Fig. S7), and localizations were well spatially separated (<1 molecule/μm2) as in standard low-density single-molecule experiments (for instance, single-particle tracking or in vitro single-molecule kinetics). Furthermore, in accordance with experiments, molecules could stay on for multiple frames (the duration of their emission was given by the absolute value of a normal distribution with a mean of one frame and an SD of three frames). Finally, because their location was randomly determined, molecules could appear at the same location as a previous molecule (like molecules 1 and 4 in Fig. 1); in these cases, a simpler algorithm would not remove the background accurately.
As a first measure of performance, we analyzed the ability of SMALL-LABS to accurately detect single molecules. The Jaccard index is the ratio of the cardinality (the number of elements in a set) of the intersection between the set of simulated molecules, S, and the set of detected molecules, D, to the cardinality of the union of S and D (11):
The false-positive and false-negative rates, FP and FN, respectively, can be similarly expressed:
The detection results for the simulated molecules (after false-positive filtering of the accurate background-subtracted data in step 5) of the three movies are presented in Table 1. In all three cases, SMALL-LABS performs well, as evidenced by a high Jaccard index and low false-positive and false-negative error rates. In particular, the FP rate does not increase upon addition in Fig. 2 b of the NP background, which is identical in appearance to the molecules. Furthermore, in the case of the laser spot background (Fig. 2 c), molecule detection without background removal would be extremely limited, leading to a substantial increase in the FN rate, whereas most molecules are correctly identified after accurate background subtraction by SMALL-LABS. To illustrate one need addressed by the SMALL-LABS algorithm, we compared the detection results for the comprehensive ThunderSTORM single-molecule image analysis package (28) to the results from our code, which implements the SMALL-LABS algorithm (Table S5). We find that SMALL-LABS successfully removes the backgrounds in all three cases to significantly reduce the false-positive and false-negative rates relative to ThunderSTORM alone (Figs. S4–S6; Supporting Materials).
Table 1.
Detection Results
| Background | Jaccard | FP Rate | FN Rate |
|---|---|---|---|
| Dark counts | 0.903 | 0.014 | 0.086 |
| Dark counts + NPs | 0.905 | 0.001 | 0.089 |
| Dark counts + NPs + laser spot | 0.878 | 0.016 | 0.100 |
The results show the Jaccard index, false-positive (FP) error rate, and false-negative (FN) error rate for single molecules in the different simulated movies of Fig. 2.
In addition to validating the ability of SMALL-LABS to detect molecules (Table 1), we also analyzed the performance of SMALL-LABS in measuring some relevant characteristics of the simulated molecules. Table 2 indicates how accurately SMALL-LABS enables the intensity and super-resolved position (as determined by a least-squares Gaussian fit) of the background-subtracted molecule images of each molecule in each movie to be measured. The mean (μ) and SD (σ) of the error distributions for each measured quantity for all molecules in each movie are tabulated in Table 2. Full distributions and further details are given in Supporting Materials. In all three movies, SMALL-LABS performs well (Figs. S8–S10; Table 2): all error distributions are centered near μ = 0 and have small σ. Furthermore, the error distributions are fairly insensitive to the nature of the background: there is little change in the statistics between the three movies. Importantly, many approximate background removal approaches introduce a bias (μ ≠ 0) in these measured quantities, with especially large biases for intensity measurements (Supporting Materials), whereas SMALL-LABS does not introduce any such systematic biases.
Table 2.
Error Distribution Characteristics
| Background | x Position Error (nm)a | y Position Error (nm)a | Intensity Error (%)b |
|---|---|---|---|
| Dark counts | μ = 0.165 | μ = 0.114 | μ = 0.821 |
| σ = 13.7 | σ = 13.6 | σ = 20.6 | |
| Dark counts + NPs | μ = –0.112 | μ = –0.029 | μ = 1.12 |
| σ = 13.7 | σ = 13.9 | σ = 20.5 | |
| Dark counts + NPs + laser spot | μ = –0.050 | μ = –0.185 | μ = 1.64 |
| σ = 14.3 | σ = 14.4 | σ = 21.2 |
The characteristics mean, μ, and SD, σ, for the simulated movies are shown.
x and y position error is the difference between the measured and true positions of the molecule.
The intensity percent error is {100% × (measured − true)/true} for the summed pixel intensities in the local region around the molecule.
Validating SMALL-LABS with live-cell single-molecule tracking
To validate SMALL-LABS and demonstrate its scope, we imaged single fluorescent proteins in living bacteria cells under optimal single-molecule tracking conditions (Fig. 3 a; Video S1) and in conditions that preclude traditional single-molecule detection (Fig. 3 b; Video S2). We imaged Bacillus subtilis strains natively expressing the DNA polymerase PolC fused to the photoactivatable fluorescent protein PAmCherry as the sole source of PolC (strain JWS213). PolC is one of the two replicative DNA polymerases in B. subtilis and has been characterized in our previous work (29). To produce the high background in Fig. 3 b, a constant 15-W/cm2, 488-nm laser illumination generated a strong autofluorescent background in the cells; this background was further complicated by its slow decay over time. By stochastically switching a small subset (1–3 molecules per cell) of the PolC-PAmCherry molecules into a fluorescent state at a time (in a single-particle tracking/ photoactivated localization microscopy experiment), we visualized the dynamics of 420 single PolC-PAmCherry molecules in 200 high-background cells (Fig. 3 b) and 200 single PolC-PAmCherry molecules in 30 low-background cells (Fig. 3 a).
Figure 3.
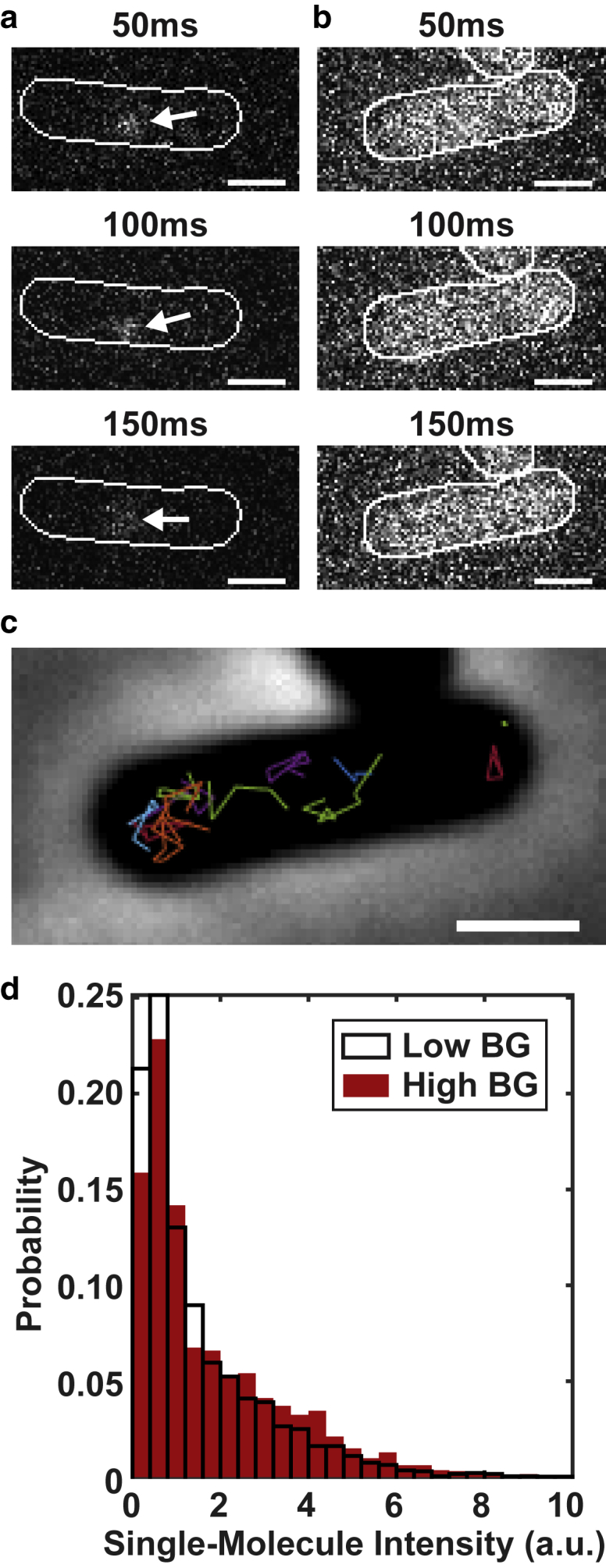
Tracking single PolC-PAmCherry molecules in living B. subtilis cells. (a) Representative raw-data images of a single PolC-PAmCherry molecule (arrow) in a B. subtilis cell are shown; the molecule is easily identifiable and can be tracked over time. (b) No PolC-PAmCherry molecules can be identified by eye in the raw-data images in high-background experimental conditions. (c) Accurate background subtraction with SMALL-LABS enables single molecules to be detected and localized from the high-background movie in (b), and trajectories are obtained (colored lines). (d) Comparison of the measured single-molecule intensities of the fluorescent protein PAmCherry in live-cell movies with low background (white) and with a high background (red) as in (a) and (b) are shown. Scale bars, 1 μm. To see this figure in color, go online.
Videos are acquired under continuous 561-nm laser excitation at a rate of 40 fps. Scale bar = 1 μm. This video corresponds to Fig. 3 a in the main text. The raw, uncompressed data corresponding to Videos S1 and S2 are provided in uncompressed TIFF format at the University of Michigan’s permanent data depository, Deep Blue. https://doi.org/10.7302/Z2CR5RKD.
A constant 15 W/cm2, 488-nm laser illumination generated a strong autofluorescent background in the cells. Videos are acquired under continuous 561-nm laser excitation at a rate of 40 fps. Scale bar = 1 μm. This video corresponds to Fig. 3 b in the main text. The raw, uncompressed data corresponding to Videos S1 and S2 are provided in uncompressed TIFF format at the University of Michigan’s permanent data depository, Deep Blue. https://doi.org/10.7302/Z2CR5RKD.
We removed the subtle background from the low-background movies with SMALL-LABS and then analyzed the subcellular single molecules with super-resolution PSF fitting. The high-background movies were analyzed with the same algorithm. Whereas the background in Fig. 3 b background is sufficiently high to make single-molecule localization essentially impossible in the raw data, after SMALL-LABS background removal, PolC-PAmCherry could be detected in these cells. Both single-molecule localization data sets were then analyzed with the same single-molecule tracking algorithm: trajectories were determined (Fig. 3 c) by optimizing all possible pairings of molecules between consecutive frames using the Hungarian algorithm (30, 31, 32). Measured diffusion coefficients for PolC-PAmCherry in the high-background cells matched our previously reported low-background measurements (29) (Fig. S11; Supporting Materials). Single-molecule intensities, measured by summing the pixel intensities around each measured molecule, yielded nearly identical distributions in the high- and low-background movies (Fig. 3 d). Both of these results show that SMALL-LABS successfully removed the background from this live-cell imaging experiment without introducing any biases to enable accurate measurements of fluorescence intensity and position in a data set that would have been impossible to analyze without background removal.
Conclusions
The SMALL-LABS algorithm, with which an arbitrary background is removed from single-molecule imaging data by separating the foreground and the background via local “on”- and “off”-frame categorization, addresses a gap in many super-resolution imaging packages and enables the detection and localization of single molecules even in the presence of obscuring backgrounds. We have benchmarked the speed of our implementation of SMALL-LABS under a variety of computer systems and tools (Supporting Materials; Table S6). In addition to improving single-molecule localization, SMALL-LABS avoids systematic biases caused by inaccurate background subtraction in the measurement of single-molecule intensities. This brightness measurement is a key metric in single-molecule fluorescence resonance energy transfer experiments, in single-molecule counting experiments, and as a single-molecule probe of the local environment. The SMALL-LABS data analysis approach requires no changes to experimental methods; in fact, it relaxes experimental constraints: with its improved accuracy and sensitivity, SMALL-LABS opens up many systems previously inaccessible to super-resolution analysis due to difficult backgrounds. Here, we have demonstrated the scope and performance of SMALL-LABS by accurately and precisely measuring simulated data under a variety of realistic background conditions and by successfully measuring and tracking single fluorescent proteins in a live-cell experiment under conditions that preclude traditional approaches.
Code availability is provided as follows. The open-source MATLAB code for implementing SMALL-LABS (GNU general public license), full documentation, and a quick-start guide with example data is provided (Data S1). Further development and expansion of the code postpublication will be hosted at https://github.com/BiteenMatlab/SMALL-LABS (https://doi.org/10.5281/zenodo.1438446).
Author Contributions
B.P.I. and J.S.B. designed the research. B.P.I. implemented the SMALL-LABS algorithm, performed the simulated experiments, and analyzed the data. Y.L. performed live-cell imaging and analyzed the data. B.P.I. and S.A.L. wrote the code. All authors discussed the results. The manuscript was written and edited by all authors. All authors read and approved the manuscript.
Acknowledgments
David Rowland provided useful coding guidance. Tiancheng (Curly) Zuo provided helpful feedback during SMALL-LABS development.
This work was supported by a National Science Foundation award (grant CHE-1807676) to J.S.B. B.P.I. was also supported by the National Science Foundation Graduate Research Fellowship Program (grant DGE-1256260).
Editor: Catherine Galbraith.
Footnotes
Supporting Materials, eleven figures, six tables, two videos, and one data file are available at http://www.biophysj.org/biophysj/supplemental/S0006-3495(19)30133-X.
Supporting Material
Code, guides, and sample data for implementing the SMALL-LABS algorithm.
References
- 1.Huang B., Bates M., Zhuang X. Super-resolution fluorescence microscopy. Annu. Rev. Biochem. 2009;78:993–1016. doi: 10.1146/annurev.biochem.77.061906.092014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thompson M.A., Lew M.D., Moerner W.E. Extending microscopic resolution with single-molecule imaging and active control. Annu. Rev. Biophys. 2012;41:321–342. doi: 10.1146/annurev-biophys-050511-102250. [DOI] [PubMed] [Google Scholar]
- 3.Joo C., Balci H., Ha T. Advances in single-molecule fluorescence methods for molecular biology. Annu. Rev. Biochem. 2008;77:51–76. doi: 10.1146/annurev.biochem.77.070606.101543. [DOI] [PubMed] [Google Scholar]
- 4.Yu J. Single-molecule studies in live cells. Annu. Rev. Phys. Chem. 2016;67:565–585. doi: 10.1146/annurev-physchem-040215-112451. [DOI] [PubMed] [Google Scholar]
- 5.Tuson H.H., Biteen J.S. Unveiling the inner workings of live bacteria using super-resolution microscopy. Anal. Chem. 2015;87:42–63. doi: 10.1021/ac5041346. [DOI] [PubMed] [Google Scholar]
- 6.Rust M.J., Bates M., Zhuang X. Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM) Nat. Methods. 2006;3:793–795. doi: 10.1038/nmeth929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Betzig E., Patterson G.H., Hess H.F. Imaging intracellular fluorescent proteins at nanometer resolution. Science. 2006;313:1642–1645. doi: 10.1126/science.1127344. [DOI] [PubMed] [Google Scholar]
- 8.Hess S.T., Girirajan T.P., Mason M.D. Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 2006;91:4258–4272. doi: 10.1529/biophysj.106.091116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sharonov A., Hochstrasser R.M. Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proc. Natl. Acad. Sci. USA. 2006;103:18911–18916. doi: 10.1073/pnas.0609643104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Small A., Stahlheber S. Fluorophore localization algorithms for super-resolution microscopy. Nat. Methods. 2014;11:267–279. doi: 10.1038/nmeth.2844. [DOI] [PubMed] [Google Scholar]
- 11.Sage D., Kirshner H., Unser M. Quantitative evaluation of software packages for single-molecule localization microscopy. Nat. Methods. 2015;12:717–724. doi: 10.1038/nmeth.3442. [DOI] [PubMed] [Google Scholar]
- 12.Cheezum M.K., Walker W.F., Guilford W.H. Quantitative comparison of algorithms for tracking single fluorescent particles. Biophys. J. 2001;81:2378–2388. doi: 10.1016/S0006-3495(01)75884-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee A., Tsekouras K., Pressé S. Unraveling the thousand word picture: an introduction to super-resolution data analysis. Chem. Rev. 2017;117:7276–7330. doi: 10.1021/acs.chemrev.6b00729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schrimpf W., Barth A., Lamb D.C. PAM: a framework for integrated analysis of imaging, single-molecule, and ensemble fluorescence data. Biophys. J. 2018;114:1518–1528. doi: 10.1016/j.bpj.2018.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen B.C., Legant W.R., Betzig E. Lattice light-sheet microscopy: imaging molecules to embryos at high spatiotemporal resolution. Science. 2014;346:1257998. doi: 10.1126/science.1257998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Greiss F., Deligiannaki M., Braun D. Single-molecule imaging in living Drosophila embryos with reflected light-sheet microscopy. Biophys. J. 2016;110:939–946. doi: 10.1016/j.bpj.2015.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tokunaga M., Imamoto N., Sakata-Sogawa K. Highly inclined thin illumination enables clear single-molecule imaging in cells. Nat. Methods. 2008;5:159–161. doi: 10.1038/nmeth1171. [DOI] [PubMed] [Google Scholar]
- 18.Axelrod D. Total internal reflection fluorescence microscopy. Methods Cell Biol. 1989;30:245–270. doi: 10.1016/s0091-679x(08)60982-6. [DOI] [PubMed] [Google Scholar]
- 19.Ha T., Tinnefeld P. Photophysics of fluorescent probes for single-molecule biophysics and super-resolution imaging. Annu. Rev. Phys. Chem. 2012;63:595–617. doi: 10.1146/annurev-physchem-032210-103340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wertz E., Isaacoff B.P., Biteen J.S. Single-molecule super-resolution microscopy reveals how light couples to a plasmonic nanoantenna on the nanometer scale. Nano Lett. 2015;15:2662–2670. doi: 10.1021/acs.nanolett.5b00319. [DOI] [PubMed] [Google Scholar]
- 21.Willets K.A., Van Duyne R.P. Localized surface plasmon resonance spectroscopy and sensing. Annu. Rev. Phys. Chem. 2007;58:267–297. doi: 10.1146/annurev.physchem.58.032806.104607. [DOI] [PubMed] [Google Scholar]
- 22.Titus E.J., Willets K.A. Accuracy of superlocalization imaging using Gaussian and dipole emission point-spread functions for modeling gold nanorod luminescence. ACS Nano. 2013;7:6258–6267. doi: 10.1021/nn4022845. [DOI] [PubMed] [Google Scholar]
- 23.Hoogendoorn E., Crosby K.C., Postma M. The fidelity of stochastic single-molecule super-resolution reconstructions critically depends upon robust background estimation. Sci. Rep. 2014;4:3854. doi: 10.1038/srep03854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Donehue J.E., Wertz E., Biteen J.S. Plasmon-Enhanced brightness and photostability from single fluorescent proteins coupled to gold nanorods. J. Phys. Chem. C. 2014;118:15027–15035. [Google Scholar]
- 25.Thompson R.E., Larson D.R., Webb W.W. Precise nanometer localization analysis for individual fluorescent probes. Biophys. J. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wertz E.A., Isaacoff B.P., Biteen J.S. Wavelength-dependent super-resolution images of dye molecules coupled to plasmonic nanotriangles. ACS Photonics. 2016;3:1733–1740. [Google Scholar]
- 27.Carattino A., Caldarola M., Orrit M. Gold nanoparticles as absolute nanothermometers. Nano Lett. 2018;18:874–880. doi: 10.1021/acs.nanolett.7b04145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ovesný M., Křížek P., Hagen G.M. ThunderSTORM: a comprehensive ImageJ plug-in for PALM and STORM data analysis and super-resolution imaging. Bioinformatics. 2014;30:2389–2390. doi: 10.1093/bioinformatics/btu202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao Y., Li Y., Biteen J.S. Single-molecule DNA polymerase dynamics at a bacterial replisome in live cells. Biophys. J. 2016;111:2562–2569. doi: 10.1016/j.bpj.2016.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kuhn H.W. The Hungarian method for the assignment problem. In: Hu M., editor. Naval Research Logistics. John Wiley & Sons; 1955. pp. 83–97. [Google Scholar]
- 31.Liao Y., Schroeder J.W., Biteen J.S. Single-molecule motions and interactions in live cells reveal target search dynamics in mismatch repair. Proc. Natl. Acad. Sci. USA. 2015;112:E6898–E6906. doi: 10.1073/pnas.1507386112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rowland D.J., Biteen J.S. Measuring molecular motions inside single cells with improved analysis of single-particle trajectories. Chem. Phys. Lett. 2017;674:173–178. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Videos are acquired under continuous 561-nm laser excitation at a rate of 40 fps. Scale bar = 1 μm. This video corresponds to Fig. 3 a in the main text. The raw, uncompressed data corresponding to Videos S1 and S2 are provided in uncompressed TIFF format at the University of Michigan’s permanent data depository, Deep Blue. https://doi.org/10.7302/Z2CR5RKD.
A constant 15 W/cm2, 488-nm laser illumination generated a strong autofluorescent background in the cells. Videos are acquired under continuous 561-nm laser excitation at a rate of 40 fps. Scale bar = 1 μm. This video corresponds to Fig. 3 b in the main text. The raw, uncompressed data corresponding to Videos S1 and S2 are provided in uncompressed TIFF format at the University of Michigan’s permanent data depository, Deep Blue. https://doi.org/10.7302/Z2CR5RKD.
Code, guides, and sample data for implementing the SMALL-LABS algorithm.