Abstract
Interferon regulatory factor 6 (IRF6) acts as a tumor suppressor and controls cell differentiation in ectodermal and craniofacial tissues by regulating expression of target genes. Haploinsufficiency of IRF6 causes Van der Woude and popliteal pterygium syndrome, 2 syndromic forms of cleft lip and palate. Around 85% of patients with Van der Woude express pits on the lower lip that continuously or intermittently drain saliva, and patients with the common cleft lip and palate have a higher prevalence of dental caries and gingivitis. This study aims to identify the role of IRF6 in development of exocrine glands, specifically the major salivary glands. Our transgenic mouse model that expresses LacZ reporter under the control of the human IRF6 enhancer element showed high expression of IRF6 in major and minor salivary glands and ducts. Immunostaining data also confirmed the endogenous expression of IRF6 in the developing ductal, serous, and mucous acinar cells of salivary glands. As such, we hypothesized that Irf6 is important for proper development of salivary glands and potentially other exocrine glands. Loss of Irf6 in mice causes an increase in the proliferation level of salivary cells, disorganized branching morphogenesis, and a lack of differentiated mucous acinar cells in submandibular and sublingual glands. Expression and localization of the acinar differentiation marker MIST1 were altered in Irf6-null salivary gland and pancreas. The RNA-Seq analysis demonstrated that 168 genes are differentially expressed and confer functions associated with transmembrane transporter activity, spliceosome, and transcriptional regulation. Furthermore, expression of genes involved in the EGF pathway—that is, Ereg, Ltbp4, Matn1, Matn3, and Tpo—was decreased at embryonic day 14.5, while levels of apoptotic proteins were elevated at postnatal day 0. In conclusion, our data report a novel role of Irf6 in exocrine gland development and support a rationale for performing exocrine functional tests for patients with IRF6-damaging mutations.
Keywords: exocrine glands, cleft lip and palate gene, saliva, mouse model, xerostomia, mucous and serous acinar cells
Introduction
Salivary glands are exocrine glands that produce saliva into the oral cavity to protect the structures of the oral mucosa and teeth, lubricate food, and initiate the process of digestion (Dodds et al. 2005; Saleh et al. 2015). The 3 major salivary glands in humans are the parotid, submandibular, and sublingual glands (Aure et al. 2015). Salivary glands consist of secretory acinar cells and branched ductal cells that carry saliva into the oral cavity. There are 2 major types of acinar cells, the serous and mucous, and one of the ways that they can be differentiated histologically is by the structure and location of their nuclei (Steinberg et al. 2005; Patel and Hoffman 2013). Serous acinar cells, which secrete watery saliva, have round nuclei that are oriented toward the center part of the cells. Conversely, mucous acinar cells, which secrete thick saliva containing mucins, have flattened nuclei located basally in the cells (Lombaert et al. 2011; Streckfus 2015). A common health problem that occurs due to a lack of saliva is dry mouth, or xerostomia, which causes significant sequelae, including problems in swallowing, infections of the oral cavity, ulcers, and dental caries (Dodds et al. 2005).
Although the role of interferon regulatory factor 6 (IRF6) during early development of the salivary gland has not been investigated yet, a recent study reported that tissue-specific deletion of Irf6 led to salivary gland dysfunction (Tamasas and Cox 2017). Moreover, pathologic studies of patients with Van der Woude syndrome reported ectopic hyperplasia in minor salivary glands (Oberoi and Vargervik 2005; Ziai et al. 2005; Souto 2008). IRF6 is a transcription factor that acts as a tumor suppressor and regulates cell cycle exit, differentiation of ectodermal tissues, and stimulation of interferons (Taniguchi et al. 2001; Ingraham et al. 2006; Moretti et al. 2010). Mutations in IRF6 cause 2 syndromic forms of orofacial clefting, Van der Woude syndrome and popliteal pterygium syndrome (Zucchero et al. 2004; Rahimov et al. 2008), and increase the risk for the common form of cleft lip and palate (Kondo et al. 2002; Vieira 2008; Fakhouri et al. 2014). While it is currently unknown if patients with Van der Woude syndrome and popliteal pterygium syndrome suffer from salivary gland disorders, some reports indicated that patients with cleft lip and palate exhibit very serious dental caries (Lam et al. 2010; Blackburn et al. 2012; Antonarakis et al. 2013). Consistent with these findings, knockout of Irf6 in oral epithelium causes a reduction in saliva production, an increase in mucus acidity, and dysplasia of the salivary gland, and these mice showed severe dental caries when placed on a high-sugar diet (Tamasas and Cox 2017). Interestingly, the minor salivary glands in patients with Van der Woude syndrome with lip pits could have nonspecific chronic inflammation with hyperplasia of epithelium (Dahllöf et al. 1989; Dissemond et al. 2004).
In mice, the Irf6-null phenotype includes skin, limb, and craniofacial abnormalities at birth (Ingraham et al. 2006), which is consistent with the spatiotemporal expression of Irf6 in the ectoderm of the facial processes and in the oral epithelium (Knight et al. 2006; Letra et al. 2012). Salivary glands are also derived from the ectoderm (Rothova et al. 2012), which is consistent with a recent finding that Irf6 is important for the salivary gland (Tamasas and Cox 2017). The expression of IRF6 is closely tied with P63 in ectoderm, which positively drives its expression (Moretti et al. 2010; Fakhouri et al. 2014). p63 is necessary for the development of ectodermal structures, including salivary glands, while Irf6 is essential for differentiation of stratified ectoderm and oral epithelium (Yang et al. 1999; Thomason et al. 2010). A previous study showed that the expression of IRF6 in oral and nasal epithelia was significantly reduced in p63-null mice at embryonic day 13.5, suggesting that p63 is a positive regulator of Irf6 expression (Moretti et al. 2010). In Sjogren’s syndrome, an immune system disorder characterized by xerostomia and dry eyes, patients exhibit shortened telomere length, associated with decreased expression of p63 as compared with non-Sjogren’s patients (Kawashima et al. 2011). Hence, the P63-IRF6 relationship merits further investigation in other organs where both factors are coexpressed (e.g., exocrine glands).
In this study, we examined the role of Irf6 in exocrine gland development, particularly in salivary glands and the pancreas. We studied the spatiotemporal expression pattern during embryonic development, characterized the pathology in Irf6-null salivary glands, and performed transcriptome analysis to identify significantly altered gene expression due to loss of Irf6. We also validated the expression of differentially expressed genes from RNA-Seq data and measured the level of candidate and marker genes at the RNA and protein level. We examined the role of Irf6 in the pancreas, an analogous exocrine gland that expresses Irf6 during development and also shares a exocrine structure similar to the salivary gland. Collectively, the data of this study show that Irf6 is critical for proper development of salivary glands and the pancreas.
Materials and Methods
Mice Handling and Generation of Irf6-Null Embryos
Animal use procedures were approved by the Animal Welfare Committee (AWC-13-055) at the University of Texas Health Science Center at Houston. Generation and genotyping of murine embryos and pups that are null for Irf6 were carried out as previously described (Ingraham et al. 2006). The age of the embryos at the desired time points in development was determined according to the presence of the copulation plug (deemed 0.5 days post coitum). Transgenic IRF6-lacZ mice were handled as previously described (Fakhouri et al. 2012).
Immunohistochemistry and Immunofluorescent Staining
Immunostaining was used to qualitatively determine the protein expression in the Irf6-null mutant salivary gland tissues as compared with wild-type (WT) tissues. Immunohistochemical (IHC) staining was performed for embryos at embryonic day 17.5 (E17.5) with ΔN-P63 (Ab735) and MIST1 (GTX31558; GeneTex) antibodies.
RNA-Seq of Irf6-Null Salivary Gland Tissues
RNA-Seq analysis was conducted on 2 male biological replicates of Irf6-null (Irf6-/-) and 2 WT (Irf6+/+) salivary glands at E14.5 to identify differentially expressed genes. The full transcriptome data sets are deposited on GEO (accession GSE102020). A subset of genes was validated on different null and WT embryos at E14.5 with quantitative real-time polymerase chain reaction (see Appendix for methods).
Bioinformatic Analysis of RNA-Seq Data
Bioinformatic analysis was performed on 168 differentially expressed genes determined by RNA-Seq analysis to identify regulatory pathways that are dependent on IRF6 expression. The online tool DAVID (Database for Annotation, Visualization and Integrated Discovery) was used to determine the gene ontology of these expressed genes so that the potential function and impact of these genes can be described across different species.
Statistical Analysis
We performed multivariate analysis of variance for the normalized expression data to determine the significance of any difference among the mean values of different genes in WT versus Irf6-null mouse tissues. We used 5 technical replicates for each biological treatment. A P value <0.05 was considered statistically significant.
Results
Expression of IRF6 during Salivary Gland Development
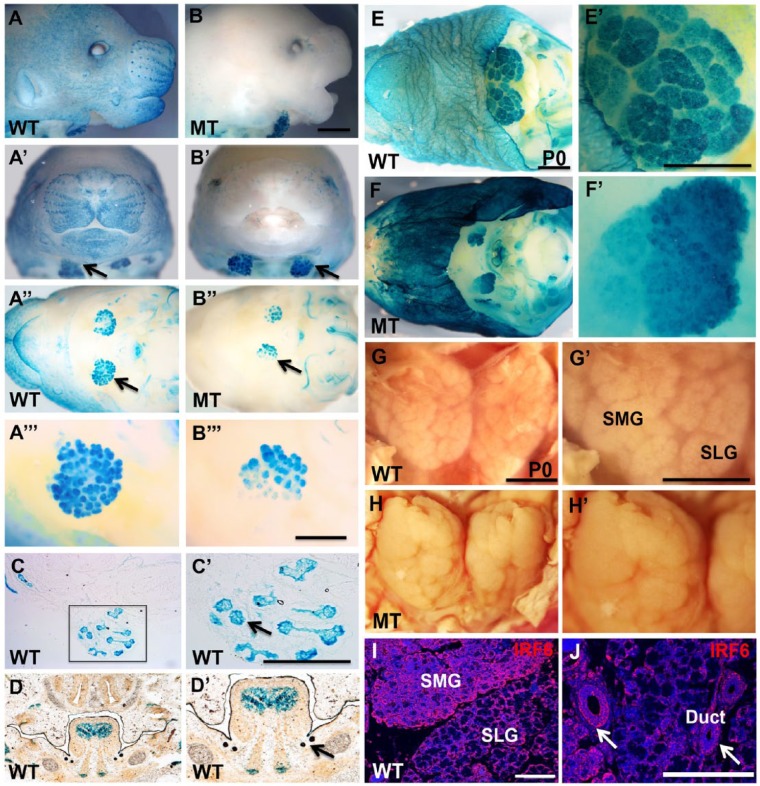
lacZ reporter gene expression driven by the IRF6 enhancer element, observed with beta-galactosidase (ßgal) staining, was noticeably different in the transgenic WT mice as compared with the same congenic background as Irf6-null alleles. For WT and Irf6-null mice, a strong ßgal staining was observed in salivary glands throughout embryonic development, including the submandibular and sublingual salivary glands (Fig. 1). At E14.5, ßgal staining of salivary gland tissues in Irf6-null murine embryos was similar to the staining in WT but was absent in the skin of Irf6-null mice while lightly visible in WT mice (Fig. 1A, B series). Coronal and transverse cryosections showed ßgal staining in acinar cells and ductal orifices of submandibular and sublingual glands (Fig. 1C, D series). At postnatal day 0 (P0), ßgal staining in salivary glands appeared similar between WT and Irf6-null pups. However, the skin of Irf6-null newborn pups was stained very dark with ßgal but was only lightly stained in WT pups (Fig. 1E, F series). Macroscopic images of dissected salivary glands at P0 showed morphologic changes in the structure and shape of Irf6-null mice as compared with WT (Fig. 1G, H series). Endogenous expression of IRF6 was confirmed and showed a wide expression in salivary glands, including ductal and acinar cells (Fig. 1I, J).
Figure 1.
Expression of IRF6 in submandibular salivary glands during development and macroscopic phenotype in Irf6-null salivary glands. Bgal staining of whole mount and coronal section was performed in wild-type (WT) and Irf6-null embryos (mutant type [MT]) at embryonic day 14.5 (A–D series) and at postnatal day 0 (P0; E, F series). The blue staining shows the expression of LacZ driven by IRF6 enhancer element in skin and submandibular glands (black arrows) of WT embryos (A–A′′′). Salivary glands but not the skin of transgenic Irf6-null littermate embryos are stained blue (black arrow; B–B′′′). Bgal staining in frozen sections of WT embryos at embryonic day 14.5 showing expression in lobes (black arrow) of submandibular salivary glands (C, C′) and in the sublingual and submandibular orifices of the ducts (black arrow; D, D′). Salivary glands and skin are stained blue in WT (E, E′), while the Bgal staining shows disorganization of salivary glands in Irf6-null newborn pup at P0 (F, F′). Dissected pair of submandibular/sublingual WT gland shows branched lobes and organized clefts in WT newborn pup (G, G′), while the salivary gland lobes in Irf6-null pup are compact and fused (H, H′). IRF6 is widely expressed in salivary glands, including the ductal (arrows) and acinar cells (I, J). Scale bars are 800 μm for all images and 200 μm for panels I and J. SLG, sublingual gland; SMG, submandibular gland.
Disruption of Branching Morphogenesis and Differentiation of Mucous Acini in Irf6-Null Salivary Glands
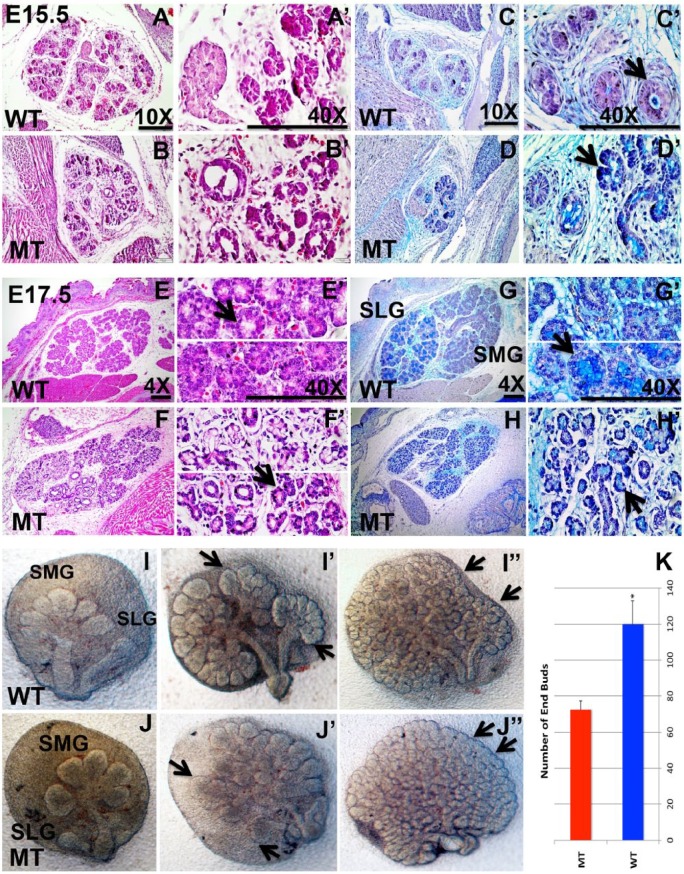
Histological studies of salivary gland sections with different staining methods were used to visualize the cell structure and organization. No difference between the WT and mutant was observed at E13.5 (data not shown). However, morphologic differences became more prominent at E15.5 (Fig. 2A–D series). We observed disruption of the lumen morphology at E15.5 (Fig. 2D series). In the salivary gland tissues of WT mice, acinar cells were differentiated into distinct serous and mucous types, where sublingual gland is composed only of mucous acini at E17.5 (Fig. 2E, G series). However, in the salivary gland tissues of the Irf6-null mutants, no SLGs were distinguishable (Fig. 2F, H series). In hematoxylin and eosin as well as the alcian blue and nuclear fast red staining, mucous acini appear to be lacking in the mutant (Fig. 3B, D series) versus WT salivary glands (Fig. 3A, C series). Alcian blue and periodic acid–Schiff staining confirmed a marked decrease in acidic mucins in Irf6-null embryos (Fig. 3F series) versus WT (Fig. 3E series). Furthermore, the acinar cells in Irf6-mutant salivary glands were disorganized and with a larger interstitial space (Fig. 2 F′, H′). These results were also supported in our salivary gland explant experiments. Salivary glands were extracted from WT and Irf6-null embryos at E13.5 and cultured for 4 d (Fig. 2I, J series). The branching of end buds was disrupted in mutant explants, and their number was reduced 2 and 4 d postincubation due to potential bud adhesion (Fig. 2J′, J′′), which is consistent with the histologic analysis. The number of end buds was significantly decreased in mutant versus WT littermate explants (Fig. 2K).
Figure 2.
Hematoxylin and eosin, alcian blue, and periodic acid–Schiff staining of submandibular glands in wild-type (WT; A, C, E, G series) and Irf6-null (mutant type [MT]) embryos (B, D, F, H series) and ex vivo culture of salivary gland explants. At embryonic day 15.5 (E15.5), histologic staining shows round ductal cells with a single circular lumen per duct (A, C series; black arrow). In comparison, the staining in Irf6-null glands shows significant ductal cell disorganization and disruption of ductal lumens (arrow in D′). At E17.5, the serous and mucous acini are detected in WT gland (black arrow in E′; E, G series), while the mucous acini (black arrow) cannot be distinguished in Irf6-null salivary glands and the interstitial spaces among the acini are wider when compared with WT (F, H series; black arrow in H′). In salivary gland explants, the sublingual gland (SLG) and submandibular gland (SMG) are formed in the WT and Irf6-null littermates at E13.5 (I, J). Explants show branched SLG and SMG after 2 d (I′, black arrow) and a remarkable number of end buds (black arrow) and branching by day 4 (I′′) of cultivation. In Irf6-null explants, the end buds of SLG and SMG seem fused by day 2 (J′, arrow) of incubation, and by day 4, many end buds appear to be coalesced or not separated (J′′, arrow). The number of end buds was counted 4 d post-incubation in 3 biological replicates, and it is significantly decreased in Irf6-null embryos (MT) versus WT littermates (WT; K). The values are the mean number of end buds, and the bars represent the standard deviation. Asterisk represents statistical significance of WT versus Irf6-null samples, P < 0.05. Scale bars are 150 μm.
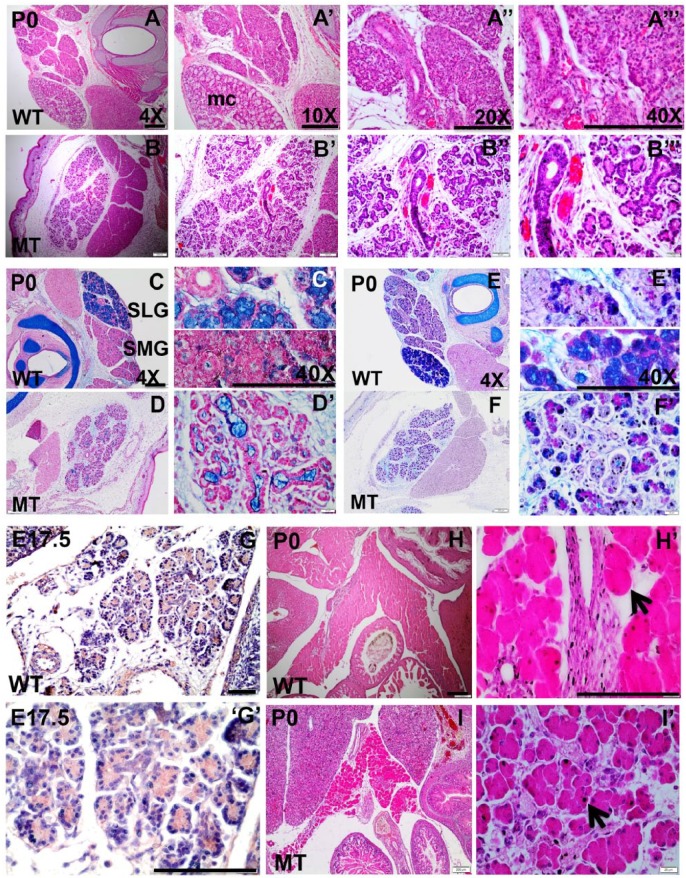
Figure 3.
Histologic studies to detect phenotypic abnormality in Irf6-null salivary gland and pancreas. Hematoxylin and eosin (H&E) staining (A, B series), alcian blue and nuclear fast red staining (C, D series), and alcian blue and periodic acid–Schiff staining (E, F series) were performed on submandibular gland (SMG) and sublingual gland (SLG) of wild-type (WT) and Irf6-null tissues at postnatal day 0 (P0). Expression of IRF6 (G series) is detected by IHC in pancreas, and H&E staining (H, I series) was performed in WT and Irf6-null pancreatic sections. H&E staining visualizes the SMG and SLG of the salivary gland in WT where SLG is recognized by the whitish cytosol of mucous acini (mc). Ducts and serous acini appear compact with single organized lumen (A′′–A′′′), while the ductal and acinar cells are disorganized in the mutant and SLG is not distinguished (B–B′′′). Alcian blue and nuclear fast red staining shows a remarkable reduction in mucin and mucous cell (mc) formation (blue-stained cells) in Irf6-null SLG and SMG (D, D′) versus WT (C, C′). Alcian blue and periodic acid–Schiff staining of SMG and SLG in WT differentiates glycoprotein substances as purple, acidic mucins as blue, neutral mucins as magenta, mixed mucous as bluish purple, and nuclei as dark blue (D, D′). Staining shows a decrease in acidic mucins in Irf6-null SMG and SLG (F, F′). IRF6 is expressed in pancreas in cytosol of acinar cells at embryonic day (E17.5; G, G′). H&E staining showed abnormal morphology, disorganized acinar cells (black arrow in I′), and wide interstitial spaces among acini in Irf6-null pancreas at P0 (I, I′) as compared with WT littermates (H, H′, black arrow). Scale bars are 150 μm.
Abnormal Acini and Disruption of MIST1 in Irf6-Null Pancreas
We performed IHC to determine if Irf6 is expressed in the pancreas. IHC staining confirmed that IRF6 is expressed in acinar cells of the pancreas (Fig. 3G, G′). In addition, we conducted histologic studies in Irf6-null pancreas at different time points. No phenotype was observed at E15.5 and E17.5 (Appendix Fig. 2A–D series). The phenotype was obvious at P0; the acinar cells had incomplete clefts, and multiple acini were clustered in a rosette-like structure as compared with WT (Fig. 3H, I series). In Irf6-null pancreatic tissues, a wide interstitial space was also observed at P0 (Fig. 3I, I′). We also performed IHC for the acinar differentiation factor MIST1 at E17.5. The level of MIST1 expression was elevated in the cytosol and nucleus of Irf6-null mutant as compared with WT (Appendix Fig. 1E, F).
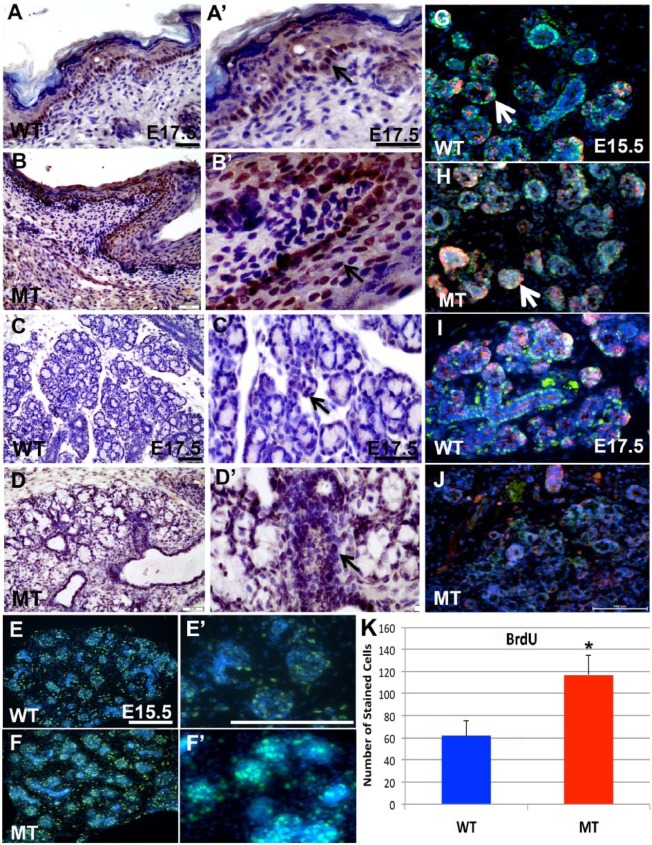
Increase Proliferation Level in Irf6-Null Salivary Glands and Elevated Expression of P63 and MIST1
The expression of P63 was observed in the basal layer of skin in WT embryos (Fig. 4A, A′), while multiple layers of the epidermis in Irf6-null embryo showed P63 expression (Fig. 4B, B′), consistent with previous reports (Ingraham et al. 2006; Thomason et al. 2010). In salivary glands, expression was detected in some acinar and ductal cells (Fig. 4C, C′). However, expression of P63 was noticeably increased in most acinar and ductal cells of Irf6-null sections (Fig. 4D, D′). The level of proliferation in salivary glands was detected with the BrdU-stained nuclei. While a good number of acinar and ductal cells were labeled in WT at E15.5 (Fig. 4E series), a significantly larger number of BrdU-labeled cells were seen in mutants (Fig. 4F series, K). Immunofluorescent staining for activated caspase 3 (Casp3) and P63 was detected in ductal and acinar cells in WT and appeared to show a difference in staining pattern from the mutant at E15.5 (Fig. 4G, H). At E17.5, the expression of P63 was detected in most ductal cells and less in acinar cells, while Casp3 was more expressed in acinar cells (Fig. 4I). Immunofluorescent staining of P63 and Casp3 was unspecific in mutants despite the large effort of modifications and optimization (Fig. 4J). Expression of differentiation factor MIST1 was investigated in acinar cells at E17.5. In WT tissues, expression of MIST1 was localized to the apical region of acini and in the nucleus of some acinar cells, while the expression in Irf6-null tissues was pronounced in the nucleus and cytosol of acinar cells (Appendix Fig. 2C, D series).
Figure 4.
Immunohistochemistry and immunofluorescent staining of wild-type (WT) and Irf6-null (mutant type [MT]) submandibular glands and skin. P63 expression was detected in the basal layer of stratified ectoderm (black arrow) in WT at embryonic day 17.5 (E17.5; A, A′), while the expression of P63 was expanded to external layers of ectoderm—namely, the spinous layer (black arrow) above the basal layer of keratinocytes (B, B′). In submandibular salivary glands, P63 was observed in the nucleus of acinar (black arrow) and ductal cells in WT (C, C′), while the expression level was increased in most of the cells (black arrow) of Irf6-null submandibular glands (D, D′). To determine proliferation level, replicated DNA in proliferative cells was labeled with BrdU, which showed even distribution of labeled green acinar and ductal cells in submandibular WT (E, E′) and Irf6-null (F, F′) salivary glands. Immunofluorescent staining was performed to determine the level of apoptotic cells based on the activated caspase 3 (Casp3) marker in WT and Irf6-null (MT) submandibular glands. In WT, P63 is expressed in the nucleus of acinar and ductal cells similar to immunohistochemistry, while Casp3 is localized to some ductal or acinar cells (G, white arrow). The expression pattern of Casp3 and P63 did not relatively change in Irf6-null at E15.5 (H, white arrow). At E17.5, expression of P63 was prominent in the ductal cells, while Casp3 is observed in more in acinar cells (I). Expression of both proteins is not detected in Irf6-null salivary glands due to probably disrupted cellular structure (J). Labeled cells with BrdU of the entire area of 20× images were counted with plug-ins of ImageJ program (K). The number of proliferative cells is significantly increased in Irf6-null embryos as compared with WT littermates at E15.5. CASP3 is shown in red, P63 in green, and DAPI in blue. The values are the mean number of stained cells with BrdU, and the bars represent the standard deviation. Asterisk represents statistical significance of Irf6-null versus WT samples, P < 0.05. Scale bars are 100 μm.
Differentially Expressed Genes and RNA-Seq Analysis
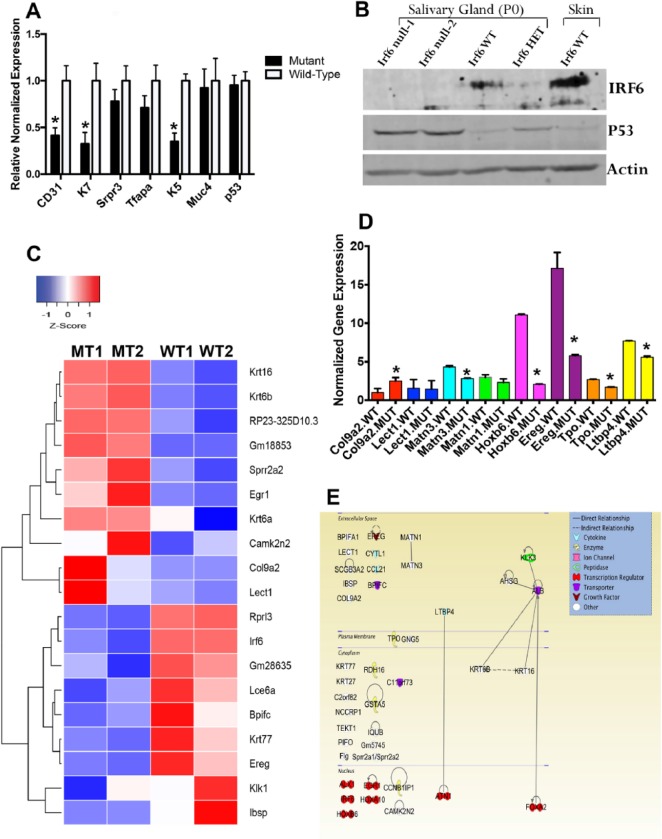
We measured the expression of cell-type markers and apoptotic factors in salivary glands. At the RNA level, markers for endothelial (CD31), ductal (K7), and progenitor (K5) cells were significantly reduced in Irf6-null tissues versus WT littermates (Fig. 5A). Immunoblot analysis showed that the expression of P53 protein was increased in Irf6-null salivary tissues as compared with Irf6-heterozygous and WT littermates at P0 (Fig. 5B). To identify differentially altered genes in an unbiased approach, RNA-Seq analysis showed that a total of 168 genes were differentially expressed in the Irf6-null mouse as compared with the WT with a false discovery rate <0.05 (Fig. 5C). The top 19 differentially expressed genes clustered closer in the dendrogram of the heat map tended to be over- or downexpressed in a similar manner within and across the different tissue samples (Fig. 5C). Eight genes were validated by reverse transcription quantitative real-time polymerase chain reaction, and 6 showed significant variation as compared with WT (Fig. 5D). There were probably global differences in the transcriptome of the 2 mutant embryos, although all embryos were male according to Appendix Table 2. The high degree of variability might reduce the number of genes that were differentially expressed as compared with WT samples (Appendix Fig. 3). The gene ontology tool DAVID was utilized to group the differentially expressed genes on the basis of their cellular functions. The data resulted in 7 groups with significant P values (Appendix Table 1). While most of these categories cover functions general to transcription factors, transmembrane transporter activity and EGF signaling pathway were most notable because they are involved in branching morphogenesis and differentiation of acinar cells. To identify enriched regulatory pathways, Ingenuity Pathway analysis was applied to all 168 differentially expressed genes and showed the genes with known function and their cellular localization and connectivity at gene-gene and protein-protein interactions (Fig. 5E).
Figure 5.
Analysis of altered gene expression in submandibular gland of wild-type (WT) and Irf6-null mice (mutant type [MT]). At embryonic day 17.5 (E17.5), normalized expression of multiple genes shows significant reduction in CD31, K7, and K5 expression, while no change is observed in mRNA of Srpr3, Tfap2a, Muc4, and p53 as compared with WT littermates (A). The values are the mean normalized expression, and the bars represent the standard deviation. Protein levels in Irf6-null, heterozygous, and WT submandibular salivary gland tissues were investigated by immunoblot assay. Skin tissue from WT pups was used as positive control and showed higher expression of IRF6 as compared with salivary glands (B). P0, postnatal day 0. The level of P53 protein was accumulated in Irf6-null versus heterozygote and WT tissues (B). Heat map of the top 19 significantly differentially expressed genes in Irf6-null embryos (MT1 and MT2) and their corresponding expression profile in WT littermate embryos (WT1 and WT2). Blue blocks represent genes that have decreased in expression, while red represents genes that have increased in expression. White represents no change in expression (C). Validation of 8 differentially expressed genes from RNA-Seq data in WT and Irf6 mutant salivary glands at E14.5 (D). The values are the mean normalized expression, and the bars represent the standard deviation. Asterisks represent statistical significance of Irf6-null samples versus WT, P < 0.05 (D). Ingenuity Pathway analysis of 41 differentially expressed genes from RNA-Seq data of Irf6-mutant SGs as compared with WT littermates was represented according to their connectivity and interactions at gene-gene and protein-protein levels (E). Several transmembrane transporter genes are involved. Five genes—that is, Ereg, Ltbp4, Matn1, Matn3, and Tpo—are involved in the EGF signaling pathway.
Discussion
The purpose of this study is to determine the role of Irf6 during development of salivary glands and the pancreas and to characterize the pathophysiology in Irf6-null tissues. While nothing is known about the function of Irf6 in early exocrine gland development, our data showed that loss of Irf6 expression led to morphologic abnormality and disrupting of expression of proliferation and differentiation markers in salivary glands and the pancreas during embryogenesis. This observation is consistent with a recent study showing that Irf6 is important for salivary gland maturation and saliva production (Tamasas and Cox 2017). The spatiotemporal expression of LacZ driven by IRF6 human enhancer recapitulates the endogenous expression of IRF6 protein, confirming the importance of this enhancer element in driving IRF6 expression in salivary glands. This observation is important because DNA mutations that disrupt the function of this element may lead to reduction in IRF6 expression during salivary gland formation. We reported that patients with Van der Woude carry damaging mutations in this enhancer element, which may explain the high incidence of dental caries in patients with cleft and palate (Dahllöf et al. 1989; Lam et al. 2010; Blackburn et al. 2012; Fakhouri et al. 2014). These data are also consistent with a recent study showing that Irf6 plays an important role in dental health and saliva production (Tamasas and Cox 2017).
To understand the importance of Irf6 in salivary gland formation, we performed histologic analysis in Irf6-null tissues to characterize morphologic changes and alteration in the structure, cell organization, and anatomy. Our data showed that lack of Irf6 led to abnormality in branching morphogenesis and cellular structure. In particular, hematoxylin and eosin staining showed obvious changes in cell types and organization. Mucous acinar cells were mostly lacking and appeared to be poorly differentiated in Irf6-null mice in submandibular and sublingual salivary glands. In WT salivary glands, the serous and mucous acinar cells were distinguishable at the E17.5 developmental stage. Furthermore, Irf6-null salivary glands lacked proper organization of the lobular structure, and the distinction of sublingual gland was difficult due to the lack of mucous acini. The phenotype in salivary glands is, to some extent, similar to skin abnormality where the last 2 layers of stratified ectoderm (the stratum corneum and stratum granulosum) were missing (Ingraham et al. 2006; Thomason et al. 2010).
We studied the expression of P63 and MIST1 in our system due to their critical role in acinar proliferation and differentiation, respectively (Botti et al. 2011; Aure et al. 2015). P63 is present in all early stages of salivary gland morphogenesis, starting from placode initiation to acinar formation. Loss of p63 expression leads to a complete lack of salivary gland formation (Ianez et al. 2010). The expression of transcription factors IRF6 and deltaN-P63 are closely regulated in single-layer and stratified epithelium (Fakhouri et al. 2012). While Irf6 is transcriptionally activated by P63, IRF6 also induces P63 proteasome-mediated degradation in human keratinocytes (Botti et al. 2011). Our immunofluorescent data showed that IRF6 was located in the cytosol of ductal and acinar cells, while ∆N-P63 was localized in the nucleus of these cell types at E15.5. Interestingly, P63 level was increased in mutant embryos according to our IHC staining. Because ∆N-P63 is a proliferative factor, this could explain the increased level of proliferation at E15.5. Noteworthy, we observed an increase in MIST1 expression in the acinar cells of salivary glands and the pancreas of Irf6-null samples. Regulation of MIST1 expression is critical for proper acinar differentiation, and our mutant tissues showed disruption of mucous acinar cell formation (Hess et al. 2016). Regulation of p53 transcription is IRF6 independent. However, loss of the IRF6 protein leads to accumulation of P53 protein at P0 due to either an increase of p53 protein stability or a decrease of degradation. This increase may explain the reduction in branching morphogenesis and the lack of mucous acini at later stages in Irf6-null salivary glands.
When we performed temporal analysis to determine the earliest time point where a change in phenotype can be detected, we found that E15.5 was the earliest time point with obvious abnormalities. Hence, we performed RNA-Seq analysis on Irf6-null and WT salivary gland tissues at E14.5. The data yielded 168 statistically differentially expressed genes with a false discovery rate <0.05. The resulting enriched gene ontology terms are associated with transmembrane transporter activity, splicesome, transcriptional regulation, and signaling pathways. Altered mRNA expression of genes involved in transmembrane transporter activity and MIST1 protein due to loss of IRF6 is interesting because it suggests that Irf6 is involved in regulating Mist1 expression that is critical for cell polarity, vesicle formation, transportation, and exocytosis. Gene ontology analysis also suggested that Irf6 regulates expression of genes involved in EGF pathway.
Preliminary exploration of the Irf6 role in pancreas development was also conducted due to the morphologic and physiologic similarities of ductal and acinar cells to those in the salivary glands (Streckfus 2015). In Irf6-null pancreas, histologic analysis showed that the acinar cells are disorganized and have a rosette-like structure with a larger spatial separation among acini as compared with their WT littermates. Interestingly, these histologic changes are to some extent similar to those that we found in Irf6-null salivary gland tissues at birth. Exploring IRF6-dependent pathways that regulate cell fate and polarity in oral epithelium and salivary gland may assist in identifying patients who are at high risk to develop long-term and severe xerostomia (Vissink et al. 2003; Konings et al. 2005; Vissink et al. 2015).
Author Contributions
K.A. Metwalli, M.A. Do, contributed to conception, design, data acquisition, and analysis, drafted the manuscript; K. Nguyen, contributed to data acquisition and analysis, drafted the manuscript; S. Mallick, K. Kin, contributed to data analysis, drafted the manuscript; N. Farokhnia, G. Jun, contributed to data analysis, critically revised the manuscript; W.D. Fakhouri, contributed to conception, design, data acquisition, analysis, and investigation, drafted and critically revised the manuscript. All authors gave final approval and agree to be accountable for all aspects of the work.
Supplementary Material
Acknowledgments
We thank Dr. Charles Streckfus for providing helpful comments and suggestions on the content of this study. We are also grateful to Angela Quispe Salcedo, Adeeb Sakkaleak, and Ishita Akhter for their excellent help with the histologic and immunofluorescent staining. We specially thank Dr. Brian Schutte for the Irf6 heterozygous mice and IRF6-lacZ transgenic mice.
Footnotes
A supplemental appendix to this article is available online.
This study was supported by start-up funding to Dr. Walid Fakhouri from the School of Dentistry at the University of Texas Health Science Center at Houston.
The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Antonarakis GS, Palaska PK, Herzog G. 2013. Caries prevalence in non-syndromic patients with cleft lip and/or palate: a meta-analysis. Caries Res. 47(5):406–413. [DOI] [PubMed] [Google Scholar]
- Aure MH, Konieczny SF, Ovitt CE. 2015. Salivary gland homeostasis is maintained through acinar cell self-duplication. Dev Cell. 33(2):231–237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn J, Ohazama A, Kawasaki K, Otsuka-Tanaka Y, Liu B, Honda K, Rountree RB, Hu Y, Kawasaki M, Birchmeier W, et al. 2012. The role of irf6 in tooth epithelial invagination. Dev Biol. 365(1):61–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botti E, Spallone G, Moretti F, Marinari B, Pinetti V, Galanti S, De Meo PD, De Nicola F, Ganci F, Castrignano T, et al. 2011. Developmental factor IRF6 exhibits tumor suppressor activity in squamous cell carcinomas. Proc Natl Acad Sci U S A. 108(33):13710–13715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahllöf G, Ussisoo-Joandi R, Ideberg M, Modeer T. 1989. Caries, gingivitis, and dental abnormalities in preschool children with cleft lip and/or palate. Cleft Palate J. 26(3):233–237. [PubMed] [Google Scholar]
- Dissemond J, Haberer D, Franckson T, Hillen U. 2004. The Van der Woude syndrome: a case report and review of the literature. J Eur Acad Dermatol Venereol. 18(5):611–613. [DOI] [PubMed] [Google Scholar]
- Dodds MW, Johnson DA, Yeh CK. 2005. Health benefits of saliva: a review. J Dent. 33(3):223–233. [DOI] [PubMed] [Google Scholar]
- Fakhouri WD, Rahimov F, Attanasio C, Kouwenhoven EN, Ferreira De Lima RL, Felix TM, Nitschke L, Huver D, Barrons J, Kousa YA, et al. 2014. An etiologic regulatory mutation in IRF6 with loss- and gain-of-function effects. Hum Mol Genet. 23(10):2711–2720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhouri WD, Rhea L, Du T, Sweezer E, Morrison H, Fitzpatrick D, Yang B, Dunnwald M, Schutte BC. 2012. MCS9.7 enhancer activity is highly, but not completely, associated with expression of Irf6 and p63. Dev Dyn. 241(2):340–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess DA, Strelau KM, Karki A, Jiang M, Azevedo-Pouly AC, Lee A-H, Deering TG, Hoang CQ, Macdonald RJ, Konieczny SF. 2016. MIST1 links secretion and stress as both target and regulator of the UPR. Mol Cell Biol. 36(23):2931–2944 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ianez RF, Buim ME, Coutinho-Camillo CM, Schultz R, Soares FA, Lourenco SV. 2010. Human salivary gland morphogenesis: myoepithelial cell maturation assessed by immunohistochemical markers. Histopathology. 57(3):410–417. [DOI] [PubMed] [Google Scholar]
- Ingraham CR, Kinoshita A, Kondo S, Yang B, Sajan S, Trout KJ, Malik MI, Dunnwald M, Goudy SL, Lovett M, et al. 2006. Abnormal skin, limb and craniofacial morphogenesis in mice deficient for interferon regulatory factor 6 (Irf6). Nat Genet. 38(11):1335–1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima M, Kawakita T, Maida Y, Kamoi M, Ogawa Y, Shimmura S, Masutomi K, Tsubota K. 2011. Comparison of telomere length and association with progenitor cell markers in lacrimal gland between Sjogren syndrome and non-Sjogren syndrome dry eye patients. Mol Vis. 17:1397–1404. [PMC free article] [PubMed] [Google Scholar]
- Knight AS, Schutte BC, Jiang R, Dixon MJ. 2006. Developmental expression analysis of the mouse and chick orthologues of IRF6: the gene mutated in Van der Woude syndrome. Dev Dyn. 235(5):1441–1447. [DOI] [PubMed] [Google Scholar]
- Kondo S, Schutte BC, Richardson RJ, Bjork BC, Knight AS, Watanabe Y, Howard E, de Lima RL, Daack-Hirsch S, Sander A, et al. 2002. Mutations in IRF6 cause Van der Woude and popliteal pterygium syndromes. Nat Genet. 32(2):285–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konings AW, Coppes RP, Vissink A. 2005. On the mechanism of salivary gland radiosensitivity. Int J Radiat Oncol Biol Phys. 62(4):1187–1194. [DOI] [PubMed] [Google Scholar]
- Lam AK, David DJ, Townsend GC, Anderson PJ. 2010. Van der Woude syndrome: dentofacial features and implications for clinical practice. Aust Dent J. 55(1):51–58. [DOI] [PubMed] [Google Scholar]
- Letra A, Fakhouri W, Fonseca RF, Menezes R, Kempa I, Prasad JL, McHenry TG, Lidral AC, Moreno L, Murray JC, et al. 2012. Interaction between IRF6 and TGFA genes contribute to the risk of nonsyndromic cleft lip/palate. PLoS One. 7(9):e45441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombaert I, Knox SM, Hoffman MP. 2011. Salivary gland progenitor cell biology provides a rationale for therapeutic salivary gland regeneration. Oral Dis. 17(5):445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moretti F, Marinari B, Lo Iacono N, Botti E, Giunta A, Spallone G, Garaffo G, Vernersson-Lindahl E, Merlo G, Mills AA, et al. 2010. A regulatory feedback loop involving p63 and IRF6 links the pathogenesis of 2 genetically different human ectodermal dysplasias. J Clin Invest. 120(5):1570–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberoi S, Vargervik K. 2005. Hypoplasia and hypodontia in Van der Woude syndrome. Cleft Palate Craniofac J. 42(5):459–466. [DOI] [PubMed] [Google Scholar]
- Patel VN, Hoffman MP. 2013. Salivary gland development: a template for regeneration. Semin Cell Dev Biol. 25–26:52–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimov F, Marazita ML, Visel A, Cooper ME, Hitchler MJ, Rubini M, Domann FE, Govil M, Christensen K, Bille C, et al. 2008. Disruption of an ap-2alpha binding site in an IRF6 enhancer is associated with cleft lip. Nat Genet. 40(11):1341–1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothova M, Thompson H, Lickert H, Tucker AS. 2012. Lineage tracing of the endoderm during oral development. Dev Dyn. 241(7):1183–1191. [DOI] [PubMed] [Google Scholar]
- Saleh J, Figueiredo MA, Cherubini K, Salum FG. 2015. Salivary hypofunction: an update on aetiology, diagnosis and therapeutics. Arch Oral Biol. 60(2):242–255. [DOI] [PubMed] [Google Scholar]
- Souto LR. 2008. Congenital bilateral lower lip pits associated with fistulae of the minor salivary glands: case report of the principal Van der Woude syndrome’s trait. Aesthetic Plast Surg. 32(1):172–174. [DOI] [PubMed] [Google Scholar]
- Steinberg Z, Myers C, Heim VM, Lathrop CA, Rebustini IT, Stewart JS, Larsen M, Hoffman MP. 2005. FGFR2b signaling regulates ex vivo submandibular gland epithelial cell proliferation and branching morphogenesis. Development. 132(6):1223–1234. [DOI] [PubMed] [Google Scholar]
- Streckfus C. 2015. Advances in salivary diagnostics. Berlin (Germany): Springer-Verlag. p. 198. [Google Scholar]
- Tamasas B, Cox TC. 2017. Massively increased caries susceptibility in an Irf6 cleft lip/palate model. J Dent Res. 96(3):315–322. [DOI] [PubMed] [Google Scholar]
- Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. 2001. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 19:623–655. [DOI] [PubMed] [Google Scholar]
- Thomason HA, Zhou H, Kouwenhoven EN, Dotto GP, Restivo G, Nguyen BC, Little H, Dixon MJ, van Bokhoven H, Dixon J. 2010. Cooperation between the transcription factors p63 and IRF6 is essential to prevent cleft palate in mice. J Clin Invest. 120(5):1561–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira AR. 2008. Unraveling human cleft lip and palate research. J Dent Res. 87(2):119–125. [DOI] [PubMed] [Google Scholar]
- Vissink A, Jansma J, Spijkervet FK, Burlage FR, Coppes RP. 2003. Oral sequelae of head and neck radiotherapy. Crit Rev Oral Biol Med. 14(3):199–212. [DOI] [PubMed] [Google Scholar]
- Vissink A, van Luijk P, Langendijk JA, Coppes RP. 2015. Current ideas to reduce or salvage radiation damage to salivary glands. Oral Dis. 21(1):e1–e10. [DOI] [PubMed] [Google Scholar]
- Yang A, Schweitzer R, Sun D, Kaghad M, Walker N, Bronsonk RT, Tabin C, Sharpe A, Caput D, Crum C, et al. 1999. P63 is essential for regenerative proliferation in limb, craniofacial and epithelial development. Nature. 398(6729):714–718. [DOI] [PubMed] [Google Scholar]
- Ziai MN, Benson AG, Djalilian HR. 2005. Congenital lip pits and Van der Woude syndrome. J Craniofac Surg. 16(5):930–932. [DOI] [PubMed] [Google Scholar]
- Zucchero TM, Cooper ME, Maher BS, Daack-Hirsch S, Nepomuceno B, Ribeiro L, Caprau D, Christensen K, Suzuki Y, Machida J, et al. 2004. Interferon regulatory factor 6 (IRF6) gene variants and the risk of isolated cleft lip or palate. N Engl J Med. 351(8):769–780. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.