Abstract
Long non-coding RNAs (lncRNAs) are transcripts longer than 200 nucleotides (nts) without obvious protein coding potential. lncRNAs act as multiple roles in biological processes of diseases, especially carcinomas. Prostate cancer associated transcript-1 (PCAT-1) is an oncogenic lncRNA that identified by RNA-Sequence in prostate cancer. High expression of PCAT-1 is observed in different types of cancers, including prostate cancer, colorectal cancer, hepatocellular cancer and gastric cancer. High expressed PCAT-1 is correlated with poor overall survival. Furthermore, PCAT-1 regulates cancer cell proliferation, apoptosis, migration and invasion. Additionally, PCAT-1 is involved in EMT and Wnt/β-catenin-signaling pathway. In this review, we focus on the implication of PCAT-1 in human cancers.
Keywords: long non-coding RNA, PCAT-1, cancer, biomarker, therapeutic target
Introduction
Cancer is a group of disease involving uncontrolled cell growth, and its high mortality and disability cause great personal suffering among patients and a huge burden to society 1. Surgical treatment, chemotherapy and radiotherapy are currently the main treatments of cancer 2, 3. However, the therapeutic effects of these primary treatments are still limited 4, 5. Therefore, early diagnostic markers and novel therapies are urgently required to cure cancer.
LncRNAs are defined as transcripts longer than 200 nucleotides (nts) without obvious protein coding potential 6, 7. The majority of lncRNAs are transcribed by RNA polymerase II and are frequently spliced and polyadenylated 8-10. Compared with protein-coding sequence, most lncRNAs tend to be weakly evolutionary constraint 11. The promoter sequence of lncRNA is strong conserved, while its transcript sequence is less conserved, indicating that lncRNA transcription itself has a greater biological consequence than the transcript sequence 11. LncRNA are involved in transcriptional regulation, epigenetic gene regulation, cellular addresses and disease, especially the growth and metastasis of cancers 11-14.
Various mutations of lncRNAs are implicated in multiple cancer types 15, 16, and lncRNAs are further considered as oncogenes or tumor suppressor genes in cancers 17, 18. For example, lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) is overexpressed in various cancers, associated with metastasis and poor prognosis, and acts as an oncogene in breast cancer by controlling the selective splicing process 19-21. Moreover, MALAT1 is an interesting tumor marker and a potential target gene for anti-metastatic therapy 20, 22-25. In contrast, lncRNA NF-KappaB Interacting LncRNA (NKILA) is a tumor suppressor and acts as a NF-κB regulator to inhibit breast cancer metastasis 26-28. Taken together, lncRNAs play important roles in cancer development, which may become diagnostic biomarkers and therapeutic targets.
Prostate cancer associated transcript-1 (PCAT-1) was originally identified in prostate cancer by large-scale RNA-Seq analyses, and functionally implicated in prostate cancer progression. The high expression of PCAT-1 has been found in several human tumor entities, and is associated with poor prognosis, including TNM stage. In addition, PCAT-1 has been implicated in regulating cancer cell proliferation, apoptosis, migration and invasion. PCAT-1 has also been shown to be involved in epithelial-mesenchymal transitions (EMT) and regulate cancer progression via the Wnt/β-catenin signaling pathway. In this review, we will focus on the implication of PCAT-1 in human cancers.
Discovery of PCAT-1
In 2011, Prensner et al. discovered 121 unannotated prostate cancer-associated ncRNA transcripts (PCATs) by high-throughput RNA-Seq from a cohort of 102 prostate tissues and cells lines. The 121 unannotated transcripts were ranked and named as PCATs according to their fold-change in tumor versus normal tissue 29, 30. PCAT-1 is significantly overexpressed in most prostate cancer tissues, particularly metastases.
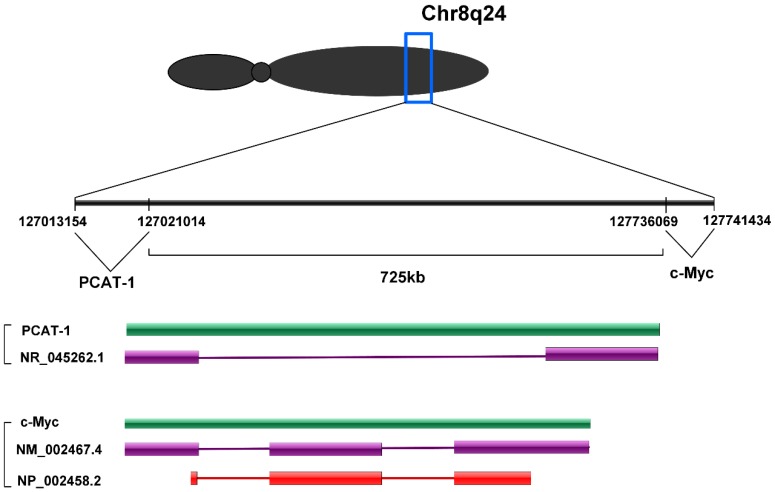
PCAT-1 is a long non-coding RNA with a length of ~1900 nt. PCAT-1 gene consists of 2 exons. Exon 1 contains a retroviral long terminal repeat (LTR) sequence and exon 2 contains sequences from the HSMAR1 mariner family transposase bisected by an AluY repeat element 30. PCAT-1 gene located at chromosome 8q24 and 725 kb upstream of the c-Myc oncogene (Fig. 1). Chr8q24 locus is an area commonly studied in prostate cancer due to frequent amplification and the presence of prostate cancer susceptibility SNP loci 31, 32. However, PCAT-1 outlier expression was not dependent on Chr8q24 amplification, as PCAT-1 highly expressing tumors often did not have 8q24 amplification and high copy number gain of 8q24 was not sufficient to upregulate PCAT-1 30.
Figure 1.
PCAT-1 is located at chromosome 8q24 and 725 kb upstream of the c-Myc oncogene.
PCAT-1 promotes cell proliferation and is upregulated in major types of cancers, including esophageal squamous cell carcinoma (ESCC) 33, bladder cancer (BC) 34, hepatocellular cancer (HCC) 35, non-small cell lung cancer (NSCL) 36, glioblastoma, multiple myeloma (MM) 37, breast cancer, colorectal cancer (CRC) 38 and gastric cancer (GC) 39. In addition, overexpression of PCAT-1 was closely associated with clinicopathological characteristics, including tumor size, lymphatic metastasis, distant metastasis, tumor-node-metastasis (TNM) stage and poor prognosis (Table 1). PCAT-1 functions predominantly as a transcriptional repressor. PCAT-1 negatively regulates the BRCA2 tumor suppressor protein and positively regulates Myc oncoprotein 40. It contains binding sites for microRNAs, and may act as a sponge for microRNAs that regulate cell growth pathways 30. Thus, current evidence suggested that PCAT-1 is an oncogenic gene and played an important role in the regulation of cancer development.
Table 1.
Functional characterization of PCAT-1 in various cancers.
| Cancer types | Expression | Effects | Related gene | Role | References |
|---|---|---|---|---|---|
| Prostate cancer | Upregulated | Proliferation, metastasis, DSB repair process | PRC2, cMyc protein, BRCA2, SUZ12, miR-3667-3p, miR-34a | Oncogene | 30, 40, 47 |
| Colorectal cancer | Upregulated | Proliferation, Cell arrest, apoptosis | Caspase-3, PARP, bax, bcl-2, cMyc protein | Oncogene | 38 |
| Hepatocellular cancer | Upregulated | Invasion, metastasis, apoptosis, TNM stage | HMGB1, mir-129-5p | Oncogene | 59, 60, 61 |
| Gastric cancer | Upregulated | Proliferation, migration, invasion, metastasis | CDKN1A | Oncogene | 39, 67 |
| Bladder cancer | Upregulated | Proliferation, apoptosis | - | Oncogene | 34 |
| Breast cancer | Upregulated | - | - | - | 91 |
| Esophageal squamous cell carcinoma | Upregulated | - | - | - | 33 |
| Non-small cell lung cancer | Upregulated | Proliferation, migration, invasion | - | Oncogene | 36 |
| Multiple myeloma (serum) | Upregulated | - | - | 37 | |
| Osteosarcoma | Upregulated | Apoptosis, metastasis | E-cadherin, N-cadherin, EZH2, p21 | Oncogene | 72, 73, 74, 75, 83 |
| Extrahepatic cholangiocarcinoma | Upregulated | Proliferation, apoptosis, metastasis | miR-122, WNT1 | Oncogene | 90 |
TNM: TNM Classification of Malignant Tumours; PRC2: Polycomb Repressive Complex 2; cMyc: MYC proto- oncogene, bHLH transcription factor; BRCA2: breast cancer 2, early onset; PARP: poly- ADP- ribose polymerase; Bax: BCL2- associated X protein; Bcl- 2: B- cell CLL/lymphoma 2 protein; HMGB1: high mobility group box 1; CDKN1A: cyclin-dependent kinase inhibitor 1A
Expression distribution and transcription regulation of PCAT-1
PCAT-1 is expressed in a wide range of different tissues and is most highly expressed in the prostate, salivary gland 41. In prostate cancer and osteosarcoma cells, PCAT-1 transcript is predominantly located in the cytoplasm and a smaller fraction is expressed in the nucleus 40. PCAT-1 might play a role in both transcriptional and post- transcriptional regulation in cancer development.
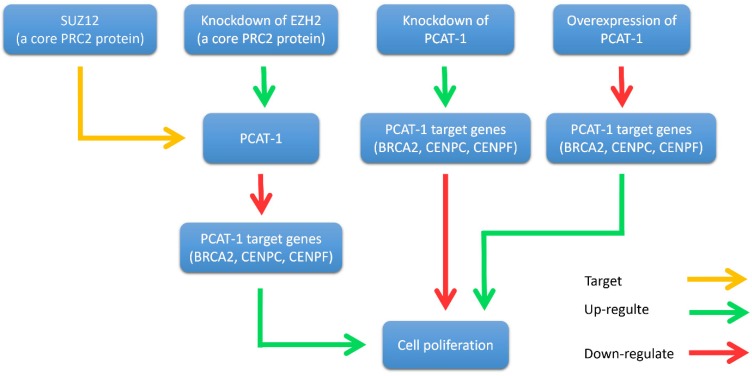
As for the mechanism of PCAT-1 expression profiles in prostate cancer, Prensner et al. found that SUZ12, a core polycomb repressive complex 2 (PRC2) protein, directly binds to the PCAT-1 promoter ~1 kb upstream of the transcription start site 30. EZH2 knockdown by short hairpin (sh)RNA or pharmacologic inhibition caused a dramatic upregulation in PCAT-1 expression levels (Fig. 3). Otherwise, not much is known about the cellular factors and pathways that lead to the strong transcription of MALAT1, especially in human cancer cells.
Figure 3.
Interplay between PCAT-1 and the polycomb inhibitory complex 2 (PRC2). PCAT-1 overexpression promotes cell proliferation, while knockdown of PCAT-1 reduces cell proliferation. EZH2 (a core PRC2 protein) knockdown up-regulates PCAT-1 expression and down-regulates PCAT-1 target genes (BRCA2, CENPC, CENPF). Similarly, PCAT-1 knockdown upregulates the target genes of PCAT-1. Another core PRC2 protein, SUZ12, binds directly to the promoter of PCAT-1.
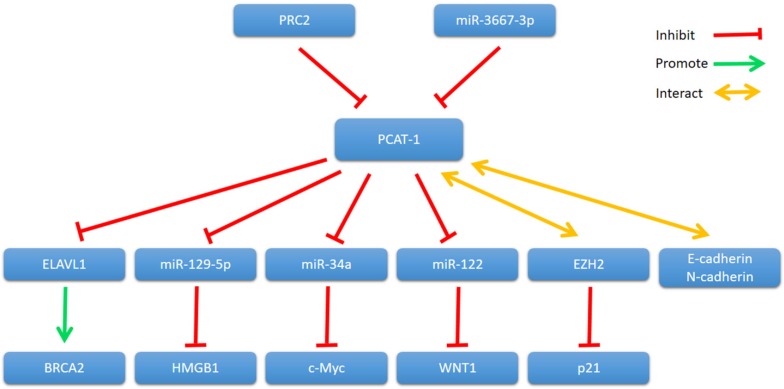
In 2011, Gene expression profiling of PCAT-1 knockdown in LNCaP cells showed that PCAT-1 modulates the transcriptional regulation of 370 genes (255 upregulated, 115 downregulated; FDR ≤ 0.01) 30. Since then, many studies have shown that PCAT-1 regulates transcription of various genes involved in cancer cell proliferation, apoptosis, migration and invasion, including BRCA2 tumor suppressor, Myc oncoprotein and so on (Fig. 2).
Figure 2.
PCAT-1 and its mechanisms of action.
PCAT-1 in human cancers
Prostate cancer
Prostate cancer is the second most common malignancy in men and the second leading cause of cancer death in the United States and Europe. It is known for a disease of older man (aged >65years), and over 10% of new cases has been diagnosed in young men (aged ≤55 years) 42. Surgery and radiotherapy are the traditional treatments to prostate cancer, but the effects of these treatments are still limited 43. Therefore, biomarkers for early diagnosis and novel targets for therapy are urgently required.
John et al. found that PCAT-1 was significantly overexpressed in most prostate cancer tissues, particularly metastases. It is well known that oncoprotein EZH2 (a core protein of PRC2) is up-regulated in tumors and contributes to a metastatic phenotype 44-46. They further investigated the relationship between PCAT-1 and EZH2 30. They discovered that the expression of PCAT-1 and EZH2 was almost mutually exclusive. Knockdown of EZH2 by shRNA or pharmacologic inhibition of EZH2 with the inhibitor 3-deazaneplanocin A (DZNep) up-regulated PCAT-1 expression levels. In addition, the expression level of three key PCAT-1 target genes (BRCA2, CENPE and CENPF) was up-regulated when PCAT-1 was knockdown in prostate cancer cells. Furthermore, another PRC2 protein SUZ12 was shown to bind directly to the promoter of PCAT-1 (Fig. 3). Further researches are required to elucidate the precise mechanism of the interplay between PCAT-1 and PRC2.
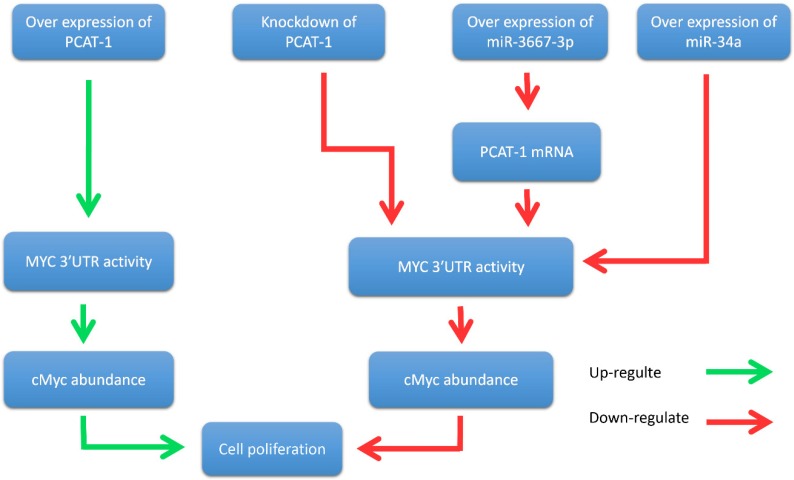
John et al. further reported that PCAT-1 posttranscriptional repressed the expression of the BRCA2 tumor suppressor, leading to downstream impairment of homologous recombination (HR) 40. In addition, Prensner et al. revealed that PCAT-1 promoted prostate cell proliferation by up-regulating the protein level of cMyc 47. It is well known that cMyc is important transcription factor that is essential for cell cycle progression 48-51. The oncogenic up-regulation of cMyc has a broad impact on cancer cell biology 52-54. Evidence showed that PCAT-1 increased MYC 3'UTR activity through interfering the regulation of MYC by microRNAs (miR-3667-3p and miR-34a), thereby promoting the proliferation of prostate tumor cells 47 (Fig. 4). In prostate cancer cells, PCAT-1 is predominantly expressed in the cytoplasm and is also expressed in a small amount in the nucleus 40. This ubiquitous subcellular localization of PCAT-1 transcripts may contribute to the function of PCAT-1 both in nucleus and cytosol. To sum up, these results suggested that PCAT-1 may function as a potential prognostic biomarker and therapeutic target in prostate cancer.
Figure 4.
PCAT-1 promotes prostate cancer cell proliferation through cMyc. Overexpression of PCAT-1 up-regulates MYC 3'UTR activity, increases cMyc abundance, and promotes prostate cancer cell proliferation. Conversely, PCAT-1 knockdown down-regulates this process and inhibites cancer cell proliferation. Overexpression of miRNA-3667-3p increases the expression level of PCAT-1 and suppresses MYC 3'UTR activity. Overexpression of miR-34a also inhibites MYC 3'UTR activity. Down-regulation of MYC 3'UTR activity reduces cMyc protein abundance and suppresses cancer cell proliferation.
Hepatocellular cancer
Hepatocellular carcinoma (HCC) is the fifth most common cancer all around the world 55, 56. Even after clinical treatment (including surgery, radiation therapy, and chemotherapy), recurrence and distant metastasis of HCC are still at high levels 57, 58. In addition, current evidence suggests that the development of HCC is associated with genetic or epigenetic mutation 59. Thus, it is urgent to seek for novel tumour biomarkers and further study the special mechanisms of HCC development.
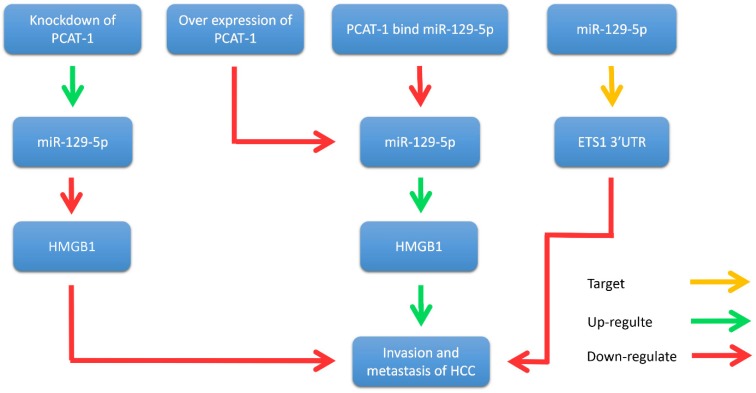
Yan et al. illustrated that PCAT-1 was not only significantly up-regulated in HCC tissues and cell lines, but also related to TNM staging, metastasis and histological grade 35. Wen et al. reported that PCAT-1 down-regulation significantly reduced the invasion and migration of HCC cell lines 60. Zhang et al. found that PCAT-1 inhibits the repressive effect of miR-129-5p and reverses the expression of high mobility group box 1 (HMGB1), a target gene of miR-129-5p 61. Importantly, HMGB1 promotes metastasis and invasion in HCC (Fig. 5). In addition, evidence revealed that PCAT-1 was an independent prognostic factor of HCC in clinic research. Overexpression of PCAT-1 in HCC was associated with more malignant phenotypes and worse prognosis. These studies suggest that PCAT-1 is a potential diagnostic and therapeutic target in HCC.
Figure 5.
PCAT-1 promotes metastasis and invasion in HCC. Overexpression of PCAT-1 down-regulates miR-129-5p, thereby increasing the expression of HMGB1 and promoting the migration and invasion of hepatoma cells. Knockdown of PCAT-1 up-regulates miR-129-5p and reduces the expression of HMGB1 protein, thereby inhibiting the migration and invasion of hepatoma cells. In addition, miR-129-5p inhibits the migration and invasion of hepatocellular carcinoma cells by directly targeting ETS1 3'UTR.
Gastric cancer
As an aggressive disease, gastric cancer (GC) is the third leading cause of cancer-related death worldwide 62, 63. Surgery and chemotherapy are the main treatments for GC, but the 5-year survival rate is still low 64-66. Therefore, it is necessary to find novel biomarkers for diagnosing GC.
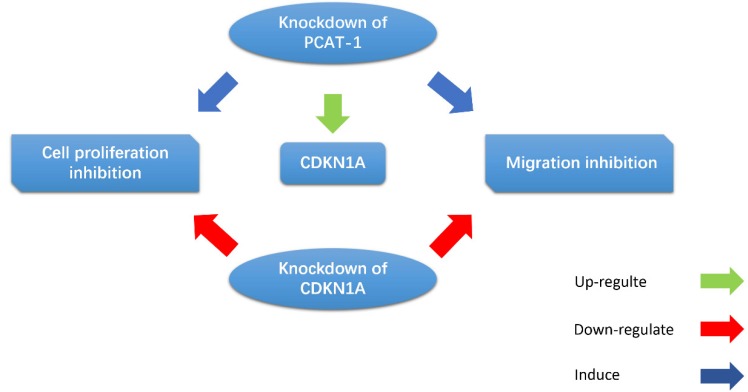
Cui et al. observed that the PCAT-1 was high-expressed in GC tissues and cell lines, and overexpression of PCAT-1 was associated with poor overall survival 39. CDKN1A acts as a regulator of cell cycle progression, and exerts tumor suppressive effects in several cancers. Bi et al. indicated that PCAT-1 promoted cell proliferation, migration and invasion in GC cells by regulating CDKN1A (Fig. 6). They found that inhibition of PCAT-1 in GC cells induced G1 arrests and apoptosis, and reduced cell migration and invasion. CDKN1A was up-regulated in PCAT-1 knockdown GC cells, and CDKN1A knockdown can rescue PCAT-1 knockdown-induced cell proliferation and migration inhibition 67. These results indicate that PCAT-1 plays an oncogenic role in the progression of GC.
Figure 6.
PCAT-1 promotes cell proliferation and migration in GC cells by regulating CDKN1A. CDKN1A was up-regulated in PCAT-1 knockdown GC cells, and CDKN1A knockdown can rescue PCAT-1 knockdown-induced cell proliferation and migration inhibition.
Cui et al. indicated that the expression level of PCAT-1 was closely correlated with the depth of invasion, TNM stage and lymph node metastasis of GC. Moreover, Kaplan-Meier analysis showed that GC patients with high PCAT-1 expression were significantly associated with lower overall survival (OS), suggesting that high expression of PCAT-1 is an independent prognostic factor for OS 39. All these results indicate that the overexpression of PCAT-1 may be used as a biomarker for GC prognosis, which has important clinical significance.
Osteosarcoma
Osteosarcoma (OS) is a malignant primary bone cancer that is diagnosed in approximately 30 new cases per million people worldwide each year 68-70. The 5-year survival rate of the patients with OS remains low 71. The major treatments for OS are surgery, chemotherapy and palliative radiotherapy, but these treatments have limited efficacy 72. Moreover, our understanding of the molecular mechanisms that underlies the development of OS remains limited. Therefore, we need to explore novel diagnostic and therapeutic targets for OS.
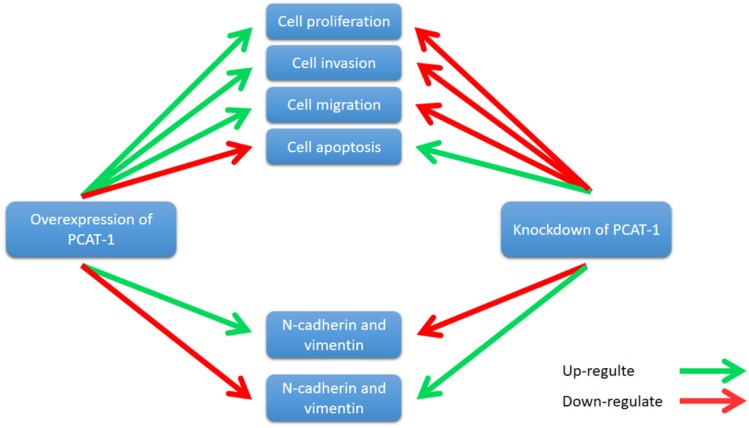
Zhang et al. reported that PCAT-1 was up-regulated in OS tissues and was correlated with several clinical features of patients with OS, including advanced clinical stage and tumor metastasis 73. High expression level of PCAT-1 predicted poor prognosis, which may be an important prognostic factor for the clinical outcomes in patients with OS 73. Moreover, knockdown of PCAT-1 promoted cell apoptosis. Cell cycle analysis showed that overexpression of PCAT-1 reduced the cell population at G0/G1 phase and increased the cell population at S phase, while knockdown of PCAT1 exerted the opposite effects. Additionally, PCAT-1 is associated with EMT by influencing the expression of EMT-related genes, including E-cadherin, N-cadherin 73. EMT is considered as an important mechanism of tumor metastasis, which promotes tumor migration, invasion and metastasis 74, 75. EMT was characterized by loss of cell polarity in epithelial cells and developing characteristics of interstitial cells, including 3 types: primary/type 1, secondary/type 2 and tertiary/type 3. Type 1 is related to embryogenesis, implantation and organogenesis; Type 2 is associated with wound healing, tissue regeneration and organ development; Type 3 regulates tumor metastasis 76-78. Importantly, the expression of some EMT-related genes changes during cancer progression and metastasis, such as mesenchymal genes (fibronectin, N-cadherin and vimentin) and epithelial genes (E-cadherin and ZO-1) 79-82. Zhang et al. observed that overexpression of PCAT-1 up-regulated the protein levels of N-cadherin and vimentin, and down-regulated the protein level of E-cadherin 73. Knockdown of PCAT-1 down-regulated the protein levels of N-cadherin and vimentin, and up-regulated the protein level of E-cadherin 73. These results showed that PCAT1 was correlated with EMT in OS (Fig. 7).
Figure 7.
PCAT-1 promotes cell proliferation, invasion, migration and EMT in osteosarcoma. Overexpression of PCAT-1 up-regulates N-cadherin and vimentin and down-regulates E-cadherin (N-cadherin and vimentin, E-cadherin are EMT-related markers). By contrast, knockdown of PCAT1 in OS cell lines exerted the opposite effect.
In addition, Huang et al found that PCAT-1 was expressed in both cytoplasm and nucleus in OS cells, although the expression level of PCAT-1 in cytoplasm was lower than that of in nucleus 83. Furthermore, they confirmed that PCAT-1 directly binds to EZH2 in OS cells by RIP assays and RNA pulldown assays. The ChIP assays revealed that EZH2 could bind to the p21 83. PCAT-1 knockdown reduced EZH2 binding to the promoter region of p21, and increased p21 expression in OS cells 83. These results suggest that PCAT-1 inhibits p21 expression through directly binding with EZH2 and the promoter region of p21 in OS cells. Moreover, cellular functional assays revealed that the functions inhibited by PCAT-1 knockdown was partly abolished by transfected si-p21 83. These results indicate that the oncogenic function of PCAT-1 is partly dependent on p21 in OS cells. To sum up, PCAT-1 has potential value as a prognostic biomarker and therapeutic target in OS.
Extrahepatic cholangiocarcinoma
Cholangiocarcinoma is a fatal cancer that can be divided into two types: intrahepatic and extrahepatic cholangiocarcinoma (ECC) 84. ECC patients often receive conventional chemotherapy or radiotherapy, but the 5-year survival rate is only 10% 85. However, the precise mechanism of ECC is still nuclear.
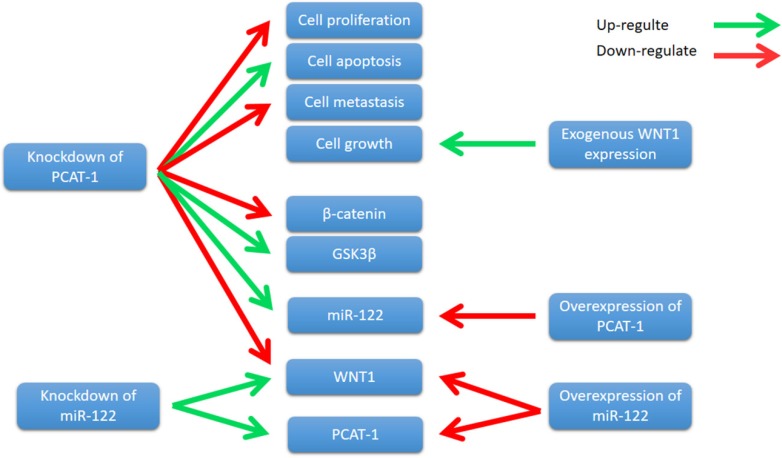
WNT1 is a member of the WNT family, which stimulates the Wnt/β- catenin-signaling pathway 86. It is well known that Wnt/β-catenin signaling has been closely associated with the initiation and progression of many cancer types 87. Wnt/β-catenin pathway mutations are frequently observed in different cancer types, such as adenomatous polyposis coli (APC) mutations in colorectal cancers and β-catenin gene mutations in colorectal cancers 88. Importantly, abnormal Wnt/β-catenin signaling directly alters a number of regulators critical for the antitumor activities of T cells, thereby promoting cancer progression 89. Zhang et al. showed that PCAT-1 was up-regulated in ECC tissue samples and cell lines. Knockdown of PCAT-1 suppressed ECC cell growth, induced apoptosis, and inhibited cell metastasis90. Moreover, PCAT-1 was associated with miR- 122, and knockdown of PCAT-1 leads to an increase in miR-122 levels, whereas PCAT-1 overexpression down-regulated miR-122 expression 90. Further study revealed that PCAT1 regulated WNT1 expression via miR-122 90. Overexpression of miR-122 significantly down-regulated the WNT1 expression, and knockdown of miR-122 up-regulated WNT1 expression90. Zhang et al. also showed that knockdown of PCAT-1 significantly reduced β-catenin levels and increased glycogen synthase kinase 3β (GSK3β), and the expression of WNT1 increased colony formation and inhibited cell apoptosis 90. These results indicate that PCAT-1 knockdown inhibits ECC cells growth by reducing Wnt/β-catenin signaling pathway activity through miR-122 (Fig. 8). In conclusion, PCAT-1 may function as a potential prognostic biomarker and therapeutic target in ECC.
Figure 8.
Knockdown of PCAT-1 inhibited cell proliferation, metastasis, and promoted cell apoptosis in ECC cell lines. PCAT-1 knockdown inhibits ECC cells growth by reducing Wnt/β-catenin signaling pathway activity through miR-122.
Other cancers
The overexpression of PCAT-1 was also observed in other major cancer tissues and cell lines. For instance, Qiao et al. revealed that PCAT-1 acted as an oncogene in colorectal cancer (CRC), and down-regulation of PCAT-1 significantly reduced the proliferation of CRC cells 38. However, our understanding of the roles of PCAT-1 in CRC is still at a preliminary stage, further experiments are required.
PCAT-1 is up-regulated in bladder cancer (BC) tissues, and PCAT-1 promotes BC cells growth and inhibits apoptosis 34. In breast cancer, PCAT-1 has been reported to be up-regulated in cancer samples compared with adjacent normal tissues 91. In esophageal squamous cell carcinoma (ESCC), the expression of PCAT-1 in tumor tissues is significantly higher than normal tissues. The high expression of PCAT-1 was closely associated with tumor invasion, clinical stage and lymph node metastasis. High expression of PCAT-1 indicates poor prognosis of ESCC 33. In non-small cell lung cancer (NSCL), PCAT-1 is considered as an oncogenic role because its expression level is up-regulated in cancer tissues and cell lines. More experiments have shown that knockdown of PCAT-1 suppressed cell proliferation, migration and invasion in NSCL, whereas up- regulation of PCAT-1 exerted the opposite effect 36. The relative expression level of PCAT-1 in serum of patients with multiple myeloma (MM) was higher than that of healthy controls, indicating that PCAT-1 can be used as an important biomarker for MM diagnosis 37.
Conclusion and future perspectives
Aberrant of PCAT-1 expression was observed in major cancer tissues and cell lines. Overexpression of PCAT-1 was closely associated with tumor size, lymphatic metastasis, distant metastasis, tumor-node- metastasis (TNM) stage and poor prognosis. PCAT-1 plays an important role in the regulation of cancer development by regulating cell proliferation, metastasis, apoptosis, cell cycle, DNA repair and homologous recombination 30, 47, 61. These results reveal that PCAT-1 acts as an oncogenic gene and plays an important role in the regulation of cancer development.
Current studies indicates that PCAT-1 is involved in EMT and Wnt/β-catenin signaling pathway 73, 90. Potential cancer therapies targeting EMT and Wnt/β-catenin signaling pathways have been investigated 86, 92. For instance, CTCs is a preliminary stage to metastatic development, and their EMT-related characteristics are emerging as novel therapeutic targets in cases of cancer chemoresistance over typical neoadjuvant therapies 92. Since Wnt/β-catenin- signaling pathway is critical for cancer, therapies based on Wnt signaling may be a novel treatment 89. Zhang et al. showed that in OS cell lines (MG-63 and U2OS), PCAT1 overexpression increased the protein levels of N-cadherin and vimentin (EMT markers), and decreased the protein level of E-cadherin, while downregulation of PCAT1 decreased the protein levels of N-cadherin and vimentin, and increased the protein level of E-cadherin 73. In the other paper, Zhang et al. reported that silencing of PCAT-1 inhibited the progression of ECC by reducing Wnt/β-catenin signaling through miR-122 repression and Wnt family member 1 (WNT1) expression 90. Since Activation of WNT signaling often promote EMT-like changes in a subset of cancers, it is possible that PCAT1 target WNT1 protein to induce EMT in ECC.
From the above researches on PCAT-1, we know a lot about the expression, regulation, and function of PCAT-1 in cancers, particular in prostate cancer, where it has been originally discovered. However, there are several questions that can be addressed to fully understand the function of PCAT-1: What is the molecular mechanism of PCAT-1 up-regulation in cancer? PCAT-1 seems to apply multiple mechanisms to fulfill its functions. How does PCAT-1 accomplish this and how is this controlled in different cancers? Which factors interact with PCAT-1 under which conditions and determine its function? Does PCAT-1 target WNT1 protein to induce EMT?
Compare to other well-known lncRANs, such as lncRNA MALAT1 93, H19 94 and HOX antisense intergenic RNA (HOTAIR) 95, our understanding of the role of PCAT-1 in cancer is still at a preliminary stage. For instance, HOTAIR promotes EMT in GC and induces cancer invasion and metastasis by regulating PRC2 target genes in breast cancer 95, 96. H19 is up-regulated in GC. Up-regulation of H19 is associated with miR-675, p53 and Isthmin1, which promote cell proliferation, migration and invasion in GC 97-99. Therefore, to detect more functions of PCAT-1, further experiments should be carried out in cancers. Moreover, more cancer samples should be used to further analyze the relationship between PCAT-1 and the progression and survival of cancer patients. The relationship between PCAT-1 and target genes can also be investigated. Notably, previous studies showed that PCAT-1 interacts with PRC2, suggesting that relevant signaling pathway should be explored in cancer.
In conclusion, PCAT-1 is considered as an oncogenic gene, and whose up-regulation is associated with clinicopathological characteristics, including tumor size, lymphatic metastasis, distant metastasis, tumor-node-metastasis (TNM) stage and poor prognosis in human cancers. PCAT-1 may serve as a potential target for cancer diagnosis and treatment, and may be a potential biomarker for cancer.
Acknowledgments
We are grateful to all authors whose names are not on the list of authors but are participating in the program. This study was supported by grants from the Shenzhen Project of Science and Technology (JCYJ20170307111748761 and 201506042).
References
- 1.Reis ES, Mastellos DC, Ricklin D, Mantovani A, Lambris JD. Complement in cancer: untangling an intricate relationship. Nature reviews Immunology; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hojman P, Gehl J, Christensen JF, Pedersen BK. Molecular Mechanisms Linking Exercise to Cancer Prevention and Treatment. Cell metabolism; 2017. [DOI] [PubMed] [Google Scholar]
- 3.Vineis P, Wild CP. Global cancer patterns: causes and prevention. Lancet. 2014;383:549–57. doi: 10.1016/S0140-6736(13)62224-2. [DOI] [PubMed] [Google Scholar]
- 4.Maresso KC, Tsai KY, Brown PH, Szabo E, Lippman S, Hawk ET. Molecular cancer prevention: Current status and future directions. CA: a cancer journal for clinicians. 2015;65:345–83. doi: 10.3322/caac.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H. et al. Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. European journal of cancer. 2013;49:1374–403. doi: 10.1016/j.ejca.2012.12.027. [DOI] [PubMed] [Google Scholar]
- 6.Bhan A, Soleimani M, Mandal SS. Long Noncoding RNA and Cancer: A New Paradigm. Cancer research. 2017;77:3965–81. doi: 10.1158/0008-5472.CAN-16-2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kwok ZH, Tay Y. Long noncoding RNAs: lincs between human health and disease. Biochemical Society transactions. 2017;45:805–12. doi: 10.1042/BST20160376. [DOI] [PubMed] [Google Scholar]
- 8.Chandra Gupta S, Nandan Tripathi Y. Potential of long non-coding RNAs in cancer patients: From biomarkers to therapeutic targets. International journal of cancer. 2017;140:1955–67. doi: 10.1002/ijc.30546. [DOI] [PubMed] [Google Scholar]
- 9.Flynn RA, Chang HY. Long noncoding RNAs in cell-fate programming and reprogramming. Cell stem cell. 2014;14:752–61. doi: 10.1016/j.stem.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knoll M, Lodish HF, Sun L. Long non-coding RNAs as regulators of the endocrine system. Nature reviews Endocrinology. 2015;11:151–60. doi: 10.1038/nrendo.2014.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–41. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 12.Schmitt AM, Chang HY. Long Noncoding RNAs in Cancer Pathways. Cancer cell. 2016;29:452–63. doi: 10.1016/j.ccell.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qi P, Du X. The long non-coding RNAs, a new cancer diagnostic and therapeutic gold mine. Modern pathology: an official journal of the United States and Canadian Academy of Pathology, Inc. 2013;26:155–65. doi: 10.1038/modpathol.2012.160. [DOI] [PubMed] [Google Scholar]
- 14.Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–307. doi: 10.1016/j.cell.2013.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang C, Li X, Zhao H, Liu H. Long non-coding RNAs: potential new biomarkers for predicting tumor invasion and metastasis. Molecular cancer. 2016;15:62. doi: 10.1186/s12943-016-0545-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huarte M. The emerging role of lncRNAs in cancer. Nature medicine. 2015;21:1253–61. doi: 10.1038/nm.3981. [DOI] [PubMed] [Google Scholar]
- 17.Serviss JT, Johnsson P, Grander D. An emerging role for long non-coding RNAs in cancer metastasis. Frontiers in genetics. 2014;5:234. doi: 10.3389/fgene.2014.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martens-Uzunova ES, Bottcher R, Croce CM, Jenster G, Visakorpi T, Calin GA. Long noncoding RNA in prostate, bladder, and kidney cancer. European urology. 2014;65:1140–51. doi: 10.1016/j.eururo.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 19.Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM. et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–41. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 20.Gutschner T, Hammerle M, Diederichs S. MALAT1 - a paradigm for long noncoding RNA function in cancer. Journal of molecular medicine. 2013;91:791–801. doi: 10.1007/s00109-013-1028-y. [DOI] [PubMed] [Google Scholar]
- 21.Wu Y, Huang C, Meng X, Li J. Long Noncoding RNA MALAT1: Insights into its Biogenesis and Implications in Human Disease. Current pharmaceutical design. 2015;21:5017–28. doi: 10.2174/1381612821666150724115625. [DOI] [PubMed] [Google Scholar]
- 22.Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW. et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–60. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F. et al. TGF-beta-induced upregulation of malat1 promotes bladder cancer metastasis by associating with suz12. Clinical cancer research: an official journal of the American Association for Cancer Research. 2014;20:1531–41. doi: 10.1158/1078-0432.CCR-13-1455. [DOI] [PubMed] [Google Scholar]
- 24.Ji Q, Zhang L, Liu X, Zhou L, Wang W, Han Z. et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. British journal of cancer. 2014;111:736–48. doi: 10.1038/bjc.2014.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fang D, Yang H, Lin J, Teng Y, Jiang Y, Chen J. et al. 17beta-estradiol regulates cell proliferation, colony formation, migration, invasion and promotes apoptosis by upregulating miR-9 and thus degrades MALAT-1 in osteosarcoma cell MG-63 in an estrogen receptor-independent manner. Biochemical and biophysical research communications. 2015;457:500–6. doi: 10.1016/j.bbrc.2014.12.114. [DOI] [PubMed] [Google Scholar]
- 26.Dijkstra JM, Alexander DB. The " NF-k B interacting long noncoding RNA" ( NKILA) transcript is antisense to cancer-associated gene PMEPA1. F1000Research. 2015;4:96. doi: 10.12688/f1000research.6400.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang W, Cui X, Chen J, Feng Y, Song E, Li J. et al. Long non-coding RNA NKILA inhibits migration and invasion of tongue squamous cell carcinoma cells via suppressing epithelial-mesenchymal transition. Oncotarget. 2016;7:62520–32. doi: 10.18632/oncotarget.11528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu X, Tang W, Yang Y, Tang L, Dai R, Pu B. et al. Long noncoding RNA NKILA enhances the anti-cancer effects of baicalein in hepatocellular carcinoma via the regulation of NF-kappaB signaling. Chemico-biological interactions. 2018;285:48–58. doi: 10.1016/j.cbi.2018.02.027. [DOI] [PubMed] [Google Scholar]
- 29.Han Y, Rand KA, Hazelett DJ, Ingles SA, Kittles RA, Strom SS, Prostate Cancer Susceptibility in Men of African Ancestry at 8q24. Journal of the National Cancer Institute; 2016. p. 108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC. et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nature biotechnology. 2011;29:742–9. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Al Olama AA, Kote-Jarai Z, Giles GG, Guy M, Morrison J, Severi G. et al. Multiple loci on 8q24 associated with prostate cancer susceptibility. Nature genetics. 2009;41:1058–60. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- 32.Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A. et al. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24. Nature genetics. 2007;39:631–7. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- 33.Shi WH, Wu QQ, Li SQ, Yang TX, Liu ZH, Tong YS. et al. Upregulation of the long noncoding RNA PCAT-1 correlates with advanced clinical stage and poor prognosis in esophageal squamous carcinoma. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:2501–7. doi: 10.1007/s13277-014-2863-3. [DOI] [PubMed] [Google Scholar]
- 34.Liu L, Liu Y, Zhuang C, Xu W, Fu X, Lv Z. et al. Inducing cell growth arrest and apoptosis by silencing long non-coding RNA PCAT-1 in human bladder cancer. Tumour biology: the journal of the International Society for Oncodevelopmental Biology and Medicine. 2015;36:7685–9. doi: 10.1007/s13277-015-3490-3. [DOI] [PubMed] [Google Scholar]
- 35.Yan TH, Yang H, Jiang JH, Lu SW, Peng CX, Que HX. et al. Prognostic significance of long non-coding RNA PCAT-1 expression in human hepatocellular carcinoma. International journal of clinical and experimental pathology. 2015;8:4126–31. [PMC free article] [PubMed] [Google Scholar]
- 36.Zhao B, Hou X, Zhan H. Long non-coding RNA PCAT-1 over-expression promotes proliferation and metastasis in non-small cell lung cancer cells. International journal of clinical and experimental medicine. 2015;8:18482–7. [PMC free article] [PubMed] [Google Scholar]
- 37.Shen X, Zhang Y, Wu X, Guo Y, Shi W, Qi J. et al. Upregulated lncRNA-PCAT1 is closely related to clinical diagnosis of multiple myeloma as a predictive biomarker in serum. Cancer biomarkers: section A of Disease markers. 2017;18:257–63. doi: 10.3233/CBM-160158. [DOI] [PubMed] [Google Scholar]
- 38.Qiao L, Liu X, Tang Y, Zhao Z, Zhang J, Feng Y. Down regulation of the long non-coding RNA PCAT-1 induced growth arrest and apoptosis of colorectal cancer cells. Life sciences. 2017;188:37–44. doi: 10.1016/j.lfs.2017.08.024. [DOI] [PubMed] [Google Scholar]
- 39.Cui WC, Wu YF, Qu HM. Up-regulation of long non-coding RNA PCAT-1 correlates with tumor progression and poor prognosis in gastric cancer. European review for medical and pharmacological sciences. 2017;21:3021–7. [PubMed] [Google Scholar]
- 40.Prensner JR, Chen W, Iyer MK, Cao Q, Ma T, Han S. et al. PCAT-1, a long noncoding RNA, regulates BRCA2 and controls homologous recombination in cancer. Cancer research. 2014;74:1651–60. doi: 10.1158/0008-5472.CAN-13-3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fagerberg L, Hallstrom BM, Oksvold P, Kampf C, Djureinovic D, Odeberg J. et al. Analysis of the human tissue-specific expression by genome-wide integration of transcriptomics and antibody-based proteomics. Mol Cell Proteomics. 2014;13:397–406. doi: 10.1074/mcp.M113.035600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Salinas CA, Tsodikov A, Ishak-Howard M, Cooney KA. Prostate cancer in young men: an important clinical entity. Nature Reviews Urology. 2014;11:317–23. doi: 10.1038/nrurol.2014.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mohler JL, Armstrong AJ, Bahnson RR, D'Amico AV, Davis BJ, Eastham JA. et al. Prostate Cancer, Version 1.2016. Journal of the National Comprehensive Cancer Network: JNCCN. 2016;14:19–30. doi: 10.6004/jnccn.2016.0004. [DOI] [PubMed] [Google Scholar]
- 44.Huang X, Yan J, Zhang M, Wang Y, Chen Y, Fu X. et al. Targeting Epigenetic Crosstalk as a Therapeutic Strategy for EZH2-Aberrant Solid Tumors. Cell. 2018;175:186–99. doi: 10.1016/j.cell.2018.08.058. e19. [DOI] [PubMed] [Google Scholar]
- 45.Zovoilis A, Cifuentes-Rojas C, Chu HP, Hernandez AJ, Lee JT. Destabilization of B2 RNA by EZH2 Activates the Stress Response. Cell. 2016;167:1788–802. doi: 10.1016/j.cell.2016.11.041. e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Prasanth SG, Prasanth KV. Easy Stress Relief by EZH2. Cell. 2016;167:1678–80. doi: 10.1016/j.cell.2016.11.051. [DOI] [PubMed] [Google Scholar]
- 47.Prensner JR, Chen W, Han S, Iyer MK, Cao Q, Kothari V. et al. The long non-coding RNA PCAT-1 promotes prostate cancer cell proliferation through cMyc. Neoplasia. 2014;16:900–8. doi: 10.1016/j.neo.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zeng J, Li HC, Tanaka T, Rabbitts TH. Selection of human single domain antibodies recognizing the CMYC protein using enhanced intracellular antibody capture. J Immunol Methods. 2015;426:140–3. doi: 10.1016/j.jim.2015.08.009. [DOI] [PubMed] [Google Scholar]
- 49.Sun L, Song L, Wan Q, Wu G, Li X, Wang Y. et al. cMyc-mediated activation of serine biosynthesis pathway is critical for cancer progression under nutrient deprivation conditions. Cell Res. 2015;25:429–44. doi: 10.1038/cr.2015.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karslioglu E, Kleinberger JW, Salim FG, Cox AE, Takane KK, Scott DK. et al. cMyc is a principal upstream driver of beta-cell proliferation in rat insulinoma cell lines and is an effective mediator of human beta-cell replication. Mol Endocrinol. 2011;25:1760–72. doi: 10.1210/me.2011-1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Heath VJ, Gillespie DA, Crouch DH. Inhibition of the terminal stages of adipocyte differentiation by cMyc. Exp Cell Res. 2000;254:91–8. doi: 10.1006/excr.1999.4736. [DOI] [PubMed] [Google Scholar]
- 52.Luo G, Li B, Duan C, Cheng Y, Xiao B, Yao F. et al. cMyc promotes cholangiocarcinoma cells to overcome contact inhibition via the mTOR pathway. Oncol Rep. 2017;38:2498–506. doi: 10.3892/or.2017.5913. [DOI] [PubMed] [Google Scholar]
- 53.Sakr HI, Chute DJ, Nasr C, Sturgis CD. cMYC expression in thyroid follicular cell-derived carcinomas: a role in thyroid tumorigenesis. Diagn Pathol. 2017;12:71. doi: 10.1186/s13000-017-0661-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Loftus RM, Assmann N, Kedia-Mehta N, O'Brien KL, Garcia A, Gillespie C. et al. Amino acid-dependent cMyc expression is essential for NK cell metabolic and functional responses in mice. Nat Commun. 2018;9:2341. doi: 10.1038/s41467-018-04719-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rinella ME. Nonalcoholic fatty liver disease: a systematic review. JAMA. 2015;313:2263–73. doi: 10.1001/jama.2015.5370. [DOI] [PubMed] [Google Scholar]
- 56.Au JS, Frenette CT. Erratum: Management of Hepatocellular Carcinoma: Current Status and Future Directions. Gut Liver. 2015;9:811. doi: 10.5009/gnl15022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waghray A, Murali AR, Menon KN. Hepatocellular carcinoma: From diagnosis to treatment. World J Hepatol. 2015;7:1020–9. doi: 10.4254/wjh.v7.i8.1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA: a cancer journal for clinicians. 2012;62:394–9. doi: 10.3322/caac.21161. [DOI] [PubMed] [Google Scholar]
- 59.Poon RT. Prevention of recurrence after resection of hepatocellular carcinoma: a daunting challenge. Hepatology. 2011;54:757–9. doi: 10.1002/hep.24569. [DOI] [PubMed] [Google Scholar]
- 60.Wen J, Xu J, Sun Q, Xing C, Yin W. Upregulation of long non coding RNA PCAT-1 contributes to cell proliferation, migration and apoptosis in hepatocellular carcinoma. Molecular medicine reports. 2016;13:4481–6. doi: 10.3892/mmr.2016.5075. [DOI] [PubMed] [Google Scholar]
- 61.Zhang D, Cao J, Zhong Q, Zeng L, Cai C, Lei L. et al. Long noncoding RNA PCAT-1 promotes invasion and metastasis via the miR-129-5p-HMGB1 signaling pathway in hepatocellular carcinoma. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2017;95:1187–93. doi: 10.1016/j.biopha.2017.09.045. [DOI] [PubMed] [Google Scholar]
- 62.Carcas LP. Gastric cancer review. Journal of carcinogenesis. 2014;13:14. doi: 10.4103/1477-3163.146506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA: a cancer journal for clinicians. 2013;63:11–30. doi: 10.3322/caac.21166. [DOI] [PubMed] [Google Scholar]
- 64.Zong L, Abe M, Seto Y, Ji J. The challenge of screening for early gastric cancer in China. Lancet. 2016;388:2606. doi: 10.1016/S0140-6736(16)32226-7. [DOI] [PubMed] [Google Scholar]
- 65.Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet. 2016;388:2654–64. doi: 10.1016/S0140-6736(16)30354-3. [DOI] [PubMed] [Google Scholar]
- 66.Ang TL, Fock KM. Clinical epidemiology of gastric cancer. Singapore Med J. 2014;55:621–8. doi: 10.11622/smedj.2014174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bi M, Yu H, Huang B, Tang C. Long non-coding RNA PCAT-1 over-expression promotes proliferation and metastasis in gastric cancer cells through regulating CDKN1A. Gene. 2017;626:337–43. doi: 10.1016/j.gene.2017.05.049. [DOI] [PubMed] [Google Scholar]
- 68.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: a cancer journal for clinicians. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 69.Bielack SS, Hecker-Nolting S, Blattmann C, Kager L. Advances in the management of osteosarcoma. F1000Research. 2016;5:2767. doi: 10.12688/f1000research.9465.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.He X, Gao Z, Xu H, Zhang Z, Fu P. A meta-analysis of randomized control trials of surgical methods with osteosarcoma outcomes. J Orthop Surg Res. 2017;12:5. doi: 10.1186/s13018-016-0500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lindsey BA, Markel JE, Kleinerman ES. Osteosarcoma Overview. Rheumatol Ther. 2017;4:25–43. doi: 10.1007/s40744-016-0050-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Durfee RA, Mohammed M, Luu HH. Review of Osteosarcoma and Current Management. Rheumatol Ther. 2016;3:221–43. doi: 10.1007/s40744-016-0046-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang X, Zhang Y, Mao Y, Ma X. The lncRNA PCAT1 is correlated with poor prognosis and promotes cell proliferation, invasion, migration and EMT in osteosarcoma. Onco Targets Ther. 2018;11:629–38. doi: 10.2147/OTT.S152063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nieto MA. Epithelial plasticity: a common theme in embryonic and cancer cells. Science. 2013;342:1234850. doi: 10.1126/science.1234850. [DOI] [PubMed] [Google Scholar]
- 75.De Craene B, Berx G. Regulatory networks defining EMT during cancer initiation and progression. Nat Rev Cancer. 2013;13:97–110. doi: 10.1038/nrc3447. [DOI] [PubMed] [Google Scholar]
- 76.Nieto MA, Huang RY, Jackson RA, Thiery JP. Emt: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- 77.Gonzalez DM, Medici D. Signaling mechanisms of the epithelial-mesenchymal transition. Sci Signal. 2014;7:re8. doi: 10.1126/scisignal.2005189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–8. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–90. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 80.Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nature reviews Molecular cell biology. 2014;15:178–96. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Santamaria PG, Moreno-Bueno G, Portillo F, Cano A. EMT: Present and future in clinical oncology. Mol Oncol. 2017;11:718–38. doi: 10.1002/1878-0261.12091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Du B, Shim JS. Targeting Epithelial-Mesenchymal Transition (EMT) to Overcome Drug Resistance in Cancer. Molecules; 2016. p. 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huang J, Deng G, Liu T, Chen W, Zhou Y. Long noncoding RNA PCAT-1 acts as an oncogene in osteosarcoma by reducing p21 levels. Biochemical and biophysical research communications. 2018;495:2622–9. doi: 10.1016/j.bbrc.2017.12.157. [DOI] [PubMed] [Google Scholar]
- 84.Alberts SR, Gores GJ, Kim GP, Roberts LR, Kendrick ML, Rosen CB. et al. Treatment options for hepatobiliary and pancreatic cancer. Mayo Clin Proc. 2007;82:628–37. doi: 10.4065/82.5.628. [DOI] [PubMed] [Google Scholar]
- 85.Skipworth JR, Olde Damink SW, Imber C, Bridgewater J, Pereira SP, Malago M. Review article: surgical, neo-adjuvant and adjuvant management strategies in biliary tract cancer. Aliment Pharmacol Ther. 2011;34:1063–78. doi: 10.1111/j.1365-2036.2011.04851.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nusse R, Clevers H. Wnt/beta-Catenin Signaling, Disease, and Emerging Therapeutic Modalities. Cell. 2017;169:985–99. doi: 10.1016/j.cell.2017.05.016. [DOI] [PubMed] [Google Scholar]
- 87.Martin PM, Stanley RE, Ross AP, Freitas AE, Moyer CE, Brumback AC. et al. DIXDC1 contributes to psychiatric susceptibility by regulating dendritic spine and glutamatergic synapse density via GSK3 and Wnt/beta-catenin signaling. Mol Psychiatry. 2018;23:467–75. doi: 10.1038/mp.2016.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.So J, Khaliq M, Evason K, Ninov N, Martin BL, Stainier DYR. et al. Wnt/beta-catenin signaling controls intrahepatic biliary network formation in zebrafish by regulating notch activity. Hepatology. 2018;67:2352–66. doi: 10.1002/hep.29752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wang B, Tian T, Kalland KH, Ke X, Qu Y. Targeting Wnt/beta-Catenin Signaling for Cancer Immunotherapy. Trends Pharmacol Sci. 2018;39:648–58. doi: 10.1016/j.tips.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 90.Zhang F, Wan M, Xu Y, Li Z, Leng K, Kang P. et al. Long noncoding RNA PCAT1 regulates extrahepatic cholangiocarcinoma progression via the Wnt/beta-catenin-signaling pathway. Biomedicine & pharmacotherapy = Biomedecine & pharmacotherapie. 2017;94:55–62. doi: 10.1016/j.biopha.2017.07.025. [DOI] [PubMed] [Google Scholar]
- 91.Sarrafzadeh S, Geranpayeh L, Ghafouri-Fard S. Expression Analysis of Long Non-Coding PCAT-1in Breast Cancer. International journal of hematology-oncology and stem cell research. 2017;11:185–91. [PMC free article] [PubMed] [Google Scholar]
- 92.Singh M, Yelle N, Venugopal C, Singh SK. EMT: Mechanisms and therapeutic implications. Pharmacol Ther. 2018;182:80–94. doi: 10.1016/j.pharmthera.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 93.Gutschner T, Hammerle M, Eissmann M, Hsu J, Kim Y, Hung G. et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer research. 2013;73:1180–9. doi: 10.1158/0008-5472.CAN-12-2850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhou X, Ye F, Yin C, Zhuang Y, Yue G, Zhang G. The Interaction Between MiR-141 and lncRNA-H19 in Regulating Cell Proliferation and Migration in Gastric Cancer. Cellular physiology and biochemistry: international journal of experimental cellular physiology, biochemistry, and pharmacology. 2015;36:1440–52. doi: 10.1159/000430309. [DOI] [PubMed] [Google Scholar]
- 95.Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ. et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–6. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Xu ZY, Yu QM, Du YA, Yang LT, Dong RZ, Huang L. et al. Knockdown of long non-coding RNA HOTAIR suppresses tumor invasion and reverses epithelial-mesenchymal transition in gastric cancer. Int J Biol Sci. 2013;9:587–97. doi: 10.7150/ijbs.6339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J. et al. Up-regulated long non-coding RNA H19 contributes to proliferation of gastric cancer cells. FEBS J. 2012;279:3159–65. doi: 10.1111/j.1742-4658.2012.08694.x. [DOI] [PubMed] [Google Scholar]
- 98.Xiang W, Ke Z, Zhang Y, Cheng GH, Irwan ID, Sulochana KN. et al. Isthmin is a novel secreted angiogenesis inhibitor that inhibits tumour growth in mice. Journal of cellular and molecular medicine. 2011;15:359–74. doi: 10.1111/j.1582-4934.2009.00961.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhuang M, Gao W, Xu J, Wang P, Shu Y. The long non-coding RNA H19-derived miR-675 modulates human gastric cancer cell proliferation by targeting tumor suppressor RUNX1. Biochemical and biophysical research communications. 2014;448:315–22. doi: 10.1016/j.bbrc.2013.12.126. [DOI] [PubMed] [Google Scholar]