Abstract
Reactive oxygen species (ROS) are involved in many important processes, including the growth, development, and responses to the environments, in rice (Oryza sativa) and Magnaporthe oryzae. Although ROS are known to be critical components in rice–M. oryzae interactions, their regulations and pathways have not yet been completely revealed. Recent studies have provided fascinating insights into the intricate physiological redox balance in rice–M. oryzae interactions. In M. oryzae, ROS accumulation is required for the appressorium formation and penetration. However, once inside the rice cells, M. oryzae must scavenge the host-derived ROS to spread invasive hyphae. On the other side, ROS play key roles in rice against M. oryzae. It has been known that, upon perception of M. oryzae, rice plants modulate their activities of ROS generating and scavenging enzymes, mainly on NADPH oxidase OsRbohB, by different signaling pathways to accumulate ROS against rice blast. By contrast, the M. oryzae virulent strains are capable of suppressing ROS accumulation and attenuating rice blast resistance by the secretion of effectors, such as AvrPii and AvrPiz-t. These results suggest that ROS generation and scavenging of ROS are tightly controlled by different pathways in both M. oryzae and rice during rice blast. In this review, the most recent advances in the understanding of the regulatory mechanisms of ROS accumulation and signaling during rice–M. oryzae interaction are summarized.
Keywords: reactive oxygen species (ROS), Magnaporthe oryzae, rice blast, disease resistance, rice-Magnaporthe oryzae interaction, NADPH oxidase OsRbohB, OsMT2b, NADP-malic enzyme2 (Os-NADP-ME2)
1. Introduction
ROS are highly reactive reduced forms of oxygen molecules, including superoxide (O2−), hydrogen peroxide (H2O2), hydroxyl radical (OH−), and singlet oxygen (O2) [1,2,3,4,5,6]. ROS are produced dependent on several classes of enzymes, including NADPH (nicotinamide adenine dinucleotide phosphate) oxidases, peroxidases, and oxidases, and cell compartments, which are mainly dominated by chloroplasts, mitochondria, and peroxisomes. The ROS generation by the enzymes and compartments has been summarized in some excellent recent reviews [6,7,8,9]. Among the ROS-generating enzymes, NADPH oxidases were best characterized in both Magnaporthe oryzae and rice. The NADPH oxidases (Nox1-Nox4, Nox5, and dual oxidase Duox) have FAD and NADPH-binding sites and an oxidase domain responsible for O2− generation [10]. In M. oryzae, there are three putative NADPH oxidases: Nox1, Nox2, and Nox3 [11]. Both Nox1 and Nox2 are known for ROS production in M. oryzae [11]. In rice, nine NADPH oxidases have been annotated. Among these NADPH oxidases, OsRbohB (a homolog of the catalytic subunit pg91phox) is well known as an important component in disease resistance against rice blast [10,12,13,14].
Rice blast is caused by the filamentous ascomycete, M. oryzae. It is one of the most serious diseases of crops and the top 10 fungal diseases in plants [15]. To establish successful infections in rice plants, M. oryzae has evolved highly sophisticated and specific infection strategies. Generally, M. oryzae utilizes highly specialized appressoria, which are produced from three-celled conidia on the rice surface, to gain entry therein [16,17]. The appressorium formation in M. oryzae requires temporal-spatial ROS production in both the germ tube tip and immature appressorium integrated with other signals. In addition, ROS production is involved in the maintenance of the F-actin network, as well as septin-dependent assemblies of the exocyst at the appressorial pore to facilitate the penetration. Once inside the rice cells, M. oryzae differentiates into invasive hyphae and then spreads to neighboring cells, resulting in the formation of lesions. During invasive growth, M. oryzae needs to elude the plant immunity, including suppressing ROS accumulation, to adapt to the host milieu [6,14,18]. On the other hand, the rice plants equip variable and remarkable defense systems to resist the potential attacks of M. oryzae. One general priming and critical event in rice immunity is the production of ROS to resist the infections of M. oryzae [2,6,14]. In the current review, the recent progresses in ROS production and signaling in both M. oryzae and rice during M. oryzae–rice interaction are summarized.
2. ROS Play Dual Roles in the Pathogenesis of M. oryzae
2.1. ROS Accumulations are Required for Infection Structure Formation and Penetration in M. oryzae
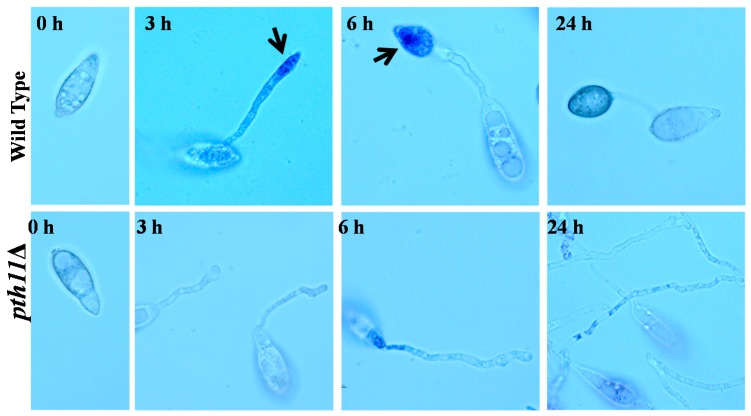
ROS are produced in the metabolic processes of M. oryzae and act as signals in a variety of developmental pathways, including the formation of infection structures. M. oryzae forms a specialized infection structure called the appressorium, which ruptures the host cuticles with high turgor, and drives a penetration peg into the underlying host cell. During appressoria morphogenesis, M. oryzae accumulates high levels of endogenous ROS in the tips of its germ tubes and immature appressoria (Figure 1) [11,19,20]. The scavenging of the ROS delays the differentiation of the appressoria, and also changes the morphology of the appressoria in M. oryzae. Recently, our studies reveal that the ROS accumulation is irregular and disrupted in the deletion mutants of PTH11 (Figure 1) and its downstream signaling components, MAGB and PMK1 [20,21]. Pth11 is an important G-protein coupled receptor required for the appressorium formation in M. oryzae [20,22]. Moreover, treatments with antioxidants induce functional appressoria differentiation in the pth11Δ strain [20], suggesting that the altered ROS accumulation is a probable cause of appressorium formation defect in pth11Δ.
Figure 1.
ROS accumulate in the tip of the germ tube and immature appressorium. Nitroblue tetrazolium (NBT) staining was performed in wild type or pth11Δ strains during the appressorium formation [20]. Arrows highlight the ROS in the tips of its germ tubes and immature appressoria. Bar = 5 μm.
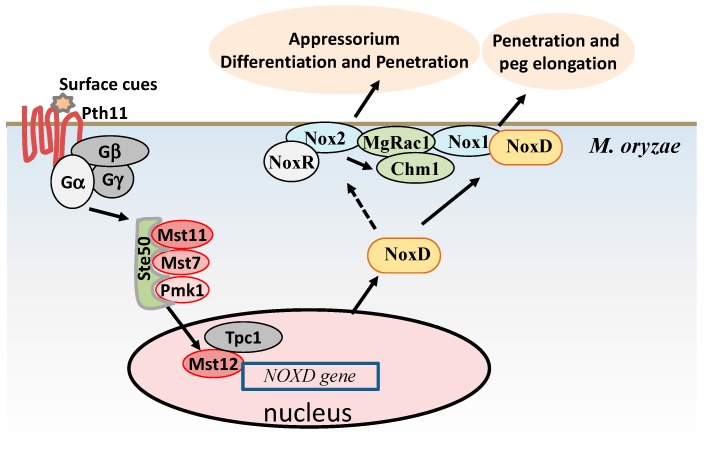
It is well known that gp91phox (Nox2) becomes active after the assembly of four cytosolic regulatory components (p47phox, p67phox, p40phox, and Rac2) with the integral membrane proteins cytochrome b558 (composed of catalytic subunit gp91phox and p22phox) in humans [23]. In M. oryzae, three putative homologs of the catalytic subunit gp91phox, Nox1, Nox2, and Nox3, have been characterized [11]. Nox1 is required for the ROS accumulations during appressorium morphogenesis and the elongation of the penetration peg [11,24]. Nox2 is necessary for the ROS accumulations in the hyphal tip and immature appressorium to form functional appressorium and the development of the penetration peg [11,24,25]. In M. oryzae, the successful infection of plants requires the maintenance of the F-actin network and septin-dependent assembly of the exocyst through the septin GTPases (sep3, Sep4, Sep5, and Sep6) at the appressorial pore. These processes require the synthesis of ROS by the Nox2-NoxR (p67phox homolog) complex to regulate the proper localization of Sep6 and organization of the exocyst complex [24,26]. Notably, Nox2 is known to regulate the localization of Chm1 kinase to possibly activate the PMK1 MAP (Mitogen-activated protein kinase) kinase pathway during the appressorium formation and penetration [26,27]. Similar to rice, the activities of both Nox1 and Nox2 may be regulated by small GTPase, MgRac1 [25]. The activity of MgRac1 is positively related to ROS accumulation in the hyphal tips [25]. Furthermore, MgRac1 directly interacts with Nox1, Nox2, and Chm1 to regulate ROS accumulation and the appressorium formation in M. oryzae [25]. In addition, the p22phox homolog, NoxD, interacts with Nox1 to play roles in the cytoskeletal re-modeling in the appressorium [28]. The expression of NOXD is directly regulated by the Tpc1 (transcription factor for polarity control 1) via an interaction with transcriptional factor Mst12 [28]. Therefore, the Tpc1 is required for NADPH oxidase dependent appressorium re-polarization through the control of the spatial and temporal regulation of the cortical F-actin during penetration [28]. All of the aforementioned findings reveal that M. oryzae needs an intricate physiological redox balance to regulate the septin assembly and formation of the F-actin network, which leads to functional appressorium formation and penetration (Figure 2).
Figure 2.
Model of NADPH oxidases-mediated appressorium formation and penetration in M. oryzae. The host surface cues are sensed by the G-protein coupled receptor, Pth11. After recognition of the cues, the Gα subunit binds to GTP (guanosine triphosphate) and dissociates from the Gβγ dimer and the induction of downstream MAPK (mitogen-activated protein kinase) cascades signaling occurs [21]. A component of the MAPK cascades signaling pathway, Mst12, interacts with the Zn(II)2Cys6 transcriptional regulator, Tpc1, to regulate the expression of the orthologue of the P22phox subunit (NOXD) of the NADPH oxidase complex. NoxD interacts with Nox1 and may be with Nox2 indirectly to control the elongation of the penetration peg. Both Nox1 and Nox2 interact with small GTPase, MgRac1, and may be regulated by MgRac1. In addition, the synthesis of ROS by the NoxR-Nox2 NADPH oxidase complex is required for the maintenance of the F-actin network and septin-dependent assembly of the exocyst at the appressorium pore to initiate plant infection.
2.2. Neutralization of the Host-Derived ROS is Required for Invasive Growth in Virulent Rice–M. Oryzae Interaction
As previously mentioned, ROS accumulation is required for functional appressorium formation and penetration in M. oryzae. However, once inside the rice cells, M. oryzae requires effective anti-oxidant defense systems to scavenge or detoxify the host-derived ROS to spread invasive hyphae by making a conclusion of the phenotypes of many mutants. In M. oryzae, secreted ROS scavenging enzymes, such as peroxidase and laccase, may contribute to the pathogenicity. The deletion mutants of a fungal-specific protein gene DES1, transcription factor MoAP1, oxidoreductase enzyme gene MoTRX2 (a target of the MoAP1), and protein phosphatase gene MoYVH1, exhibit stronger ROS accumulation, induction of defense response genes, and reduced pathogenicity [29,30,31,32]. It is believed that the ROS accumulations in the des1Δ, Moap1Δ, Motrx2Δ, and Moyvh1Δ mutants during M. oryzae infection may be related to lower production or activation levels of extracellular peroxidase and laccases [29,30,31,32]. Similarly, the deletion mutant of the soluble NSF (N-ethyl-maleimide-sensitive protein) attachment protein receptor, MoVAM7, which is required for the secretory transport of laccases, affects the ROS accumulation in the hypha and the pathogenicity [33]. Moreover, the mutants of ROS detoxifying enzymes encoding genes in M. oryzae, such as glutathione peroxidase domain-containing gene HRY1, glutathione reductase gene GTR1, and nitronate monooxygenases gene NMO2, fail to inhibit host ROS accumulation during infection [18,34,35]. Although these three enzymes likely exhibit ROS detoxifying activities, questions regarding how the fungal resident enzymes regulate the ROS accumulations in the host remain to be answered. A similar question can be raised about how the NAD (nicotinamide adenine dinucleotide)-dependent histone deacetylase MoSir2 regulates the expression of a fungal mitochondrion localized enzyme to suppress defense and the ROS accumulation in the host [36].
Moreover, there are an abundance of mutants found in M. oryzae, which fail to suppress the ROS accumulation in the host without having obvious relationships with the ROS generating or scavenging components [37,38,39,40,41,42,43,44]. Collectively, the results of previous studies have strongly implied that failures in ROS detoxification are highly likely to cause virulent defects in M. oryzae. However, this hypothesis is not supported by the observations of ROS levels using Grx1-roGFP2 in M. oryzae [19,45]. In regard to this issue, Samalova et al. proposed that M. oryzae has the ability to tolerate extreme oxidative stress, and does not undergo oxidative stress in planta [19]. One explanation may be that the function of these genes is more than just ROS detoxification in the ROS signaling of the host. Further elucidations of these mutants will provide important clues to understand the ROS generating and signaling in the virulent rice–M. oryzae interaction.
3. ROS as Key Players against M. oryzae Attacks in Rice
3.1. OsRbohB as an Important Component against Rice Blast
ROS play important roles in both the first line of defense termed as pathogen-associated molecular patterns (PAMPs) triggered immunity (PTI) and the second line of defense related to effector-triggered immunity (ETI) [2,6,9,14,46,47,48]. By the perception of the PAMPs, pattern recognition receptors (PRRs) activate a variety of immune responses, including the rapid and strong production of ROS through OsRbohB and other ROS generating components, to trigger PTI in rice [49,50]. In the second line of defense, plant intracellular immune receptors directly or indirectly recognize the specific pathogen effectors to induce ETI. In this situation, rice plants activate a series of signaling pathways, which lead to hypersensitive responses (HR). The HR activation involves the ROS burst, callose deposition, induction of the expression of pathogenesis-related protein genes, and programmed cell death [14,49]. In rice, OsRbohB is required for ROS burst in the HR during immunity against rice blast [12,13,51,52]. In addition, OsRbohB is also involved in the ROS generations, which are regulated by lesion mimic genes and plant hormones to contribute to the resistance of rice against M. oryzae [53,54].
3.1.1. Small GTPase OsRac1 Plays Dual Roles in the Induction of OsRbohB-Dependent ROS and the Suppression of ROS Scavenging during Immunity against Rice Blast
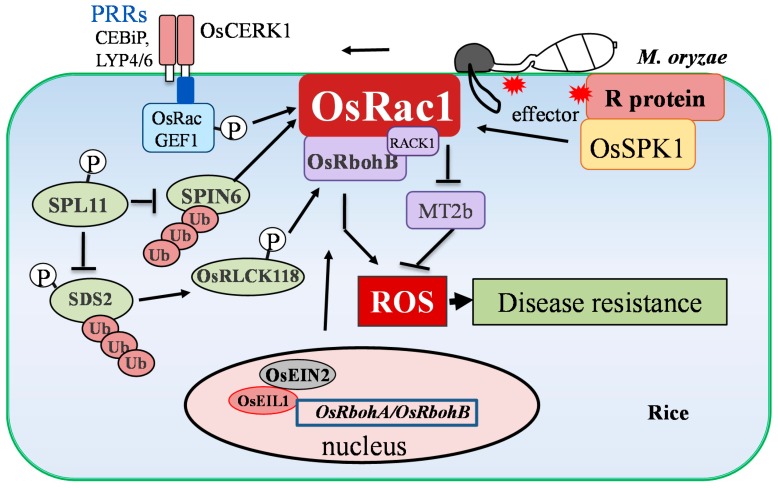
Some of the PAMPs from M. oryzae have been known to induce ROS in rice [55,56]. Chitin derived from fungi is one of the best-characterized PAMPs [56]. The perception of chitin in rice depends on a ligand induced complex between the LysM-containing proteins, CEBiP (chitin elicitor-binding protein) or LYP4/6 and OsCERK1 [57,58,59,60,61,62]. Upon the perception of chitin, the OsCERK1 forms a complex with OsRacGEF1/2 at the endoplasmic reticulum. Then, the complex is transported to the plasma membrane, where it interacts with the OsRac1 to activate a series of responses, including ROS production and expression of defense genes [63,64]. Recently, a new report suggests that the chitinase, MoChai, binds chitin to suppress the chitin-triggered plant immune response and ROS generation. Conversely, MoChai also acts as PMAP to induce PTI [48,65]. It is unknown whether the MoChai is involved in the OsRac1-mediated signaling. In addition, OsRac1 may directly interacts with several disease resistance (R) proteins (including Pit, Pib, Pi9, and Pita), which recognize the effectors from the M. oryzae and trigger ETI [51,66,67,68]. It has been noted that inductions of the HR and ROS burst by Pit are dependent on the OsRac1 [66,67]. Furthermore, recent studies showed that Pit or Pia activates OsRac1 by GEF OsSPK1 (a DOCK family guanine nucleotide exchange factor) on the plasma membrane [68]. All of the aforementioned results demonstrate that OsRac1 can be activated by both PTI and ETI to induce ROS accumulations during rice blast (Figure 3).
Figure 3.
OsRbohB as an important component against rice blast. Small GTPase, OsRac1, can be activated by both pathogen-associated molecular patterns (PAMPs) triggered immunity (PTI) and effector-triggered immunity (ETI) to induce OsRbohB-dependent ROS production and suppress ROS scavenging. By sensing the PAMPs or effectors, the OsRac1 is activated to induce ROS accumulation by NADPH oxidase OsRbohB activation and suppress ROS scavenging by down regulating the expression of OsMT2b. In addition, Spl11 (Spotted leaf 11) regulates the activity of OsRbohB through SDS2 (SPL11 cell-death suppressor) and OsRLCK118 to suppress ROS production. Moreover, the ethylene signaling components, OsEIN2, and its downstream transcription factor, OsEIL1, could regulate the expression of OsRbohA/OsRbohB to regulate ROS generation during rice–M. oryzae interaction.
It has been shown that OsRac1 regulates ROS production through its interaction with NADPH oxidase, OsRbohB [10,12,13,52]. Constitutively active OsRac1 induces ROS accumulation by regulating OsRbohB activity and enhances the resistance of rice to M. oryzae [10,12,51,69]. Meanwhile, the OsRac1-interacted protein, RACK1A, which plays a role in the resistance against rice blast infection, is also known to interact with OsRbohB [52]. The OsRbohB-knockdown rice plants increase the susceptibility to virulent M. oryzae isolates, suggesting that OsRbohB is essential for basal resistance [13]. However, it remains to be seen whether knockdown OsRbohB compromises the R gene mediated disease resistance. In addition to inducing ROS burst, OsRac1 down regulates the metallothionein gene (OsMT2b) to suppress ROS scavenging [70]. The OsMT2b has superoxide- and hydroxyl radical-scavenging activities and functions as a negative regulator of rice blast resistance [70]. Taken together, these findings provide fascinating evidence that OsRac1 plays dual roles in the induction of ROS production and the suppression of ROS scavenging (Figure 3).
3.1.2. Rice Blast Resistance Pathways Involved in spotted leaf 11 (spl11) could be Integrated with ROS Signaling via OsRbohB
ROS generation must be tightly controlled to avoid detrimental effects on rice plants. It must be produced in appropriate amounts, with the correct localization, and at the right time. Abnormal accumulation of ROS may cause defects in the rice, such as lesion-mimics and spots. Over the last two decades, many lesion-mimic mutants in rice have been identified with an abnormal accumulation of ROS and programmed cell death [71,72,73,74,75,76,77,78,79,80,81,82,83,84]. The majority of these lesion-mimic mutants have displayed enhanced disease resistance, including resistance against rice blast [72,74,76,77,78,79,80,83]. However, spontaneous lesions in rice plants with increased ROS accumulation and susceptibility to M. oryzae infection have been observed, suggesting that highly accumulated ROS in rice may not directly kill M. oryzae during infection [85].
SPL11 encodes an E3 ubiquitin ligase [86], which is one of best characterized lesion-mimic mutant genes found in rice. It has been determined that the spl11 mutant accumulates increased H2O2 and confers resistance to rice blast [73,87]. Spl11 interacts with a Rho GTPase activating protein, SPIN6, to ubiquitinate and degrade SPIN6 through the 26S proteasome pathway [53]. RNAi silencing or knockout of the SPIN6 gene elevates the chitin- and flg22-mediated defense responses and the resistance to rice blast [53]. Moreover, SPIN6 functions as a GTPase-activating protein (GAP) towards the OsRac1-associated defense response [53]. Recently, it has been found that Spl11 regulates the activity of the OsRbohB through the SDS2 and OsRLCK118 [88]. SDS2, encoding S-domain receptor-like kinase, is an Spl11 cell-death suppressor, as well as a positive regulator of resistance to rice blast [88,89]. SDS2 interacts with and phosphorylates OsRLCK118, which positively regulate rice blast resistance by phosphorylating the OsRbohB to stimulate the generation of ROS [88]. These results highlight that the pathways involved in the spl11 mutant could be integrated with the ROS signaling via the OsRbohB (Figure 3). In addition, detailed analyses between ROS production and other lesion-mimic mutant genes would be expected.
3.1.3 Plant Hormones May Regulate ROS Accumulation through OsRbohA/OsRbohB during Rice-M. oryzae Interaction
Other possible regulators of ROS generation (for example, plant hormones) during resistance against rice blast have also been investigated during recent years. Plant hormones, such as salicylic acids (SA), jasmonates (JAs), and ethylene (ET), are known as signals of diverse array of defense responses in rice [4,90]. Treating rice plants with SA or methyl-jasmonate (MeJA) induces ROS accumulation and enhances the resistance against rice blast [91,92]. The SA-deficient NahG rice plants, which have greatly reduced SA due to the overexpression of the bacterial nahG gene, increase the levels of superoxide and H2O2 and the susceptibility to the virulent isolate of M. oryzae [85]. Also, it has been shown that the ethylene (ET) biosynthesis is important for rice plants’ resistance to rice blast [93,94]. Recently, the ethylene signaling components, OsEIN2, and its downstream transcription factor, OsEIL1, have been determined to positively regulate ROS accumulation and disease resistance to rice blast [54]. Interestingly, the OsEIL1 directly binds to the promoters of OsRbohA/OsRbohB [54], suggesting that plant hormones may possibly regulate ROS accumulation through OsRbohA/OsRbohB during rice–M. oryzae interaction (Figure 3).
3.2. NADP-Malic Enzyme 2 Regulates ROS Production during M. oryzae Infection
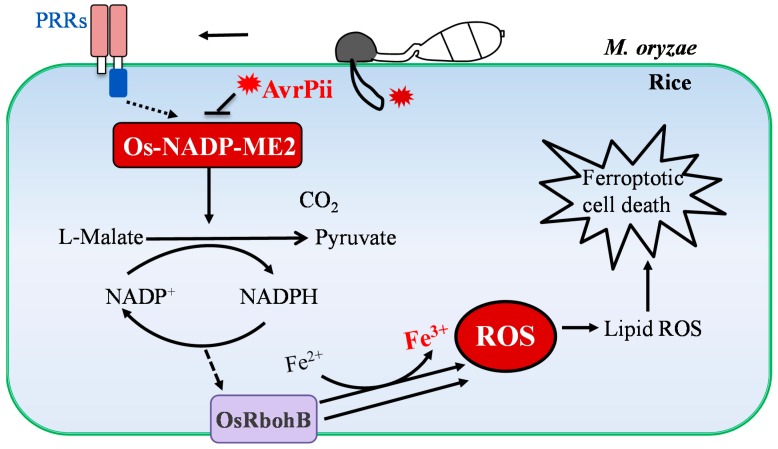
In general, evidence suggests that avirulent pathogens induce a strong ROS burst and HR cell death [2,14]. The most recent study has highlighted that the accumulation of ferric ions and ROS-dependent ferroptotic cell death occur during avirulent M. oryzae infection [95]. Suppressing iron-dependent ROS accumulation and lipid peroxidation with ferroptosis inhibitors and the NADPH oxidase inhibitor abolish the HR cell death during avirulent M. oryzae infection [95]. In contrast, the HR cell death is induced in virulent M. oryzae infection by erastin, which can trigger iron-dependent ROS burst, the accumulation of ferric ions, and glutathione depletion [95]. Notably, chitin fails to induce the accumulation of ferric ions. The accumulation of ROS and ferric ions in the HR cell death is dependent on the NADP-malic enzyme 2 (Os-NADP-ME2) (Figure 4) [95,96]. Knock out of the Os-NADP-ME2 gene in a resistant rice line disrupts the immunity against M. oryzae, suggesting that Os-NADP-ME2 plays an important role in the exclusion of pathogens. Furthermore, recent research has shown that Os-NADP-ME2 is involved in AvrPii-triggered ROS inhibition [96]. The interaction of Os-NADP-ME2 and AvrPii suppresses the malic enzyme activity of Os-NADP-ME2 and NADPH production to attenuate the host ROS burst in virulent M. oryzae infection [96]. As indicated above, the Os-NADP-ME2 may directly regulate the ROS production during immunity. It would be interesting to determine the virulence functions of the AvrPii, as well as the functions of the Os-NADP-ME2 in Pii-mediated disease resistance. Additionally, Os-NADP-ME2 is also required for chitin-triggered ROS burst [95,96]. A detailed correlation between the PAMPs-induced ROS production and Os-NADP-ME2 activity would be expected. Furthermore, it is worthwhile to clarify whether the Os-NADP-ME2 regulates ROS production during immunity through OsRbohB.
Figure 4.
NADP-malic enzyme 2 regulates ROS production during M. oryzae infection. Os-NADP-ME2 is involved in the AvrPii-triggered ROS inhibition, chitin-triggered ROS burst, and ROS-dependent ferroptotic cell death. The malic enzyme activity of Os-NADP-ME2 is activated by chitin-treatment, while it can be suppressed by AvrPii. The activated Os-NADP-ME2 catalyzes malic acid into pyruvic acid, which generates NADPH, and subsequently produces ROS to induce the ROS-dependent ferroptotic cell death in rice.
3.3. Other ROS Accumulation Mechanisms during Rice–M. Oryzae Interaction
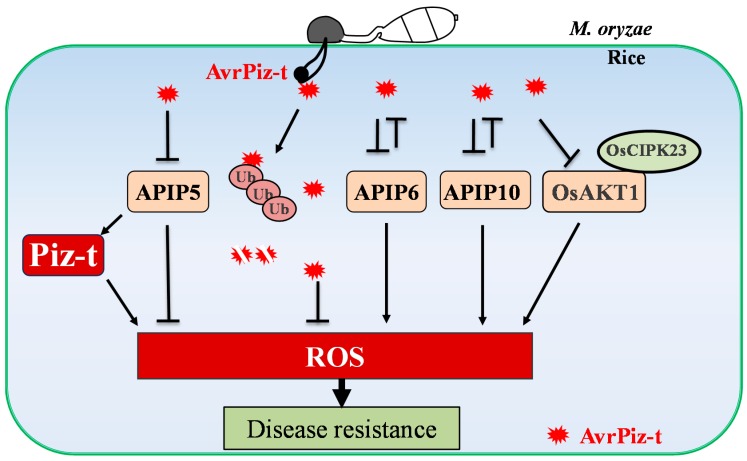
Another best-characterized ROS related signaling pathway in rice is the Piz-t mediated disease resistance. The R protein Piz-t recognizes the cognate avirulence factor, AvrPiz-t, which is a secreted 108 amino acids polypeptide, by indirect interaction [97,98]. It is known that AvrPiz-t functions to suppress flg22- and chitin-induced ROS generation, and also enhances the susceptibility of rice plants to M. oryzae [99]. Recent studies have shown that the AvrPiz-t targets proteins in rice, including APIP5, APIP6, APIP10, APIP12, and OsAKT1, to modulate the host defense responses (Figure 5) [99,100,101,102,103]. Both the APIP6 and APIP10 are RING E3 ubiquitin ligase, which ubiquitinate AvrPiz-t during infection [99,100]. Conversely, AvrPiz-t suppresses the ubiquitin ligase activities of the APIP6 and APIP10. Silencing APIP6 or APIP10 reduces flg22-induced ROS production, and enhances the susceptibility of rice plants to M. oryzae [99,100]. APIP5, which is a bZIP-type transcriptional factor, interacts with both AvrPiz-t and Piz-t. AvrPiz-t attenuates the transcriptional activity and protein accumulation of the APIP5, while Piz-t stabilizes the APIP5 to prevent necrosis during the necrotrophic stage. Silencing APIP5 leads to ROS accumulation, cell death, and enhanced resistance to rice blast [101]. AvrPiz-t also interacts with OsAKT1, which is a rice plasma-membrane-localized K+ channel protein, and suppresses the OsAKT1-mediated K+ currents [103]. OsAKT1, with the cytoplasmic kinase, OsCIPK23, plays a positive role in K+ absorption, chitin-induced ROS accumulation, and resistance against M. oryzae [103]. It has become apparent that the accumulation of ROS is clearly important in AvrPiz-t triggered immunity. However, at the present time, very little is actually known about how AvrPiz-t suppresses ROS production or how AvrPiz-t interacted proteins regulate ROS accumulation. One possibility has been mentioned that the OsMT2b is downregulated in APIP5 RNAi transgenic plants [70,101]. It will be important to clarify the relationship between AvrPiz-t and its interacted proteins and the ROS generating components or ROS scavengers during immunity in rice plants.
Figure 5.
AvrPiz-t and AvrPiz-t targeted proteins regulate ROS accumulation during immunity. APIP5, APIP6, and APIP10 are AvrPiz-t interacting proteins. OsAKT1, which interacts with AvrPiz-t, is a rice plasma-membrane-localized K+ channel protein. Silencing APIP6, APIP10, or OsAKT1 reduces the PMAP-induced ROS production. In contrast, silencing APIP5 leads to ROS accumulation.
In addition, there are a few of other genes, such as calcium-dependent protein kinase, OsCPK10, DICER-like (DCL) ribonuclease, as well as AGC kinase, OsOxi1, and its interacted protein, OsPti1a, which have been demonstrated to regulate ROS accumulation in the resistance of rice against M. oryzae [104,105,106,107]. However, the mechanisms that these genes regulate ROS accumulation remain largely unknown at this time. The most recent report has shown that the rice miRNA, miR398b, could target Cu/Zn-Superoxidase Dismutasel (CSD1), CSD2, and SODX to boost H2O2 accumulation and enhance resistance to M. oryzae [108]. These reports suggest that rice plants could regulate ROS accumulation by multiple ways to contribute to resistance against M. oryzae.
4. Conclusions
It is known that M. oryzae needs an intricate ROS accumulation during the appressorium formation and penetration, as well as the neutralization of host-derived ROS during in planta growth. However, there are still many gaps in our knowledge of ROS regulations in M. oryzae. Most importantly, how M. oryzae suppresses the host-derived ROS accumulation remains largely unclear. Also, whether ROS are related to the biotrophy-necrotrophy switch of rice blast has not yet been confirmed.
On the other hand, there is no doubt that ROS production plays an important role in rice against M. oryzae. However, little is known about where the ROS originates from during rice blast. As mentioned above, three enzymes (NADPH oxidase OsRbohB, metallothionein OsMT2b, and NADP-malic enzyme 2 Os-NADP-ME2) are highly regulated and are responsible for ROS accumulation during rice blast infection. However, with the exception of those enzymes, the functions of other ROS generating or scavenging enzymes during rice immunity remain unclear. In addition, barely anything is currently known about the spatio-temporal generation and accumulation of ROS in the rice cells during rice–M. oryzae interaction. The detection or measurement of ROS in local and systemic tissues of rice is one of the bottlenecks for analyzing the role of ROS during rice blast. Luminol-chemiluminescence assay by using rice protoplast and DAB (3,3’-Diaminobenzidine) staining are the most common methods to determine ROS in rice. However, it is difficult to use these methods to track ROS generation and accumulation during rice–M. oryzae interaction. It might be helpful to use reporters, such as the luciferase reporter, roGFP, or the Hyper sensor, for living cell imaging to answer this important question [19,47,109]. Another important question is why the ROS burst actually occurs. It seems that ROS produced by rice are not a sufficient toxic line of defense to M. oryzae even in avirulent interactions or transgenic rice plants with spontaneous lesions [19,85]. It is possible that ROS play a dominant role in signaling, rather than in ROS toxicity. If this is true, it would be important to determine the downstream events of ROS signaling during rice–M. oryzae interaction. Moreover, it will be interesting to address the mechanisms that determine the ROS signal specificity in PTI and ETI.
Acknowledgments
This project was supported by the Chinese Academy of Agricultural Sciences under the “Elite Youth” program, the Agricultural Sciences and Technologies Innovation Program. We thank Prof. Yan Liang, Yoji Kawano, Yuese Ning, and Jun Liu for helpful discussion and suggestions.
Funding
This research was funded by Zhejiang Provincial Natural Science Foundation of China, grant number “LQ19C140004 and LQ19C130007” and Key project of Zhejiang province, grant number “2019C02018”.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Kotchoni S.O., Gachomo E.W. The reactive oxygen species network pathways:an essential prerequisite for perception of pathogen attack and the acquired disease resistance in plants. J. Biosci. 2006;31:389–404. doi: 10.1007/BF02704112. [DOI] [PubMed] [Google Scholar]
- 2.Averyanov A. Oxidative burst and plant disease resistance. Front. Biosci. 2009;1:142–152. doi: 10.2741/E14. [DOI] [PubMed] [Google Scholar]
- 3.Nathan C., Cunningham-Bussel A. Beyond oxidative stress: An immunologist’s guide to reactive oxygen species. Nat. Rev. Immunol. 2013;13:349–361. doi: 10.1038/nri3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang D.L., Yang Y., He Z. Roles of plant hormones and their interplay in rice immunity. Mol. Plant. 2013;6:675–685. doi: 10.1093/mp/sst056. [DOI] [PubMed] [Google Scholar]
- 5.Gilroy S., Suzuki N., Miller G., Choi W.G., Toyota M., Devireddy A.R., Mittler R. A tidal wave of signals: Calcium and ROS at the forefront of rapid systemic signaling. Trends Plant Sci. 2014;19:623–630. doi: 10.1016/j.tplants.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 6.Camejo D., Guzman-Cedeno A., Moreno A. Reactive oxygen species, essential molecules, during plant-pathogen interactions. Plant Physiol. Biochem. 2016;103:10–23. doi: 10.1016/j.plaphy.2016.02.035. [DOI] [PubMed] [Google Scholar]
- 7.Tripathy B.C., Oelmuller R. Reactive oxygen species generation and signaling in plants. Plant Signal. Behav. 2012;7:1621–1633. doi: 10.4161/psb.22455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shapiguzov A., Vainonen J.P., Wrzaczek M., Kangasjarvi J. ROS-talk—How the apoplast, the chloroplast, and the nucleus get the message through. Front. Plant Sci. 2012;3:292. doi: 10.3389/fpls.2012.00292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mittler R. ROS Are Good. Trends Plant Sci. 2017;22:11–19. doi: 10.1016/j.tplants.2016.08.002. [DOI] [PubMed] [Google Scholar]
- 10.Oda T., Hashimoto H., Kuwabara N., Akashi S., Hayashi K., Kojima C., Wong H.L., Kawasaki T., Shimamoto K., Sato M., et al. Structure of the N-terminal regulatory domain of a plant NADPH oxidase and its functional implications. J. Biol. Chem. 2010;285:1435–1445. doi: 10.1074/jbc.M109.058909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egan M.J., Wang Z.Y., Jones M.A., Smirnoff N., Talbot N.J. Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc. Natl. Acad. Sci. USA. 2007;104:11772–11777. doi: 10.1073/pnas.0700574104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kosami K., Ohki I., Nagano M., Furuita K., Sugiki T., Kawano Y., Kawasaki T., Fujiwara T., Nakagawa A., Shimamoto K., et al. The crystal structure of the plant small GTPase OsRac1 reveals its mode of binding to NADPH oxidase. J. Biol. Chem. 2014;289:28569–28578. doi: 10.1074/jbc.M114.603282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagano M., Ishikawa T., Fujiwara M., Fukao Y., Kawano Y., Kawai-Yamada M., Shimamoto K. Plasma Membrane Microdomains Are Essential for Rac1-RbohB/H-Mediated Immunity in Rice. Plant Cell. 2016;28:1966–1983. doi: 10.1105/tpc.16.00201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jwa N.S., Hwang B.K. Convergent Evolution of Pathogen Effectors toward Reactive Oxygen Species Signaling Networks in Plants. Front. Plant Sci. 2017;8:1687. doi: 10.3389/fpls.2017.01687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dean R., Van Kan J.A., Pretorius Z.A., Hammond-Kosack K.E., Di Pietro A., Spanu P.D., Rudd J.J., Dickman M., Kahmann R., Ellis J., et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012;13:414–430. doi: 10.1111/j.1364-3703.2011.00783.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hamer J.E., Howard R.J., Chumley F.G., Valent B. A mechanism for surface attachment in spores of a plant pathogenic fungus. Science. 1988;239:288–290. doi: 10.1126/science.239.4837.288. [DOI] [PubMed] [Google Scholar]
- 17.Kou Y., Naqvi N.I. Surface sensing and signaling networks in plant pathogenic fungi. Semin. Cell Dev. Biol. 2016;57:84–92. doi: 10.1016/j.semcdb.2016.04.019. [DOI] [PubMed] [Google Scholar]
- 18.Marroquin-Guzman M., Hartline D., Wright J.D., Elowsky C., Bourret T.J., Wilson R.A. The Magnaporthe oryzae nitrooxidative stress response suppresses rice innate immunity during blast disease. Nat. Microbiol. 2017;2:17054. doi: 10.1038/nmicrobiol.2017.54. [DOI] [PubMed] [Google Scholar]
- 19.Samalova M., Meyer A.J., Gurr S.J., Fricker M.D. Robust anti-oxidant defences in the rice blast fungus Magnaporthe oryzae confer tolerance to the host oxidative burst. New Phytol. 2014;201:556–573. doi: 10.1111/nph.12530. [DOI] [PubMed] [Google Scholar]
- 20.Kou Y., Tan Y.H., Ramanujam R., Naqvi N.I. Structure-function analyses of the Pth11 receptor reveal an important role for CFEM motif and redox regulation in rice blast. New Phytol. 2017;214:330–342. doi: 10.1111/nph.14347. [DOI] [PubMed] [Google Scholar]
- 21.Park G., Xue C., Zhao X., Kim Y., Orbach M., Xu J.R. Multiple upstream signals converge on the adaptor protein Mst50 in Magnaporthe grisea. Plant Cell. 2006;18:2822–2835. doi: 10.1105/tpc.105.038422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.DeZwaan T.M., Carroll A.M., Valent B., Sweigard J.A. Magnaporthe grisea pth11p is a novel plasma membrane protein that mediates appressorium differentiation in response to inductive substrate cues. Plant Cell. 1999;11:2013–2030. doi: 10.1105/tpc.11.10.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nauseef W.M. Nox enzymes in immune cells. Semin. Immunopathol. 2008;30:195–208. doi: 10.1007/s00281-008-0117-4. [DOI] [PubMed] [Google Scholar]
- 24.Ryder L.S., Dagdas Y.F., Mentlak T.A., Kershaw M.J., Thornton C.R., Schuster M., Chen J., Wang Z., Talbot N.J. NADPH oxidases regulate septin-mediated cytoskeletal remodeling during plant infection by the rice blast fungus. Proc. Natl. Acad. Sci. USA. 2013;110:3179–3184. doi: 10.1073/pnas.1217470110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen J., Zheng W., Zheng S., Zhang D., Sang W., Chen X., Li G., Lu G., Wang Z. Rac1 is required for pathogenicity and Chm1-dependent conidiogenesis in rice fungal pathogen Magnaporthe grisea. PLoS Pathog. 2008;4:e1000202. doi: 10.1371/journal.ppat.1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gupta Y.K., Dagdas Y.F., Kershaw M.J., Littlejohn G.R., Martinez-Rocha A.L., Menke F., Ryder L.S., Sklenar J., Talbot N.J. Septin-Dependent Assembly of the Exocyst Is Essential for Plant Infection by Magnaporthe oryzae. Plant Cell. 2015;27:3277–3289. doi: 10.1105/tpc.15.00552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li L., Xue C., Bruno K., Nishimura M., Xu J.R. Two PAK kinase genes, CHM1 and MST20, have distinct functions in Magnaporthe grisea. Mol. Plant-Microbe Interact. 2004;17:547–556. doi: 10.1094/MPMI.2004.17.5.547. [DOI] [PubMed] [Google Scholar]
- 28.Galhano R., Illana A., Ryder L.S., Rodriguez-Romero J., Demuez M., Badaruddin M., Martinez-Rocha A.L., Soanes D.M., Studholme D.J., Talbot N.J., et al. Tpc1 is an important Zn(II)2Cys6 transcriptional regulator required for polarized growth and virulence in the rice blast fungus. PLoS Pathog. 2017;13:e1006516. doi: 10.1371/journal.ppat.1006516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chi M.H., Park S.Y., Kim S., Lee Y.H. A novel pathogenicity gene is required in the rice blast fungus to suppress the basal defenses of the host. PLoS Pathog. 2009;5:e1000401. doi: 10.1371/journal.ppat.1000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo M., Chen Y., Du Y., Dong Y., Guo W., Zhai S., Zhang H., Dong S., Zhang Z., Wang Y., et al. The bZIP transcription factor MoAP1 mediates the oxidative stress response and is critical for pathogenicity of the rice blast fungus Magnaporthe oryzae. PLoS Pathog. 2011;7:e1001302. doi: 10.1371/journal.ppat.1001302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang J., Yin Z., Tang W., Cai X., Gao C., Zhang H., Zheng X., Wang P., Zhang Z. The thioredoxin MoTrx2 protein mediates reactive oxygen species (ROS) balance and controls pathogenicity as a target of the transcription factor MoAP1 in Magnaporthe oryzae. Mol. Plant Pathol. 2017;18:1199–1209. doi: 10.1111/mpp.12484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu X., Yang J., Qian B., Cai Y., Zou X., Zhang H., Zheng X., Wang P., Zhang Z. MoYvh1 subverts rice defense through functions of ribosomal protein MoMrt4 in Magnaporthe oryzae. PLoS Pathog. 2018;14:e1007016. doi: 10.1371/journal.ppat.1007016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dou X., Wang Q., Qi Z., Song W., Wang W., Guo M., Zhang H., Zhang Z., Wang P., Zheng X. MoVam7, a conserved SNARE involved in vacuole assembly, is required for growth, endocytosis, ROS accumulation, and pathogenesis of Magnaporthe oryzae. PLoS ONE. 2011;6:e16439. doi: 10.1371/journal.pone.0016439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang K., Czymmek K.J., Caplan J.L., Sweigard J.A., Donofrio N.M. HYR1-mediated detoxification of reactive oxygen species is required for full virulence in the rice blast fungus. PLoS Pathog. 2011;7:e1001335. doi: 10.1371/journal.ppat.1001335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fernandez J., Wilson R.A. Characterizing roles for the glutathione reductase, thioredoxin reductase and thioredoxin peroxidase-encoding genes of Magnaporthe oryzae during rice blast disease. PLoS ONE. 2014;9:e87300. doi: 10.1371/journal.pone.0087300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fernandez J., Marroquin-Guzman M., Nandakumar R., Shijo S., Cornwell K.M., Li G., Wilson R.A. Plant defence suppression is mediated by a fungal sirtuin during rice infection by Magnaporthe oryzae. Mol. Microbiol. 2014;94:70–88. doi: 10.1111/mmi.12743. [DOI] [PubMed] [Google Scholar]
- 37.Liu X., Cai Y., Zhang X., Zhang H., Zheng X., Zhang Z. Carbamoyl Phosphate Synthetase Subunit MoCpa2 Affects Development and Pathogenicity by Modulating Arginine Biosynthesis in Magnaporthe oryzae. Front. Microbiol. 2016;7:2023. doi: 10.3389/fmicb.2016.02023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li Y., Zhu J., Hu J., Meng X., Zhang Q., Zhu K., Chen X., Chen X., Li G., Wang Z., et al. Functional characterization of electron-transferring flavoprotein and its dehydrogenase required for fungal development and plant infection by the rice blast fungus. Sci. Rep. 2016;6:24911. doi: 10.1038/srep24911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chen X.L., Shen M., Yang J., Xing Y., Chen D., Li Z., Zhao W., Zhang Y. Peroxisomal fission is induced during appressorium formation and is required for full virulence of the rice blast fungus. Mol. Plant Pathol. 2017;18:222–237. doi: 10.1111/mpp.12395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L., Wang J., Chen H., Chai R., Zhang Z., Mao X., Qiu H., Jiang H., Wang Y., Sun G. Pex14/17, a filamentous fungus-specific peroxin, is required for the import of peroxisomal matrix proteins and full virulence of Magnaporthe oryzae. Mol. Plant Pathol. 2017;18:1238–1252. doi: 10.1111/mpp.12487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fan G., Zhang K., Huang H., Zhang H., Zhao A., Chen L., Chen R., Li G., Wang Z., Lu G.D. Multiprotein-bridging factor 1 regulates vegetative growth, osmotic stress, and virulence in Magnaporthe oryzae. Curr. Genet. 2017;63:293–309. doi: 10.1007/s00294-016-0636-9. [DOI] [PubMed] [Google Scholar]
- 42.Pan Y., Pan R., Tan L., Zhang Z., Guo M. Pleiotropic roles of O-mannosyltransferase MoPmt4 in development and pathogenicity of Magnaporthe oryzae. Curr. Genet. 2018;65:223–239. doi: 10.1007/s00294-018-0864-2. [DOI] [PubMed] [Google Scholar]
- 43.Qian B., Liu X., Jia J., Cai Y., Chen C., Zhang H., Zheng X., Wang P., Zhang Z. MoPpe1 partners with MoSap1 to mediate TOR and cell wall integrity signalling in growth and pathogenicity of the rice blast fungus Magnaporthe oryzae. Environ. Microbiol. 2018;20:3964–3979. doi: 10.1111/1462-2920.14421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang S., Liu X., Li L., Yu R., He J., Zhang H., Zheng X., Wang P., Zhang Z. The ArfGAP protein MoGlo3 regulates the development and pathogenicity of Magnaporthe oryzae. Environ. Microbiol. 2017;19:3982–3996. doi: 10.1111/1462-2920.13798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Donofrio N.M., Wilson R.A. Redox and rice blast: New tools for dissecting molecular fungal-plant interactions. New Phytol. 2014;201:367–369. doi: 10.1111/nph.12623. [DOI] [PubMed] [Google Scholar]
- 46.Torres M.A. ROS in biotic interactions. Physiol. Plant. 2010;138:414–429. doi: 10.1111/j.1399-3054.2009.01326.x. [DOI] [PubMed] [Google Scholar]
- 47.Miller G., Schlauch K., Tam R., Cortes D., Torres M.A., Shulaev V., Dangl J.L., Mittler R. The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal. 2009;2:ra45. doi: 10.1126/scisignal.2000448. [DOI] [PubMed] [Google Scholar]
- 48.Peng Y., van Wersch R., Zhang Y. Convergent and Divergent Signaling in PAMP-Triggered Immunity and Effector-Triggered Immunity. Mol. Plant-Microbe Interact. 2018;31:403–409. doi: 10.1094/MPMI-06-17-0145-CR. [DOI] [PubMed] [Google Scholar]
- 49.Liu W., Liu J., Triplett L., Leach J.E., Wang G.L. Novel insights into rice innate immunity against bacterial and fungal pathogens. Annu. Rev. Phytopathol. 2014;52:213–241. doi: 10.1146/annurev-phyto-102313-045926. [DOI] [PubMed] [Google Scholar]
- 50.Sang Y., Macho A.P. Analysis of PAMP-Triggered ROS Burst in Plant Immunity. Methods Mol. Biol. 2017;1578:143–153. doi: 10.1007/978-1-4939-6859-6_11. [DOI] [PubMed] [Google Scholar]
- 51.Ono E., Wong H.L., Kawasaki T., Hasegawa M., Kodama O., Shimamoto K. Essential role of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. USA. 2001;98:759–764. doi: 10.1073/pnas.98.2.759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakashima A., Chen L., Thao N.P., Fujiwara M., Wong H.L., Kuwano M., Umemura K., Shirasu K., Kawasaki T., Shimamoto K. RACK1 functions in rice innate immunity by interacting with the Rac1 immune complex. Plant Cell. 2008;20:2265–2279. doi: 10.1105/tpc.107.054395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu J., Park C.H., He F., Nagano M., Wang M., Bellizzi M., Zhang K., Zeng X., Liu W., Ning Y., et al. The RhoGAP SPIN6 associates with SPL11 and OsRac1 and negatively regulates programmed cell death and innate immunity in rice. PLoS Pathog. 2015;11:e1004629. doi: 10.1371/journal.ppat.1004629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang C., Li W., Cao J., Meng F., Yu Y., Huang J., Jiang L., Liu M., Zhang Z., Chen X., et al. Activation of ethylene signaling pathways enhances disease resistance by regulating ROS and phytoalexin production in rice. Plant J. Cell Mol. Biol. 2017;89:338–353. doi: 10.1111/tpj.13388. [DOI] [PubMed] [Google Scholar]
- 55.Fang Y.L., Peng Y.L., Fan J. The Nep1-like protein family of Magnaporthe oryzae is dispensable for the infection of rice plants. Sci. Rep. 2017;7:4372. doi: 10.1038/s41598-017-04430-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Boller T., Felix G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 57.Hayafune M., Berisio R., Marchetti R., Silipo A., Kayama M., Desaki Y., Arima S., Squeglia F., Ruggiero A., Tokuyasu K., et al. Chitin-induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich-type dimerization. Proc. Natl. Acad. Sci. USA. 2014;111:E404–E413. doi: 10.1073/pnas.1312099111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kaku H., Nishizawa Y., Ishii-Minami N., Akimoto-Tomiyama C., Dohmae N., Takio K., Minami E., Shibuya N. Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl. Acad. Sci. USA. 2006;103:11086–11091. doi: 10.1073/pnas.0508882103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shimizu T., Nakano T., Takamizawa D., Desaki Y., Ishii-Minami N., Nishizawa Y., Minami E., Okada K., Yamane H., Kaku H., et al. Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. Cell Mol. Biol. 2010;64:204–214. doi: 10.1111/j.1365-313X.2010.04324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kouzai Y., Mochizuki S., Nakajima K., Desaki Y., Hayafune M., Miyazaki H., Yokotani N., Ozawa K., Minami E., Kaku H., et al. Targeted gene disruption of OsCERK1 reveals its indispensable role in chitin perception and involvement in the peptidoglycan response and immunity in rice. Mol. Plant-Microbe Interact. 2014;27:975–982. doi: 10.1094/MPMI-03-14-0068-R. [DOI] [PubMed] [Google Scholar]
- 61.Liu B., Li J.F., Ao Y., Qu J., Li Z., Su J., Zhang Y., Liu J., Feng D., Qi K., et al. Lysin motif-containing proteins LYP4 and LYP6 play dual roles in peptidoglycan and chitin perception in rice innate immunity. Plant Cell. 2012;24:3406–3419. doi: 10.1105/tpc.112.102475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kishimoto K., Kouzai Y., Kaku H., Shibuya N., Minami E., Nishizawa Y. Perception of the chitin oligosaccharides contributes to disease resistance to blast fungus Magnaporthe oryzae in rice. Plant J. Cell Mol. Biol. 2010;64:343–354. doi: 10.1111/j.1365-313X.2010.04328.x. [DOI] [PubMed] [Google Scholar]
- 63.Akamatsu A., Wong H.L., Fujiwara M., Okuda J., Nishide K., Uno K., Imai K., Umemura K., Kawasaki T., Kawano Y., et al. An OsCEBiP/OsCERK1-OsRacGEF1-OsRac1 module is an essential early component of chitin-induced rice immunity. Cell Host Microbe. 2013;13:465–476. doi: 10.1016/j.chom.2013.03.007. [DOI] [PubMed] [Google Scholar]
- 64.Akamatsu A., Uno K., Kato M., Wong H.L., Shimamoto K., Kawano Y. New insights into the dimerization of small GTPase Rac/ROP guanine nucleotide exchange factors in rice. Plant Signal. Behav. 2015;10:e1044702. doi: 10.1080/15592324.2015.1044702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Han Y., Song L., Peng C., Liu X., Liu L., Zhang Y., Wang W., Zhou J., Wang S., Ebbole D.J., et al. A Magnaporthe Chitinase Interacts with a Rice Jacalin-related Lectin to Promote Host Colonization. Plant Physiol. 2019 doi: 10.1104/pp.18.01594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kawano Y., Akamatsu A., Hayashi K., Housen Y., Okuda J., Yao A., Nakashima A., Takahashi H., Yoshida H., Wong H.L., et al. Activation of a Rac GTPase by the NLR family disease resistance protein Pit plays a critical role in rice innate immunity. Cell Host Microbe. 2010;7:362–375. doi: 10.1016/j.chom.2010.04.010. [DOI] [PubMed] [Google Scholar]
- 67.Kawano Y., Fujiwara T., Yao A., Housen Y., Hayashi K., Shimamoto K. Palmitoylation-dependent membrane localization of the rice resistance protein pit is critical for the activation of the small GTPase OsRac1. J. Biol. Chem. 2014;289:19079–19088. doi: 10.1074/jbc.M114.569756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang Q., Li Y., Ishikawa K., Kosami K.I., Uno K., Nagawa S., Tan L., Du J., Shimamoto K., Kawano Y. Resistance protein Pit interacts with the GEF OsSPK1 to activate OsRac1 and trigger rice immunity. Proc. Natl. Acad. Sci. USA. 2018;115:E11551–E11560. doi: 10.1073/pnas.1813058115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kawasaki T., Henmi K., Ono E., Hatakeyama S., Iwano M., Satoh H., Shimamoto K. The small GTP-binding protein rac is a regulator of cell death in plants. Proc. Natl. Acad. Sci. USA. 1999;96:10922–10926. doi: 10.1073/pnas.96.19.10922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wong H.L., Sakamoto T., Kawasaki T., Umemura K., Shimamoto K. Down-regulation of metallothionein, a reactive oxygen scavenger, by the small GTPase OsRac1 in rice. Plant Physiol. 2004;135:1447–1456. doi: 10.1104/pp.103.036384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ueno M., Shibata H., Kihara J., Honda Y., Arase S. Increased tryptophan decarboxylase and monoamine oxidase activities induce Sekiguchi lesion formation in rice infected with Magnaporthe grisea. Plant J. Cell Mol. Biol. 2003;36:215–228. doi: 10.1046/j.1365-313X.2003.01875.x. [DOI] [PubMed] [Google Scholar]
- 72.Zeng L.R., Qu S., Bordeos A., Yang C., Baraoidan M., Yan H., Xie Q., Nahm B.H., Leung H., Wang G.L. Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell. 2004;16:2795–2808. doi: 10.1105/tpc.104.025171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kojo K., Yaeno T., Kusumi K., Matsumura H., Fujisawa S., Terauchi R., Iba K. Regulatory mechanisms of ROI generation are affected by rice spl mutations. Plant Cell Physiol. 2006;47:1035–1044. doi: 10.1093/pcp/pcj074. [DOI] [PubMed] [Google Scholar]
- 74.Yamaguchi T., Kuroda M., Yamakawa H., Ashizawa T., Hirayae K., Kurimoto L., Shinya T., Shibuya N. Suppression of a phospholipase D gene, OsPLDbeta1, activates defense responses and increases disease resistance in rice. Plant Physiol. 2009;150:308–319. doi: 10.1104/pp.108.131979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Qiao Y., Jiang W., Lee J., Park B., Choi M.S., Piao R., Woo M.O., Roh J.H., Han L., Paek N.C., et al. SPL28 encodes a clathrin-associated adaptor protein complex 1, medium subunit micro 1 (AP1M1) and is responsible for spotted leaf and early senescence in rice (Oryza sativa) New Phytol. 2010;185:258–274. doi: 10.1111/j.1469-8137.2009.03047.x. [DOI] [PubMed] [Google Scholar]
- 76.Babu R., Jiang C.J., Xu X., Kottapalli K.R., Takatsuji H., Miyao A., Hirochika H., Kawasaki S. Isolation, fine mapping and expression profiling of a lesion mimic genotype, spl(NF4050-8) that confers blast resistance in rice. TAG. 2011;122:831–854. doi: 10.1007/s00122-010-1490-7. [DOI] [PubMed] [Google Scholar]
- 77.Zhu X., Yin J., Liang S., Liang R., Zhou X., Chen Z., Zhao W., Wang J., Li W., He M., et al. The Multivesicular Bodies (MVBs)-Localized AAA ATPase LRD6-6 Inhibits Immunity and Cell Death Likely through Regulating MVBs-Mediated Vesicular Trafficking in Rice. PLoS Genet. 2016;12:e1006311. doi: 10.1371/journal.pgen.1006311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhou Q., Zhang Z., Liu T., Gao B., Xiong X. Identification and Map-Based Cloning of the Light-Induced Lesion Mimic Mutant 1 (LIL1) Gene in Rice. Front. Plant Sci. 2017;8:2122. doi: 10.3389/fpls.2017.02122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Wang S., Lei C., Wang J., Ma J., Tang S., Wang C., Zhao K., Tian P., Zhang H., Qi C., et al. SPL33, encoding an eEF1A-like protein, negatively regulates cell death and defense responses in rice. J. Exp. Bot. 2017;68:899–913. doi: 10.1093/jxb/erx001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu Q., Ning Y., Zhang Y., Yu N., Zhao C., Zhan X., Wu W., Chen D., Wei X., Wang G.L., et al. OsCUL3a Negatively Regulates Cell Death and Immunity by Degrading OsNPR1 in Rice. Plant Cell. 2017;29:345–359. doi: 10.1105/tpc.16.00650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Chen Z., Chen T., Sathe A.P., He Y., Zhang X.B., Wu J.L. Identification of a Novel Semi-Dominant Spotted-Leaf Mutant with Enhanced Resistance to Xanthomonas oryzae pv. oryzae in Rice. Int. J. Mol. Sci. 2018;19:3766. doi: 10.3390/ijms19123766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xiao G., Zhou J., Lu X., Huang R., Zhang H. Excessive UDPG resulting from the mutation of UAP1 causes programmed cell death by triggering reactive oxygen species accumulation and caspase-like activity in rice. New Phytol. 2018;217:332–343. doi: 10.1111/nph.14818. [DOI] [PubMed] [Google Scholar]
- 83.Lee D., Lee G., Kim B., Jang S., Lee Y., Yu Y., Seo J., Kim S., Lee Y.H., Lee J., et al. Identification of a Spotted Leaf Sheath Gene Involved in Early Senescence and Defense Response in Rice. Front. Plant Sci. 2018;9:1274. doi: 10.3389/fpls.2018.01274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ruan B., Hua Z., Zhao J., Zhang B., Ren D., Liu C., Yang S., Zhang A., Jiang H., Yu H., et al. OsACL-A2 negatively regulates cell death and disease resistance in rice. Plant Biotechnol. J. 2018 doi: 10.1111/pbi.13058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yang Y., Qi M., Mei C. Endogenous salicylic acid protects rice plants from oxidative damage caused by aging as well as biotic and abiotic stress. Plant J. Cell Mol. Biol. 2004;40:909–919. doi: 10.1111/j.1365-313X.2004.02267.x. [DOI] [PubMed] [Google Scholar]
- 86.Vega-Sanchez M.E., Zeng L., Chen S., Leung H., Wang G.L. SPIN1, a K homology domain protein negatively regulated and ubiquitinated by the E3 ubiquitin ligase SPL11, is involved in flowering time control in rice. Plant Cell. 2008;20:1456–1469. doi: 10.1105/tpc.108.058610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yin Z., Chen J., Zeng L., Goh M., Leung H., Khush G.S., Wang G.L. Characterizing rice lesion mimic mutants and identifying a mutant with broad-spectrum resistance to rice blast and bacterial blight. Mol. Plant-Microbe Interact. 2000;13:869–876. doi: 10.1094/MPMI.2000.13.8.869. [DOI] [PubMed] [Google Scholar]
- 88.Fan J., Bai P., Ning Y., Wang J., Shi X., Xiong Y., Zhang K., He F., Zhang C., Wang R., et al. The Monocot-Specific Receptor-like Kinase SDS2 Controls Cell Death and Immunity in Rice. Cell Host Microbe. 2018;23:498–510.e5. doi: 10.1016/j.chom.2018.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Shirsekar G.S., Vega-Sanchez M.E., Bordeos A., Baraoidan M., Swisshelm A., Fan J., Park C.H., Leung H., Wang G.L. Identification and characterization of suppressor mutants of spl11-mediated cell death in rice. Mol. Plant-Microbe Interact. 2014;27:528–536. doi: 10.1094/MPMI-08-13-0259-R. [DOI] [PubMed] [Google Scholar]
- 90.Spoel S.H., Dong X. Making sense of hormone crosstalk during plant immune responses. Cell Host Microbe. 2008;3:348–351. doi: 10.1016/j.chom.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 91.Li Y., Zhang Z., Nie Y., Zhang L., Wang Z. Proteomic analysis of salicylic acid-induced resistance to Magnaporthe oryzae in susceptible and resistant rice. Proteomics. 2012;12:2340–2354. doi: 10.1002/pmic.201200054. [DOI] [PubMed] [Google Scholar]
- 92.Li Y., Nie Y., Zhang Z., Ye Z., Zou X., Zhang L., Wang Z. Comparative proteomic analysis of methyl jasmonate-induced defense responses in different rice cultivars. Proteomics. 2014;14:1088–1101. doi: 10.1002/pmic.201300104. [DOI] [PubMed] [Google Scholar]
- 93.Singh M.P., Lee F.N., Counce P.A., Gibbons J.H. Mediation of partial resistance to rice blast through anaerobic induction of ethylene. Phytopathology. 2004;94:819–825. doi: 10.1094/PHYTO.2004.94.8.819. [DOI] [PubMed] [Google Scholar]
- 94.Iwai T., Miyasaka A., Seo S., Ohashi Y. Contribution of ethylene biosynthesis for resistance to blast fungus infection in young rice plants. Plant Physiol. 2006;142:1202–1215. doi: 10.1104/pp.106.085258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Dangol S., Chen Y., Hwang B.K., Jwa N.S. Iron- and Reactive Oxygen Species-Dependent Ferroptotic Cell Death in Rice-Magnaporthe oryzae Interactions. Plant Cell. 2018 doi: 10.1105/tpc.18.00535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Singh R., Dangol S., Chen Y., Choi J., Cho Y.S., Lee J.E., Choi M.O., Jwa N.S. Magnaporthe oryzae Effector AVR-Pii Helps to Establish Compatibility by Inhibition of the Rice NADP-Malic Enzyme Resulting in Disruption of Oxidative Burst and Host Innate Immunity. Mol. Cells. 2016;39:426–438. doi: 10.14348/molcells.2016.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Li W., Wang B., Wu J., Lu G., Hu Y., Zhang X., Zhang Z., Zhao Q., Feng Q., Zhang H., et al. The Magnaporthe oryzae avirulence gene AvrPiz-t encodes a predicted secreted protein that triggers the immunity in rice mediated by the blast resistance gene Piz-t. Mol. Plant-Microbe Interact. 2009;22:411–420. doi: 10.1094/MPMI-22-4-0411. [DOI] [PubMed] [Google Scholar]
- 98.Zhou B., Qu S., Liu G., Dolan M., Sakai H., Lu G., Bellizzi M., Wang G.L. The eight amino-acid differences within three leucine-rich repeats between Pi2 and Piz-t resistance proteins determine the resistance specificity to Magnaporthe grisea. Mol. Plant-Microbe Interact. 2006;19:1216–1228. doi: 10.1094/MPMI-19-1216. [DOI] [PubMed] [Google Scholar]
- 99.Park C.H., Chen S., Shirsekar G., Zhou B., Khang C.H., Songkumarn P., Afzal A.J., Ning Y., Wang R., Bellizzi M., et al. The Magnaporthe oryzae effector AvrPiz-t targets the RING E3 ubiquitin ligase APIP6 to suppress pathogen-associated molecular pattern-triggered immunity in rice. Plant Cell. 2012;24:4748–4762. doi: 10.1105/tpc.112.105429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Park C.H., Shirsekar G., Bellizzi M., Chen S., Songkumarn P., Xie X., Shi X., Ning Y., Zhou B., Suttiviriya P., et al. The E3 Ligase APIP10 Connects the Effector AvrPiz-t to the NLR Receptor Piz-t in Rice. PLoS Pathog. 2016;12:e1005529. doi: 10.1371/journal.ppat.1005529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wang R., Ning Y., Shi X., He F., Zhang C., Fan J., Jiang N., Zhang Y., Zhang T., Hu Y., et al. Immunity to Rice Blast Disease by Suppression of Effector-Triggered Necrosis. Curr. Biol. 2016;26:2399–2411. doi: 10.1016/j.cub.2016.06.072. [DOI] [PubMed] [Google Scholar]
- 102.Tang M., Ning Y., Shu X., Dong B., Zhang H., Wu D., Wang H., Wang G.L., Zhou B. The Nup98 Homolog APIP12 Targeted by the Effector AvrPiz-t is Involved in Rice Basal Resistance Against Magnaporthe oryzae. Rice. 2017;10:5. doi: 10.1186/s12284-017-0144-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Shi X., Long Y., He F., Zhang C., Wang R., Zhang T., Wu W., Hao Z., Wang Y., Wang G., et al. The fungal pathogen Magnaporthe oryzae suppresses innate immunity by modulating a host potassium channel. PLoS Pathog. 2018;14:e1006878. doi: 10.1371/journal.ppat.1006878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Matsui H., Yamazaki M., Kishi-Kaboshi M., Takahashi A., Hirochika H. AGC kinase OsOxi1 positively regulates basal resistance through suppression of OsPti1a-mediated negative regulation. Plant Cell Physiol. 2010;51:1731–1744. doi: 10.1093/pcp/pcq132. [DOI] [PubMed] [Google Scholar]
- 105.Bundo M., Coca M. Calcium-dependent protein kinase OsCPK10 mediates both drought tolerance and blast disease resistance in rice plants. J. Exp. Bot. 2017;68:2963–2975. doi: 10.1093/jxb/erx145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Liu Q., Li X., Yan S., Yu T., Yang J., Dong J., Zhang S., Zhao J., Yang T., Mao X., et al. OsWRKY67 positively regulates blast and bacteria blight resistance by direct activation of PR genes in rice. BMC Plant Biol. 2018;18:257. doi: 10.1186/s12870-018-1479-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Salvador-Guirao R., Baldrich P., Tomiyama S., Hsing Y.I., Okada K., San Segundo B. OsDCL1a activation impairs phytoalexin biosynthesis and compromises disease resistance in rice. Ann. Bot. 2018;123:79–93. doi: 10.1093/aob/mcy141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Li Y., Cao X.L., Zhu Y., Yang X.M., Zhang K.N., Xiao Z.Y., Wang H., Zhao J.H., Zhang L.L., Li G.B., et al. Osa-miR398b boosts H2O2 production and rice blast disease-resistance via multiple superoxide dismutases. New Phytol. 2019 doi: 10.1111/nph.15678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Huang K., Caplan J., Sweigard J.A., Czymmek K.J., Donofrio N.M. Optimization of the HyPer sensor for robust real-time detection of hydrogen peroxide in the rice blast fungus. Mol. Plant Pathol. 2017;18:298–307. doi: 10.1111/mpp.12392. [DOI] [PMC free article] [PubMed] [Google Scholar]