Abstract
Aptamers are short, single-stranded DNA, RNA, or synthetic XNA molecules that can be developed with high affinity and specificity to interact with any desired targets. They have been widely used in facilitating discoveries in basic research, ensuring food safety and monitoring the environment. Furthermore, aptamers play promising roles as clinical diagnostics and therapeutic agents. This review provides update on the recent advances in this rapidly progressing field of research with particular emphasis on generation of aptamers and their applications in biosensing, biotechnology and medicine. The limitations and future directions of aptamers in target specific delivery and real-time detection are also discussed.
Keywords: aptamer, systematic evolution of ligands by exponential enrichment (SELEX), diagnostics, therapeutics, biosensor, nanorocket
1. Introduction
Target-specific therapy was discovered by the inventor of chemotherapy, Paul Ehrlich [1]. The ensuing development of hybridoma technology for generating monoclonal antibodies, which realized Ehrlich’s vision [2], along with the therapeutic success of antibodies led to the development of a number of novel antibody drugs. The examples of these drugs include trastuzumab targeting receptor tyrosine-protein kinase 2 ERBB2 implicated in ovarian and breast tumors [3], anti-CD20 chimeric mAb rituximab developed for treating B-cell non-Hodgkin lymphoma [4], and Vedolizumab inhibiting integrin α4β7, which plays an important role in ulcerative colitis and Crohn’s disease [5].
The idea to use nucleic acids to recognize protein targets only emerged later as a result of HIV research. It was shown that the trans-acting response element (TAR)-containing RNA sequences can inhibit HIV replication with high affinity and specificity [6].
Aptamers are short, single-stranded oligonucleotides (DNA or RNA) that bind to targets with high affinity and specificity by folding into tertiary structures [7,8]. Aptamers have been extensively used in basic research, to ensure food safety and to monitor the environment. Furthermore, aptamers have promising role in clinical diagnostics and as therapeutic agents. Although aptamers recognize and bind targets of interest just like antibodies, they have a number of advantages, such as shorter generation time, lower costs of manufacturing, no batch-to-batch variability, higher modifiability, better thermal stability and higher target potential ranging from ions to live animals (Table 1). A more thorough comparison of aptamers and antibodies was reviewed by Zhou and Rossi [9]. Our review provides an update on recent advances in the field of aptamers with particular focus on the methods of generating novel aptamers and the applications of aptamers in biosensing, biotechnology and biomedicine.
Table 1.
Comparison between aptamers and antibodies. A comparison of critical features of aptamers shows how aptamers can supplement monoclonal antibodies.
| Aptamers | Antibodies | |
|---|---|---|
| Stability | Withstand repeated rounds of denaturation/renaturation. Temperature resistant: stable at room temperature. Long shelf life (several years). Can be lyophilized. Degradable by nucleases. Resistant to proteases. |
Easily denatured. Temperature sensitive and require refrigeration to avoid denaturation. Limited shelf life. Must be refrigerated for storage and transport. Degradable by proteases. Resistant to nucleases. |
| Synthesis | In vitro SELEX takes only 2–8 weeks. No batch-to-batch variation. Cheap to synthesize. |
Produced in vivo. More than 6 months. Batch-to-batch variations. Laborious and expensive. |
| Target potential | From ions and small molecules to whole cells and live animals. | Targets must cause a strong immune response for antibodies to be produced. |
| Size | Small molecules. | Relatively large by comparison. |
| Modifiability | Aptamers can readily and easily be modified without affinity loss. | Modifications often lead to reduced activity. |
| Affinity | High and increased in multivalent aptamers. | Dependent on the number of epitopes on the antigen. |
| Specificity | Single point mutations identifiable. | Different antibodies might bind the same antigen. |
| Tissue uptake/kidney filtration | Fast. | Slow. |
2. Generation of Aptamers
A number of approaches were developed in recent years to generate aptamers more reliably and efficiently. These include the systematic evolution of ligands by exponential Enrichment (SELEX) approach and its variations, such as immunoprecipitation-coupled SELEX (IP-SELEX), capture-SELEX, cell-SELEX, capillary electrophoresis-SELEX (CE-SELEX), atomic force microscopy-SELEX (AFM-SELEX), and artificially expanded genetic information system-SELEX (AEGIS-SELEX), which are herein reviewed. The key aspects, advantages, and disadvantages of each SELEX method [9,10,11,12] are highlighted in Table 2.
Table 2.
Key aspects, advantages, and disadvantages of the currently used SELEX methods.
| Method | Key Aspects | Advantages | Disadvantages |
|---|---|---|---|
| IP-SELEX | Includes immunoprecipitation. | Selects aptamers against proteins under normal physiological conditions. Increased affinity and specificity. | More time-consuming than standard SELEX. |
| Capture-SELEX | Oligonucleotide library is immobilized on a support instead of the targets to identify aptamers against small soluble molecules. | Suitable for the selection of aptamers against small molecules. Immobilization of the target not required. Used for the discovery of structure-switching aptamers. | Some oligonucleotides from the library might be not released/selected. |
| Cell-SELEX | Utilizes whole live cells as targets for selection of aptamers. | Prior knowledge of the target not required. Aptamers are selected against molecules in their native state. Many potential targets available on the cell surface. Protein purification not required. | Suitable for cell surface targets. Requires high level of technical expertise. Costly. Time consuming. Post SELEX identification of the target required. |
| CE-SELEX | Involves separation of ions based on electrophoretic mobility. | Fast. Only few (1–4) rounds of selection required. Reduced non-specific binding. Target immobilization not required. | Not suitable for small molecules. Expensive equipment. |
| M-SELEX | Combines SELEX with a microfluidic system. | Rapid. Very efficient (only small amounts of reagents needed). Applicable to small molecules. Automatable. | Low purity/recovery of aptamers. Target immobilization required. |
| AFM-SELEX | Employs AFM to create three-dimensional image of the sample surface. | Able to isolate high affinity aptamers. Fast (only 3–4 rounds required). | Expensive equipment required. Immobilization of target and aptamers required. |
| AEGIS-SELEX | Utilizes libraries with the artificially expanded genetic code. | High specificity of the selected aptamers. | Poor recognition of the unnatural bases by natural DNA polymerases. |
| Animal-SELEX | Aptamers are selected directly within live animal models. | Selected aptamers bind the targets in their natural environment. Prior knowledge of the target not required. Minimal optimization needed. | Time consuming (many rounds required). |
2.1. Systematic Evolution of Ligands by EXponential Enrichment (SELEX)
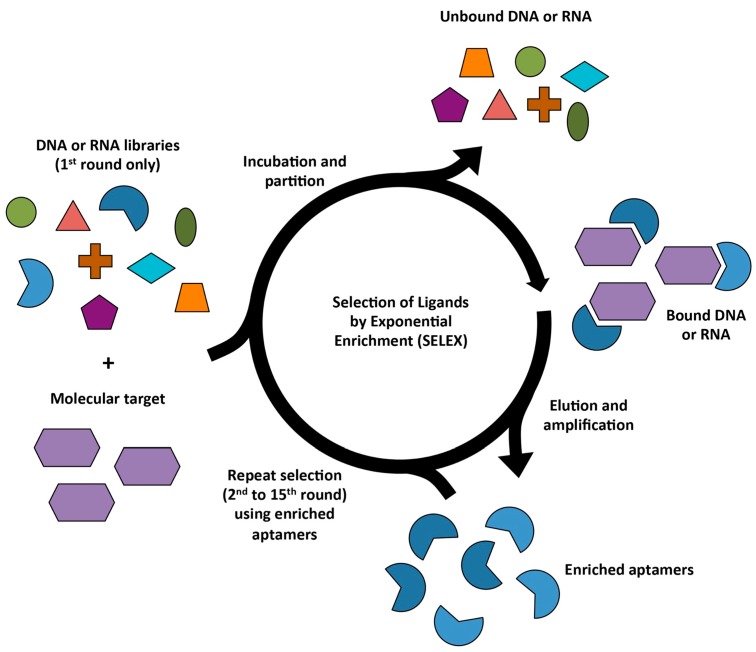
SELEX is a standard method of generating aptamers, where the target of interest is first incubated with a pool of 1014~1016 single-stranded random oligonucleotides. Oligonucleotides in the SELEX library typically consist of 40~100 nucleotides, which harbor a random region in the middle and fixed sequences on both ends. Subsequently, the target-binding oligonucleotides are separated from the unbound ones. The bound DNA oligomers are then eluted and amplified by PCR. After several rounds of selection, the resulting DNA sequences (aptamers) with high affinity and specificity are enriched in the pool and sequenced (Figure 1) [7,8]. The most common modifications toward traditional SELEX method aimed to simplify the SELEX procedure or to generate improved aptamers include changing binding conditions, platform on which the selection is performed, type of target, library design and immobilization matrix. Besides DNA libraries, RNA libraries have also been successfully used for SELEX [9,13,14]. The differences in the majority of RNA SELEX protocols compared to DNA SELEX include the requirement of the protection of RNA from RNAases, amplification by T7 RNA polymerase and reverse transcription step before PCR. Consequently, the 5′-primer used for RNA SELEX usually encodes a promoter for T7 RNA polymerase [13].
Figure 1.
Schematic representation of SELEX (Systematic Evolution of Ligands by EXponential Enrichment). The starting single-stranded DNA or RNA library (1014~1016 random oligonucleotides) is composed of sequences 20~100 nucleotides in length with a random region in the middle flanked by fixed primer sequences. After incubation with the target of interest, the bound oligonucleotides are partitioned from unbound sequences and amplified by PCR. The resulting enriched DNA pool is used for the next round of selection.
2.2. Immunoprecipitation-Coupled SELEX (IP-SELEX)
The affinity and specificity of aptamers largely depend on their targets’ three-dimensional structures. As the three-dimensional targets of artificial, recombinant proteins can differ from their native conformation, it is suggested that the in vitro selection of aptamers might not be the best approach to select aptamers with the highest affinity and specificity under the standard physiological conditions [15]. IP-SELEX was developed to address this issue. Although IP-SELEX is comparable to the traditional SELEX method, it includes immunoprecipitation step to pull down the desirable targets in their native form. This leads to the enrichment for aptamers that retain the ability to recognize proteins under the normal physiological conditions. In the first round of IP-SELEX selection, cells harboring the protein of interest are incubated with ssDNA library in the suitable buffer and condition. Subsequently, cells are lysed, and the target protein is immunoprecipitated using antibody-coated beads. Following several rounds of washing, the protein-aptamer complexes are eluted from the beads and DNA sequences are PCR amplified for the next round of selection. The process of selection for the oligonucleotides with the high specificity and affinity against a desired target is repeated, thus leading to their final enrichment in the aptamer pool. The selected aptamers are then identified by sequencing [16]. IP-SELEX has been successfully used for the identification of anti-CD8 aptamers [17,18].
2.3. Capture-SELEX
In comparison to most SELEX methods, which require immobilization of targets on a solid surface, Capture-SELEX is used for a soluble small molecule targets. Capture-SELEX utilizes a special DNA library harboring fixed domain in the center flanked by two random domains, each of which is flanked by primer-binding sequences. As the fixed domain in the center is designed to be complementary to a biotinylated antisense oligonucleotide, this allows immobilization of the DNA library onto avidin-coated beads by hybridization. Aptamers that are able to form complexes with their targets detach from beads and solubilize, which allows their collection and PCR amplification [19]. Capture-SELEX strategy has been used successfully to select both DNA and RNA aptamers against small organic molecules in solution [20]. Furthermore, Capture-SELEX has been used recently to select aptamers against furaneol [21], penicillin [22], fluoroquinolone antibiotics [23], tobramycin, and other aminoglycoside antibiotics [24,25].
2.4. Cell-SELEX
In Cell-SELEX, live cells are used to select aptamers [26]. Cell-SELEX is considered to have practical applications particularly in oncology, as this approach can select aptamers specific for the cancer cell targets. Employing Cell-SELEX, aptamer-based probes have been developed for a number of cancers, including cervical cancer [27,28], ovarian cancer [29,30], liver cancer [31], prostate cancer [32], breast cancer [33,34], glioma, [35], colorectal carcinoma [36], and lung carcinoma [37]. A number of cell-SELEX variants have been developed in the last years. These include cell-internalization SELEX where aptamers are transported intracellularly and internalized by the analyzed cells, 3D cell-SELEX which involves development of 3D laboratory cell cultures to develop aptamers and cross-over SELEX which utilizes both the cells and the purified protein to increase the selection efficiency [38,39] (Table 2). Ligand-guided selection (LIGS) is another interesting variant of Cell-SELEX which selects aptamers specifically binding known cell surface proteins [11]. LIGS is therefore particularly useful for the generation of aptamers with high specificity and has been utilized also for the selection of aptamers with high specificity against membrane-bound immunoglobulin M (mIgM) [40,41,42]. Furthermore, flow cytometry has been successfully integrated into the Cell-SELEX approach to isolate aptamer-bound cells and eliminate dead cells [15,43]. The main disadvantage of Cell-SELEX and its variants is that the whole process is time consuming and requires a certain level of technical expertise [11] (Table 2). The main advantages of cell-SELEX and its variants are the abundance of molecules on the cell surface with the potential to become targets and their native conformation which is important for diagnostic and therapeutic applications [31,44]. Furthermore, as the whole cells are applied to the selection, there is no need to have the prior knowledge of the biomarkers. In cell-SELEX, the successful selection leads to the generation of aptamers against unknown biomarkers which can be subsequently identified by purification and analysis of the aptamers [11] (Table 2).
2.5. Capillary Electrophoresis-SELEX (CE-SELEX)
CE-SELEX exploits capillary electrophoresis, an analytical technique for separating ions based on their electrophoretic mobility, to generate aptamers with high affinity and specificity in as few as one to four rounds of selection compared to more than 15 selection rounds required for traditional SELEX [12,45]. Capillary electrophoresis in CE-SELEX separates bound DNA molecules from unbound ones in a solution, thus eliminating linker and kinetic bias by stationary support preparation and washing. CE-SELEX was first used to generate aptamers against neuropeptide Y and IgE [46,47]. A number of aptamers have been successfully generated employing CE-SELEX, including those against alpha-fetoprotein and activated protein C [48,49]. Furthermore, a fraction collection approach has been integrated into CE-SELEX for the partition of a bound DNA–target complex in order to generate the target specific binding aptamers in a single round of selection [50]. Recently, CE-SELEX has been successfully used to generate aptamers against glypican-3, a tumor biomarker for the early diagnosis of hepatocellular carcinoma [51]. However, capillary electrophoresis is restricted to the selection against high molecular weight targets. In addition the diversity of the libraries that are handled in CE-SELEX is largely reduced compared to SELEX (Table 2).
2.6. Microfluidic-SELEX (M-SELEX)
M-SELEX based on microfluidics system capable of handling small volumes of fluid was employed to obtain high affinity aptamers against diverse protein targets recently, including influenza A nucleoprotein (infA NP), ovarian cancer and cardiovascular biomarkers [52,53,54,55]. M-SELEX is a universal and automatable approach for rapid generation of aptamers with high affinity and specificity at the microscale level. The most notable improvements to M-SELEX include a micro-magnetic separation device to increase the efficiency of separation [56,57], sol-gel technique [58,59], bead-based acoustophoresis technique [60], and microarray-integrated microfluidic chip technique [12,61].
2.7. Atomic Force Microscopy-SELEX (AFM-SELEX)
AFM-SELEX exploits a high resolution atomic force microscopy (AFM) with cantilever as a probe to measure the weak force between the sample surface and the probe to create the three-dimensional images of the sample surface. AFM-SELEX approach utilizing AFM dynamic affinity force measurement has been developed recently to obtain aptamers with increased affinity [62]. Furthermore, AFM-SELEX has been also successfully applied to develop aptamers against human serum albumin recently [63]. In this study, aptamers against human serum albumin could be selected in the fourth round, based on the measuring of the DNA-duplex interactions by AFM [63].
2.8. Artificially Expanded Genetic Information System-SELEX (AEGIS-SELEX)
AEGIS-SELEX utilizes modified libraries with the artificially expanded genetic code. This includes incorporation of hydrophobic base 7-(2-thienyl) imidazo (4,5-b) pyridine (Ds) nucleotides into a random natural nucleotides sequence library to obtain aptamers with the increased affinity [64]. AEGIS-SELEX has also been used to incorporate four natural and two synthetic nucleotides (commonly referred to as P and Z) [65]. Rearranging donor and acceptor of hydrogen bond on nucleobases by incorporating unnatural base pairs was shown to significantly increase the functional diversity of aptamers. Furthermore, AEGIS-SELEX has been used successfully for the laboratory evolution of artificially expanded DNA to generate aptamers targeting toxic form of Bacillus anthracis protective antigen [66]. Current applications of AEGIS-SELEX are limited mainly by the poor recognition of the unnatural base by the naturally occurring DNA polymerases. However, this obstacle can be overcome by the direct evolution of polymerases able to recognize unnatural bases [67].
2.9. Animal-SELEX
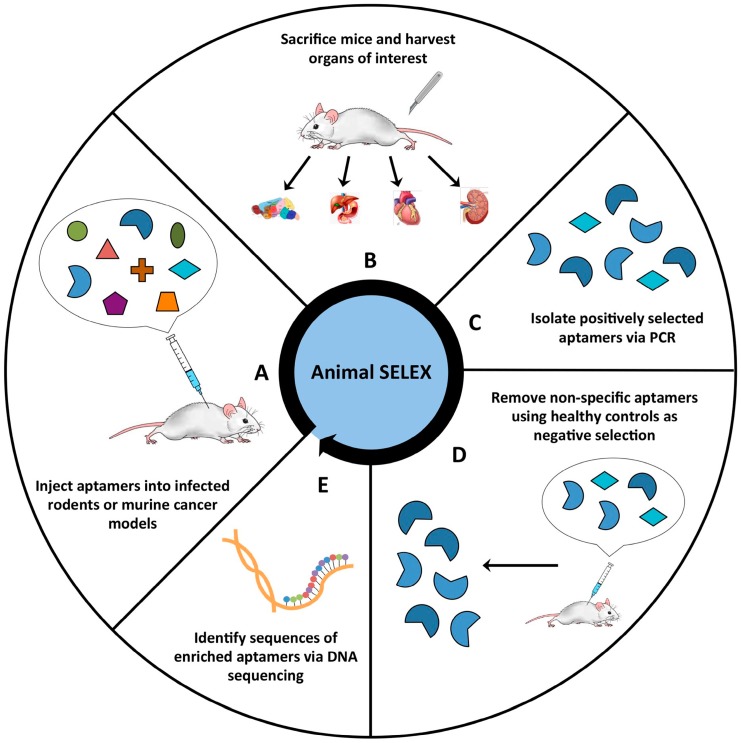
In the whole animal in vivo SELEX, mouse cancer models or pathogen-infected mice can serve as a positive target. Here, aptamer libraries are first injected into the target mice (Figure 2A) and, following inoculation, the organs of interest harvested (Figure 2B). Next, the selected aptamers are isolated and amplified by PCR (Figure 2C). After selection, counter selection can be introduced by inoculating the aptamer pool into the healthy mouse tissues (Figure 2D). The resulting sequences of the disease-specific aptamers with high affinity and specificity to target tissues can be enriched and identified by sequencing (Figure 2E). Aptamers penetrating the blood–brain barrier (BBB) were successfully developed using this selection strategy against brain tissue from mice [68].
Figure 2.
Flowchart of animal SELEX. Animal SELEX can be used to generate aptamers specific to target tissues. (A) Aptamer libraries are first injected into the target mice. (B) After inoculation, the organs of interest are harvested. (C) The selected aptamers are isolated and amplified by PCR. (D) After rounds of selection, counter selection can be performed by inoculating aptamer pool into the healthy mouse tissues. (E) The aptamer sequences with high affinity and specificity to the target tissues of interest are selected and identified by sequencing.
Animal-SELEX was employed recently to identify bone targeting aptamer in a mouse model with prostate cancer bone metastasis [69], Toll-like receptor 4 (TLR4) blocking aptamers for use as acute stroke treatment [70], aptamers with the potential to be used as biomarkers for neurological disorders [71]. Furthermore, animal-SELEX in a murine model of lymphoma has been used recently to screen DNA aptamers with homing specificity to lymphoma bone marrow involvement [72].
3. Applications of Aptamers
Analogically to monoclonal antibodies, aptamers can specifically recognize and bind to their target [73]. Therefore, following their isolation, aptamers can be utilized for molecular recognition of their targets. Consequentially, aptamers have a number of diagnostic and therapeutic applications, such as biosensors and target inhibitors. Due to simple preparation, easy modification, and stability, aptamers have been used in the diverse areas within molecular biology, biotechnology, and biomedicine.
3.1. Aptamers as Diagnostics
The high affinity and specificity of aptamers make them ideal diagnostic agents with the potential to replace conventional antibodies in clinical diagnosis, environmental protection, and food safety. Like monoclonal antibodies, aptamers can be used for the molecular recognition of their respective targets. Aptamers have been successfully used for pathogen recognition, cancer recognition, monitoring environmental contamination, and as stem cell markers.
3.1.1. Pathogen Recognition
The fluorescence resonance energy transfer (FRET)-aptamers were developed as a novel high-throughput screening tool against Escherichia coli outer membrane proteins to detect enterotoxaemia E. coli (ETEC) K88 [74]. Furthermore, aptamers were utilized to detect surface proteins of Campylobacter jejuni [75]. In addition to using purified bacterial proteins as targets, the whole bacterium-based SELEX procedure was applied to detect E. coli [76], Lactobacillus acidophilus [76], Staphylococcus aureus [77], the virulent strain of Mycobacterium tuberculosis [78], Vibrio parahemolyticus [79], Shigella sonnei [78], and C. jejuni [80]. This led to development of aptamers with increased affinity and specificity. SELEX-based approaches can be also used to generate molecular probes for detecting viral infections, such as vaccinia virus [81], herpes simplex virus [82], hepatitis C virus [83,84], hepatitis B virus [83,84], human immunodeficiency virus [85], influenza virus [86], and Severe Acute Respiratory Syndrome (SARS) coronavirus [87]. Furthermore, SELEX has been used successfully to generate aptamers for the detection of a number of parasites, such as Trypanosoma spp. [88], Leishmania spp. [89], Plasmodium spp. [90,91,92,93,94,95,96,97,98,99], Cryptosporidium parvum [100], Entamoeba histolytica [101]. A more thorough overview of the recent advances on aptamers as diagnostics of protozoan parasites was reviewed by Ospina-Villa et al. [73].
3.1.2. Cancer Recognition
Development of aptamers for a reliable and timely cancer diagnosis and prognosis evaluation is of the highest importance. To address this issue, aptamers have been developed for the detection of a number of cancer-related biomarkers [102], including multiple tumor-related proteins in living cancer cells, such as MUC1 (mucin 1), HER2 (human epidermal growth factor receptor 2), and estrogen receptor [102]. Aptamers for the detection of the MCF-7 breast cancer cells [103] and leukemia CCRF-CEM cells were also developed recently [104]. In addition, aptamers have been successfully used for the detection of a number of tumor-related soluble biomarkers, including carcinoembryonic antigen (CEA), prostate specific antigen (PSA) [105,106]. Fluorescently labeled aptamers showed high detection rates of the metastatic tumor tissues [107]. Furthermore, aptamers were successfully used also for the in vivo imaging of lymphoma, adenocarcinoma, leukemia, glioblastoma and other cancer types [102,108]. The current developments in the application of aptamers for the molecular recognition of cancer have been summarized in a recent review by Sun et al. [109].
3.1.3. Stem Cell Recognition
Progress in developing aptamers against stem cells has been slow. There are only a few aptamers against stem cells markers such as cancer stem cells (CSCs), including cancer cell surface biomarkers: Epithelial cell adhesion molecule (EpCAM), CD133, CD117, and CD44 [110], and those for mouse embryonic stem cells [111].
3.1.4. Monitoring Environmental Contamination
Aptamers have the potential to monitor and to minimize the environmental pollutants and the resulting illnesses. Antibiotics, heavy metals, toxins, and pathogens can be toxic to nervous, endocrine, and reproduction systems. Antibiotics used for farm animals may accumulate in the animal tissues and transmit to human upon ingestion. To address this issue, aptamers have been developed against some antibiotics, such as chloramphenicol [112] and tetracycline [113].
Furthermore, aptamers for a number of environmental toxins have been developed recently. These include aptamers against ochratoxin A (OTA) [114], bacterial endotoxins [115], and bisphenol A [116]. Aptamers against mercury [117,118], arsenic [119,120,121], copper [122], and lead [123] have also been generated to identify heavy metal contamination.
Aptamers for the detection of various pesticides in the environment have been also developed recently, such as fungicide carbendazim [124], acetamiprid and atrazine [125,126], and chlorpyriphos [127].
In addition, aptamers have been generated against various types of herbicides [128] and insecticides [129], which may cause reproductive damage in humans. Abraham et al. reported on the identification and characterization of an ssDNA aptamer against herbicide atrazine, recently [130].
3.2. Aptamers Used in Biosensors
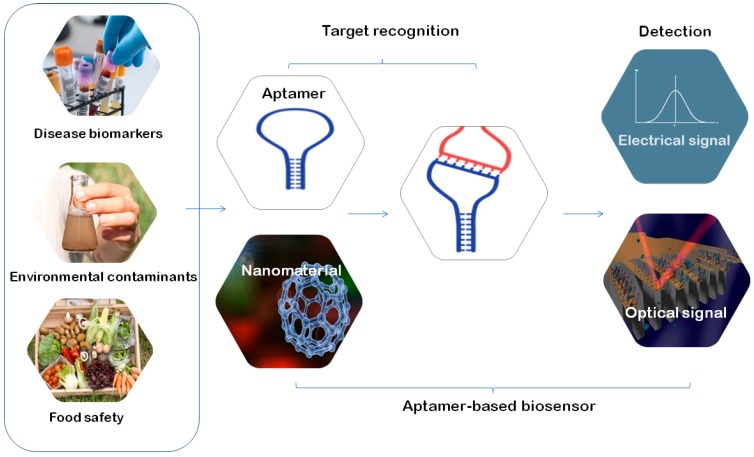
The ease of which different aptamer structures can be generated together with their ability to bind specific targets by forming stable tertiary structures has led to widespread applications of aptamers in biosensors [123,131,132,133] (Figure 3).
Figure 3.
Aptamers used in biosensors. Aptamer-based biosensors are used to detect disease biomarkers, monitor environmental contaminants, or to ensure food safety. Aptamers can be further enhanced by different nanomaterials or biomaterials. The signal of detection in the most of the recently developed aptamer-based biosensors is based on electric or optic/fluorescent signal.
Besides monitoring environmental contamination described above, examples of the recently developed biosensors include the following: (1) Aptamer-based biosensors to target disease biomarkers, such as platelet-derived growth factor BB (PDGF-BB), a cancer related protein, to help early diagnosis and prognosis of cancer development [134]. (2) Biosensors for the azole class of antifungal drugs with application in the therapeutic drug monitoring of patients with invasive fungal infections [135]. (3) Biosensors harboring DNA aptamers targeting B-lactoglobulin for the detection of milk allergen [124]. (4) Highly sensitive biosensors for the detection of human epidermal growth factor receptor 2 with application in real-time detection of breast cancer cells [136]. (5) Biosensors for the detection of bisphenol A [137].
Biosensors can be enhanced by using different nanomaterials (Figure 3). A good example of this is the graphene-based biosensor targeting B-globulin for the detection of milk allergen [124] and the biosensor for the detection of human epidermal growth factor receptor 2 [136] described above. In the latter, ultrafine graphene nanomesh, a continuous 2D graphene nanostructure with a high density of holes punched in the basal plane, has been created to improve the signal on/off ratio [136]. Biosensors can be further enhanced by using biomaterials, such as antibodies to form a "sandwich" structure. In the biosensor developed by Wiedman et al. for the azole class of antifungal drugs, two aptamers were combined to form a "sandwich" structure [135].
Currently, change in the biosensor’s electric signal (current and resistance) upon binding to analytes is the major principle of detection, while the electrochemical impedance spectroscopy (EIS) and cyclic voltammetry (CV) are usually used to monitor aptamer–ligand complexes occurring on electrode surface [133]. DNA-based electrochemical biosensors have been widely used for the detection of heavy metal ions, including mercury, lead, and copper [138]. Fluorescence-labeled aptamer probes have also been widely used in biosensors (Figure 3). A novel label-free aptasensor for acetamiprid through fluorescence resonance energy transfer (FRET) involves the conformational change taking place after the aptamer binds to acetamiprid and its effect on the stability of the gold nanoparticles in solution. In this biosensor, dual-colored Au NCs (gold nanoclusters) excitable by single-wavelength excitation were used as energy donors to achieve simultaneous detection of multiple tumor markers [139]. Another recent example of the fluorescent biosensor is the biosensor for the detection of bisphenol A based on the adsorption ability of a nano-sized iron metal–organic framework (Fe-MIL-88B–NH2) to a DNA aptamer [137].
To further enhance the affinity of detection, recent studies have focused on three aspects: reconstructing aptamers, modifying electrodes and exploiting the inter-substance interactions. It has been demonstrated that removing the non-binding domain of aptamers could increase the binding affinity and specificity of the aptamer-target complex by improving the density of the binding domains [140]. Furthermore, the rolling circle amplification increases the length of the aptamer, thus increasing the number of binding domains. Electrochemical roughening approach also enhanced the signaling of electrochemical sensors by increasing the microscopic surface area of gold electrodes [141]. This simple method does not require pretreatment compared to the conventional electrochemical deposition to increase the electrode surface [141]. Signal amplification was also achieved via synergetic catalysis using DNAzyme-decorated AuPd nanoparticles. The signal was amplified by synergistic catalysis of G-quadruplex/hemin/HRP/AuPd/poly(o-phenylenediamine) bioconjugates, thus greatly enhancing the sensitivity of the biosensor [142]. Jeddi and Saiz presented the first approach to predict three-dimensional structures of aptamers from sequences by focusing explicitly on ssDNA hairpins [143]. Different 3D aptamer configurations resulted in different affinities, thus highlighting the role of 3D configuration of aptamers in their sensitivity, specificity, and reliability.
3.3. Aptamers as Therapeutics
Due to their ability to compete with small molecules and protein ligands and to inhibit their targets [144], aptamers are considered to be promising therapeutics. Furthermore, aptamers can activate the function of the target receptors or act as carriers for the delivery of therapeutic agents to the target cells or tissue [9]. For example, aptamers have the potential to act as antiviral agents. Previous work suggests that RNA aptamers against a synthetic derivative of gp120 can neutralize HIV-1 [145]. Furthermore, RNA aptamers against RIG-I can inhibit Newcastle disease virus (NDV), vesicular stomatitis virus (VSV), and influenza virus replication [144]. RNA aptamers able to efficiently inhibit protease and helicase activities were developed which target hepatitis C virus (HCV) NS3 helicase domain, thus inhibiting HCV [146,147,148]. Other aptamers in discovery and preclinical stages include those targeting cancer, such as those binding different growth factors (e.g. VEGF, bFGF, PDGF, KGF), NOX-A12 RNA aptamer binding the chemokine ligand CXCL12 [149] and NAS-24 DNA aptamer targeting vimentin involved in maintaining cell shape, cytoplasm integrity, and cytoskeleton stability [150]. Furthermore, aptamers in discovery and preclinical stages include those against bacterial infections, such as those caused by E. coli, S. aureus, M. tuberculosis, and Salmonella spp. [151,152]. A number of aptamers for immune disorders are in the preclinical stages too, including ssDNA aptamer BC007 targeting beta1-adrenoreceptor autoantibodies [153]. Furthermore, aptamers in the preclinical stages of development include those targeting neurodegenerative diseases, immune disorders, and inflammation [151].
A number of aptamers have entered clinical trials, such as for ocular diseases [154], haematologic diseases [155], and cancer [156]. Pegaptanib, a vascular endothelial growth factor (VEGF)-specific aptamer, was approved for therapeutic use for age-related macular degeneration. Pegaptanib blocks VEGF which plays a key role in pathological angiogenesis, for example, in ocular neovascular diseases, such as age-related macular degeneration (AMD) and diabetic macular oedema [157]. AS1411, a G-rich DNA aptamer against nucleolin, is undergoing phase II clinical trial for acute myeloid leukemia. The anti-tumor activity of AS1411 stems from its ability to bind cell surface nucleolin, thereby inhibiting DNA synthesis, preventing cell growth signaling, and inducing apoptosis [156]. AS1411 was shown to be efficient against a variety of cancer cells, including lung cancer [156], colorectal cancer [158], breast cancer [159], and hepatocellular carcinoma [160]. NOX-E36, l-RNA aptamer with 3′-PEG against chemokine (C-C motif) ligand 2, is in phase I clinical trial for Type 2 diabetes and diabetic nephropathy. NOX-E36 binds and neutralizes human chemokine CCL2 and related chemokines. This prevents infiltration of pro-inflammatory cells into the kidney and allows resolving of the existing inflammation over time [161,162]. Aptamers as therapeutics, including those in clinical trials, have been summarized in Table 3. The current list of the therapeutic aptamers in clinical trials which is being constantly updated can be found at the Clinical Trials website of the NIH U.S. National Library of Medicine (https://clinicaltrials.gov/) and in the recent review by Ismail and Alshaer [151].
Table 3.
Aptamer as therapeutics.
| Target | Aptamers | References |
|---|---|---|
| VEGF-165 | SL (2)-B (DNA), RNV66 (DNA) | [166] |
| Nucleolin | FCL-II | [167] |
| CXCL12 | NOX-A12 | [168] |
| EGFR | TuTu2231, KDI130 | [32] |
| Vimentin | NAS-24 | [150] |
| E-selectin | ESTA | [169] |
| PD-1 | MP7 | [170] |
| CTLA-4 | AptCTLA-4 | [171] |
| C5a | AON-D21 I-aptamer | [172] |
| CD44/EpCAM | CD44-EpCAM aptamer | [173] |
| Thrombin | Anti-Thrombin aptamer | [174] |
Aptamers have been used successfully for the delivery of a variety of therapeutic reagents into the target cells and tissues [9]. Aptamer-based delivery systems include the aptamer-therapeutic oligonucleotide conjugates [163], aptamer-drug conjugates [164], and aptamer-decorated nanomaterials [9,165]. Examples of aptamers used for targeted drug delivery are shown in Table 4.
Table 4.
Aptamers for the targeted drug delivery.
| Target Name | Aptamer | Selection Target | Delivery/Application |
|---|---|---|---|
| Epidermal growth factor receptor (EGFR) | RNA | Purified extracellular domain of EGFR | Nanoparticle delivery |
| Immunoglobin heavy chain (IGHM) | DNA | Cell | Micelle nanoparticles for drug delivery |
| Mucin1 (MUC-1) | DNA | Recombinant peptides | Photodynamic therapy, radionuclide delivery |
| Prostate-specific membrane antigen (PSMA) | RNA | Purified extracellular domain of PSMA | siRNA delivery, Chemotherapeutic drug delivery |
| Protein tyrosine kinase-7 (PTK7) | DNA | Cell | Chemotherapeutic drug delivery |
4. Conclusions and Future Directions
As described in this review, aptamers have a number of biotechnologically and medically-relevant applications; however, the main limitations for their in vivo therapeutic applications are target specific delivery. Cell-SELEX and whole animal in vivo SELEX can be employed to address these issues.
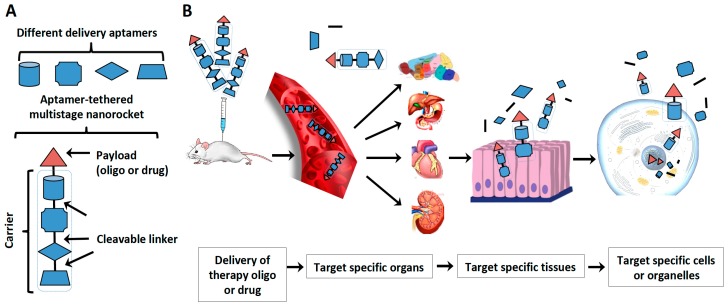
Target site-specific delivery is a crucial problem in the therapeutic use of oligonucleotides, which can be overcome by employing aptamers in the form of a “nanorocket”. An aptamer-tethered multistage “nanorocket” is a complex of two or more aptamers, each of which contains its own functional oligonucleotide. Serial stages of aptamers are mounted on top of another to build a carrier “nanorocket” for payload drug, allowing delivery into selected tissues, cell types or even subcellular organelles (Figure 4A). To design and engineer the multistage “nanorocket”, whole animal-SELEX, tissue-SELEX, cell-SELEX and fractionation-SELEX can be used. This involves selection of the oligonucleotides pool for different targets. After incubation and isolation of the fraction of interest, such as specific animal tissues, cell types or subcellular fractions, those containing the binding complex (targets and oligonucleotide sequences) are partitioned from the unbound sequences and amplified by PCR. The process can be repeated until the pool is enriched for target-specific sequences. To increase the specificity, counter selection can be introduced.
Figure 4.
Aptamer-tethered multistage “nanorocket” for target-specific delivery. (A) An aptamer-tethered multistage “nanorocket” is a complex of two or more aptamers, each of which contains its own functional oligonucleotide. Serial stages of aptamers are mounted on top of another to build a carrier “nanorocket” for the target specific recognition and delivery. With appropriate linkages, each stage of aptamer “nanorocket” can freely rotate and perform its function. (B) The selected aptamers can be used in different stages of “nanorockets”, designed for tissue penetration, cell target recognition and cellular internalization. After reaching target, the recognition and delivering stages of the “nanorockets” can be cleaved and degraded by the cell leaving only the cargo oligos. Aptamers in multi-stage “nanorockets” can be used as tissue-, cell-type-, and cellular compartment-specific delivery systems.
Prior knowledge of the delivery mechanism, binding targets and delivery pathway through the tissue or cell surface barrier is unnecessary. Crucial are the types of tissues, cells and organelles used in the process, as they will be specifically recognized and targeted by the generated aptamers. The selected aptamers can be used in different stages of “nanorockets”, designed for tissue penetration, cell target recognition and cellular internalization. With appropriate linkages, each aptamer stage can freely rotate and perform its function. With cytosolic cleavable linkers, such as N-succinimidyl-4-(2-pyridyldithio) pentanoate (SPP) and N-succinimidyl-4-(2-pyridyldithio) butyrate (SPDB) or acid-cleavable hydrazone linkers for lysosome-specific release, the recognition and delivering stages of the “nanorockets” can be removed at the desired time point. After reaching the target, the recognition and delivering stages of the “nanorockets” can be cleaved and degraded by the cell leaving only the cargo oligos chemically modified to be resistant to the degradation. Modified aptamers in multi-stage “nanorockets” can be used as tissue-, cell-type-, and cellular compartment-specific delivery systems (Figure 4B).
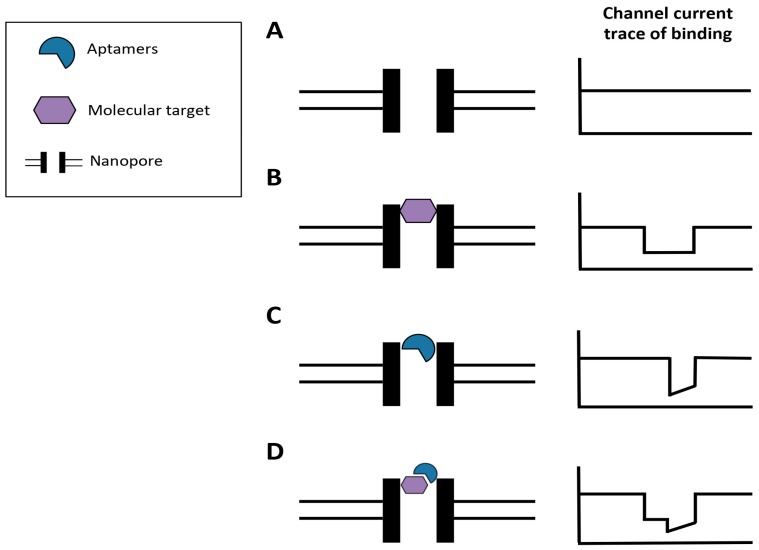
In term of diagnostics, a number of analytic devices were introduced to in the past years to improve performance and simplify the diagnostic procedure. AFM, microfluidic, and capillary electrophoresis were employed to couple with aptamers for high specific detection. AFM permits assessing of aptamer binding affinities during interaction by visualizing aptamer-target complexes and is considered a step forward towards real-time aptamer monitoring. In the future, a nanopore sensor (nano-scale pore with voltage applied across it) can be incorporated into the procedure, thus allowing real-time observation of binding. Single-molecule detection can be achieved by measuring the current disruption caused by molecules electrophoretically driven through the pore. The aptamer-protein target binding can be characterized by ion blockade level, and the binding evaluated in real-time (Figure 5).
Figure 5.
Flowchart of nanopore-based aptamer assaying. Nanopore sensors can be used to detect aptamer binding and selection in a real-time. Aptamer and target interaction can be detected by measuring the current disruption caused by molecules electrophoretically driven through the pore. (A) An ionic current is passed through the nanopore. The current changes as molecular target (B), aptamer (C), or aptamer–target complex (D) passes through the nanopore.
Aptamers are cheap to manufacture, thermally stable and non-immunogenic. They specifically target harmful cells or tissues with minimal toxicity to the healthy ones, and can be chemically modified to facilitate their visualization, absorption, and delivery. Due to their specificity, stability, and versatility, aptamers embody the future of diagnostics and therapy.
Acknowledgments
We would like to thank all researchers who contributed to the analysis of aptamers and to the members of our labs for the constructive discussions.
Funding
The project financially supported by the Natural Science Foundation of Shenzhen City (Project number JCYJ20170307150444573).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Strebhardt K., Ullrich A. Paul Ehrlich’s magic bullet concept: 100 years of progress. Nat. Rev. Cancer. 2008;8:473–480. doi: 10.1038/nrc2394. [DOI] [PubMed] [Google Scholar]
- 2.Köhler G., Milstein C. Continuous cultures of fused cells secreting antibody of predefined specificity. Nature. 1975;256:495–497. doi: 10.1038/256495a0. [DOI] [PubMed] [Google Scholar]
- 3.Hudziak R.M., Lewis G.D., Winget M., Fendly B.M., Shepard H.M., Ullrich A. p185HER2 monoclonal antibody has antiproliferative effects in vitro and sensitizes human breast tumor cells to tumor necrosis factor. Mol. Cell. Biol. 1989;9:1165–1172. doi: 10.1128/MCB.9.3.1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Gaetano N., Cittera E., Nota R., Vecchi A., Grieco V., Scanziani E., Botto M., Introna M., Golay J. Complement activation determines the therapeutic activity of rituximab in vivo. J. Immunol. 2003;171:1581–1587. doi: 10.4049/jimmunol.171.3.1581. [DOI] [PubMed] [Google Scholar]
- 5.Sandborn W.J., Feagan B.G., Rutgeerts P., Hanauer S., Colombel J.F., Sands B.E., Lukas M., Fedorak R.N., Lee S., Bressler B., et al. Vedolizumab as induction and maintenance therapy for Crohn’s disease. N. Engl. J. Med. 2013;369:711–721. doi: 10.1056/NEJMoa1215739. [DOI] [PubMed] [Google Scholar]
- 6.Sullenger B.A., Gallardo H.F., Ungers G.E., Gilboa E. Analysis of trans-acting response decoy RNA-mediated inhibition of human immunodeficiency virus type 1 transactivation. J. Virol. 1991;65:6811–6816. doi: 10.1128/jvi.65.12.6811-6816.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ellington A.D., Szostak J.W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990;346:818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- 8.Tuerk C., Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 9.Zhou J., Rossi J. Aptamers as targeted therapeutics: Current potential and challenges. Nat. Rev. Drug Discov. 2017;16:181–202. doi: 10.1038/nrd.2016.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bayat P., Nosrati R., Alibolandi M., Rafatpanah H., Abnous K., Khedri M., Ramezani M. SELEX methods on the road to protein targeting with nucleic acid aptamers. Biochimie. 2018;154:132–155. doi: 10.1016/j.biochi.2018.09.001. [DOI] [PubMed] [Google Scholar]
- 11.Kaur H. Recent developments in cell-SELEX technology for aptamer selection. Biochim. Biophys. Acta Gen. Subj. 2018;1862:2323–2329. doi: 10.1016/j.bbagen.2018.07.029. [DOI] [PubMed] [Google Scholar]
- 12.Zhuo Z., Yu Y., Wang M., Li J., Zhang Z., Liu J., Wu X., Lu A., Zhang G., Zhang B. Recent Advances in SELEX Technology and Aptamer Applications in Biomedicine. Int. J. Mol. Sci. 2017;18:2142. doi: 10.3390/ijms18102142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vorobyeva M.A., Davydova A.S., Vorobjev P.E., Venyaminova A.G. Key Aspects of Nucleic Acid Library Design for in Vitro Selection. Int. J. Mol. Sci. 2018;19:470. doi: 10.3390/ijms19020470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Randrianjatovo-Gbalou I., Rosario S., Sismeiro O., Varet H., Legendre R., Coppée J.Y., Huteau V., Pochet S., Delarue M. Enzymatic synthesis of random sequences of RNA and RNA analogues by DNA polymerase theta mutants for the generation of aptamer libraries. Nucleic Acids Res. 2018;46:6271–6284. doi: 10.1093/nar/gky413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mayer G., Ahmed M.S., Dolf A., Endl E., Knolle P.A., Famulok M. Fluorescence-activated cell sorting for aptamer SELEX with cell mixtures. Nat. Protoc. 2010;5:1993–2004. doi: 10.1038/nprot.2010.163. [DOI] [PubMed] [Google Scholar]
- 16.Chang Y.C., Kao W.C., Wang W.Y., Yang R.B., Peck K. Identification and characterization of oligonucleotides that inhibit Toll-like receptor 2-associated immune responses. FASEB J. 2009;23:3078–3088. doi: 10.1096/fj.09-129312. [DOI] [PubMed] [Google Scholar]
- 17.Wang C.W., Chung W.H., Cheng Y.F., Ying N.W., Peck K., Chen Y.T., Hung S.I. A new nucleic acid-based agent inhibits cytotoxic T lymphocyte-mediated immune disorders. J. Allergy Clin. Immunol. 2013;132:713–722.e11. doi: 10.1016/j.jaci.2013.04.036. [DOI] [PubMed] [Google Scholar]
- 18.Mercier M.C., Dontenwill M., Choulier L. Selection of Nucleic Acid Aptamers Targeting Tumor Cell-Surface Protein Biomarkers. Cancers. 2017;9:69. doi: 10.3390/cancers9060069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nutiu R., Li Y. In vitro selection of structure-switching signaling aptamers. Angew. Chem. Int. Ed. Engl. 2005;44:1061–1065. doi: 10.1002/anie.200461848. [DOI] [PubMed] [Google Scholar]
- 20.Lauridsen L.H., Doessing H.B., Long K.S., Nielsen A.T. A Capture-SELEX Strategy for Multiplexed Selection of RNA Aptamers Against Small Molecules. Methods Mol. Biol. 2018;1671:291–306. doi: 10.1007/978-1-4939-7295-1_18. [DOI] [PubMed] [Google Scholar]
- 21.Komarova N., Andrianova M., Glukhov S., Kuznetsov A. Selection, Characterization, and Application of ssDNA Aptamer against Furaneol. Molecules. 2018;23:3159. doi: 10.3390/molecules23123159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paniel N., Istamboulié G., Triki A., Lozano C., Barthelmebs L., Noguer T. Selection of DNA aptamers against penicillin G using Capture-SELEX for the development of an impedimetric sensor. Talanta. 2017;162:232–240. doi: 10.1016/j.talanta.2016.09.058. [DOI] [PubMed] [Google Scholar]
- 23.Reinemann C., Freiin von Fritsch U., Rudolph S., Strehlitz B. Generation and characterization of quinolone-specific DNA aptamers suitable for water monitoring. Biosens. Bioelectron. 2016;77:1039–1047. doi: 10.1016/j.bios.2015.10.069. [DOI] [PubMed] [Google Scholar]
- 24.Nikolaus N., Strehlitz B. DNA-aptamers binding aminoglycoside antibiotics. Sensors. 2014;14:3737–3755. doi: 10.3390/s140203737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spiga F.M., Maietta P., Guiducci C. More DNA-Aptamers for Small Drugs: A Capture-SELEX Coupled with Surface Plasmon Resonance and High-Throughput Sequencing. ACS Comb. Sci. 2015;17:326–333. doi: 10.1021/acscombsci.5b00023. [DOI] [PubMed] [Google Scholar]
- 26.Sefah K., Shangguan D., Xiong X., O’Donoghue M.B., Tan W. Development of DNA aptamers using Cell-SELEX. Nat. Protoc. 2010;5:1169–1185. doi: 10.1038/nprot.2010.66. [DOI] [PubMed] [Google Scholar]
- 27.Graham J.C., Zarbl H. Use of cell-SELEX to generate DNA aptamers as molecular probes of HPV-associated cervical cancer cells. PLoS ONE. 2012;7:e36103. doi: 10.1371/journal.pone.0036103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang J., Gao T., Luo Y., Wang Z., Zhang Y., Pei R. In Vitro Selection of a DNA Aptamer by Cell-SELEX as a Molecular Probe for Cervical Cancer Recognition and Imaging. J. Mol. Evol. 2019 doi: 10.1007/s00239-019-9886-8. [DOI] [PubMed] [Google Scholar]
- 29.Hung L.Y., Wang C.H., Hsu K.F., Chou C.Y., Lee G.B. An on-chip Cell-SELEX process for automatic selection of high-affinity aptamers specific to different histologically classified ovarian cancer cells. Lab Chip. 2014;14:4017–4028. doi: 10.1039/C4LC00587B. [DOI] [PubMed] [Google Scholar]
- 30.He J., Wang J., Zhang N., Shen L., Wang L., Xiao X., Wang Y., Bing T., Liu X., Li S., et al. In vitro selection of DNA aptamers recognizing drug-resistant ovarian cancer by cell-SELEX. Talanta. 2019;194:437–445. doi: 10.1016/j.talanta.2018.10.028. [DOI] [PubMed] [Google Scholar]
- 31.Rong Y., Chen H., Zhou X.F., Yin C.Q., Wang B.C., Peng C.W., Liu S.P., Wang F.B. Identification of an aptamer through whole cell-SELEX for targeting high metastatic liver cancers. Oncotarget. 2016;7:8282–8294. doi: 10.18632/oncotarget.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang Y., Luo Y., Bing T., Chen Z., Lu M., Zhang N., Shangguan D., Gao X. DNA aptamer evolved by cell-SELEX for recognition of prostate cancer. PLoS ONE. 2014;9:e100243. doi: 10.1371/journal.pone.0100243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang K., Sefah K., Tang L., Zhao Z., Zhu G., Ye M., Sun W., Goodison S., Tan W. A novel aptamer developed for breast cancer cell internalization. ChemMedChem. 2012;7:79–84. doi: 10.1002/cmdc.201100457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li W.M., Zhou L.L., Zheng M., Fang J. Selection of Metastatic Breast Cancer Cell-Specific Aptamers for the Capture of CTCs with a Metastatic Phenotype by Cell-SELEX. Mol. Ther. Nucleic Acids. 2018;12:707–717. doi: 10.1016/j.omtn.2018.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wu Q., Wang Y., Wang H., Wu L., Zhang H., Song Y., Zhu Z., Kang D., Yang C. DNA aptamers from whole-cell SELEX as new diagnostic agents against glioblastoma multiforme cells. Analyst. 2018;143:2267–2275. doi: 10.1039/C8AN00271A. [DOI] [PubMed] [Google Scholar]
- 36.Maimaitiyiming Y., Yang C., Wang Y., Hussain L., Naranmandura H. Selection and characterization of novel DNA aptamer against colorectal carcinoma Caco-2 cells. Biotechnol. Appl. Biochem. 2019 doi: 10.1002/bab.1737. [DOI] [PubMed] [Google Scholar]
- 37.Shi H., Cui W., He X., Guo Q., Wang K., Ye X., Tang J. Whole cell-SELEX aptamers for highly specific fluorescence molecular imaging of carcinomas in vivo. PLoS ONE. 2013;8:e70476. doi: 10.1371/journal.pone.0070476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thiel W.H., Thiel K.W., Flenker K.S., Bair T., Dupuy A.J., McNamara J.O., Miller F.J., Giangrande P.H. Cell-internalization SELEX: Method for identifying cell-internalizing RNA aptamers for delivering siRNAs to target cells. Methods Mol. Biol. 2015;1218:187–199. doi: 10.1007/978-1-4939-1538-5_11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iaboni M., Fontanella R., Rienzo A., Capuozzo M., Nuzzo S., Santamaria G., Catuogno S., Condorelli G., de Franciscis V., Esposito C.L. Targeting Insulin Receptor with a Novel Internalizing Aptamer. Mol. Ther. Nucleic Acids. 2016;5:e365. doi: 10.1038/mtna.2016.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zümrüt H.E., Batool S., Van N., George S., Bhandari S., Mallikaratchy P. Structural optimization of an aptamer generated from Ligand-Guided Selection (LIGS) resulted in high affinity variant toward mIgM expressed on Burkitt’s lymphoma cell lines. Biochim. Biophys. Acta Gen. Subj. 2017;1861:1825–1832. doi: 10.1016/j.bbagen.2017.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zumrut H.E., Ara M.N., Fraile M., Maio G., Mallikaratchy P. Ligand-Guided Selection of Target-Specific Aptamers: A Screening Technology for Identifying Specific Aptamers Against Cell-Surface Proteins. Nucleic Acid Ther. 2016;26:190–198. doi: 10.1089/nat.2016.0611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zumrut H.E., Ara M.N., Maio G.E., Van N.A., Batool S., Mallikaratchy P.R. Ligand-guided selection of aptamers against T-cell Receptor-cluster of differentiation 3 (TCR-CD3) expressed on Jurkat.E6 cells. Anal. Biochem. 2016;512:1–7. doi: 10.1016/j.ab.2016.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kim J.W., Kim E.Y., Kim S.Y., Byun S.K., Lee D., Oh K.J., Kim W.K., Han B.S., Chi S.W., Lee S.C., et al. Identification of DNA aptamers toward epithelial cell adhesion molecule via cell-SELEX. Mol. Cells. 2014;37:742–746. doi: 10.14348/molcells.2014.0208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Civit L., Taghdisi S.M., Jonczyk A., Haßel S.K., Gröber C., Blank M., Stunden H.J., Beyer M., Schultze J., Latz E., et al. Systematic evaluation of cell-SELEX enriched aptamers binding to breast cancer cells. Biochimie. 2018;145:53–62. doi: 10.1016/j.biochi.2017.10.007. [DOI] [PubMed] [Google Scholar]
- 45.Mosing R.K., Bowser M.T. Isolating aptamers using capillary electrophoresis-SELEX (CE-SELEX) Methods Mol. Biol. 2009;535:33–43. doi: 10.1007/978-1-59745-557-2_3. [DOI] [PubMed] [Google Scholar]
- 46.Mendonsa S.D., Bowser M.T. In vitro selection of aptamers with affinity for neuropeptide Y using capillary electrophoresis. J. Am. Chem. Soc. 2005;127:9382–9383. doi: 10.1021/ja052406n. [DOI] [PubMed] [Google Scholar]
- 47.Mendonsa S.D., Bowser M.T. In vitro evolution of functional DNA using capillary electrophoresis. J. Am. Chem. Soc. 2004;126:20–21. doi: 10.1021/ja037832s. [DOI] [PubMed] [Google Scholar]
- 48.Dong L., Tan Q., Ye W., Liu D., Chen H., Hu H., Wen D., Liu Y., Cao Y., Kang J., et al. Screening and Identifying a Novel ssDNA Aptamer against Alpha-fetoprotein Using CE-SELEX. Sci. Rep. 2015;5:15552. doi: 10.1038/srep15552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hamedani N.S., Müller J. Capillary Electrophoresis for the Selection of DNA Aptamers Recognizing Activated Protein C. Methods Mol. Biol. 2016;1380:61–75. doi: 10.1007/978-1-4939-3197-2_5. [DOI] [PubMed] [Google Scholar]
- 50.Luo Z., Zhou H., Jiang H., Ou H., Li X., Zhang L. Development of a fraction collection approach in capillary electrophoresis SELEX for aptamer selection. Analyst. 2015;140:2664–2670. doi: 10.1039/C5AN00183H. [DOI] [PubMed] [Google Scholar]
- 51.Dong L., Zhou H., Zhao M., Gao X., Liu Y., Liu D., Guo W., Hu H., Xie Q., Fan J., et al. Phosphorothioate-Modified AP613-1 Specifically Targets GPC3 when Used for Hepatocellular Carcinoma Cell Imaging. Mol. Ther. Nucleic Acids. 2018;13:376–386. doi: 10.1016/j.omtn.2018.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahmad K.M., Oh S.S., Kim S., McClellen F.M., Xiao Y., Soh H.T. Probing the limits of aptamer affinity with a microfluidic SELEX platform. PLoS ONE. 2011;6:e27051. doi: 10.1371/journal.pone.0027051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Leblebici P., Leirs K., Spasic D., Lammertyn J. Encoded particle microfluidic platform for rapid multiplexed screening and characterization of aptamers against influenza A nucleoprotein. Anal. Chim. Acta. 2019;1053:70–80. doi: 10.1016/j.aca.2018.11.055. [DOI] [PubMed] [Google Scholar]
- 54.Hung L.Y., Fu C.Y., Wang C.H., Chuang Y.J., Tsai Y.C., Lo Y.L., Hsu P.H., Chang H.Y., Shiesh S.C., Hsu K.F., et al. Microfluidic platforms for rapid screening of cancer affinity reagents by using tissue samples. Biomicrofluidics. 2018;12:054108. doi: 10.1063/1.5050451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sinha A., Gopinathan P., Chung Y.D., Lin H.Y., Li K.H., Ma H.P., Huang P.C., Shiesh S.C., Lee G.B. An integrated microfluidic platform to perform uninterrupted SELEX cycles to screen affinity reagents specific to cardiovascular biomarkers. Biosens. Bioelectron. 2018;122:104–112. doi: 10.1016/j.bios.2018.09.040. [DOI] [PubMed] [Google Scholar]
- 56.Lou X., Qian J., Xiao Y., Viel L., Gerdon A.E., Lagally E.T., Atzberger P., Tarasow T.M., Heeger A.J., Soh H.T. Micromagnetic selection of aptamers in microfluidic channels. Proc. Natl. Acad. Sci. USA. 2009;106:2989–2994. doi: 10.1073/pnas.0813135106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cho M., Xiao Y., Nie J., Stewart R., Csordas A.T., Oh S.S., Thomson J.A., Soh H.T. Quantitative selection of DNA aptamers through microfluidic selection and high-throughput sequencing. Proc. Natl. Acad. Sci. USA. 2010;107:15373–15378. doi: 10.1073/pnas.1009331107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim S., Kim Y., Kim P., Ha J., Kim K., Sohn M., Yoo J.S., Lee J., Kwon J.A., Lee K.N. Improved sensitivity and physical properties of sol-gel protein chips using large-scale material screening and selection. Anal. Chem. 2006;78:7392–7396. doi: 10.1021/ac0520487. [DOI] [PubMed] [Google Scholar]
- 59.Bae H., Ren S., Kang J., Kim M., Jiang Y., Jin M.M., Min I.M., Kim S. Sol-gel SELEX circumventing chemical conjugation of low molecular weight metabolites discovers aptamers selective to xanthine. Nucleic Acid Ther. 2013;23:443–449. doi: 10.1089/nat.2013.0437. [DOI] [PubMed] [Google Scholar]
- 60.Park J.W., Lee S.J., Ren S., Lee S., Kim S., Laurell T. Acousto-microfluidics for screening of ssDNA aptamer. Sci. Rep. 2016;6:27121. doi: 10.1038/srep27121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liu X., Li H., Jia W., Chen Z., Xu D. Selection of aptamers based on a protein microarray integrated with a microfluidic chip. Lab Chip. 2016;17:178–185. doi: 10.1039/C6LC01208F. [DOI] [PubMed] [Google Scholar]
- 62.Miyachi Y., Shimizu N., Ogino C., Kondo A. Selection of DNA aptamers using atomic force microscopy. Nucleic Acids Res. 2010;38:e21. doi: 10.1093/nar/gkp1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Takenaka M., Okumura Y., Amino T., Miyachi Y., Ogino C., Kondo A. DNA-duplex linker for AFM-SELEX of DNA aptamer against human serum albumin. Bioorg. Med. Chem. Lett. 2017;27:954–957. doi: 10.1016/j.bmcl.2016.12.080. [DOI] [PubMed] [Google Scholar]
- 64.Kimoto M., Yamashige R., Matsunaga K., Yokoyama S., Hirao I. Generation of high-affinity DNA aptamers using an expanded genetic alphabet. Nat. Biotechnol. 2013;31:453–457. doi: 10.1038/nbt.2556. [DOI] [PubMed] [Google Scholar]
- 65.Sefah K., Yang Z., Bradley K.M., Hoshika S., Jiménez E., Zhang L., Zhu G., Shanker S., Yu F., Turek D., et al. In vitro selection with artificial expanded genetic information systems. Proc. Natl. Acad. Sci. USA. 2014;111:1449–1454. doi: 10.1073/pnas.1311778111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Biondi E., Lane J.D., Das D., Dasgupta S., Piccirilli J.A., Hoshika S., Bradley K.M., Krantz B.A., Benner S.A. Laboratory evolution of artificially expanded DNA gives redesignable aptamers that target the toxic form of anthrax protective antigen. Nucleic Acids Res. 2016;44:9565–9577. doi: 10.1093/nar/gkw890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen T., Romesberg F.E. Directed polymerase evolution. FEBS Lett. 2014;588:219–229. doi: 10.1016/j.febslet.2013.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cheng C., Chen Y.H., Lennox K.A., Behlke M.A., Davidson B.L. In vivo SELEX for Identification of Brain-penetrating Aptamers. Mol. Ther. Nucleic Acids. 2013;2:e67. doi: 10.1038/mtna.2012.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen L., He W., Jiang H., Wu L., Xiong W., Li B., Zhou Z., Qian Y. In vivo SELEX of bone targeting aptamer in prostate cancer bone metastasis model. Int. J. Nanomed. 2019;14:149–159. doi: 10.2147/IJN.S188003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fernández G., Moraga A., Cuartero M.I., García-Culebras A., Peña-Martínez C., Pradillo J.M., Hernández-Jiménez M., Sacristán S., Ayuso M.I., Gonzalo-Gobernado R., et al. TLR4-Binding DNA Aptamers Show a Protective Effect against Acute Stroke in Animal Models. Mol. Ther. 2018;26:2047–2059. doi: 10.1016/j.ymthe.2018.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lecocq S., Spinella K., Dubois B., Lista S., Hampel H., Penner G. Aptamers as biomarkers for neurological disorders. Proof of concept in transgenic mice. PLoS ONE. 2018;13:e0190212. doi: 10.1371/journal.pone.0190212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Mai J., Li X., Zhang G., Huang Y., Xu R., Shen Q., Lokesh G.L., Thiviyanathan V., Chen L., Liu H., et al. DNA Thioaptamer with Homing Specificity to Lymphoma Bone Marrow Involvement. Mol. Pharm. 2018;15:1814–1825. doi: 10.1021/acs.molpharmaceut.7b01169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ospina-Villa J.D., López-Camarillo C., Castañón-Sánchez C.A., Soto-Sánchez J., Ramírez-Moreno E., Marchat L.A. Advances on Aptamers against Protozoan Parasites. Genes. 2018;9:584. doi: 10.3390/genes9120584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bruno J.G., Carrillo M.P., Phillips T., Andrews C.J. A novel screening method for competitive FRET-aptamers applied to E. coli assay development. J. Fluoresc. 2010;20:1211–1223. doi: 10.1007/s10895-010-0670-9. [DOI] [PubMed] [Google Scholar]
- 75.Bruno J.G., Phillips T., Carrillo M.P., Crowell R. Plastic-adherent DNA aptamer-magnetic bead and quantum dot sandwich assay for Campylobacter detection. J. Fluoresc. 2009;19:427–435. doi: 10.1007/s10895-008-0429-8. [DOI] [PubMed] [Google Scholar]
- 76.Hamula C.L., Zhang H., Guan L.L., Li X.F., Le X.C. Selection of aptamers against live bacterial cells. Anal. Chem. 2008;80:7812–7819. doi: 10.1021/ac801272s. [DOI] [PubMed] [Google Scholar]
- 77.Cao X., Li S., Chen L., Ding H., Xu H., Huang Y., Li J., Liu N., Cao W., Zhu Y., et al. Combining use of a panel of ssDNA aptamers in the detection of Staphylococcus aureus. Nucleic Acids Res. 2009;37:4621–4628. doi: 10.1093/nar/gkp489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chen F., Zhou J., Luo F., Mohammed A.B., Zhang X.L. Aptamer from whole-bacterium SELEX as new therapeutic reagent against virulent Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 2007;357:743–748. doi: 10.1016/j.bbrc.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 79.Duan N., Wu S., Chen X., Huang Y., Wang Z. Selection and Identification of a DNA Aptamer Targeted to Vibrio parahemolyticus. J. Agric. Food Chem. 2012;60:4034–4038. doi: 10.1021/jf300395z. [DOI] [PubMed] [Google Scholar]
- 80.Dwivedi H.P., Smiley R.D., Jaykus L.A. Selection and characterization of DNA aptamers with binding selectivity to Campylobacter jejuni using whole-cell SELEX. Appl. Microbiol. Biotechnol. 2010;87:2323–2334. doi: 10.1007/s00253-010-2728-7. [DOI] [PubMed] [Google Scholar]
- 81.Labib M., Zamay A.S., Muharemagic D., Chechik A.V., Bell J.C., Berezovski M.V. Aptamer-based viability impedimetric sensor for viruses. Anal. Chem. 2012;84:1813–1816. doi: 10.1021/ac203412m. [DOI] [PubMed] [Google Scholar]
- 82.Gopinath S.C., Hayashi K., Kumar P.K. Aptamer that binds to the gD protein of herpes simplex virus-1 and efficiently inhibits viral entry. J. Virol. 2012;86:6732–6744. doi: 10.1128/JVI.00377-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Fukuda K., Vishinuvardhan D., Sekiya S., Kakiuchi N., Shimotohno K., Kumar P.K., Nishikawa S. Specific RNA aptamers to NS3 protease domain of hepatitis C virus. Nucleic Acids Symp. Ser. 1997;37:237–238. [PubMed] [Google Scholar]
- 84.Kumar P.K., Machida K., Urvil P.T., Kakiuchi N., Vishnuvardhan D., Shimotohno K., Taira K., Nishikawa S. Isolation of RNA aptamers specific to the NS3 protein of hepatitis C virus from a pool of completely random RNA. Virology. 1997;237:270–282. doi: 10.1006/viro.1997.8773. [DOI] [PubMed] [Google Scholar]
- 85.Boiziau C., Dausse E., Yurchenko L., Toulmé J.J. DNA aptamers selected against the HIV-1 trans-activation-responsive RNA element form RNA-DNA kissing complexes. J. Biol. Chem. 1999;274:12730–12737. doi: 10.1074/jbc.274.18.12730. [DOI] [PubMed] [Google Scholar]
- 86.Gopinath S.C., Kawasaki K., Kumar P.K. Selection of RNA-aptamer against human influenza B virus. Nucleic Acids Symp. Ser. 2005;49:85–86. doi: 10.1093/nass/49.1.85. [DOI] [PubMed] [Google Scholar]
- 87.Jang K.J., Lee N.R., Yeo W.S., Jeong Y.J., Kim D.E. Isolation of inhibitory RNA aptamers against severe acute respiratory syndrome (SARS) coronavirus NTPase/Helicase. Biochem. Biophys. Res. Commun. 2008;366:738–744. doi: 10.1016/j.bbrc.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Nagarkatti R., de Araujo F.F., Gupta C., Debrabant A. Aptamer based, non-PCR, non-serological detection of Chagas disease biomarkers in Trypanosoma cruzi infected mice. PLoS Negl. Trop. Dis. 2014;8:e2650. doi: 10.1371/journal.pntd.0002650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Guerra-Pérez N., Ramos E., García-Hernández M., Pinto C., Soto M., Martín M.E., González V.M. Molecular and Functional Characterization of ssDNA Aptamers that Specifically Bind Leishmania infantum PABP. PLoS ONE. 2015;10:e0140048. doi: 10.1371/journal.pone.0140048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cheung Y.W., Dirkzwager R.M., Wong W.C., Cardoso J., D’Arc Neves Costa J., Tanner J.A. Aptamer-mediated Plasmodium-specific diagnosis of malaria. Biochimie. 2018;145:131–136. doi: 10.1016/j.biochi.2017.10.017. [DOI] [PubMed] [Google Scholar]
- 91.Tang M.S.L., Shiu S.C., Godonoga M., Cheung Y.W., Liang S., Dirkzwager R.M., Kinghorn A.B., Fraser L.A., Heddle J.G., Tanner J.A. An aptamer-enabled DNA nanobox for protein sensing. Nanomedicine. 2018;14:1161–1168. doi: 10.1016/j.nano.2018.01.018. [DOI] [PubMed] [Google Scholar]
- 92.Godonoga M., Lin T.Y., Oshima A., Sumitomo K., Tang M.S., Cheung Y.W., Kinghorn A.B., Dirkzwager R.M., Zhou C., Kuzuya A., et al. A DNA aptamer recognising a malaria protein biomarker can function as part of a DNA origami assembly. Sci. Rep. 2016;6:21266. doi: 10.1038/srep21266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Jain P., Chakma B., Singh N.K., Patra S., Goswami P. Aromatic Surfactant as Aggregating Agent for Aptamer-Gold Nanoparticle-Based Detection of Plasmodium Lactate Dehydrogenase. Mol. Biotechnol. 2016;58:497–508. doi: 10.1007/s12033-016-9946-x. [DOI] [PubMed] [Google Scholar]
- 94.Geldert A., Zhang X., Zhang H., Lim C.T. Enhancing the sensing specificity of a MoS. Analyst. 2017;142:2570–2577. doi: 10.1039/C7AN00640C. [DOI] [PubMed] [Google Scholar]
- 95.Frith K.A., Fogel R., Goldring J.P.D., Krause R.G.E., Khati M., Hoppe H., Cromhout M.E., Jiwaji M., Limson J.L. Towards development of aptamers that specifically bind to lactate dehydrogenase of Plasmodium falciparum through epitopic targeting. Malar. J. 2018;17:191. doi: 10.1186/s12936-018-2336-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Fraser L.A., Kinghorn A.B., Dirkzwager R.M., Liang S., Cheung Y.W., Lim B., Shiu S.C., Tang M.S.L., Andrew D., Manitta J., et al. A portable microfluidic Aptamer-Tethered Enzyme Capture (APTEC) biosensor for malaria diagnosis. Biosens. Bioelectron. 2018;100:591–596. doi: 10.1016/j.bios.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 97.Dirkzwager R.M., Kinghorn A.B., Richards J.S., Tanner J.A. APTEC: Aptamer-tethered enzyme capture as a novel rapid diagnostic test for malaria. Chem. Commun. 2015;51:4697–4700. doi: 10.1039/C5CC00438A. [DOI] [PubMed] [Google Scholar]
- 98.Choi S.J., Ban C. Crystal structure of a DNA aptamer bound to PvLDH elucidates novel single-stranded DNA structural elements for folding and recognition. Sci. Rep. 2016;6:34998. doi: 10.1038/srep34998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Wang W.X., Cheung Y.W., Dirkzwager R.M., Wong W.C., Tanner J.A., Li H.W., Wu Y. Specific and sensitive detection of Plasmodium falciparum lactate dehydrogenase by DNA-scaffolded silver nanoclusters combined with an aptamer. Analyst. 2017;142:800–807. doi: 10.1039/C6AN02417C. [DOI] [PubMed] [Google Scholar]
- 100.Iqbal A., Labib M., Muharemagic D., Sattar S., Dixon B.R., Berezovski M.V. Detection of Cryptosporidium parvum Oocysts on Fresh Produce Using DNA Aptamers. PLoS ONE. 2015;10:e0137455. doi: 10.1371/journal.pone.0137455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ospina-Villa J.D., Dufour A., Weber C., Ramirez-Moreno E., Zamorano-Carrillo A., Guillen N., Lopez-Camarillo C., Marchat L.A. Targeting the polyadenylation factor EhCFIm25 with RNA aptamers controls survival in Entamoeba histolytica. Sci. Rep. 2018;8:5720. doi: 10.1038/s41598-018-23997-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zheng C.Y., Pestilli F., Rokem A. Deconvolution of High Dimensional Mixtures via Boosting, with Application to Diffusion-Weighted MRI of Human Brain. Adv. Neural Inf. Process. Syst. 2014;27:2699–2707. [PMC free article] [PubMed] [Google Scholar]
- 103.Wang K., Fan D., Liu Y., Wang E. Highly sensitive and specific colorimetric detection of cancer cells via dual-aptamer target binding strategy. Biosens. Bioelectron. 2015;73:1–6. doi: 10.1016/j.bios.2015.05.044. [DOI] [PubMed] [Google Scholar]
- 104.Ye X., Shi H., He X., Wang K., He D., Yan L., Xu F., Lei Y., Tang J., Yu Y. Iodide-Responsive Cu-Au Nanoparticle-Based Colorimetric Platform for Ultrasensitive Detection of Target Cancer Cells. Anal. Chem. 2015;87:7141–7147. doi: 10.1021/acs.analchem.5b00943. [DOI] [PubMed] [Google Scholar]
- 105.Zhang F., Li S., Cao K., Wang P., Su Y., Zhu X., Wan Y. A Microfluidic Love-Wave Biosensing Device for PSA Detection Based on an Aptamer Beacon Probe. Sensors. 2015;15:13839–13850. doi: 10.3390/s150613839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Yang X., Zhuo Y., Zhu S., Luo Y., Feng Y., Xu Y. Selectively assaying CEA based on a creative strategy of gold nanoparticles enhancing silver nanoclusters’ fluorescence. Biosens. Bioelectron. 2015;64:345–351. doi: 10.1016/j.bios.2014.09.029. [DOI] [PubMed] [Google Scholar]
- 107.Li X., An Y., Jin J., Zhu Z., Hao L., Liu L., Shi Y., Fan D., Ji T., Yang C.J. Evolution of DNA aptamers through in vitro metastatic-cell-based systematic evolution of ligands by exponential enrichment for metastatic cancer recognition and imaging. Anal. Chem. 2015;87:4941–4948. doi: 10.1021/acs.analchem.5b00637. [DOI] [PubMed] [Google Scholar]
- 108.Wu X., Zhao Z., Bai H., Fu T., Yang C., Hu X., Liu Q., Champanhac C., Teng I.T., Ye M., et al. DNA Aptamer Selected against Pancreatic Ductal Adenocarcinoma for in vivo Imaging and Clinical Tissue Recognition. Theranostics. 2015;5:985–994. doi: 10.7150/thno.11938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sun H., Tan W., Zu Y. Aptamers: Versatile molecular recognition probes for cancer detection. Analyst. 2016;141:403–415. doi: 10.1039/C5AN01995H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ababneh N., Alshaer W., Allozi O., Mahafzah A., El-Khateeb M., Hillaireau H., Noiray M., Fattal E., Ismail S. In vitro selection of modified RNA aptamers against CD44 cancer stem cell marker. Nucleic Acid Ther. 2013;23:401–407. doi: 10.1089/nat.2013.0423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Iwagawa T., Ohuchi S.P., Watanabe S., Nakamura Y. Selection of RNA aptamers against mouse embryonic stem cells. Biochimie. 2012;94:250–257. doi: 10.1016/j.biochi.2011.10.017. [DOI] [PubMed] [Google Scholar]
- 112.Burke D.H., Hoffman D.C., Brown A., Hansen M., Pardi A., Gold L. RNA aptamers to the peptidyl transferase inhibitor chloramphenicol. Chem. Biol. 1997;4:833–843. doi: 10.1016/S1074-5521(97)90116-2. [DOI] [PubMed] [Google Scholar]
- 113.Kim Y.J., Kim Y.S., Niazi J.H., Gu M.B. Electrochemical aptasensor for tetracycline detection. Bioprocess Biosyst. Eng. 2010;33:31–37. doi: 10.1007/s00449-009-0371-4. [DOI] [PubMed] [Google Scholar]
- 114.Cruz-Aguado J.A., Penner G. Determination of ochratoxin a with a DNA aptamer. J. Agric. Food Chem. 2008;56:10456–10461. doi: 10.1021/jf801957h. [DOI] [PubMed] [Google Scholar]
- 115.Kim S.E., Su W., Cho M., Lee Y., Choe W.S. Harnessing aptamers for electrochemical detection of endotoxin. Anal. Biochem. 2012;424:12–20. doi: 10.1016/j.ab.2012.02.016. [DOI] [PubMed] [Google Scholar]
- 116.Jo M., Ahn J.Y., Lee J., Lee S., Hong S.W., Yoo J.W., Kang J., Dua P., Lee D.K., Hong S., et al. Development of single-stranded DNA aptamers for specific Bisphenol a detection. Oligonucleotides. 2011;21:85–91. doi: 10.1089/oli.2010.0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zeng G., Zhang C., Huang D., Lai C., Tang L., Zhou Y., Xu P., Wang H., Qin L., Cheng M. Practical and regenerable electrochemical aptasensor based on nanoporous gold and thymine-Hg. Biosens. Bioelectron. 2017;90:542–548. doi: 10.1016/j.bios.2016.10.018. [DOI] [PubMed] [Google Scholar]
- 118.Li L., Li B., Qi Y., Jin Y. Label-free aptamer-based colorimetric detection of mercury ions in aqueous media using unmodified gold nanoparticles as colorimetric probe. Anal. Bioanal. Chem. 2009;393:2051–2057. doi: 10.1007/s00216-009-2640-0. [DOI] [PubMed] [Google Scholar]
- 119.Cui L., Wu J., Ju H. Label-free signal-on aptasensor for sensitive electrochemical detection of arsenite. Biosens. Bioelectron. 2016;79:861–865. doi: 10.1016/j.bios.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 120.Kim M., Um H.J., Bang S., Lee S.H., Oh S.J., Han J.H., Kim K.W., Min J., Kim Y.H. Arsenic removal from Vietnamese groundwater using the arsenic-binding DNA aptamer. Environ. Sci. Technol. 2009;43:9335–9340. doi: 10.1021/es902407g. [DOI] [PubMed] [Google Scholar]
- 121.Oroval M., Coll C., Bernardos A., Marcos M.D., Martínez-Máñez R., Shchukin D.G., Sancenón F. Selective Fluorogenic Sensing of As(III) Using Aptamer-Capped Nanomaterials. ACS Appl. Mater. Interfaces. 2017;9:11332–11336. doi: 10.1021/acsami.6b15164. [DOI] [PubMed] [Google Scholar]
- 122.Chen Z., Li L., Mu X., Zhao H., Guo L. Electrochemical aptasensor for detection of copper based on a reagentless signal-on architecture and amplification by gold nanoparticles. Talanta. 2011;85:730–735. doi: 10.1016/j.talanta.2011.04.056. [DOI] [PubMed] [Google Scholar]
- 123.Mishra G.K., Sharma V., Mishra R.K. Electrochemical Aptasensors for Food and Environmental Safeguarding: A Review. Biosensors. 2018;8:28. doi: 10.3390/bios8020028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Eissa S., Zourob M. In vitro selection of DNA aptamers targeting β-lactoglobulin and their integration in graphene-based biosensor for the detection of milk allergen. Biosens. Bioelectron. 2017;91:169–174. doi: 10.1016/j.bios.2016.12.020. [DOI] [PubMed] [Google Scholar]
- 125.Hu W., Chen Q., Li H., Ouyang Q., Zhao J. Fabricating a novel label-free aptasensor for acetamiprid by fluorescence resonance energy transfer between NH2-NaYF4: Yb, Ho@SiO2 and Au nanoparticles. Biosens. Bioelectron. 2016;80:398–404. doi: 10.1016/j.bios.2016.02.001. [DOI] [PubMed] [Google Scholar]
- 126.Madianos L., Tsekenis G., Skotadis E., Patsiouras L., Tsoukalas D. A highly sensitive impedimetric aptasensor for the selective detection of acetamiprid and atrazine based on microwires formed by platinum nanoparticles. Biosens. Bioelectron. 2018;101:268–274. doi: 10.1016/j.bios.2017.10.034. [DOI] [PubMed] [Google Scholar]
- 127.Xu G., Huo D., Hou C., Zhao Y., Bao J., Yang M., Fa H. A regenerative and selective electrochemical aptasensor based on copper oxide nanoflowers-single walled carbon nanotubes nanocomposite for chlorpyrifos detection. Talanta. 2018;178:1046–1052. doi: 10.1016/j.talanta.2017.08.086. [DOI] [PubMed] [Google Scholar]
- 128.Sinha J., Reyes S.J., Gallivan J.P. Reprogramming bacteria to seek and destroy an herbicide. Nat. Chem. Biol. 2010;6:464–470. doi: 10.1038/nchembio.369. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 129.Fan L., Zhao G., Shi H., Liu M., Li Z. A highly selective electrochemical impedance spectroscopy-based aptasensor for sensitive detection of acetamiprid. Biosens. Bioelectron. 2013;43:12–18. doi: 10.1016/j.bios.2012.11.033. [DOI] [PubMed] [Google Scholar]
- 130.Abraham K.M., Roueinfar M., Ponce A.T., Lussier M.E., Benson D.B., Hong K.L. In Vitro Selection and Characterization of a Single-Stranded DNA Aptamer Against the Herbicide Atrazine. ACS Omega. 2018;3:13576–13583. doi: 10.1021/acsomega.8b01859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Wang K.Y., Krawczyk S.H., Bischofberger N., Swaminathan S., Bolton P.H. The tertiary structure of a DNA aptamer which binds to and inhibits thrombin determines activity. Biochemistry. 1993;32:11285–11292. doi: 10.1021/bi00093a004. [DOI] [PubMed] [Google Scholar]
- 132.Zhang W., Liu Q.X., Guo Z.H., Lin J.S. Practical Application of Aptamer-Based Biosensors in Detection of Low Molecular Weight Pollutants in Water Sources. Molecules. 2018;23:344. doi: 10.3390/molecules23020344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Zhou W., Huang P.J., Ding J., Liu J. Aptamer-based biosensors for biomedical diagnostics. Analyst. 2014;139:2627–2640. doi: 10.1039/c4an00132j. [DOI] [PubMed] [Google Scholar]
- 134.Hong L., Zhou F., Shi D., Zhang X., Wang G. Portable aptamer biosensor of platelet-derived growth factor-BB using a personal glucose meter with triply amplified. Biosens. Bioelectron. 2017;95:152–159. doi: 10.1016/j.bios.2017.04.023. [DOI] [PubMed] [Google Scholar]
- 135.Wiedman G.R., Zhao Y., Mustaev A., Ping J., Vishnubhotla R., Johnson A.T.C., Perlin D.S. An Aptamer-Based Biosensor for the Azole Class of Antifungal Drugs. mSphere. 2017;2:e00274-17. doi: 10.1128/mSphere.00274-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yang Y., Yang X., Zou X., Wu S., Wan D., Cao A., Liao L., Yuan Q., Duan X. Ultrafine Graphene Nanomesh with Large On/Off Ratio for High-Performance Flexible Biosensors. Adv. Funct. Mater. 2017;27:1604096. doi: 10.1002/adfm.201604096. [DOI] [Google Scholar]
- 137.Chen M.-L., Chen J.-H., Ding L., Xu Z., Wen L., Wang L.-B., Cheng Y.-H. Study of the detection of bisphenol A based on a nano-sized metal–organic framework crystal and an aptamer. Anal. Methods. 2017;9:906–909. doi: 10.1039/C6AY03151J. [DOI] [Google Scholar]
- 138.Saidur M.R., Aziz A.R., Basirun W.J. Recent advances in DNA-based electrochemical biosensors for heavy metal ion detection: A review. Biosens. Bioelectron. 2017;90:125–139. doi: 10.1016/j.bios.2016.11.039. [DOI] [PubMed] [Google Scholar]
- 139.Xu S., Feng X., Gao T., Wang R., Mao Y., Lin J., Yu X., Luo X. A novel dual-functional biosensor for fluorometric detection of inorganic pyrophosphate and pyrophosphatase activity based on globulin stabilized gold nanoclusters. Anal. Chim. Acta. 2017;958:22–29. doi: 10.1016/j.aca.2016.12.026. [DOI] [PubMed] [Google Scholar]
- 140.Alhadrami H.A., Chinnappan R., Eissa S., Rahamn A.A., Zourob M. High affinity truncated DNA aptamers for the development of fluorescence based progesterone biosensors. Anal. Biochem. 2017;525:78–84. doi: 10.1016/j.ab.2017.02.014. [DOI] [PubMed] [Google Scholar]
- 141.Arroyo-Currás N., Scida K., Ploense K.L., Kippin T.E., Plaxco K.W. High Surface Area Electrodes Generated via Electrochemical Roughening Improve the Signaling of Electrochemical Aptamer-Based Biosensors. Anal. Chem. 2017;89:12185–12191. doi: 10.1021/acs.analchem.7b02830. [DOI] [PubMed] [Google Scholar]
- 142.Wang X., Sun D., Tong Y.A., Zhong Y., Chen Z. A voltammetric aptamer-based thrombin biosensor exploiting signal amplification via synergetic catalysis by DNAzyme and enzyme decorated AuPd nanoparticles on a poly(o-phenylenediamine) support. Microchim. Acta. 2017;184:1791–1799. doi: 10.1007/s00604-017-2160-0. [DOI] [Google Scholar]
- 143.Jeddi I., Saiz L. Three-dimensional modeling of single stranded DNA hairpins for aptamer-based biosensors. Sci. Rep. 2017;7:1178. doi: 10.1038/s41598-017-01348-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hwang S.Y., Sun H.Y., Lee K.H., Oh B.H., Cha Y.J., Kim B.H., Yoo J.Y. 5′-Triphosphate-RNA-independent activation of RIG-I via RNA aptamer with enhanced antiviral activity. Nucleic Acids Res. 2012;40:2724–2733. doi: 10.1093/nar/gkr1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mufhandu H.T., Gray E.S., Madiga M.C., Tumba N., Alexandre K.B., Khoza T., Wibmer C.K., Moore P.L., Morris L., Khati M. UCLA1, a Synthetic Derivative of a gp120 RNA Aptamer, Inhibits Entry of Human Immunodeficiency Virus Type 1 Subtype C. J. Virol. 2012;86:4989–4999. doi: 10.1128/JVI.06893-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Nishikawa F., Kakiuchi N., Funaji K., Fukuda K., Sekiya S., Nishikawa S. Inhibition of HCV NS3 protease by RNA aptamers in cells. Nucleic Acids Res. 2003;31:1935–1943. doi: 10.1093/nar/gkg291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Umehara T., Fukuda K., Nishikawa F., Sekiya S., Kohara M., Hasegawa T., Nishikawa S. Designing and analysis of a potent bi-functional aptamers that inhibit protease and helicase activities of HCV NS3. Nucleic Acids Symp. Ser. 2004;48:195–196. doi: 10.1093/nass/48.1.195. [DOI] [PubMed] [Google Scholar]
- 148.Urvil P.T., Kakiuchi N., Zhou D.M., Shimotohno K., Kumar P.K., Nishikawa S. Selection of RNA aptamers that bind specifically to the NS3 protease of hepatitis C virus. Eur. J. Biochem. 1997;248:130–138. doi: 10.1111/j.1432-1033.1997.t01-1-00130.x. [DOI] [PubMed] [Google Scholar]
- 149.Hoellenriegel J., Zboralski D., Maasch C., Rosin N.Y., Wierda W.G., Keating M.J., Kruschinski A., Burger J.A. The Spiegelmer NOX-A12, a novel CXCL12 inhibitor, interferes with chronic lymphocytic leukemia cell motility and causes chemosensitization. Blood. 2014;123:1032–1039. doi: 10.1182/blood-2013-03-493924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Zamay T.N., Kolovskaya O.S., Glazyrin Y.E., Zamay G.S., Kuznetsova S.A., Spivak E.A., Wehbe M., Savitskaya A.G., Zubkova O.A., Kadkina A., et al. DNA-aptamer targeting vimentin for tumor therapy in vivo. Nucleic Acid Ther. 2014;24:160–170. doi: 10.1089/nat.2013.0471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ismail S.I., Alshaer W. Therapeutic aptamers in discovery, preclinical and clinical stages. Adv. Drug Deliv. Rev. 2018;134:51–64. doi: 10.1016/j.addr.2018.08.006. [DOI] [PubMed] [Google Scholar]
- 152.Pan Q., Yan J., Liu Q., Yuan C., Zhang X.L. A single-stranded DNA aptamer against mannose-capped lipoarabinomannan enhances anti-tuberculosis activity of macrophages through downregulation of lipid-sensing nuclear receptor peroxisome proliferator-activated receptor γ expression. Microbiol. Immunol. 2017;61:92–102. doi: 10.1111/1348-0421.12470. [DOI] [PubMed] [Google Scholar]
- 153.Wallukat G., Müller J., Haberland A., Berg S., Schulz A., Freyse E.J., Vetter R., Salzsieder E., Kreutz R., Schimke I. Aptamer BC007 for neutralization of pathogenic autoantibodies directed against G-protein coupled receptors: A vision of future treatment of patients with cardiomyopathies and positivity for those autoantibodies. Atherosclerosis. 2016;244:44–47. doi: 10.1016/j.atherosclerosis.2015.11.001. [DOI] [PubMed] [Google Scholar]
- 154.Gragoudas E.S., Adamis A.P., Cunningham E.T., Feinsod M., Guyer D.R. VEGF Inhibition Study in Ocular Neovascularization Clinical Trial Group. Pegaptanib for neovascular age-related macular degeneration. N. Engl. J. Med. 2004;351:2805–2816. doi: 10.1056/NEJMoa042760. [DOI] [PubMed] [Google Scholar]
- 155.Rusconi C.P., Scardino E., Layzer J., Pitoc G.A., Ortel T.L., Monroe D., Sullenger B.A. RNA aptamers as reversible antagonists of coagulation factor IXa. Nature. 2002;419:90–94. doi: 10.1038/nature00963. [DOI] [PubMed] [Google Scholar]
- 156.Bates P.J., Laber D.A., Miller D.M., Thomas S.D., Trent J.O. Discovery and development of the G-rich oligonucleotide AS1411 as a novel treatment for cancer. Exp. Mol. Pathol. 2009;86:151–164. doi: 10.1016/j.yexmp.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Ng E.W., Shima D.T., Calias P., Cunningham E.T., Guyer D.R., Adamis A.P. Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat. Rev. Drug Discov. 2006;5:123–132. doi: 10.1038/nrd1955. [DOI] [PubMed] [Google Scholar]
- 158.Alibolandi M., Taghdisi S.M., Ramezani P., Hosseini Shamili F., Farzad S.A., Abnous K., Ramezani M. Smart AS1411-aptamer conjugated pegylated PAMAM dendrimer for the superior delivery of camptothecin to colon adenocarcinoma in vitro and in vivo. Int. J. Pharm. 2017;519:352–364. doi: 10.1016/j.ijpharm.2017.01.044. [DOI] [PubMed] [Google Scholar]
- 159.Wang Y., Chen X., Tian B., Liu J., Yang L., Zeng L., Chen T., Hong A., Wang X. Nucleolin-targeted Extracellular Vesicles as a Versatile Platform for Biologics Delivery to Breast Cancer. Theranostics. 2017;7:1360–1372. doi: 10.7150/thno.16532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Cho Y., Lee Y.B., Lee J.H., Lee D.H., Cho E.J., Yu S.J., Kim Y.J., Kim J.I., Im J.H., Oh E.J., et al. Modified AS1411 Aptamer Suppresses Hepatocellular Carcinoma by Up-Regulating Galectin-14. PLoS ONE. 2016;11:e0160822. doi: 10.1371/journal.pone.0160822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Kulkarni O., Pawar R.D., Purschke W., Eulberg D., Selve N., Buchner K., Ninichuk V., Segerer S., Vielhauer V., Klussmann S., et al. Spiegelmer inhibition of CCL2/MCP-1 ameliorates lupus nephritis in MRL-(Fas)lpr mice. J. Am. Soc. Nephrol. 2007;18:2350–2358. doi: 10.1681/ASN.2006121348. [DOI] [PubMed] [Google Scholar]
- 162.Kulkarni O., Eulberg D., Selve N., Zöllner S., Allam R., Pawar R.D., Pfeiffer S., Segerer S., Klussmann S., Anders H.J. Anti-Ccl2 Spiegelmer permits 75% dose reduction of cyclophosphamide to control diffuse proliferative lupus nephritis and pneumonitis in MRL-Fas(lpr) mice. J. Pharmacol. Exp. Ther. 2009;328:371–377. doi: 10.1124/jpet.108.142711. [DOI] [PubMed] [Google Scholar]
- 163.Shu D., Li H., Shu Y., Xiong G., Carson W.E., Haque F., Xu R., Guo P. Systemic Delivery of Anti-miRNA for Suppression of Triple Negative Breast Cancer Utilizing RNA Nanotechnology. ACS Nano. 2015;9:9731–9740. doi: 10.1021/acsnano.5b02471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Gijs M., Aerts A., Impens N., Baatout S., Luxen A. Aptamers as radiopharmaceuticals for nuclear imaging and therapy. Nucl. Med. Biol. 2016;43:253–271. doi: 10.1016/j.nucmedbio.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 165.Liang C., Guo B., Wu H., Shao N., Li D., Liu J., Dang L., Wang C., Li H., Li S., et al. Aptamer-functionalized lipid nanoparticles targeting osteoblasts as a novel RNA interference-based bone anabolic strategy. Nat. Med. 2015;21:288–294. doi: 10.1038/nm.3791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Gantenbein A.R., Sarikaya H., Riederer F., Goadsby P.J. Postoperative hemicrania continua-like headache—A case series. J. Headache Pain. 2015;16:526. doi: 10.1186/s10194-015-0526-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Fan X., Sun L., Li K., Yang X., Cai B., Zhang Y., Zhu Y., Ma Y., Guan Z., Wu Y., et al. The Bioactivity of d-/l-Isonucleoside- and 2′-Deoxyinosine-Incorporated Aptamer AS1411s Including DNA Replication/MicroRNA Expression. Mol. Ther. Nucleic Acids. 2017;9:218–229. doi: 10.1016/j.omtn.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zboralski D., Hoehlig K., Eulberg D., Frömming A., Vater A. Increasing Tumor-Infiltrating T Cells through Inhibition of CXCL12 with NOX-A12 Synergizes with PD-1 Blockade. Cancer Immunol. Res. 2017;5:950–956. doi: 10.1158/2326-6066.CIR-16-0303. [DOI] [PubMed] [Google Scholar]
- 169.Morita Y., Kamal M., Kang S.A., Zhang R., Lokesh G.L., Thiviyanathan V., Hasan N., Woo S., Zhao D., Leslie M., et al. E-selectin Targeting PEGylated-thioaptamer Prevents Breast Cancer Metastases. Mol. Ther. Nucleic Acids. 2016;5:e399. doi: 10.1038/mtna.2016.103. [DOI] [PubMed] [Google Scholar]
- 170.Prodeus A., Abdul-Wahid A., Fischer N.W., Huang E.H., Cydzik M., Gariépy J. Targeting the PD-1/PD-L1 Immune Evasion Axis With DNA Aptamers as a Novel Therapeutic Strategy for the Treatment of Disseminated Cancers. Mol. Ther. Nucleic Acids. 2015;4:e237. doi: 10.1038/mtna.2015.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Huang B.T., Lai W.Y., Chang Y.C., Wang J.W., Yeh S.D., Lin E.P., Yang P.C. A CTLA-4 Antagonizing DNA Aptamer with Antitumor Effect. Mol. Ther. Nucleic Acids. 2017;8:520–528. doi: 10.1016/j.omtn.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ajona D., Ortiz-Espinosa S., Moreno H., Lozano T., Pajares M.J., Agorreta J., Bértolo C., Lasarte J.J., Vicent S., Hoehlig K., et al. A Combined PD-1/C5a Blockade Synergistically Protects against Lung Cancer Growth and Metastasis. Cancer Discov. 2017;7:694–703. doi: 10.1158/2159-8290.CD-16-1184. [DOI] [PubMed] [Google Scholar]
- 173.Zheng J., Zhao S., Yu X., Huang S., Liu H.Y. Simultaneous targeting of CD44 and EpCAM with a bispecific aptamer effectively inhibits intraperitoneal ovarian cancer growth. Theranostics. 2017;7:1373–1388. doi: 10.7150/thno.17826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Waters B., Lillicrap D. The molecular mechanisms of immunomodulation and tolerance induction to factor VIII. J. Thromb. Haemost. 2009;7:1446–1456. doi: 10.1111/j.1538-7836.2009.03538.x. [DOI] [PubMed] [Google Scholar]