Abstract
The antioxidant defense system acts to maintain the equilibrium between the production of reactive oxygen species (ROS) and the elimination of toxic levels of ROS in plants. Overproduction and accumulation of ROS results in metabolic disorders and can lead to the oxidative destruction of the cell. Several stress factors cause ROS overproduction and trigger oxidative stress in crops and weeds. Recently, the involvement of the antioxidant system in weed interference and herbicide treatment in crops and weeds has been the subject of investigation. In this review, we address ROS production and plant mechanisms of defense, alterations in the antioxidant system at transcriptional and enzymatic levels in crops induced by weed interference, and herbicide exposure in crops and weeds. We also describe the mechanisms of action in herbicides that lead to ROS generation in target plants. Lastly, we discuss the relations between antioxidant systems and weed biology and evolution, as well as the interactive effects of herbicide treatment on these factors.
Keywords: reactive oxygen species (ROS), oxidative stress, herbicide treatment, herbicide resistance, weed evolution
1. Introduction
In the field, plant growth, development, and reproduction are affected to various degrees by biotic and abiotic factors. Plant stress is most often the result of combinations of biotic and abiotic factors; interactions are complex but need to be understood since they affect plant performance [1]. Stress conditions often induce the overproduction of reactive oxygen species (ROS), which are toxic molecules that can lead to the oxidative destruction of cells. Plants have cellular antioxidant machinery, including enzymatic and non-enzymatic antioxidants that control ROS levels and maintain cellular homeostasis [2,3,4].
The enzymatic and non-enzymatic responses to abiotic stress are widely studied in crops. One gap in our knowledge is the dynamic effects of weed interference on crops and changes rendered by herbicide exposure, especially at the molecular level. Antioxidant response to stress caused by weed interference and herbicide treatment in crops and weeds have been recent subjects of investigation [5,6,7,8,9,10,11,12,13,14,15]. Both weed interference and herbicide treatment stimulate the synthesis of antioxidant molecules in plants in response to stress [10,11,15,16]. Alterations in enzymatic antioxidants such as superoxide dismutase, catalase, ascorbate peroxidase, and peroxidase have been documented in crops and weeds in response to weed interference [11,13,14,17] and herbicide exposure [1,5,18,19,20] together with the synthesis of ROS and causing lipid peroxidation. Also, antioxidant-related gene expression and protein synthesis appear to be responsive to those stressors [8,9,12,16].
The increase in the synthesis of several antioxidant defense compounds by crops in response to weed interference could result in crop energy drains and yield losses. Weeds have a high genetic variation that allows them to survive under different stress conditions [21]. In this way, the evolution of herbicide resistance in weeds is a threat to sustainable agriculture, and their mechanisms have been widely studied. In this review, we focus on ROS production in crops under weed interference and in both crops and weeds after herbicide exposure, as well as antioxidant defense systems against ROS. Also, we discuss molecular approaches that may be used to understand responses to ROS better and increase plant adaptation.
2. The Basis of ROS Production and Plant Mechanisms of Defense
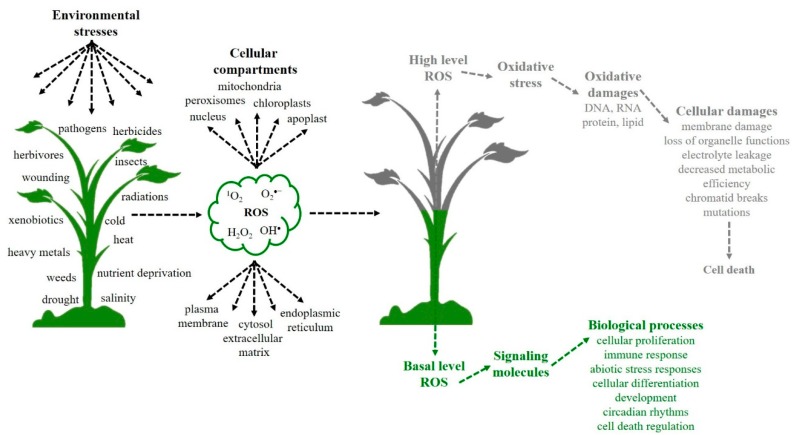
ROS such as singlet oxygen (1O2), superoxide radical (O2•−), hydrogen peroxide (H2O2), and hydroxyl radical (OH•) are partially reduced or excited forms of atmospheric oxygen and considered to be toxic molecules [3]. These molecules have different levels of reactivity, sites of production, and the potential to cross biological membranes [3]. ROS are mainly formed in organelles with high electron flux such as chloroplasts, mitochondria, and peroxisomes [3,4], but also in plant cells in the plasma membrane, cell wall, cytosol, apoplast, endoplasmic reticulum, nucleus, and extracellular matrix (Figure 1) [3,4,22].
Figure 1.
Environmental stressors that can lead to increased generation of reactive oxygen species (ROS) in different cell compartments and the biological consequences. Environmental stressors result in ROS production in different cellular compartments. High-level ROS production results in oxidative stress, oxidative and cellular damages, and even cell death. The basal level of ROS production serves as a signaling molecule and may be involved in various important biological processes.
In plants, ROS at a basal cellular level is important for signaling molecules capable of regulating diverse metabolic pathways [3,4,23]. Also, these molecules work by activating and controlling gene expression in response to stress [24] and are involved in critical plant biological processes like photosynthesis [2,3]. Thus, the basal level of ROS in cells is essential for plant life [3] and cellular processes. However, high concentrations of ROS can be fatal [2,4,22,25].
The stress generated by biotic and abiotic factors generally induces the overproduction of ROS [2,26]. In response to ROS overproduction, the plant’ defense system acts to scavenge toxic molecules and restore cellular homeostasis. However, when ROS overproduction exceeds the capacity of the antioxidant scavenging system, ROS accumulation occurs [3]. The over-accumulated ROS in plant cells results in lipidic peroxidation (LPO) and membrane damage [2]. LPO is a complex process, which is initiated by hydrogen abstraction or the addition of an oxygen radical. This reaction results in oxidative damage of polyunsaturated fatty acids (PUFA). The initiation step involves transition metal complexes such as Fe and Cu. The O2•− and H2O2 can initiate the reactions. However, OH• is sufficiently reactive and can initiate LPO by the abstraction of a hydrogen atom, in an unsaturated fatty acyl chain of a PUFA residue. In aerobic conditions, fatty acids can be oxygenated to give rise to a lipid peroxyl group (ROO•), which can then be propagated via a peroxidation chain reaction by abstracting a hydrogen atom from adjacent PUFA side chains. The resulting lipid hydroperoxide can decompose into several reactive products such as aldehydes (malonyl dialdehyde), alkanes, lipid alkoxyl radicals, lipid epoxides, and alcohols [2]. Therefore, LPO is considered as the most destructive process known to occur in living organisms altering membrane fluidity and permeability, damaging membrane proteins, inactivating receptors as well as other enzymes and ion channels [2,4,27]. After LPO, there can be damage to nucleic acids including chromatid breaks and other mutations, loss of organelle function, reduction in metabolic efficiency, electrolyte leakage, and subsequently, cell death [2,4] (Figure 1). If plants can survive to LPO, a decrease in crop biomass yield is almost always certain [2,3,4,25].
The ROS scavenging antioxidant-defense machinery includes enzymatic and non-enzymatic components which serve to balance the production and detoxification of ROS (Figure 2) [2].
Figure 2.
Key enzymatic and non-enzymatic antioxidants and the reactions catalyzed. (a) When ROS is overproduced, the enzymatic and non-enzymatic antioxidants act to scavenge the toxic molecules and restore the redox balance. (b) Reactions catalyzed by enzymatic antioxidants as well as the functions of the non-enzymatic antioxidants are shown.
Enzymatic antioxidants include superoxide dismutase (SOD), catalase (CAT), ascorbate peroxidase (APX), glutathione peroxidase (GPX), peroxidase (POD), peroxiredoxin (PRX), monodehydroascorbate reductase (MDHAR), dehydroascorbate reductase (DHAR), glutathione reductase (GR), and glutathione-S-transferase (GST) [2,4,22]. SODs are enzymes that catalyze the dismutation of O2•− to H2O2. Thus, SODs enzymes are at the frontline in the defense against ROS. CAT enzymes catalyze the dismutation of two H2O2 molecules to water and O2. These enzymes are localized mainly in peroxisomes. APXs play a key role catalyzing the conversion of H2O2 into H2O using the ascorbate as a specific electron donor. APXs are distributed in chloroplasts, mitochondria, peroxisomes, and the cytosol, coding different isoforms. GPXs catalyze the reduction of H2O2 or organic hydroperoxides to H2O. GPXs are nonheme thiol peroxidases and in plants are localized to mitochondria, chloroplasts, and cytosol. PODs are involved in H2O2 detoxification. Also, PODs can perform a second cyclic reaction, the hydroxylic reaction, this is distinct from the peroxidative reaction. Further, these enzymes are involved in the biosynthesis of lignin and defense against biotic stresses by consuming H2O2. MDHARs catalyze the regeneration of ascorbic acid (ASC) from the monodehydroascorbate radical using NAD(P)H as an electron donor. Thus, MDHARs play an essential role in the antioxidant system to maintain the ascorbate pool. DHARs are thiol enzymes that maintain the ascorbate in reduced form. DHARs catalyze the reduction of dehydroascorbate to ascorbate using glutathione (GSH) as a reducing substrate. GR enzymes catalyze the reduction of oxidized glutathione (GSSG) to reduced GSH. GRs are a NAD(P)H-dependent enzyme that protects cells against oxidative damage [2,4,25,26,27,28]. Also, GSTs are isozymes known to protect cells against chemical-induced toxicity. These enzymes catalyze the conjugation of GSH to a variety of electrophilic and hydrophobic substrates [29]. Accordingly, the action of the cellular antioxidant machinery is essential to control excess ROS to protect plant cells from oxidative damage and to restore the redox homeostasis.
Non-enzymatic antioxidants include ASC, glutathione, tocopherol, flavonoids, proline, and phenolic compounds [2,4]. It is important to highlight the role played by non-enzymatic compounds, that are substances found in all cellular compartments and may act directly in the detoxification of ROS and radicals or reduce substrates for antioxidant enzymes [2,3]. ASC is the most abundant, powerful, and water-soluble antioxidant which acts in all tissues, serving as a scavenger for O2•−, OH•, and 1O2, and can reduce H2O2 to H2O via the APX reaction. Glutathione is considered a powerful intracellular antioxidant localized in all cell compartments [2]. It is a potential scavenger of 1O2, H2O2, and OH•. GSH acts in regenerating ASC via the ASC-GSH cycle; these have redox potential to interact with numerous components and pathways [2,4,26]. Also, carotenoids serve to quench 1O2 and flavonoids as ROS scavengers [2]. Tocopherols serve to protect the membrane from lipid peroxidation, detoxify lipid peroxides, and quench 1O2 [2]. Proline is an osmoprotectant that has been considered to be a powerful antioxidant [4]. Proline can act as an inhibitor of lipid peroxidation and ROS scavenger [2,4]. Phenolic compounds play roles as antioxidants through their ability to donate electrons or hydrogen atoms to ROS [2,4]. Thus, the widespread biochemical activity of non-enzymatic antioxidants can prevent, reduce, or eliminate ROS damage in different plant tissues and cell compartments.
3. Modulation of Enzymes and Antioxidant Genes Induced by Weed Interference and Herbicide Exposure
Plants are immobile organisms that are adapted to cope with suites of environmental stresses present in their native environments. Crops retain some adaptations to deal with stress, but cultivation and crops are largely artificial constructs. One prominent stress in crops comes from weed interference and then also the subsequent application of herbicides to control weeds in modern agriculture. Certainly, these stresses are not completely understood, but play critical roles in crop management, crop development, and reproduction, and often lead to yield loss and reduced farmer profits [26].
3.1. Crop and Weed Interference
Weed interference is the most important biotic stress in crop production [30,31]. The level of interference varies with the crop and weed species, growing season, plant spatial distribution and density, the period of coexistence, edaphoclimatic conditions, and potential allelopathy [30,31,32]. Competition is the most common type of interference exerted from weeds on crops, which is established since at least one environmental resource is limited [31]. Competition can occur among plants of different species (interspecific) and the same species (intraspecific—e.g., high crop density). When the competition is established, the species with higher competitive capacity and ability get an advantage to access the environmental resources and grow faster than others, resulting in yield losses to those less competitive [30,32].
Studies have demonstrated alterations in ROS production in crops as a response to weed interference, as well as changes in enzymatic activity and gene expression of antioxidant components [7,11,13,14,33]. Current research is needed to tie weed interference with crop oxidative stress more closely. Especially important is quantifying the dynamic expression of antioxidant genes as well as changes in enzyme activity and compounds as a means of coping with ROS. Finally, it is important to assess the energetics of crops as they cope with oxidative stress and the effects on yield.
Despite documented alterations in oxidative stress and antioxidant systems, the literature remains divided on the response of crops to weed interference. In soybean and bean, Piasecki et al. [13,14] showed that H2O2 levels, APX, and CAT activity decreased with the increase in volunteer corn density, while SOD activity increased commensurate with weed density. Another study documented that the interference of wild poinsettia (Euphorbia heterophylla) on soybean did not result in any cellular damage or change in SOD, CAT, and APX enzyme activity in soybean [34]. Darmanti et al. [10] studied soybean under purple nutsedge (Cyperus rotundus L.) competition and found reduced activities of SOD, CAT, and APX in soybean.
On the other hand, studies conducted with soybean and wheat under the interference of Italian ryegrass (Lolium multiflorum) resulted in oxidative damage and enhanced of the activity of SOD, CAT, and APX enzyme [11,17]. The interference of ryegrass with maize seedlings increased the H2O2 level and the antioxidant gene expression of ZmGST1, ZmSOD2, ZmAPX2, and ZmCAT3 [7]. These same authors suggested that the changes which occurred in response to weed interference resulted in a physiological cost to the crop, which contributes to yield loss. Gal et al. [33] studied the interference of ryegrass on soybean and found an increase in H2O2 content and LPO with a concomitant reduction in flavonoid content in soybean. Also, the transcript levels of the antioxidant genes apx, cat, sod, and gpx increased [33], demonstrating that biochemical and molecular mechanisms were altered in soybean under weed interference.
After stress signals emanate from weed interferences, crop plants activate their antioxidant defense mechanisms that deal with ROS and restore cellular homeostasis [13,14,33]; these defenses certainly have energetic costs vis-à-vis yield. For example, soybean yield is decreased when volunteer maize is a competitor [13]. Although some studies show the involvement of the crop antioxidant system with weed interference, specific details on how it is initiated are lacking and might be related to light conditions [13] and allelopathic compounds exuded from weeds [35]. A consequence of far-red-enriched (FR-E) light is the generation of ROS [7]. Therefore, it is proposed that the FR-E light reflected from neighboring weeds increases the production of 1O2 which initiates the formation of H2O2 via ascorbate and disrupts thiol-modulated chloroplast enzymes. This triggers a physiological event that impacts both photosynthesis and carbon partitioning [36]. Allelochemicals stimulate the production of ROS by blocking the electron-carrying chain: electrons become free and react easily with O2 to form superoxide [37]. Thus, triggering ROS production and activation of the antioxidant-mediated defense [35] may result in damage to DNA, proteins and cellular membranes. In maize, allelochemical stress was applied by treatment with walnut husk wash water, which possesses allelopathy and phytotoxic effects. The treatment increased H2O2 content and changed the activity of CAT, SOD, and APX enzyme in maize. In this way, CAT activity increased by 85% in maize roots after 3 h [38]. Furthermore, 4-day juglone treatment (allelochemical) stimulated the expression of the glutathione transferase (GstI) gene in maize seedlings [39].
Studies of crops under oxidative stress caused by weed interference generally do not assess the yield components and yield. Thus, the understanding of the effective costs for the crop yield as a response to weed interference is limited with regards to ROS. Multiple stresses, ROS dynamics, and crop responses and defenses need to be better understood in relevant field studies as crops proceed from seed to seed. Modern management practices, including herbicide and other pesticide treatments, should also be factored into the stress–yield equation.
3.2. Herbicide Treatment
Herbicides, by definition, cause abiotic stress on plants. Globally, herbicides are the predominant method for controlling weeds in modern crop production, contributing to protecting the crop yield and economic profit [40]. Despite their inherent selectivity mechanisms that facilitate crop production, herbicides can cause some phytotoxicity to crop plants and cause reductions in leaf area index (LAI), shoot dry weight (SDW), plant height, and alterations in plant metabolism by generating ROS [41]. Most of the perturbations caused by herbicide treatment in plants are related to ROS generation and consequent oxidative stress [41].
Numerous herbicide modes of action (MOA) have been commercialized for application to agricultural fields. The herbicide MOA is the physiological and biochemical process (step-by-step) related to herbicide treatment. Each herbicide MOA has a specific target site (TS) which is referred to as a mechanism of action (MA). The target site herbicide is usually an enzyme/protein which is inhibited by herbicides at the molecular level. After herbicide TS inhibition in susceptible plants, at least a vital biological process is interrupted causing a sequence of secondary effects which ultimately leads to plant death [41]. For example, in susceptible plants, glyphosate MA is the inhibition of the enzyme 5-enolpyruvylshikimate 3-phosphate synthase (EPSPS). On the other hand, glyphosate MOA involves processes which occur after EPSPS inhibition, such as the inhibition of the the shikimic acid pathway and biosynthesis of aromatic amino acids (phenylalanine, tyrosine, and tryptophan), accumulation of shikimic acid and reducing power (NADPH+H), ROS generation, oxidative stress, and susceptible plant death from seven to 15 days after treatment [9,42,43].
The MOA for some herbicides induces the generation of ROS in plants as secondary effects after the specific TS is sufficiently inhibited. In this case, the oxidative stress generated is responsible for an important part of cellular and tissue damage. For example, glyphosate action produces ROS as a secondary consequence of the inhibition of the shikimic acid pathway. After the blockage of the shikimic acid pathway, occurs the reducing power accumulation in the chloroplasts. Also, the lack of tyrosine inhibits the synthesis of plastoquinone, which is an electron acceptor in the photosynthetic electron transport chain in the photosystem II (PSII). The non-regeneration of plastoquinone in the PSII interrupts the electron transport, leading to an energy accumulation [41]. Therefore, both processes, reducing power accumulation and PSII blockage, lead to ROS production, cell damage, and plant death [41].
The summary of the major MA according to the Herbicide Resistance Action Committee (HRAC) and whether their actions lead to ROS production in some action step [41] are presented in Table 1. From 21 known groups of herbicides classified by the mode of action described in Table 1, 15, i.e., 71%, causes ROS overproduction after their target site inhibition. Nine of the 15 MA groups (~60%) kill plants by direct ROS production (C1, C2, C3, D, E, F1, F2, F3, and H), whereas the other six, i.e., ~40%, cause ROS production as a secondary effect (B, G, I, M, O, and P) (Table 1). Only six MA herbicides (~29%) (A, K1, K2, K3, L, and N) are not documented to produce oxidative stress in any phase of their action (Table 1) [41]. Also, the use of herbicide-active ingredients from this last group is lower when compared to A, B, and G groups (ACCase, ALS, and EPSPS, respectively) that are the most used herbicides [44].
Table 1.
Description of the herbicide mechanisms of action (MA) and their effects on treated plants related to the production of reactive oxygen species.
| HRAC Group a | Herbicide Mechanism of Action (MA) | Biological Process Committed | Herbicide Chemical Family | Herbicide Active Ingredient b | ROS Production c |
|---|---|---|---|---|---|
| A | Inhibition of acetyl-CoA carboxylase (ACCase) | Fatty acid biosynthesis | Aryloxyphenoxy-propionate “FOPS”, Cyclohexanedione “DIMs”, Phenylpyrazoline “DEN” | benzoylprop-ethyl, diclofop-methyl, haloxyfop-methyl, cyhalofop, clethodim, setoxydim, pinoxaden | No |
| B | Inhibition of acetohydroxyacid synthase (AHAS, ALS) | Amino acid biosynthesis (Leu, Ile, Val) | Sulfonylurea, Imidazolinone, Triazolopyrimidine, Pyrimidinyl(thio)benzoate | metsulfuron-methyl, chlorimuron-ethyl, nicosulfuron, imazapyr, imazethapyr, flumetsulam, cloransulam-methyl, diclosulam, flucarbazone-sodium, pyritiobac | Yes |
| C1, C2, C3 | Inhibition of photosystem II protein D1 (psbA) | Photosynthesis (electron transfer) | Triazine, Triazinone, Triazolinone, Uracil, Pyridazinone, Phenyl-carbamate, Urea, Amide, Nitrile, Benzothiadiazinone, Phenyl-pyridazine | ametryne, atrazine, simazine, hexazinone, metribuzin, amicarbazone, bromacil, pyrazon, desmedipham, chlorotoluron, diuron, linuron, propanil, bromoxynil, ioxynil, bentazon, pyridate | Yes |
| D | Diversion of the electrons transferred by the photosystem I ferredoxin (Fd) | Photosynthesis (electron transfer) | Bipyridylium | diquat, paraquat | Yes |
| E | Inhibition of protoporphyrinogen oxidase (PPO) | Photosynthesis (heme biosyn- thesis for chlorophyll) | Diphenylether, Phenylpyrazole, N-phenylphthalimide, Thiadiazole, Oxadiazole, Triazolinone, Oxazolidinedione, Pyrimidindione | acifluorfen-Na, fomesafen, lactofen, oxyfluorfen, pyraflufen-ethyl, flumioxazin, fluthiacet-methyl, oxadiazon, azafenidin, pentoxazone, butafenacil, flufenpyr-ethyl | Yes |
| F1, F2, F3 | Inhibition of phytoene desaturase (PDS) or 4-hydroxyphenylpyruvate dioxygenase (4-HPPD) or of an unknown protein | Photosynthesis (carotenoid biosynthesis) | Pyridazinone, Pyridinecarboxamide, Triketone, Isoxazole, Pyrazole, Triazole, Isoxazolidinone, Urea, Diphenylether | norflurazon, diflufenican, fluridone, mesotrione, isoxaflutole, pyrazoxyfen, amitrole, clomazone, fluometuron, aclonifen | Yes |
| G | Inhibition of 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) | Amino acid biosynthesis (Phe, Trp, Tyr) | Glycine | glyphosate | Yes |
| H | Inhibition of glutamine synthase | Amino acid biosynthesis (Gln) | Phosphinic acid | glufosinate-ammonium | Yes |
| I | Inhibition of dihydropteroate synthase | Tetrahydrofolate biosynthesis | Carbamate | asulam | Yes |
| K1, K2 | Enhancement of tubulin depolymerization | Microtubule polymerization | Dinitroaniline, Phosphoroamidate, Pyridine, Benzamide, Benzoic acid, Carbamate | oryzalin, pendimethalin, trifluralin, amiprophos-methyl, dithiopyr, propyzamide, DCPA, carbetamide | No |
| K3 | Inhibition of fatty acid synthase (FAS) | Fatty acid biosynthesis | Chloroacetamide, Acetamide, Oxyacetamide, Tetrazolinone | acetochlor, alachlor, metolachlor, napropamide, flufenacet, fentrazamide, anilofos | No |
| L | Inhibition of cellulose-synthase | Cell wall biosynthesis | Nitrile, Benzamide, Triazolocarboxamide, Quinoline carboxylic acid | dichlobenil, isoxaben, flupoxam, quinclorac | No |
| M | Uncoupling of oxidative phosphorylation | ATP biosynthesis | Dinitrophenol | dinoseb, dinoterb | Yes |
| N | Inhibition of fatty acid elongase | Fatty acid biosynthesis | Thiocarbamate, Phosphorodithioate, Benzofuran, Chloro-Carbonic-acid | butylate, cycloate, EPTC, bensulide, ethofumesate, TCA, dalapon | No |
| O | Stimulation of transport inhibitor response protein 1 (TIR1) | Regulation of auxin-responsive genes | Phenoxy-carboxylic-acid, Benzoic acid, Pyridine carboxylic acid, Quinoline carboxylic acid | 2,4-D, MCPA, dicamba, clopyralid, fluroxypyr, picloram, triclopyr, quinclorac, quinmerac, benazolin-ethyl | Yes |
| P | Inhibition of auxin transport | Long-range hormone signaling | Phthalamate, Semicarbazone | naptalam, diflufenzopyr-Na | Yes |
a The 21 known groups of herbicides classified by mechanims of action (MA) according to the global Herbicide Resistance Action Committee (HRAC; http://www.hracglobal.com, accessed on: 28 January 2019. Also, there are other MAs, but they are not completely understood as the biochemical processes were not described. Please see the HRAC website to check all MA. b Not all active herbicide ingredients are presented, please see the HRAC website to check all of them. c Indicates the ROS production in some phase of the herbicide action [41,45].
After the active herbicide reaches and inhibits the TS, a series of stress events are initiated by the signaling of plant defense systems against perturbations. The response time for oxidative stress occurrence and visible plant damage varies with the herbicide mode of action, type of herbicide and formulation, plant species, development stage, and environmental conditions [41]. For example, paraquat (photosynthesis inhibitor—PSI) damage could be observed from 2 hours after treatment under light conditions, and susceptible plants die from 3 to 7 days after. On the other hand, the first visible plant symptoms from glyphosate and plant death for susceptible species may occur around 5 days and 7 to 15 days, respectively (Table 1).
The plant antioxidant defense system must act quickly and efficiently to cope with ROS produced by herbicides, especially those which produce ROS directly (Table 1). However, herbicides which produce ROS as a secondary effect may have a lag time between treatment to creating oxidative stress. Thus, as these herbicides cause a wide range of perturbations, likely the antioxidant system acts as a complement to the resistance process. The complexity of all processes involved in plant defense against herbicide action related to NTSR is still poorly understood.
4. Coevolution of Herbicide Resistance and Antioxidant Systems
Weed evolution has been hastened by human action in the last two to three decades, mainly through the intensive use of herbicides [40,46,47]. According to Heap [48], globally, there are 497 unique cases of weeds resistant to herbicides including 23 of 26 known herbicide’s sites of action. These cases comprise 255 species (148 dicots and 107 monocots), reported in 92 crops in 70 countries. Also, weeds have evolved resistance to around 90% of the known herbicide sites of action.
Herbicide resistance occurs when a plant biotype survives and reproduces after exposure to a normally lethal dose of herbicide [49]. According to Yuan et al. [49] and Délye [47], in weeds there are two primary mechanisms of herbicide resistance: (1) alterations in the herbicide target-site (target-site resistance—TSR) such as mutation and overexpression; and (2) any other alterations which do not involve the herbicide target-site (non-target site resistance—NTSR), such as herbicide absorption, translocation, metabolism, and compensation or protection [44]. In general, TSR resistance is monogenic, and as herbicides are developed to target specific enzymes or proteins, a simple point mutation could alter the enzyme structure and confer the resistance, making the herbicide treatment ineffective. On the other hand, NTSR is a complex polygenic adaptation to herbicides which involves a multi-step process [44,49]. NTSR is considered the predominant type of resistance and comprises a very complex molecular process which remains to be fully comprehended [43]. This type of resistance is a serious threat for agriculture because it can confer cross-resistance to various modes of action herbicides (multiple herbicide resistance), including those not yet discovered [44,49].
The NTSR can be caused by a plant detoxification process that follows a four-phase: I—detoxification; II—conjugation; III—transport; and IV—degradation [44,47,49]. In the first phase, detoxification involves the oxidation process carried out by P450 monooxygenases or oxidases with various functions. In the second phase, conjugation of xenobiotic occurs by the addition of thiols or sugars, or directly by glutathione S-transferases and glycosyltransferases. The third phase comprises the transport of the conjugated molecule into the vacuole or extracellular space by ABC transporters, which are the most known group of transporters. Finally, the fourth phase involves degradation of the conjugated molecule [44,47,49].
Antioxidant systems and protection against oxidative damage caused by herbicide action plays an important role in NTSR [9,44,50,51,52]. This system could act to directly scavenge ROS or complement other herbicide resistance mechanisms in a complex set of coordinated processes [9]. In this way, as ~70% of MA herbicides involve ROS production and oxidative stress (Table 1), this type of resistance can cause a multi-herbicide resistance, representing a threat to sustainable agriculture and worries scientists, herbicides developers, and farmers.
Research has shown modulations in the scavenging activity of ROS within the antioxidant defense machinery in plants exposed to the herbicide. In general, the antioxidant enzyme activities, ROS levels, and LPO are elevated. However, in some cases, high herbicide concentrations decrease the antioxidant enzyme activities. Table 2 shows the percentage values to variations in antioxidants enzyme, ROS, and LPO (Table 2).
Table 2.
Alterations in antioxidant enzyme activities, ROS level, and lipid peroxidation in different species after herbicide treatment.
| Species | Herbicide Concentration | Time | Tissue | * Antioxidants Enzyme | * ROS Level | * Lipid Peroxidation | References |
|---|---|---|---|---|---|---|---|
| Triticum aestivum L. | Chlorotoluron 0, 5, 10, 15, 20 25 mg kg−1 |
10 days | Roots Leaves |
CAT ↑ 5 mg kg−1 (80%); ↑ 10 mg kg−1 (35%); ↑ 15 mg kg−1 (5%); ↓ 20 mg kg−1 (11%); ↓ 25 mg kg−1 (23%); SOD ↑ 5 mg kg−1 (100%); ↑ 10 mg kg−1 (200%); ↑ 15 mg kg−1 (300%); ↑ 20 mg kg−1 (430%); ↑ 25 mg kg−1 (500%); APX ↑ 5 mg kg−1 (160%); ↑ 10 mg kg−1 (260%); ↑ 15 mg kg−1 (80%); ↑ 20 mg kg−1 (70%); ↑ 25 mg kg−1 (40%); POD ↑ 5 mg kg−1 (88%); ↑ 10 mg kg−1 (233%); ↑ 15 mg kg−1 (210%); ↑ 20 mg kg−1 (188%); ↑ 25 mg kg−1 (133%); CAT ↓ 5 mg kg−1 (17%); ↓ 10 mg kg−1 (23%); ↓ 15 mg kg−1 (35%); ↓ 20 mg kg−1 (41%); ↓ 25 mg kg−1 (47%); SOD ↑ 5 mg kg−1 (4%); ↑ 10 mg kg−1 (60%); ↑ 15 mg kg−1 (180%); ■ 20 mg kg−1 (0%); ↓ 25 mg kg−1 (4%); APX ↑ 5 mg kg−1 (100%); ↑ 10 mg kg−1 (300%); ↑ 15 mg kg−1 (75%); ↑ 20 mg kg−1 (50%); ↑ 25 mg kg−1 (25%); POD ■ |
nd †H2O2 ↑ †O2•− ↑ |
■ 5 mg kg−1 (0%) ↑ 10 mg kg−1 (50%) ↑ 15 mg kg−1 (40%) ↑ 20 mg kg−1 (35%) ■ 25 mg kg−1 (0%) ↑ 5 mg kg−1 (125%) ↑ 10 mg kg−1 (225%) ↑ 15 mg kg−1 (150%) ↑ 20 mg kg−1 (50%) ↑ 25 mg kg−1 (25%) |
[19] |
| Triticum aestivum L. | Prometryne 0, 4, 8, 12, 16, 20, 24 mg kg−1 |
10 days | Roots Leaves |
CAT ↓ 4 mg kg−1 (20%); ↓ 8 mg kg−1 (24%); ↓ 12 mg kg−1 (37%); ↓ 16 mg kg−1 (42%); ↓ 20 mg kg−1 (48%); ↓ 24 mg kg−1 (55%); SOD ↑ 4 mg kg−1 (14%); ↑ 8 mg kg−1 (52%); ↑ 12 mg kg−1 (45%); ■ 16 mg kg−1 (0%); ■ 20 mg kg−1 (0%); ↓ 24 mg kg−1 (37%); APX ■ 4 mg kg−1 (0%); ↑ 8 mg kg−1 (23%); ↑ 12 mg kg−1 (44%); ↑ 16 mg kg−1 (16%); ■ 20 mg kg−1 (0%); ↓ 24 mg kg−1 (25%); POD ↑ 4 mg kg−1 (58%); ↑ 8 mg kg−1 (76%); ↑ 12 mg kg−1 (66%); ↑ 16 mg kg−1 (58%); ↑ 20 mg kg−1 (23%); ■ 24 mg kg−1 (0%); GST ↑ 4 mg kg−1 (50%); ↑ 8 mg kg−1 (64%); ↑ 12 mg kg−1 (57%); ↑ 16 mg kg−1 (42%); ↑ 20 mg kg−1 (21%); ■ 24 mg kg−1 (0%); CAT ↑ 4 mg kg−1 (21%); ↑ 8 mg kg−1 (50%); ↑ 12 mg kg−1 (42%); ↑ 16 mg kg−1 (21%); ■ 20 mg kg−1 (0%); ↓ 24 mg kg−1 (30%); SOD ■ 4 mg kg−1 (0%); ↑ 8 mg kg−1 (57%); ↑ 12 mg kg−1 (68%); ↑ 16 mg kg−1 (47%); ↑ 20 mg kg−1 (0%); ■ 24 mg kg−1 (0%); APX ↑ 4 mg kg−1 (16%); ↑ 8 mg kg−1 (36%); ↑ 12 mg kg−1 (56%); ↑ 16 mg kg−1 (66%); ↑ 20 mg kg−1 (43%); ↑ 24 mg kg−1 (26%); POD ■ 4 mg kg−1 (0%); ↑ 8 mg kg−1 (37%); ↑ 12 mg kg−1 (43%); ↑ 16 mg kg−1 (18%); ■ 20 mg kg−1 (0%); ■ 24 mg kg−1 (0%); GST ■ 4 mg kg−1 (0%); ↑ 8 mg kg−1 (100%); ↑ 12 mg kg−1 (116%); ↑ 16 mg kg−1 (50%); ↑ 20 mg kg−1 (16%); ↓ 24 mg kg−1 (33%); |
nd |
↑ 4 mg kg−1 (250%) ↑ 8 mg kg−1 (450%) ↑ 12 mg kg−1 (400%) ↑ 16 mg kg−1 (325%) ↑ 20 mg kg−1 (275%) ↑ 24 mg kg−1 (250%) ↑ 4 mg kg−1 (140%) ↑ 8 mg kg−1 (260%) ↑ 12 mg kg−1 (200%) ↑ 16 mg kg−1 (140%) ↑ 20 mg kg−1 (135%) ↑ 24 mg kg−1 (135%) |
[5] |
|
Brassica napus L. Brassica rapa L. |
ZJ0273 0, 100, 500, 1000 mg L−1 |
7 days 14 days 28 days 7 days 14 days 28 days |
Leaves | SOD ↓ 100 mg L−1 (5%); ↓ 500 mg L−1 (22%); ↓ 1000 mg L−1 (38%); SOD ↓ 100 mg L−1 (3%); ↓ 500 mg L−1 (17%); ↓ 1000 mg L−1 (28%); SOD ↓ 100 mg L−1 (8%); ↓ 500 mg L−1 (21%); ↓ 1000 mg L−1 (33%); POD ↓ 100 mg L−1 (5%); ↓ 500 mg L−1 (42%); ↓ 1000 mg L−1 (55%); POD ↓ 100 mg L−1 (6%); ↓ 500 mg L−1 (33%); ↓ 1000 mg L−1 (47%); POD ↓ 100 mg L−1 (1%); ↓ 500 mg L−1 (3%); ↓ 1000 mg L−1 (29%); SOD ↓ 100 mg L−1 (10%); ↓ 500 mg L−1 (22%); ↓ 1000 mg L−1 (34%); SOD ↓ 100 mg L−1 (2%); ↓ 500 mg L−1 (17%); ↓ 1000 mg L−1 (29%); SOD ↓ 100 mg L−1 (1%); ↓ 500 mg L−1 (8%); ↓ 1000 mg L−1 (15%); POD ↓ 100 mg L−1 (9%); 500 mg L−1 (36%); ↓ 1000 mg L−1 (49%); POD ↓ 100 mg L−1 (5%); ↓ 500 mg L−1 (21%); ↓ 1000 mg L−1 (45%); POD ↓ 100 mg L−1 (1%); ↓ 500 mg L−1 (8%); ↓ 1000 mg L−1 (24%); |
nd | (7) ↑ 100 mg L−1 (9%) ↑ 500 mg L−1 (53%) ↑ 1000 mg L−1 (58%) (14) ■ 100 mg L−1 (0%) ↑ 500 mg L−1 (32%) ↑ 1000 mg L−1 (44%) (28) ↓ 100 mg L−1 (9%) ■ 500 mg L−1 (0%) ↑ 1000 mg L−1 (1%) (7) ↑ 100 mg L−1 (32%) ↑500 mg L−1 (86%) ↑ 1000 mg L−1 (101%) (14) ↑ 100 mg L−1 (25%) ↑ 500 mg L−1 (45%) ↑ 1000 mg L−1 (63%) (28) ■ 100 mg L−1 (0%) ↑ 500 mg L−1 (11%) ↑ 1000 mg L−1 (19%) |
[53] |
| Oryza sativa L. | Fluroxypyr 0, 0.05, 0.1, 0.2, 0.4, 0.8 mg L−1 |
6 days | Roots Leaves |
CAT ■ SOD ↑ 0.05 mg L−1 (18%); ↑ 0.1 mg L−1 (20%); ↑ 0.2 mg L−1 (32%); ↑ 0.4 mg L−1 (22%); ↑ 0.8 mg L−1 (13%); APX ↑ 0.05 mg L−1 (10%); ↑ 0.1 mg L−1 (15%); ↑ 0.2 mg L−1 (10%); ■ 0.4 mg L−1 (0%); ↓ 0.8 mg L−1 (10%); POD ↑ 0.05 mg L−1 (50%); ↑ 0.1 mg L−1 (57%); ↑ 0.2 mg L−1 (90%); ↑ 0.4 mg L−1 (93%); ↑ 0.8 mg L−1 (110%); CAT ■ 0.05 mg L−1 (0%); ↑ 0.1 mg L−1 (15%); ■ 0.2 mg L−1 (0%); ↓ 0.4 mg L−1 (10%); ↓ 0.8 mg L−1 (30%); SOD ↑ 0.05 mg L−1 (20%); ↑ 0.1 mg L−1 (35%); ↑ 0.2 mg L−1 (40%); ↑ 0.4 mg L−1 (35%); ↑ 0.8 mg L−1 (30%); APX ■ POD ■ 0.05 mg L−1 (0%); ■ 0.1 mg L−1 (0%); ■ 0.2 mg L−1 (0%); ↑ 0.4 mg L−1 (45%); ↑ 0.8 mg L−1 (55%); |
†H2O2 ↑ †O2•− ↑ |
↑ 0.05 mg L−1 (17%) ↑ 0.1 mg L−1 (25%) ↑ 0.2 mg L−1 (45%) ↑ 0.4 mg L−1 (40%) ↑ 0.8 mg L−1 (17%) ■ 0.05 mg L−1 (0%) ↑ 0.1 mg L−1 (10%) ↑ 0.2 mg L−1 (13%) ↑ 0.4 mg L−1 (22%) ↑ 0.8 mg L−1 (9%) |
[54] |
| Zea mays L. | Clethodim 0, 50, 100, 200, 500, 1000 ppm |
21 days | Leaves | CAT ↓ 50 ppm (57%); ↓ 100 ppm (47%); ↓ 200 ppm (23%); ↓ 500 ppm (15%); ↓ 1000 ppm (9%); SOD ↓ 50 ppm (13%); ↓ 100 ppm (25%); ↓ 200 ppm (35%); ↓ 500 ppm (35%); ↓ 1000 ppm (32%); APX ↑ 50 ppm (90%); ↑ 100 ppm (175%); ↑ 200 ppm (82%); ↑ 500 ppm (75%); ↑ 1000 ppm (17%); POD ↑ 50 ppm (92%); ↑ 100 ppm (77%); ↑ 200 ppm (180%); ↑ 500 ppm (123%); ↑ 1000 ppm (190%); |
H2O2↑ 50 ppm (1%) ↑ 100 ppm (23%) ↑ 200 ppm (36%) ↑ 500 ppm (50%) ↑ 1000 ppm (63%) |
↑ 50 ppm (45%) ↓ 100 ppm (7%) ↑ 200 ppm (67%) ↑ 500 ppm (120%) ↑ 1000 ppm (182%) |
[55] |
| Oryza sativa L. | Atrazine 0, 0.05, 0.1, 0.2, 0.4, 0.8 mg L−1 |
6 days | Roots Leaves |
CAT ■ 0.05 mg L−1 (0%); ↑ 0.1 mg L−1 (25%); ↑ 0.2 mg L−1 (25%); ↑ 0.4 mg L−1 (25%); ■ 0.8 mg L−1 (0%); SOD ■ 0.05 mg L−1 (0%); ↑ 0.1 mg L−1 (60%); ↑ 0.2 mg L−1 (75%); ↑ 0.4 mg L−1 (150%); ↑ 0.8 mg L−1 (95%); APX ■ 0.05 mg L−1 (0%); ↑ 0.1 mg L−1 (25%); ↑ 0.2 mg L−1 (65%); ↑ 0.4 mg L−1 (70%); ↑ 0.8 mg L−1 (25%); POD ■ 0.05 mg L−1 (0%); ■ 0.1 mg L−1 (0%); ↑ 0.2 mg L−1 (65%); ↑ 0.4 mg L−1 (85%); ↑ 0.8 mg L−1 (125%); GST ■ 0.05 mg L−1 (0%); ■ 0.1 mg L−1 (0%); ↓ 0.2 mg L−1 (50%); ↓ 0.4 mg L−1 (58%); ↓ 0.8 mg L−1 (50%); GR ■ 0.05 mg L−1 (0%); ↑ 0.1 mg L−1 (25%); ↑ 0.2 mg L−1 (100%); ↑ 0.4 mg L−1 (50%); ↑ 0.8 mg L−1 (40%); CAT ↑ 0.05 mg L−1 (50%); ↑ 0.1 mg L−1 (100%); ↑ 0.2 mg L−1 (125%); ↑ 0.4 mg L−1 (150%); ↑ 0.8 mg L−1 (200%); SOD ■ 0.05 mg L−1 (0%); ↑ 0.1 mg L−1 (40%); ↑ 0.2 mg L−1 (50%); ↑ 0.4 mg L−1 (140%); ↑ 0.8 mg L−1 (300%); APX ■ 0.05 mg L−1 (0%); ■ 0.1 mg L−1 (0%); ↑ 0.2 mg L−1 (40%); ↑ 0.4 mg L−1 (45%); ■ 0.8 mg L−1 (0%); POD ■ 0.05 mg L−1 (0%); ↑ 0.1 mg L−1 (40%); ↑ 0.2 mg L−1 (45%); ↑ 0.4 mg L−1 (360%); ↑ 0.8 mg L−1 (540%); GST ■ 0.05 mg L−1 (0%); ■ 0.1 mg L−1 (0%); ↑ 0.2 mg L−1 (50%); ↑ 0.4 mg L−1 (50%); ↑ 0.8 mg L−1 (40%); GR ■ 0.05 mg L−1 (0%); ↑ 0.1 mg L−1 (50%); ↑ 0.2 mg L−1 (95%); ↑ 0.4 mg L−1 (150%); ↑ 0.8 mg L−1 (115%); |
nd †H2O2 ↑ †O2•− ↑ |
■ 0.05 mg L−1 (0%) ↑ 0.1 mg L−1 (22%) ↑ 0.2 mg L−1 (33%) ↑ 0.4 mg L−1 (45%) ↑ 0.8 mg L−1 (22%) ■ 0.05 mg L−1 (0%) ↑ 0.1 mg L−1 (25%) ↑ 0.2 mg L−1 (25%) ↑ 0.4 mg L−1 (37%) ↑ 0.8 mg L−1 (25%) |
[18] |
| Triticum aestivum L. | Simetryne 0, 0.8, 1.6, 3.2, 4.8, 6.4, 8.0 mg kg−1 |
7 days | Roots Leaves |
CAT ↑ 0.8 mg kg−1 (43%); ↑ 1.6 mg kg−1 (73%); ↑ 3.2 mg kg−1 (15%); ↓ 4.8 mg kg−1 (30%); ↓ 6.4 mg kg−1 (45%); ↓ 8.0 mg kg−1 (70%); SOD ↑ 0.8 mg kg−1 (25%); ↑ 1.6 mg kg−1 (65%); ↑ 3.2 mg kg−1 (105%); ↑ 4.8 mg kg−1 (60%); ↑ 6.4 mg kg−1 (40%); ↑ 8.0 mg kg−1 (20%); APX ↑ 0.8 mg kg−1 (50%); ↑ 1.6 mg kg−1 (90%); ↑ 3.2 mg kg−1 (135%); ↑ 4.8 mg kg−1 (120%); ↑ 6.4 mg kg−1 (65%); ↑ 8.0 mg kg−1 (50%); POD ↑ 0.8 mg kg−1 (10%); ↑ 1.6 mg kg−1 (50%); ↑ 3.2 mg kg−1 (100%); ↑ 4.8 mg kg−1 (80%); ↑ 6.4 mg kg−1 (30%); ■ 8.0 mg kg−1 (0%); GST ↑ 0.8 mg kg−1 (20%); ↑ 1.6 mg kg−1 (25%); ↑ 3.2 mg kg−1 (75%); ↑ 4.8 mg kg−1 (55%); ↑ 6.4 mg kg−1 (30%); ↑ 8.0 mg kg−1 (20%); GR ↑ 0.8 mg kg−1 (45%); ↑ 1.6 mg kg−1 (90%); ↑ 3.2 mg kg−1 (170%); ↑ 4.8 mg kg−1 (150%); ↑ 6.4 mg kg−1 (140%); ↑ 8.0 mg kg−1 (95%); CAT ↑ 0.8 mg kg−1 (25%); ↑ 1.6 mg kg−1 (85%); ↑ 3.2 mg kg−1 (150%); ↑ 4.8 mg kg−1 (100%); ↑ 6.4 mg kg−1 (50%); ↓ 8.0 mg kg−1 (25%); SOD ↑ 0.8 mg kg−1 (40%); ↑ 1.6 mg kg−1 (110%); ↑ 3.2 mg kg−1 (195%); ↑ 4.8 mg kg−1 (145%); ↑ 6.4 mg kg−1 (100%); ↑ 8.0 mg kg−1 (50%); APX ↑ 0.8 mg kg−1 (20%); ↑ 1.6 mg kg−1 (45%); ↑ 3.2 mg kg−1 (100%); ↑ 4.8 mg kg−1 (90%); ↑ 6.4 mg kg−1 (65%); ↑ 8.0 mg kg−1 (45%); POD ■ 0.8 mg kg−1 (0%); ↑ 1.6 mg kg−1 (15%); ↑ 3.2 mg kg−1 (35%); ↑ 4.8 mg kg−1 (15%); ↑ 6.4 mg kg−1 (10%); ■ 8.0 mg kg−1 (0%); GST ↑ 0.8 mg kg−1 (10%); ↑ 1.6 mg kg−1 (25%); ↑ 3.2 mg kg−1 (50%); ↑ 4.8 mg kg−1 (70%); ↓ 6.4 mg kg−1 (25%); ↓ 8.0 mg kg−1 (35%); GR ■ 0.8 mg kg−1 (0%); ↑ 1.6 mg kg−1 (10%); ↑ 3.2 mg kg−1 (25%); ↑ 4.8 mg kg−1 (10%); ↑ 6.4 mg kg−1 (5%); ■ 8.0 mg kg−1 (0%); |
†H2O2 ↑ †O2•− ↑ |
↑ 0.8 mg kg−1 (20%) ↑ 1.6 mg kg−1 (20%) ↑ 3.2 mg kg−1 (45%) ↑ 4.8 mg kg−1 (25%) ↑ 6.4 mg kg−1 (20%) ■ 8.0 mg kg−1 (0%) ↑ 0.8 mg kg−1 (15%) ↑ 1.6 mg kg−1 (35%) ↑ 3.2 mg kg−1 (45%) ↑ 4.8 mg kg−1 (35%) ↑ 6.4 mg kg−1 (15%) ↑ 8.0 mg kg−1 (10%) |
[56] |
| Phaseolus vulgaris L. | Prometryn 0, 10, 100, 500 µM |
21 days | Roots Leaves |
CAT ↑ 10 µM (35%); ↑ 100 µM (40%); ■ 500 µM (0%); APX ↑ 10 µM (35%); ↑ 100 µM (70%); ↓ 500 µM (22%); GST ↑ 10 µM (8%); ↑ 100 µM (15%); ↓ 500 µM (11%); CAT ↑ 10 µM (30%); ↑ 100 µM (100%); ↓ 500 µM (25%); APX ↑ 10 µM (20%); ↑ 100 µM (42%); ↓ 500 µM (49%); GST ↑ 10 µM (55%); ↑ 100 µM (110%); ↓ 500 µM (18%); |
nd nd |
nd ■ 10 µM (0%) ↑ 100 µM (80%) ↑ 500 µM (148%) |
[20] |
|
Oryza sativa L. (ZJ 88) Oryza sativa L. (XS 134) |
2,4-D 0.8 kg a.i. ha−1 |
15 days | Roots | CAT ↑ 0.8 kg a.i. ha−1 (15%); SOD ↑ 0.8 kg a.i. ha−1 (79%); APX ↑ 0.8 kg a.i. ha−1 (15%); POD ↓ 0.8 kg a.i. ha−1 (7%); CAT ↑ 0.8 kg a.i. ha−1 (19%); SOD ↑ 0.8 kg a.i. ha−1 (32%); APX ↑ 0.8 kg a.i. ha−1 (54%); POD ↑ 0.8 kg a.i. ha−1 (2%); |
H2O2 ↑ 0.8 kg a.i. ha−1 (59%) O2•− ↑ 0.8 kg a.i. ha−1 (29%) H2O2 ↑ 0.8 kg a.i. ha−1 (22%) O2•− ↑ 0.8 kg a.i. ha−1 (19%) |
↑ 0.8 kg a.i. ha−1 (214%) ↑ 0.8 kg a.i. ha−1 (121%) |
[57] |
| Brassica napus L. | Metazachlor 0, 0.2, 0.4 mM |
14 days 28 days |
Leaves | CAT ↑ 0.2 Mm (80%); ↑ 0.4 mM (25%); SOD ↑ 0.2 Mm (30%); ↑ 0.4 mM (25%); APX ↑ 0.2 Mm (42%); ↑ 0.4 mM (35%); POD ↑ 0.2 Mm (170%); ↑ 0.4 mM (130%); GR ↑ 0.2 Mm (42%); ↑ 0.4 mM (83%); CAT ↑ 0.2 Mm (107%); ↑ 0.4 mM (175%); SOD ↑ 0.2 Mm (15%); ↑ 0.4 mM (68%); APX ↑ 0.2 Mm (42%); ↑ 0.4 mM (65%); POD ↑ 0.2 Mm (22%); ↑ 0.4 mM (220%); GR ↑ 0.2 Mm (30%); ↑ 0.4 mM (63%); |
nd | ↑ 0.2 mM (10%) ↑ 0.4 mM (23%) ↑ 0.2 mM (40%) ↑ 0.4 mM (43%) |
[58] |
|
Setaria italica L. (Jingu 21) Setaria italica L. (Zhangzagu 3) Setaria italica L. (Zhangzagu 5) Setaria italica L. (Zhangzagu 10) |
Fluroxypyr 0, 0.5, 1, 2, 4 L a.i. ha−1 |
15 days | Leaves | CAT ↑ 0.5 L a.i. ha−1 (138%); ↑ 1 L a.i. ha−1 (480%); ↑ 2 L a.i. ha−1 (265%); ↑ 4 L a.i. ha−1 (65%); SOD ↑ 0.5 L a.i. ha−1 (75%); ↑ 1 L a.i. ha−1 (98); ↑ 2 L a.i. ha−1 (75%); ↑ 4 L a.i. ha−1 (75%;); APX ↑ 0.5 L a.i. ha−1 (72%); ↑ 1 L a.i. ha−1 (300%); ↑ 2 L a.i. ha−1 (155%); ↑ 4 L a.i. ha−1 (163%); POD ↑ 0.5 L a.i. ha−1 (80%); ↑ 1 L a.i. ha−1 (213%); ↑ 2 L a.i. ha−1 (200%); ↑ 4 L a.i. ha−1 (185%); GR ↑ 0.5 L a.i. ha−1 (57%); ↑ 1 L a.i. ha−1 (255%); ↑ 2 L a.i. ha−1 (150%); ↑ 4 L a.i. ha−1 (100%); CAT ↑ 0.5 L a.i. ha−1 (110%); ↑ 1 L a.i. ha−1 (210%); ↑ 2 L a.i. ha−1 (222%); ↑ 4 L a.i. ha−1 (115%); SOD ↑ 0.5 L a.i. ha−1 (575%); ↑ 1 L a.i. ha−1 (673%); ↑ 2 L a.i. ha−1 (718%); ↑ 4 L a.i. ha−1 (520%); APX ↑ 0.5 L a.i. ha−1 (65%); ↑ 1 L a.i. ha−1 (212%); ↑ 2 L a.i. ha−1 (243%); ↑ 4 L a.i. ha−1 (118%); POD ↑ 0.5 L a.i. ha−1 (100%); ↑ 1 L a.i. ha−1 (110%); ↑ 2 L a.i. ha−1 (180%); ↑ 4 L a.i. ha−1 (185%); GR ↑ 0.5 L a.i. ha−1 (32%); ↑ 1 L a.i. ha−1 (142%); ↑ 2 L a.i. ha−1 (272%); ↑ 4 L a.i. ha−1 (97%); CAT ↑ 0.5 L a.i. ha−1 (412%); ↑ 1 L a.i. ha−1 (370%); ↑ 2 L a.i. ha−1 (311%); ↑ 4 L a.i. ha−1 (435%); SOD ↑ 0.5 L a.i. ha−1 (72%); ↑ 1 L a.i. ha−1 (140%); ↑ 2 L a.i. ha−1 (227%); ↑ 4 L a.i. ha−1 (125%); APX ↑ 0.5 L a.i. ha−1 (15%); ↑ 1 L a.i. ha−1 (20%); ↑ 2 L a.i. ha−1 (92%); ↑ 4 L a.i. ha−1 (70%); POD ↑ 0.5 L a.i. ha−1 (28%); ↑ 1 L a.i. ha−1 (83%); ↑ 2 L a.i. ha−1 (90%); ↑ 4 L a.i. ha−1 (125%); GR ↑ 0.5 L a.i. ha−1 (18%); ↑ 1 L a.i. ha−1 (105%); ↑ 2 L a.i. ha−1 (295%); ↑ 4 L a.i. ha−1 (25%); CAT ↑ 0.5 L a.i. ha−1 (293%); ↑ 1 L a.i. ha−1 (320%); ↑ 2 L a.i. ha−1 (430%); ↑ 4 L a.i. ha−1 (110%); SOD ↑ 0.5 L a.i. ha−1 (138%); ↑ 1 L a.i. ha−1 (202%); ↑ 2 L a.i. ha−1 (230%); ↑ 4 L a.i. ha−1 (140%); APX ↑ 0.5 L a.i. ha−1 (113%); ↑ 1 L a.i. ha−1 (163%); ↑ 2 L a.i. ha−1 (345%); ↑ 4 L a.i. ha−1 (235%); POD ↑ 0.5 L a.i. ha−1 (88%); ↑ 1 L a.i. ha−1 (235%); ↑ 2 L a.i. ha−1 (670%); ↑ 4 L a.i. ha−1 (740%); GR ↑ 0.5 L a.i. ha−1 (13%); ↑ 1 L a.i. ha−1 (62%); ↑ 2 L a.i. ha−1 (195%); ↑ 4 L a.i. ha−1 (109%); |
H2O2 ↑ 0.5 L a.i. ha−1 (70%) ↑ 1 L a.i. ha−1 (130%) ↑ 2 L a.i. ha−1 (160%) ↑ 4 L a.i. ha−1 (182%) O2•− ↑ 0.5 L a.i. ha−1 (3%) ↑ 1 L a.i. ha−1 (10%) ↑ 2 L a.i. ha−1 (15%) ↑ 4 L a.i. ha−1 (28%) H2O2 ↑ 0.5 L a.i. ha−1 (2%) ↑ 1 L a.i. ha−1 (10%) ↑ 2 L a.i. ha−1 (30%) ↑ 4 L a.i. ha−1 (60%) O2•− ■ 0.5 L a.i. ha−1 (0%) ↑ 1 L a.i. ha−1 (2%) ↑ 2 L a.i. ha−1 (2%) ↑ 4 L a.i. ha−1 (3%) H2O2 ↑ 0.5 L a.i. ha−1 (42%) ↑ 1 L a.i. ha−1 (55%) ↑ 2 L a.i. ha−1 (55%) ↑ 4 L a.i. ha−1 (60%) O2•− ■ 0.5 L a.i. ha−1 (0%) ■ 1 L a.i. ha−1 (0%) ■ 2 L a.i. ha−1 (0%) ↑ 4 L a.i. ha−1 (10%) H2O2 ↑ 0.5 L a.i. ha−1 (1%) ↑ 1 L a.i. ha−1 (12%) ↑ 2 L a.i. ha−1 (13%) ↑ 4 L a.i. ha−1 (80%) O2•− ↑ 0.5 L a.i. ha−1 (5%) ↑ 1 L a.i. ha−1 (10%) ↑ 2 L a.i. ha−1 (10%) ↑ 4 L a.i. ha−1 (13%) |
↑ 0.5 L a.i. ha− (35%) ↑ 1 L a.i. ha−1 (52%) ↑ 2 L a.i. ha−1 (62%) ↑ 4 L a.i. ha−1 (80%) ↑ 0.5 L a.i. ha−1 (10%) ↑ 1 L a.i. ha−1 (37%) ↑ 2 L a.i. ha−1 (52%) ↑ 4 L a.i. ha−1 (65%) ■ 0.5 L a.i. ha−1 (0%) ↑ 1 L a.i. ha−1 (7%) ↑ 2 L a.i. ha−1 (20%) ↑ 4 L a.i. ha−1 (25%) ↑ 0.5 L a.i. ha−1 (20%) ↑ 1 L a.i. ha−1 (37%) ↑ 2 L a.i. ha−1 (65%) ↑ 4 L a.i. ha−1 (80%) |
[59] |
* Approximate percentage values relative to controls for antioxidants enzyme, ROS and lipid peroxidation; Lipid peroxidation—determined by measuring the concentration of malondialdehyde as thiobarbituric acid reactive substances; † Histochemical analysis of ROS; ↑ Increase in roots and leaves with the application of herbicide in concentrations cited; ↓ Decrease in roots and leaves with the application of herbicide in concentrations cited; ■ Unchanged; nd, not determined. SOD—superoxide dismutase; CAT—catalase; APX—ascorbate peroxidase; POD—peroxidase; GR—glutathione reductase; GST—glutathione-S-transferase; O2•−—superoxide radical; H2O2—hydrogen peroxide; ZJ0273—Propyl 4-(2-(4,6-dimethoxypyrimidin-2-yloxy)benzylamino)benzoate; 2,4-D—2,4-dichlorophenoxyacetic acid.
In multiple herbicide-resistant (MHR) wild oats (Avena fatua), redox-related enzymes with elevated expression were identified, suggesting that these plants exhibit a high capacity for redox maintenance since these plants are resistant to 11 herbicides from five different MA [12]. Also, the authors [12] described that the transcriptional regulation of MHR was similar to those presented by phenotypes tolerant to abiotic stress. For goosegrass (Eleusine indica L. (Gaertn)), the analysis of the differentially expressed genes revealed that most ROS pathway genes were up-regulated in resistant and susceptible biotypes after the application of paraquat [8]. In wheat, prometryne and symetrine-induced oxidative stress and increased transcript abundance of Cu/Zn-SOD, GR, APX, and GST genes in leaves and roots [5,6,56]. The Cu/Zn-SOD expression in leaves increased 2.6-fold with 12 mg kg−1 prometryne [5], and Cu/Zn-SOD and GST genes in leaves with 8 mg kg−1 prometryne increased 7.6 and 4.4-fold, respectively [6]. Also, the effects of simetryne on transcript abundance of Cu/Zn-SOD, GR, APX, and GST genes showed an increase of 2.1, 1.3, 1.6, and 1.4-fold, respectively [56]. Furthermore, in rice, atrazine treatment resulted in OsGST3, OsGST4, OsAPX2, OsAPX3, OsGR1, and OsGR3 genes up-regulation [18]. These results indicate that antioxidants gene can be highly regulated by herbicides at the molecular level in plants. Thus, crops are capable of activating antioxidants mechanisms to alleviate herbicide-induced stress. In rice, atrazine treatment resulted in OsGST3, OsGST4, OsAPX2, OsAPX3, OsGR1, and OsGR3 genes up-regulation [18]. Maroli et al. [9] demonstrated the potential role of antioxidant systems in glyphosate resistance in Palmer amaranth (Amaranthus palmeri) using metabolic profile and enzyme analyses. Harre et al. [15] documented the involvement of antioxidant enzymes in the glyphosate resistance in Ambrosia trifida.
In rice, the over-expression of the OsGSTL2 gene improved glyphosate and chlorsulfuron tolerance [60]. Transgenic plants contained higher levels of GST activities, 2.1, 1.8 and 2.4-fold, which were enough to provide tolerance to herbicides. Further, in transgenic plants, the GPX activity was higher, 1.6, 1.5 and 1.7-fold of that detected in non-transformed plants; the GPX enzyme, therefore, can help degrade superoxide levels [60]. Transgenic potato overexpressing Arabidopsis cytosolic AtDHAR1 gene exhibited increased DHAR activity up to 4.5 times, as well as higher tolerance to methyl viologen herbicide [61]. These plants, when subjected to herbicide, exhibited less ion leakage, greater chlorophyll contents, less accumulation of H2O2, and less severe visual injury symptoms [61], demonstrating the power of the antioxidant system.
5. Transcriptomic/Proteomic Approaches Helping to Clarify the Antioxidant Response to Herbicides in Plants
Transcriptomics and proteomic approaches have helped to clarify the genetic regulation in response to herbicide treatment [9,44,47]. Transcriptomic and proteomic approaches permit the characterization of the expression levels of genes or gene families and proteins for important biochemical pathways in response to herbicide treatment. Thus, in these studies the expression levels of metabolic pathways may change during the organism life cycle and tissue, and according to the environmental conditions.
RNA-sequencing is the preferred method these days to identify transcripts that are differentially expressed in a genome in target tissues under environmental treatments. Differential expression of genes related to antioxidant pathways following herbicide treatment of weeds is important to characterize in species with evolved herbicide resistance (Table 3).
Table 3.
Antioxidant-related genes differentially expressed identified from RNA-Seq studies performed for weed herbicide resistance investigation after herbicide treatment.
| Weed Species | Herbicide Resistance | ROS Scavenging Pathway Genes | Reference |
|---|---|---|---|
| Avena fatua | Pinoxaden Flucarbazone |
GST, SOD | [12] |
| Alopecurus aequalis Sobol | Mesosulfuron-methyl | GST, POD | [62] |
| Apera spica-venti | Sensitive | GST | [63] |
| Brachypodium hybridum | Pinoxaden | GST, POD | [64] |
| Eleusine indica L. | Glyphosate | GST | [65] |
| Lolium spp. | Pyroxsulam; Iodosulfuron+mesosulfuron |
GST | [66] |
| Beckmannia syzigachne | Fenoxaprop-P-ethyl | GST, POD | [67] |
| Descurainia sophia L. | Tribenuron-methyl | GST, POD | [68] |
| Alopecurus myosuroides L. | Iodosulfuron+mesosulfuron | GST, POD | [69] |
| Euphorbia esula | Glyphosate | GST | [70] |
| Eleusine indica L. | Paraquat | GLR, MDAR, GR, POD, GST, CAT, Trx | [8] |
| Lolium rigidum | Diclofop-methyl | GST | [71] |
GST—glutathione-S-transferase; POD—peroxidase; GLR—glutaredoxin; MDAR—monodehydroascorbate reductase; GR—glutathione reductase; CAT—catalase; Trx—thioredoxin.
These studies have shown that several antioxidants genes have altered expression in weeds after herbicide treatment. The data shows how antioxidant defense mechanisms were activated in response to herbicide, thus working to restore redox homeostasis. Considering all aspects indicated above, the data suggests that these weedy species exhibit a high capacity for redox maintenance. The synergistic transcriptome/proteomic combination provides a complete representation of the plant phenotype [12]. Accordingly, the proteomic analyses of the rice plants treated with glyphosate herbicide demonstrated that antioxidant proteins APX, GST, PRX, and SOD were accumulated. Further, the transcript levels of the genes GST, PRX, and APX increased [16], suggesting that the herbicide generated oxidative stress.
6. Concluding Remarks
ROS overproduction occurs in plants exposed to environmental stresses. ROS are potential toxic molecules that are damaging to cell components and can lead to cell death. The role of antioxidant response systems is to modulate ROS during periods of normal growth or in response to stress to regulate cellular redox homeostasis.
Weed interference is consistently a biotic stressor in crops; both weed interference and herbicide treatment can lead to ROS accumulation in crops as indicated by increased O2•− and H2O2 levels. Various crop species and genotypes vary in these responses in space and time, as well as by degree of interference and herbicide treatment. Lipid peroxidation responses from ROS overproduction are a pernicious effect that commonly causes reduced crop biomass and yield.
Herbicides remain as the primary tool for implementing weed management in the major agronomic crops of the world. Of the 21 herbicide mechanisms of action we described, nearly 43% cause direct ROS production and an additional 29% are indirectly involved in increased ROS in plants. Weeds may have increased the propensity to evolve antioxidant defenses compared with crops. Weedy biotypes with herbicide resistance, especially NTSR to multiple herbicides, appear to have enhanced mechanisms to deal with ROS. It is critical to better understand the interplay of evolved antioxidant responses at the molecular level in weeds.
To this end, transcriptomic and proteomic approaches are beginning to illuminate which antioxidant defense mechanisms are activated in response to ROS and their roles to maintain redox homeostasis. Reverse genetics studies in crops that alter antioxidant enzyme profiles for stress tolerance are an important approach for crop improvement and to improve our understanding of basic cellular mechanisms. Moreover, the application of exogenous protectants such as plant nutrients, antioxidants, osmolytes, phytohormones, signaling molecules, and others have been employed and may contribute to mitigating the toxic effects of a high ROS level through increasing the antioxidant defenses in crops.
Acknowledgments
The authors are grateful to Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) for financial support, PNPD/CAPES for the scholarship of Andréia Caverzan. Also, the authors thank Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) for the scholarship of Cristiano Piasecki. We also appreciate USDA Hatch funding to CNS.
Abbreviations
| 1O2 | singlet oxygen |
| O2•− | superoxide radical |
| H2O2 | hydrogen peroxide |
| OH• | hydroxyl radical |
| SOD | superoxide dismutase |
| CAT | catalase |
| APX | ascorbate peroxidase |
| GPX | glutathione peroxidase |
| POD | peroxidase |
| PRX | peroxiredoxin |
| MDHAR DHAR |
monodehydroascorbate reductase dehydroascorbate reductase |
| GR | glutathione reductase |
| GST | glutathione-S-transferase |
| ASC | ascorbic acid |
| GSH | glutathione |
| Trx | thioredoxin |
Funding
This research received no external funding.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Pan D., Li Q.X., Lin Z., Chen C., Tang W., Pan C., Tan H., Zeng D. Interactions between salicylic acid and antioxidant enzymes tilting the balance of H2O2 from photorespiration in non-target crops under halosulfuron-methyl stress. Pestic. Biochem. Physiol. 2017;143:214–223. doi: 10.1016/j.pestbp.2017.09.007. [DOI] [PubMed] [Google Scholar]
- 2.Gill S.S., Tuteja N. Reactive oxygen species and antioxidant machinery in abiotic stresses tolerance in crop plants. Plant Physiol. Biochem. 2010;48:909–930. doi: 10.1016/j.plaphy.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 3.Mittler R. ROS are good. Trends Plant Sci. 2017;22:11–19. doi: 10.1016/j.tplants.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 4.Czarnocka W., Karpiński S. Friend or foe? Reactive oxygen species production, scavenging and signaling in plant response to environmental stresses. Free Radic. Biol. Med. 2018;122:4–20. doi: 10.1016/j.freeradbiomed.2018.01.011. [DOI] [PubMed] [Google Scholar]
- 5.Jiang L., Yang H. Prometryne-induced oxidative stress and impact on antioxidant enzymes in wheat. Ecotoxicol. Environ. Saf. 2009;72:1687–1693. doi: 10.1016/j.ecoenv.2009.04.025. [DOI] [PubMed] [Google Scholar]
- 6.Jiang L., Ma L., Sui Y., Han S.Q., Wu Z.Y., Feng Y.X., Yang H. Effect of manure compost on the herbicide prometryne bioavailability to wheat plants. J. Hazard. Mater. 2010;184:337–344. doi: 10.1016/j.jhazmat.2010.08.041. [DOI] [PubMed] [Google Scholar]
- 7.Afifi M., Swanton C. Early physiological mechanisms of weed competition. Weed Sci. 2012;60:542–551. doi: 10.1614/WS-D-12-00013.1. [DOI] [Google Scholar]
- 8.An J., Shen X., Ma Q., Yang C., Liu S., Chen Y. Transcriptome profiling to discover putative genes associated with paraquat resistance in goosegrass (Eleusine indica L.) PLoS ONE. 2014;9:e99940. doi: 10.1371/journal.pone.0099940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maroli A.S., Nandula V., Dayan F.E., Duke E., Gerard P., Tharayil N. Metabolic profiling and enzyme analyses indicate a potential role of antioxidant systems in complementing glyphosate resistance in an Amaranthus palmeri biotype. J. Agric. Food Chem. 2015;63:9199–9209. doi: 10.1021/acs.jafc.5b04223. [DOI] [PubMed] [Google Scholar]
- 10.Darmanti S., Santosa L.H., Dewi K., Nugroho L.H. Antioxidative defenses of soybean [Glycine max (L.) Merr. cv. Grobogan] against purple nutsedge (Cyperus rotundus L.) interference during drought stress. J. Anim. Plant Sci. 2016;26:225–232. [Google Scholar]
- 11.Agostinetto D., Tarouco C.P., Nohatto M.A., Oliveira C., Fraga D.S. Metabolic activity of wheat and ryegrass plants in competition. Planta Daninha. 2017;35:e017155463. doi: 10.1590/s0100-83582017350100044. [DOI] [Google Scholar]
- 12.Keith B.K., Burns E.E., Bothner B., Carey C.C., Mazurie A.J., Hilmer J.K., Biyiklioglu S., Budak H., Dyer W.E. Intensive herbicide use has selected for constitutively elevated levels of stress-responsive mRNAs and proteins in multiple herbicide-resistant Avena fatua L. Pest. Manag. Sci. 2017;73:2267–2281. doi: 10.1002/ps.4605. [DOI] [PubMed] [Google Scholar]
- 13.Piasecki C., Rizzardi M.A., Schons J., Caverzan A., Chavarria G. Does the interference of GR® volunteer corn alter stress metabolism on soybean? Planta Daninha. 2018;36:e018171955. doi: 10.1590/s0100-83582018360100018. [DOI] [Google Scholar]
- 14.Piasecki C., Rizzardi M.A., Schons J., Caverzan A., Oliveira C. Interference of volunteer corn on stress metabolism and yield of dry bean. Planta Daninha. 2018;36:e018176669. doi: 10.1590/s0100-83582018360100112. [DOI] [Google Scholar]
- 15.Harre N.T., Nie H., Jiang Y., Young B.G. Differential antioxidant enzyme activity in rapid-response glyphosate-resistant Ambrosia trifida. Pest. Manag. Sci. 2018;74:2125–2132. doi: 10.1002/ps.4909. [DOI] [PubMed] [Google Scholar]
- 16.Ahsan N., Lee D.G., Lee K.W., Alam I., Lee S.H., Bahk J.D., Lee B.H. Glyphosate-induced oxidative stress in rice leaves revealed by proteomic approach. Plant Physiol. Biochem. 2008;46:1062–1070. doi: 10.1016/j.plaphy.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 17.Agostinetto D., Oliveira C., Langaro A.C., Nohatto M.A., Manica-Berto R. Change in physiological features in ryegrass biotypes in competition with soybean due resistance to glyphosate. Planta Daninha. 2016;34:517–526. doi: 10.1590/s0100-83582016340300012. [DOI] [Google Scholar]
- 18.Zhang J.J., Lu Y.C., Zhang J.J., Tan L.R., Yang H. Accumulation and toxicological response of atrazine in rice crops. Ecotoxicol. Environ. Saf. 2014;102:105–112. doi: 10.1016/j.ecoenv.2013.12.034. [DOI] [PubMed] [Google Scholar]
- 19.Song N.H., Yin X.L., Chen G.F., Yang H. Biological responses of wheat (Triticum aestivum) plants to the herbicide chlorotoluron in soils. Chemosphere. 2007;68:1779–1787. doi: 10.1016/j.chemosphere.2007.03.023. [DOI] [PubMed] [Google Scholar]
- 20.Boulahia K., Carol P., Planchais S., Abrous-Belbachir O. Phaseolus vulgaris L. seedlings exposed to prometryn herbicide contaminated soil trigger an oxidative stress response. J. Agric. Food Chem. 2016;64:3150–3160. doi: 10.1021/acs.jafc.6b00328. [DOI] [PubMed] [Google Scholar]
- 21.Neve P., Vila-Aiub M., Roux F. Evolutionary-thinking in agricultural weed management. New Phytol. 2009;184:783–793. doi: 10.1111/j.1469-8137.2009.03034.x. [DOI] [PubMed] [Google Scholar]
- 22.Mittler R. Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 2002;7:405–410. doi: 10.1016/S1360-1385(02)02312-9. [DOI] [PubMed] [Google Scholar]
- 23.Suzuki N., Koussevitzky S., Mittler R., Miller G. ROS and redox signaling in the response of plants to abiotic stress. Plant Cell Environ. 2012;35:259–270. doi: 10.1111/j.1365-3040.2011.02336.x. [DOI] [PubMed] [Google Scholar]
- 24.Das K., Roychoudhury A. Reactive oxygen species (ROS) and response of antioxidants as ROS-scavengers during environmental stress in plants. Front. Plant Sci. 2014;2:1–13. doi: 10.3389/fenvs.2014.00053. [DOI] [Google Scholar]
- 25.Scandalios J.G. The rise of ROS. Trends Biochem. Sci. 2002;27:483–486. doi: 10.1016/S0968-0004(02)02170-9. [DOI] [PubMed] [Google Scholar]
- 26.Caverzan A., Casassola A., Brammer S.P. Abiotic and Biotic Stress in Plants-Recent Advances and Future Perspectives. InTech; Rijeka, Croatia: 2016. Reactive oxygen species and antioxidant enzymes involved in plant tolerance to stress; pp. 463–480. [Google Scholar]
- 27.Foyer C.H., Noctor G. Redox homeostasis and antioxidant signaling: A metabolic interface between stress perception and physiological responses. Plant Cell. 2005;17:1866–1875. doi: 10.1105/tpc.105.033589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Anjum N.A., Gill S.S., Gill R., Hasanuzzaman M., Duarte A.C., Pereira E., Ahmad I., Tuteja R., Tuteja N. Metal/metalloid stress tolerance in plants: Role of ascorbate, its redox couple, and associated enzymes. Protoplasma. 2014;251:1265–1283. doi: 10.1007/s00709-014-0636-x. [DOI] [PubMed] [Google Scholar]
- 29.Kumar S., Trivedi P.K. Glutathione S-Transferases: Role in Combating Abiotic Stresses Including Arsenic Detoxification in Plants. Front. Plant Sci. 2018;7:751. doi: 10.3389/fpls.2018.00751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ramos L.R.M., Pitelli R.A. Effects of different weed control periods on the productivity of corn crop. Pesq. Agropecu. Bras. 1998;29:1523–1531. [Google Scholar]
- 31.Radosevich S.R., Holt J.S., Ghersa C.M. Ecology of Weeds and Invasive Plants: Relationship to Agriculture and Natural Resource Management. 3rd ed. John Wiley & Sons; Hoboken, NJ, USA: 2007. [Google Scholar]
- 32.Barroso A.A.M., Yamauti M.S., Alves P.L.C.A. Interference between weed species and two bean cultivars in two times of sowing. Bragantia. 2010;69:609–616. doi: 10.1590/S0006-87052010000300012. [DOI] [Google Scholar]
- 33.Gal J., Afifi M., Lee E., Lukens L., Swanton C.J. Detection of neighboring weeds alters soybean seedling roots and nodulation. Weed Sci. 2015;63:888–900. doi: 10.1614/WS-D-15-00039.1. [DOI] [Google Scholar]
- 34.Ulguim A.R., Agostinetto D., Oliveira C., Ruchel Q., Silva J.D.G., Vargas L., Avila L.A. Does competition between soybeans and Wild Poinsettia with low-level resistance or susceptibility to glyphosate affect physiology and secondary metabolism? Semina. 2017;38:1133–1144. doi: 10.5433/1679-0359.2017v38n3p1133. [DOI] [Google Scholar]
- 35.Rockenbach A.P., Rizzardi M.A., Nunes A.L., Bianchi M.A., Caverzan A., Schneider T. Interference between weeds and crop: Changes in secondary metabolism. Rev. Bras. Herbic. 2018;17:59–70. doi: 10.7824/rbh.v17i1.527. [DOI] [Google Scholar]
- 36.McKenzie-Gopsill A. Ph.D. Thesis. University of Guelph; Guelph, ON, Canada: 2016. Morphological, Molecular and Physiological Mechanisms of Early Season Weed Competition in Soybean (Glycine max (L.) Merr.) [Google Scholar]
- 37.De Almeida D.G., Zucoloto M., Zetun M.C., Coelho I., Sobreir F.M. Oxidative stress in vegetable cells mediated by allelochemicals. Revista Facultad Nacional de Agronomía Medellín. 2008;61:4237–4247. [Google Scholar]
- 38.Ciniglia C., Mastrobuoni F., Scortichini M., Petriccione M. Oxidative damage and cell-programmed death induced in Zea mays L. by allelochemical stress. Ecotoxicology. 2015;24:926–937. doi: 10.1007/s10646-015-1435-7. [DOI] [PubMed] [Google Scholar]
- 39.Sytykiewicz H. Expression patterns of glutathione transferase gene (GstI) in maize seedlings under juglone-induced oxidative stress. Int. J. Mol. Sci. 2011;12:7982–7995. doi: 10.3390/ijms12117982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Heap I. Global perspective of herbicide-resistant weeds. Pest Manag. Sci. 2014;70:1306–1315. doi: 10.1002/ps.3696. [DOI] [PubMed] [Google Scholar]
- 41.Cobb A.H., Reade P.H.R. Herbicides and Plant Physiology. 2nd ed. John Wiley & Sons; New York, NY, USA: 2010. 286p [Google Scholar]
- 42.Servaites J.C., Tucci M.A., Geiger D.R. Glyphosate effects on carbon assimilation, ribulose bisphosphate carboxylase activity, and metabolite levels in sugar beet leaves. Plant Physiol. 1987;85:370–374. doi: 10.1104/pp.85.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duke S.O., Baerson S.R., Rimando A.M. Herbicides: Glyphosate. In: Plimmer J.R., Gammon D.W., Ragsdale N.N., editors. Encyclopedia of Agrochemicals. John Wiley & Sons; New York, NY, USA: 2007. [Google Scholar]
- 44.Délye C., Jasieniuk M., Le Corre V. Deciphering the evolution of herbicide resistance in weeds. Trends Genet. 2013;29:649–658. doi: 10.1016/j.tig.2013.06.001. [DOI] [PubMed] [Google Scholar]
- 45.Grossmann K., Niggeweg R., Christiansen N., Looser R., Ehrhardt T. The herbicide saflufenacil (KixorTm) is a new inhibitor of protoporphyrinogen IX oxidase activity. Weed Sci. 2010;58:1–9. doi: 10.1614/WS-D-09-00004.1. [DOI] [Google Scholar]
- 46.Jasieniuk M., Brûlé-Babel A.L., Morrison I.N. The evolution and genetics of herbicide resistance in weeds. Weed Sci. 1996;44:176–193. doi: 10.7202/706069ar. [DOI] [Google Scholar]
- 47.Délye C. Unraveling the genetic bases of non-target-site-based resistance (NTSR) to herbicides: A major challenge for weed science in the forthcoming decade. Pest Manag. Sci. 2013;69:176–187. doi: 10.1002/ps.3318. [DOI] [PubMed] [Google Scholar]
- 48.Heap I. The International Survey of Herbicide Resistant Weeds. [(accessed on 1 February 2019)]; Available online: www.weedscience.org.
- 49.Yuan J.S., Tranel P.J., Stewart C.N., Jr. Non-target-site herbicide resistance: A family business. Trends Plant Sci. 2007;12:6–13. doi: 10.1016/j.tplants.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 50.Cummins I., Cole D.J., Edwards R. A role for glutathione transferases functioning as glutathione peroxidases in resistance to multiple herbicides in black-grass. Plant J. 1999;18:285–292. doi: 10.1046/j.1365-313X.1999.00452.x. [DOI] [PubMed] [Google Scholar]
- 51.Cummins I., Bryant D.N., Edwards R. Safener responsiveness and multiple herbicide resistance in the weed black-grass (Alopecurus myosuroides) Plant Biotechnol. J. 2009;7:807–820. doi: 10.1111/j.1467-7652.2009.00445.x. [DOI] [PubMed] [Google Scholar]
- 52.Cummins I., Wortley D.J., Sabbadin F., He Z., Coxon C.R., Straker H.E., Sellars J.D., Knight K., Edwards L., Hughes D., et al. Key role for a glutathione transferase in multiple-herbicide resistance in grass weeds. Proc. Natl. Acad. Sci. USA. 2013;110:5812–5817. doi: 10.1073/pnas.1221179110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jin Z.L., Zhang F., Ahmed Z.I., Rasheed M., Naeem M.S., Ye Q.F., Zhou W.J. Differential morphological and physiological responses of two oilseed Brassica species to a new herbicide ZJ0273 used in rapeseed fields. Pest. Biochem. Physiol. 2010;98:1–8. doi: 10.1016/j.pestbp.2010.04.002. [DOI] [Google Scholar]
- 54.Wu G.L., Cui J., Tao L., Yang H. Fluroxypyr triggers oxidative damage by producing superoxide and hydrogen peroxide in rice (Oryza sativa) Ecotoxicology. 2010;19:124–132. doi: 10.1007/s10646-009-0396-0. [DOI] [PubMed] [Google Scholar]
- 55.Radwan D.E.M. Salicylic acid-induced alleviation of oxidative stress caused by clethodim in maize (Zea mays L.) leaves. Pest. Biochem. Physiol. 2012;102:182–188. doi: 10.1016/j.pestbp.2012.01.002. [DOI] [Google Scholar]
- 56.Jiang L., Yang Y., Jia L.X., Lin J.L., Liu Y., Pan B., Lin Y. Biological responses of wheat (Triticum aestivum) plants to the herbicide simetryneinsoils. Ecotoxicol. Environ. Saf. 2016;127:87–94. doi: 10.1016/j.ecoenv.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 57.Islam F., Farooq M.A., Gill R.A., Wang J., Yang C., Ali B., Wang G.X., Zhou W. 2,4-D attenuates salinity-induced toxicity by mediating anatomical changes, antioxidant capacity and cation transporters in the roots of rice cultivars. Sci. Rep. 2017;7:10443. doi: 10.1038/s41598-017-09708-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vercampt H., Koleva L., Vassilev A., Vangronsveld J., Cuypers A. Short-term phytotoxicity in Brassica napus (L.) in response to pre-emergently applied metazachlor: A microcosm study. Environ. Toxicol. Chem. 2017;36:59–70. doi: 10.1002/etc.3538. [DOI] [PubMed] [Google Scholar]
- 59.Guo M.J., Wang Y.G., Yuan X.Y., Dong S.Q., Wen Y.Y., Song X.E., Guo P.Y. Responses of the antioxidant system to fluroxypyr in foxtail millet (Setaria italica L.) at the seedling stage. J. Integr. Agric. 2018;17:554–565. doi: 10.1016/S2095-3119(17)61808-2. [DOI] [Google Scholar]
- 60.Hu T. A glutathione s-transferase confers herbicide tolerance in rice. Crop Breed. Appl. Biotechnol. 2014;14:76–81. doi: 10.1590/1984-70332014v14n2a14. [DOI] [Google Scholar]
- 61.Eltayeb A.E., Yamamoto S., Habora M.E.E., Yin L., Tsujimoto H., Tanaka K. Transgenic potato overexpressing Arabidopsis cytosolic AtDHAR1 showed higher tolerance to herbicide, drought and salt stresses. Breed. Sci. 2011;61:3–10. doi: 10.1270/jsbbs.61.3. [DOI] [Google Scholar]
- 62.Zhao N., Li W., Bai S., Guo W., Yuan G., Wang F., Liu W., Wang J. Transcriptome profiling to identify genes involved in mesosulfuron-methyl resistance in Alopecurus aequalis. Front. Plant Sci. 2017;8:1–16. doi: 10.3389/fpls.2017.01391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Babineau M., Mahmood K., Mathiassen S.K., Kudsk P., Kristensen M. De novo transcriptome assembly analysis of weed Apera spica-venti from seven tissues and growth stages. BMC Genom. 2017;18:128. doi: 10.1186/s12864-017-3538-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Matzrafi M., Shaar-Moshe L., Rubin B., Peleg Z. Unraveling the transcriptional basis of temperature-dependent pinoxaden resistance in Brachypodium hybridum. Front. Plant Sci. 2017;8:1–11. doi: 10.3389/fpls.2017.01064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen J., Huang H., Wei S., Huang Z., Wang X., Zhang C. Investigating the mechanisms of glyphosate resistance in goosegrass (Eleusine indica (L.) Gaertn.) by RNA sequencing technology. Plant J. 2017;89:407–415. doi: 10.1111/tpj.13395. [DOI] [PubMed] [Google Scholar]
- 66.Duhoux A., Carrère S., Duhoux A., Délye C. Transcriptional markers enable identification of rye-grass (Lolium sp.) plants with non-target-site-based resistance to herbicides inhibiting acetolactate-synthase. Plant Sci. 2015;257:22–36. doi: 10.1016/j.plantsci.2017.01.009. [DOI] [PubMed] [Google Scholar]
- 67.Pan L., Gao H., Xia W., Zhang T., Dong L. Establishing a herbicide-metabolizing enzyme library in Beckmannia syzigachne to identify genes associated with metabolic resistance. J. Exp. Bot. 2016;67:1745–1757. doi: 10.1093/jxb/erv565. [DOI] [PubMed] [Google Scholar]
- 68.Yang Q., Deng W., Li X., Yu Q., Bai L., Zheng M. Target-site and non-target-site based resistance to the herbicide tribenuron-methyl in flixweed (Descurainia sophia L.) BMC Genom. 2016;17:551–564. doi: 10.1186/s12864-016-2915-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gardin J.A.C., Gouzy J., Carrère S., Délye C. ALOMYbase, a resource to investigate non-target-site-based resistance to herbicides inhibiting acetolactate-synthase (ALS) in the major grass weed Alopecurus myosuroides (black-grass) BMC Genom. 2015;16:590. doi: 10.1186/s12864-015-1804-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Doğramacı M., Foley M.E., Horvath D.P., Hernandez A.G., Khetani R.S., Fields C.J., Keating K.M., Mikel M.A., Anderson J.V. Glyphosate’s impact on vegetative growth in leafy spurge identifies molecular processes and hormone cross-talk associated with increased branching. BMC Genom. 2015;16:395. doi: 10.1186/s12864-015-1627-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gaines T.A., Lorentz L., Figge A., Herrmann J., Maiwald F., Ott M.C., Han H., Busi R., Yu Q., Powles S.B., et al. RNA-Seq transcriptome analysis to identify genes involved in metabolism-based diclofop resistance in Lolium rigidum. Plant J. 2014;78:865–876. doi: 10.1111/tpj.12514. [DOI] [PubMed] [Google Scholar]