Abstract
Flower color is a charming phenotype with very important ornamental and commercial values. Anthocyanins play a critical role in determining flower color pattern formation, and their biosynthesis is typically regulated by R2R3-MYB transcription factors (TFs). Paeonia suffruticosa is a famous ornamental plant with colorful flowers. However, little is known about the R2R3-MYB TFs that regulate anthocyanin accumulation in P. suffruticosa. In the present study, two R2R3-MYB TFs, namely, PsMYB114L and PsMYB12L, were isolated from the petals of P. suffruticosa ‘Shima Nishiki’ and functionally characterized. Sequence analysis suggested that PsMYB114L contained a bHLH-interaction motif, whereas PsMYB12L contained two flavonol-specific motifs (SG7 and SG7-2). Subsequently, the in vivo function of PsMYB114L and PsMYB12L was investigated by their heterologous expression in Arabidopsis thaliana and apple calli. In transgenic Arabidopsis plants, overexpression of PsMYB114L and of PsMYB12L caused a significantly higher accumulation of anthocyanins, resulting in purple-red leaves. Transgenic apple calli overexpressing PsMYB114L and PsMYB12L also significantly enhanced the anthocyanins content and resulted in a change in the callus color to red. Meanwhile, gene expression analysis in A. thaliana and apple calli suggested that the expression levels of the flavonol synthase (MdFLS) and anthocyanidin reductase (MdANR) genes were significantly downregulated and the dihydroflavonol 4-reductase (AtDFR) and anthocyanin synthase (AtANS) genes were significantly upregulated in transgenic lines of PsMYB114L. Moreover, the expression level of the FLS gene (MdFLS) was significantly downregulated and the DFR (AtDFR/MdDFR) and ANS (AtANS/MdANS) genes were all significantly upregulated in transgenic lines plants of PsMYB12L. These results indicate that PsMYB114L and PsMYB12L both enhance anthocyanin accumulation by specifically regulating the expression of some anthocyanin biosynthesis-related genes in different plant species. Together, these results provide a valuable resource with which to further study the regulatory mechanism of anthocyanin biosynthesis in P. suffruticosa and for the breeding of tree peony cultivars with novel and charming flower colors.
Keywords: P. suffruticosa, R2R3-MYB, overexpression, anthocyanin, transcriptional regulation
1. Introduction
Paeonia suffruticosa is a very popular ornamental flowering plant that was first cultivated more than 1600 years ago in China and is currently distributed worldwide. This species is in the Paeoniaceae family and has been named ‘the king of flowers’ for its showy and colorful flowers [1]. Approximately 1500 cultivars of P. suffruticosa with a variety of flower colors have been produced by breeders worldwide [2]. Among the many flower colors of this species, most fit into two clusters: monochrome color (red, pink, white, purple, black, blue, green, and yellow) and double color. Cultivars with a double-color phenotype are rarer and more sought after, and thus have great ornamental and commercial value [3]. Among them, P. suffruticosa ‘Shima Nishiki’, a well-known chimeric cultivar, was selected from the bud mutation of P. suffruticosa ‘Taiyoh’. ‘Shima Nishiki’ usually has red and pink petals on the same flower, and this aesthetically pleasing double-color phenotype can be stably inherited [4]. Therefore, the ‘Shima Nishiki’ cultivar is regarded as an important experimental material with which to study the molecular regulatory mechanism of flower color and in the breeding of new cultivars [5].
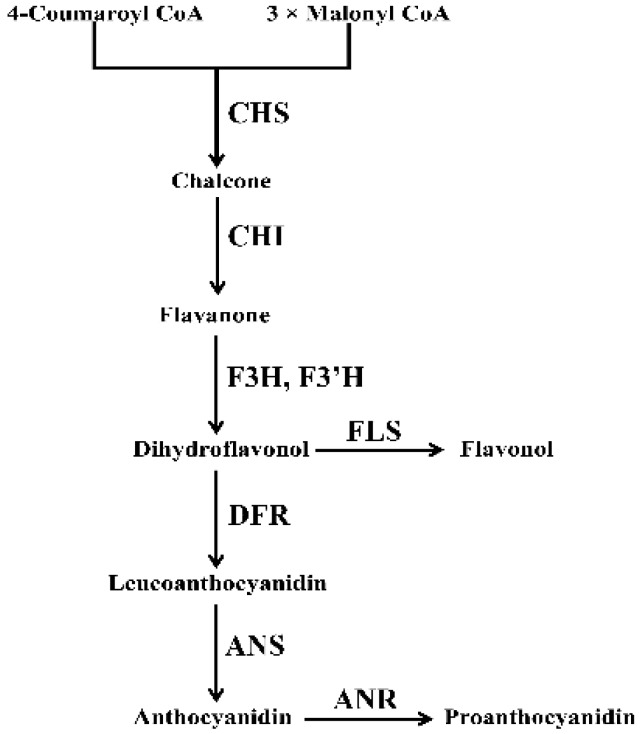
Anthocyanins are important soluble flavonoid compounds that are widely distributed in the leaves, flowers, fruits, seeds and other tissues of many plants [6]. Anthocyanin composition and concentration are usually closely related to flower color intensity [7,8]. The anthocyanin biosynthetic pathway is well known to be highly conserved in many ornamental plants [9,10,11,12,13,14]. Anthocyanin biosynthesis and accumulation are usually regulated by a series of structural genes and regulatory genes [15,16]. The structural genes encode enzymes associated with anthocyanin biosynthesis, including chalcone synthase (CHS), chalcone isomerase (CHI), flavanone 3-hydroxylase (F3H), flavonoid 3′-hydroxylase (F3′H), dihydroflavonol 4-reductase (DFR) anthocyanin synthase (ANS), Flavonol synthase (FLS), and anthocyanidin reductase (ANR) [17,18,19] (Figure 1). Among them, FLS is a dedicated enzyme involved in flavonol biosynthesis, and ANR is a key enzyme for proanthocyanidin biosynthesis. The regulatory genes can be divided into three families R2R3-MYB, bHLH, and WD40 [20,21,22] and they usually form a regulatory complex to activate the expression of anthocyanin biosynthetic genes [23,24,25].
Figure 1.
A general schematic diagram of the metabolic pathway related to anthocyanin biosynthesis. CHS, chalcone synthase; CHI, chalcone isomerase; F3H, flavanone 3-hydroxylase; F3′H, flavonoid 3′-hydroxylase; DFR, dihydroflavonol 4-reductase; ANS, anthocyanidin synthase; FLS, flavonol synthase; ANR, anthocyanidin reductase.
Many structural genes have been characterized and cloned in P. suffruticosa [12,26,27,28]. In the MYB-bHLH-WDR (MBW) complex, R2R3-MYB transcription factors (TFs) usually play critical roles in anthocyanin biosynthesis and accumulation [29,30]. Many R2R3-MYB TFs involved in anthocyanin biosynthesis have been isolated and characterized from various plants, including Arabidopsis thaliana [24], Zea mays [31], Vitis vinifera [32], Malus crabapple [33], Petunia hybrida [34], Antirrhinum majus [29], Dendranthema morifolium [35] and Phalaenopsis aphrodite [14]. In P. suffruticosa, most previous studies were focused primarily on the preliminary investigation of R2R3-MYB TFs based on transcriptome sequencing and qRT-PCR analyses [12,36,37,38], whereas whether and how R2R3-MYB TFs control anthocyanin biosynthesis and accumulation in P. suffruticosa are almost unknown.
In the present study, two novel R2R3-MYB TFs, namely, PsMYB114L and PsMYB12L, were cloned in P. suffruticosa. Subsequently, the expression patterns of PsMYB114L and PsMYB12L were determined at five developmental stages in P. suffruticosa ‘Shima Nishiki’. Furthermore, the function of these two TFs was further verified by heterologous expression in Arabidopsis and apple calli. These results will provide valuable insights into understanding the putative roles of PsMYB114L and PsMYB12L in regulating anthocyanin biosynthesis in P. suffruticosa.
2. Results
2.1. Cloning and Analysis of the PsMYB114L and PsMYB12L Genes
Based on the functional annotation and gene expression analysis of transcriptome sequencing data in P. suffruticosa ‘Shima Nishiki’ [39], we filtered two MYB unigenes exhibiting relatively high expression differences between the red and pink petals as the targeted genes of this study.
The full-length cDNA sequences of the two novel MYB genes were obtained with PCR amplification. By conducting GenBank BLAST searches of the amino acid sequences of these two genes, we found that these genes have the highest homology with transcription factor MYB114-like [Quercus suber] and transcription factor MYB12-like [Juglans regia], respectively. Therefore, we named these genes PsMYB114L and PsMYB12L. Sequencing results revealed that PsMYB114L (Figure S1A,B) and PsMYB12L (Figure S1C,D) contained an open reading frame (ORF) of 600 and 1140 bp encoding 199 and 379 amino acids and that their predicted proteins had a molecular mass of 22.81 and 42.61 kDa and a theoretical isoelectric point (pI) of 8.53 and 4.86, respectively.
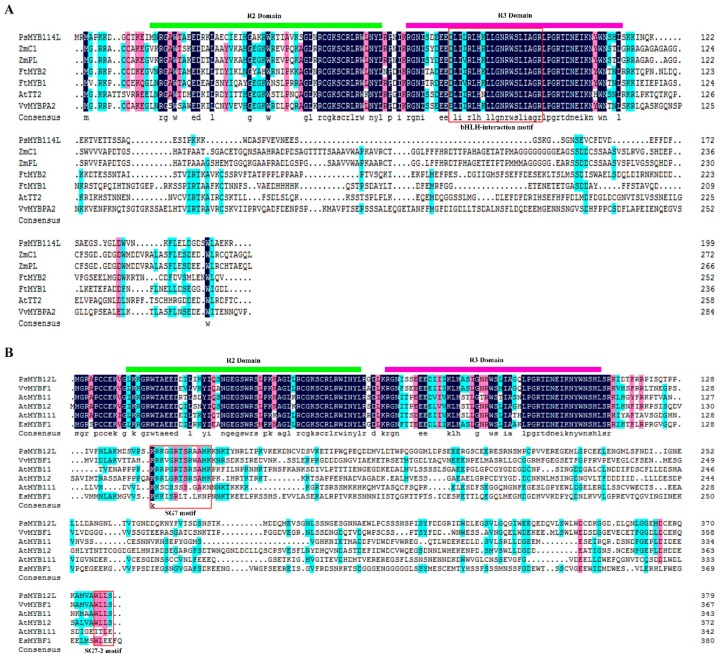
Multiple sequence alignment of amino acids revealed that PsMYB114L and PsMYB12L, belonging to the SANT superfamily (which typically consists of tandem repeats of three alpha-helices arranged in a helix-turn-helix motif, with each alpha helix containing a bulky aromatic residue), and other known R2R3-MYB TFs related to anthocyanin biosynthesis contained a highly conserved R2R3 DNA-binding domain. The presence of this conserved domain means that PsMYB114L and PsMYB12L are also R2R3-MYB TFs and may perform similar functions in regulating anthocyanin biosynthesis. Furthermore, PsMYB114L had a bHLH-interaction motif ([D/E]Lx2[R/K]x3Lx6Lx3R) in the R3 domain at the N terminus and did not have any conserved motifs at the C terminus (Figure 2A). Moreover, PsMYB12L did not have any bHLH-interaction motifs at the N terminus, whereas it contained two flavonol-specific motifs [40], namely, SG7 ([K/R][R/x][R/K]xGRT[S/x][R/G]xx[M/x]K) and SG7-2 ([W/x][L/x]LS), at the C terminus (Figure 2B).
Figure 2.
Amino acid sequence alignment analysis of the PsMYB114L (A) and PsMYB12L (B) genes with other known R2R3-MYB TFs. The green and pink long lines indicate the R2 and R3 domain, respectively. The red boxes show the conserved bHLH-interaction, SG7 and SG7-2 motifs. The NCBI GenBank accession numbers of these sequences are as follows: ZmC1, Zea mays, AF320613.3; ZmPL, Zea mays, NM_001112415.1; FtMYB2, Fagopyrum tataricum, JF313346.1; FtMYB1, Fagopyrum tataricum, JF313344.1; AtTT2, Arabidopsis thaliana, NM_122946.3; VvMYBPA2, Vitis vinifera, NM_001281024.1; VvMYBF1, Vitis vinifera, FJ948477.2; AtMYB11, Arabidopsis thaliana, NM_116126.3; AtMYB12, Arabidopsis thaliana, NM_130314.4; AtMYB111, Arabidopsis thaliana, NM_124310.3; EsMYBF1, Epimedium sagittatum, KU365320.1.
To better evaluate the phylogenetic relationships of PsMYB114L, PsMYB12L and 16 other known MYB TFs related to the regulation of anthocyanin biosynthesis, a phylogenetic tree was constructed based on the amino acid sequences of these 18 MYB TFs from different species using the neighbor-joining method. The phylogenetic analysis indicated that these 18 MYB TFs were classified into four groups (Flavonol, Anthocyanin, Anthocyanin/Proanthocyanidin and Proanthocyanidin) based on their specific roles in the flavonoid biosynthesis pathway. Among them, PsMYB114L had the closest phylogenetic relationship with ZmC1 and ZmPL, which are involved in regulating anthocyanin biosynthesis, whereas PsMYB12L belongs to a subgroup of MYB proteins that includes VvMYBF1, EsMYBF1, AtMYB11, AtMYB12 and AtMYB111, which regulate flavonol synthesis and had the closest phylogenetic relationship with VvMYBF1 (Figure 3).
Figure 3.
Phylogenetic analysis of the PsMYB114L and PsMYB12L genes with R2R3-MYB TFs from other species. The NCBI GenBank accession numbers of these sequences are as follows: FaMYB1, Fragaria x ananassa, AF401220.1; VvMYBPA1, Vitis vinifera, NM_001281231.1; VvMYB5b, Vitis vinifera, NM_001280925.1; AtMYB5, Arabidopsis thaliana, AF401220.1; GtMYB3, Gentiana triflora, AB289445.1; ZmC1, Zea mays, AF320613.3; ZmPL, Zea mays, NM_001112415.1; FtMYB2, Fagopyrum tataricum, JF313346.1; FtMYB1, Fagopyrum tataricum, JF313344.1; AtTT2, Arabidopsis thaliana, NM_122946.3; VvMYBPA2, Vitis vinifera, NM_001281024.1; VvMYBF1, Vitis vinifera, FJ948477.2; AtMYB11, Arabidopsis thaliana, NM_116126.3; AtMYB12, Arabidopsis thaliana, NM_130314.4; AtMYB111, Arabidopsis thaliana, NM_124310.3; EsMYBF1, Epimedium sagittatum, KU365320.1.
2.2. Subcellular Localization of PsMYB114L and PsMYB12L
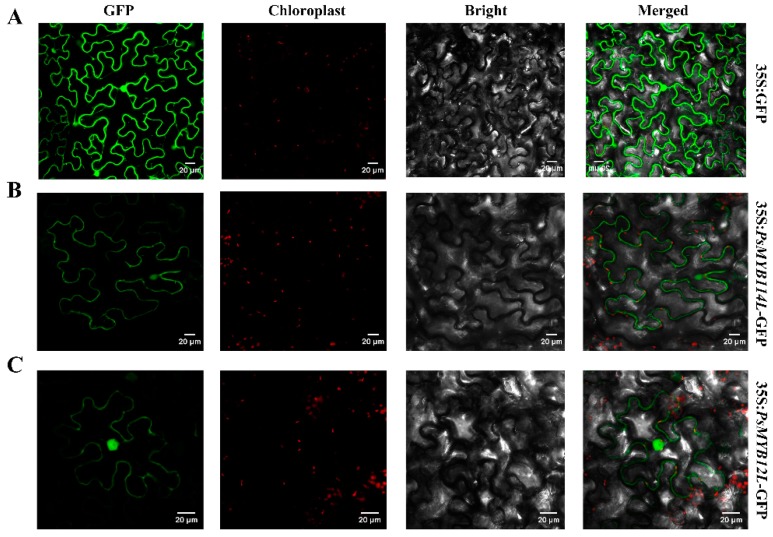
To examine the subcellular localization of PsMYB114L and PsMYB12L, the recombinant vector (PsMYB114L-GFP/PsMYB12L-GFP) and the control vector (pCAMBIA1301-GFP) were introduced into the tobacco leaves. Our results were basically consistent with those of previous studies [41,42]. The green fluorescent protein (GFP) fluorescence of the control vector was clearly distributed throughout the entire cell (Figure 4A), and the PsMYB114L-GFP/PsMYB12L-GFP vector displayed a strong fluorescence signal in the nucleus and cytoplasm of tobacco cells (Figure 4B,C). Therefore, we speculated that the two R2R3-MYB TFs (PsMYB114L/PsMYB12L) were simultaneously localized and functioned in the nucleus and cytoplasm.
Figure 4.
Subcellular localization analysis of the PsMYB114L and PsMYB12L genes. (A) Control vector (pCAMBIA1301-GFP) expressed in epidermal cells of tobacco leaves. (B) Recombinant vector (PsMYB114L-GFP) expressed in epidermal cells of tobacco leaves. (C) Recombinant vector (PsMYB12L-GFP) expressed in epidermal cells of tobacco leaves. White lines at the bottom right of the picture represent 20 μm in the respective pixel. GFP, GFP fluorescence; Chloroplast, Chloroplast fluorescence; Bright, Bright field; Merged, Superposition of bright field and fluorescence.
2.3. Expression Patterns of PsMYB114L and PsMYB12L in P. suffruticosa ‘Shima Nishiki’
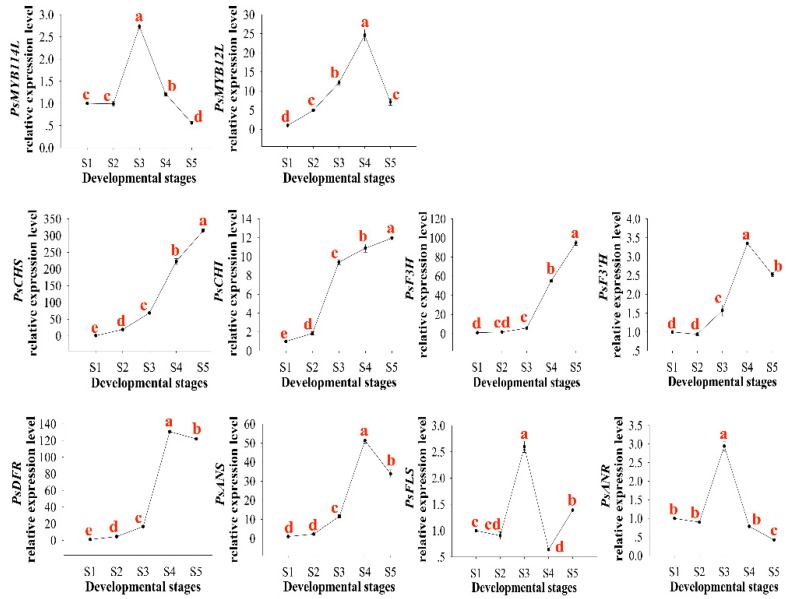
qRT-PCR analysis was conducted to survey the expression patterns of PsMYB114L and PsMYB12L in P. suffruticosa ‘Shima Nishiki’ (Figure 5). Petal samples of this cultivar were collected at five developmental stages (Figure S2). The expression levels of the PsMYB114L gene peaked at S3 and then decreased from S3 to S5, whereas the PsMYB12L gene exhibited the highest expression at S4. Furthermore, the expression levels of the eight anthocyanin biosynthesis-related genes (PsCHS, PsCHI, PsF3H, PsF3′H, PsDFR, PsANS, PsFLS, and PsANR) were analyzed. Among these genes, PsF3′H, PsDFR, and PsANS showed a trend similar to that of PsMYB12L, whereas PsFLS and PsANR showed a trend similar to that of PsMYB114L.
Figure 5.
The expression patterns of the PsMYB114L gene, PsMYB12L gene and anthocyanin biosynthesis-related structural genes in P. suffruticosa ‘Shima Nishiki’. S1, flower bud emerging stage; S2 small bell-like flower-bud stage; S3, large bell-like flower-bud stage; S4, bell-like flower-bud extending stage; S5, color exposing stage. Different lowercase letters indicate significant differences at p < 0.05.
2.4. Overexpression of PsMYB114L and PsMYB12L in Arabidopsis
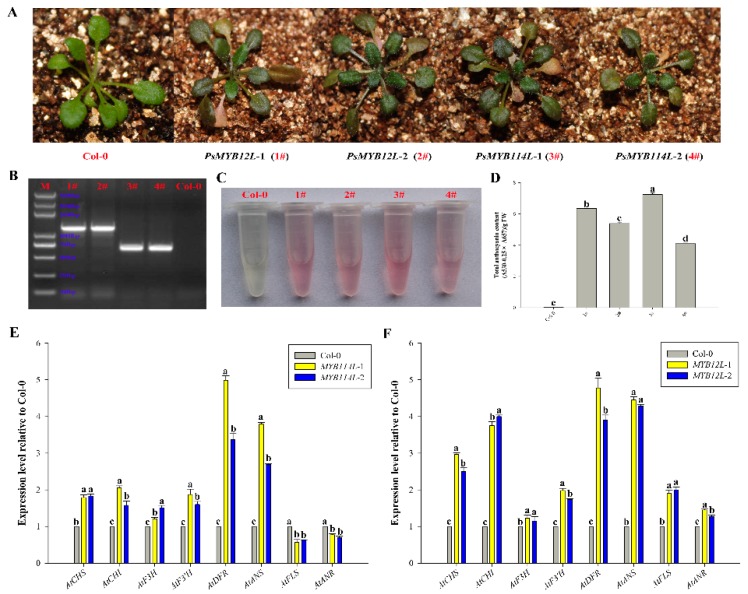
To characterize the functions of PsMYB114L and PsMYB12L, these two genes under the expression of the 35S promoter were genetically transformed into Arabidopsis. Phenotypic investigations of the transgenic lines of PsMYB114L and PsMYB12L revealed that their leaves were much deeper in color than those of Col-0 and showed a purple-red color (Figure 6A). Meanwhile, these transgenic lines of the two genes were confirmed by PCR analysis (Figure 6B). Furthermore, the total anthocyanin content results indicated that the transgenic lines of PsMYB114L and PsMYB12L produced much more anthocyanin than Col-0 (Figure 6C,D).
Figure 6.
Overexpression analysis of the PsMYB114L and PsMYB12L genes in Arabidopsis. (A) Comparison of leaf colors in transgenic Arabidopsis plants and Col-0. (B) Results of positive PCR detection in transgenic Arabidopsis plants. (C) Anthocyanin extraction solutions for transgenic Arabidopsis plants and Col-0. (D) Total anthocyanin content in transgenic Arabidopsis plants and Col-0. (E) Expression analysis of anthocyanin biosynthesis-related genes in transgenic Arabidopsis plants of PsMYB114L and Col-0. (F) Expression analysis of anthocyanin biosynthesis-related genes in transgenic Arabidopsis plants of PsMYB12L and Col-0. Col-0, Arabidopsis thaliana ecotype Columbia; 1# and 2#, two transgenic lines of the PsMYB12L gene; 3# and 4#, two transgenic lines of the PsMYB114L gene. Different lowercase letters indicate significant differences at p < 0.05.
Additionally, the expression levels of anthocyanin biosynthesis-related genes (AtCHS, AtCHI, AtF3H, AtF3′H, AtDFR, AtANS, AtFLS, and AtANR) in the Col-0 and the transgenic Arabidopsis plants of PsMYB114L and PsMYB12L were analyzed with qRT-PCR experiments. Compared with the Col-0, overexpression of PsMYB114L upregulated the expression of most of the genes (AtCHS, AtCHI, AtF3H, AtF3′H, AtDFR, and AtANS) in transgenic PsMYB114L plants; among them, both of the AtDFR/AtANS genes showed a relatively high difference between the Col-0 and transgenic plants, whereas AtFLS and AtANR were downregulated in transgenic PsMYB114L plants (Figure 6E).
For PsMYB12L overexpression in Arabidopsis, the expression levels of all eight genes were upregulated in transgenic PsMYB12L plants. Among them, the four genes (AtCHS, AtCHI, AtDFR, and AtANS) all showed a relatively high difference between the Col-0 and transgenic plants (Figure 6F).
2.5. Overexpression of PsMYB114L and PsMYB12L in Apple Calli
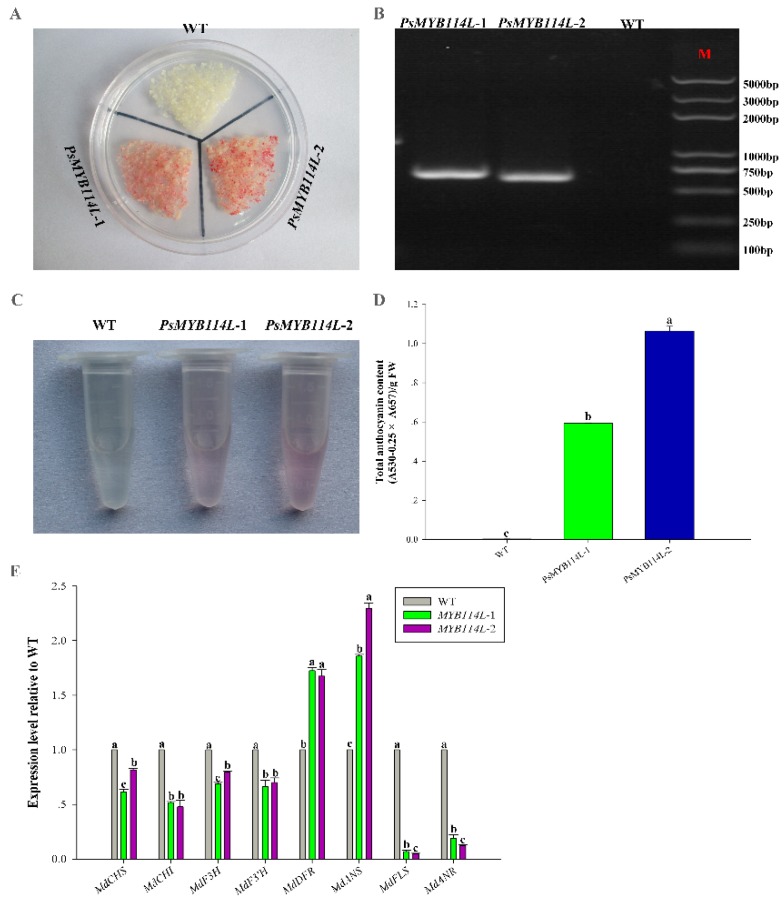
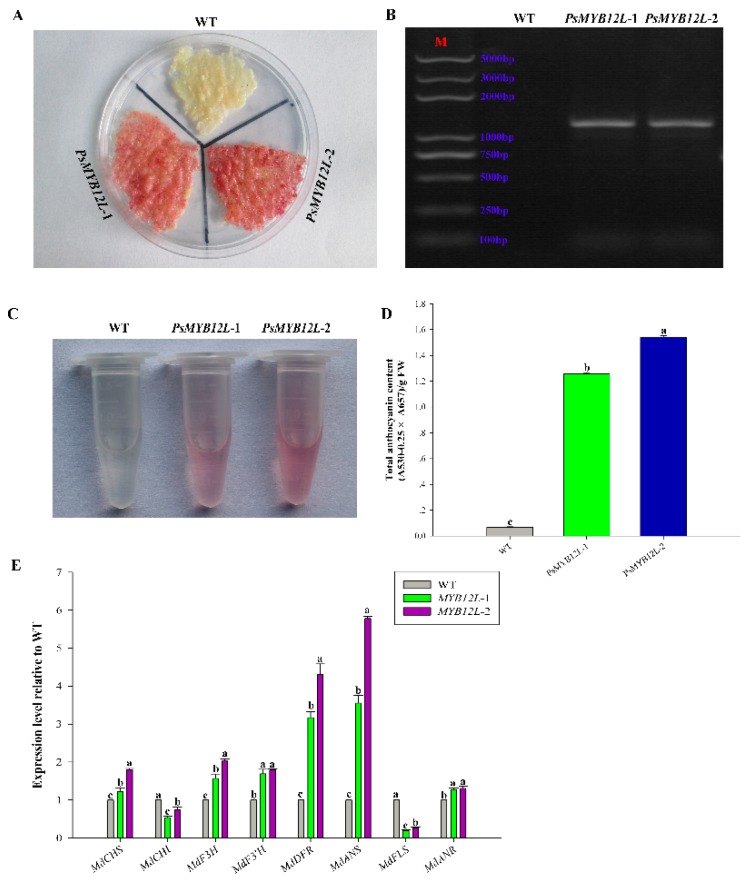
For further functional validation, the two genes (PsMYB114L and PsMYB12L) were ectopically expressed in the calli of ‘Orin’ apple. Interestingly, after light and low-temperature treatments, the WT had almost no phenotypic changes, but an especially obvious color change was observed in the transgenic lines of PsMYB114L and PsMYB12L (Figure 7A and Figure 8A). PCR amplification confirmed that these transgenic apple calli carry PsMYB114L and PsMYB12L (Figure 7B and Figure 8B). With regard to the total anthocyanin content, the transgenic lines of PsMYB114L and PsMYB12L all accumulated markedly higher amounts of anthocyanins than did the WT (Figure 7C,D and Figure 8C,D).
Figure 7.
Overexpression analysis of the PsMYB114L gene in apple calli. (A) Colors observed in transgenic apple calli and the WT. (B) Results of positive PCR detection in transgenic apple calli. (C) Anthocyanin extraction solutions for transgenic apple calli and the WT. (D) Total anthocyanin content in transgenic apple calli and the WT. (E) Expression analysis of anthocyanin biosynthesis-related genes in transgenic apple calli and the WT. WT, Wild-type ‘Orin’ apple calli; PsMYB114L-1 and PsMYB114L-2, two transgenic lines of the PsMYB114L gene. Different lowercase letters indicate significant differences at p < 0.05.
Figure 8.
Overexpression analysis of the PsMYB12L gene in apple calli. (A) Colors observed in transgenic apple calli and the WT. (B) Results of positive PCR detection in transgenic apple calli. (C) Anthocyanin extraction solutions for transgenic apple calli and the WT. (D) Total anthocyanin content in transgenic apple calli and the WT. (E) Expression analysis of anthocyanin biosynthesis-related genes in transgenic apple calli and the WT. PsMYB12L-1 and PsMYB12L-2, two transgenic lines of the PsMYB12L gene. Different lowercase letters indicate significant differences at p < 0.05.
Additionally, the expression levels of anthocyanin biosynthesis-related genes (MdCHS, MdCHI, MdF3H, MdF3′H, MdDFR, MdANS, MdFLS, and MdANR) in the WT and the transgenic lines of PsMYB114L and PsMYB12L were analyzed by qRT-PCR. Compared with the WT, overexpression of PsMYB114L downregulated the expression of most of the genes, specifically, MdCHS, MdCHI, MdF3H, MdF3′H, MdFLS, and MdANR, and upregulated the expression of MdDFR and MdANS in transgenic PsMYB114L calli (Figure 7E).
For PsMYB12L overexpression, the expression levels of most genes, including MdCHS, MdF3H, MdF3′H, MdDFR, MdANS, and MdANR, were upregulated, but those of MdCHI and MdFLS were downregulated in transgenic PsMYB12L calli (Figure 8E).
3. Discussion
Flower color is a very important trait in many ornamental plants and has a close association with their ornamental and commercial value. Many prior studies have shown that anthocyanins are a key factor influencing flower color [43,44,45]. R2R3-MYB TFs comprise one of the largest gene families in plants and play key roles in regulating anthocyanin accumulation by activating the expression of structural genes involved in the anthocyanin biosynthetic pathway [46,47]. However, the role of R2R3-MYB TFs in regulating flower color in P. suffruticosa has seldom been functionally verified. Therefore, determining how certain R2R3-MYB TFs regulate anthocyanin production in P. suffruticosa would aid in breeding improved cultivars with desirable flower colors.
In the present study, two novel R2R3-MYB TFs (PsMYB114L and PsMYB12L) possibly involved in anthocyanin biosynthesis were successfully cloned and characterized from the petals of P. suffruticosa ‘Shima Nishiki’ and found to contain full-length cDNA of 600 and 1140 bp encoding 199 and 379 amino acids, respectively. The amino acid sequence alignment between PsMYB114L/PsMYB12L and other known R2R3-MYB TFs involved in anthocyanin regulation indicated that the R2R3 domain distributions of these R2R3-MYB TFs were highly similar, but a bHLH-interaction motif ([D/E]Lx2[R/K]x3Lx6Lx3R) existed in the R3 domain of PsMYB114L, whereas PsMYB12L did not contain this motif for interaction with bHLH proteins. In Arabidopsis, based on a similar function, 125 TFs of R2R3-MYB gene-family members were classified into more than 25 subgroups [48]. Furthermore, many previous studies demonstrated that subgroup 7 [49,50], characterized by both the SG7 ([K/R][R/x][R/K]xGRT[S/x][R/G]xx[M/x]K) and SG7-2 ([W/x][L/x]LS) motifs, specifically regulated flavonol biosynthesis. PsMYB12L contained these two motifs (SG7 and SG7-2) at the C terminus of the protein, but PsMYB114L lacked these two motifs.
Phylogenetic analysis indicated that PsMYB12L and 5 flavonol-regulating R2R3-MYB TFs (VvMYBF1, EsMYBF1, and AtMYB11/12/111) belonging to subgroup 7 [30,51] were clustered together, and PsMYB114L and certain R2R3-MYB TFs belonging to subgroup 5 (AtTT2, ZmC1, VvMYBPA2, etc.) [52,53,54,55] had relatively higher homology. Based on the motif analysis of amino acid sequences and phylogenetic analysis, PsMYB114L might regulate anthocyanin production by combinatorially interacting with a basic helix-loop-helix (bHLH) factor [25,56,57]. PsMYB12L might independently regulate the expression of anthocyanin biosynthesis-related genes without the MBW complex [58].
In addition, we conducted further ectopic transgenic studies by overexpressing PsMYB114L/PsMYB12L in Arabidopsis and apple calli. In contrast to the green-colored leaves of the Col-0 A. thaliana ecotype and the white-colored WT apple calli, the leaves of these transgenic Arabidopsis plants turned purple-red and the transgenic calli of PsMYB114L and PsMYB12L were red, which was in agreement with their remarkably higher anthocyanin content. The color and total anthocyanin content analyses of Arabidopsis and apple calli indicated that these two R2R3-MYB TFs contribute to anthocyanin accumulation in transgenic lines.
Subsequently, qRT-PCR analysis of seven anthocyanin biosynthesis-related genes (MdCHS, MdCHI, MdF3H, MdF3′H, MdDFR, MdANS, MdFLS, and MdANR) was further performed in Arabidopsis and apple calli. In terms of PsMYB114L, the qRT-PCR results in Arabidopsis showed that the expression levels of AtDFR and AtANS were significantly upregulated, whereas AtFLS and AtANR were downregulated to a certain extent compared with the levels in the Col-0. Furthermore, the qRT-PCR results in apple calli showed that the expression levels of MdDFR and MdANS were upregulated to a certain extent, whereas MdFLS and MdANR (especially MdFLS) were significantly downregulated compared with the levels in the WT. Meanwhile, based on the results of expression patterns of PsMYB114L in P. suffruticosa ‘Shima Nishiki’, we have known that PsMYB114L have a positive correlation with PsFLS and PsANR. By comparing these three qRT-PCR results in Arabidopsis, apple calli, and P. suffruticosa, we found differences in the expression patterns of some anthocyanin biosynthesis-related genes. Previous studies have showed that many R2R3-MYB TFs usually regulate flavonoid biosynthesis by interacting with the promoter of the targeted structural genes [55,58]. For promoter region, in general, the sequence of the same structural gene in different plant species also differs greatly. Therefore, it is possible that the same MYB TFs performed different regulatory mechanisms of flavonoid biosynthesis in different species [59]. Dihydroflavonol is the direct substrate for two key genes (FLS and DFR) in the flavonoid biosynthetic pathways, and these two genes usually show a competitive interaction in producing colored anthocyanidin and colorless flavonols [60]. In this study, the strong upregulation of AtDFR and AtANS may have played key roles in activating the branch of the anthocyanin biosynthesis, resulting in purple-red leaves in the transgenic Arabidopsis plants of PsMYB114L, whereas the strong downregulation of MdFLS would inhibit the branch of the flavonol biosynthesis, resulting in the production of anthocyanins and a red-colored phenotype in the transgenic calli of PsMYB114L. Furthermore, because PsMYB114L has a bHLH-interaction motif, it may form an MBW complex and contribute to anthocyanin accumulation by regulating the expression of these key genes (AtDFR, AtANS, MdFLS, and MdANR) in Arabidopsis and apple calli.
With regard to PsMYB12L, the qRT-PCR results showed that the expression levels of the four genes (AtCHS, AtCHI, AtDFR and AtANS) were all significantly upregulated in the transgenic Arabidopsis plants. Moreover, the expression levels of MdDFR and MdANS were significantly upregulated in the transgenic calli of PsMYB12L, but MdFLS was significantly downregulated. Meanwhile, based on the results of expression patterns of PsMYB12L in P. suffruticosa ‘Shima Nishiki’, we have known that PsMYB12L have a positive correlation with PsDFR, PsANS, and PsF3′H. By comparing these three qRT-PCR results in Arabidopsis, apple calli, and P. suffruticosa, we can found that the two key anthocyanin biosynthesis-related genes (DFR and ANS) showed a very similar expression pattern. We considered that PsMYB12L should be a specific transcriptional regulator on DFR and ANS genes in these three species. Furthermore, we also found differences in the expression patterns of the FLS gene in Arabidopsis and apple calli, and considered that the expression difference of the FLS gene is likely caused by the promoter sequence specificity of this gene in these two species [61]. Based on the motif analysis of PsMYB12L, we speculated that the TF may be a flavonol-specific MYB regulator. Many flavonol-specific MYB TFs have been isolated and functionally verified in various plants, such as A. thaliana, Vitis vinifera, and Epimedium sagittatum [50,62,63]. Furthermore, many flavonol-specific MYB TFs negatively regulate anthocyanin accumulation by inducing higher expression of the FLS gene. By overexpressing AtMYB12 in tobacco, the expression of NtCHS, NtCHI, and NtFLS was specifically activated; moreover, the flowers of the transgenic plants were paler in color than their wild-type counterparts [64]. Ectopic expression analysis of EsMYBF1 in transgenic tobacco indicated that NtCHS, NtCHI, NtF3H, and NtFLS were upregulated but NtDFR and NtANS were significantly downregulated, and the accumulation of anthocyanins in transgenic tobacco flowers was also remarkably decreased [63]. A study on the overexpression of PpMYB15 in tobacco showed that it can significantly activate the expression of NtCHS, NtCHI, NtF3H, and NtFLS, while it had no effects on the expression of NtDFR and NtANS, resulting in pale-pink or pure white flowers in transgenic tobacco plants [40]. Compared with the expression of anthocyanin biosynthesis-related genes documented in the abovementioned studies, in this study AtCHS/MdCHS, AtCHI, AtF3H/MdF3H, and AtFLS had a somewhat similar expression pattern and MdFLS, AtDFR/MdDFR and AtANS/MdANS exhibited the opposite pattern. However, the lower expression of the MdFLS gene and the higher expression of AtDFR/MdDFR and AtANS/MdANS were consistent with the significantly higher anthocyanin accumulation in transgenic lines of PsMYB12L. Beacuse PsMYB12L has the flavonol-specific motif and lacks the bHLH-interaction motif, it alone enhances anthocyanin production by regulating the expression of these key genes (AtDFR/MdDFR, AtANS/MdANS, and MdFLS) independently of bHLH cofactors in Arabidopsis and apple calli.
4. Materials and Methods
4.1. Plant Materials
The tree peony cultivar P. suffruticosa ‘Shima Nishiki’ was grown in the experimental nursery of Forestry College, Shandong Agricultural University, Tai’an, Shandong, China. Flower samples were collected at five early flower-bud developmental stages (flower bud emerging stage (S1), small bell-like flower-bud stage (S2), large bell-like flower-bud stage (S3), bell-like flower-bud extending stage (S4), and color exposing stage (S5)) (Figure S2) [65]. All these samples were immediately frozen in liquid nitrogen and then stored at –80 °C for further experiments.
The A. thaliana ecotype Columbia (Col-0) was used for genetic transformation and phenotypic analysis in the present study. The plants were grown under a 16 h light/ 8 h dark photoperiod at 23 °C/21 °C
Furthermore, calli of the wild type (WT) of ‘Orin’ apple were subcultured on Murashige and Skoog (MS) medium with 1.5 mg L−1 6-benzyl adenine (6-BA) and 0.5 mg L−1 2,4-dichlorophenoxyacetic acid (2,4-D) at room temperature (24 °C) in a continuous dark environment at 15-day intervals [66]. Subsequently, the calli were used for genetic transformation and phenotypic analysis.
4.2. Total RNA Extraction and cDNA Synthesis
Total RNA was extracted from all samples according to instructions of the EASY Spin Plant RNA Rapid Extraction Kit (Aidlab Biotech, Beijing, China). The purity and concentration of all RNA samples were assessed using a Nanodrop 2000C spectrophotometer (Thermo Fisher Scientific, Wilmington, Delaware, DE, USA), and RNA quality was detected using 1 % agarose gels. Furthermore, cDNA was synthesized with 1 µg of total RNA using 5× All-In-One RT MasterMix (with an AccuRT Genomic DNA Removal Kit) (ABM, Vancouver, BC, Canada).
4.3. Cloning of the PsMYB114L and PsMYB12L Genes in P. suffruticosa
In this study, based on the transcriptome sequencing data of P. suffruticosa ‘Shima Nishiki’ in our laboratory, two R2R3-MYB transcription factors were filtered by analyzing the functional annotations of MYB unigenes and performing gene expression analysis.
The cDNA of the ‘Shima Nishiki’ cultivar’s petals was used as the template. The full-length coding sequence (CDS) of the PsMYB114L (MK518073) and PsMYB12L (MK518074) genes was amplified using PCR. The complete 5′ CDS of the PsMYB114L and PsMYB12L genes was identified from the transcriptome sequencing data of P. suffruticosa ‘Shima Nishiki’. The cDNA 3′ end sequence of these candidate genes was obtained using nested PCR technology using PsMYB114L-1-F/PsMYB114L-2-F and PsMYB12L-1-F/PsMYB12L-2-F as forward primers (Table S1), respectively, and B26 was used as the common reverse primer. The full-length cDNA of the PsMYB114L and PsMYB12L genes was amplified with the forward primers PsMYB114L-F1/PsMYB12L-F1 and the reverse primers PsMYB114L-R1/PsMYB12L-R1 (Table S1). The PCR program of gene amplification was as follows: initial denaturation at 95 °C for 1 min, followed by 30 cycles of 98 °C for 10 s, 60 °C for 15 s and 68 °C for 60 s. The PCR products were purified and cloned into the pTOPO-Blunt Simple vector for sequencing.
4.4. Subcellular Localization
The full-length cDNA without the termination codon of PsMYB114L/PsMYB12L was amplified with special primers (PsMYB114L-GFPF/PsMYB12L-GFPF and PsMYB114L-GFPR/PsMYB12L-GFPR) (Table S1) with restriction sites (Xba I and Kpn I) and subcloned into the pCAMBIA1301-GFP vector between the Xba I and Kpn I sites to create the PsMYB114L-GFP/PsMYB12L-GFP fusion construct. The recombinant vectors (PsMYB114L-GFP/PsMYB12L-GFP) and control vector (pCAMBIA1301-GFP) were then introduced into tobacco leaves by agroinfiltration. These infiltrated plants were grown for over 72 h in a growth chamber, and the GFP fluorescence of samples was observed under a Nikon C2-ER confocal laser scanning microscope (Nikon, Tokyo, Japan) [67].
4.5. Overexpression Vector Construction
The full-length cDNA of the PsMYB114L and PsMYB12L genes from the petals of P. suffruticosa ‘Shima Nishiki’ was amplified using recombinant primers (PsMYB114L-F2/PsMYB12L-F2 and PsMYB114L-R2/PsMYB12L-R2) (Table S1) with restriction sites (Spe I and BstE II). Based on the predesigned vector construction procedure, the pCAMBIA1304 empty vector and the pTOPO-Blunt Simple vector containing the target genes (PsMYB114L and PsMYB12L) with restriction sites were double digested separately between the Spe I and BstE II sites and then recombined (Figure S3A–C). Subsequently, the two recombinant vectors pCAMBIA1304-PsMYB114L (Figure S3D) and pCAMBIA1304-PsMYB12L (Figure S3E) were verified successfully by PCR and sequencing with the forward vector validation primer 1304Ve-F and the reverse primers PsMYB114L-R2/PsMYB12L-R2 (Table S1). These two overexpression constructs were also introduced into Agrobacterium tumefaciens strain GV3101 using the freeze-thaw method.
4.6. Stable Transformation of Arabidopsis
The transformation of Arabidopsis was performed using the floral dip transformation method [68]. An A. tumefaciens infection solution (OD600 = 0.8–1.2) containing 5 % sucrose and 0.01 % Silwet L-77 was prepared to infect inflorescences, and the infection time per inflorescence was 15 s. Subsequently, these plants were transferred to a dark treatment for 24 h. These steps were repeated twice more according to the growth state of the plant. Mature T1 seeds were harvested, surface sterilized, and then sown on MS medium with 30 mg L−1 hygromycin B to screen for positive transformants. The resistant seedlings were transplanted into soil and then placed in a light incubator (16 h light/8 h dark at 23 °C/21 °C). When these transgenic Arabidopsis plants had grown to a certain size, they were further verified with gene-specific primers by PCR.
4.7. Stable Transformation of Apple Calli
To transform apple calli, 15-day-old WT apple calli were incubated with A. tumefaciens infection solution that carried pCAMBIA1304-PsMYB114L/pCAMBIA1304-PsMYB12L for 20 min, and the apple calli were then cocultured on MS medium supplemented with 0.5 mg L−1 2,4-D and 1.5 mg L−1 6-BA for 2 days at 24 °C in the dark. Subsequently, the apple calli were washed three times with sterile water and transferred to a selective medium that contained 15 mg L−1 hygromycin B for transgene selection. The transgenic apple calli were cocultured in the selective medium containing appropriate concentrations of an antibiotic and transferred to a light incubator with constant light (photon flux density of ~100 μmol s−1 m−2) and low-temperature (15 °C) treatments for phenotypic observation [69,70].
4.8. Measurement of Total Anthocyanin Content
Total anthocyanin were extracted from the rosette leaves of 25-day-old Arabidopsis plants and apple calli cultured for 7 days. Anthocyanin extraction was performed using a methanol–HCl method [71]. Approximately 0.1 g of each sample was incubated in 5 mL of 0.1 % acidic methanol solution (CH3OH:HCl:H2O = 70:0.1:29.9, v/v/v) overnight in the dark at 4 °C. The absorbance of each extract was measured at 530 and 657 nm with a UV-1600 spectrophotometer (SHIMADZU, Kyoto, Japan). The total anthocyanin content was calculated using the following equation: QTotal Anthocyanin = (A530 − 0.25 × A657) × FM−1. There were three biological replicates for each sample.
4.9. Quantitative Real-Time PCR (qRT-PCR) Analysis
qRT-PCR was performed to analyze the expression levels of anthocyanin biosynthesis-related genes in all plant materials in this study. The qRT-PCR experiments were conducted using SYBR® Premix Ex Taq™ (Tli RNaseH Plus) (TaKaRa, Kyoto, Japan) on a Bio-Rad CFX96™ Real-Time system (Bio-Rad, Hercules, CA, USA) with three biological replicates according to the manufacturer’s instructions. The PCR conditions were as follows: 95 °C for 30 s, 40 cycles of 95 °C for 5 s and 60 °C for 30 s and then a dissociation stage at 95 °C for 10 s, 65 °C for 5 s and 95 °C for 5 s. The Psubiquitin gene, AtActin2 gene and MdActin gene were used as internal controls to normalize the expression levels in P. suffruticosa, A. thaliana and Malus domestica, respectively. All gene-specific primers used in this study are shown in Table S1 [39,66]. The relative expression levels of genes were calculated using the 2−ΔΔCt method [72].
4.10. Sequence and Statistical Analysis
Multiple sequence alignment was performed using DNAMAN 8.0 software (Lynnon Biosoft, San Ramon, CA, USA). Homology search of sequences was carried out using the GenBank BLAST. Phylogenetic tree construction of sequences was performed using MEGA 5.0 software (Arizona State University, Tempe, AZ, USA) with the bootstrap values from 1000 replicates. Primers were designed using Primer Premier 5.0 software (PREMIER Biosoft International, Palo Alto, CA, USA). All experiments were repeated three times, and the data are expressed as the mean ± standard error. Variance analyses were performed using SPSS software ver. 17.0 (SPSS Inc., Chicago, IL, USA). p-values of < 0.05 were considered statistically significant.
5. Conclusions
In conclusion, two novel R2R3-MYB TFs, namely PsMYB114L and PsMYB12L, were successfully cloned from the petals of P. suffruticosa ‘Shima Nishiki’ and functionally characterized by heterologous expression in Arabidopsis and apple calli. Based on the above results, we preliminarily demonstrated the potential functional roles of PsMYB114L and PsMYB12L in regulating anthocyanin biosynthesis. These results provide a valuable resource for further understanding the molecular regulatory mechanisms of anthocyanin biosynthesis and accumulation in P. suffruticosa and breeding improved cultivars of P. suffruticosa with desirable flower colors in the future.
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/20/5/1055/s1. Table S1. Primers used in this study. Figure S1. Full-length cDNA amplification of the PsMYB114L and PsMYB12L genes. Figure S2. Flowers of P. suffruticosa ‘Shima Nishiki’ at five developmental stages. Figure S3. Construction of the recombinant expression vectors of pCAMBIA1304-PsMYB114L and pCAMBIA1304-PsMYB12L.
Author Contributions
X.Y. and L.Z. conceived and designed the research. X.Z. and Z.X. participated in the specific design of the study. X.Z. and Z.X. performed the experiments and the data analysis, and drafted the manuscript. M.Z., X.H., and S.Q. contributed analysis tools and helped analyze the data. All authors contributed to manuscript revision and approved the final version.
Funding
This research was funded by Forestry Science and Technology Innovation Projects of Shandong Province (Grant Number LYCX06-2018-32).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Zhang J.J., Shu Q.Y., Liu Z.A., Ren H.X., Wang L.S., De Keyser E. Two EST-derived marker systems for cultivar identification in tree peony. Plant Cell Rep. 2012;31:299–310. doi: 10.1007/s00299-011-1164-1. [DOI] [PubMed] [Google Scholar]
- 2.Shi Q.Q., Zhou L., Wang Y., Li K., Zheng B.Q., Miao K. Transcriptomic analysis of Paeonia delavayi wild population flowers to identify differentially expressed genes involved in purple-red and yellow petal pigmentation. PLoS ONE. 2015;10:e0135038. doi: 10.1371/journal.pone.0135038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhao D.Q., Tao J. Recent advances on the development and regulation of flower color in ornamental plants. Front. Plant Sci. 2015;6:261. doi: 10.3389/fpls.2015.00261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang X.P., Zhao M.Y., Guo J., Zhao L.Y., Xu Z.D. Anatomical and biochemical analyses reveal the mechanism of double-color formation in Paeonia suffruticosa ‘Shima Nishiki’. 3 Biotech. 2018;8:420. doi: 10.1007/s13205-018-1459-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noman A., Aqeel M., Deng J.M., Khalid N., Sanaullah T., He S.H. Biotechnological advancements for improving loral attributes in ornamental plants. Front. Plant Sci. 2017;8:530. doi: 10.3389/fpls.2017.00530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li C.H., Qiu J., Yang G.S., Huang S.R., Yin J.M. Isolation and characterization of a R2R3-MYB transcription factor gene related to anthocyanin biosynthesis in the spathes of Anthurium andraeanum (Hort.) Plant Cell Rep. 2016;35:2151–2165. doi: 10.1007/s00299-016-2025-8. [DOI] [PubMed] [Google Scholar]
- 7.Grotewold E. The genetics and biochemistry of floral pigments. Annu. Rev. Plant Biol. 2006;57:761–780. doi: 10.1146/annurev.arplant.57.032905.105248. [DOI] [PubMed] [Google Scholar]
- 8.Miyagawa N., Miyahara T., Okamoto M., Hirose Y., Sakaguchi K., Hatano S., Ozeki Y. Dihydroflavonol 4-reductase activity is associated with the intensity of flower colors in delphinium. Plant Biotechnol. 2015;32:249–255. doi: 10.5511/plantbiotechnology.15.0702b. [DOI] [Google Scholar]
- 9.Zhang J.L., Pan D.R., Zhou Y.F., Wang Z.C., Hua S.M., Hou L.L., Sui F.F. Cloning and expression of genes involved in anthocyanins synthesis in ornamental sunflower. Acta Hortic. Sin. 2009;36:73–80. [Google Scholar]
- 10.Chen S.M., Li C.H., Zhu X.R., Deng Y.M., Sun W., Wang L.S., Chen F.D., Zhang Z. The identification of flavonoids and the expression of genes of anthocyanin biosynthesis in the chrysanthemum flowers. Biol. Plant. 2012;56:458–464. doi: 10.1007/s10535-012-0069-3. [DOI] [Google Scholar]
- 11.Zhao D.Q., Tao J., Han C.X., Ge J.T. Flower color diversity revealed by differential expression of flavonoid biosynthetic genes and flavonoid accumulation in herbaceous peony (Paeonia lactiflora Pall.) Mol. Biol. Rep. 2012;39:11263–11275. doi: 10.1007/s11033-012-2036-7. [DOI] [PubMed] [Google Scholar]
- 12.Zhang C., Wang W.N., Wang Y.J., Gao S.L., Du D.N., Fu J.X., Dong L. Anthocyanin biosynthesis and accumulation in developing flowers of tree peony (Paeonia suffruticosa) ‘Luoyang Hong’. Postharvest Biol. Technol. 2014;97:11–22. doi: 10.1016/j.postharvbio.2014.05.019. [DOI] [Google Scholar]
- 13.Shi S.G., Yang M., Zhang M., Wang P., Kang Y.X., Liu J.J. Genome-wide transcriptome analysis of genes involved in flavonoid biosynthesis between red and white strains of Magnolia sprengeri pamp. BMC Genom. 2014;15:706. doi: 10.1186/1471-2164-15-706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsu C.C., Chen Y.Y., Tsai W.C., Chen W.H., Chen H.H. Three R2R3-MYB transcription factors regulate distinct floral pigmentation patterning in Phalaenopsis spp. Plant Physiol. 2015;168:175–191. doi: 10.1104/pp.114.254599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakatsuka T., Nishihara M., Mishiba K., Yamamura S. Temporal expression of flavonoid biosynthesis-related genes regulates flower pigmentation in gentian plants. Plant Sci. 2005;168:1309–1318. doi: 10.1016/j.plantsci.2005.01.009. [DOI] [Google Scholar]
- 16.Jaakola L. New insights into the regulation of anthocyanin biosynthesis in fruits. Trends Plant Sci. 2013;18:477–483. doi: 10.1016/j.tplants.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka Y., Nakamura N., Togami J. Altering flower color in transgenic plants by RNAi-mediated engineering of flavonoid biosynthetic pathway. Methods Mol. Biol. 2008;442:245–257. doi: 10.1007/978-1-59745-191-8_17. [DOI] [PubMed] [Google Scholar]
- 18.Petroni K., Tonelli C. Recent advances on the regulation of anthocyanin synthesis in reproductive organs. Plant Sci. 2011;181:219–229. doi: 10.1016/j.plantsci.2011.05.009. [DOI] [PubMed] [Google Scholar]
- 19.Tian J., Chen M.C., Zhang J., Li K.T., Song T.T., Zhang X., Yao Y.C. Characteristics of dihydrofavonol 4-reductase gene promoters from different leaf colored Malus crabapple cultivars. Hortic. Res. 2017;4:17070. doi: 10.1038/hortres.2017.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hichri I., Barrieu F., Bogs J., Kappel C., Delrot S., Lauvergeat V. Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J. Exp. Bot. 2011;62:2465–2483. doi: 10.1093/jxb/erq442. [DOI] [PubMed] [Google Scholar]
- 21.Huang Y.J., Song S., Allan A.C., Liu X.F., Yin X.R., Xu C.J., Chen K.S. Differential activation of anthocyanin biosynthesis in Arabidopsis and tobacco over-expressing an R2R3 MYB from Chinese bayberry. Plant Cell Tiss. Org. Cult. 2013;1113:491–499. doi: 10.1007/s11240-013-0291-5. [DOI] [Google Scholar]
- 22.Schaart J.G., Dubos C., Romero De La Fuente I., van Houwelingen A.M.M.L., de Vos R.C.H., Jonker H.H., Xu W.J., Routaboul J.M., Lipinec L., Bovy A.G. Identification and characterization of MYB-bHLH-WD40 regulatory complexes controlling proanthocyanidin biosynthesis in strawberry (Fragaria × ananassa) fruits. New Phytol. 2013;197:454–467. doi: 10.1111/nph.12017. [DOI] [PubMed] [Google Scholar]
- 23.Koes R., Verweij W., Quattrocchio F. Flavonoids: A colorful model for the regulation and evolution of biochemical pathways. Trends Plant Sci. 2005;10:236–242. doi: 10.1016/j.tplants.2005.03.002. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalez A., Zhao M.Z., Leavitt J.M., Lloyd A.M. Regulation of the anthocyanin biosynthetic pathway by the TTG1/bHLH/Myb transcriptional complex in Arabidopsis seedlings. Plant J. 2008;53:814–827. doi: 10.1111/j.1365-313X.2007.03373.x. [DOI] [PubMed] [Google Scholar]
- 25.Albert N.W., Davies K.M., Lewis D.H., Zhang H.B., Montefiori M., Brendolise C., Boase M.R., Ngo H., Jameson P.E., Schwinn K.E. A conserved network of transcriptional activators and repressors regulates anthocyanin pigmentation in eudicots. Plant Cell. 2014;24:962–980. doi: 10.1105/tpc.113.122069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou L., Wang Y., Peng Z. Molecular characterization and expression analysis of chalcone synthase gene during flower development in tree peony (Paeonia suffruticosa) Afr. J. Biotechnol. 2011;10:1275–1284. [Google Scholar]
- 27.Zhou L., Wang Y., Peng Z. Cloning and expression analysis of dihydroflavonol 4-reductase gene PsDFR1 from tree peony (Paeonia suffruticosa) Plant Physiol. J. 2011;47:885–892. [Google Scholar]
- 28.Zhou L., Wang Y., Ren L., Shi Q.Q., Zheng B.Q., Miao K., Guo X. Overexpression of PsCHI1, a homologue of the chalcone isomerase gene from tree peony (Paeonia suffruticosa), reduces the intensity of flower pigmentation in transgenic tobacco. Plant Cell Tiss. Org. Cult. 2014;116:285–295. doi: 10.1007/s11240-013-0403-2. [DOI] [Google Scholar]
- 29.Schwinn K., Venail J., Shang Y.J., Mackay S., Alm V., Butelli E., Oyama R., Bailey P., Davies K., Martin C. A small family of MYB-regulatory genes controls foral pigmentation intensity and patterning in the Genus Antirrhinum. Plant Cell. 2006;18:831–851. doi: 10.1105/tpc.105.039255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dubos C., Stracke R., Grotewold E., Weisshaar B., Martin C., Lepiniec L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010;15:573–581. doi: 10.1016/j.tplants.2010.06.005. [DOI] [PubMed] [Google Scholar]
- 31.Carey C.C., Strahle J.T., Selinger D.A., Chandler V.L. Mutations in the pale aleurone color1 regulatory gene of the Zea mays anthocyanin pathway have distinct phenotypes relative to the functionally similar TRANSPARENT TESTA GLABRA1 gene in Arabidopsis thaliana. Plant Cell. 2004;16:450–464. doi: 10.1105/tpc.018796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walker A.R., Lee E., Bogs J., McDavid D.A.J., Thomas M.R., Robinson S.P. White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 2007;49:772–785. doi: 10.1111/j.1365-313X.2006.02997.x. [DOI] [PubMed] [Google Scholar]
- 33.Tian J., Peng Z., Zhang J., Song T.T., Wan H.H., Zhang M.L., Yao Y.C. McMYB10 regulates coloration via activating McF3′H and later structural genes in ever-red leaf crabapple. Plant Biotechnol. J. 2015;13:948–961. doi: 10.1111/pbi.12331. [DOI] [PubMed] [Google Scholar]
- 34.Albert N.W., Lewis D.H., Zhang H.B., Schwinn K.E., Jameson P.E., Davies K.M. Members of an R2R3-MYB transcription factor family in Petunia are developmentally and environmentally regulated to control complex floral and vegetative pigmentation patterning. Plant J. 2011;65:771–784. doi: 10.1111/j.1365-313X.2010.04465.x. [DOI] [PubMed] [Google Scholar]
- 35.Zhu L., Shan H., Chen S.M., Jiang J.F., Gu C.S., Zhou G.Q., Chen Y., Song A.P., Chen F.D. The heterologous expression of the chrysanthemum R2R3-MYB transcription factor CmMYB1 alters lignin composition and represses flavonoid synthesis in Arabidopsis thaliana. PLoS ONE. 2013;8:e65680. doi: 10.1371/journal.pone.0065680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang Y.X., Zhang L., Gai S.P., Liu C.Y., Lu S. Cloning and expression analysis of the R2R3-PsMYB1 gene associated with bud dormancy during chilling treatment in the tree peony (Paeonia suffruticosa) Plant Growth Regul. 2015;75:667–676. doi: 10.1007/s10725-014-9968-y. [DOI] [Google Scholar]
- 37.Gao L.X., Yang H.X., Liu H.F., Yang J., Hu Y.H. Extensive transcriptome changes underlying the flower color intensity variation in Paeonia ostii. Front. Plant Sci. 2016;6:1205. doi: 10.3389/fpls.2015.01205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi Q.Q., Li L., Zhang X.X., Luo J.R., Li X., Zhai L.J., He L.X., Zhang Y.L. Biochemical and comparative transcriptomic analyses identify candidate genes related to variegation formation in Paeonia rockii. Molecules. 2017;22:1364. doi: 10.3390/molecules22081364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X.P., Zhao L.Y., Xu Z.D., Yu X.Y. Transcriptome sequencing of Paeonia suffruticosa ‘Shima Nishiki’to identify differentially expressed genes mediating double-color formation. Plant Physiol. Biochem. 2018;123:114–124. doi: 10.1016/j.plaphy.2017.12.009. [DOI] [PubMed] [Google Scholar]
- 40.Cao Y.L., Xie L.F., Ma Y.Y., Ren C.H., Xing M.Y., Fu Z.S., Wu X.Y., Yin X.R., Xu C.J., Li X. PpMYB15 and PpMYBF1 Transcription factors are involved in regulating flavonol biosynthesis in peach fruit. J. Agric. Food Chem. 2019;67:644–652. doi: 10.1021/acs.jafc.8b04810. [DOI] [PubMed] [Google Scholar]
- 41.Gong B.H., Yi J., Sui J.J., Wu J., Wu Z., Cheng Y.H., Wu C.Y., Liu C., Yi M.F. Cloning and expression analysis of LlHsfA1 from Lilium longiforum. Acta Hortic. Sin. 2014;41:1400–1408. [Google Scholar]
- 42.Ding K., Ma P.D., Jia Y.Y., Pei T.L., Bai Z.Q., Liang Z.S. Subcellular localization and transactivation analysis of three R2R3-MYB in Salvia miltiorrhiza Bunge. Acta Agric. Boreali-Occidentalis Sin. 2018;27:586–594. [Google Scholar]
- 43.Moreau C., Ambrose M.J., Turner L., Hill L., Noel Ellis T.H., Hofer J.M.I. The b gene of pea encodes a defective flavonoid 3′,5′-hydroxylase and confers pink flower color. Plant Physiol. 2012;159:759–768. doi: 10.1104/pp.112.197517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davies K.M., Albert N.W., Schwinn K.E. From landing lights to mimicry: The molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct. Plant Biol. 2012;39:619–638. doi: 10.1071/FP12195. [DOI] [PubMed] [Google Scholar]
- 45.Li X., Lu M., Tang D.Q., Shi Y.M. Composition of carotenoids and flavonoids in narcissus cultivars and their relationship with flower color. PLoS ONE. 2015;10:e0142074. doi: 10.1371/journal.pone.0142074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang Q., Hao R.J., Xu Z.D., Yang W.R., Wang J., Cheng T.R., Pan H.T., Zhang Q.X. Isolation and functional characterization of a R2R3-MYB regulator of Prunus mume anthocyanin biosynthetic pathway. Plant Cell. Tiss. Org. Cult. 2017;131:417–429. doi: 10.1007/s11240-017-1294-4. [DOI] [Google Scholar]
- 47.Feng K., Xu Z.S., Que F., Liu J.X., Wang F., Xiong A.S. An R2R3-MYB transcription factor, OjMYB1, functions in anthocyanin biosynthesis in Oenanthe javanica. Planta. 2018;247:301–315. doi: 10.1007/s00425-017-2783-8. [DOI] [PubMed] [Google Scholar]
- 48.Stracke R., Werber M., Weisshaar B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001;4:447–456. doi: 10.1016/S1369-5266(00)00199-0. [DOI] [PubMed] [Google Scholar]
- 49.Stracke R., Ishihara H., Huep G., Barsch A., Mehrtens F., Niehaus K., Weisshaar B. Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 2007;50:660–677. doi: 10.1111/j.1365-313X.2007.03078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Czemmel S., Stracke R., Weisshaar B., Cordon N., Harris N.N., Walker A.R., Robinson S.P., Bogs J. The grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol. 2009;151:1513–1530. doi: 10.1104/pp.109.142059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schwinn K.E., Ngo H., Kenel F., Brummell D.A., Albert N.W., McCallum J.A., Pither-Joyce M., Crowhurst R.N., Eady C., Davies K.M. The onion (Allium cepa L.) R2R3-MYB gene MYB1 regulates anthocyanin biosynthesis. Front. Plant Sci. 2016;7:1865. doi: 10.3389/fpls.2016.01865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Paz-Ares J., Wienand U., Peterson P.A., Saedler H. Molecular cloning of the c locus of Zea mays: A locus regulating the anthocyanin pathway. EMBO J. 1986;5:829–834. doi: 10.1002/j.1460-2075.1986.tb04291.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nesi N., Jond C., Debeaujon I., Caboche M., Lepiniec L. The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell. 2001;13:2099–2114. doi: 10.1105/TPC.010098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Terrier N., Torregrosa L., Ageorges A., Vialet S., Verriès C., Cheynier V., Romieu C. Ectopic expression of VvMybPA2 promotes proanthocyanidin biosynthesis in grapevine and suggests additional targets in the pathway. Plant Physiol. 2009;149:1028–1041. doi: 10.1104/pp.108.131862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gu Z.Y., Zhu J., Hao Q., Yuan Y.W., Duan Y.W., Men S.Q., Wang Q.Y., Hou Q.Z., Liu Z.A., Shu Q.Y., et al. A novel R2R3-MYB transcription factor contributes to petal blotch formation by regulating organ-specific expression of PsCHS in tree peony (Paeonia suffruticosa) Plant Cell Physiol. 2018:pcy232. doi: 10.1093/pcp/pcy232. [DOI] [PubMed] [Google Scholar]
- 56.Goff S.A., Cone K.C., Chandler V.L. Functional analysis of the transcriptional activator encoded by the maize B-gene: Evidence for a direct functional interaction between two classes of regulatory proteins. Genes Dev. 1992;6:864–875. doi: 10.1101/gad.6.5.864. [DOI] [PubMed] [Google Scholar]
- 57.Zimmermann I.M., Heim M.A., Weisshaar B., Uhrig J.F. Comprehensive identifcation of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J. 2004;40:22–34. doi: 10.1111/j.1365-313X.2004.02183.x. [DOI] [PubMed] [Google Scholar]
- 58.Wang N., Xu H.F., Jiang S.H., Zhang Z.Y., Lu N.L., Qiu H.R., Qu C.Z., Wang Y.C., Wu S.J., Chen X.S. MYB12 and MYB22 play essential roles in proanthocyanidin and flavonol synthesis in red-fleshed apple (Malus sieversii f. niedzwetzkyana) Plant J. 2017;90:276–292. doi: 10.1111/tpj.13487. [DOI] [PubMed] [Google Scholar]
- 59.Liu C.Y., Long J.M., Zhu K.J., Liu L.L., Yang W., Zhang H.Y., Li L., Xu Q., Deng X.X. Characterization of a Citrus R2R3-MYB transcription factor that regulates the flavonol and hydroxycinnamic acid biosynthesis. Sci. Rep. 2016;6:25352. doi: 10.1038/srep25352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Davies K.M., Schwinn K.E., Deroles S.C., Manson D.G., Lewis D.H., Bloor S.J., Bradley J.M. Enhancing anthocyanin production by altering competition for substrate between flavonol synthase and dihydroflavonol 4-reductase. Euphytica. 2003;131:259–268. doi: 10.1023/A:1024018729349. [DOI] [Google Scholar]
- 61.Wang F.B., Kong W.L., Wong G., Fu L.F., Peng R.H., Li Z.J., Yao Q.H. AtMYB12 regulates flavonoids accumulation and abiotic stress tolerance in transgenic Arabidopsis thaliana. Mol. Genet. Genom. 2016;291:1545–1559. doi: 10.1007/s00438-016-1203-2. [DOI] [PubMed] [Google Scholar]
- 62.Mehrtens F., Kranz H., Bednarek P., Weisshaar B. The Arabidopsis transcription factor MYB12 is a flavonol-specific regulator of phenylpropanoid biosynthesis. Plant Physiol. 2005;138:1083–1096. doi: 10.1104/pp.104.058032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang W.J., Khaldun A.B.M., Chen J.J., Zhang C.J., Lv H.Y., Yuan L., Wang Y. A R2R3-MYB transcription factor regulates the flavonol biosynthetic pathway in a traditional Chinese medicinal plant, Epimedium sagittatum. Front. Plant Sci. 2016;7:1089. doi: 10.3389/fpls.2016.01089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Luo J., Butelli E., Hill L., Parr A., Niggeweg R., Bailey P., Weisshaar B., Martin C. AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: Expression in fruit results in very high levels of both types of polyphenol. Plant J. 2008;56:316–326. doi: 10.1111/j.1365-313X.2008.03597.x. [DOI] [PubMed] [Google Scholar]
- 65.Ren X.X., Wang S.L., Xue J.Q., Zhu F.Y., Liu C.J., Zhang X.X. Molecular cloning and expression analysis of cryptochrome gene PsCRY2 in tree peony. Acta Hortic. Sin. 2015;42:2229–2236. doi: 10.1016/j.hpj.2016.10.003. [DOI] [Google Scholar]
- 66.An J.P., An X.H., Yao J.F., Wang X.N., You C.X., Wang X.F., Hao Y.J. BTB protein MdBT2 inhibits anthocyanin and proanthocyanidin biosynthesis by triggering MdMYB9 degradation in apple. Tree Physiol. 2018;38:1578–1587. doi: 10.1093/treephys/tpy063. [DOI] [PubMed] [Google Scholar]
- 67.Verma A., Lee C., Morriss S., Odu F., Kenning C., Rizzo N., Spollen W.G., Lin M., McRae A.G., Givan S.A., et al. The novel cyst nematode effector protein 30D08 targets host nuclear functions to alter gene expression in feeding sites. New Phytol. 2018;219:697–713. doi: 10.1111/nph.15179. [DOI] [PubMed] [Google Scholar]
- 68.Clough S.J., Bent A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 69.An J.P., Liu X., Li H.H., You C.X., Wang X.F., Hao Y.J. Apple RING E3 ligase MdMIEL1 inhibits anthocyanin accumulation by ubiquitinating and degrading MdMYB1 protein. Plant Cell Physiol. 2017;58:1953–1962. doi: 10.1093/pcp/pcx129. [DOI] [PubMed] [Google Scholar]
- 70.An J.P., Qu F.J., Yao J.F., Wang X.N., You C.X., Wang X.F., Hao Y.J. The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic. Res. 2017;4:17023. doi: 10.1038/hortres.2017.56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zhao D.Q., Jiang Y., Ning C.L., Meng J.S., Lin S.S., Ding W., Tao J. Transcriptome sequencing of a chimaera reveals coordinated expression of anthocyanin biosynthetic genes mediating yellow formation in herbaceous peony (Paeonia lactilora Pall.) BMC Genom. 2014;15:689. doi: 10.1186/1471-2164-15-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.