Abstract
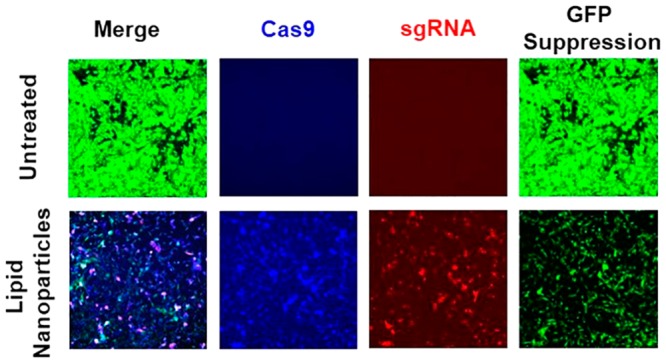
CRISPR/Cas9 system is a promising approach for gene editing in gene therapy. Effective gene editing requires safe and efficient delivery of CRISPR/Cas9 system in target cells. Several new multifunctional pH-sensitive amino lipids were designed and synthesized with modification of the amino head groups for intracellular delivery of CRISPR/Cas9 system. These multifunctional pH-sensitive amino lipids exhibited structurally dependent formulation of stable nanoparticles with the DNA plasmids of CRISPR/Cas9 system with the sizes ranging from 100 to 200 nm. The amino lipid plasmid DNA nanoparticles showed pH-sensitive hemolysis with minimal hemolytic activity at pH 7.4 and increased hemolysis at acidic pH (pH = 5.5, 6.5). The nanoparticles exhibited low cytotoxicity at an N/P ratio of 10. Expression of both Cas9 and sgRNA of the CRISPR/Cas9 system was in the range from 4.4% to 33%, dependent on the lipid structure in NIH3T3-GFP cells. The amino lipids that formed stable nanoparticles with high expression of both Cas9 and sgRNA mediated high gene editing efficiency. ECO and iECO mediated more efficient gene editing than other tested lipids. ECO mediated up to 50% GFP suppression based on observations with confocal microscopy and nearly 80% reduction of GFP mRNA based on RT-PCR measurement in NIH3T3-GFP cells. The multifunctional pH-sensitive amino lipids have the potential for efficient intracellular delivery of CRISPR/Cas9 for effective gene editing.
Introduction
CRISPR (clustered regulatory interspaced short palindromic repeats)/Cas9 (CRISPR associated protein 9) system has the potential to become a revolutionary and powerful gene editing method, and presents a promising approach for gene therapy.1−5 CRISPR/Cas9 system acts as an adaptive immune system in bacteria and archaea. It relies on integration of foreign DNA fragments into CRISPR loci and subsequent transcription and processing of these RNA transcripts into short CRISPR RNAs (crRNAs), which in turn anneal to a trans-activating crRNA (tracrRNA) and direct sequence-specific silencing of foreign nucleic acids by Cas proteins.6 Recent studies have shown that a synthetic single guide RNA (sgRNA) consisting of a fusion of crRNA and tracrRNA can direct Cas9 endonuclease-mediated cleavage of target DNA, which enables sequence-specific DNA editing. Cas9 nuclease is guided by programmable sgRNA, recognizes the genomic sequence with a 3′ protospacer adjacent motif (PAM) sequence, and cleaves the recognized DNA. Following DNA cleavage, double-strand break repair mechanisms allow mutagenesis or insertion/deletion (INDEL) mutations.7,8
CRISPR/Cas9 has the potential to treat autosomal dominant diseases, such as Huntington's disease and retinitis pigmentosa, which are caused by mutations in one of a pair of autosomal chromosomes. CRISPR/Cas9 can facilitate allele specific genome editing by selectively targeting and permanently inactivating the mutant allele while leaving the normal allele functionally intact.9,10 The effectiveness of CRISPR/Cas9 for allele specific editing and disease phenotype alleviation has been demonstrated in animal models.3,9−11 For therapeutic applications, both Cas9 nuclease and sgRNA can be incorporated in DNA plasmids and expressed in the same cell for gene editing after intracellular delivery. Therefore, codelivery of Cas9 and sgRNA is a critical step for therapeutic applications of CRISPR/Cas9 systems in treating autosomal dominant diseases.
Various delivery strategies of nucleic acid based therapeutics, including both viral and nonviral approaches, have been tested for the delivery of CRISPR/Cas9 system.2,12−14 Although successful gene editing has been induced by both viral and nonviral delivery systems, the available delivery systems suffer from various limitations, e.g., immunogenicity and random genome integration for viral systems15 and low transcription efficiency for nonviral systems.16−18 We have previously reported a class of multifunctional pH-sensitive amino lipids, including (1-aminoethyl)iminobis[N-(oleoylcysteinyl-1-aminoethyl)propionamide] (ECO), as simple and smart carriers for cytosolic delivery of plasmid DNA and siRNA for gene replacement therapies and gene regulation.19−22 The multifunctional amino lipids have a protonatable amino headgroup, two cysteinyl residues, and two distant lipid tails. They can form stable self-assembly nanoparticles with nucleic acids by electrostatic complexation with the headgroup, hydrophobic condensation of the lipid tails and disulfide bond cross-linking of the cysteinyl residues without any helper lipids. The multifunctional lipids mediate highly efficient cytosolic delivery of therapeutic DNA and siRNA through pH-sensitive amphiphilic endosomal escape and reductive cytosolic release (PERC).23
In this work, a series of new multifunctional pH-sensitive amino lipids were designed and synthesized based on the PERC approach for intracellular delivery of CRISPR/Cas9 system. In these lipid structures, a new headgroup 2,2′,2″-triaminotriethylamine was used to replace the moiety of 3,3′-[(2-aminoethyl)imino]bis[N-(2-aminoethyl)propanamide] in the previous design. Lysine and histidine were also introduced to assess the effects of the side chains with amino groups of different pKas. These new isotypic ECO lipids were synthesized and characterized for cytosolic delivery of CRISPR/Cas9 and gene editing. The intracellular transfection efficiency and gene knockdown efficacy of the lipids were assessed with a CRISPR/Cas9 system targeting GFP sequence.
Results
Synthesis of Multifunctional pH-Sensitive Amino Lipids
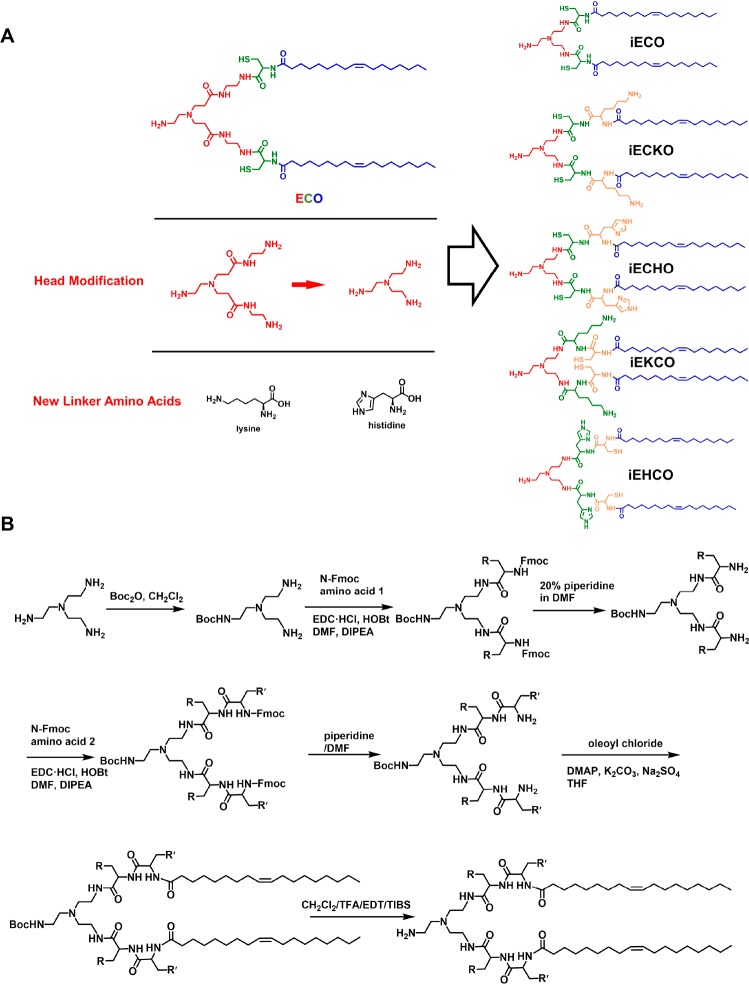
The structures and abbreviated names of the newly designed multifunctional pH-sensitive lipids are shown in Figure 1A. 2,2′,2″-Triaminotriethylamine was used to design the protonatable ethylene diamine headgroup with Y-shaped branches for conjugating cysteinyl residues with different combination of histidinyl or lysinyl functional residues, and two oleoyl tails. The designed amino lipids were synthesized following liquid phase protocols, which are shown in Figure 1B. The final products were purified by flash chromatography and characterized by MALDI-TOF mass spectrometry and 1H NMR spectroscopy.
Figure 1.
Design and synthesis of isotypic ECO derivatives with modifications of the headgroup and new amino acid functional linkers. (A) Chemical structures of ECO isotypic derivatives. (B) Synthetic scheme of ECO isotypic derivatives.
Formulation of Lipid CRISPR/Cas9 Plasmid Nanoparticles
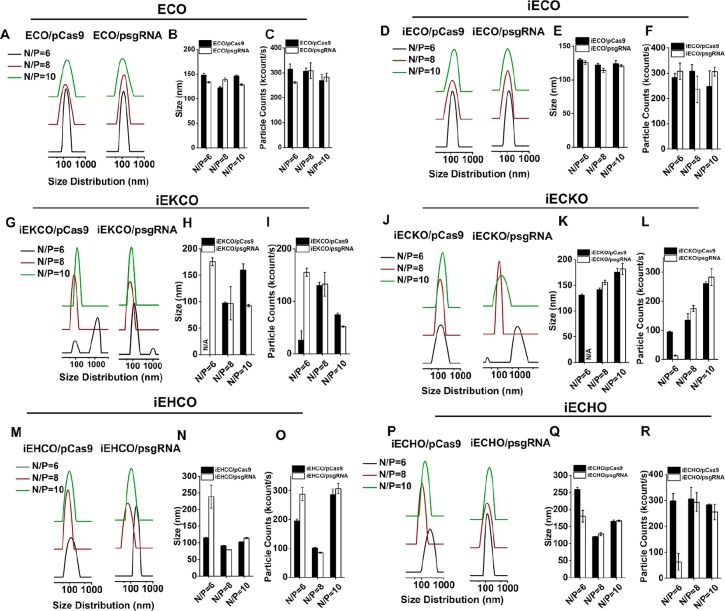
The amino lipids were used to formulate nanoparticles with a plasmid DNA expressing sgRNAs (psgRNA) (9.4 kb) and a plasmid DNA expressing Cas9 (pCas9) (13.6 kb), respectively, through self-assembly for gene editing (Figure 2). The nanoparticles were formulated by simply mixing the stock solutions of the lipids and plasmid DNA stock solutions at N/P ratios of 6, 8, and 10. The nanoparticles were characterized by dynamic light scattering (DLS) (Figure 3). All amino lipids, including ECO, formed nanoparticles with both plasmids, which had a narrow size distribution between 100 and 150 nm at the tested N/P ratios, except for iEKCO, iECKO, iEHCO, and iECHO at the N/P of 6. The lipids iEKCO, iECKO, iEHCO, and iECHO appeared unable to form stable nanoparticles with both psgRNA and pCas9 at the N/P ratio of 6. Large nanoparticles or aggregations were detected for the lipids at the low N/P ratio. iEHCO and iECHO could form stable nanoparticles at N/P = 8 and 10 with good particle counts. Among these lipids, ECO and iECO were able to formulate stable nanoparticles with size distributions in the range of 100–200 nm and high nanoparticle counts within the range of 200–350 kilocounts at all N/P ratios.
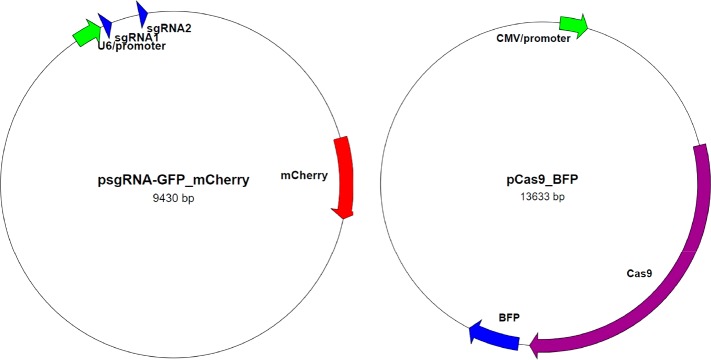
Figure 2.
Plasmid maps of CRISPR/Cas9 system that targets GFP gene. The system includes a plasmid expressing two sgRNAs targeting GFP sequence and mCherry reporter, and a plasmid expressing Cas9 nuclease and BFP reporter.
Figure 3.
DLS measurements of the nanoparticles formed by the multifunctional pH-sensitive lipids with psgRNA and pCas9. The size distribution of nanoparticles formulated between (A) ECO, (D) iECO, (G) iEKCO, (J) iECKO, (M) iEHCO, (P) iECHO, and psgRNA or pCas9. The average sizes and particle counts of nanoparticles formulated with (B,C) ECO, (E,F) iECO, (H,I) iEKCO, (K,L) iECKO, (H,O) iEHCO, (Q,R) iECHO, and the plasmids. N/A = no reliable readouts due to formation of unstable and varied nanoparticles.
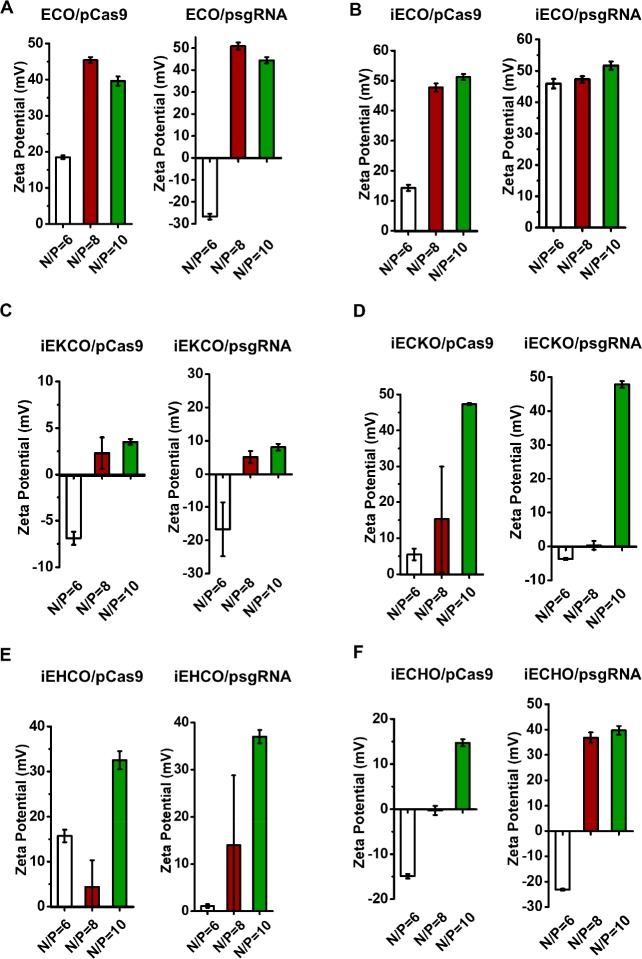
The zeta potential of the nanoparticles varied with the N/P ratios, as shown in Figure 4. In general, an increase in zeta potential was observed with the increase of N/P ratios for all carriers. At N/P ratio of 6, the nanoparticles formed with both pCas9 and psgRNA have either negative zeta potentials or slightly positive zeta potentials except for iECO, which was able to form nanoparticles with psgRNA that had a zeta potential larger than +30 mV (45.9 ± 1.5 mV) at this N/P ratio. At N/P ratio of 8, ECO and iECO formed nanoparticles with both pCas9 and psgRNA with zeta potentials higher than +30 mV. Other nanoparticles formed by iEKCO, iECKO, iEHCO, and iECHO had zeta potentials in the range of 0–15 mV except for iECHO/sgRNA nanoparticles. At N/P ratio of 10, ECO, iECO, iECKO, and iEHCO were able to form nanoparticles with both plasmids demonstrating zeta potentials over +30 mV. However, iEKCO still formed nanoparticles with slightly positive zeta potentials (<+10 mV) for both plasmids. iECHO/psgRNA nanoparticles had a zeta potential of 39.7 ± 1.7 mV, while iECHO/pCas9 nanoparticles had a zeta potential of 14.7 ± 0.8 mV.
Figure 4.
DLS zeta potential measurements of the nanoparticles formed by the multifunctional pH-sensitive lipids with psgRNA and pCas9 at N/P ratios of 6, 8, and 10.
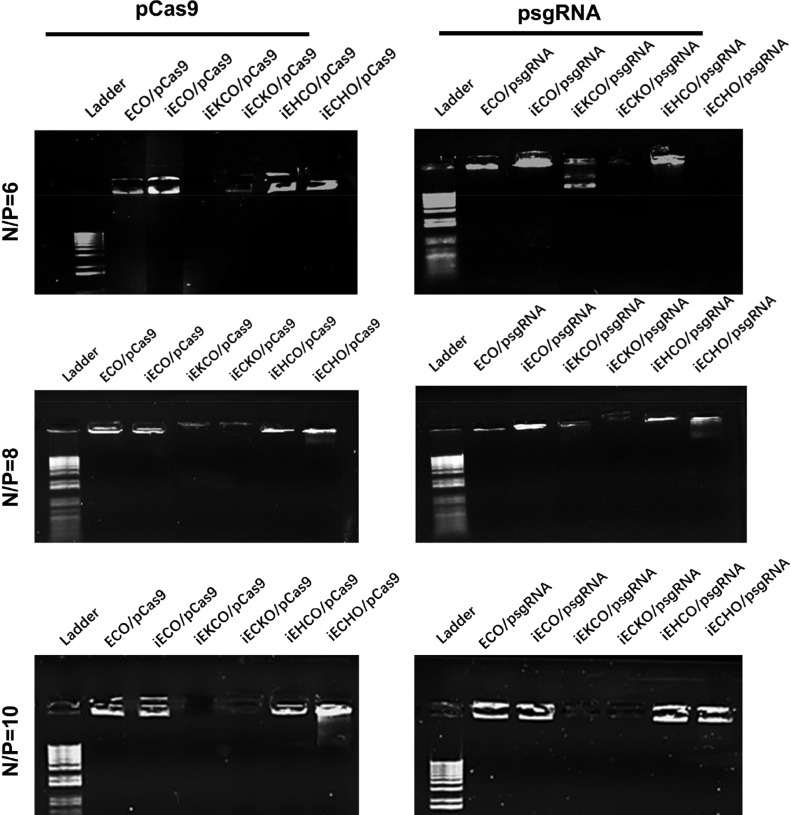
The encapsulation and stability of the nanoparticles were further evaluated with agarose gel electrophoresis (Figure 5). At N/P ratio of 6, good encapsulation and stability were observed for ECO, iECO, and iEHCO with both DNA plasmids. For iEKCO, iECKO and iECHO, low encapsulation and stability were shown by faint and unclear bands. At N/P ratio of 8 and 10, ECO, iECO, and iEHCO, and iECHO were able to encapsulate plasmids of CRISPR/Cas9 system and the nanoparticles showed good stability, indicated by the bright bands at the top. iECKO and iEKCO did not show efficient encapsulation at all tested N/P ratios, where faint bands were also observed.
Figure 5.
Agarose gel electrophoresis showing the encapsulation and stability of the nanoparticles formed by the multifunctional pH-sensitive lipids with psgRNA and pCas9 at N/P ratios of 6, 8, and 10.
pH-Dependent Hemolytic Activities
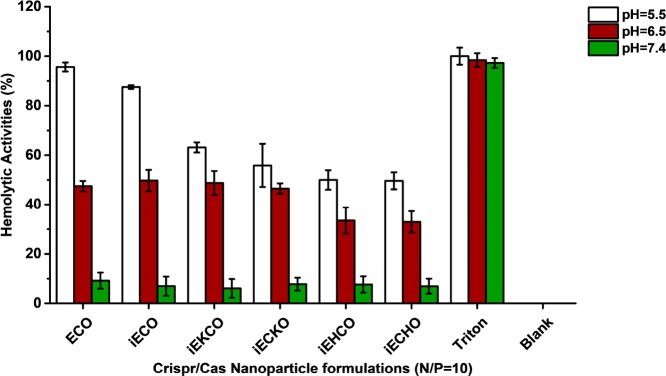
Membrane disruptive capabilities of the nanoparticles formulated by amino lipids and plasmid DNA were evaluated using hemolysis assay. Figure 6 shows that nanoparticles of all new carriers exhibited pH-sensitive hemolytic activity at N/P ratio of 10. At pH = 7.4, all of the amino lipid carriers have the same minimal hemolytic activities that were less than 10%. At pH = 6.5, increase in hemolytic activity was observed for all the carriers, and even more so at pH = 5.5, the late endosomal pH. However, ECO and iECO exhibited a greater increase in hemolytic activity than the other carriers at this pH.
Figure 6.
pH-dependent hemolytic activities of all carriers at N/P ratio of 10. Rat blood cells were diluted 1:50 in PBS and incubated with each formulation at pH = 7.4, 6.5, and 5.4 for 2 h at 37 °C. Triton X-100 (1% v/v) was implemented as a positive control. Blank was rat blood cells incubated with PBS solutions. Data were subtracted by the average of blank readouts and normalized to positive control.
Gene Editing Efficiency with CRISPR/Cas9 Mediated by ECO
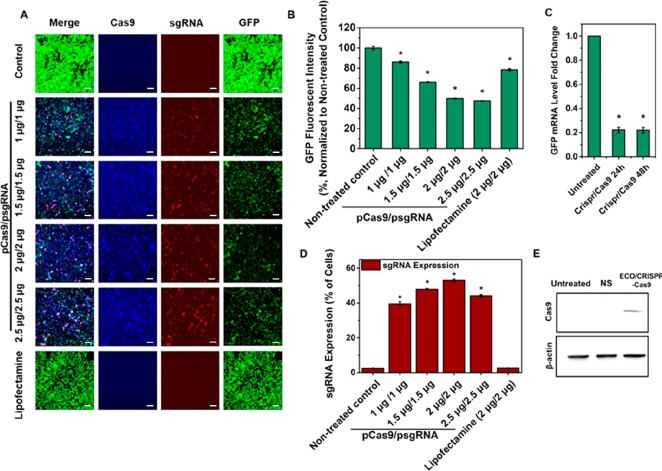
The gene editing efficiency of ECO was first evaluated with the GFP targeting CRISPR/Cas9 system, since ECO was well characterized in plasmid DNA delivery in our previous work.21,22 NIH3T3-GFP cells were transfected with ECO/psgRNA and ECO/pCas9 nanoparticles at a particle ratio of 1:1, with doses of 1, 1.5, 2.0, and 2.5 μg/well for each plasmid for 8 h. As the dose increased, higher expression levels of Cas9 (Blue) and sgRNA (Red) were observed under confocal microscope 72 h after transfection (Figure 7A). Consequently, substantial reduction of green fluorescence intensity was observed in the confocal images of the treated cells. The silencing efficiency of the ECO/CRISPR/Cas9 system was quantitatively determined using flow cytometry. The silencing efficiency increased with the dose of the system and plateaued at the dose over 2 μg-DNA/well. Approximately 50% reduction of GFP fluorescence intensity were observed with the doses of 2.0 and 2.5 μg/well (Figure 7B), while Lipofectamine 2000 resulted in a 28.6% reduction of GFP fluorescence intensity at 2 μg/well. The GFP knockdown effect was also confirmed by qRT-PCR tests (Figure 7C). At the dose of 2 μg/well, about 78% reduction in GFP mRNA expression was observed at 24 and 48 h after CRISPR/Cas9 treatments. The expression of sgRNA determined based on mCherry reporter gene increased from 39% to 53% with dose increase from 1 to 2.0 μg/well (Figure 7D), while much lower sgRNA expression was observed for Lipofectamine 2000. The expression of Cas9 was also verified with Western blot at 2 μg/well (Figure 7E). A clear Cas9 band was observed from the blot compared with nontreated control and nonspecific control at 72 h. The results suggested that ECO can effectively deliver both Cas9 and sgRNA and mediate efficient GFP knockdown with the CRISPR/Cas9 system.
Figure 7.
ECO mediated GFP silence with the CRISPR/Cas9 system. (A) In vitro transfection of ECO/psgRNA and ECO/pCas9 (particle ratio 1:1) in NIH3T3-GFP cells, with the dose of each plasmid at 1, 1.5, 2, and 2.5 μg/well in the transfection media. Cas9 expression (blue), gRNA expression (red), and GFP knockdown (green) were imaged by confocal microscopy. (B) Quantitative flow cytometry measurements of GFP knockdown efficiency represented the percentage of GFP fluorescent intensity normalized to nontreated control. (C) GFP knockdown efficiency in mRNA levels measured by qRT-PCR. Cells were treated with ECO/CRISPR/Cas9 nanoparticles at a dose of 2 μg/well and mRNA levels were evaluated at 24 and 48 h. (D) Quantitative flow cytometry measurements of sgRNA expression represented by percentage of cells with mCherry expression. (E) Western blot showing Cas9 protein expression 72 h after CRISPR/Cas9 treatments by ECO at a dose of 2 μg/well. (NS = nonspecific control, β-actin expression as inner control). (Error bars = ± std. * p < 0.05 relative to untreated control). Scale bars = 200 μm.
Gene Editing Efficiency Mediated by the New Amino Lipids
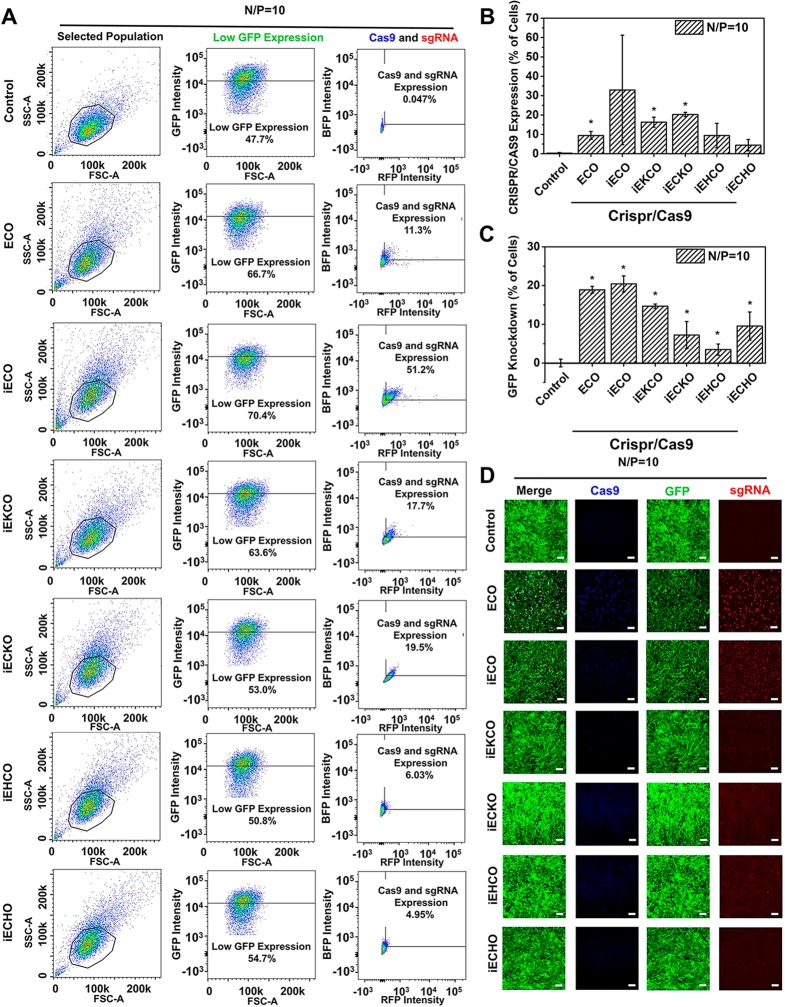
Gene editing efficiency mediated by the multifunctional amino lipids was evaluated in NIH3T3-GFP cells at N/P ratio of 10 and a dose of 1 μg/well for each plasmid in the transfection media. Flow cytometry was used to assess the cell population change after CRISPR/Cas9 treatment mediated by the amino lipids. Cell population was selected in SSC (side-scattered light) and FSC (forward-scattered light) plot. Results of one treatment from each lipid are represented in Figure 8A. After treatments, increases of cell population with reduced GFP expression were observed for all the amino lipids, which were summarized in Figure 8B. Averaged GFP knockdown levels ranging from 3.5% to 20% were observed for the amino lipids. Cell populations with expression of both Cas9 and sgRNA were also shown in the flow cytometry results and summarized in Figure 8C. iECO demonstrated up to 51% CRISPR/Cas9 expression and the highest average GFP knockdown efficiency of 20%. ECO demonstrated an average CRISPR/Cas9 expression level of 11.3% and a GFP knockdown efficiency of 19%. iEKCO and iECKO had averaged CRISPR/Cas9 expression levels of approximately 18% and 20%, with averaged GFP knockdown efficiency of 15% and 7.2%, respectively. iEHCO and iECHO demonstrated average CRISPR/Cas9 expression levels of 9.4% and 4.4%, with average GFP knockdown efficiency of 3.5% and 9.5%, respectively. These results were verified by confocal images shown in Figure 8D. Clear reduction in GFP fluorescence intensity was observed for ECO and iECO, with high Cas9 (Blue) and sgRNA (Red) expression. Reductions in GFP fluorescence and CRISPR/Cas9 expressions were also observed for other amino lipids, which were not as high as ECO and iECO. Interestingly, although iEKCO, iECKO, iEHCO, and iECHO demonstrated comparable average CRISPR/Cas9 expression level as to ECO, GFP knockdown efficiency was not as high as ECO. The higher editing efficiency of ECO was attributed to higher expression levels of both Cas9 and sgRNA in each cell of the CRISPR/Cas9 expressing cell population mediated by ECO than iEKCO, iECKO, iEHCO, and iECHO (Figure 8A).
Figure 8.
In vitro CRISPR/Cas9 transfection and gene editing using the multifunctional amino lipids in NIH3T3-GFP cells. (A) Flow cytometry of NIH3T3 cells from one treatment group transfected with CRISPR/Cas9 (SSC for Side-scattered light, FSC for Forward-scattered light, B525 for GFP, UV440 for BFP and YG610 for RFP). Cell population was gated in SSC and FSC plot and was used for the following fluorescence analysis. Low GFP expression was gated in the control group by selecting 47.7% cells with low GFP fluorescence intensity. The low GFP expression cell population was then analyzed RFP (sgRNA) and BFP (Cas9) expression. (B) Quantitative flow cytometry measurements of Cas9 and sgRNA expression in low GFP expression cell population. (C) Quantitative flow cytometry measurements of GFP knockdown represented by increased percentage of low GFP expression cell population. (D) Confocal images of GFP knockdown in NIH3T3 cells after CRISPR/Cas9 treatments. For each carrier, the lipid/psgRNA and lipid/pCas9 nanoparticle ratio was 1:1, with the dose of each plasmid at 1 μg, in the transfection media. Cas9 expression (blue), gRNA expression (red), and GFP knockdown (green) were imaged 72 h after transfection. (Error bars = ± std. * p < 0.05 relative to untreated control). Scale bars = 200 μm.
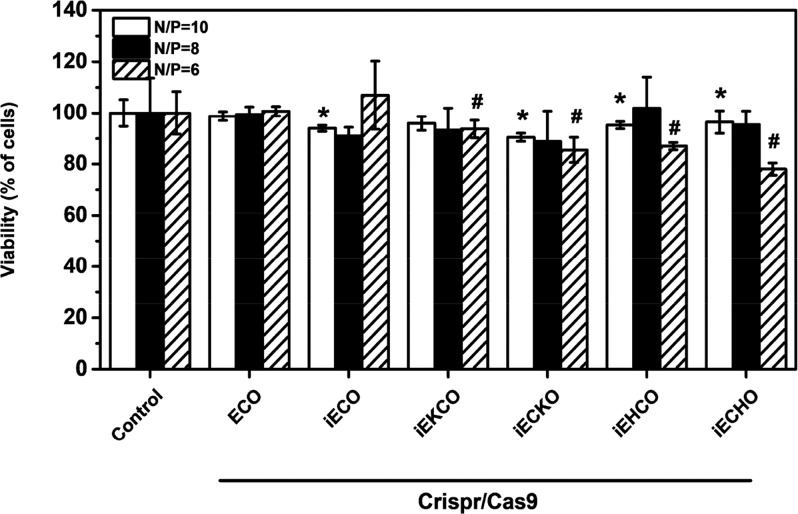
The cell viability after in vitro transfection of the nanoparticles of CRISPR/Cas9 system with the amino lipids was evaluated using MTT assay, results are shown in Figure 9. The nanoparticles demonstrated excellent cell viability, which was more than 75% across all N/P ratios. Especially for ECO and iECO plasmid DNA nanoparticles, over 90% viability was observed for all tested N/P ratios. It appears that the lipids that the nanoparticles with poor stability and DNA encapsulation, especially at the N/P = 6, exhibited a moderate cytotoxicity.
Figure 9.
Cell viability of in vitro transfection of CRISPR/Cas9 system using amino lipids in NIH3T3-GFP cells by an MTT assay of cytotoxicity 72 h after transfection. Nanoparticles had plasmid concentrations of 1 μg/well for each plasmid. For each carrier, the lipid/psgRNA and lipid/pCas9 nanoparticle ratio was 1:1. (Error bars = ± std. *, # p < 0.05 relative to untreated control).
Discussion
In this work, we synthesized and tested several new multifunctional pH-sensitive amino lipids for CRISRPR/Cas9 delivery. Multifunctional pH-sensitive amino lipids were designed to form stable nanoparticles with nucleic acids of different sizes and to facilitate efficient cytosolic delivery using the PERC mechanism. Cytosolic release of the gene cargo is facilitated by pH-sensitive amphiphilic endosomal membrane destabilization in the acidic endosomal compartment (pH = 5–6) and dissociation of the nanoparticles by reduction of the disulfide bonds from cysteine functional linkers in the cytoplasm.23 In this study, the headgroup for new amino lipids was simplified to 2,2′,2″-triaminotriethylamine, which has three aminoethyl branches, one as protonatable ethylene diamine headgroup and two for conjugation of lipid groups. Amino acids with side chains containing amino groups (lysine and histidine) were also introduced in the structure. When iECO with new headgroup was compared to ECO, no significant difference was observed between the amino lipid head groups in nanoparticle formation with the plasmids. Nanoparticles formulated by ECO and iECO with CRISPR/Cas9 plasmids demonstrated similar sizes and zeta potentials at N/P ratios of 8 and 10. The DNA nanoparticles of both ECO and iECO showed similar pH-sensitive hemolysis at N/P = 10. The results indicate the change of the headgroup does not affect the formation of nanoparticles with plasmid DNA and the new amino lipids.
Histidinyl and lysyl residues were also introduced in the new amino lipids to evaluate the effects of additional amino groups of different pKas on nanoparticle formation with plasmid DNA and their gene editing efficiency. The extra protonatable amino groups after introduction of these amino acids residues could enhance the formulation of stable nanoparticles with nucleic acids and their endosomal escape with a reduced amount of amino lipids. Imidazole group in histidine is known to act as endogenous buffers, which are commonly used as modifying groups to improve the endosomal escape of several nonviral gene carriers.24−26 With a pKa around 6, the imidazole group is a weak base that can acquire a cationic charge when the pH of the environment drops below 6 in the endosomal compartments after cellular uptake. Protonation of the imidazole groups could induce fusogenic activity of gene carriers.27 Lysine has a primary amine of a pKa value of 10.7, which remains protonated at physiological and endosomal pH. Introduction of lysinyl residues could potentially increase the electrostatic interactions with nucleic acids to form stable nanoparticles with reduced amount of the lipids.28−31 The amino lipids with histidinyl or lysyl residues formed stable nanoparticles at a high N/P ratio of 10 and were not able to consistently to form stable nanoparticles at the N/P ratios of 6 and 8, as compared to ECO and iECO. Their pH-sensitive hemolytic activities, an indicator for pH-sensitive cell membrane disruption and endosome escape, were not as efficient as ECO and iECO. The relatively low effectiveness of the lysine and histidine modified amino lipids as compared to ECO and iECO at the same N/P ratio could be attributed to the low concentration of the modified lipids, 50% of that of ECO and iECO, used in the nanoparticle formation at the same N/P ratio. The observation suggests that the concentration of the amino lipids is also critical to form stable nanoparticles with plasmid DNA.
Efficient gene editing requires the delivery and expression of both Cas9 and sgRNA in the same cells. The previously reported ECO mediated efficient delivery and expression of both Cas9 and sgRNA and significant gene editing efficiency. Dose dependent knockdown was also shown with ECO with the GFP-targeting CRISPR/Cas9 system. ECO mediated higher gene editing efficiency than Lipofectamine 2000, a commercially available lipid delivery system for nucleic acids. However, the efficacy of CRISPR/Cas9 system plateaued at a high concentration, possible due to a saturation effect. More CRISPR/Cas9 expression might not translate to more gene editing.
The tested amino lipids demonstrated structurally dependent nanoparticle formation with plasmid DNA expressing CRISPR/Cas9, pH-sensitive hemolysis, gene expression, and gene editing efficiency. ECO and iECO exhibited higher efficiency to form stable nanoparticles of relatively high zeta potential with the DNA plasmids and more effective pH-sensitive hemolysis, and resulted in higher gene transfection and gene editing than other lipids. Although iECO induced more CRISPR/Cas9 expression in cells than ECO, both ECO and iECO mediated similar gene editing efficiency at N/P = 10. The amino lipids containing histidinyl or lysinyl residues did not show improved intracellular expression of the CRISPR/Cas9 system and GFP knockdown efficiency as compared ECO and iECO at the same N/P ratio, possibly due to low overall lipid concentration in the nanoparticle formation. Further detailed studies are needed to explore the optimal conditions, e.g., the concentration of the amino lipids in correlation with N/P ratio, for highly efficient transfection of CRISPR/Cas9 system and gene editing with minimal toxic side effects from the delivery system. Nevertheless, the multifunctional pH-sensitive amino lipids, especially ECO and iECO, are promising carriers for CRISPR/Cas9 system mediated gene editing. The lipids have the potential to be used in gene therapy with CRISPR/Cas9 system, especially for treating autosomal dominant diseases.
Conclusion
The multifunctional pH-sensitive amino lipids were modified and evaluated for intracellular delivery of CRISPR/Cas9 system and gene editing. The modified amino lipids exhibited structurally dependent nanoparticle formation, pH-sensitive hemolysis, and gene editing with the DNA plasmids expressing CRISPR/Cas9 system. ECO and iECO formed stable nanoparticles with the DNA plasmids, and exhibited effective pH-sensitive hemolysis, high expression of the CRISPR/Cas9 system, and high gene editing efficiency. A saturation effect for gene editing with the CRISPR/Cas9 system was observed for ECO when the dose of DNA plasmids increased. The nanoparticles of the amino lipids and CRISPR/Cas9 plasmids exhibited low cytotoxicity. The multifunctional pH-sensitive amino lipids have the promise for intracellular delivery of CRISPR/Cas9 to mediate effective gene editing for gene therapy.
Materials and Methods
All reagents purchased from the vendors were used without further purification unless otherwise stated. Anhydrous N,N-diisopropylethylamine (DIPEA) was purchased from Alfa Aesar (Ward Hill, MA). Trifluoroacetic acid (TFA) was purchased from Oakwood Products, Inc. (West Columbia, SC). All the amino acids used in the chemical synthesis were purchased from Novabiochem (Darmstadt, Germany). Ethylenediamine (EDA), piperidine, 1,2-ethanedithiol (EDT), triisobutylsilane (TIBS), oleoyl chloride, and N-(3-(dimethylamino)propyl)-N′-ethylcarbodiimide hydrochloride (EDC) were purchased from Sigma-Aldrich Corporation (St. Louis, MO). Hydroxybenzotriazole (HOBt) was purchased from Peptides International Inc. (Louisville, KY). Fmoc-His(Trt)-OH and Fmoc-Cys (Trt)-OH were purchased from EMD Biosciences (San Diego, CA). Sodium carbonate, potassium carbonate, methylene chloride (DCM), N,N-dimethylformamide (DMF), chloroform, methanol, and tetrahydrofuran (THF) were purchased from Fisher Scientific (Hampton, NH). Reagents for cell culturing, including fetal bovine serum, streptomycin, and penicillin were from Invitrogen (Carlsbad, CA).
Synthesis of the Multifunctional Amino Lipids
Carrier ECO was prepared and purified according to our previous report.32 The synthetic route of ECO isotypic derivatives were shown in Figure 1B, and is described here with the synthesis of iEHCO as an example. tert-Butyl 2-(bis(2-aminoethyl)amino)ethylcarbamate (BocTren) was prepared with a good yield (72%) as reported previously.33 Then, BocTren reacted with Fmoc-His(Trt)-OH to provide ipEH (2,2′,2″-triaminotriethylamine conjugated with two protected histidinyl residues) in the presence of EDC·HCl, HOBt, and DIPEA.34−36 To a DMF (40 mL) solution of BocTren (246 mg, 1.0 mmol) and Fmoc-His(Trt)-OH (1859 mg, 3.0 mmol), 1-hydroxybenzotriazol hydrate (HOBt·H2O, 306.2 mg, 2.0 mmol), 1-(3-(dimethylamino)propyl)-3-ethylcarbodiimide hydrochloride (EDC·HCl, 383.4 mg, 2.0 mmol), and diisopropylethylamine (DIPEA, 348 μL, 2.0 mmol) were successively added. The mixture was stirred for 9 h at room temperature under N2 atmosphere, and the solvent was evaporated to dryness. The residue was dissolved in methylene chloride and washed twice with 5% aqueous sodium carbonate. The organic extract was dried over anhydrous Na2SO4 and filtered off. Solvent was evaporated to dryness, and the residue was purified by recrystallization from methylene chloride/diethyl ether twice. The raw product ipEH was directly used to next step reaction.
A solution of ipEH (363 mg, 0.25 mmol) in DMF (10 mL) containing 20% piperidine was stirred under N2 atmosphere at room temperature for 2 h to remove Fmoc protection and the reaction was monitored by mass spectrometry. Once ipEH was consumed, the solution was evaporated to dryness, washed with 5% aqueous sodium carbonate, and extracted with methylene chloride. The organic extract was dried over anhydrous Na2SO4, filtered off, and evaporated to dryness. The residue was purified on silica gel using a gradient eluent (chloroform to chloroform/methanol (v/v, 30/1)). The product (ipEH-NH2) was obtained in a yield of 78%. 1H NMR (500 MHz, CDCl3, δ ppm): 7.93, 7.33, 7.12, 6.66, 6.00, 4.67, 3.64, 3.24, 3.12, 3.07, 2.71, 2.56, 1.38. 13C NMR (126 MHz, CDCl3, δ ppm): 142.42, 138.48, 129.74, 128.05, 119.43, 55.82, 53.68, 36.85, 28.46. Electrospray ionization mass spectrometry (Thermo Finnigan LCQ Deca XP LCMSMS System) (Thermo Fisher Scientific, Waltham, MA): m/z = 1027 (M+Na+, m/z), 1005 (M+H+) measured; 1005.28 calculated.
Intermediate ipEH-NH2 (2,2′,2″-triaminotriethylamine conjugated with two histidinyl residues) reacted with Fmoc-Cys(Trt)-OH to provide ipEHC in the presence of EDC·HCl, HOBt and DIPEA. To a DMF (40 mL) solution of ipEH-NH2 (503 mg, 0.5 mmol) and Fmoc-Cys(Trt)-OH (879 mg, 1.5 mmol), 1-hydroxybenzotriazol hydrate (HOBt·H2O, 153 mg, 1.0 mmol), 1-(3-(dimethylamino)propyl)-3-ethylcarbodiimide hydrochloride (EDC·HCl, 192 mg, 1.0 mmol), and diisopropylethylamine (DIPEA, 192 μL, 1.0 mmol) were successively added. The mixture was stirred for 9 h at room temperature under N2 atmosphere, and the solvent was evaporated to dryness. The residue was dissolved in methylene chloride and washed twice with 5% aqueous sodium carbonate. The organic extract was dried over anhydrous Na2SO4 and filtered off. Solvent was evaporated to dryness, and the residue was purified by recrystallization from methylene chloride/diethyl ether twice. The raw product ipEHC was directly used to next step reaction.
A solution of ipEHC (535 mg, 0.25 mmol) in DMF (10 mL) containing 20% piperidine was stirred under N2 atmosphere at room temperature for 2 h to remove the Fmoc protection and the reaction was monitored by MS. Once no ipEHC was comsumed, the solution was evaporated to dryness, washed with 5% aqueous sodium carbonate, and extracted with methylene chloride. The organic extract was dried over anhydrous Na2SO4, filtered off, and evaporated to dryness. The product ipEHC-NH2 was purified from recrystallization of methylene chloride/hexane twice and direct used into next step reaction.
To a THF (20 mL) solution of ipEHC-NH2 (ipEH conjugated with two cysteinyl residues) (170 mg, 0.1 mmol), 4-(dimethylamino)pyridine (12.2 mg, 0.1 mmol), sodium sulfate (340 mg), and potassium carbonate (340 mg) was successively added. After cooling to 0 °C, a THF (10 mL) solution of oleoyl chloride (90.3 mg, 0.3 mmol) was added dropwise. The reaction was monitored by MS and once no iEHC remained, the salts were filtered off and evaporated to dryness. The residue was washed with water and centrifuged to separate the supernatant and precipitate. The precipitate was lyophilized to provide ipEHCO.
To a methylene chloride (3 mL) solution of ipEHCO (100 mg, 0.045 mmol), a freshly prepared cocktail solution of trifluoroacetic acid/water/1,2-ethanedithiol/triisobutylsilane (1.5 mL/75 μL/75 μL/150 μL) was added to remove the protecting groups in ice/water bath. The solution was stirred for 1 h monitored by MS. Once no ipEHCO remained, the reaction solution was evaporated to dryness and purified via Biotage flash chromatography (Biotage, Charlotte, NC) (eluents: water and methanol) to provide pure EHCO in a yield of 20%. 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.93 (s, 2H), 8.25 (m, 4H), 8.17 (b, 2H), 8.07 (b, 2H), 7.31 (m, 2H), 5.26 (m, 4H), 4.54 (m, 4H), 3.13 (m, 8H), 2.93 (m, 4H), 2.80 (m, 8H), 2.11 (m, 4H), 1.91 (m, 10H), 1.41 (b, 6H), 1.17 (b, 38H), 0.79 (t, 6H). MALDI-TOF MS (M + H+, m/z): 1155.84, measured; 1155.79, calculated.
Other amino lipid carriers were synthesized similarly with characteristic structural information listed as the following. Intermediate ipEC-NH2 for iECO, iECHO, and iECKO: yield: 72%; 1H NMR (500 MHz, CDCl3, δ ppm): 7.42 (b, 2H), 7.34 (m, 12H), 7.19 (m, 12H), 7.12 (m, 12H), 5.46 (t, 1H), 3.08 (m, 8H), 2.61 (m, 2H), 2.41 (m, 8H), 1.33 (s, 9H); 13C NMR (126 MHz, CDCl3, δ ppm): 175.77, 172.67, 155.94, 144.60, 129.60, 128.00, 126.82, 66.95, 54.06, 53.56, 53.14, 38.66, 37.37, 36.96, 28.53; ESI-MS (m/z, M+H+): 936.44, calculated; 937.24, measured.
iECO: yield: 40%: 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.22 (m, 4H), 5.33 (m, 4H), 3.20 (m, 4H), 2.83 (m, 4H), 2.18 (b, 6H), 1.98 (b, 8H), 1.49 (b, 8H), 1.25 (b, 46H), 0.87 (t, J = 5.5 Hz, 6H); MALDI-TOF MS (m/z, M+H+): 881.67, calculated; 881.82, measured.
iECHO: yield: 32%; 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.94 (s, 2H), 8.80 (b, 2H), 8.18 (b, 4H), 7.73 (b, 2H), 7.31 (b, 2H), 5.32 (m, 4H), 4.56 (m, 4H), 3.18 (m, 10H), 2.74 (m, 10h), 2.07 (M, 12H), 1.41 (m, 8H), 1.23 (b, 38H), 0.85 (t, J = 6.0 Hz, 6H); MALDI-TOF MS (m/z, M + H)+: 1155.79, calculated; 1155.60, measured.
Intermediate ipEK-NH2: yield: 81%; 1H NMR (500 MHz, CDCl3, δ ppm): 7.73 (b 2H), 5.54 (b, 1H), 4.75 (b, 2H), 3.39 (b, 2H), 3.31 (m, 4H), 3.12 (m, 6H), 2.61 (m, 6), 1.84 (m, 2H), 1.76 (b, 4H), 1.53 (m, 6H), 1.45 (s, 27H); 13C NMR (126 MHz, CDCl3, δ, ppm): 175.20, 156.08, 55.31, 54.01, 53.56, 40.21, 38.72, 37.16, 34.74, 29.94, 28.38, 23.02; MALDI-TOF MS (m/z, M + Na+): 725.49, calculated; 725.56, measured. (m/z, M+H+): 703.51 calculated; 703.54, measured.
iEKCO: yield: 35%. 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.08 (m, 4H), 7.79 (m, 2H), 5.34 (m, 4H), 4.31 (m, 4H), 3.13 (m, 10H), 2.77 (m, 6H), 2.14 (m, 6H), 1.99 (m, 6H), 1.25 (m, 62H), 0.87 (t, J = 5.5 Hz, 6H); MALDI-TOF MS (m/z, M+H+): 1137.86, calculated; 1138.13, measured.
iECKO: yield: 39%: 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.83 (m, 4H), 7.67 (m, 2H), 5.37 (m, 4H), 4.23 (m, 4H), 3.20 (m, 8H), 2.76 (m, 6H), 2.11 (m, 6H), 1.95 (m, 6H), 1.51 (m, 18H), 1.20 (b, 46H), 0.86 (t, J = 7.0 Hz, 6H); MALDI-TOF MS (m/z, M+H+): 1137.86, calculated; 1137.83, measured.
Cell Culture
NIH3T3 cell line with stable GFP expression was prepared as previously described.37 The NIH3T3-GFP cells were cultured in DMEM and supplemented with 10% fetal bovine serum, 100 μg/mL streptomycin, and 100 units/mL penicillin (all reagents were from Invitrogen). Cells were maintained in a humidified incubator at 37 °C and 5% CO2.
Nanoparticle Formulation and Characterization
CRISPR/Cas9 plasmids were purchased from the Addgene repository. (Addgene plasmids #78535 and 78547).38 Two sgRNAs were selected to target eGFP sequence (sgRNA1: GAGCTGGACGGCGACGTAAACGG; sgRNA2: CAGAACACCCCCATCGGCGACGG). The amino lipid carriers were dissolved in ethanol at a stock concentration of 2.5 mM, while the plasmids of CRISPR/Cas9 system were reconstituted in nuclease-free water at 0.5 μg/μL. Nanoparticles are formulated by mixing the amino lipids with plasmid DNA for 30 min in nuclease-free water at prespecified N/P ratios. The size and zeta potential of the nanoparticles were analyzed using an Anton Paar Litesizer 500 instrument (Anton Paar USA Inc., Ashhland, VA) in nuclease free water.
The encapsulation of CRISPR/cas9 plasmids in the nanoparticles was assessed by gel electrophoresis. Lipid/plasmid DNA nanoparticles (4 μL) and 4 μL of loading dye (Promega, Madison, WI) and 16 μL nuclease free water were mixed. The mixture (20 μL) was loaded onto a 0.7% agarose gel containing ethidium bromide. The gel was submerged in 0.5× Tris/Borate/EDTA (TBE) buffer and run at 100 V for 25 min. Plasmid DNA bands were visualized using GelDoc XRS (Bio-Rad, Hercules, CA).
Evaluation of pH-Sensitive Membrane Disruption
Red blood cells (RBCs) (Innovative Research Inc., Novi, MI) were diluted 1:50 in PBS solutions of pH = 5.4, 6.5, or 7.4. The solution of nanoparticles (100 μL, pH = 5.4, 6.5, or 7.4) at N/P ratio of 10 was incubated with 100 μL of diluted RBCs at 37 °C for 2 h and tested in triplicates. Nanoparticles were formulated so that the final amine concentration for all the samples was 150 μM after mixing with the RBCs. The absorbance of the supernatant of each sample was measured on a SpectraMax spectrophotometer (Molecular Devices, San Jose, CA) at a wavelength of 540 nm to determine the amount of hemoglobin released from the RBCs, due to membrane destabilization. PBS and 1% (v/v) Triton-X100 were used as negative and positive controls, respectively. The background absorption of the RBCs treated with PBS treatment was subtracted from the solutions treated with the samples. The percentage of hemolysis of each sample was calculated from the average of the released hemoglobin relative to the hemoglobin release from the positive control with 1% (v/v) Triton-X100 surfactant during the 2-h incubation period.
In Vitro Transfection
NIH3T3-GFP cells were seeded either onto 12-well plates at a density of 5 × 104 cells per well or confocal dish at a density of 1 × 105 cells and allowed to grow for 24 h at 37 °C. Transfections were conducted in 10% serum media with the ECO/pCas9 and ECO/psgRNA nanoparticles at plasmid concentrations of 1, 1.5, 2, and 2.5 μg/well for each plasmid. The nanoparticles were incubated with NIH3T3 cells for 8 h at 37 °C. Then, the transfection media was replaced with fresh serum-containing media (10% serum) and cells were allowed to grow until 24, 48, or 72 h. Lipofectamine 2000 particles were formulated with pCas9 and psgRNA at a concentration of 2 μg/well for each plasmid according to the protocol from the vendor. The expression of Cas9 (BFP), sgRNA (mCherry), and GFP knockdown efficiency were evaluated with an Olympus FV1000 confocal microscope (Olympus, Center Valley, PA). The quantitative analysis was performed with flow cytometry. The cells were harvested by treatment with 0.25% trypsin containing 0.26 mmol EDTA (Invitrogen, Carlsbad, CA), collected by centrifugation at 1500 rpm for 5 min, resuspended in 500 μL of PBS containing 4% paraformaldehyde, and finally passed through a 35 μm cell strainer (BD Biosciences, Franklin Lakes, NJ). Cas9 expression, gRNA expression, and GFP knockdown were quantified by the fluorescence intensity measurement for a total of 10,000 cells per sample using a BD FACSCalibur flow cytometer (BD Biosciences, Franklin Lakes, NJ). The other lipids were similarly tested the plasmid concentration of 1 μg/well for each plasmid. Each sample was conducted in triplicate.
Cytotoxicity
Cytotoxicity of the nanoparticles of the amino lipids was investigated using MTT colorimetric assay (Invitrogen, Carlsbad, CA). NIH3T3-GFP cells were seeded onto 12-well plates at a density of 5 × 104 cells per well. For each carrier, the lipid/psgRNA and lipid/pCas9 nanoparticle ratio was 1:1 and nanoparticles had concentrations of 1 μg/well for each plasmid, which were incubated with NIH3T3 cells for 8 h at 37 °C. Then, the transfection media was replaced with fresh serum-containing media (10% serum) and cells were allowed to grow until 72 h. NIH3T3-GFP cells were incubated with 100 μL MTT reagent for 4 h, followed by an additional 4 h incubation with 1 mL SDS-HCl solution to dissolve formazan crystals formed by the reduction of MTT by NAD(P)H-dependent enzymes in the cells. The absorbance of each sample was measured at 570 nm using a SpectraMax microplate reader (Molecular Devices, San Jose, CA). Cellular viability was calculated by averaging the signal intensities over three replicates and then normalizing the results relative to the negative control data.
qRT-PCR
NIH3T3-GFP cells were treated with the lipid CRISPR/Cas9 nanoparticles for 8 h and the transfection media was replaced with fresh media containing 10% serum. Cells were allowed to grow until 24 or 48 h. Total RNA was extracted from cells using the RNeasy Plus Mini Kit (Qiagen, Germantown, MD), according to manufacturer’s instructions. Reverse transcription was performed using the miScript II RT Kit (Qiagen) and qPCR was performed using the SyBr Green PCR Master Mix (Applied Biosystems, Foster City, CA). Gene expression was analyzed by the 2–ΔΔCt method with 18S expression as the control. The following primer sequences were used: mCherry, Fwd 5′-GAACGGCCACGAGTTCGAGA-3′ and Rev 5′-CTTGGAGCCGTACATGAACTGAGG-3′; eGFP, Fwd 5′-ACGTAAACGGCCACAAGTTC-3′ and Rev 5′-AAGTCGTGCTGCTTCATGTG-3′; 18S (rRNA), Fwd 5′-TCAAGAACGAAAGTCGGAGG-3′ and Rev 5′-GGACATCTAAGGGCATCACA-3′.
Western Blot
Similarly, NIH3T3-GFP cells were treated with CRISPR/Cas9 nanoparticles for 8 h and the transfection media was replaced with fresh media containing 10% serum. Then, cells were allowed to grow for 72 h. Total cellular protein was extracted as a reported method.39 Protein extracts (40 μg) were separated by SDS-PAGE, transferred onto nitrocellulose membrane and immunoblotted with primary antibodies overnight. Anti-Cas9 and anti-β-actin (loading control) from Cell Signaling Technology (Danvers, MA) were used to stain the proteins. The membrane was visualized using an AlphaImager imaging system (Bio-Rad, Hercules, CA). Protein expression were determined by analyzing the bands on the membranes.
Acknowledgments
This project was supported by the NIH grant R01 CA194518. Z.R.L. is an M. Frank Rudy and Margaret Domiter Rudy Professor of Biomedical Engineering.
Glossary
Abbreviations
- CRISPR
clustered regulatory interspaced short palindromic repeats
- Cas9
CRISPR associated protein 9
- sgRNA
single guide RNA
- PAM
protospacer adjacent motif sequence
- PERC
pH-sensitive amphiphilic endosomal escape and reductive cytosolic release
- SSC
Side-scattered light
- FSC
Forward-scattered light
Author Contributions
† Da Sun and Zhanhu Sun contributed equally to this work.
The authors declare no competing financial interest.
References
- Millington-Ward S.; Chadderton N.; O’Reilly M.; Palfi A.; Goldmann T.; Kilty C.; Humphries M.; Wolfrum U.; Bennett J.; Humphries P.; et al. (2011) Suppression and Replacement Gene Therapy for Autosomal Dominant Disease in a Murine Model of Dominant Retinitis Pigmentosa. Mol. Ther. 19, 642–649. 10.1038/mt.2010.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin H.; Song C.-Q.; Dorkin J. R.; Zhu L. J.; Li Y.; Wu Q.; Park A.; Yang J.; Suresh S.; Bizhanova A.; et al. (2016) Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat. Biotechnol. 34, 328. 10.1038/nbt.3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakondi B.; Lv W.; Lu B.; Jones M. K.; Tsai Y.; Kim K. J.; Levy R.; Akhtar A. A.; Breunig J. J.; Svendsen C. N.; et al. (2016) In Vivo CRISPR/Cas9 Gene Editing Corrects Retinal Dystrophy in the S334ter-3 Rat Model of Autosomal Dominant Retinitis Pigmentosa. Mol. Ther. 24, 556–563. 10.1038/mt.2015.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandal P. K.; Ferreira L. M. R.; Collins R.; Meissner T. B.; Boutwell C. L.; Friesen M.; Vrbanac V.; Garrison Brian S.; Stortchevoi A.; Bryder D.; et al. (2014) Efficient Ablation of Genes in Human Hematopoietic Stem and Effector Cells using CRISPR/Cas9. Cell Stem Cell 15, 643–652. 10.1016/j.stem.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gori J. L.; Hsu P. D.; Maeder M. L.; Shen S.; Welstead G. G.; Bumcrot D. (2015) Delivery and Specificity of CRISPR/Cas9 Genome Editing Technologies for Human Gene Therapy. Hum. Gene Ther. 26, 443–451. 10.1089/hum.2015.074. [DOI] [PubMed] [Google Scholar]
- Mali P.; Yang L.; Esvelt K. M.; Aach J.; Guell M.; DiCarlo J. E.; Norville J. E.; Church G. M. (2013) RNA-Guided Human Genome Engineering via Cas9. Science 339, 823. 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L.; Ran F. A.; Cox D.; Lin S.; Barretto R.; Habib N.; Hsu P. D.; Wu X.; Jiang W.; Marraffini L. (2013) Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 339, 819. 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu P. D.; Lander E. S.; Zhang F. (2014) Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 157, 1262–1278. 10.1016/j.cell.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christie K. A.; Courtney D. G.; DeDionisio L. A.; Shern C. C.; De Majumdar S.; Mairs L. C.; Nesbit M. A.; Moore C. B. T. (2017) Towards personalised allele-specific CRISPR gene editing to treat autosomal dominant disorders. Sci. Rep. 7, 16174. 10.1038/s41598-017-16279-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X.; Tao Y.; Lamas V.; Huang M.; Yeh W.-H.; Pan B.; Hu Y.-J.; Hu J. H.; Thompson D. B.; Shu Y.; et al. (2017) Treatment of autosomal dominant hearing loss by in vivo delivery of genome editing agents. Nature 553, 217. 10.1038/nature25164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W.; Mookherjee S.; Chaitankar V.; Hiriyanna S.; Kim J.-W.; Brooks M.; Ataeijannati Y.; Sun X.; Dong L.; Li T.; et al. (2017) Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal degeneration in mice. Nat. Commun. 8, 14716. 10.1038/ncomms14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt R. J.; Chen S.; Zhou Y.; Yim M. J.; Swiech L.; Kempton H. R.; Dahlman J. E.; Parnas O.; Eisenhaure T. M.; Jovanovic M.; et al. (2014) CRISPR-Cas9 Knockin Mice for Genome Editing and Cancer Modeling. Cell 159, 440–455. 10.1016/j.cell.2014.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang C.; Mei M.; Li B.; Zhu X.; Zu W.; Tian Y.; Wang Q.; Guo Y.; Dong Y.; Tan X. (2017) A non-viral CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA and pcsk9 in vivo. Cell Res. 27, 440. 10.1038/cr.2017.16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senís E.; Fatouros C.; Große S.; Wiedtke E.; Niopek D.; Mueller A.-K.; Börner K.; Grimm D. (2014) CRISPR/Cas9-mediated genome engineering: An adeno-associated viral (AAV) vector toolbox. Biotechnol. J. 9, 1402–1412. 10.1002/biot.201400046. [DOI] [PubMed] [Google Scholar]
- Thomas C. E.; Ehrhardt A.; Kay M. A. (2003) Progress and problems with the use of viral vectors for gene therapy. Nat. Rev. Genet. 4, 346. 10.1038/nrg1066. [DOI] [PubMed] [Google Scholar]
- Mahato R. I.; Rolland A.; Tomlinson E. (1997) Cationic Lipid-Based Gene Delivery Systems: Pharmaceutical Perspectives. Pharm. Res. 14, 853–859. 10.1023/A:1012187414126. [DOI] [PubMed] [Google Scholar]
- Li L.; He Z.-Y.; Wei X.-W.; Gao G.-P.; Wei Y.-Q. (2015) Challenges in CRISPR/CAS9 Delivery: Potential Roles of Nonviral Vectors. Hum. Gene Ther. 26, 452–462. 10.1089/hum.2015.069. [DOI] [PubMed] [Google Scholar]
- Yin H.; Kanasty R. L.; Eltoukhy A. A.; Vegas A. J.; Dorkin J. R.; Anderson D. G. (2014) Non-viral vectors for gene-based therapy. Nat. Rev. Genet. 15, 541. 10.1038/nrg3763. [DOI] [PubMed] [Google Scholar]
- Sun D.; Schur R. M.; Lu Z.-R. (2017) A novel nonviral gene delivery system for treating Leber’s congenital amaurosis. Ther. Delivery 8, 823–826. 10.4155/tde-2017-0072. [DOI] [PubMed] [Google Scholar]
- Malamas A. S.; Gujrati M.; Kummitha C. M.; Xu R.; Lu Z.-R. (2013) Design and evaluation of new pH-sensitive amphiphilic cationic lipids for siRNA delivery. J. Controlled Release 171, 296–307. 10.1016/j.jconrel.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D.; Maeno H.; Gujrati M.; Schur R.; Maeda A.; Maeda T.; Palczewski K.; Lu Z.-R. (2015) Self-Assembly of a Multifunctional Lipid With Core–Shell Dendrimer DNA Nanoparticles Enhanced Efficient Gene Delivery at Low Charge Ratios into RPE Cells. Macromol. Biosci. 15, 1663–1672. 10.1002/mabi.201500192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun D.; Sahu B.; Gao S.; Schur R. M.; Vaidya A. M.; Maeda A.; Palczewski K.; Lu Z.-R. (2017) Targeted Multifunctional Lipid ECO Plasmid DNA Nanoparticles as Efficient Non-viral Gene Therapy for Leber’s Congenital Amaurosis. Mol. Ther.--Nucleic Acids 7, 42–52. 10.1016/j.omtn.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujrati M.; Vaidya A.; Lu Z.-R. (2016) Multifunctional pH-Sensitive Amino Lipids for siRNA Delivery. Bioconjugate Chem. 27, 19–35. 10.1021/acs.bioconjchem.5b00538. [DOI] [PubMed] [Google Scholar]
- Midoux P.; Monsigny M. (1999) Efficient Gene Transfer by Histidylated Polylysine/pDNA Complexes. Bioconjugate Chem. 10, 406–411. 10.1021/bc9801070. [DOI] [PubMed] [Google Scholar]
- Shigeta K.; Kawakami S.; Higuchi Y.; Okuda T.; Yagi H.; Yamashita F.; Hashida M. (2007) Novel histidine-conjugated galactosylated cationic liposomes for efficient hepatocyte-selective gene transfer in human hepatoma HepG2 cells. J. Controlled Release 118, 262–270. 10.1016/j.jconrel.2006.12.019. [DOI] [PubMed] [Google Scholar]
- Yang S. R.; Lee H. J.; Kim J.-D. (2006) Histidine-conjugated poly(amino acid) derivatives for the novel endosomolytic delivery carrier of doxorubicin. J. Controlled Release 114, 60–68. 10.1016/j.jconrel.2006.05.016. [DOI] [PubMed] [Google Scholar]
- Midoux P.; Pichon C.; Yaouanc J.-J.; Jaffrès P.-A. (2009) Chemical vectors for gene delivery: a current review on polymers, peptides and lipids containing histidine or imidazole as nucleic acids carriers. Br. J. Pharmacol. 157, 166–178. 10.1111/j.1476-5381.2009.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. E.; Rice K. G. (2007) Peptide-guided gene delivery. AAPS J. 9, E18–E29. 10.1208/aapsj0901003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toncheva V.; Wolfert M. A.; Dash P. R.; Oupicky D.; Ulbrich K.; Seymour L. W.; Schacht E. H. (1998) Novel vectors for gene delivery formed by self-assembly of DNA with poly(l-lysine) grafted with hydrophilic polymers. Biochim. Biophys. Acta, Gen. Subj. 1380, 354–368. 10.1016/S0304-4165(98)00004-X. [DOI] [PubMed] [Google Scholar]
- Choi J. S.; Lee E. J.; Choi Y. H.; Jeong Y. J.; Park J. S. (1999) Poly(ethylene glycol)-block-poly(l-lysine) Dendrimer: Novel Linear Polymer/Dendrimer Block Copolymer Forming a Spherical Water-Soluble Polyionic Complex with DNA. Bioconjugate Chem. 10, 62–65. 10.1021/bc9800668. [DOI] [PubMed] [Google Scholar]
- Singh R.; Pantarotto D.; McCarthy D.; Chaloin O.; Hoebeke J.; Partidos C. D.; Briand J.-P.; Prato M.; Bianco A.; Kostarelos K. (2005) Binding and Condensation of Plasmid DNA onto Functionalized Carbon Nanotubes: Toward the Construction of Nanotube-Based Gene Delivery Vectors. J. Am. Chem. Soc. 127, 4388–4396. 10.1021/ja0441561. [DOI] [PubMed] [Google Scholar]
- Malamas A. S.; Gujrati M.; Kummitha C. M.; Xu R.; Lu Z. R. (2013) Design and evaluation of new pH-sensitive amphiphilic cationic lipids for siRNA delivery. J. Controlled Release 171, 296–307. 10.1016/j.jconrel.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon J. M.; Lambert T. N.; Smith B. D.; Beatty A. M.; Ugrinova V.; Brown S. N. (2002) Structure/Activity Study of Tris(2-aminoethyl)amine-Derived Translocases for Phosphatidylcholine. J. Org. Chem. 67, 2168–2174. 10.1021/jo016416s. [DOI] [PubMed] [Google Scholar]
- Okuro K.; Kinbara K.; Tsumoto K.; Ishii N.; Aida T. (2009) Molecular glues carrying multiple guanidinium ion pendants via an oligoether spacer: stabilization of microtubules against depolymerization. J. Am. Chem. Soc. 131, 1626–7. 10.1021/ja800491v. [DOI] [PubMed] [Google Scholar]
- Sun Z.; Gilles A.; Kocsis I.; Legrand Y. M.; Petit E.; Barboiu M. (2016) Squalyl Crown Ether Self-Assembled Conjugates: An Example of Highly Selective Artificial K Channels. Chem. - Eur. J. 22, 2158–2164. 10.1002/chem.201503979. [DOI] [PubMed] [Google Scholar]
- Sun Z. H.; Albrecht M.; Raabe G.; Pan F. F.; Rauber C. (2015) Solvent-dependent enthalpic versus entropic anion binding by biaryl substituted quinoline based anion receptors. J. Phys. Chem. B 119, 301–6. 10.1021/jp510796f. [DOI] [PubMed] [Google Scholar]
- Golczak M.; Maeda A.; Bereta G.; Maeda T.; Kiser P. D.; Hunzelmann S.; von Lintig J.; Blaner W. S.; Palczewski K. (2008) Metabolic Basis of Visual Cycle Inhibition by Retinoid and Nonretinoid Compounds in the Vertebrate Retina. J. Biol. Chem. 283, 9543–9554. 10.1074/jbc.M708982200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulido-Quetglas C.; Aparicio-Prat E.; Arnan C.; Polidori T.; Hermoso T.; Palumbo E.; Ponomarenko J.; Guigo R.; Johnson R. (2017) Scalable Design of Paired CRISPR Guide RNAs for Genomic Deletion. PLoS Comput. Biol. 13, e1005341 10.1371/journal.pcbi.1005341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidya A.; Mao Z.; Tian X.; Spencer B.; Seluanov A.; Gorbunova V. (2014) Knock-In Reporter Mice Demonstrate that DNA Repair by Non-homologous End Joining Declines with Age. PLoS Genet. 10, e1004511 10.1371/journal.pgen.1004511. [DOI] [PMC free article] [PubMed] [Google Scholar]