Abstract
Periodontitis is caused by dysbiotic subgingival bacterial communities that may lead to increased bacterial invasion into gingival tissues. Although shifts in community structures associated with transition from health to periodontitis have been well characterized, the nature of bacteria present within the gingival tissue of periodontal lesions is not known. To characterize microbiota within tissues of periodontal lesions and compare them with plaque microbiota, gingival tissues and subgingival plaques were obtained from 7 patients with chronic periodontitis. A sequencing analysis of the 16S rRNA gene revealed that species richness and diversity were not significantly different between the 2 groups. However, intersubject variability of intratissue communities was smaller than that of plaque communities. In addition, when compared with the plaque communities, intratissue communities were characterized by decreased abundance of Firmicutes and increased abundance of Fusobacteria and Chloroflexi. In particular, Fusobacterium nucleatum and Porphyromonas gingivalis were highly enriched within the tissue, composing 15% to 40% of the total bacteria. Furthermore, biofilms, as visualized by alcian blue staining and atomic force microscopy, were observed within the tissue where the degradation of connective tissue fibers was prominent. In conclusion, very complex bacterial communities exist in the form of biofilms within the gingival tissue of periodontal lesions, which potentially serve as a reservoir for persistent infection. This novel finding may prompt new research on therapeutic strategies to treat periodontitis.
Keywords: periodontitis, gingiva, microbial ecology, metagenomics, bacteria, Fusobacterium nucleatum
Introduction
The human body is colonized by a large number of microorganisms, and the human microbiome is a part of the human physiology. The advent of next-generation sequencing technologies has enabled us to characterize not only the composition and function of the human microbiome but also variations across anatomic sites, time, and individuals (Human Microbiome Project Consortium 2012). Accumulating evidence suggests that many diseases are associated with altered resident bacterial communities, leading to dysbiosis (Frank et al. 2011; DeGruttola et al. 2016).
Periodontitis is the best example of a disease caused by dysbiosis of the human microbiome, specifically subgingival communities. The altered subgingival microbiome induces gingival inflammation that accompanies immune cell infiltration, leading to the destruction of the connective tissue and alveolar bone (Darveau 2010; Cekici et al. 2014). Changes in subgingival bacterial communities characterizing the transition from health to periodontitis include 1) increases in total bacterial load and richness; 2) shifts from gram-positive aerobic cocci-dominated communities to predominantly strict anaerobic, gram-negative, and motile organism-dominated ones; and 3) shifts in the core microbiome through an increase in low-abundance community members, including “established” periodontal pathogens and newly identified periodontitis-associated taxa (Marsh 1994; Abusleme et al. 2013; Galimanas et al. 2014).
Of the bacteria that increase in periodontitis, several species—including Fusobacterium nucleatum, Prevotella intermedia, Treponema denticola, and Porphyromonas gingivalis—have an ability to invade the gingival tissue through transcellular and/or paracellular pathways (Ji et al. 2015). The presence of bacteria, including the aforementioned species, in the epithelium and connective tissue of periodontal lesions has been reported by electron microscopy, immunofluorescence, immunohistochemistry, and in situ hybridization (Frank 1980; Saglie et al. 1982; Pekovic and Fillery 1984; Saglie et al. 1988; Kim et al. 2010). Higher levels of bacteria were detected in periodontal lesions than in healthy sites (Choi et al. 2014). In murine models of periodontitis, the amounts of bacteria within gingival tissues were positively correlated with T-cell infiltration and alveolar bone loss (Choi et al. 2013; Baek et al. 2017). Therefore, the bacteria present within tissues seem to be important for the progression of periodontitis.
However, the characteristics of bacterial communities present within the gingival tissue of periodontal lesions have not been studied yet. This study aimed to characterize bacterial communities located within gingival tissues in comparison with subgingival plaque communities obtained from periodontal lesions.
Materials and Methods
Study Samples
Seven subjects were recruited from among adult patients with chronic periodontitis who needed surgery after undergoing scaling and root debridement at the Anam Hospital of Korea University. This study was approved by the Institutional Review Board for Human Subjects of the Korea University Anam Hospital (No. ED14038). Informed consent was obtained from all subjects. Exclusion criteria included medication history of antibiotics or anti-inflammatory agents within 6 wk, ongoing medication of phenytoin or cyclosporine, pregnancy/breast-feeding, uncontrolled diabetes, and other systemic diseases that could influence the prognosis of periodontitis. Subgingival plaque and gingival tissue samples were obtained during a modified Widman flap surgery. The probing depth, clinical attachment level, and bleeding on probing were recorded from 6 sites of each tooth in the surgical site (Appendix Table).
From each patient, a subgingival plaque sample was taken with a sterile paper point (Caulk-Dentsply) from a bleeding on probing–positive site showing probing depth ≥5 mm after removal of the supragingival plaque and stored at –80 °C until use. From the same surface (buccal or lingual) of the tooth from which the subgingival plaque was sampled, gingival tissue samples for sequencing analysis were obtained and then stored in a sterile tube at –80 °C. Gingival tissue samples for histology were obtained from the opposite surface of the tooth from which the subgingival plaque was sampled or from the adjacent tooth and transferred into a tube containing zinc fixative (BD Bioscience).
The sections of gingival biopsies obtained from the periodontal lesions of 5 patients in a previous study (Choi et al. 2014) were additionally used for histologic studies. The mean probing depth and clinical attachment level of the sampled sites were 5.3 ± 0.7 mm and 6.4 ± 1.0 mm, respectively.
Preparation of Bacterial DNA Samples
Bacterial genomic DNA was isolated from the subgingival plaque and gingival tissues with a commercial kit for soil bacteria (MO BIO Laboratories). The gingival tissues were first washed and incubated with 1 mL of PBS containing lysozyme (300 μg/mL) and antibiotics (penicillin, streptomycin, and gentamicin) at 37 °C for 1 h to damage the cell walls of surface bacteria. Subsequently, bacterial DNA on the surface of tissues was digested with DNase I. After heat inactivation of DNase I and washing, the tissues were homogenized and subjected to bacterial DNA extraction. After DNase I treatment, bacterial DNA was not detected in the wash solution (Appendix Fig. 1A). When the degree of potential contamination with plaque bacteria was estimated with additional plaque samples, treatment with lysozyme and DNase I removed >99% of bacterial DNA (Appendix Fig. 1B).
Estimation of Total Bacterial Loads
Because sequencing of the bacterial 16S rRNA gene does not give absolute measurements, total bacterial loads in plaque and tissue samples were estimated by real-time polymerase chain reaction (PCR) with universal primers targeting the 16S rRNA gene and a standard curve. Details of the real-time PCR method are described in the Appendix.
Analysis of Bacterial Communities
DNA fragments including the V3-to-V4 hypervariable regions of the bacterial 16S rRNA gene were amplified by PCR as previously described (Lee and Eom 2016); then, the PCR products were sequenced with an Illumina MiSeq Sequencing System at ChunLab Inc. Processing and analysis of sequences were performed as previously described with the CLcommunity software provided by ChunLab Inc. (Kim et al. 2016). For the uncharacterized phylotypes mentioned in this article, oral taxon number was assigned by searching against the Human Oral Microbiome Database (http://www.homd.org/). The sequence data are available in the NCBI Sequence Read Archive under BioProject accession SRP106728.
Hematoxylin and Eosin Stain, Alcian Blue Stain, In Situ Hybridization, and Image Analysis
Fixed gingival tissues were embedded with paraffin. Serial 4-μm-thick sections were subjected to hematoxylin and eosin staining, alcian blue staining, and in situ hybridization of P. gingivalis and F. nucleatum with a species-specific probe for each. For alcian blue staining, deparaffinized sections were sequentially treated with 0.1N hydrochloric acid (HCl) and 1% alcian blue (pH 1.0) solutions and then counterstained with nuclear fast red solution.
Preparation of P. gingivalis–specific probe and in situ hybridization were performed according to a previously described method (Choi et al. 2015). A 313-bp DNA fragment of the F. nucleatum 16S rRNA gene was amplified by PCR with the forward (5′-AACTTAGGTTTGGGTGGCGG-3′) and reverse (5′-TGCTGGATCAGACTCTTGGT-3′) primers and labeled with digoxigenin with a commercial kit (Roche Applied Science). The specificity of the digoxigenin-labeled probe for F. nucleatum was confirmed by dot blotting with the genomic DNA samples of 6 bacterial species (Appendix Fig. 2).
Three areas with varying degrees of alcian blue staining were chosen from each sample, and anatomically corresponding areas in the in situ hybridized sections were photographed at ×200 magnification. The signal intensities of alcian blue staining, F. nucleatum, and P. gingivalis were analyzed with ImageJ software (National Institute of Mental Health).
Atomic Force Microscopy
Sections of gingival tissues were examined by atomic force microscopy (AFM) after deparaffinization, staining with alcian blue, and drying. The images were taken with the noncontact mode of an XE-100 atomic force microscope (Park Systems). A PPP-NCHR probe (NanoSensors) with a resonance frequency of 330 kHz and a force constant of 42 N/m was applied to analyze the morphology. The typical scan rate was kept at 0.1 Hz.
Statistical Analysis
The Wilcoxon signed-rank test was used to compare various parameters between the subgingival plaque and intratissue communities. The Kruskal-Wallis test, followed by Mann-Whitney U test, was used to determine differences in relative abundance among the 5 F. nucleatum subspecies. Spearman’s rank correlation test was used to determine the correlation between 2 measured parameters. All statistical analyses were performed with SPSS Statistics 22 software (IBM). Significance was set at P < 0.05.
Results
Intratissue Bacterial Communities Were as Complex as the Subgingival Plaque Communities but Had Altered Composition
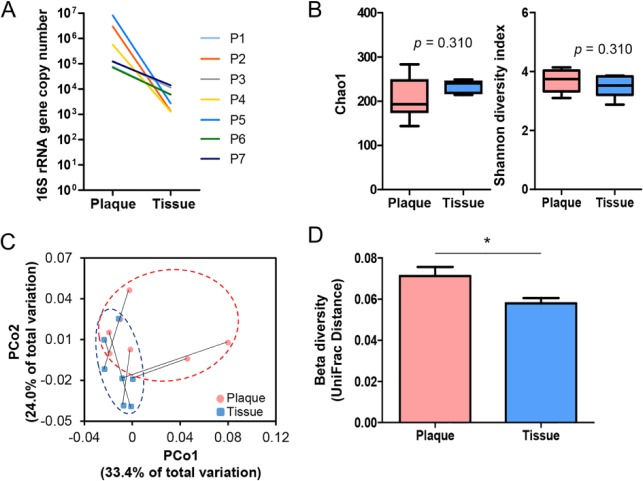
Total bacterial loads estimated by real-time PCR of the 16S rRNA gene were lower in intratissue than those in subgingival plaque samples by 1 to 4 orders of magnitude (Fig. 1A). From the 7 subgingival plaque and 7 intratissue bacterial communities, a total of 261,690 filtered sequences (13,668 to 19,962 per sample) with an average length of 417 bp were obtained, which presented ≥99.7% Good’s coverage for each community. The species richness determined by Chao 1 index (210.1 ± 48.0 vs. 232.8 ± 13.9, P = 0.310) and Shannon diversity (3.7 ± 0.4 vs. 3.5 ± 0.6, P = 0.310) were not different between the 2 sample sites (Fig. 1B). To compare the degree of phylogenic distances among communities, UniFrac-based principal coordinates analysis was performed. In the principal coordinates analysis plot, the tissue samples were partially separated from the plaque samples and clustered in a smaller area (Fig. 1C), presenting a significantly decreased UniFrac distance (0.072 ± 0.004 vs. 0.059 ± 0.003), which is an index of intragroup intersubject variability (Fig. 1D).
Figure 1.
Alpha and beta diversities of subgingival plaque and intratissue communities within periodontal lesions. Subgingival plaque and gingival tissue samples were obtained from 7 patients with chronic periodontitis. Bacterial genomic DNA was prepared as described in the Materials and Methods section and subjected to MiSeq sequencing of the 16S rRNA gene. (A) Total bacterial load in each sample was estimated by real-time polymerase chain reaction with universal primers targeting the bacterial 16S rRNA gene. The total bacterial load was expressed as the 16S rRNA gene copy number in the total DNA obtained from each sample. P, patient. (B) The Chao1 and Shannon index are expressed with box and whisker plots (P value by 2-tailed Wilcoxon signed-rank test). (C) The principal coordinates analysis (PCoA) plot was generated with weighted UniFrac metric. Samples from the same subject are connected with a solid line. (D) The intersubject UniFrac distances of the subgingival plaque and intratissue communities were obtained with a weighted metric. *P < 0.05 by 2-tailed Wilcoxon signed-rank test.
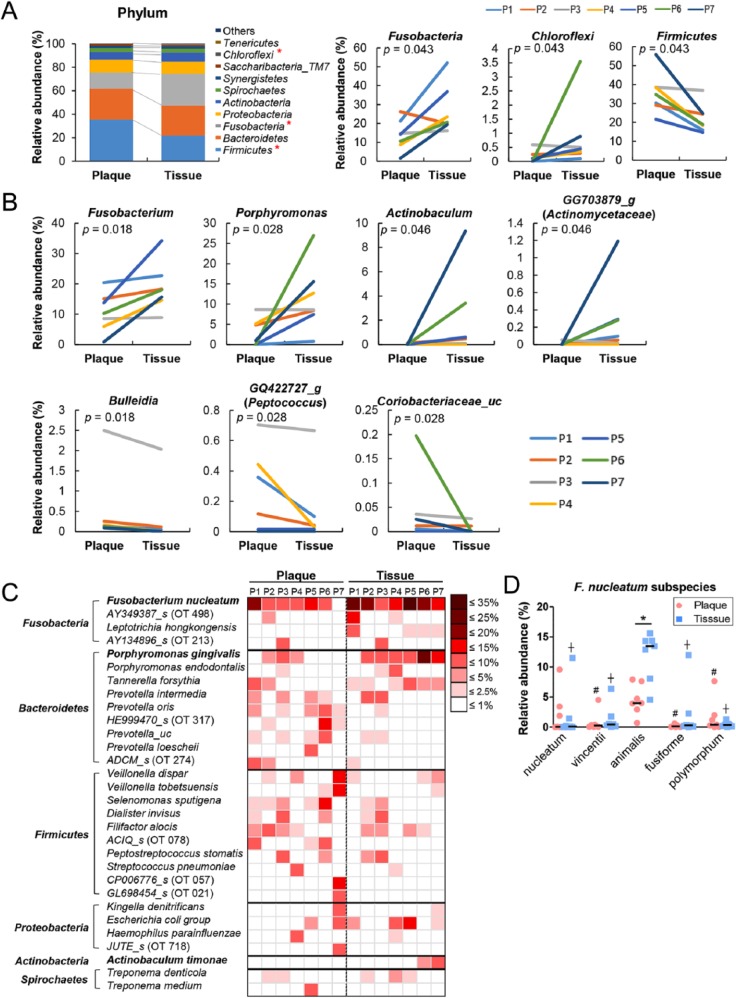
To investigate differences in the composition of bacterial communities, the relative abundance of each taxon between the 2 groups was compared from phylum to subspecies. Although a total of 14 and 17 phyla were identified from the subgingival plaque and tissue samples, respectively, only 8 phyla—Firmicutes, Bacteroidetes, Fusobacteria, Proteobacteria, Actinobacteria, Spirochaetes, Synergistetes, and Saccharibacteria_ TM7—were detected in all samples. Differences in the bacterial composition between the plaque and tissue communities were evident. When compared with the subgingival plaque communities, the tissue communities harbored significantly increased abundances of Fusobacteria and Chloroflexi but decreased abundance of Firmicutes (Fig. 2A). At the genus level, the relative abundance of Fusobacterium, Porphyromonas, Actinobaculum, and GG703879_g (Actinomycetaceae family) was significantly increased in the tissue, whereas the abundance of Bulleidia, GQ422727_g (Peptococcaceae family) and Coriobacteriaceae_uc was decreased (Fig. 2B). At the species level, 24 species/phylotypes were differently distributed between the 2 groups (Table). Among these species, F. nucleatum and P. gingivalis were highly enriched in the tissue communities, composing 15% to 40% of the total bacteria (Fig. 2C). Among the 5 F. nucleatum subspecies, the abundance of F. nucleatum subsp. animalis was higher than that of the other subspecies in plaque and tissue and was significantly increased in the tissue samples (Fig. 2D).
Figure 2.
Differences in bacterial composition between the subgingival plaque and intratissue communities. The relative abundance of each taxon between the subgingival plaque and intratissue communities was compared. P, patient. (A) The members of top 10 phyla are shown (left panel). Three phyla were differently distributed between the 2 communities (right panel). (B) Seven genera were differently distributed between the 2 communities (P value by 2-tailed Wilcoxon signed-rank test). (C) A heat map was generated for the species/phylotypes whose relative abundance was >2.5% in any sample. (D) The relative abundance of Fusobacterium nucleatum subspecies is shown. #P < 0.05 compared with animalis subspecies in plaques. †P < 0.05 compared with animalis subspecies in tissues by Kruskal-Wallis test, followed by Mann-Whitney U test. *P < 0.05 by 2-tailed Wilcoxon signed-rank test.
Table.
Relative Abundance of Species/Phylotype Differently Distributed between Subgingival Plaque and Gingival Tissues.
| Plaque (n = 7)a | Tissue (n = 7)a | P Valueb | |
|---|---|---|---|
| Fusobacterium nucleatum group | 8.46 (0.78 to 19.52) | 16.03 (8.55 to 32.67) | 0.018 |
| Porphyromonas gingivalis | 0.83 (0 to 6.26) | 6.32 (0.61 to 23.54) | 0.018 |
| Actinobaculum timonae | 0 (0 to 0.11) | 0.43 (0 to 8.72) | 0.046 |
| Porphyromonas_uc | 0.15 (0.01 to 0.66) | 0.96 (0.08 to 3.35) | 0.043 |
| Fusobacterium_uc | 0.45 (0.03 to 0.69) | 0.74 (0.32 to 1.34) | 0.028 |
| APVJ_s (Chloroflexi OT 439) | 0 (0 to 0.56) | 0.42 (0.10 to 3.37) | 0.043 |
| JQ473872_s (Bacteroidetes OT 336) | 0.06 (0 to 0.19) | 0.45 (0.02 to 0.64) | 0.018 |
| GG703879_s (Actinobacteria OT 848) | 0 (0 to 0.05) | 0.07 (0 to 1.06) | 0.046 |
| Corynebacterium durum | 0 (0 to 0.18) | 0.03 (0 to 0.23) | 0.028 |
| Leptotrichia hofstadii | 0 (0 to 0.03) | 0.02 (0 to 0.28) | 0.028 |
| Cardiobacterium_uc | 0 (0 to 0.02) | 0.04 (0 to 0.10) | 0.046 |
| Lautropia_f_uc_s | 0 (0 to 0.01) | 0.02 (0 to 0.03) | 0.028 |
| GQ422712_s (Firmicutes OT 093) | 0 (0 to 0.03) | 0.01 (0 to 0.04) | 0.043 |
| Cardiobacteriaceae_uc_s | 0 (0 to 0) | 0.01 (0 to 0.05) | 0.028 |
| AM420132_s (TM7 OT 346) | 0.37 (0.08 to 0.91) | 0.09 (0 to 0.53) | 0.028 |
| Neisseria bacilliformis | 0.01 (0 to 1.57) | 0 (0 to 0.13) | 0.043 |
| AY005448_s (TM7 OT 349) | 0.01 (0 to 0.93) | 0 (0 to 0.24) | 0.043 |
| Prevotella buccae | 0.11 (0.03 to 0.53) | 0 (0 to 0.07) | 0.018 |
| Johnsonella ignava | 0.03 (0 to 0.49) | 0 (0 to 0.21) | 0.043 |
| Saccharimonas_uc | 0.05 (0.01 to 0.28) | 0.02 (0 to 0.24) | 0.018 |
| Bulleidia_uc | 0.01 (0.01 to 0.12) | 0 (0 to 0.13) | 0.028 |
| Atopobium parvulum | 0.02 (0 to 0.10) | 0.01 (0 to 0.03) | 0.043 |
| Coriobacteriaceae_uc_s | 0.01 (0 to 0.20) | 0 (0 to 0.03) | 0.043 |
| JX010937_s (Synergistetes OT 359) | 0.01 (0 to 0.11) | 0 (0 to 0.03) | 0.043 |
Relative abundance expressed as median percentage (minimum to maximum.)
Two-tailed Wilcoxon signed-rank test.
Bacterial Aggregates in the Form of Biofilms Were Observed within Gingival Tissues
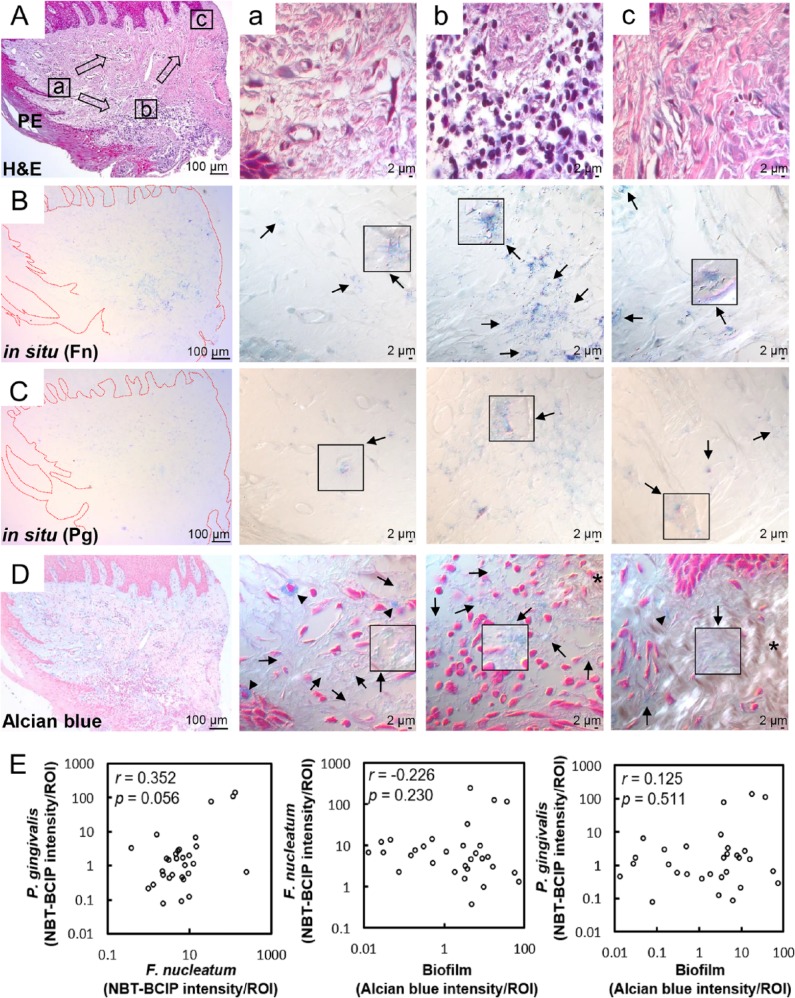
The distribution of the 2 highly abundant species F. nucleatum and P. gingivalis in gingival tissues was examined by in situ hybridization with specific probes. Both species were detected throughout the section from the pocket epithelium, connective tissue, and oral epithelium of all gingival biopsies but were particularly abundant at the area of inflammatory infiltration (Fig. 3A–C).
Figure 3.
Distribution of Fusobacterium nucleatum, Porphyromonas gingivalis, and biofilm within the gingival tissue. (A) Tissue sections were stained with hematoxylin and eosin (H&E). Three areas (a, b, c) were examined under high magnification (×1,000) with differential interference contrast microscopy. Arrows indicate the potential directions of infection spread. PE, pocket epithelium. (B, C) Tissue sections were in situ hybridized with F. nucleatum– and P. gingivalis–specific probes, respectively. Arrows, bacterial signals; insets, areas with biofilm-like structure are magnified. (D) Tissue sections were stained with 1% alcian blue for acid mucopolysaccharide and counterstained with nuclear fast red. Arrows, biofilm-like structures; arrowheads, mast cells; asterisk, area with intact connective tissue fibers. (E) Correlation plots between bacterial signals and biofilm formation (r and P values by Spearman’s rank correlation test). ROI, region of interest. See Appendix Figure 3 for DIC images taken by confocal microscopy.
Because the bacteria were often observed as aggregates of different sizes, we hypothesized that they form biofilms. To confirm this possibility, alcian blue staining was performed to visualize polysaccharides in the extracellular polymeric substances (EPSs) of biofilms. At low magnification, weak alcian blue staining was observed in almost half the dense connective tissue along the pocket epithelium, where abundant white spaces were observed in the hematoxylin and eosin–stained section. At high magnification, cobweb-like structures with bead-like bacterial clusters (arrows in Fig. 3D) were readily observed in free spaces formed by degradation of connective tissue fibers but were rarely observed in the areas where fibers were relatively intact (asterisk in Fig. 3D).
To determine associations between the biofilm and F. nucleatum or P. gingivalis, 3 areas with varying degrees of alcian blue staining were chosen from each sample, and the intensities of alcian blue staining and the signals of F. nucleatum and P. gingivalis were measured. Although the amounts of F. nucleatum and P. gingivalis tended to be positively correlated, suggesting coexistence of the 2 species, the amount of neither species was associated with the amount of biofilm (Fig. 3E).
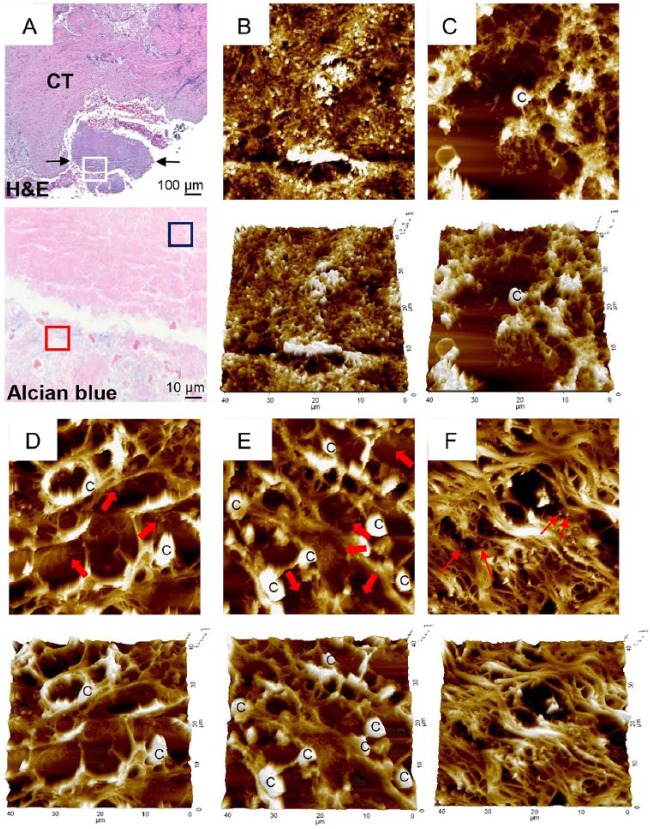
The presence of biofilms within gingival tissues was further verified by AFM. First, a piece of plaque biofilm coembedded with tissue was examined. The central area of it was not stained with alcian blue, and it revealed tightly packed bacterial cells under AFM. The periphery of the plaque biofilm was stained with alcian blue, and it presented the cobweb-like structures with bacterial clusters under AFM (Fig. 4A–C). Similar cobweb-like structures were also observed within tissues where collagen fibers were severely degraded (Fig. 4D, E). In the areas where the fibers were intact, scattered bacterial cells adhering to collagen fibers were observed (Fig. 4F).
Figure 4.
Atomic force microscopy examination of biofilm. (A) A piece of plaque biofilm coembedded with tissue was located in a hematoxylin and eosin (H&E)–stained section (between 2 arrows in top panel). CT, connective tissue. Area corresponding to the white-boxed region in the H&E-stained section was examined in the serial section stained with alcian blue under high magnification (×1,000, bottom panel). Two typical areas were chosen based on alcian blue staining and examined by atomic force microscopy: (B) blue- and (C) red-boxed areas from panel A. Areas a–c (D–F, respectively) from Figure 3A were examined by atomic force microscopy. Thick arrows indicate biofilm-like structures within tissue. Thin arrows indicate scattered bacterial cells. C, eukaryotic cell.
Discussion
Periodontitis involves polymicrobial infection resulting from a homeostatic imbalance in the subgingival microbiome, and the shifts in the subgingival microbiota from health to periodontitis are relatively well established. We now report that very complex bacterial communities exist and can form biofilms within gingival tissues of periodontal lesions.
The presence of heterogeneous bacteria within gingival tissues or buccal epithelial cells has been reported for decades (Saglie et al. 1982; Rudney et al. 2005). However, the complexity and composition of the intratissue bacterial communities were first revealed in this study. In the periodontal pockets, bacteria exist as biofilms composed of hundreds of species rather than as individual planktonic cells. Therefore, bacteria that invade tissues are also likely to be in the form of multispecies aggregates and reflect the composition of plaque community. Actually, bacterial aggregates of a corncob-like structure penetrating into intercellular spaces of the pocket epithelium had been demonstrated by scanning electron microscopy (Saglie et al. 1982). The intratissue bacterial communities would eventually differentiate from the parent community as a result of adaptation to the new environment. The significantly decreased intersubject variability in the tissue communities suggests that the environment within the tissue is more uniform than that in periodontal pockets.
The validity of the current study depends on whether bacteria on the tissue surface were completely removed during sample preparation. Our preliminary experiment suggested that contamination of the tissue samples with bacteria on the tissue surface cannot be excluded. However, in situ hybridization revealed relatively few bacteria on the surface of tissues that were embedded after a simple wash procedure, while substantial levels of bacteria were observed throughout the tissues. Furthermore, 1 of the established periodontal pathogens (P. gingivalis) was rarely detected in the plaque samples treated with lysozyme and DNase I (Appendix Fig. 1B) but highly enriched within the tissues, which may be attributed to its immune-evasive characteristics and ability to survive within macrophages (Wang et al. 2007; Hajishengallis 2011). Therefore, the differences in bacterial composition between the plaque and tissue communities seem to be valid.
As a bridge between the early and late colonizers in dental biofilm, F. nucleatum is the most prevalent taxon in health and periodontitis, but its abundance is increased in periodontal lesions (Socransky and Haffajee 2005; Abusleme et al. 2013). Coinfection with F. nucleatum and P. gingivalis has been shown to synergistically enhance pathogenicity, alveolar bone loss, and soft tissue destruction (Metzger et al. 2009; Polak et al. 2009). Furthermore, P. gingivalis enhanced biofilm formation by F. nucleatum, whereas F. nucleatum, a species with higher tolerance to oxygen, supported the growth of P. gingivalis in an aerated environment (Diaz et al. 2002; Saito et al. 2008). Therefore, enrichment of P. gingivalis and F. nucleatum in the tissue communities suggests that these 2 species also interact within tissues to facilitate survival in the new environment. Indeed, the distribution of P. gingivalis and F. nucleatum examined by in situ hybridization revealed a tendency for them to coexist.
In addition to P. gingivalis and F. nucleatum, Chloroflexi OT 439—a phylotype reported to be a member of core-microbiome in periodontitis (Abusleme et al. 2013)—was overrepresented in tissues. This OT 439 was recently cultured in the presence of a helper strain F. nucleatum (Vartoukian et al. 2016), which is consistent with our finding. Tannerella forsythia and T. denticola, other established periodontal pathogens, and Filifactor alocis, a new candidate periodontal pathogen, were also observed in substantial amounts in the tissue communities (Fig. 2C), although the abundances of these species were not different between the locations. The species/phylotypes that are present in substantial amounts or overrepresented within gingival tissues may have a more important role in the pathogenesis of periodontitis.
The most interesting finding in this study was the presence of biofilms within gingival tissues, which was confirmed by alcian blue staining and AFM. Bacterial plaques located on the resorbed surface of alveolar bone obtained from a patient with juvenile periodontitis were presented earlier by scanning electron microscopy (Carranza et al. 1983). Biofilm-like structures were also reported in kidney abscesses of BALB/c mice caused by infection with Staphylococcus aureus (Cheng et al. 2009).
Although F. nucleatum and P. gingivalis were detected in the areas of the biofilm, the degree of biofilm formation within the tissues was not associated with the amounts of either F. nucleatum or P. gingivalis. Other species may play a role in biofilm formation within the tissues. For example, Actinobaculum timonae, which was overrepresented in the tissues, was isolated from the duodenal biopsy of a patient with enteritis catheter-related bacteremia, a biofilm disease (Drancourt et al. 2004). Otherwise, as shown in the plaque biofilm (Fig. 4A), the degree of biofilm formation based on the EPS staining may not necessarily correlate with the number of bacteria: only the area with a loose structure, not the area highly packed with bacterial cells, was stained with alcian blue. Similarly, the EPS of the biofilm within tissue was readily observed only in the areas where collagen breakdown was evident. Collagen breakdown in gingival tissues has been attributed to inflammatory cytokines and matrix metalloproteinases secreted by inflammatory cells (Seguier et al. 2000; Seguier et al. 2001; Ejeil et al. 2003). The diverse functions of EPSs include accumulation and stabilization of enzymes (Flemming and Wingender 2010); therefore, EPSs may facilitate collagen breakdown by concentrating enzymes of bacterial and host origins.
In Figure 3A, bacterial infection seemed to have spread from area a to b and c along the apical migration of the junctional epithelium during progression of periodontitis (Bosshardt and Lang 2005). In the initial stage of periodontitis, the junctional epithelium and the primary focus of infection must have been located near area a, which migrated to near area b. We speculate that the massive immune cell infiltration seen in area b had already passed area a and cleared the infected bacteria but not completely, probably due to the protection afforded by biofilms. Area c may represent a region where the bacterial infection spread, but few immune cells had arrived yet. Although the triggering cue for biofilm formation in gingival tissues is not currently known, biofilms would definitely assist the bacteria to resist host immune responses.
Clinical implications for the presence of biofilms in the gingival tissue are not clear yet. Biofilm is one of the best examples of bacterial persistence and thus may be the cause of chronic inflammation. Alternatively, it may be beneficial by limiting the systemic spread of bacteria. Although the immune response to bacterial infection usually accompanies tissue damage, the damaged tissue is repaired once the infection is cleared (Garlet 2010). As long as biofilms persist within gingival tissue, however, the immune response would not be switched from inflammation to the reparative phase, hampering the repair of destroyed gingival tissues. According to the current paradigm, the mechanical treatment of periodontitis is focused on the elimination of subgingival biofilm (Drisko 2014). Whether the removal of biofilm-containing gingival tissues improves long-term prognosis merits further study.
One of the limitations of this study is the small sample size. Although we identified differences between subgingival and intratissue communities through parallel comparison, more samples, including those from periodontally healthy subjects, need to be studied in the future.
In conclusion, bacterial communities present within the gingival tissues of periodontal lesions are as complex as subgingival plaque communities and can form biofilm. These findings may provide a novel insight into the pathogenesis of periodontitis and prompt new research on therapeutic strategies to treat periodontitis.
Author Contributions
K. Baek, contributed to design, data acquisition, analysis, and interpretation, drafted and critically revised the manuscript; S. Ji, contributed to data acquisition, critically revised the manuscript; Y. Choi, contributed to conception, design, and data interpretation, critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplementary Material
Acknowledgments
We acknowledge professor Hyun-Man Kim at the Department of Cell and Developmental Biology for providing histologic expertise.
Footnotes
A supplemental appendix to this article is available online.
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (2016-929358 and 2016R1E1A1A01942402).
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Abusleme L, Dupuy AK, Dutzan N, Silva N, Burleson JA, Strausbaugh LD, Gamonal J, Diaz PI. 2013. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 7(5):1016–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baek KJ, Choi YS, Kang CK, Choi Y. 2017. The proteolytic activity of Porphyromonas gingivalis is critical in a murine model of periodontitis. J Periodontol. 88(2):218–224. [DOI] [PubMed] [Google Scholar]
- Bosshardt DD, Lang NP. 2005. The junctional epithelium: from health to disease. J Dent Res. 84(1):9–20. [DOI] [PubMed] [Google Scholar]
- Carranza FA, Jr, Saglie R, Newman MG, Valentin PL. 1983. Scanning and transmission electron microscopic study of tissue-invading microorganisms in localized juvenile periodontitis. J Periodontol. 54(10):598–617. [DOI] [PubMed] [Google Scholar]
- Cekici A, Kantarci A, Hasturk H, Van Dyke TE. 2014. Inflammatory and immune pathways in the pathogenesis of periodontal disease. Periodontol 2000. 64(1):57–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng AG, Kim HK, Burts ML, Krausz T, Schneewind O, Missiakas DM. 2009. Genetic requirements for Staphylococcus aureus abscess formation and persistence in host tissues. FASEB J. 23(10):3393–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Kim YC, Baek KJ, Choi Y. 2015. In situ detection of bacteria within paraffin-embedded tissues using a digoxin-labeled DNA probe targeting 16S rRNA. J Vis Exp. 99:e52836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi YS, Kim YC, Ji S, Choi Y. 2014. Increased bacterial invasion and differential expression of tight-junction proteins, growth factors, and growth factor receptors in periodontal lesions. J Periodontol. 85(8):e313–e322. [DOI] [PubMed] [Google Scholar]
- Choi YS, Kim YC, Jo AR, Ji S, Koo KT, Ko Y, Choi Y. 2013. Porphyromonas gingivalis and dextran sulfate sodium induce periodontitis through the disruption of physical barriers in mice. Eur J Inflamm. 11(2):419–431. [Google Scholar]
- Darveau RP. 2010. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 8(7):481–490. [DOI] [PubMed] [Google Scholar]
- DeGruttola AK, Low D, Mizoguchi A, Mizoguchi E. 2016. Current understanding of dysbiosis in disease in human and animal models. Inflamm Bowel Dis. 22(5):1137–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz PI, Zilm PS, Rogers AH. 2002. Fusobacterium nucleatum supports the growth of Porphyromonas gingivalis in oxygenated and carbon-dioxide-depleted environments. Microbiology. 148(Pt 2):467–472. [DOI] [PubMed] [Google Scholar]
- Drancourt M, Berger P, Raoult D. 2004. Systematic 16S rRNA gene sequencing of atypical clinical isolates identified 27 new bacterial species associated with humans. J Clin Microbiol. 42(5):2197–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drisko CL. 2014. Periodontal debridement: still the treatment of choice. J Evid Based Dent Pract. 14(Suppl):33–41. [DOI] [PubMed] [Google Scholar]
- Ejeil AL, Gaultier F, Igondjo-Tchen S, Senni K, Pellat B, Godeau G, Gogly B. 2003. Are cytokines linked to collagen breakdown during periodontal disease progression? J Periodontol. 74(2):196–201. [DOI] [PubMed] [Google Scholar]
- Flemming HC, Wingender J. 2010. The biofilm matrix. Nat Rev Microbiol. 8(9):623–633. [DOI] [PubMed] [Google Scholar]
- Frank DN, Zhu W, Sartor RB, Li E. 2011. Investigating the biological and clinical significance of human dysbioses. Trends Microbiol. 19(9):427–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank RM. 1980. Bacterial penetration in the apical pocket wall of advanced human periodontitis. J Periodontal Res. 15(6):563–573. [DOI] [PubMed] [Google Scholar]
- Galimanas V, Hall MW, Singh N, Lynch MD, Goldberg M, Tenenbaum H, Cvitkovitch DG, Neufeld JD, Senadheera DB. 2014. Bacterial community composition of chronic periodontitis and novel oral sampling sites for detecting disease indicators. Microbiome. 2:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garlet GP. 2010. Destructive and protective roles of cytokines in periodontitis: a re-appraisal from host defense and tissue destruction viewpoints. J Dent Res. 89(12):1349–1363. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G. 2011. Immune evasion strategies of Porphyromonas gingivalis. J Oral Biosci. 53(3):233–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Human Microbiome Project Consortium. 2012. Structure, function and diversity of the healthy human microbiome. Nature. 486(7402):207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji S, Choi YS, Choi Y. 2015. Bacterial invasion and persistence: critical events in the pathogenesis of periodontitis? J Periodontal Res. 50(5):570–585. [DOI] [PubMed] [Google Scholar]
- Kim YC, Ko Y, Hong SD, Kim KY, Lee YH, Chae C, Choi Y. 2010. Presence of Porphyromonas gingivalis and plasma cell dominance in gingival tissues with periodontitis. Oral Dis. 16(4):375–381. [DOI] [PubMed] [Google Scholar]
- Kim YJ, Choi YS, Baek KJ, Yoon SH, Park HK, Choi Y. 2016. Mucosal and salivary microbiota associated with recurrent aphthous stomatitis. BMC Microbiol. 16(Suppl 1):57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SY, Eom YB. 2016. Analysis of microbial composition associated with freshwater and seawater. Biomed Sci Lett. 22(4):150–159. [Google Scholar]
- Marsh PD. 1994. Microbial ecology of dental plaque and its significance in health and disease. Adv Dent Res. 8(2):263–271. [DOI] [PubMed] [Google Scholar]
- Metzger Z, Lin YY, DiMeo F, Ambrose WW, Trope M, Arnold RR. 2009. Synergistic pathogenicity of Porphyromonas gingivalis and Fusobacterium nucleatum in the mouse subcutaneous chamber model. J Endod. 35(1):86–94. [DOI] [PubMed] [Google Scholar]
- Pekovic DD, Fillery ED. 1984. Identification of bacteria in immunopathological mechanisms of human periodontal diseases. J Periodontal Res. 19(4):329–351. [DOI] [PubMed] [Google Scholar]
- Polak D, Wilensky A, Shapira L, Halabi A, Goldstein D, Weiss EI, Houri-Haddad Y. 2009. Mouse model of experimental periodontitis induced by Porphyromonas gingivalis/Fusobacterium nucleatum infection: bone loss and host response. J Clin Periodontol. 36(5):406–410. [DOI] [PubMed] [Google Scholar]
- Rudney JD, Chen R, Sedgewick GJ. 2005. Actinobacillus actinomycetemcomitans, Porphyromonas gingivalis, and Tannerella forsythensis are components of a polymicrobial intracellular flora within human buccal cells. J Dent Res. 84(1):59–63. [DOI] [PubMed] [Google Scholar]
- Saglie FR, Marfany A, Camargo P. 1988. Intragingival occurrence of Actinobacillus actinomycetemcomitans and Bacteroides gingivalis in active destructive periodontal lesions. J Periodontol. 59(4):259–265. [DOI] [PubMed] [Google Scholar]
- Saglie R, Newman MG, Carranza FA, Jr, Pattison GL. 1982. Bacterial invasion of gingiva in advanced periodontitis in humans. J Periodontol. 53(4):217–222. [DOI] [PubMed] [Google Scholar]
- Saito Y, Fujii R, Nakagawa KI, Kuramitsu HK, Okuda K, Ishihara K. 2008. Stimulation of Fusobacterium nucleatum biofilm formation by Porphyromonas gingivalis. Oral Microbiol Immunol. 23(1):1–6. [DOI] [PubMed] [Google Scholar]
- Seguier S, Godeau G, Brousse N. 2000. Collagen fibers and inflammatory cells in healthy and diseased human gingival tissues: a comparative and quantitative study by immunohistochemistry and automated image analysis. J Periodontol. 71(7):1079–1085. [DOI] [PubMed] [Google Scholar]
- Seguier S, Gogly B, Bodineau A, Godeau G, Brousse N. 2001. Is collagen breakdown during periodontitis linked to inflammatory cells and expression of matrix metalloproteinases and tissue inhibitors of metalloproteinases in human gingival tissue? J Periodontol. 72(10):1398–1406. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD. 2005. Periodontal microbial ecology. Periodontol 2000. 38:135–187. [DOI] [PubMed] [Google Scholar]
- Vartoukian SR, Adamowska A, Lawlor M, Moazzez R, Dewhirst FE, Wade WG. 2016. In vitro cultivation of “unculturable” oral bacteria, facilitated by community culture and media supplementation with siderophores. PLoS One. 11(1):e0146926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Shakhatreh MA, James D, Liang S, Nishiyama S, Yoshimura F, Demuth DR, Hajishengallis G. 2007. Fimbrial proteins of Porphyromonas gingivalis mediate in vivo virulence and exploit TLR2 and complement receptor 3 to persist in macrophages. J Immunol. 179(4):2349–2358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.