Rad51-mediated homologous recombination is the major mechanism for repairing DNA double-strand break (DSB) repair in cancer cells. Thus, regulating Rad51 activity could be an attractive target. The sequential assembly and disassembly of Rad51 to the broken DNA ends depend on reversible protein-protein interactions. Here, we discovered that a dynamic interaction with molecular chaperone Hsp90 is one such regulatory event that governs the recruitment of Rad51 onto the damaged DNA. We uncovered that Rad51 associates with Hsp90, and upon DNA damage, this complex dissociates to facilitate the loading of Rad51 onto broken DNA. In a mutant where such dissociation is incomplete, the occupancy of Rad51 at the broken DNA is partial, which results in inefficient DNA repair. Thus, it is reasonable to propose that any small molecule that may alter the dynamics of the Rad51-Hsp90 interaction is likely to impact DSB repair in cancer cells.
KEYWORDS: DNA repair, Hsp90, homologous recombination, Hsp90-Rad51 interaction, Rad51 recruitment to chromatin, molecular chaperone
ABSTRACT
DNA damage-induced Rad51 focus formation is the hallmark of homologous recombination-mediated DNA repair. Earlier, we reported that Rad51 physically interacts with Hsp90, and under the condition of Hsp90 inhibition, it undergoes proteasomal degradation. Here, we show that the dynamic interaction between Rad51 and Hsp90 is crucial for the DNA damage-induced nuclear function of Rad51. Guided by a bioinformatics study, we generated a single mutant of Rad51, which resides at the N-terminal domain, outside the ATPase core domain. The mutant with an E to L change at residue 108 (Rad51E108L) was predicted to bind more strongly with Hsp90 than the wild-type (Rad51WT). A coimmunoprecipitation study demonstrated that there exists a distinct difference between the in vivo associations of Rad51WT-Hsp90 and of Rad51E108L-Hsp90. We found that upon DNA damage, the association between Rad51WT and Hsp90 was significantly reduced compared to that in the undamaged condition. However, the mutant Rad51E108L remained tightly associated with Hsp90 even after DNA damage. Consequently, the recruitment of Rad51E108L to the double-stranded broken ends was reduced significantly. The E108L-rad51 strain manifested severe sensitivity toward methyl methanesulfonate (MMS) and a complete loss of gene conversion efficiency, a phenotype similar to that of the Δrad51 strain. Previously, some of the N-terminal domain mutants of Rad51 were identified in a screen for a Rad51 interaction-deficient mutant; however, our study shows that Rad51E108L is not defective either in the self-interaction or its interaction with the members of the Rad52 epistatic group. Our study thus identifies a novel mutant of Rad51 which, owing to its greater association with Hsp90, exhibits a severe DNA repair defect.
IMPORTANCE Rad51-mediated homologous recombination is the major mechanism for repairing DNA double-strand break (DSB) repair in cancer cells. Thus, regulating Rad51 activity could be an attractive target. The sequential assembly and disassembly of Rad51 to the broken DNA ends depend on reversible protein-protein interactions. Here, we discovered that a dynamic interaction with molecular chaperone Hsp90 is one such regulatory event that governs the recruitment of Rad51 onto the damaged DNA. We uncovered that Rad51 associates with Hsp90, and upon DNA damage, this complex dissociates to facilitate the loading of Rad51 onto broken DNA. In a mutant where such dissociation is incomplete, the occupancy of Rad51 at the broken DNA is partial, which results in inefficient DNA repair. Thus, it is reasonable to propose that any small molecule that may alter the dynamics of the Rad51-Hsp90 interaction is likely to impact DSB repair in cancer cells.
INTRODUCTION
Whenever cells are exposed to DNA-damaging agents, the family of DNA repair proteins must relocate to the nucleus and be recruited to the damaged chromatins to elicit a DNA damage response and to ensure efficient repair of damaged DNA (1–3). These groups of proteins include DNA damage signaling proteins (Mre11, ATM, ATR, and DNA-PKcs), cell cycle checkpoint effectors (Chk1 and Chk2), and DNA processing enzymes (Mre11, ExoI, Sae2, Rad51, Rad52, Rad54, BRCA1/2, BLM, Ku70/80, ligase IV, etc.) (4). The sequential assembly and disassembly of DNA repair proteins at DNA broken ends depend on reversible protein-protein interactions. Rad51, a central player of homology-directed double-strand break (DSB) repair, remains in the cytoplasm under normal conditions. DNA damage leads to the redistribution of Rad51 from the cytoplasm to the nucleus and its loading onto the broken ends of DNA. It is reasonable to propose that insufficient recruitment of Rad51 onto the chromatin is likely to have a severe impact on homologous recombination (HR) efficiency. Earlier reports demonstrated that in a human cell line, BRCA1 promotes the localization of BRCA2 to damage foci through the BRCA2 binding protein PALB2 (5–8). BRCA2 interacts with RAD51 and promotes RAD51 assembly onto single-stranded DNA (ssDNA) (9–11). However, BRCA2 is absent in lower eukaryotes, where HR is the predominant pathway for DNA repair. It is reported that in Saccharomyces cerevisiae, Rad52 promotes Rad51 filament assembly (12) by interacting with RPA. Rad52 is thought to replace RPA bound to ssDNA with Rad51 or provide a seeding site within the RPA-bound ssDNA for subsequent binding of Rad51 (13).
Our previous study revealed that Rad51 is a direct client of Hsp90 and is dependent upon Hsp90 for its maturity and activity (14). Apart from merely providing maturity to the client proteins, Hsp90 also assists in the translocation of proteins to different cellular compartments (15). Previous reports have established that the Hsp90 chaperone machinery not only escorts steroid hormone receptors (SHRs) to the nucleus but is also responsible for the recycling of the receptor on chromatin and stabilizing the DNA-binding properties of the receptor (16). Two cochaperones of Hsp90, p23 and Bag-1L, are found to modulate steroid hormone receptor function by controlling receptor binding to chromatin (16).
Our earlier study demonstrated that the charged linker deletion mutant of yHsp90 (Δ211-259hsp82) inhibits effective Rad51 focus formation in the nucleus upon DNA damage (14). This finding was positively correlated with severe methyl methanesulfonate (MMS) sensitivity (comparable to that for the Δrad51 strain) and with the complete loss of Rad51-dependent gene targeting function. We demonstrated that the charged linker deletion (Δ211-259hsp82) mutant strain is strikingly different than the wild-type strain in the distribution of Rad51 foci upon MMS treatment. Although there was only a 20% overall reduction in the Rad51 focus formation, the number of nuclei having multiple foci was drastically reduced in the mutant strain. This clearly indicates that in mutant nuclei, effective Rad51 levels may be low. Since the charged linker region is responsible for providing structural flexibility between amino and carboxyl-terminal domains of Hsp90 (17), an optimum interaction between Rad51 and Hsp90 may be compromised in the mutant. Hence, we hypothesize that effective Hsp90 and Rad51 interaction may be crucial for nuclear function of Rad51. To prove this, we utilized a bioinformatics approach to design a point mutant with an E to L change at residue 108 (Rad51E108L), which has a stronger affinity toward Hsp90. Our data reveal that there exists a dynamic equilibrium between the association of wild-type Rad51 (Rad51WT) and Hsp90 under a normal condition and dissociation under DNA-damaging conditions. In the case of Rad51E108L, due to tighter association, the interaction between Hsp90 and mutant Rad51 becomes irreversible; hence, even under DNA-damaging conditions, the mutant Rad51 protein does not proficiently dissociate from Hsp90. As a result, the mutant Rad51E108L is not recruited to the broken DNA ends as efficiently as wild-type Rad51. Hence, the E108L-rad51 strain shows extreme sensitivity toward DNA-damaging agents and poor gene conversion activity. This study points out that the DNA damage-induced reversible protein-protein interaction between Rad51 and Hsp90 plays a critical role in Rad51 function.
RESULTS
Generation of RAD51 mutant strain based on the molecular docking studies between yHsp90 and Rad51.
Earlier studies in our lab demonstrated that yHsp90 and Rad51 can physically interact (14). Unlike other chaperones, there is no specific binding pocket present in Hsp90 through which it binds to the client proteins. Hence, in order to understand the point of contacts between yHsp90 and Rad51, we employed a bioinformatics approach. To that end, Rad51 proteins (PDB identifier [ID] 1SZP) having various combinations of monomers, dimers, and hexamers were allowed to dock with yHsp90 (PDB ID 2CG9) using the fully automated web-based program ClusPro 2.0 (18), which employs the improved fast Fourier transform (FFT)-based rigid docking program PIPER (19). Thirty models of the protein-protein complex for each type of interaction, namely, balanced, electrostatic favored, hydrophobic favored, and van der Waal's plus electrostatic, were generated for each docking. It was found that a hydrophobic-favored interaction showed the lowest energy scores; hence, the corresponding protein complex model with the largest cluster was chosen. The surface view of the three-dimensional structure of Rad51 displays a characteristic pocket in each of the monomers into which the yHsp90 is found to dock. The docked complex models showed that the N-terminal residue of the Rad51 E chain, Glu 108 (1.88 Å), has the shortest bond distance with yHsp90 C-terminal residues. We conducted a multiple-sequence alignment of Rad51 (Fig. 1A) and found that E108, which is predicted to have the strongest association with Hsp90, is evolutionarily conserved. In Rad51, the amino acid residue E108 is present in the N-terminal domain of Rad51, which lies outside its catalytic domain (Fig. 1B). To explore whether the Hsp90 and Rad51 association mediates Rad51 nuclear function under DNA-damaging conditions, one approach may be the generation of a Rad51 mutant with a reduced affinity for Hsp90. However, as Rad51 is a client of Hsp90, we reasoned that any mutant of Rad51 that fails to interact with Hsp90 due to a low affinity would be unstable in the cell. Hence, we designed a strong-affinity mutant to establish our hypothesis. By in silico mutation, we created four single mutants of Rad51 where the glutamic acid at the 108th position was replaced by neutral residues (glycine, alanine, leucine, and isoleucine). Table 1 displays a comparison of the parameters of yHsp90 docking with the wild-type and mutant Rad51 based on ClusPro results. Our study shows that the mutant Rad51E108L and Hsp90 docked complex results in a maximum increase in cluster size of 139 compared to 71 for the wild type. This implies a greater probability of the receptor-ligand complex being found in that specific conformation. Furthermore, there is a decrease in the energy score from −1,407.2 to −1,512.6 between the wild-type and Rad51E108L mutant, respectively, which points to an increased stability of the protein complex. The rad51 mutant was subsequently cloned into a yeast 2µ expression vector pTA (20) having the GPD promoter. As the Rad51 and Hsp90 interaction is essential for the stability of Rad51, we determined the stability of Rad51 mutant proteins by Western blot analysis. For this, we generated yeast strains NRY1, NRY2, and TSY17 by transforming empty vector (pTA), pTA-RAD51, and pTA-E108L-rad51 vectors into a null rad51 yeast strain. The steady-state level of the mutant Rad51 was comparable to that of the wild type (Fig. 1C).
FIG 1.

Generation of RAD51 mutant strain. (A) Multiple sequence alignment of Rad51 (N-terminal domain) protein sequences of S. cerevisiae (yeast) with Homo sapiens (human), Mus musculus (mouse), Gallus gallus (bird), Danio rerio (zebrafish), Caenorhabditis elegans (nematode), and Drosophila melanogaster (fruit fly). The conserved glutamic acid residues among various organisms are represented by the red box. (B) Schematic representation of Rad51 domains demonstrating boundaries of N-terminal, ATPase (AD), and C-terminal domains along with the Walker A and Walker B motifs. The star depicts the approximate location of E108 in the N-terminal domain of Rad51. (C) Western blot was performed using protein extracts from wild-type, Δrad51, and E108L-rad51 strains. Actin was used as a loading control.
TABLE 1.
ClusPro results depicting cluster sizes and energy scores of yHsp90 (2CG9A) with wild-type and mutant Rad51
| Rad51 (1SZP ABCDEF) strain | Hydrophobic-favored interaction |
|
|---|---|---|
| Cluster size | Energy score | |
| Wild-type (E108) | 71 | −1,407.2 |
| E108G | 117 | −1,534.0 |
| E108A | 117 | −1,518.4 |
| E108I | 113 | −1,543.3 |
| E108L | 139 | −1,512.6 |
Rad51E108L shows a stronger association with Hsp90 than the wild-type Rad51.
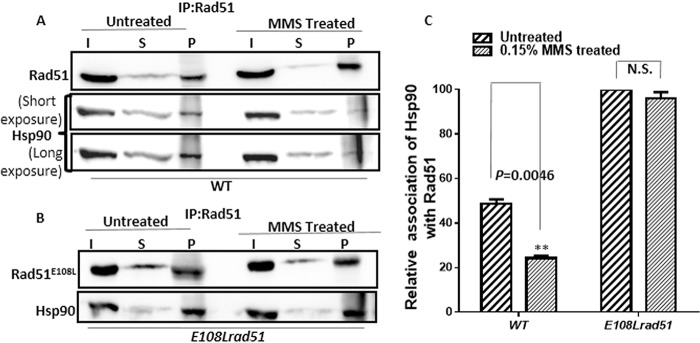
To investigate the interaction between Rad51 and yHsp90, we performed coimmunoprecipitation experiments under normal as well as MMS treatment conditions. To capture a detectable association between yHsp90 and Rad51, we overexpressed both yHSP90 and RAD51 (or its mutant version) from two 2µ vectors, each having a GPD promoter. The yHsp90-Rad51 complex was coimmunoprecipitated from the whole-cell extract with an anti-Rad51 antibody, followed by detection on a Western blot using an anti-Hsp82 antibody (Fig. 2A and B). Under normal conditions, in the wild-type strain, a small fraction of Hsp90 was associated with Rad51, whereas, in the case of the mutant strain, a significantly larger fraction of Hsp90 was associated with Rad51. Quantification of the several experimental repeats showed that the relative association between Hsp90 and Rad51E108L was almost double the association found between Hsp90 and Rad51WT. This signifies a stronger association of Hsp90 with Rad51E108L than with Rad51WT. In the presence of MMS, Hsp90 and Rad51 association was reduced in the wild-type strain. On the other hand, in the E108L-rad51 strain, even in the presence of MMS, there was no detectable reduction in the association between Hsp90 and Rad51E108L. We repeated this experiment three times and calculated the relative association of Hsp90 with Rad51 in the presence and absence of MMS. Our analysis shows that approximately 50% dissociation of the Rad51WT-Hsp90 complex occurs upon MMS treatment, whereas no significant dissociation of the Rad51E108L-Hsp90 complex was observed under similar conditions (Fig. 2C). Thus, from this experiment, we conclude that there is a dynamic equilibrium between Rad51-Hsp90 complexes: in the presence of DNA damage, the equilibrium is shifted toward the dissociation of Rad51-Hsp90. However, this dynamic interaction is absent in the E108L-rad51 strain, and the complex remains in the associated form even in the presence of the DNA-damaging agent.
FIG 2.
Rad51E108L shows a stronger association with Hsp90 than the wild-type Rad51. (A) Western blot showing coimmunoprecipitation of Rad51 with Hsp90 from whole-cell extracts of wild-type strain and cells treated with 0.15% MMS for 2 h. I, input; S, supernatant; P, pellet. (B) Western blot showing coimmunoprecipitation of Rad51E108L with Hsp90 from whole-cell extracts of E108L-rad51 mutant strain untreated and treated with 0.15% MMS for 2 h. Immunoprecipitation (IP) was performed using an anti-Rad51 antibody. An anti-Hsp90 antibody was used for Western blotting. (C) Relative association of Hsp90 with Rad51 was calculated from at least three independent experiments, and standard deviations are plotted for both wild-type and mutant strains. P values were calculated using the two-tailed Student’s t test. **, P = 0.0046; N.S., not significant.
HO-induced Rad51 recruitment to the broken DNA ends is compromised in the E108L-rad51 strain.
During homologous recombination-mediated DNA repair, Rad51 is recruited to the ssDNA overhangs. It searches for the homologous DNA and, once found, facilitates the repair by performing a strand exchange reaction. The recruitment of Rad51 to the broken ends is the hallmark of DNA repair. Our previous observations suggest that the E108L-rad51 mutant is defective in dissociating from Hsp90 upon DNA damage. This defect may cause inadequate recruitment of Rad51 mutants to the broken DNA. To study the recruitment of mutant Rad51 to the DSB, we employed chromatin immunoprecipitation (ChIP) assays. To that end, we used NA14 strains (21) harboring null rad51. We modified the NA14 strain and generated three strains, namely, TSY20, TSY21, and TSY22, where native RAD51 is knocked out, and into those backgrounds, the empty plasmid, wild-type RAD51, and the mutant rad51 were transformed, respectively. These strains have a cassette inserted in chromosome V with two copies of URA3, separated by 3 kb, of which one ura3 copy is inactivated by the insertion of an HO endonuclease restriction site (Fig. 3A). The KANMX gene is incorporated within the two URA3 genes. HO endonuclease is expressed in the strain by a galactose inducible promoter. A double-strand break (DSB) is generated in the ura3 gene upon induction of HO endonuclease. We pulled down the Rad51-bound DNA segments from uninduced and HO-induced samples and subsequently compared the recruitment of mutant Rad51 protein to the donor URA3 locus (22). This experiment was repeated three times, and representative data from one of these are presented (Fig. 3B). To ensure the specificity of Rad51 recruitment to the broken locus, we probed its recruitment at the ACT1 locus, which does not contain an HO cut site. We did not detect any band at the ACT1 locus. We quantified the extent of recruitment of Rad51 proteins by measuring the ratio of amplification in the pellet sample with respect to the amplification observed in the input. To confirm the specificity of Rad51 recruitment to the DSB, we performed ChIP with IgG, which does not result in any amplification with the precipitated sample (Fig. 3B). Although there was no recruitment of Rad51 in the HO-uninduced condition, upon HO induction, the recruitment of Rad51E108L was only 40% of that for the wild type (Fig. 3C). To ensure that the defect in the recruitment of the mutant Rad51 to the DSB was not due to the inefficiency of galactose-induced DSB, we probed the HO endonuclease recognition site in the presence and absence of HO induction. To that end, we amplified the HO site flanking the ura3 region using a forward primer, which is 20 bp upstream of the HO site, and a reverse primer, which is complementary to the middle part of KANMX gene. We observed the amplification of the target region in a galactose-untreated sample; however, after 1 h of galactose induction, the amplicon disappeared, indicating the successful generation of DSBs in all the strains (Fig. 3D). Overall, from these experiments, we conclude that the effective concentration of the Rad51E108L mutant at broken DNA ends is less than that of the wild-type Rad51.
FIG 3.
HO-induced Rad51 recruitment to the broken DNA ends is severely compromised in E108L-rad51 mutant. (A) Schematic diagram of a cassette incorporated in the strain used for studying gene conversion efficiency. It harbors two copies of URA3, one of which is mutated by the insertion of an HO endonuclease site. Induction with galactose creates single DSB in the mutated ura3, repair of which takes place in either a Rad51-dependent or Rad51-independent manner. KANMX cassette will be retained only if repair happens via the Rad51-dependent manner. (B) Chromatin immunoprecipitation (ChIP) of strains expressing wild-type Rad51 and E108L-rad51. Gel image showing one of the representative PCR products of input and precipitated samples using URA3 donor-specific primer and ACT1-specific primer. Immunoprecipitation was performed using anti-Rad51 and IgG antibodies. Input represents the total amount of DNA in the sample. (C) Each set was repeated three times, and the band intensities of the recruited samples upon HO induction were quantified using ImageJ software; comparative recruitment of Rad51 and Rad51E108L is plotted with respect to the input. Error bars indicate standard deviations (SDs); n = 3 (P values were calculated using the two-tailed Student's t test). **, P < 0.01. (D) Semiquantitative reverse transcriptase PCR (RT-PCR), representing the amplification of DNA around the DSB site in ura3 before and after HO endonuclease induction. Lower intensity of band in HO-induced sample indicates the DSB generation in strains having wild-type Rad51 and E108L-rad51. Actin was used as a loading control.
Mutation at the E108 position of Rad51 sensitizes the cells to MMS and renders them deficient in gene conversion.
In S. cerevisiae, homologous recombination is the preferred pathway for repairing DSBs, in which Rad51 plays a central role. To understand the effect of rad51 mutation, we performed the return-to-growth assay upon DNA damage. This was conducted by exposing the strains to 0.03% MMS (methyl methanesulfonate) for 2 h. Subsequently, treated and untreated cells were serially diluted by 10-fold as presented in Fig. 4A and spotted on selective medium. We observed that the E108L-rad51 strain showed a slow growth phenotype compared to that of the wild type and Δrad51 strains. The survivability of the cells was positively correlated with the efficiency of DNA repair. We observed that E108L-rad51 cells were highly sensitive to MMS-induced DNA damage, similar to that observed in Δrad51 cells. The mechanism of homologous recombination involves repairing the DSBs by utilizing a homologous sequence from the genome. If the genome contains repetitive sequences and a double-strand break is created in any one of the repeats, it can be repaired by gene conversion, which is Rad51 dependent. We examined the gene conversion efficiency of the Rad51 mutant in the yeast strain NA14 (21). The DSB can be repaired by either of the two HR pathways (gene conversion or single strand annealing), and the repair products are easily distinguishable. If repaired by the Rad51-dependent gene conversion pathway, the strain behaves as G418 sulfate resistant; if it is repaired by the Rad51-independent single-strand annealing (SSA) pathway, the strain will be G418 sulfate sensitive (Fig. 3A) (21). The percent gene conversion was scored by growing cells on G418 sulfate-containing plates after galactose induction. Our experimental data indicate that there was no significant change in the gene conversion (GC) efficiency of the wild type (near 40%). However, the GC score for the E108L-rad51 mutant (10.5%) was comparable to that of the Δrad51 strain (7%) (Fig. 4B). Overall, we conclude from our experimental data that the E108L-rad51 mutant behaved as a complete loss-of-function mutant of Rad51 in our assay.
FIG 4.
Mutation at E108 position of Rad51 sensitizes the cells to MMS and renders them deficient in gene conversion. (A) Pictorial representation of return-to-growth assay upon MMS treatment. Cells were spotted after serial dilution of treated and untreated cells for wild-type and mutant strains. First lane for each strain shows untreated and second lane shows treated cells. (B) Graph showing the percentages of gene conversion. Cells were spread on galactose-containing plates and subsequently obtained colonies were patched on G418 sulfate plates. Percentage was determined by calculating the number of colonies grown on G418 sulfate plate versus number of colonies obtained on galactose plate. Error bars indicate SDs; n = 3; P values were calculated using the two-tailed Student’s t test. ****, P < 0.0001; N.S., not significant.
Rad51E108L can form homodimers and interacts efficiently with the Rad52 epistasis group of proteins.
It has been established that to execute the nuclear function, Rad51 interacts with itself. Also, Rad52 and Rad54 modulate the catalytic activity of Rad51 via direct physical interaction. We wanted to test whether Rad51E108L has any defect in self-association or association with Rad52 and Rad54. To that end, we used a yeast two-hybrid assay to measure the protein-protein interaction between Rad51E108L and the Rad52 epistasis group. Figure 5 (top) shows the results with wild-type Rad51, which acts as a positive control in our study. The bottom of Fig. 5 shows that Rad51E108L interacted efficiently with itself as well as with Rad52 and Rad54. We verified that Rad52 and Rad54 do not cause self-activation of a reporter gene (data not shown). No growth in a triple-drop-out plate for the strains PMY11 and PMY14 indicated that there was no self-activation for the indicative strains.
FIG 5.

Rad51E108L can form homodimers and bind efficiently to the Rad52 epistasis group of proteins. Yeast two-hybrid analysis depicting the interaction of RAD51/rad51 mutants with Rad52 epistasis group. Various strains harboring bait and prey vectors are represented on the left. Cells of each strain were grown to an OD600 of 0.5 and serially diluted before spotting. To monitor the interaction between proteins, diluted cells were spotted on medium lacking Leu and Ura (left panel) as well as on medium lacking Leu, Ura, and Ade. Homodimerization as well as interaction of Rad51 (positive control) (I) and Rad51E108L (II) with Rad52 and Rad54 was unaltered.
DISCUSSION
Rad51 protein, which facilitates homologous strand exchange, is the central player for HR in mammalian cells. Disruption of this gene is associated with embryonic lethality in mice (23). It is reported that haploinsufficiency of this gene is linked with defects in human neurodevelopment (24, 25). The Rad51 focus formation in response to DNA damage is one of the regulatory events in HR.
Previously, we established that besides providing stability to the Rad51 protein, Hsp90 also controls its nuclear function, i.e., DNA damage-induced focus formation. Taking that study further, we show that the dynamic interaction between Hsp90 and Rad51 can influence the nuclear function of Rad51. We are reporting for the first time that DNA damage triggers the dissociation of Rad51 and Hsp90, which could be a prerequisite for the nuclear function of Rad51. Due to a stronger association with Hsp90, the Rad51E108L protein probably remains locked with Hsp90; hence, the recruitment of Rad51E108L to the broken DNA ends, even at a very high MMS concentration (0.15%), is considerably defective. This is evident by 10.5% GC efficiency and complete loss of cell survivability in the E108L-rad51 mutant cells under DNA-damaging conditions. Thus, our study shows that there is a positive correlation between the extent of Hsp90-Rad51 dissociation after DNA damage and Rad51 nuclear activity. It appears that in the case of E108L-rad51, a major portion of the Hsp90 pool is associated with Rad51, which might result in an insufficient availability of free Hsp90 for other cellular functions. This is supported by our observation that the E108L-rad51 mutant strain showed a slow growth phenotype compared to that of the wild-type strain. However, it is possible that the constitutive form of yHsp90, namely, Hsc82, might be sufficient for the essential cellular function of Hsp90, ensuring the survivability of the mutant strain.
A defect in recruitment to the damaged DNA may result from a defect in DNA binding or defects in its interactions with other nuclear proteins. An earlier report showed that glycine at the 103rd position of Rad51 is crucial for DNA binding (26). Another report showed that valine at 328, proline at 339, and isoleucine at 345 are also involved in DNA binding (27). Although there is no report available regarding the DNA binding capacity of the mutant used in our study, we do not anticipate any defect in DNA binding, as the mutant was recruited to the chromatin DNA albeit at lesser extent, probably due to the lesser availability of free Rad51E108L proteins. In the case of the Rad51E108L mutant, despite its apparent defect in reversible dissociation from Hsp90 under DNA-damaging conditions, its 40% recruitment confirms that it is not defective in DNA binding.
In our study, we expressed RAD51 and E108L-rad51 from episomal plasmids in a Δrad51 background and compared their phenotypes. Thus, it is important to ensure that the observed phenotypes were not due to overexpression. In an earlier study, it was observed that overexpression of Rad51 does not have any effect on MMS sensitivity or repair of a single DSB in wild-type cells. However, it sensitizes Δsrs2 and Δku70 strains toward MMS (28). It was also observed that a high level of Rad51 reduces the frequency of but does not eliminate HR (28). In our study, the steady-state levels of Rad51WT and Rad51E108L were comparable. Thus, the severe DNA repair defects observed in the E108L-rad51 strain compared to that in RAD51 cells are not due to overexpression but rather to the point mutation.
It did not escape our notice that nearly 50% less recruitment of Rad51 in the E108L-rad51 strain had a profound effect on DNA repair. It is not unexpected, as our earlier study demonstrated that an only 20% reduction of Rad51 focus formation in the Δ211-259hsp82 strain led to severe sensitivity to MMS and UV treatment (14). These findings prompted us to conclude that 20% to 50% less occupancy of Rad51 at the broken DNA ends is sufficient to perturb DSB repair.
The E108 residue of Rad51 that is in close proximity to Hsp90 resides outside the ATPase domain of Rad51 and is evolutionary conserved. The N-terminal domain of Rad51 is implicated in the monomer-monomer interaction as well as the interactions with the members of the Rad52 epistasis group (27, 29). Although the mutation is present in the N-terminal domain, it was not previously identified in Rad51 interaction-deficient mutants (30). The yeast two-hybrid assay confirms that the ability of Rad51E108L for self-association as well as for associations with Rad52 and Rad54 are comparable to that of wild-type Rad51. As Rad51 recruitment to the broken DNA ends is an upstream event, the defect will be dominant over any other defects. Thus, the drastic phenotype found in the E108L-rad51 strain is likely to be one of the primary causes for the loss-of-function phenotype in the mutant strain.
It is known that Hsp90 shows a variable degree of association with its clients. Hsp90 clients such as kinases are primarily associated with Hsp90 through transient interactions, and once chaperoned, they are readily released from Hsp90 as functional proteins. On the other hand, clients such as steroid hormone receptors remain associated with Hsp90 to maintain their functional forms. Also, the extent of association between Hsp90 and its client can alter the cellular function of its client. For example, the single point mutations in the epidermal growth factor receptor (EGFRL858R) and B-Raf kinase (B-RafV600E) promote tumor formation. It was observed that these point mutants have enhanced levels of association with Hsp90 compared to those of their wild-type counterparts (31, 32). While binding with its clients, Hsp90 exhibits specificity toward the hydrophobic residues of proteins (33). The incorporation of leucine at the 108th position of Rad51 increases the hydrophobic stretch on Rad51 (107 to 113 amino acids). We speculate that such an increase in hydrophobicity might result in a tighter binding between Hsp90 and mutant Rad51 protein.
Collectively, our work establishes the importance of Hsp90 in the HR pathway, where it appears to regulate the stability and functions of Rad51. Increasing lines of evidence suggest that the functions of several DNA repair proteins, such as BRCA1, BRCA2, Chk1, DNA-PKcs, FANCA, and the Mre11/Rad50/NBS, are likely to be dependent on Hsp90 (34). A recent report showed that overexpression of Hsp90 leads to genomic instability through a negative regulation of the checkpoint kinase RAD53 (22). Our work along with these reports embarks on the relationship of Hsp90 with DNA repair. Currently, DNA repair along with the Hsp90 inhibitor is being targeted in many cancer studies. Understanding the detailed regulation of HR will be beneficial for further knowledge in the field.
There are many reports which show that in response to various signals, Hsp90/Hsp82 gets posttranslational modifications (PTMs), and such PTMs help the release of the client protein (35–37). Currently, it is not known whether such PTM of Hsp90 occurs due to MMS treatment and that causes the decrease in association between Rad51WT and Hsp90. It is also unclear how the stronger association between Rad51E108L and Hsp90 was not overcome during the DNA damage response (DDR). These questions are interesting but beyond the scope of this report, and future studies might unravel the mechanism underlying the dissociation of Rad51 from Hsp90 upon DNA damage.
MATERIALS AND METHODS
Plasmids.
The sequences of all the primers used in this paper are tabulated in Table 2. The RAD51 mutant (E108L-rad51) was cloned in 2µ yeast expression vector pTA (20) between the BamH1 and Pst1 restriction sites to generate the pTA-E108L-rad51 plasmid. pTA-RAD51 was used as a positive control in our study (20). Full-length RAD51 and E108L-rad51 were subcloned into prey vector pGADC1 and bait vector pGBDUC1 from pTA-RAD51 and pTA-E108L-rad51, respectively. Thus, the plasmids pGADC1/RAD51, pGBDUC1/RAD51, pGADC1/E108L-rad51, and pGBDUC1/E108L-rad51 were generated. Full-length RAD52 was amplified using the OSB330/OSB331 primer set and cloned into pGBDUC1 vector between EcoRI and SalI restriction sites to create the pGBDUC1/RAD52 plasmid. To generate the pGBDUC1/RAD54 plasmid, RAD54 was amplified using the OSB332/OSB333 primer set and cloned into pGBDUC1 vector between EcoRI and SalI restriction sites.
TABLE 2.
Primer list
| Primer | Sequence (5′→3′) | Purpose |
|---|---|---|
| OMKB90 | GGATCCATGTCTCAAGTTCAAGAAC | Forward primer to amplify full-length RAD51 |
| OMKB88 | CTGCAGCTACTCGTCTTCTTCTC | Reverse primer to amplify full-length RAD51 |
| OMKB149 | GTCGACCTCGTCTTCTTCTCTGG | Reverse primer used to clone E108L-rad51 into pET22b vector |
| OSB305 | CTCGGATCCATGTCTCAAGTTCAAGAACAAC | Forward primer used to amplify full-length rad51 mutants |
| OSB293 | GTCGTCGACCTCGTCTTCTTCTCTGGGG | Reverse primer used to amplify full-length rad51 mutants |
| OSB315 | AGTGGGCTTCACACTGCTTTGGCGGTAGCA | Forward primer to create rad51 E108L mutation |
| OSB314 | TCTGGGAGCATATGCTACCGCCAAAGCAGTG | Reverse primer to create rad51 E108L mutation |
| OSB278 | CATGCAAGGGCTCCCTAGC | Forward primer used to amplify URA3 region for ChIP |
| OSB279 | CAACCAATCGTAACCTTCATCT | Reverse primer used to amplify URA3 region for ChIP |
| OSB289 | GTTAGTTGAAGCATTAGGTCC | Forward primer used to confirm HO digestion |
| KanB1 | TGTACGGGCGACAGTCACAT | Reverse primer used to confirm HO digestion |
| OSB21 | GACGGATCCATGGCTAGTGAAACTTTTGAATTTC | Forward primer to amplify full-length hsp82 |
| OSB22 | CGGGTCGACCTAATCTACCTCTTCCATTTCGG | Reverse primer to amplify full-length hsp82 |
| OSB16 | TGACCAAACTACTTACAACTCC | Forward primer to amplify 307 bp of 3′ end of ACT1 |
| OSB14 | TTAGAAACACTTGTGGTGAACG | Reverse primer to amplify ACT1 |
| OSB330 | CATGAATTCATGAATGAAATTATGGATATCGATG | Forward primer to amplify RAD52 |
| OSB331 | CATGTCGACTCAAGTAGGCTTGCGTGCATG | Reverse primer to amplify RAD52 |
| OSB332 | CATGAATTCATGGCAAGACGCAGATTACC | Forward primer to amplify RAD54 |
| OSB333 | CATGTCGACTCAATGTGAAATATATTGAAATGC | Reverse primer to amplify RAD54 |
Site-directed mutagenesis.
Point mutations were introduced in RAD51 by using the splice overlap extension (SOE) PCR technique. A primer set was designed to incorporate the required mutation in RAD51 at the desired location. Yeast genomic DNA was used as a template, and the full-length gene was amplified in two segments in order to insert the point mutation. For amplifying the first and second segments to generate the E108L-rad51 mutation, primer sets OSB305/OSB314 and OSB315/OSB293 were used, respectively. Full-length RAD51 containing the E108L mutation was then amplified by using the first two segments along with primer set OMKB90/OMKB88. The rad51 mutant was then cloned into the pTA 2µ yeast expression vector using the sites BamH1 and PstI. After successful cloning, the pTA-E108L-rad51 construct was sequenced to confirm the desired mutation. To create the E108L-rad51 mutant, we changed the codon GAA to TTG.
Yeast strains.
The strains used in this study are tabulated in Table 3. LS402 Δrad51 was transformed with empty vector (pTA), pTA-RAD51, and pTA-E108L-rad51 to generate NRY1, NRY2, and TSY17, respectively. For the gene conversion assay, pTA-RAD51 and pTA-E108L-rad51 were transformed into NA14 Δrad51 (21) to generate TSY21 and TSY22. For a negative control, the NA14 Δrad51 strain was transformed with pTA empty vector to generate TSY20. To perform the yeast two-hybrid analysis, empty pGADC1 and pGBDUC1 vectors were transformed into a pJ694a parent strain to generate the PMY3 yeast strain. To study the interaction of wild-type Rad51 with itself and with Rad52 and Rad54 proteins, PMY8, PMY9, and PMY10 were created by transforming prey-RAD51 plus bait-RAD51, prey-RAD51 plus bait-RAD52, and prey-RAD51 plus bait-RAD54 constructs, respectively, into the pJ694a strain. Similarly, to study the interaction of Rad51E108L with itself and with Rad52 and Rad54, strains TSY10, PMY12, and PMY13 were generated by transforming prey-E108L-rad51 plus bait-E108L-rad51, prey-E108L-rad51 plus bait-RAD52, and prey-E108L-rad51 plus bait-RAD54 constructs, respectively. Strains PMY4, PMY7, PMY14, and PMY11 were utilized as controls. These strains were generated by transforming empty prey plus bait-RAD51, prey-RAD51 plus empty bait, empty prey plus bait-E108L-rad51, and prey-E108L-rad51 plus empty bait vectors, respectively, into the pJ694a strain.
TABLE 3.
Yeast strains
| Strain | Genotype | Source or reference |
|---|---|---|
| NRY1 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 [phi+] RAD51::LEU2 pTA | 20 |
| NRY2 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 [phi+] RAD51::LEU2 pTA-RAD51 | 20 |
| TSY17 | MATa leu2-3,112 trp1-1 can1-100 ura3-1 ade2-1 his3-11,15 [phi+] RAD51::LEU2 pTA-E108L-rad51 | This study |
| TSY20 | MATa inc ura3-HOcs lys2::ura3-HOcs-inc ade3::GALHO ade2-1 leu2-3,112 his3-11,15 trp1-1 can1-100 RAD51::LEU2 pTA | This study |
| TSY21 | MATa inc ura3-HOcs lys2::ura3-HOcs-inc ade3::GALHO ade2-1 leu2-3,112 his3-11,15 trp1-1 can1-100 RAD51::LEU2 pTA-RAD51 | This study |
| TSY22 | MATa inc ura3-HOcs lys2::ura3-HOcs-inc ade3::GALHO ade2-1 leu2-3,112 his3-11,15 trp1-1 can1-100 RAD51::LEU2 pTA-E108L-rad51 | This study |
| PMY3 | MATa trpl-901 leu2-3,112 ura3-52 his3-200 ga14Δ ga180Δ LYS2::GALl-HIS3 GAL2-ADE2 met2::GAL7-lacZ pGADC1 pGBDUC1 | This study |
| PMY8 | MATa trpl-901 leu2-3,112 ura3-52 his3-200 ga14Δ ga180Δ LYS2::GALl-HIS3 GAL2-ADE2 met2::GAL7-lacZ pGADC1/RAD51 pGBDUC1/RAD51 | This study |
| PMY9 | MATa trpl-901 leu2-3,112 ura3-52 his3-200 ga14Δ ga180Δ LYS2::GALl-HIS3 GAL2-ADE2 met2::GAL7-lacZ pGADC1/RAD51 pGBDUC1/RAD52 | This study |
| PMY10 | MATa trpl-901 leu2-3,112 ura3-52 his3-200 ga14Δ ga180Δ LYS2::GALl-HIS3 GAL2-ADE2 met2::GAL7-lacZ pGADC1/ScRAD51 pGBDUC1/RAD54 | This study |
| TSY10 | MATa trpl-901 leu2-3,112 ura3-52 his3-200 ga14Δ ga180Δ LYS2::GALl-HIS3 GAL2-ADE2 met2::GAL7-lacZ pGADC1/E108L-rad51 pGBDUC1/E108L-rad51 | This study |
| PMY12 | MATa trpl-901 leu2-3,112 ura3-52 his3-200 ga14Δ ga180Δ LYS2::GALl-HIS3 GAL2-ADE2 met2::GAL7-lacZ pGADC1/E108L-rad51 pGBDUC1/RAD52 | This study |
| PMY13 | MATa trpl-901 leu2-3,112 ura3-52 his3-200 ga14Δ ga180Δ LYS2::GALl-HIS3 GAL2-ADE2 met2::GAL7-lacZ pGADC1/E108L-rad51 pGBDUC1/RAD54 | This study |
| PMY4 | MATa trpl-901 leu2-3,112 ura3-52 his3-200 ga14Δ ga180Δ LYS2::GALl-HIS3 GAL2-ADE2 met2::GAL7-lacZ pGADC1 pGBDUC1/RAD51 | This study |
| PMY7 | MATa trpl-901 leu2-3,112 ura3-52 his3-200 ga14Δ ga180Δ LYS2::GALl-HIS3 GAL2-ADE2 met2::GAL7-lacZ pGADC1/RAD51 pGBDUC1 | This study |
| PMY14 | MATa trpl-901 leu2-3,112 ura3-52 his3-200 ga14Δ ga180Δ LYS2::GALl-HIS3 GAL2-ADE2 met2::GAL7-lacZ pGADC1 pGBDUC1/E108L-rad51 | This study |
| PMY11 | MATa trpl-901 leu2-3,112 ura3-52 his3-200 ga14Δ ga180Δ LYS2::GALl-HIS3 GAL2-ADE2 met2::GAL7-lacZ pGADC1/E108L-rad51 pGBDUC1 | This study |
Yeast two-hybrid analysis.
Yeast two hybrid analysis was performed as described earlier (20). The strains PMY3, PMY8, PMY9, PMY10, TSY10, PMY12, PMY13, PMY4, PMY7, PMY14, and PMY11 were grown in SC-Ura-Leu medium until logarithmic phase. They were then diluted serially as shown in Fig. 5 and spotted on SC-uracil (Ura)-Leu and SC-Ura-Leu-adenine (Ade) medium. The plates were kept at 30°C for 3 to 4 days. The strain PMY3 was used as the negative control in our study.
MMS sensitivity assay.
NRY1, NRY2, and TSY17 were tested for DNA damage sensitivity. All strains were grown in tryptophan dropout synthetic medium overnight at 30°C. The next day, a secondary culture was grown to an optical density at 600 nm (OD600) of 0.5 at 30°C. The culture was then divided into two sets. One set of cells was treated with 0.03% (vol/vol) of methyl methanesulfonate (MMS) (Sigma-Aldrich) and grown at 30°C for 2 h, and the other set was continuously grown at 30°C for 2 h without MMS. After that, the cells were serially diluted as mentioned, spotted on selective medium, and incubated at 30°C for 2 to 3 days.
Gene conversion assay.
TSY20, TSY21, and TSY22 strains were generated by transforming pTA (empty vector), pTA-RAD51 and pTA-E108L-rad51, respectively, into the NA14 Δrad51 strain. The transformed cells were initially patched on a plate containing glycerol as a sole carbon source. Next, equal numbers of cells were counted and spread on two different plates, one containing glycerol and other containing galactose as a carbon source, and incubated at 30°C for 3 to 5 days. Cells which survived on galactose plates were then patched on another plate containing G418 sulfate and incubated at 30°C for 36 h in order to determine the percentage gene conversion. Cells grown on G418 sulfate-containing plates utilize the Rad51-mediated gene conversion pathway for repair as they retain KANMX6. The ratio of the number of cells grown on the G418 sulfate plate to the number of cells grown on the galactose plate was calculated to determine the percent gene conversion. The assay was performed more than 3 times, and the mean values were plotted using GraphPad Prism.
Chromatin immunoprecipitation.
TSY21 and TSY22 were grown in the selective medium to an OD600 of 0.3 in the presence of 3% glycerol. Half of the batch of cells was then treated with 3% galactose for 3 h, and other half continued to grow in glycerol medium. The ChIP assay was performed as described earlier (38). One microgram anti-Rad51 antibody was added to the sample to precipitate Rad51-bound DNA fragments. Recruitment of Rad51 was then monitored by PCR with 30 cycles using primer set OSB278/OSB279 in a reaction mixture volume of 50 µl using the immunoprecipitate and input DNA samples. Samples were subjected to electrophoresis on 2% agarose. For control, ChIP was performed with rabbit IgG antibody. To verify whether a double-stranded break (DSB) was generated by HO digestion in the assay strain, we used OSB289 as a forward primer, which is complementary to the 20 bp upstream of HO cut site (HOcs), and a reverse primer (KanB1) which is complementary to the KANMX gene. We amplified full-length ACT1 using OSB14 and OSB16 as a normalization control.
Western blotting.
Western blottin was performed to check Rad51 levels in NRY1, NRY2, and TSY17 strains. Protein samples were loaded on an SDS polyacrylamide gel. A polyvinylidene difluoride (PVDF) membrane was used for the transfer as described earlier (39). The primary antibodies used were mouse anti-Act1 (Abcam), rabbit anti-Rad51 (Santa Cruz), and mouse anti-Hsp82 (Calbiochem) at 1:5,000 dilutions. For subcellular fractionation, we used anti-Pgk1 antibody (Novus Biologicals) and mouse anti-Nsp1 antibody (Abcam) at 1:3,000 and 1:5,000 dilutions, respectively. For secondary antibodies, horseradish peroxide-conjugated anti-rabbit antibody (Promega) and anti-mouse antibody (Santa Cruz Biotechnology Inc., CA, USA) were used at 1:10,000 dilutions. The Western blots were developed using a chemiluminescent detection system (Pierce). Every experiment was repeated at least 3 times, and band intensities were quantified by using Image J software. Mean relative densities were plotted using GraphPad prism.
Protein-protein docking.
The protein sequence of Rad51 with entry P25454 and the ATP-dependent molecular chaperone yHsp90 (Hsp82) with entry P02829 of Saccharomyces cerevisiae (strain ATCC 204508/S288c) are publicly available from the central repository of protein sequence and function, UniProt (Universal Protein Resource). The three-dimensional (3D) structures of Rad51 (PDB ID 1SZP) and yHsp90 (PDB ID 2CG9) were retrieved from the RCSB protein data bank. Protein-protein docking was conducted using a fully automated web-based program ClusPro 2.0, which employs an improved fast Fourier transform (FFT)-based rigid docking program PIPER. The program output is a short list of putative complexes ranked according to their clustering properties (18). Biovia Discovery Studio Visualizer is utilized for visualization and analysis of protein complexes. For mutation studies, the sequence of Rad51 protein retrieved from PDB (1SZP ABCDEF) was viewed in the sequence viewer of Biovia Discovery Studio software. The critical amino acids to be mutated were selected in all six chains and replaced. The sulfate ions were removed, and the structure of the protein generated was subjected to clean geometry and energy minimization before using for protein-protein docking. The amino acid Glu108 (E108) was mutated with four different amino acids, namely, leucine (E108L), alanine (E108A), glycine (E108G), and isoleucine (E108I), in chains A, B, C, D, E, and F to generate single mutant hexamers. The mutated Rad51 proteins were again subjected to protein-protein interaction with yHsp90 2CG9A. Protein-protein docking similar to that of the wild type was repeated with the mutant protein against Hsp90 using the online tool ClusPro.
Coimmunoprecipitation.
Wild-type and E108L-rad51 cells harboring yHsp90 overexpression plasmid (under GPD promoter; 2µ vector) were grown to an OD600 of 0.5. Ten milliliters of each culture was harvested, resuspended in 1 ml spheroplast buffer (50 mM Tris-HCl [pH 8], 25 mM HEPES [pH 7.4], 0.2% Casamino Acids, 0.2% yeast nitrogen base [YNB], 1% glucose, 18.2% sorbitol) containing dithiothreitol (DTT) and lyticase, and incubated at 30°C for 90 min. Subsequently, glass beads were added and the cells were intermittently vortexed and incubated on ice six times for a period of 30 s each. An anti-Rad51 antibody was added to the supernatant for overnight incubation at 4°C. Protein A agarose (25%; Calbiochem) was added, and the mixture was incubated for 2 h at room temperature. The beads were then spun down for 15 s at 1,000 rpm, and the pellet was washed 3 times with NETNS buffer (20 mM Tris-HCl [pH 8], 1 mM EDTA, 1 M NaCl, 0.5% [vol/vol] NP-40 with protease inhibitor) and twice with NETN buffer (20 mM Tris-HCl [pH 8], 1 mM EDTA, 100 mM NaCl, 0.5% [vol/vol] NP-40 with protease inhibitor). The bound protein was eluted with 4× Laemmli buffer by boiling for 10 min and was further spun down, and the supernatant was collected and used for Western blotting. The proteins in the supernatant were precipitated using 20% trichloroacetic acid, eluted using 4 × SDS loading dye containing dithiothreitol (DTT) and Tris (pH 8.8), and boiled for 10 min. The sample was spun down and the proteins in the supernatant were used for Western blotting. After the coimmunoprecipitation, the relative association of Hsp90 with Rad51 was calculated for each experiment using the following formula: relative association of Hsp90 with Rad51 = (Hsp90 in the pellet/Hsp90 in the input) ÷ (Rad51 in the pellet/Rad51 in the input).
ACKNOWLEDGMENTS
The work was supported by the Council of Scientific and Industrial Research (India), [37(1669)/16/EMR-II] to S.B. T.S. and W.T. were supported by senior research fellowships from the University Grants Commission, India, and N.F. was supported by a junior research fellowship from the Council for Scientific and Industrial Research (CSIR), India. P.M. was supported by an MSc thesis grant, Department of Biotechnology, India.
We declare no conflict of interest.
REFERENCES
- 1.Polo SE, Jackson SP. 2011. Dynamics of DNA damage response proteins at DNA breaks: a focus on protein modifications. Genes Dev 25:409–433. doi: 10.1101/gad.2021311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tembe V, Henderson BR. 2007. Protein trafficking in response to DNA damage. Cell Signal 19:1113–1120. doi: 10.1016/j.cellsig.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 3.Reynolds P, Botchway SW, Parker AW, O’Neill P. 2013. Spatiotemporal dynamics of DNA repair proteins following laser microbeam induced DNA damage–when is a DSB not a DSB? Mutat Res 756:14–20. doi: 10.1016/j.mrgentox.2013.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ciccia A, Elledge SJ. 2010. The DNA damage response: making it safe to play with knives. Mol Cell 40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, Liu X, Jasin M, Couch FJ, Livingston DM. 2006. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell 22:719–729. doi: 10.1016/j.molcel.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 6.Sy SMH, Huen MSY, Chen J. 2009. PALB2 is an integral component of the BRCA complex required for homologous recombination repair. Proc Natl Acad Sci U S A 106:7155–7160. doi: 10.1073/pnas.0811159106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang F, Ma J, Wu J, Ye L, Cai H, Xia B, Yu X. 2009. PALB2 links BRCA1 and BRCA2 in the DNA-damage response. Curr Biol 19:524–529. doi: 10.1016/j.cub.2009.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang F, Fan Q, Ren K, Andreassen PR. 2009. PALB2 functionally connects the breast cancer susceptibility proteins BRCA1 and BRCA2. Mol Cancer Res 7:1110–1118. doi: 10.1158/1541-7786.MCR-09-0123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen RB, Carreira A, Kowalczykowski SC. 2010. Purified human BRCA2 stimulates RAD51-mediated recombination. Nature 467:678–683. doi: 10.1038/nature09399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu J, Doty T, Gibson B, Heyer WD. 2010. Human BRCA2 protein promotes RAD51 filament formation on RPA-covered single-stranded DNA. Nat Struct Mol Biol 17:1260–1262. doi: 10.1038/nsmb.1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thorslund T, McIlwraith MJ, Compton SA, Lekomtsev S, Petronczki M, Griffith JD, West SC. 2010. The breast cancer tumor suppressor BRCA2 promotes the specific targeting of RAD51 to single-stranded DNA. Nat Struct Mol Biol 17:1263–1265. doi: 10.1038/nsmb.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shinohara A, Ogawa T. 1998. Stimulation by Rad52 of yeast Rad51-mediated recombination. Nature 391:404–407. doi: 10.1038/34943. [DOI] [PubMed] [Google Scholar]
- 13.Song B, Sung P. 2000. Functional interactions among yeast Rad51 recombinase, Rad52 mediator, and replication protein A in DNA strand exchange. J Biol Chem 275:15895–15904. doi: 10.1074/jbc.M910244199. [DOI] [PubMed] [Google Scholar]
- 14.Suhane T, Laskar S, Advani S, Roy N, Varunan S, Bhattacharyya D, Bhattacharyya S, Bhattacharyya MK. 2015. Both charged linker region and ATPase domain of Hsp90 are essential for Rad51-dependent DNA repair. Eukaryot Cell 14:64–77. doi: 10.1128/EC.00159-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pratt WB, Toft DO. 2003. Regulation of signaling protein function and trafficking by the hsp90/hsp70-based chaperone machinery 1. Exp Biol Med (Maywood) 228:111–133. doi: 10.1177/153537020322800201. [DOI] [PubMed] [Google Scholar]
- 16.Cato L, Neeb A, Brown M, Cato AC. 2014. Control of steroid receptor dynamics and function by genomic actions of the cochaperones p23 and Bag-1L. Nucl Recept Signal 12:e005. doi: 10.1621/nrs.12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jahn M, Rehn A, Pelz B, Hellenkamp B, Richter K, Rief M, Buchner J, Hugel T. 2014. The charged linker of the molecular chaperone Hsp90 modulates domain contacts and biological function. Proc Natl Acad Sci U S A 111:17881–17886. doi: 10.1073/pnas.1414073111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Comeau SR, Gatchell DW, Vajda S, Camacho CJ. 2004. ClusPro: a fully automated algorithm for protein–protein docking. Nucleic Acids Res 32:W96–W99. doi: 10.1093/nar/gkh354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozakov D, Brenke R, Comeau SR, Vajda S. 2006. PIPER: an FFT‐based protein docking program with pairwise potentials. Proteins 65:392–406. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- 20.Roy N, Bhattacharyya S, Chakrabarty S, Laskar S, Babu SM, Bhattacharyya MK. 2014. Dominant negative mutant of Plasmodium Rad51 causes reduced parasite burden in host by abrogating DNA double‐strand break repair. Mol Microbiol 94:353–366. doi: 10.1111/mmi.12762. [DOI] [PubMed] [Google Scholar]
- 21.Agmon N, Pur S, Liefshitz B, Kupiec M. 2009. Analysis of repair mechanism choice during homologous recombination. Nucleic Acids Res 37:5081–5092. doi: 10.1093/nar/gkp495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khurana N, Laskar S, Bhattacharyya MK, Bhattacharyya S. 2016. Hsp90 induces increased genomic instability toward DNA-damaging agents by tuning down RAD53 transcription. Mol Biol Cell 27:2463–2478. doi: 10.1091/mbc.E15-12-0867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tsuzuki T, Fujii Y, Sakumi K, Tominaga Y, Nakao K, Sekiguchi M, Matsushiro A, Yoshimura Y, Morita T. 1996. Targeted disruption of the Rad51 gene leads to lethality in embryonic mice. Proc Natl Acad Sci U S A 93:6236–6240. doi: 10.1073/pnas.93.13.6236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Depienne C, Bouteiller D, Méneret A, Billot S, Groppa S, Klebe S, Charbonnier-Beaupel F, Corvol JC, Saraiva JP, Brueggemann N, Bhatia K, Cincotta M, Brochard V, Flamand-Roze C, Carpentier W, Meunier S, Marie Y, Gaussen M, Stevanin G, Wehrle R, Vidailhet M, Klein C, Dusart I, Brice A, Roze E. 2012. RAD51 haploinsufficiency causes congenital mirror movements in humans. Am J Hum Genet 90:301–307. doi: 10.1016/j.ajhg.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gallea C, Popa T, Hubsch C, Valabregue R, Brochard V, Kundu P, Schmitt B, Bardinet E, Bertasi E, Flamand-Roze C, Alexandre N, Delmaire C, Méneret A, Depienne C, Poupon C, Hertz-Pannier L, Cincotta M, Vidailhet M, Lehericy S, Meunier S, Roze E. 2013. RAD51 deficiency disrupts the corticospinal lateralization of motor control. Brain 136:3333–3346. doi: 10.1093/brain/awt258. [DOI] [PubMed] [Google Scholar]
- 26.Zhang XP, Lee KI, Solinger JA, Kiianitsa K, Heyer WD. 2005. Gly-103 in the N-terminal domain of Saccharomyces cerevisiae Rad51 protein is critical for DNA binding. J Biol Chem 280:26303–26311. doi: 10.1074/jbc.M503244200. [DOI] [PubMed] [Google Scholar]
- 27.Fortin GS, Symington LS. 2002. Mutations in yeast Rad51 that partially bypass the requirement for Rad55 and Rad57 in DNA repair by increasing the stability of Rad51-DNA complexes. EMBO J 21:3160–3170. doi: 10.1093/emboj/cdf293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paffett KS, Clikeman JA, Palmer S, Nickoloff JA. 2005. Overexpression of Rad51 inhibits double-strand break-induced homologous recombination but does not affect gene conversion tract lengths. DNA Repair (Amst) 4:687–698. doi: 10.1016/j.dnarep.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 29.Donovan JW, Milne GT, Weaver DT. 1994. Homotypic and heterotypic protein associations control Rad51 function in double-strand break repair. Genes Dev 8:2552–2562. doi: 10.1101/gad.8.21.2552. [DOI] [PubMed] [Google Scholar]
- 30.Krejci L, Damborsky J, Thomsen B, Duno M, Bendixen C. 2001. Molecular dissection of interactions between Rad51 and members of the recombination-repair group. Mol Cell Biol 21:966–976. doi: 10.1128/MCB.21.3.966-976.2001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Grbovic OM, Basso AD, Sawai A, Ye Q, Friedlander P, Solit D, Rosen N. 2006. V600E B-Raf requires the Hsp90 chaperone for stability and is degraded in response to Hsp90 inhibitors. Proc Natl Acad Sci U S A 103:57–62. doi: 10.1073/pnas.0609973103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shimamura T, Lowell AM, Engelman JA, Shapiro GI. 2005. Epidermal growth factor receptors harboring kinase domain mutations associate with the heat shock protein 90 chaperone and are destabilized following exposure to geldanamycins. Cancer Res 65:6401–6408. doi: 10.1158/0008-5472.CAN-05-0933. [DOI] [PubMed] [Google Scholar]
- 33.Karagöz GE, Rüdiger SG. 2015. Hsp90 interaction with clients. Trends Biochem Sci 40:117–125. doi: 10.1016/j.tibs.2014.12.002. [DOI] [PubMed] [Google Scholar]
- 34.Pennisi R, Ascenzi P, di Masi A. 2015. Hsp90: a new player in DNA repair? Biomolecules 5:2589–2618. doi: 10.3390/biom5042589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kekatpure VD, Dannenberg AJ, Subbaramaiah K. 2009. HDAC6 modulates Hsp90 chaperone activity and regulates activation of aryl hydrocarbon receptor signaling. J Biol Chem 284:7436–7445. doi: 10.1074/jbc.M808999200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Kovacs JJ, Murphy PJ, Gaillard S, Zhao X, Wu JT, Nicchitta CV, Yoshida M, Toft DO, Pratt WB, Yao TP. 2005. HDAC6 regulates Hsp90 acetylation and chaperone-dependent activation of glucocorticoid receptor. Mol Cell 18:601–607. doi: 10.1016/j.molcel.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 37.Robbins N, Leach MD, Cowen LE. 2012. Lysine deacetylases Hda1 and Rpd3 regulate Hsp90 function thereby governing fungal drug resistance. Cell Rep 2:878–888. doi: 10.1016/j.celrep.2012.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laskar S, Sheeba K, Bhattacharyya MK, Nair AS, Dhar P, Bhattacharyya S. 2015. Heat stress-induced Cup9-dependent transcriptional regulation of SIR2. Mol Cell Biol 35:437–450. doi: 10.1128/MCB.01046-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Laskar S, Bhattacharyya MK, Shankar R, Bhattacharyya S. 2011. HSP90 controls SIR2 mediated gene silencing. PLoS One 6:e23406. doi: 10.1371/journal.pone.0023406. [DOI] [PMC free article] [PubMed] [Google Scholar]