Abstract
Aldehyde Oxidase (AO) is an enzyme involved in the metabolism of aldehydes and N-containing heterocyclic compounds. Many drug compounds contain heterocyclic moieties, and AO metabolism has lead to failure of several late-stage drug candidates. Therefore, it is important to take AO-mediated metabolism into account early in the drug discovery process, and thus, to have fast and reliable models to predict the site of metabolism (SOM). We have collected a dataset of 78 substrates of human AO with a total of 89 SOMs and 347 non-SOMs and determined atomic descriptors for each compound. The descriptors comprise NMR shielding and ESP charges from density functional theory (DFT), NMR chemical shift from ChemBioDraw, and Gasteiger charges from RDKit. Additionally, atomic accessibility was considered using 2D-SASA and relative span descriptors from SMARTCyp. Finally, stability of the product, the metabolite, was determined with DFT and also used as a descriptor. All descriptors have AUC larger than 0.75. In particular, descriptors related to the chemical shielding and chemical shift (AUC = 0.96) and ESP charges (AUC = 0.96) proved to be good descriptors. We recommend two simple methods to identify the SOM for a given molecule: 1) use ChemBioDraw to calculate the chemical shift or 2) calculate ESP charges or chemical shift using DFT. The first approach is fast but somewhat difficult to automate, while the second is more time-consuming, but can easily be automated. The two methods predict correctly 93% and 91%, respectively, of the 89 experimentally observed SOMs.
Keywords: Aldehyde oxidase, Drug metabolism, Sites of metabolism, Density functional theory, Chemical shielding, ESP charges, Solvent accessible surface area
Graphical Abstract
1. Introduction
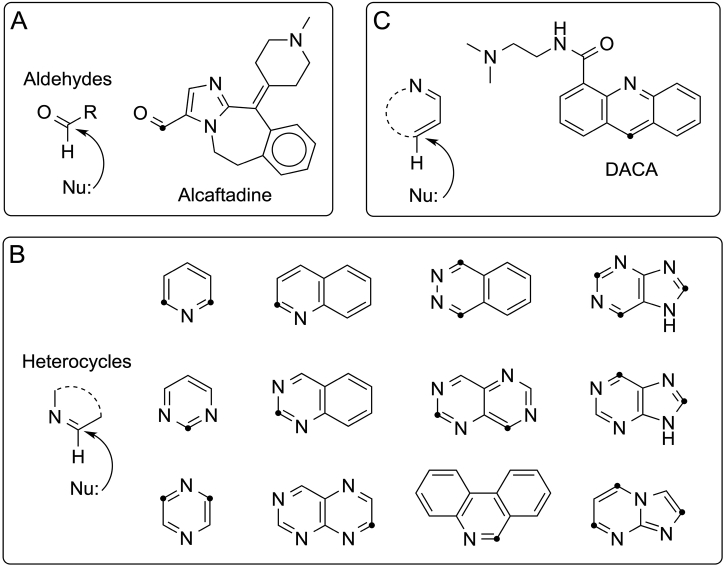
Aldehyde Oxidase (AO) enzymes metabolize different chemical functionalities, including aldehydes although this chemical fragment is not often present in drug compounds (cf. Fig. 1A) [1]. Aldehydes can be a result of a biotransformation by other drug metabolizing enzymes, such as the cytochrome P450s (CYPs), and can be subsequently oxidized to a carboxylic acid by AO. However, AO plays an important role in the oxidation of aromatic azaheterocyclic groups to oxoheterocycles, e.g. of pyridines, diazines, purines or benzimidazoles (cf. Fig. 1B). AO can also reduce N- and S-oxides and hydrolyze amides [2,3]. Since chemical groups like these ones are often present in drug-like compounds, e.g. because azaheterocyclic rings have been introduced to avoid CYP metabolism, there has lately been a lot of attention to AO metabolism since a number of compounds have been discontinued in clinical trials due to too rapid clearance or toxicity [1,[4], [5], [6], [7]]. Thus, it is highly relevant to be able to predict AO metabolism. One approach, frequently used by many predictive methods (SMARTCyp [8,9], StarDrop [10], FAME2 [11]), is to predict where a compound potentially will be metabolized if being a substrate (site-of-metabolism, SOM) and, thereby, indirectly also identify the possible metabolite(s).
Fig. 1.
The mechanism of AO mediated metabolism involves a nucleophilic attack on the electron-deficient carbon atom. Potential SOMs are marked by a dot. A: Alcaftadine is one of the few registered drug compounds being an aldehyde; B: Examples on heterocyclic rings systems present in drug compounds. See Fig. S1 in Supplementary Material for the structures of the actual drug compounds; C: DACA, an example on an unusual AO substrate.
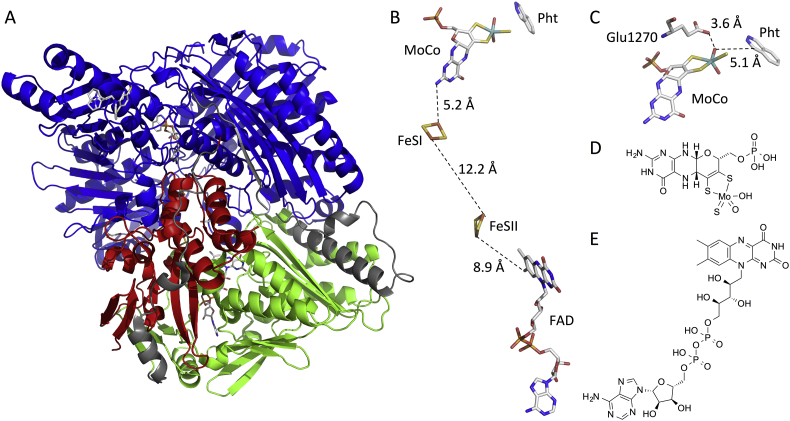
Recently, three human AO structures have been determined (PDB entries 5EPG [12], 4UHW and 4UHX [13]), which allow a more detailed analysis of the molecular processes associated with AO metabolism. The AO enzyme is a 150 kDa protein comprising three domains, a small N-terminal domain containing two [2Fe-2S] centers, a reductive flavin domain and an oxidative molybdenum domain (cf. Fig. 2). The [2Fe-2S] centers are probably responsible for the electron flow from the flavin to molybdenum sites [[12], [13], [14]]. Oxidation of N-containing heterocycles takes place at the molybdenum site, where the molybdenum cofactor (MoCo), activated by a glutamate residue (Glu1270), acts as a nucleophile attacking an electron-deficient carbon atom next to the hetero-atom (cf. Fig. 2) [[15], [16], [17]]. The nucleophilic attack is rate-limiting and has the lowest activation barrier on electron deficient C atoms [17].
Fig. 2.
A: Ribbon representation of the human AO crystal structure (PDF entry 4UHX [18]). The threee domains are: N terminal or 2Fe-2S domain (Ala4-Lys166, red), FAD domain (Gln231-Asp538, green) and MoCo domain (Asp555-Val1336, blue). The linker regions between the domains are colored grey. B: Colour-coded stick models of prostetic groups (MoCo, FeSI, FeSII and FAD) and phthalazine (Pht) and distances between them. B: Close-up showing the contacts between MoCo, Glu1270 and phthalazine. D and E: 2D structures of MoCo and FAD. Colour coding and domain definitions adapted from Coelho et al. [13,18].
Only a few methods for prediction of AO metabolism have been reported. Torres et al. used density functional theory (DFT) methods to calculate the tetrahedral intermediate for the reaction leading to potential metabolites, and in more than 90% of the cases, the intermediate with the lowest energy relative to the initial substrate corresponded to the experimentally found metabolite [19]. Jones et al. calculated the relative heats of formation by DFT methods for eight AO substrates and in combination with a steric model for the accessibility to the AO active site, they were able to develop a model for in vivo as well as in vitro clearance [16]. Xu et al. used the energy and accessibility descriptors by Jones et al. to develop a decision three model for identifying and ranking possible SOMs in compounds with multiple SOMs [20]. Montefiori et al. studied different reaction mechanisms for AO mediated metabolism of a series of 4-quinazolinones by DFT methods, and showed that the lowest activation energy in all cases corresponded to the experimentally observed SOMs [17].
All these methods for prediction of SOM are based on DFT calculations and, accordingly, time-consuming for larger compounds and/or series of compounds. Therefore, to avoid the time-consuming DFT calculations to identify the transition states, it would be desirable to use other descriptors as predictor for whether the reaction would occur [2,19]. Montefiori et al. also showed that ESP charges correlate with the computed activation energies which makes it an attractive descriptor for the prediction of site of metabolism (SOM) [17]. Thus, it has been our aim to develop faster methods for accurate AO SOM prediction, fast enough to be useful in drug development projects in the pharmaceutical industry.
In this work, we have developed models for prediction of the AO mediated SOMs in heterocyclic compounds containing the -CH=N- moiety (cf. Fig. 1B) using electronic descriptors of substrate atoms. We used the two DFT calculated descriptors from Montefiori et al. with the best correlation to the activation energies, i.e. the stability of the generated product and the ESP charges [17]. In addition, we used NMR shielding constants determined at the DFT level and chemical shifts from empirical models, since these also reflect the atomic electron density. To address the accessibility, descriptors like those from SMARTCyp that are based on 2D structures and thus fast to determine were used [9,21]. A data set comprising 78 compounds with a total of 89 SOMs and 347 non-SOMs were collected to evaluate the performance of our models.
2. Results and Discussion
2.1. Atomic Descriptors
A set of atomic descriptors related to the charge distribution, stability of the product, accessibility or whether an aromatic or sp2-hybridized atom is next to an aromatic or sp2-hybridized N atom were determined. For each descriptor, the area under the curve (AUC) has been calculated in two different ways: 1) considering all the atoms of the dataset, checking if both the SOMs and non-SOMs were correctly predicted, or 2) per molecule, checking if at least one of the SOMs was in the top n rank with n being equal to the number of SOMs in the molecule. In the first case, the AUC values calculated considering all the atoms are biased by the large number of non-SOMs relative to SOMs (347 and 89, respectively) and may not be a relevant measure for the ability to predict the SOMs. In the second case, a correct prediction requires that the predicted SOM is identical to the experimentally observed SOM if the molecule only has one experimentally observed SOM, and is in the top two if the molecule has two experimentally observed SOMs and so on. This way to determine the performance has previously been used by us [8,9] as well as by other groups [10,11,22,23] in evaluating and comparing methods for predicting cytochrome P450 mediated SOMs.
The descriptors with the best AUC are the NMR shielding and chemical shift and the ESP charges (AUC = 0.96, cf. Table 1), which can be related to the charge at the potential SOMs and thus, the reactivity of the molecule towards AO. The atomic accessibility, on the other hand, is not directly predictive for this set of compounds. The limited effect of atomic accessibility on the prediction of the SOM may be related to the fact that the substrates generally are small. For larger substrates, these descriptors may be more important. The AUC for the top N position in the molecule follows the same trend as the one for all the atoms, with a few exceptions. The chemical shift calculated with ChemBioDraw (AUC = 0.90) is slightly better than DFT calculated chemical shielding and ESP charges (cf. Table 1). The ESP charges, however, are more predictive than the empirical Gasteiger charges. The "C-next-to-N" rule-of-thumb with an aromatic C atom next to an N atom in a heterocyclic ring has the best performance in the AUC calculated per molecule. This is due to the fact that this is a binary descriptor, and as such the atom is either classified as a SOM or not and, accordingly, there is no ranking of the atoms. This is also seen in Table 2 where the true positive rate (sensitivity) for C-next-to-N is higher (0.99) than for the chemical shielding, shift and ESP charges (0.89–0.91). However, the true negative rate (specificity) is lower considering the aromatic C atoms next to N (0.88). It is also reflected in the accuracy (0.90 for C-next-to-N compared to 0.95–0.96 for shielding, shift and ESP charges) and particularly the low precision of 0.65 due to a large false positive rate. Thus, although the correct SOM is almost always identified considering the presence of an aromatic C atom next to an N atom, it is difficult to choose the correct one if there are more of the same type of C atom. On the other hand, descriptors like chemical shielding; chemical shift and ESP charges that showed only a slightly lower performance in the AUC compared to the C-next-to-N rule-of-thumb are more predictive considering specificity, precision and accuracy.
Table 1.
AUC values for the different descriptors for our dataset comprising 78 substrates (see Fig. S1 in Supplementary Material for 2D structures of the compounds).
| AUC (all values)i | AUC (top n)j | |
|---|---|---|
| Chemical shieldinga | 0.96 | 0.88 |
| Chemical shiftb | 0.96 | 0.90 |
| ESP chargesc | 0.96 | 0.88 |
| Gasteiger chargesd | 0.91 | 0.67 |
| Product stabilitye | 0.79 | 0.59 |
| Relative spanf | 0.62 | 0.22 |
| 2D-SASAg | 0.80 | 0.72 |
| C-next-to-Nh | 0.93 | 0.99 |
Determined at the B3LYP/6-31G* level.
Determined with ChemBioDraw.
Determined at the B3LYP/6-31G* level.
Determined with RDKit.
Determined at the B3LYP/6-31G* level.
From SMARTCyp.
From SMARTCyp.
Aromatic C atom next to N atom.
AUC (all values) evaluates the descriptor with both the SOMs and the non SOMs, thus giving us 89 (SOMs) and 347 (non SOMs) data points to evaluate the performance.
AUC (top n) evaluates the SOMs (thus 89 SOMs) where n refers to the number of SOMs in each molecule (n = 1 or 2).
Table 2.
Classification of SOMs. The total number of SOMs and non-SOMs is 89 and 347, respectively.
| TPa | FPb | Sensc | Specd | Prece | Accf | |
|---|---|---|---|---|---|---|
| Chemical shielding | 79 | 9 | 0.90 | 0.97 | 0.90 | 0.96 |
| Chemical shift | 80 | 8 | 0.91 | 0.98 | 0.91 | 0.96 |
| ESP charges | 78 | 10 | 0.89 | 0.97 | 0.89 | 0.95 |
| Gasteiger charges | 60 | 28 | 0.68 | 0.92 | 0.68 | 0.87 |
| Product stability | 55 | 33 | 0.63 | 0.91 | 0.63 | 0.85 |
| Relative span | 20 | 68 | 0.23 | 0.81 | 0.23 | 0.69 |
| 2D-SASA | 64 | 24 | 0.73 | 0.93 | 0.73 | 0.89 |
| C-next-to-N | 87 | 47 | 0.99 | 0.88 | 0.65 | 0.90 |
True positives (TP: atoms correctly classified as SOMs).
False positives (FP: atoms wrongly classified as SOMs).
Sensitivity (Sens): TP/(TP + FN).
Specificity (Spec): TN/(TN + FP).
Precision (Prec): TP/(TP + FP).
Accuracy (Acc): (TP + TN)/(TP + FP + FN + TN).
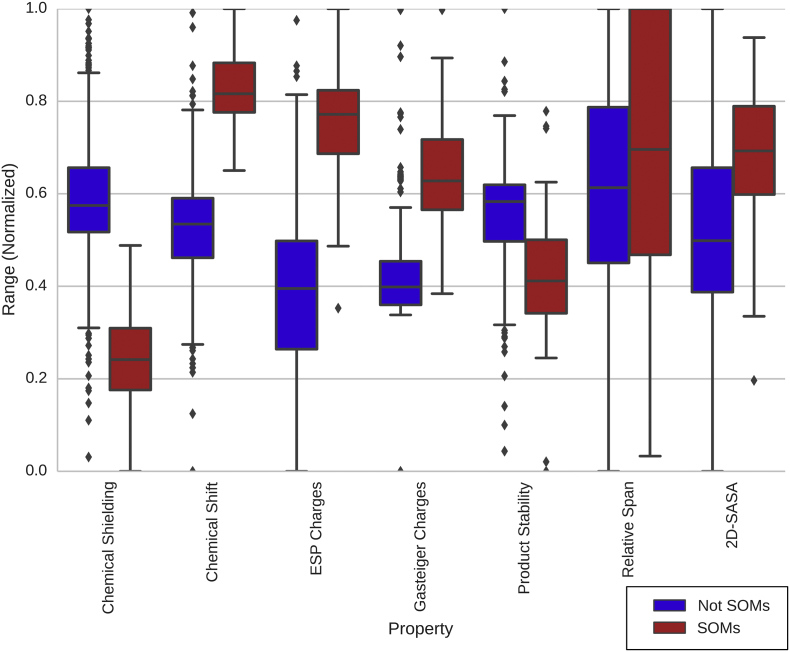
In Fig. 3, it can be seen that the SOMs have large chemical shifts and ESP charges and thus low chemical shielding. In addition, there is a clear separation between these properties of the SOMs and non-SOMs with only a few outliers. The SOMs are characterized by positive ESP charges with 50% of the data in the range of 0.27–0.47 (cf. Table S1). This agrees well with the chemical shielding of the SOMs mostly having low values in the range of 43–51, or high chemical shifts from 122 to 132 (Table S1). The accessibility descriptors have a less clear separation between the SOMs and non-SOMs as also indicated by the statistical data in Tables 1 and 2. Nevertheless, there is a tendency that the SOMs are more accessible than the non-SOMs.
Fig. 3.
Box plot of calculated descriptors. All descriptors have been normalized between 0 and 1 for easier comparison using the following formula: Normalized value = (Value - min of all the descriptors)/(max of all descriptors - min of all descriptors). The binary C-next-to-N descriptor has been omitted from the figure. The absolute values are found in Table S1 in Supplementary Material.
2.2. Examples of AO SOMs
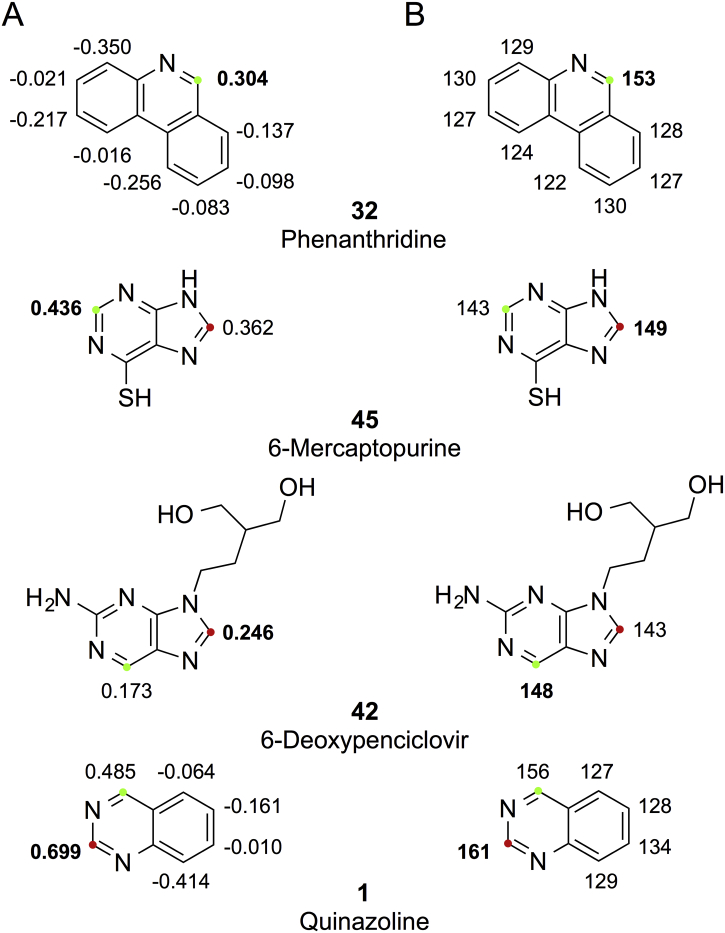
In Fig. 4, we report chemical shifts and ESP charges for four representative molecules to compare the two best methods. Considering either chemical shifts or ESP charges, the most electrophilic carbon atoms in these compounds are unambiguously identified as the carbon atoms next to the nitrogen atoms and, thereby, as the potential SOMs for AO mediated metabolism. Both methods predict the correct carbon atom as the primary SOM for the majority of the 78 compounds (e.g. compound 32 in Fig. 4). Several of the compounds contain two experimentally observed SOMs, and we see a few examples where one or both methods have difficulties identifying the correct relative ranking of the primary and secondary SOMs, although both the primary and secondary SOMs are clearly distinguishable from the remaining C atoms by both methods. For example, the ranking of the primary and secondary SOM is opposite for 6-mercaptopurine (45) and 6-deoxypenciclovir (42); and for quinazoline (1) both methods predict the secondary SOM as primary SOM (Fig. 4).
Fig. 4.
ESP charges (A) and chemical shifts (B) for four representative molecules. The values for predicted primary SOMs are in bold, the experimentally observed primary and secondary SOMs are shown by green and red dots, respectively.
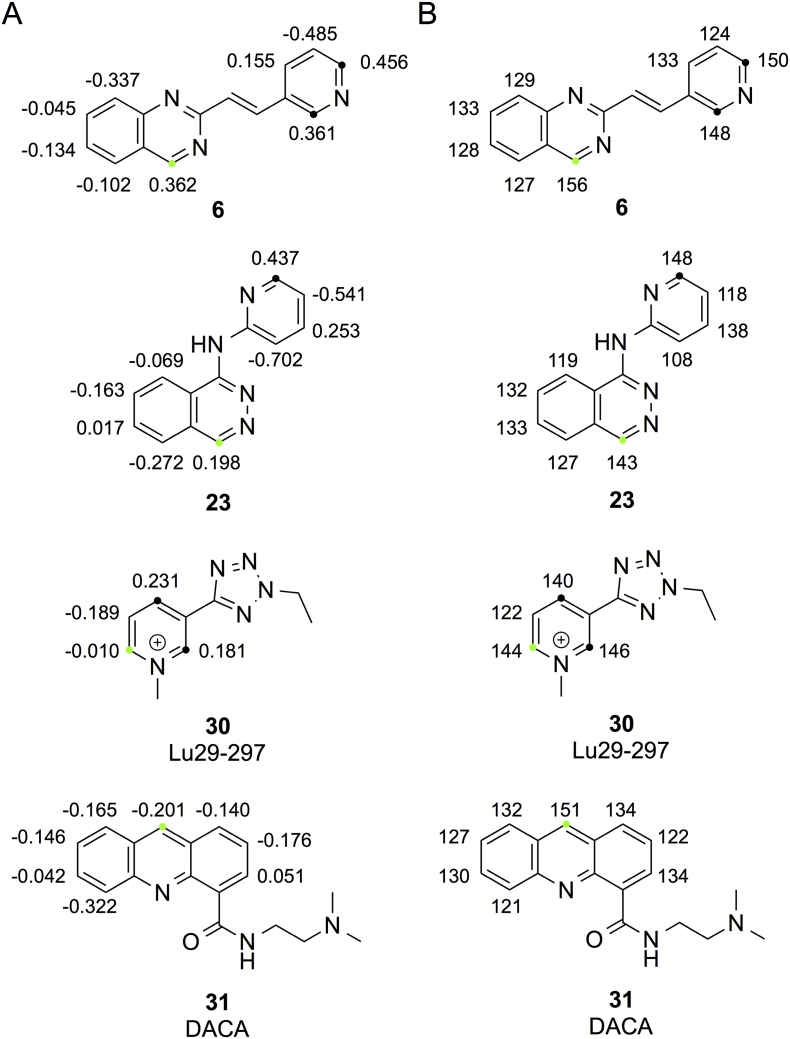
We also see a few examples where one or both of the methods predict the primary SOM to correspond to a site not observed experimentally. For example, in compound 6 (cf. Fig. 5) chemical shifts correctly predict SOM to be in the quinazoline moiety, whereas ESP charges predict the pyridine moiety to be the most reactive [24]. In compound 23 (cf. Fig. 5) the experimental SOM is in the phthalazine moiety, whereas both chemical shifts and ESP charges indicate that the six-membered ring is more prone to nucleophilic attack [24].
Fig. 5.
ESP charges (A) and chemical shifts (B) of 6, 23, 30 and 31. Experimentally observed SOMs are marked by green dots, whereas SOMs not observed experimentally are marked by black dots.
The reasons for the discrepancies between the experimentally identified and predicted SOMs and between the two predictive methods may have several reasons. We have generated one likely 3D conformation for each compound, but we have not performed a conformational analysis of the compounds and, accordingly, conformational effects may have an effect on the predicted values of the chemical shifts and ESP charges. The potential SOMs in the molecules are not equally accessible (cf. Table S2, Supplementary Material), but we have not been able to establish a correlation between reactivity and accessibility for the present dataset as we have previously done for CYP mediated metabolism [25,26]. The effect of the protein, i.e. the shape of the active site, has not been considered and the generated 3D structures may therefore not be identical to the bioactive conformations of the compounds. Coelho et al. noticed increased mobility (altered orientations or poor electron density) of several hydrophobic residues at the entrance to the active site when phtalazine binds to AO and suggested that the binding process involves an induced-fit mechanism [13].
Finally, a few atypical AO substrates present in our dataset should be mentioned. Lu29-297 (30), a CYP metabolite of alvameline, containing a positively charged pyridinium ring, is further metabolized by AO at the C atom next to the positive nitrogen atom and opposite to the tetrazole substituent (cf. Fig. 5) [27]. This compound is the only charged compound in our dataset and, accordingly, we do not have enough basis to warrant whether charged compounds should be treated independently or analogously to the neutral compounds, as we have done in this study. DACA (31) (cf. Figs. 1C and 5) is another example on an atypical AO substrate by being metabolized at C9 to the 9(10H)-acridone metabolite [28]. We have previously by DFT calculations shown that metabolism at C9 is associated with the lowest transition state, although C9 is not placed next to a N atom [17]. It is notable that considering the chemical shift values, C9 is correctly identified as the preferred SOM.
3. Conclusions
We have collected a set of AO substrates consisting of 78 substrates with 89 SOMs and 347 non-SOMs. For this dataset, a set of atomic descriptors related to the charge distribution or accessibility was determined. In particular, descriptors related to the chemical shift and shielding (AUC = 0.96) and ESP charges (AUC = 0.96) prove to be good descriptors. The rule-of-thumb of a C atom next to a N atom in a heterocyclic ring is good at predicting SOMs (AUC = 0.93), but has a large false positive rate. The stability of the products shows a weaker performance (AUC = 0.79). For this set of compounds, the atomic accessibility descriptors do not yield high predictions rates, probably because most of the compounds are relatively small. For larger compounds, it could be important to include the accessibility, e.g. in case of sterically hindered atoms. We propose two simple methods to identify the SOM for a given compound: (1) Use ChemBioDraw to calculate the chemical shift which is very fast and reliable for compounds with good parameters, but somewhat difficult to automate. (2) Calculate the ESP charges or NMR chemical shielding using a DFT program. This is more time-consuming, but easier to automate and independent of whether empirical parameters exist. ChemBioDraw-calculated chemical shifts and DFT-calculated ESP charges predict correctly 83 (93%) and 81 (91%), respectively, of the 89 experimentally observed SOMs.
4. Methods
A dataset comprising 78 substrates of human AO with experimentally determined SOMs were collected from the literature. Only compounds tested against human AO (hAO) were included (see Fig. S1 in Supplementary Material and Montefiori.sdf for 2D and 3D structures, respectively). The SOMs were taken from the literature and references to the original papers can be found in the Supplementary Material (Montifiori.csv). In total, the dataset contains 89 SOMs and 347 non-SOMs where the potential SOMs are all the aromatic C atoms. Aldehydes, amides or N- and S-oxides are not included.
The KNIME Analytics Platform (version 2.12.2; www.knime.org) [29] with the Schrodinger (www.schrodinger.com) and RDKit nodes (Open-source cheminformatics, http://www.rdkit.org) was used to optimize structures, determine the atomic descriptors and structures of the possible products.
The structure of substrates and possible products were prepared with LigPrep [30,31] (Epik version 3.5014), followed by a MCMM conformational search with MacroModel using the OPLS3 force field [[32], [33], [34], [35]]. Each structure was subsequently optimized with Jaguar [36], using the B3LYP/6-31G** basis set with the exception of bromine for which we used LACVP. The DFT optimized structures were used to calculate the atomic properties (chemical shielding, ESP charges). The energies of the substrates and products were extracted and used to determine product stabilities. Gasteiger charges [37] were determined using the RDKit node. The 2D-SASA and span2end values were determined with SMARTCyp [8,9]. ChemBioDraw [38] was used to determine the chemical shift for the carbon atoms in the compounds. Fig. S1 was produced by the Mona program [39].
Acknowledgements
This work was supported by the European Union via the Advanced Research Infrastructure for Analytical Research in ADME profiling (ARIADME) [607517].
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.csbj.2019.03.003.
Appendix A. Supplementary data
Supplementary material 1
CSV file with calculated data for SOMs and non-SOMs in the 78 compounds.
SDF file with coordinates of the 78 compounds and experimental SOMs.
References
- 1.Pryde D.C., Dalvie D., Hu Q.Y., Jones P., Obach R.S., Tran T.D. Aldehyde oxidase: an enzyme of emerging importance in drug discovery. J Med Chem. 2010;53:8441–8460. doi: 10.1021/jm100888d. [DOI] [PubMed] [Google Scholar]
- 2.Lepri S., Ceccarelli M., Milani N., Tortorella S., Cucco A., Valeri A. Structure–metabolism relationships in human-AOX: chemical insights from a large database of aza-aromatic and amide compounds. Proc Natl Acad Sci U S A. 2017;114 doi: 10.1073/pnas.1618881114. [E3178-E87] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sodhi J.K., Wong S., Kirkpatrick D.S., Liu L., Khojasteh S.C., Hop C.E. A novel reaction mediated by human aldehyde oxidase: amide hydrolysis of GDC-0834. Drug Metab Dispos. 2015;43:908–915. doi: 10.1124/dmd.114.061804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Akabane T., Tanaka K., Irie M., Terashita S., Teramura T. Case report of extensive metabolism by aldehyde oxidase in humans: pharmacokinetics and metabolite profile of FK3453 in rats, dogs, and humans. Xenobiotica. 2011;41:372–384. doi: 10.3109/00498254.2010.549970. [DOI] [PubMed] [Google Scholar]
- 5.Infante J.R., Rugg T., Gordon M., Rooney I., Rosen L., Zeh K. Unexpected renal toxicity associated with SGX523, a small molecule inhibitor of MET. Invest New Drugs. 2013;31:363–369. doi: 10.1007/s10637-012-9823-9. [DOI] [PubMed] [Google Scholar]
- 6.Lolkema M.P., Bohets H.H., Arkenau H.-T., Lampo A., Barale E., de Jonge M.J.A. The c-met tyrosine kinase inhibitor JNJ-38877605 causes renal toxicity through species-specific insoluble metabolite formation. Clin Cancer Res. 2015;21:2297–2304. doi: 10.1158/1078-0432.CCR-14-3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang X., Liu H.H., Weller P., Zheng M., Tao W., Wang J. In silico and in vitro pharmacogenetics: aldehyde oxidase rapidly metabolizes a p38 kinase inhibitor. Pharmacogenomics J. 2011;11:15–24. doi: 10.1038/tpj.2010.8. [DOI] [PubMed] [Google Scholar]
- 8.Rydberg P., Gloriam D.E., Olsen L. The SMARTCyp cytochrome P450 metabolism prediction server. Bioinformatics. 2010;26:2988–2989. doi: 10.1093/bioinformatics/btq584. [DOI] [PubMed] [Google Scholar]
- 9.Rydberg P., Gloriam D.E., Zaretzki J., Breneman C., Olsen L. SMARTCyp: a 2D method for prediction of cytochrome P450-mediated drug metabolism. ACS Med Chem Lett. 2010;1:96–100. doi: 10.1021/ml100016x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tyzack J.D., Hunt P.A., Segall M.D. Predicting regioselectivity and lability of cytochrome P450 metabolism using quantum mechanical simulations. J Chem Inf Model. 2016;56:2180–2193. doi: 10.1021/acs.jcim.6b00233. [DOI] [PubMed] [Google Scholar]
- 11.Sicho M., Kops C.D., Stork C., Svozil D., Kirchmair J. FAME 2: simple and effective machine learning model of cytochrome P450 Regioselectivity. J Chem Inf Model. 2017;57:1832–1846. doi: 10.1021/acs.jcim.7b00250. [DOI] [PubMed] [Google Scholar]
- 12.Foti A., Hartmann T., Coelho C., Santos-Silva T., Romão M.J., Leimkühler S. Optimization of the expression of human aldehyde oxidase for investigations of single-nucleotide polymorphisms. Drug Metab Dispos. 2016;44:1277. doi: 10.1124/dmd.115.068395. [DOI] [PubMed] [Google Scholar]
- 13.Coelho C., Foti A., Hartmann T., Santos-Silva T., Leimkuehler S., Romao M.J. Structural insights into xenobiotic and inhibitor binding to human aldehyde oxidase. Nat Chem Biol. 2015;11:779–783. doi: 10.1038/nchembio.1895. [DOI] [PubMed] [Google Scholar]
- 14.Terao M., Romao M.J., Leimkuehler S., Bolis M., Fratelli M., Coelho C. Structure and function of mammalian aldehyde oxidases. Arch Toxicol. 2016;90:753–780. doi: 10.1007/s00204-016-1683-1. [DOI] [PubMed] [Google Scholar]
- 15.Alfaro J.F., Jones J.P. Studies on the mechanism of aldehyde oxidase and xanthine oxidase. J Org Chem. 2008;73:9469–9472. doi: 10.1021/jo801053u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones J.P., Korzekwa K.R. Predicting intrinsic clearance for drugs and drug candidates metabolized by aldehyde oxidase. Mol Pharm. 2013;10:1262–1268. doi: 10.1021/mp300568r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montefiori M., Jørgensen F.S., Olsen L. Aldehyde oxidase: reaction mechanism and prediction of site of metabolism. ACS Omega. 2017;2:4237–4244. doi: 10.1021/acsomega.7b00658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coelho C., Mahro M., Trincao J., Carvalho A.T., Ramos M.J., Terao M. The first mammalian aldehyde oxidase crystal structure: insights into substrate specificity. J Biol Chem. 2012;287:40690–40702. doi: 10.1074/jbc.M112.390419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Torres R.A., Korzekwa K.R., McMasters D.R., Fandozzi C.M., Jones J.P. Use of density functional calculations to predict the regioselectivity of drugs and molecules metabolized by aldehyde oxidase. J Med Chem. 2007;50:4642–4647. doi: 10.1021/jm0703690. [DOI] [PubMed] [Google Scholar]
- 20.Xu Y., Li L., Wang Y., Xing J., Zhou L., Zhong D. Aldehyde oxidase mediated metabolism in drug-like molecules: a combined computational and experimental study. J Med Chem. 2017;60:2973–2982. doi: 10.1021/acs.jmedchem.7b00019. [DOI] [PubMed] [Google Scholar]
- 21.Rydberg P., Rostkowski M., Gloriam D.E., Olsen L. The contribution of atom accessibility to site of metabolism models for cytochromes P450. Mol Pharm. 2013;10:1216–1223. doi: 10.1021/mp3005116. [DOI] [PubMed] [Google Scholar]
- 22.Ford K.A., Ryslik G., Sodhi J., Halladay J., Diaz D., Dambach D. Computational predictions of the site of metabolism of cytochrome P450 2D6 substrates: comparative analysis, molecular docking, bioactivation and toxicological implications. Drug Metab Rev. 2015;47:291–319. doi: 10.3109/03602532.2015.1047026. [DOI] [PubMed] [Google Scholar]
- 23.Zaretzki J., Matlock M., Swamidass S.J. XenoSite: accurately predicting CYP-mediated sites of metabolism with neural networks. J Chem Inf Model. 2013;53:3373–3383. doi: 10.1021/ci400518g. [DOI] [PubMed] [Google Scholar]
- 24.Beedham C., Critchley D.J.P., Rance D.J. Substrate specificity of human liver aldehyde oxidase toward substituted quinazolines and phthalazines: a comparison with hepatic enzyme from Guinea pig, rabbit, and baboon. Arch Biochem Biophys. 1995;319:481–490. doi: 10.1006/abbi.1995.1320. [DOI] [PubMed] [Google Scholar]
- 25.Bonomo S., Jørgensen F.S., Olsen L. Dissecting the cytochrome P450 1A2- and 3A4-mediated metabolism of Aflatoxin B1 in ligand and protein contributions. Chemistry. 2017;23:2884–2893. doi: 10.1002/chem.201605094. [DOI] [PubMed] [Google Scholar]
- 26.Leth R., Ercig B., Olsen L., Jørgensen F.S. Both reactivity and accessibility are important in cytochrome P450 metabolism: a combined DFT and MD study of Fenamic acids in BM3 mutants. J Chem Inf Model. 2019;59:743–753. doi: 10.1021/acs.jcim.8b00750. [DOI] [PubMed] [Google Scholar]
- 27.Christensen E.B., Andersen J.B., Pedersen H., Jensen K.G., Dalgaard L. Metabolites of [(14)C]-5-(2-ethyl-2H-tetrazol-5-yl)-1-methyl-1,2,3, 6-tetrahydropyridine in mice, rats, dogs, and humans. Drug Metab Dispos. 1999;27:1341–1349. [PubMed] [Google Scholar]
- 28.Barr J.T., Jones J.P. Evidence for substrate-dependent inhibition profiles for human liver aldehyde oxidase. Drug Metab Dispos. 2012;41:24. doi: 10.1124/dmd.112.048546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berthold M.R., Cebron N., Dill F., Gabriel T.R., Kötter T., Meinl T. KNIME: the Konstanz information miner. In: Preisach C., Burkhardt H., Schmidt-Thieme L., Decker R., editors. Data analysis, machine learning and applications: proceedings of the 31st annual conference of the Gesellschaft für Klassifikation eV. Springer Berlin Heidelberg; Berlin, Heidelberg: 2008. pp. 319–326. Albert-Ludwigs-Universität Freiburg, March 7–9, 2007. [Google Scholar]
- 30.Shelley J.C., Cholleti A., Frye L.L., Greenwood J.R., Timlin M.R., Uchimaya M. Epik: a software program for pKa prediction and protonation state generation for drug-like molecules. J Comput Aided Mol Des. 2007;21:681–691. doi: 10.1007/s10822-007-9133-z. [DOI] [PubMed] [Google Scholar]
- 31.Greenwood J.R., Calkins D., Sullivan A.P., Shelley J.C. Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J Comput Aided Mol Des. 2010;24:591–604. doi: 10.1007/s10822-010-9349-1. [DOI] [PubMed] [Google Scholar]
- 32.Shivakumar D., Williams J., Wu Y., Damm W., Shelley J., Sherman W. Prediction of absolute solvation free energies using molecular dynamics free energy perturbation and the OPLS force field. J Chem Theory Comput. 2010;6:1509–1519. doi: 10.1021/ct900587b. [DOI] [PubMed] [Google Scholar]
- 33.Harder E., Damm W., Maple J., Wu C., Reboul M., Xiang J.Y. OPLS3: a force field providing broad coverage of drug-like small molecules and proteins. J Chem Theory Comput. 2015;12:281–296. doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- 34.Jorgensen W.L., Maxwell D.S., Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118:11225–11236. [Google Scholar]
- 35.Jorgensen W.L., Tirado-Rives J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 36.Bochevarov A.D., Harder E., Hughes T.F., Greenwood J.R., Braden D.A., Philipp D.M. Jaguar: a high-performance quantum chemistry software program with strengths in life and materials sciences. Int J Quantum Chem. 2013;113:2110–2142. [Google Scholar]
- 37.Gasteiger J., Marsili M. Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron. 1980;36:3219–3228. [Google Scholar]
- 38.ChemBioDraw, CambridgeSoft corporation (Perkin Elmer), (2014) In: Elmer CCP, [editor].
- 39.Hilbig M., Rarey M. MONA 2: a light cheminformatics platform for interactive compound library processing. J Chem Inf Model. 2015;55:2071–2078. doi: 10.1021/acs.jcim.5b00292. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material 1
CSV file with calculated data for SOMs and non-SOMs in the 78 compounds.
SDF file with coordinates of the 78 compounds and experimental SOMs.