Abstract
Members of the Hedgehog (Hh) family of signaling proteins are powerful regulators of developmental processes in many organisms and have been implicated in many human disease states. Here we report the results of a genome-wide RNA interference screen in Drosophila melanogaster cells for new components of the Hh signaling pathway. The screen identified hundreds of potential new regulators of Hh signaling, including many large protein complexes with pleiotropic effects, such as the coat protein complex I (COPI) complex, the ribosome and the proteasome. We identified the multimeric protein phosphatase 2A (PP2A) and two new kinases, the D. melanogaster orthologs of the vertebrate PITSLRE and cyclin-dependent kinase-9 (CDK9) kinases, as Hh regulators. We also identified a large group of constitutive and alternative splicing factors, two nucleoporins involved in mRNA export and several RNA-regulatory proteins as potent regulators of Hh signal transduction, indicating that splicing regulation and mRNA transport have a previously unrecognized role in Hh signaling. Finally, we showed that severalof these genes have conserved roles in mammalian Hh signaling.
The Hh signaling pathway is atypical in that its mode of action seems to be unique at almost every step1–3. Hh proteins undergo a complex maturation process involving an autocatalytic cleavage and the covalent addition of both a cholesterol and a palmitic acid moiety. Hh binds to its receptor, the multiple-pass transmembrane protein Patched (Ptc), which relays the signal to a large intracellular complex consisting of the kinesin-like protein Costal2 (Cos2), the serinethreonine kinase Fused (Fu) and the zinc-finger protein Cubitus interruptus (Ci)1–3. This intracellular complex binds both micro-tubules and intracellular vesicles containing the seven-pass trans-membrane protein Smoothened (Smo), an essential positive regulator of Hh signaling1–3. In the absence of Hh stimulation, Ptc exerts a repressive effect on the complex and on Smo, preventing activation of both elements. Binding of Hh to Ptc relieves this repression, allowing the complex to become hyperphosphorylated and allowing Smo to activate downstream components of the pathway. The complex interplay between these factors ultimately results in precise regulation of Ci. In the absence of Hh, Ci is processed into an N-terminal fragment that seems to repress the transcription of Hh target genes. Efficient processing of Ci requires the cAMP-dependent protein kinase-1 (PKA-C1), casein kinase Iα (CKIα) and Shaggy (GSK-3β) kinases in addition to Supernumerary limbs (Slimb), a component of a multimeric ubiquitin ligase. Hh stimulation inhibits this processing to generate full-length Ci and also activates this full-length Ci. The full-length, activated form of Ci can then act as a transcriptional activator, increasing the transcription of Hh target genes such as ptc1–3.
RNA interference (RNAi) is a technique that uses double-stranded RNA (dsRNA) or small interfering RNA (siRNA) to potently and specifically degrade the mRNA, and ultimately diminish the protein, encoded by a gene of interest4. Large RNAi libraries, including many genome-wide libraries, have been established and used to screen whole animals (in the case of Caenorhabditis elegans) and cell lines (from vertebrates and D. melanogaster) for genes affecting particular pathways or processes5–9. To obtain a complete picture of the genes required for Hh signaling, we carried out a genome-wide RNAi screen for new components of the Hh pathway using D. melanogaster cells and a dsRNA library previously developed in our laboratory5. We identified a broad range of new genes involved in Hh signaling. In accordance with previous results, we found that ribosomal genes and members of the COPI complex are strong regulators of Hh signaling9. We also found a large number of new candidate Hh regulators, including PP2A, two new kinases, lipid synthesis proteins, proteasomal components and components of ubiquitin-ligase complexes. We also identified a set of splicing genes, many of which are involved in alternative splicing, as potent regulators of Hh signaling. Finally, we showed that some of these genes have conserved roles in vertebrate Hh signaling.
RESULTS
Genome-wide RNAi screen for Hh signaling factors
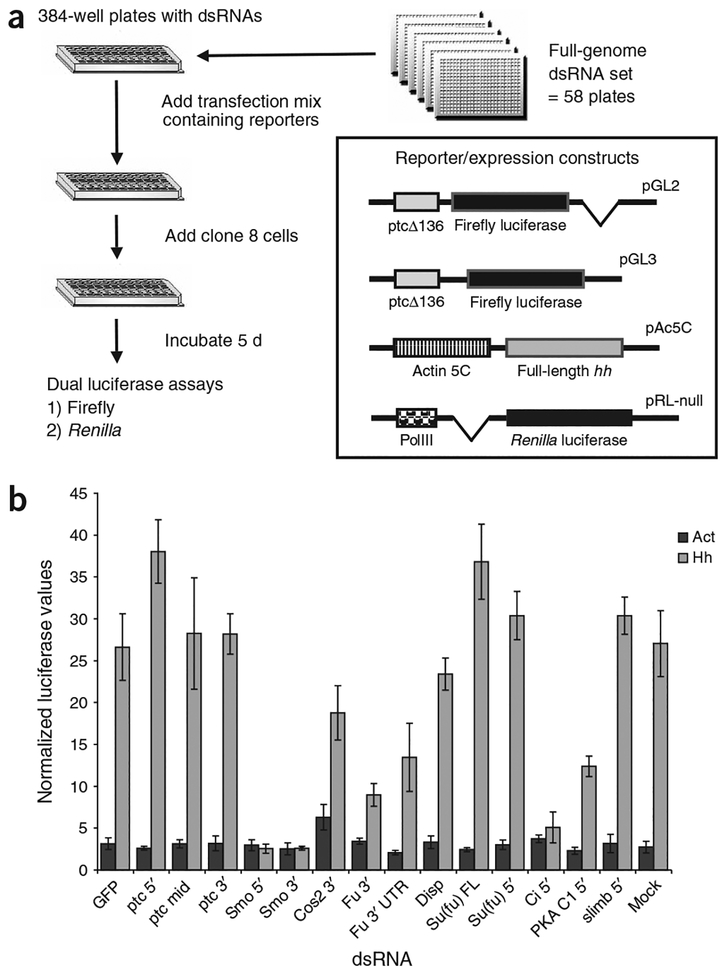
Because our dsRNA library is divided into 384-well tissue-culture plates, we needed a robust Hh signaling assay that could be used in this format. To that end, we developed a transfection-based dual-luciferase assay using a previously described Hh reporter in which a portion of the ptc promoter, called ptcΔ136, drives expression of firefly luciferase10 (Fig. 1a). The assay was verified before screening by testing the effects of dsRNAs targeted against known components of the Hh pathway on the normalized luciferase signal. None of the test dsRNAs, including known negative regulators, caused any substantial increase in basal reporter activity, with the previously noted exception of cos dsRNA, which roughly doubled basal reporter activity9 (Fig. 1b). The lack of any ectopic increase in reporter activity when known negative regulators were knocked down convinced us that a genome-wide screen for dsRNAs that increase basal reporter activity was not worthwhile in the context of our assay. But RNAi directed against most of the known members of the Hh pathway in the presence of Hh stimulation had the expected effect of either reducing or increasing reporter activity, indicating that our assay should be able to identify regulators of Hh signaling in a genome-wide assay (Fig. 1b).
Figure 1.
Assay design and validation of the primary screen for new components of Hh signaling. (a) Outline of screen design and schematic representation of constructs used in the primary screen. Relevant portions of the vectors shown are shown in inset. Left boxes are promoter fragments and right boxes are coding regions; parent vectors are indicated at the upper right of each construct. (b) Verification of Hh assay in 384-well plate format. Clone 8 cells were assayed using the ptcΔ136-GL2 reporter and either Act5C-Hh–expressing vector (Hh) or empty Act5C vector (Act). Normalized luciferase values were averaged from 4 wells in a 384-well plate. The dsRNA used to treat the cells is indicated. smo and ci dsRNAs eliminated most reporter activity and resulted in a sixfold difference in normalized signal between cells treated with GFP dsRNA and cells treated with smo or ci dsRNAs. Treatment with dsRNA against the fu coding region reduced reporter signal by ~75%, whereas dsRNAs against the 5′ untranslated region (UTR) of fu reduced signal by 50%. Error bars in this figure and all subsequent figures indicate two standard deviations.
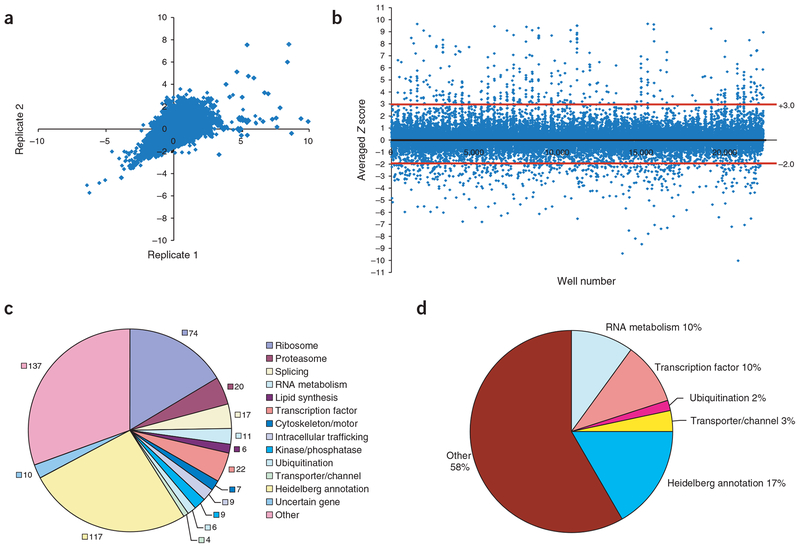
The RNAi screen was done in duplicate to identify genes that either reduce or increase Hh reporter activity in D. melanogaster clone 8 cells. Analysis of the Z scores of replicate plates showed that the assay was highly reproducible, with most points falling along a diagonal through the origin (Fig. 2a). Wells were scored as hits if the average of the Z scores from the two replicates was less than −2.0 or greater than +3.0 (scoring rationale is given in Supplementary Methods online); most wells fell within these thresholds (Fig. 2b). Of the ~21,000 dsRNAs screened, we identified 509 hits using these criteria; 449 reduced reporter activity, suggesting that they function as positive regulators of Hh signaling, and 60 increased reporter activity, suggesting that they act as negative regulators (Supplementary Tables 1 and 2 online). Notably, we identified nine of the previously known components of the Hh signaling pathway: smo, ci, cos, fu, Pka-C1 and combgap were identified as positive regulators of Hh signaling, whereas Su(fu), slimb and ptc were identified as negative regulators (Supplementary Tables 1 and 2). Our assay did not identify sgg, rasp, dlp, disp or CkIα as hits. Because most known Hh signaling genes were identified in our screen, we concluded that our assay can identify legitimate Hh signaling factors.
Figure 2.
Primary screen results. (a) Correlation of replicate Z scores for the first five primary screening plates. The data in a and b were filtered to remove values greater than +10.0, which correspond to the artificially high normalized values found in some edge wells. (b) Scatter plot of all averaged Z scores from the primary screen. Red lines indicate −2.0 and +3.0 averaged Z score thresholds. Each screening plate contained four control wells, one of which contained smo dsRNA. These smo control wells formed the majority of the data points with Z scores near −5.0. (c) Functional classification of the 449 genes with Z scores less than −2.0 when reduced by RNAi in the Hh signaling assay. These genes are likely to function as positive regulators of Hh signaling. Numbers of genes in each category are indicated. (d) Functional classification of the 60 genes with Z scores greater than +3.0 when reduced by RNAi in the Hh signaling assay. These genes are likely to function as negative regulators of Hh signaling.
The screen identified many components of known macromolecular complexes and functional groups defined by the FlyBase annotations of the targeted genes. The distribution of all the positive and negative hits to assignable complexes or functional groups is shown in Figure 2c,d. Most of the components of several large protein complexes were identified as positive regulators of Hh signaling (Fig. 2c). Many other genes belonging to a variety of functional groups were also identified in the screen, and many have orthologs in other phyla. The human, mouse, yeast and nematode orthologs of our hits are listed in Supplementary Table 3 online. We discuss some of these functional groups in more detail below.
Secondary assays
To confirm our initial results and ascertain how the identified genes function in Hh signaling, we produced 96-well secondary screening plates containing dsRNAs from 255 of the hits identified in the primary screen (Supplementary Methods). We then used these plates to carry out a series of secondary assays designed to identify genuine Hh regulators and to begin to determine where each regulator acts on the pathway.
The first series of assays was designed to control for the effects of the reporter and expression constructs (Supplementary Fig. 1 online and Supplementary Methods). Because the original experimental firefly and control Renilla reporters contained introns, we suspected that many of the RNA regulatory and splicing genes identified as hits may have been affecting the splicing or RNA stability of the reporter mRNAs and not directly affecting Hh signaling. To test this possibility, we assayed the secondary plates using an experimental reporter lacking the intron and found that 80% of the dsRNAs tested passed this assay (Supplementary Table 4 online). Next, we reversed the control and experimental promoters, using the ptcΔ136 experimental promoter to drive Renilla luciferase and the RNA polymerase III (PolIII) control promoter to drive firefly luciferase. If a dsRNA scored solely because of its differential effects on one luciferase mRNA versus another, that score would be greatly reduced or reverse sign in this assay. Almost 60% of the hits passed this assay (Supplementary Table 4). Many splicing factors passed both of these assays, indicating that they are legitimate Hh signaling factors. We then carried out a secondary assay to control for changes that might have been caused by differences in the regulation of the PolIII promoter fragment driving our control Renilla construct and the actin promoter construct driving the full-length Hh. When Hh was driven by the PolIII promoter, 65% of the dsRNAs rescored as hits (Supplementary Table 4). Overall, 90 positive and 6 negative regulators passed all three secondary screens and a retest of the primary screen, confirming the robustness of our assays and the large number of new genes that might regulate Hh signaling. Finally, we tested how our candidate genes affected basal transcription from the ptcΔ136 reporter to determine whether any of them could induce reporter activity to a level similar to that seen with Hh stimulation. Although the percentage changes in basal reporter activity caused by application of a particular dsRNA were often similar to those seen in the Hh-stimulated state, none of the dsRNAs activated the reporter to a normalized level near the stimulated state (comparing the plate means and percent changes in the presence or absence of Hh; Supplementary Table 4).
The zinc-finger protein Ci is a key downstream regulator of Hh signaling. Therefore, we sought to order the screen hits relative to ci to ascertain their epistatic relationships with ci. To do this, we examined reporter activity when a construct expressing full-length (activating) Ci instead of a construct expressing Hh was used to transfect clone 8 cells. Ci expression stimulated ptcΔ136 reporter activity to a level approximately half of that induced by Hh expression (Supplementary Fig. 1). We considered ci to be epistatic to candidate genes if the difference between their Hh and Ci scores was greater than 40 percentage points. Consistent with our previous understanding of Hh signaling, ci was epistatic to fu and cos using these criteria. When all the secondary screening plates were screened with ci and hits with substantially lowered cell viability were discarded, we found 27 hits whose scores were changed by 40 percentage points or greater when Ci was compared with Hh (Table 1 and Supplementary Table 4). A pattern emerged from this comparison: RNAi against cos substantially decreased reporter activity when Hh was expressed but increased reporter activity when Ci was expressed in clone 8 cells. We therefore examined our list for genes with substantial shifts in reporter activity and found that the genes microtubule star (mts) and CG1874 also changed from reducing to increasing reporter activity when Ci was used as the stimulus instead of Hh. These genes may have a role in Hh signaling similar to that of cos.
Table 1.
Epistasis of ci with candidate Hh regulators
| Locus tag | Gene | Hh score | Ci score | Difference (Hh − Ci) |
|---|---|---|---|---|
| CG1708 | cos | −73 | 61 | −134 |
| CG7109 | mts | −62 | 42 | −105 |
| CG3412 | slimb | 36 | 127 | −91 |
| CG1135 | CG1135 | 76 | 153 | −77 |
| CG18041 | CG18041 | 45 | 121 | −76 |
| CG6292 | CycT | −77 | −7 | −70 |
| CG6551 | fu | −73 | −8 | −65 |
| CG1874 | CG1874 | −25 | 38 | −63 |
| CG9282 | RpL24 | −70 | −8 | −62 |
| CG11561 | Smo | −94 | −34 | −60 |
| CG2184 | Mlc2 | −55 | −4 | −52 |
| CG3193 | Crn | −72 | −22 | −50 |
| CG17489 | RpL5 | −65 | −16 | −50 |
| CG14813 | δ COP | −67 | −17 | −50 |
| CG1475 | RpL13A | −83 | −37 | −47 |
| CG6699 | β′COP | −73 | −29 | −44 |
| CG1821 | RpL31 | −78 | −35 | −44 |
| CG6625 | Snap | −52 | −8 | −44 |
| CG10198 | Nup98 | −59 | −16 | −43 |
| CG5179 | Cdk9 | −63 | −20 | −43 |
| CG7961 | αCOP | −65 | −23 | −42 |
| CG1528 | γCOP | −57 | −16 | −41 |
| CG5931 | CG5931 | −64 | −23 | −41 |
| CG7757 | CG7757 | −48 | −8 | −40 |
| CG7923 | Fad2 | −48 | −8 | −40 |
| CG32955 | CG32955 | −58 | −19 | −40 |
Candidate Hh signaling factors identified in the screen to which ci is epistatic. ci epistasis was determined as a difference of 40 percentage points or greater between the Hh and Ci reporter scores. Gray shading indicates genes that had a Hh/Ci profile similar to that of cos, in that they shifted from reducing the reporter score in the Hh assay to increasing it in the Ci assay. Bold type indicates ribosomal genes. Genes were discarded if they reduced the control Renilla values to less than 75% of the plate average or if they were not rescored as hits in the Hh secondary assay.
Large protein complexes involved in Hh signaling
It has been difficult to identify all the components of Hh signaling using genetic screens and biochemical methods. One reason for this may be that the missing components are pleiotropic genes. We therefore expected that our screen might identify genes or groups of genes with multiple functions. Indeed, our screen identified several groups of genes that affect many aspects of cellular metabolism but also seem to have roles in Hh signaling.
Foremost among these were the genes encoding ribosomal proteins. Of the 93 cytoplasmic ribosomal subunits, we identified 74 that reduce Hh signaling when targeted by RNAi (Supplementary Table 1). This is unlikely to have been caused by a sharp reduction in the cells’ translational ability, as reduction of ribosomal proteins by RNAi results in less than a twofold reduction in protein translation in D. melanogaster S2 cells6. Furthermore, a screen for genes affecting Wingless signaling done with the same control reporter and in the same cell type (clone 8) identified no ribosomal genes11. This apparent requirement for ribosomal components in Hh signaling could reflect a consistent need for a short-lived Hh signaling component(s). A reduction in overall translation efficiency through loss of individual ribosomal components would greatly reduce this factor’s abundance in the cell relative to other Hh components and so impair Hh signaling. As the ribosome can interact with alternative splicing and translation factors to influence the generation of alternative protein isoforms, the ribosome may also interact with these factors to generate new protein isoforms required for Hh signaling12,13. In any case, our data indicate that the Hh signaling pathway is relatively more sensitive to ribosomal perturbation than the Wnt signaling pathway.
In addition to the ribosomal components, our screen identified 20 members of the 26S proteasome, all of which act as positive regulators of Hh signaling (Supplementary Table 1). This result was somewhat unexpected because slimb, Roc1a and lin19 (also called Cul1) all increase Hh reporter activity and full-length Ci accumulation when mutated in D. melanogaster, indicating that they act as negative regulators of Hh signaling14–16. The proteins encoded by slimb,Roc1a, lin19 and skpA are components of a tetrameric ubiquitin ligase, called the Skp1/Cul1/F-box (SCF) complex, that is involved in targeting proteins to the proteasome for degradation17. Therefore, the opposing effects of RNAi reduction of proteasomal components and RNAi reduction of one of the proteasome-targeting complexes suggests that the proteasome and its targeting systems may have a more complex interaction within the Hh pathway than previously realized. It is possible that the SCF complex targets a positive regulator of Hh signaling, such as Ci, for proteasomal degradation, and a negative regulator of Hh signaling downstream of the SCF target is also degraded by the proteasome. Thus, loss of the SCF complex would increase Hh signaling, whereas loss of the proteasome would decrease Hh signaling. This model suggests that there is another ubiquitin ligase complex involved in Hh signaling, which acts on the downstream negative regulator. Our screen identified several new ubiquitin and ubiquitin ligase components that could have this role, including Ubiquitin-conjugating enzyme (UbcD6), CG11700 (a ubiquitin-like protein), Ubiquitin-63E, Effete and CG32676 (another ubiquitin-like protein; Supplementary Tables 1 and 4). Although slimb was identified, however, Roc1a, lin19 and skpA were not identified as Hh regulators in our screen. This may be the result of an inherent difficulty in knocking down these genes, or the SCF complex may act on Hh signaling by maintaining a certain threshold level of Ci below which slimb function has little effect but above which slimb has a more substantial effect. Indeed, loss of slimb by RNAi greatly increased reporter activity in the Ci overexpression assays compared with the Hh assays (Supplementary Table 4).
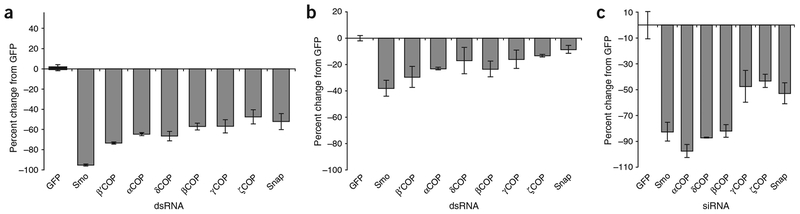
We also identified six of the seven core subunits of the COPI vesicular transport complex as positive regulators of Hh signaling (Fig. 3a). COPI components seem to act upstream of Ci (Fig. 3b). To show that the COPI components have a conserved role in Hh signaling, we showed that the COPI components reduce reporter activity in an assay of mouse soluble Sonic hedgehog (ShhN; Fig. 3c). Notably, siRNAs directed against some COPI subunits, such as αCOP and δCOP, reduced signaling by more than 85%, making them more potent regulators of Shh signaling than Smo in this assay (Fig. 3c and Supplementary Note online).
Figure 3.
Vesicular trafficking genes are important in Hh signaling. (a) COP and Snap dsRNAs reduce Hh signaling by 40% or more in secondary assays. Scores were taken from the Hh (GL3) secondary assay and are presented as the average percent change from a GFP dsRNA control. dsRNAs targeting smo were included as a control. (b) Effects of COP and Snap dsRNAs on activation of reporter by Ci. Scores were taken from Ci secondary assay. dsRNAs and scale are the same as in a. (c) Effects of mouse Cop and Snap siRNAs on soluble Shh signaling. RNAi of mouse Copa (ortholog of αCOP), in particular, caused a greater reduction in reporter activity than even siRNA targeting mouse Smo.
Identification of new kinases involved in Hh signaling
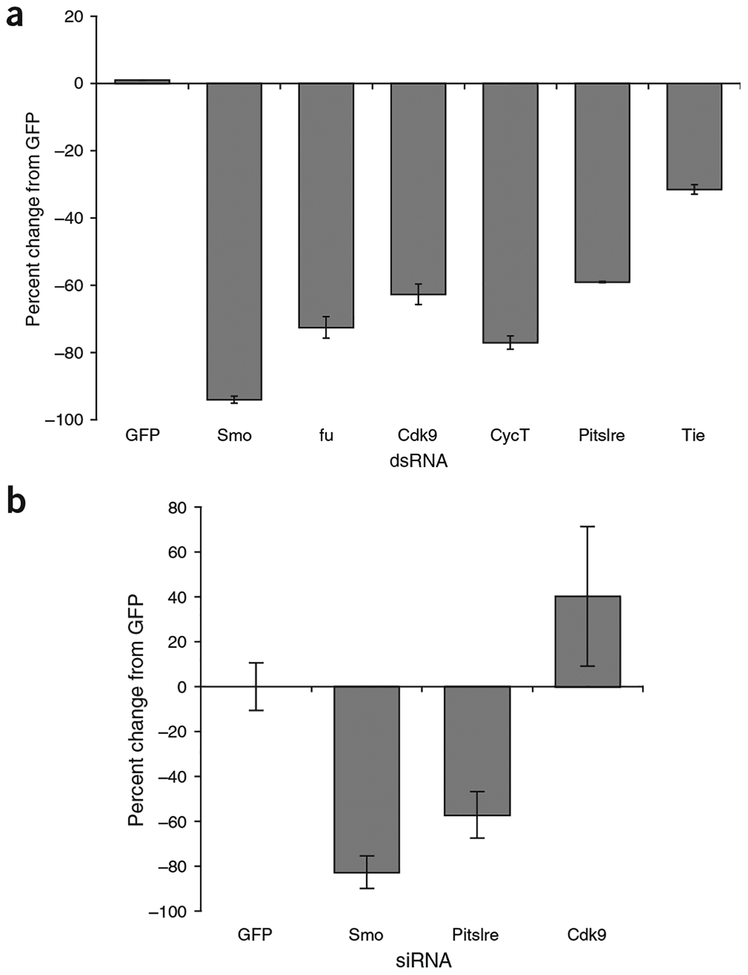
In addition to the members of large functional groups, we also identified particular genes of certain functional classes. We identified five protein kinases, all of which reduced Hh signaling when targeted by RNAi (Fig. 4a). Two of these, Fu and PKA-C1, had previously been identified as Hh pathway members. The remaining three kinases, all of which reduced basal reporter activity, were Cdk9 and Pitslre, the D. melanogaster orthologs of the vertebrate CDK9 and PITSLRE (or CDK11) kinases, and Tie-like receptor tyrosine kinase. The Tie-like receptor tyrosine kinase did not pass two of the four secondary screens and was not considered further. Cdk9 is involved in regulation of transcription and stress responses and can phosphorylate the C-terminal domain of RNA polymerase II18,19. Cdk9 is associated with an unusual cyclin, Cyclin T (CycT), in a complex called P-TEFb20. We also identified CycT in our screen as a strong positive regulator of Hh signaling (Fig. 4a). Pitslre is a kinase that functions in both transcription and splicing, in addition to a role in cell-cycle regulation21–23. Pitlsre physically associates with splicing components and transcription factors, especially those involved in transcriptional elongation, and regulates the phosphorylation of SR-family splicing factors, two of which we identified in our screen.
Figure 4.
Identification of new kinases affecting Hh signaling. (a) Reduction of Pitslre and Cdk9/CycT by RNAi substantially reduce Hh signaling, as assayed by ptcΔ136-GL3 reporter activity. dsRNAs targeting smo and fu were included as controls. (b) Effect of siRNAs targeting mouse Pitslre and Cdk9 on ShhN signaling in NIH3T3 cells. Cdk9 RNAi had variable effects on ShhN signaling but consistently increased reporter activity.
To determine whether either of these two kinases has a more universal role in Hh signaling, we tested their effects on ShhN (a form of Shh protein containing only the secreted, N-terminal region) signaling in mouse NIH3T3 cells (Fig. 4b). We found that Cdk9 siRNA consistently increased reporter activity, although the variability was too high to determine whether the changes were statistically significant. Pitslre siRNA, however, consistently reduced Shh signaling by ~50%. Therefore, Pitslre seems to have a conserved and previously unrecognized role in Hh signaling in both flies and vertebrates. Whether it affects Hh signaling through its effects on transcription or splicing, or perhaps through a yet unrecognized role, is not yet known.
RNAi against Cdk9 or CycT had little effect on induction of reporter activity by Ci overexpression, whereas RNAi against Pitslre did, suggesting that the Cdk9-CycT complex acts upstream of Ci, whereas Pitslre acts downstream of Ci (Table 1). As Fu and Cos are hyperphosphorylated in response to Hh signaling, we also examined whether Cdk9 or Pitslre could be phosphorylating Fu or Cos. Hh stimulation normally causes ~50% of Fu and most of Cos to migrate more slowly on polyacrylamide gels. But neither Cdk9 nor Pitslre RNAi had an effect on the phosphorylation of Fu or Cos isolated from Hh-expressing cells, indicating that Cdk9 and Pitslre act in parallel to or downstream of fu and cos (Supplementary Fig. 2 online).
Role of mRNA splicing, transport and regulation in Hh signaling
One of the more unexpected results of our screen was the large number of splicing and RNA-regulatory proteins that modulated Hh signaling. Almost all the genes annotated as splicing and RNA-regulatory factors function as positive regulators of Hh signaling (Table 2). Most of the splicing factors repeated as hits in all the secondary assays (although most also reduced basal reporter activity), and some were very strong positive regulators of Hh signaling, reducing reporter activity by more than 75% (Fig. 5a). crooked neck (crn) was a prominent hit in the splicing factor group, and all the dsRNAs directed against it in our library resulted in strong decreases in reporter activity. crn was originally isolated as a gene involved in the morphogenesis of the D. melanogaster embryo and was subsequently shown to be a component of the splicing machinery24–26. crn has been implicated in the regulation of alternative splicing of the D. melanogaster genes Ubx and Adar27,28. Mouse crn siRNAs reduced ShhN signaling by 50% (Fig. 5b), indicating that the gene has a phylogenetically conserved role in vertebrate Hh signaling.
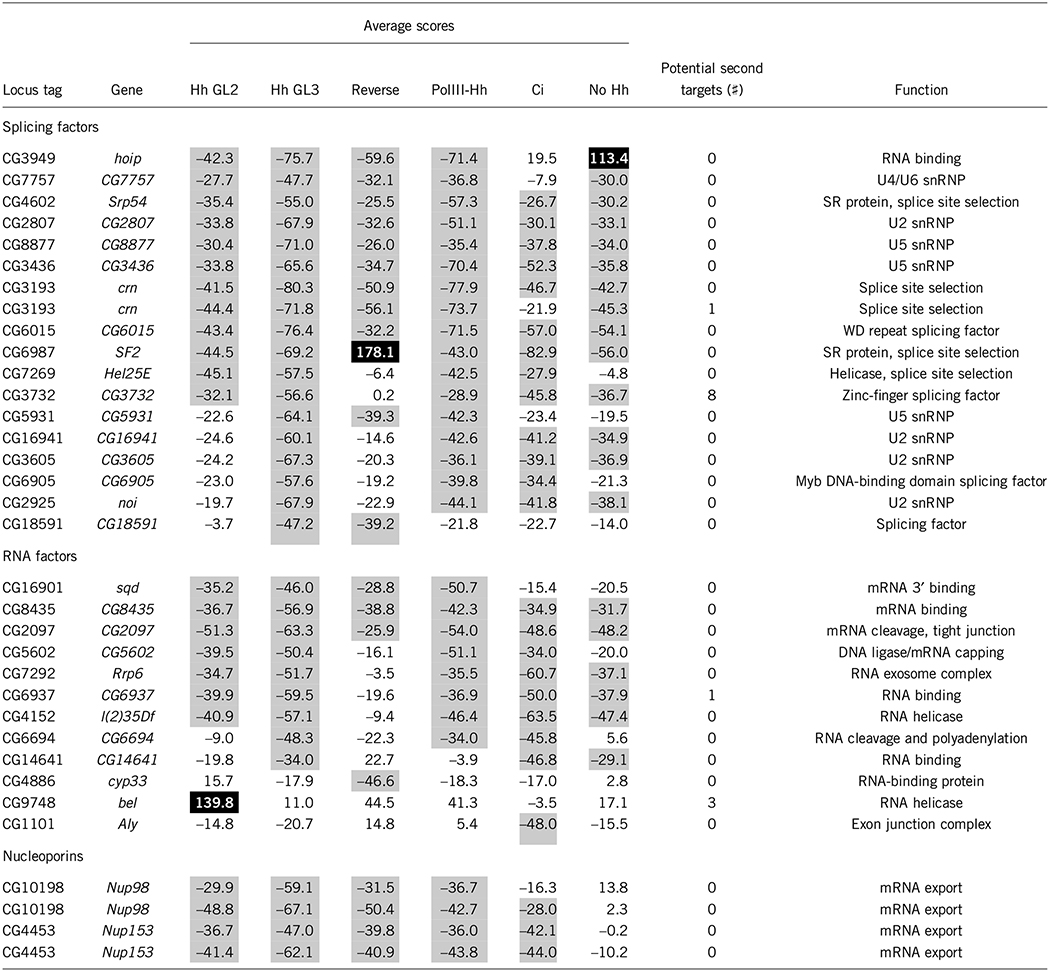
Table 2.
Splicing components, RNA-binding factors and nucleoporins affecting Hh signaling
Putative molecular functions were obtained from the FlyBase gene annotations. Hh assays using the ptcΔ136-GL2–and ptcΔ136-GL3–derived reporters are indicated; the PolIII-Hh, Ci and no-Hh assays all used the ptcΔ136-GL3 reporter. Scores for each gene in the indicated secondary assay are percent change from averaged Gfp dsRNA controls. Gray shading indicates secondary scores lower than −25%; black shading indicates those higher than +50%. Nup98 is represented twice because two different dsRNAs directed against it were found in the dsRNA library. Nup153 is represented twice because the same dsRNA was found in two different wells in the dsRNA library.
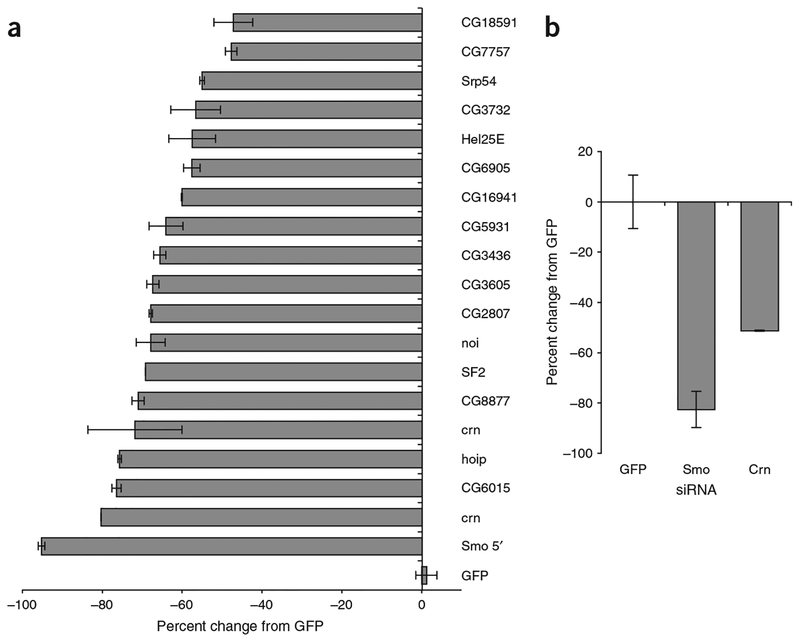
Figure 5.
Splicing factors have a role in Hh signaling. (a) Degree to which splicing factors modulate Hh signaling. Scores were taken from the Hh (GL3) secondary assay and are presented as the average percent change from a GFP dsRNA control. crn, a gene involved in splicing in Drosophila and humans, is a strong positive regulator of the Hh signaling pathway. All three dsRNAs representing this gene in the dsRNA library were identified in the primary screen, although only two were tested in secondary assays. (b) siRNA against mouse Crnkl1 (ortholog of crn) reduced ShhN signaling by almost 50%.
The identification of splicing and RNA-regulatory factors as Hh signaling components might simply suggest that splicing is required to generate most mRNAs, including those in the Hh pathway, but we do not believe that this is the case. Many of the splicing factors we identified are not involved in the core splicing machinery. Of the splicing factors we identified that are known to belong to certain core splicing complexes, there were three components of U5, four components of U2 and a single U4/U6 component. If splicing in general were being affected and abrogating Hh signaling, then we should have identified more of the essential splicing factors, including components of the U1 and U2 AF complexes29,30. In addition, a recent study showed that RNAi reduction of some the spliceosome’s core components (including the U2 component CG16941, which we identified as a hit) can modulate alternative splice site selection in D. melanogaster27. Therefore, it seems more likely that the splicing genes we identified are involved in some sort of regulated alternative splicing event within the Hh pathway (Supplementary Note).
In the case of the RNA-regulatory proteins, three of the hits passed all four secondary assays (Table 2). One of these hits was the gene squid, which encodes an RNA transport protein whose best-characterized role is in mRNA transport in the D. melanogaster oocyte31,32. Another hit in this category was CG2097, the D. melanogaster ortholog of the vertebrate gene Sympk, whose protein acts as a scaffold on which several components of the polyadenylation machinery assemble in both the nucleus and the cytoplasm33.
Further evidence for the involvement of mRNA regulation in Hh signaling came from the identification of two nucleoporins as hits. These nucleoporins, Nup98 and Nup153, were identified as hits that reduced Hh signaling. Both scored in all four secondary assays (Table 2), indicating that they are legitimate Hh signaling components. In the absence of Hh stimulation, Nup98 and Nup153 RNAi had no effect on basal reporter activity, implying that they are only required when Hh signaling is activated (Supplementary Table 4). Both Nup98 and Nup153 have vital roles in mRNA export in vertebrate cells, and neither appears to play any part in nuclear import34–36. The finding that nucleoporins involved in mRNA export would be the only nuclear transport factors identified in our screen is unexpected, as the current view of Hh signaling calls for full-length Ci to be imported into the nucleus upon Hh stimulation. If nuclear import of Ci has a key role in Hh signaling, we would have expected to identify at least one nuclear import factor in the screen.
Regulation of Hh signaling by PP2A
Phosphorylation is associated with the activities of at least five components of the Hh pathway: Fu, Cos, Smo, Su(fu) and Ci37–42. Little is known about the kinases that phosphorylate Su(fu) and Fu, but at least two sites in Cos are phosphorylated by Fu, and several kinases are involved in phosphorylating Ci and Smo, including PKA-C1, CkIα and Sgg41,43. But no phosphatase has been implicated in Hh signaling, and a previous RNAi screen did not identify any phosphatases involved in Hh signaling9.
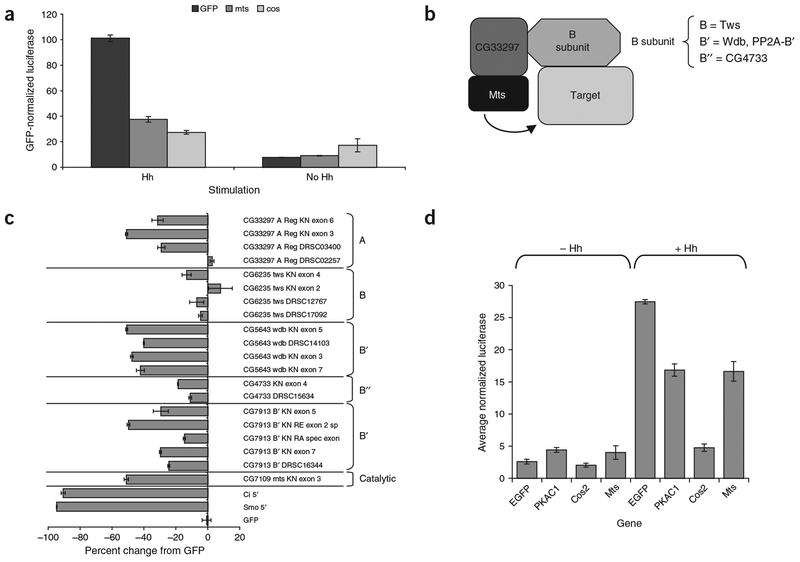
Our screen identified mts, which encodes the D. melanogaster PP2A catalytic subunit, as a gene that substantially reduced Hh signaling when targeted by RNAi (Fig. 6a). PP2A is a multimeric enzyme that consists at minimum of the catalytic subunit, a regulatory A subunit (encoded by CG33297 in D. melanogaster) and a B subunit principally involved in substrate selection (Fig. 6b)44. The B-subunit family in D. melanogaster is represented by the gene twins (tws), the B′ family by the genes widerborst (wdb) and PP2A-B′, and the B″ family by CG4733. We obtained and tested all the PP2A component dsRNAs from our dsRNA library and generated and tested addi tional, distinct dsRNAs to these components (Supplementary Note and Supplementary Methods). In addition to confirming the mts result, we found that both the original-library dsRNA and three new, unique dsRNAs targeting wdb all reduced Hh signaling by ~50% (Fig. 6c). This indicated that Wdb is likely to be the B subunit that targets Mts to its substrate in the Hh signaling pathway. This hypothesis is in agreement with recent findings from Xenopus laevis, where the wdb ortholog encoding B56e has been found to regulate Hh signaling (J. Yang, personal communication). In addition, some PP2A-B′ amplicons cause a reduction in reporter activity averaging ~30%, indicating that they may have a partially redundant role in targeting PP2A to its Hh pathway substrate.
Figure 6.
PP2A and its regulatory B subunits regulate Hh signaling. (a) Effect of GFP, mts and cos dsRNA on reporter activity in cells expressing Hh or empty vector. All values were taken from the Hh (GL3) secondary assay and were normalized such that Hh-stimulated GFP dsRNA was 100. (b) Presumed structure of D. melanogaster PP2A holoenzyme, based on data from vertebrate PP2A studies. The catalytic subunit is bound to a regulatory A subunit. This minimal dimer is bound to a regulatory B subunit that is involved in targeting the PP2A to its substrates or proper subcellular location. The names of the four D. melanogaster B subunits are indicated. (c) Effects of dsRNAs targeting the catalytic, A and B subunits of PP2A on Hh signaling. dsRNA against the A subunit had mixed results, with one amplicon having no effect and the others reducing Hh signaling by 30–50% relative to GFP dsRNA. None of the four twins dsRNAs has an effect on reporter activity. dsRNAs against wdb resulted in a 40–50% reduction in reporter activity. dsRNAs against PP2A-B′ give mixed results depending on the particular dsRNA. Amplicons from the Drosophila RNAi Screening Center are indicated by DRSC amplicon numbers. RNAi against ci and smo was included as controls. (d) Overexpression of mts and Pka-C1 results in matching phenotypes. Equal amounts of each expression construct were cotransfected with reporters. Overexpression of both mts and Pka-C1 in the absence of Hh stimulation resulted in small but reproducible increases in reporter activity.
To determine whether PP2A acts on Cos, we examined whether overexpression of cos and mts results in similar phenotypes. When overexpressed in Hh-stimulated clone 8 cells, cos completely abrogated Hh signaling, reducing it to near uninduced levels, whereas over-expression of mts reduced Hh signaling by 40% (Fig. 6d). Thus, Mts and Cos have different overexpression profiles and do not seem to regulate Hh signaling in the same way. We then compared the overexpression phenotype of mts with those of cos and 14 other hits from the screen, including the fu, Cdk9 and Pka-C1 kinases. Over-expressing cos in uninduced cells further reduced background signaling, whereas mts overexpression doubled reporter activity, although these levels were still very low compared with the Hh-activated state (Fig. 6d). Of the 18 other genes tested, only Pka-C1 overexpression had an effect on Hh reporter activity similar to that of mts: doubling of reporter activity in the Hh-uninduced state and a 50% reduction of activity in the Hh-stimulated state (Fig. 6d and data not shown). It is therefore possible that PKA-C1 and Mts act on similar substrates. Because several studies have identified Ci as a substrate of PKA-C1, Mts could also be acting on Ci, perhaps removing inhibitory phosphates in response to Hh stimulation.
DISCUSSION
In this study, we identified a wide range of new components with potential roles in Hh signaling. Among these new components are members of protein complexes that affect multiple cellular processes and would be difficult to place in any one signaling pathway using classical genetic techniques. The sensitivity of cell-based RNAi assays, however, allows us to dissect out the roles of these genes in particular signaling pathways in isolation from complicating tissue-specific interactions and cell competition effects. Our screen allowed us to group the ribosome, proteasome, COPI complex and PP2A phosphatase as important regulators of Hh signaling, none of which had been identified as Hh regulators in vivo. Notably, some of the components we identified in the screen had already been implicated in aspects of Hh signaling. For instance, the gene encoding eRF1, a translational regulator, was identified in a screen for modifiers of a gain-of-function smo allele, and polyhomeotic and additional sex combs have both been shown to modify ectopic hh expression phenotypes45,46.
Our results open many new avenues for investigation of Hh signaling. In particular, elucidation of the Hh pathway substrates affected by PP2A will be important in defining the role of dephosphorylation in Hh signaling. Finally, the paradigm of Hh signaling would change substantially if further investigation determines that alternative splicing and mRNA regulation do have vital roles in Hh signaling.
METHODS
DNA constructs and cloning.
The ptcΔ136 luciferase reporter in the pGL2 vector (ptcΔ136-GL2) was a gift from P. Beachy (Johns Hopkins University). The ptcΔ136-GL3 reporter was generated by excising the ptcΔ136 promoter as a MluI-HindIII fragment from ptcΔ136-pGL2 and ligating it into the MluI and HindIII sites of pGL3-basic (Promega), which has no 3′ intron. The pAct5C-Hh construct was generated by PCR of full-length hh from a cDNA (gift fromT. Kornberg; UCSF) with a 5′ BamHI site and a 3′ stop followed by a NotI site. This PCR product was then ligated into the pActin5C vector47 and verified by sequencing. The PolIII–Renilla luciferase control construct was generated by PCR amplification of a fragment (base pairs 43,224–43,389 of scaffold AE003823) of the promoter of the D. melanogaster RNA PolIII 128 subunit (RpIII128) with a 5′ BglII and a 3′ SpeI site and ligation of this fragment into the BglII and SpeI sites of pRL-null (Promega). The ptcΔ136-Renilla construct was made by excision of a fragment containing the ptcΔ136 promoter region with SacI and HindIII and subsequent ligation into the SacI and HindIII sites of a form of pRL-null with the NsiI and PstI sites excised and ligated together. The PolIII-luciferase construct was made by PCR of the same fragment of the PolIII promoter described above but with a 5′ BglII site and a 3′ HindIII site. This fragment was then ligated into the BglII and HindIII sites of pGL3-basic.
The pAct5C-Mts and pAct5C-PKA-C1 overexpression constructs were constructed by PCR amplification of a full-length fragment of each gene containing a 5′ BamHI site and a 3′ NotI site. These fragments were restriction-digested and then ligated into a subcloning vector containing three tandem hemagglutinin (HA) epitope tags C-terminal to the cloning site. The 3HA-tagged genes were then excised using EcoRV and XbaI and ligated into the EcoRV and XbaI sites of pActin5C. The pAct5C-Ci construct was made by PCR amplification of the full-length open reading frame of ci with 5′ BamHI and 3′ NotI sites. This fragment was digested and ligated into the BamHI and NotI sites of a modified form of pBluescript. Most of this ci PCR fragment (3,948 bp) was then excised by digestion with MluI and BsaBI and replaced by a MluI-BsaBI fragment taken from the original clone, and the ends were sequenced. The ci fragment was then excised by digestion with BamHI and XbaI and ligated into the BamHI and XbaI sites of pAct5C.
dsRNA.
For D. melanogaster dsRNAs not obtained from the Drosophila RNAi Screening Center, fragments of the genes were amplified by PCR from cDNAs or genomic DNA using T7-tailed oligonucleotides (Supplementary Methods). PCR products obtained from the screening collection were reamplified by PCR using T7 primers. The resulting PCR products were then transcribed using a T7 Megascript kit (Ambion) in accordance with the manufacturer’s instructions, purified using filter plates (MANU03050; Millipore) and quantified by spectrophotometry. Some of the dsRNAs used in Figure 1b were produced by T7 and T3 transcription (Ambion) of full-length clones inserted into pBluescript KS+ (Stratagene), followed by annealing and plate purification. Purified, quantified dsRNAs were stored in sealed deep-well plates at −80 °C. Transfection of dsRNA into clone 8 cells is described in Supplementary Methods. Bathing RNAi of S2-Hh cells was done as described5.
Insect cell culture.
Clone 8 cells were maintained in Shields and Sang M3 medium with 2.0% fetal bovine serum, 2.5% fly extract, 1× penicillin/streptomycin and 0.0125 IU ml−1 insulin. Cells were split at a 1:10 dilution 2–3 d before the assays were done. S2-Hh cells were maintained in Schneider medium with 5.0% fetal bovine serum and 1× penicillin/streptomycin.
Primary and secondary screening.
The primary and secondary screens are described in Supplementary Methods.
Overexpression analysis.
For overexpression of ci, 100 ng of pAct5C-Ci was used in place of pAct5c-Hh. For overexpression of mts, Pka-C1 and cos, 100 ng of the respective pAct5C vectors containing the 3HA-tagged version of each gene was mixed with a master mix containing 125 ng of ptcΔ136-pGL3 experimental reporter, 50 ng of the PolIII-Renilla control reporter and 50 ng of either pAct5C-Hh or pAct5C. The mix was used to transfect clone 8 cells using Effectene (Qiagen). The cells were incubated at 25 °C for 5 d and then assayed for luciferase activity as described in Supplementary Methods.
Mammalian cell culture.
Mouse NIH3T3 cells were seeded on gelatinized 24-well plates at 25,000 cells cm−2 in standard medium with 10% calf serum approximately 18 h before transfection. For the Shh assay, mouse NIH3T3 cells were transfected with 300 ng of ptcΔ36-GL3 luciferase reporter, 100 ng of pSV-b-galactosidase normalizing vector (Promega) and ~100 ng of the experimental or control siRNA pool using Lipofectamine 2000 (Invitrogen). ShhN protein (Shh2I; Biogen)48 was added to the appropriate wells at a concentration of 200 ng ml−1, and the cells were incubated in transfection medium. After 2 d, the wells were changed to low-serum medium (0.5% calf serum) with 200 ng ml−1 ShhN and incubated for an additional 2 d.
Cells were collected in trypsin-EDTA and split into two tubes. Cells in one tube were lysed with Dual Glo luciferase buffer (Promega), and luciferase activity was assayed 10 min later on a plate-reading luminometer. Cells in the other tube were assayed with BetaFluor reaction buffer (Novagen) to quantify β-galactosidase activity. Reactions were incubated at 37 °C for 3 h and measured by fluorimeter. Luciferase values were normalized to β-galactosidase values and expressed as a percentage of Hh stimulation.
siRNA preparation.
PCR primer pairs with T7 polymerase binding sites were designed for each specific gene (Supplementary Methods). The reactions used cDNA from whole mouse embryos at embryonic day 12.5 as a template. The resulting amplicons (200–400 bp) were transcribed with T7 polymerase to generate dsRNA, treated with DNase I (Ambion) and purified on a P-30 column (Bio-Rad). Approximately 1 μg of purified dsRNA was added to 1 unit of Dicer enzyme (Invitrogen) and incubated overnight at 37 °C. Reactions were purified using the manufacturer’s suggested protocol with sequential purification on a G-25 column (Amersham Biosciences) and using a Microcon YM100 filter (Millipore) to remove the remaining uncleaved dsRNA.
Mammalian ortholog selection.
The sequences of all D. melanogaster genes were compared to their mouse orthologs using Tblastn. The top scores were compared against known information using the National Center for Biotechnology Information’s HomoloGene database and data from previous studies. In cases where an ortholog could not be identified, a putative ortholog was assigned if there was an extended region of amino acid homology greater than 45%.
Immunoblotting.
Polyacrylamide gels were run in accordance with standard protocols and transferred using a semi-dry transfer apparatus (E&K Scientific Products). Immunoblotting was also done using standard protocols in 5% milk block. Fu was detected using a rabbit polyclonal antiserum38, and Cos was detected using a mouse monoclonal antiserum (5D6)49. Infrared-conjugated secondary antibodies were used to detect the primary antisera, and the membranes were probed using an Odyssey machine (Li-Cor).
URL.
The National Center for Biotechnology Information’s HomoloGene database is available at http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=homologene.
Supplementary Material
ACKNOWLEDGMENTS
We thank R. Dasgupta, S. Armknecht, K. Kerr, S. Talala, J. Murphy, I. Flockhart,M. Booker, N. Ramadan, B. Mathey-Prevot and the entire staff of the Drosophila RNAi Screening Center at Harvard Medical School for assistance throughout the project; the Harvard Institute of Chemistry and Cell Biology screening center and its staff for use of equipment and advice; A. Kiger, M. Boutros and the Heidelberg Screening Consortium for their work in establishing the dsRNA collection; K. Richards, K. Kerr, L. Hrdlicka and C. Villalta for technical assistance; P. Bradley, H. Agaisse, R. Zhou and the rest of the Perrimon labfor advice; P. Beachy for the ptcΔ136 reporter construct; T. Kornberg and P. Aza-Blanc for the Hh and Ci plasmids; and P. Wolfe for assistance in identifying mammalian orthologs. This work was supported in part by a National Institutes of Health postdoctoral fellowship (K.N.), the Howard Hughes Medical Institute (N.P.), National Institutes of Health grants (T.-Y.L., N.P. and A.P.M.) and the Helen Hay Whitney Foundation (S.A.V.).
Footnotes
Note: Supplementary information is available on the Nature Genetics website.
COMPETING INTERESTS STATEMENT
The authors declare competing financial interests; see the Nature Genetics website for details.
References
- 1.Ogden SK, Ascano M Jr., Stegman MA & Robbins DJ Regulation of Hedgehog signaling: a complex story. Biochem. Pharmacol 67, 805–814 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nybakken K & Perrimon N Hedgehog signal transduction: recent findings. Curr. Opin. Genet. Dev 12, 503–511 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Lum L & Beachy PA The Hedgehog response network: sensors, switches, and routers. Science 304, 1755–1759 (2004). [DOI] [PubMed] [Google Scholar]
- 4.Mello CC & Conte D Jr. Revealing the world of RNA interference. Nature 431, 338–342 (2004). [DOI] [PubMed] [Google Scholar]
- 5.Boutros M et al. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science 303, 832–835 (2004). [DOI] [PubMed] [Google Scholar]
- 6.Cherry S et al. Genome-wide RNAi screen reveals a specific sensitivity of IRES-containing RNA viruses to host translation inhibition. Genes Dev. 19, 445–452 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foley E & O’Farrell PH Functional dissection of an innate immune response by a genome-wide RNAi screen. PLoS Biol. 2, E203 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kiger AA et al. A functional genomic analysis of cell morphology using RNA interference. J. Biol 2, 27 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lum L et al. Identification of Hedgehog pathway components by RNAi in Drosophila cultured cells. Science 299, 2039–2045 (2003). [DOI] [PubMed] [Google Scholar]
- 10.Chen CH et al. Nuclear trafficking of Cubitus interruptus in the transcriptional regulation of Hedgehog target gene expression. Cell 98, 305–316 (1999). [DOI] [PubMed] [Google Scholar]
- 11.DasGupta R, Kaykas A, Moon RT & Perrimon N Functional genomic analysis of the Wnt-wingless signaling pathway. Science 308, 826–833 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Lejeune F & Maquat LE Mechanistic links between nonsense-mediated mRNA decay and pre-mRNA splicing in mammalian cells. Curr. Opin. Cell Biol. 17, 309–315 (2005). [DOI] [PubMed] [Google Scholar]
- 13.Pyronnet S, Pradayrol L & Sonenberg N Alternative splicing facilitates internal ribosome entry on the ornithine decarboxylase mRNA. Cell. Mol. Life Sci. 62, 1267–1274 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noureddine MA, Donaldson TD, Thacker SA & Duronio RJ Drosophila Roc1a encodes a RING-H2 protein with a unique function in processing the Hh signal transducer Ci by the SCF E3 ubiquitin ligase. Dev. Cell 2, 757–770 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Ou CY, Lin YF, Chen YJ & Chien CT Distinct protein degradation mechanisms mediated by Cul1 and Cul3 controlling Ci stability in Drosophila eye development. Genes Dev. 16, 2403–2414 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang J & Struhl G Regulation of the Hedgehog and Wingless signalling pathways by the F-box/WD40-repeat protein Slimb. Nature 391, 493–496 (1998). [DOI] [PubMed] [Google Scholar]
- 17.Deshaies RJ SCF and Cullin/Ring H2-based ubiquitin ligases. Annu. Rev. Cell Dev. Biol 15, 435–467 (1999). [DOI] [PubMed] [Google Scholar]
- 18.de Falco G & Giordano A CDK9 (PITALRE): a multifunctional cdc2-related kinase.J. Cell. Physiol 177, 501–506 (1998). [DOI] [PubMed] [Google Scholar]
- 19.De Luca A, De Falco M, Baldi A & Paggi MG Cyclin T: three forms for different roles in physiological and pathological functions. J. Cell. Physiol 194, 101–107 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Garriga J & Grana X Cellular control of gene expression by T-type cyclin/CDK9 complexes. Gene 337, 15–23 (2004). [DOI] [PubMed] [Google Scholar]
- 21.Hu D, Mayeda A, Trembley JH, Lahti JM & Kidd VJ CDK11 complexes promote pre-mRNA splicing. J. Biol. Chem 278, 8623–8629 (2003). [DOI] [PubMed] [Google Scholar]
- 22.Lahti JM, Xiang J & Kidd VJ The PITSLRE protein kinase family. Prog. Cell Cycle Res. 1, 329–338 (1995). [DOI] [PubMed] [Google Scholar]
- 23.Trembley JH et al. PITSLRE p110 protein kinases associate with transcription complexes and affect their activity. J. Biol. Chem 277, 2589–2596 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Zhang K, Smouse D & Perrimon N The crooked neck gene of Drosophila contains a motif found in a family of yeast cell cycle genes. Genes Dev. 5, 1080–1091 (1991). [DOI] [PubMed] [Google Scholar]
- 25.Raisin-Tani S & Leopold P Drosophila crooked-neck protein co-fractionates in a multiprotein complex with splicing factors. Biochem. Biophys. Res. Commun 296, 288–292 (2002). [DOI] [PubMed] [Google Scholar]
- 26.Chung S, McLean MR & Rymond BC Yeast ortholog of the Drosophila crooked neck protein promotes spliceosome assembly through stable U4/U6.U5 snRNP addition. RNA 5, 1042–1054 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park JW, Parisky K, Celotto AM, Reenan RA & Graveley BR Identification of alternative splicing regulators by RNA interference in Drosophila. Proc. Natl. Acad. Sci. USA 101, 15974–15979 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Burnette JM, Hatton AR & Lopez AJ Trans-acting factors required for inclusion of regulated exons in the Ultrabithorax mRNAs of Drosophila melanogaster. Genetics 151, 1517–1529 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Black DL Mechanisms of alternative pre-messenger RNA splicing. Annu. Rev. Biochem 72, 291–336 (2003). [DOI] [PubMed] [Google Scholar]
- 30.Hastings ML & Krainer AR Pre-mRNA splicing in the new millennium. Curr. Opin. Cell Biol. 13, 302–309 (2001). [DOI] [PubMed] [Google Scholar]
- 31.Kelley RL Initial organization of the Drosophila dorsoventral axis depends on an RNA-binding protein encoded by the squid gene. Genes Dev. 7, 948–960 (1993). [DOI] [PubMed] [Google Scholar]
- 32.Lall S et al. Squid hnRNP protein promotes apical cytoplasmic transport and localization of Drosophila pair-rule transcripts. Cell 98, 171–180 (1999). [DOI] [PubMed] [Google Scholar]
- 33.Barnard DC, Ryan K, Manley JL & Richter JD Symplekin and xGLD-2 are required for CPEB-mediated cytoplasmic polyadenylation. Cell 119, 641–651 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Bastos R, Lin A, Enarson M & Burke B Targeting and function in mRNA export of nuclear pore complex protein Nup153. J. Cell Biol. 134, 1141–1156 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Powers MA, Forbes DJ, Dahlberg JE & Lund E The vertebrate GLFG nucleoporin, Nup98, is an essential component of multiple RNA export pathways. J. Cell Biol. 136, 241–250 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ullman KS, Shah S, Powers MA & Forbes DJ The nucleoporin nup153 plays a critical role in multiple types of nuclear export. Mol. Biol. Cell 10, 649–664 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Therond PP, Knight JD, Kornberg TB & Bishop JM Phosphorylation of the fused protein kinase in response to signaling from hedgehog. Proc. Natl. Acad. Sci. USA 93, 4224–4228 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Robbins DJ et al. Hedgehog elicits signal transduction by means of a large complex containing the kinesin-related protein costal2. Cell 90, 225–234 (1997). [DOI] [PubMed] [Google Scholar]
- 39.Wang QT & Holmgren RA The subcellular localization and activity of Drosophila cubitus interruptus are regulated at multiple levels. Development 126, 5097–5106 (1999). [DOI] [PubMed] [Google Scholar]
- 40.Nybakken KE, Turck CW, Robbins DJ & Bishop JM Hedgehog-stimulated phosphorylation of the kinesin-related protein Costal2 is mediated by the serine/threonine kinase fused. J. Biol. Chem 277, 24638–24647 (2002). [DOI] [PubMed] [Google Scholar]
- 41.Price MA & Kalderon D Proteolysis of the Hedgehog signaling effector Cubitus interruptus requires phosphorylation by Glycogen Synthase Kinase 3 and Casein Kinase 1. Cell 108, 823–835 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Denef N, Neubuser D, Perez L & Cohen SM Hedgehog induces opposite changes in turnover and subcellular localization of patched and smoothened. Cell 102, 521–531 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Jia J et al. Shaggy/GSK3 antagonizes Hedgehog signalling by regulating Cubitus interruptus. Nature 416, 548–552 (2002). [DOI] [PubMed] [Google Scholar]
- 44.Janssens V & Goris J Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J 353, 417–439 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Randsholt NB, Maschat F & Santamaria P Polyhomeotic controls engrailed expression and the hedgehog signaling pathway in imaginal discs. Mech. Dev 95, 89–99 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Collins RT & Cohen SM A genetic screen in Drosophila for identifying novel components of the hedgehog signaling pathway. Genetics 170, 173–184 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bond BJ & Davidson N The Drosophila melanogaster actin 5C gene uses two transcription initiation sites and three polyadenylation sites to express multiple mRNA species. Mol. Cell. Biol 6, 2080–2088 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor FR et al. Enhanced potency of human Sonic hedgehog by hydrophobic modification. Biochemistry 40, 4359–4371 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Ascano M Jr., Nybakken KE, Sosinski J, Stegman MA & Robbins DJ The carboxyl-terminal domain of the protein kinase fused can function as a dominant inhibitor of hedgehog signaling. Mol. Cell. Biol 22, 1555–1566 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.