 A cooperativity framework to describe and interpret small-molecule stabilization of protein–protein interactions (PPI) is presented, which allows elucidating structure–activity relationships regarding cooperativity and intrinsic affinity.
A cooperativity framework to describe and interpret small-molecule stabilization of protein–protein interactions (PPI) is presented, which allows elucidating structure–activity relationships regarding cooperativity and intrinsic affinity.
Abstract
A cooperativity framework to describe and interpret small-molecule stabilization of protein–protein interactions (PPI) is presented. The stabilization of PPIs is a versatile and emerging therapeutic strategy to target specific combinations of protein partners within the protein interactome. Currently, the potency of PPI stabilizers is typically expressed by their apparent affinity or EC50. Here, we propose that the effect of a PPI stabilizer be best described involving the cooperativity factor, α, between the stabilizer and binding partners in addition to the intrinsic affinity, KDII, of the stabilizer for one of the apo-proteins. By way of illustration, we combine fluorescence polarization measurements with thermodynamic modeling to determine the α and KDII for the PPI stabilization of 14-3-3 and TASK3 by fusicoccin-A (FC-A) and validate our approach by studying other PPI-partners of 14-3-3 proteins. Finally, we characterize a library of different stabilizer compounds, and perform structure–activity relationship studies in which molecular changes could be attributed to either changes in cooperativity or intrinsic affinity. Such insights should aid in the development of more effective protein–protein stabilizer drugs.
Introduction
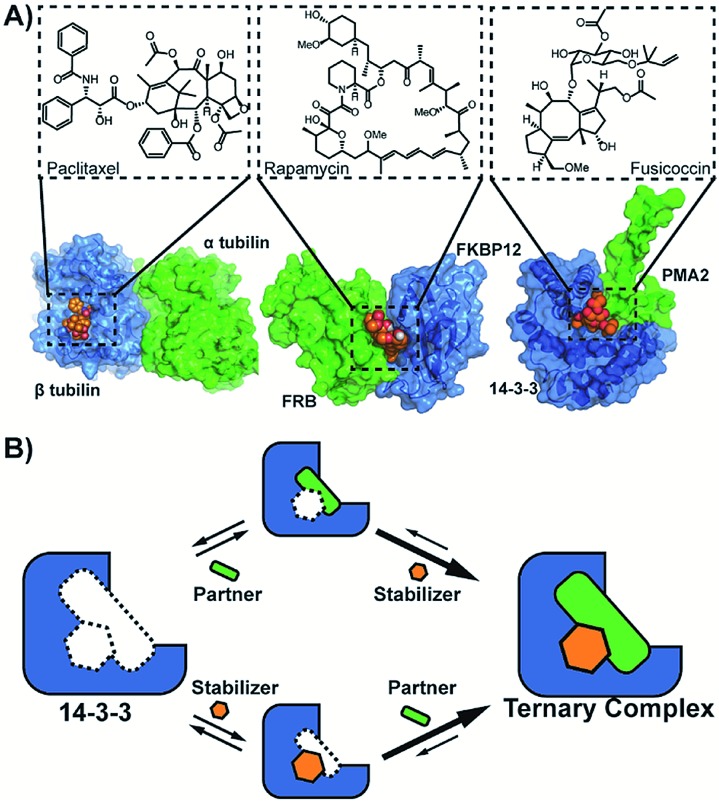
The stabilization of protein–protein interactions (PPI) using small molecules is an emerging and versatile strategy in drug development.1–3 PPI stabilizers target a specific combination of protein partners within the interactome, thereby increasing the stability of the resulting protein complex (molecular glues) (Fig. 1A).1 The direct stabilization of PPIs with small-molecules is conceptually challenging since it requires the simultaneous targeting of more than one protein within the complex.2 While several promising examples of PPI stabilizers have been reported, including Tafamidis, Rapamycin and Fusicoccin A,3 their discovery typically relied on serendipity.4–8 Therefore, rational approaches are urgently needed to assist in the development of PPI stabilizers.
Fig. 1. (A) Examples of natural compounds that stabilize protein–protein interactions. (pdb: 1JFF,10; 1FAP,11; 2O98.12 (B) Cooperativity scheme for PPI stabilization involving the sequential addition of PPI partner and stabilizer or vice versa.
The potency of a PPI stabilizer is typically expressed as a change in apparent affinity between the protein partners, either in the presence of a fixed concentration of stabilizer compound or as an EC50 of a dose-response similar to PPI inhibition.9 However, a single EC50 value does not capture the multitude of binding events in operation within the ternary complex of partner proteins and stabilizer molecule (Fig. 1B). The EC50-values depend for example on the relative concentrations of proteins and stabilizer compound. This would tend to complicate medchem optimization of stabilizer drugs. An objective framework is needed to quantify the activity of PPI stabilizers and enable subsequent structure–activity relationship studies for drug development. Here, we propose the use of cooperativity as a fundamental, objective metric to enable such comparisons of PPI stabilizers.
Cooperativity, commonly expressed through a cooperativity factor α, is a widespread phenomenon which strengthens binding within biological multicomponent systems.13,14 In GPCR research, cooperativity is used to characterize the activity of allosteric modulators15,16 and it has as well been used to describe the activity of chimeric compounds.17 To the best of our knowledge, cooperativity has yet to be established for PPI-stabilization, likely, in part, because it is more labour intensive to measure compared to direct binding measurements such as EC50 by FP and KD by ITC for example. Also, the typical low intrinsic affinity of small molecule stabilizers for either one of the individual protein partners limits facile detection or running of simple screening assays. The correlated intrinsic affinities and the cooperativity factor in PPI-stabilization require to be determined with the support of a thermodynamic model.
PPIs involving the 14-3-3 hub proteins are widely studied. 14-3-3 are intrinsic dimers that interact with several hundred protein partners, typically through short phosphorylated motifs.18 Since many of these interactions are involved in human disease, 14-3-3 proteins have been extensively studied as drug targets,19 making 14-3-3 a useful platform to explore new types of PPI modulation, for example by natural products, stapled peptides, or supramolecular ligands and for more fundamental studies.20,21 Examples of 14-3-3 PPI partners of fundamental importance or susceptible for small molecule modulation include the cystic fibrosis transmembrane conductance regulator CFTR,22 the regulatory-associated protein of mTOR Raptor controlling cell growth,23 the bacterial ADP-ribosyltransferase toxin of Pseudomonas aeruginosa ExoS,24 and the cancer relevant targets C-Raf, ERα and p53.25–27 Here we report a theoretical framework in which the activity of PPI stabilizers for such 14-3-3 mediated PPIs can be quantified and objectively compared in a straightforward manner using two parameters, viz. the cooperativity between the stabilizer and binding partner, α, and the intrinsic affinity of the stabilizer for the apo-protein, KDII. As a proof of principle, we applied this concept to revise PPI stabilization of the neuropathology relevant TWIK-related acid sensitive K+-channel 3 (TASK3) and hub protein 14-3-3, by the natural product fusicoccin A (FC-A).28 Subsequently, we broaden the scope of our approach by performing binding studies of 14-3-3 proteins and other binding partners. Finally, we perform a structure–activity relationship study on a small library of FC-A analogues in terms of their intrinsic affinity and cooperativity.
Results and discussion
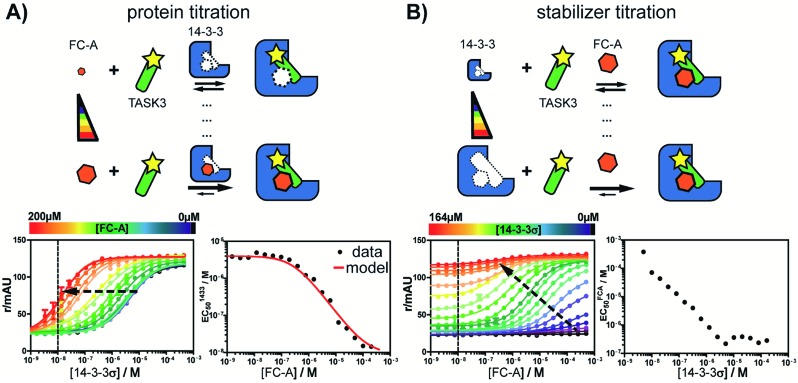
First, the affinity of TASK3 toward 14-3-3 was measured by 2D-fluorescence polarization titrations, varying the concentration of both 14-3-3 protein and stabilizer compound, FC-A (Fig. 2). In the first experiment, 14-3-3σ was titrated to 10 nM FITC-labelled TASK3 peptide at varying (0 to 200 μM) constant concentrations of FC-A (Fig. 2A). In the absence of FC-A, 14-3-3 binds to the labelled TASK3 epitope with an EC50 of 4 μM, in agreement with prior studies.28 Upon addition of increasing constant concentrations of FC-A, this EC50 gradually decreased over two orders of magnitude to 0.014 μM. Similarly, when titrating FC-A to 10 nM of labelled TASK3 (Fig. 2B), the EC50 value is also strongly dependent on the concentration of 14-3-3 protein. Here, the dynamic range of the S-curve was observed to decrease at higher 14-3-3 protein concentrations, due to the intrinsic binding affinity of the labelled peptide to the 14-3-3 protein. The data from these two experiments highlight the dependency of EC50 values on our assay conditions and the need, therefore, for additional metrics, independent of the relative concentrations, to enable the activities of analogue stabilizers to be objectively compared.
Fig. 2. Concentration dependence of fusicoccin-A (FC-A) stabilization of the TASK3/14-3-3σ PPI interaction measured by 2D fluorescence polarization. (A) Titration of 14-3-3 protein to FITC-labelled TASK3 peptide (10 nM, dashed line) against varying fixed concentrations of FC-A, between 0 and 200 μM (left panel). The EC50's of 14-3-3 to TASK3, for each FC-A concentration is compared to the EC50 values as calculated by the model (red line, right panel). (B) Titration of FC-A to FITC-labelled TASK3 peptide (10 nM, dashed line) against varying fixed concentrations of 14-3-3 between 0 and 164 μM (left panel). The EC50 of the FC-A titration depends on the 14-3-3 concentration in the assay (right panel). Here, the measured EC50 values were not compared to a calculated model fit, as the titration data at both low and high 14-3-3 concentrations prohibit clear determination of the EC50 values.
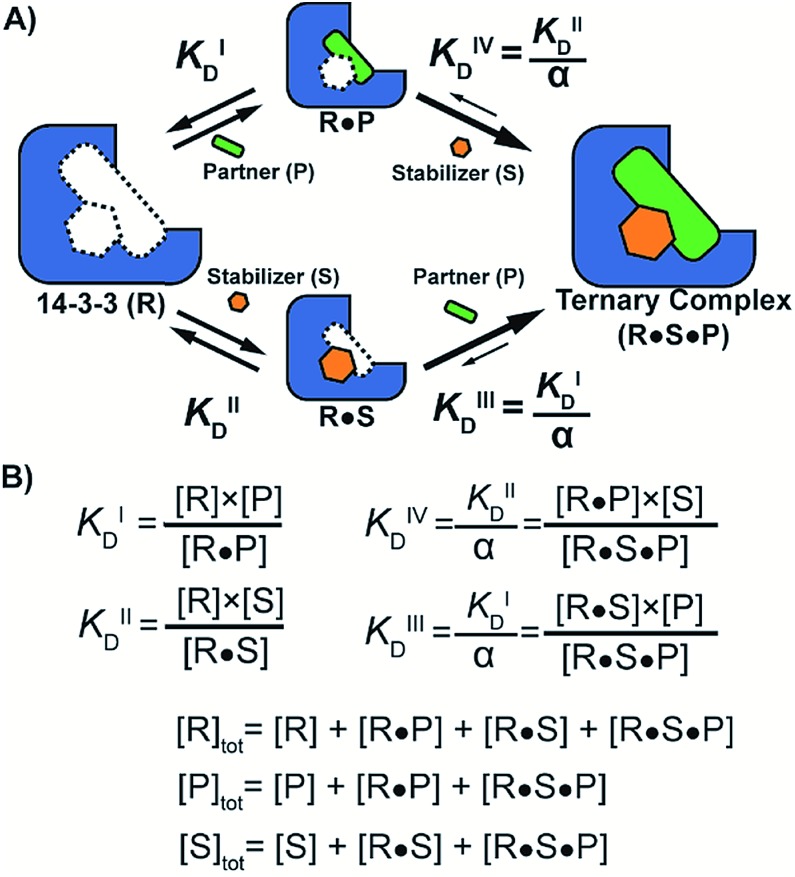
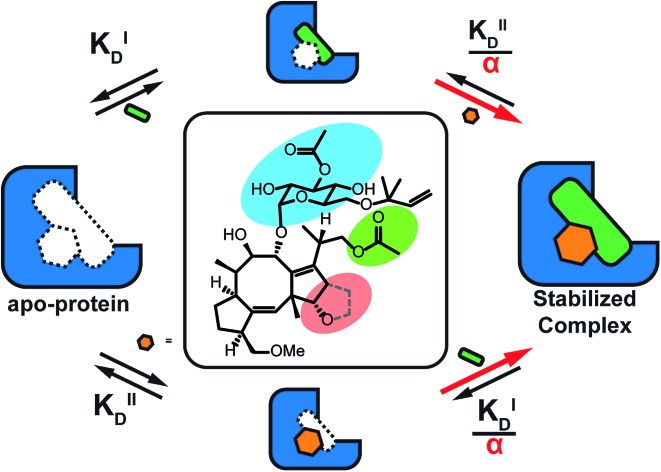
To quantify the degree of stabilization, the titration data was analysed using a cooperativity model (Fig. 3A) based on the sequential binding of protein (or representative peptide) partners (R and P in blue and green) and stabilizer molecule (S in orange). The binding partner binds to the target protein with KDI and in the presence of a stabilizer this affinity is altered to KDI/α. Similarly, the stabilizer binds with an intrinsic affinity KDII and an enhanced affinity KDII/α when the partner is already bound to the target protein. To provide analytical solutions for such a ternary equilibrium system, a semi-numerical thermodynamic model based on mass-action laws and mass-balance equations was established (Fig. 3B). These expressions can be rewritten into three interdependent master equations, which can be solved in a straightforward numerical manner (see ESI† for derivation).29–31
Fig. 3. (A) Cooperativity scheme of PPI stabilization involving sequential binding of protein (or representative peptide) partners (blue and green) and stabilizer molecule (orange). The binding partner binds to the target protein with KDI and in the presence of a stabilizer this affinity is altered to KDI/α. Similarly, the stabilizer binds with an intrinsic affinity KDII and an enhanced affinity KDII/α when the partner is already bound to the target protein. (B) Mass action laws and mass balance equations.
Analysis of the experimental data depicted in Fig. 2 with the cooperativity model resulted in an intrinsic affinity (KDII) of 0.3 mM for FC-A to the apo 14-3-3σ protein and an α-factor of 1 × 103. The intrinsic affinity (KDII) is in the same range as previously reported using indirect measurements.32 The resulting cooperativity factor, in contrast, is significantly larger than previously determined via classical 2D-plot analysis of the change in EC50 value at different relative concentrations of stabilizer.28 This observation highlights the importance of objective metrics for the characterization of PPI stabilizers. Fig. 2A (right bottom panel), for example, depicts the relationship between EC50 and the stabilizer (FC-A) concentration: the EC50 gradually decreases with increasing concentration of stabilizer. The EC50 values, as predicted by the cooperativity model (in red), highly correlate with the experimental data.
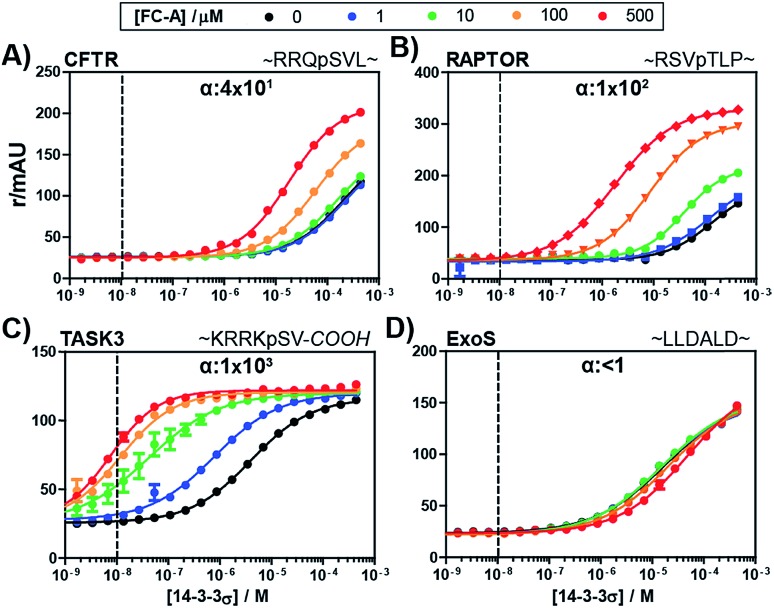
To evaluate the broader applicability of our cooperativity model, we applied it to several other 14-3-3-binding partners, each with different affinities and FC-A sensitivities.18,33 14-3-3σ was titrated to each binding partner at five different concentrations of FC-A to obtain the stabilization landscape and extract the corresponding α-factors (a selection is depicted in Fig. 4, see ESI Fig. 1† for a complete view). For some of the PPIs, the apparent affinity increased upon increasing concentrations of FC-A. For example, in the case of the CFTR-sequence the EC50 decreased from 174 μM to 2 μM in the presence of 500 μM FC-A (Fig. 4A). Fitting of the various PPIs resulted in a range of α-factors. The obtained intrinsic affinity (KDII) of FC-A for the 14-3-3 protein is, as expected, similar for all measured PPIs (around 0.3 mM). A KDII of 0.3 mM was also measured in the case where FC-A functioned as a PPI inhibitor of 14-3-3/ExoS and the cooperativity of the system is in effect negative (α ≤ 1) (Fig. 4D). In the absence of, or in case of very low cooperativity (α ∼ 1) the KDII could not be determined based on that individual data set alone (ESI Fig. 1†). The cooperativity of stabilizer and partner binding to 14-3-3 results in differentiated dose responses for different PPI pairs. For example, at least 100 μM FC-A is required to stabilize the 14-3-3 binding of CFTR-peptide (Fig. 4A, yellow line) while at a dose of 1 μM FC-A, the binding of the TASK3 peptide to 14-3-3 is already stabilized 20-fold (Fig. 4C, blue line). Even though the intrinsic affinity of FC-A for the apo 14-3-3 protein is of course the same and independent of the partner, stabilization of the TASK3/14-3-3 interaction occurs at a much lower stabilizer concentration due to the higher cooperativity factor in comparison to the other partners. This data makes an important point with respect to PPI stabilization, and demonstrates the value of characterizing PPI stabilizers using cooperativity. This difference in cooperativity provides a basis for selectivity in PPI stabilization. Especially for PPIs with high α-factor values, the small molecule stabilizers can act at much lower concentrations than their intrinsic affinity for one of the binding partners alone due to the mutual enhancement of stabilizer and peptide binding. The variation in cooperativity factors between the different 14-3-3 PPIs stabilized by FC-A can be explained by studying previously published crystal structure data (See ESI Fig. 2). For example, the TASK3 protein binds such that the terminal valine residue makes a van der Waals contact with the stabilizer molecule. By contrast, the binding mode of the ExoS peptide overlaps with the FC-A-pocket, which prevents simultaneous binding of stabilizer and peptide, and thus explains the absence of stabilization (and function as inhibitor).
Fig. 4. Fluorescence polarization assays of 14-3-3σ interacting with multiple FITC-labelled binding partners (10 nM, indicated by the dashed vertical line) at various concentration of FC-A between 0 and 500 μM. The cooperativity factor α, is obtained though data-fitting according to the model depicted in Fig. 3. (A) CFTR-sequence (B) RAPTOR-sequence. (C) TASK3-sequence. (D) ExoS-sequence.
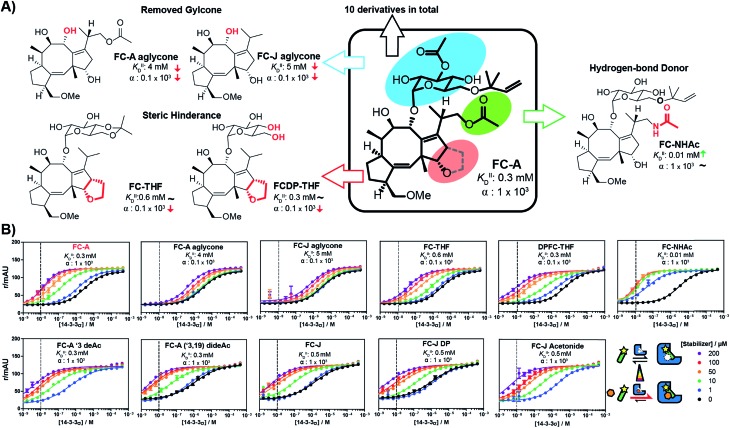
With the methodology to determine the intrinsic affinity and cooperativity factor established, we used our approach to perform a structure–acitivity relationship study of FC-A derivatives for the stabilization of the 14-3-3/TASK3 PPI. To this end, we compiled a library of eleven fusicoccin analogues, among them FC-A and FC-J and semi-synthetic derivatives (Fig. 5 & iESI†).28,34,35 We observed that the different FC analogues each stabilized the 14-3-3/TASK3 interaction differently (Fig. 5). The α-factor and KDII of each FC analogue was determined by fitting our numerical model to the corresponding 2D-FP titration of 14-3-3σ to the labelled TASK3 (summarized in Fig. 5B). With the use of our model, a simple structure–activity relationship of PPI stabilization can thus be obtained. As previously, FC-A binds to the apo form of 14-3-3σ with a KDII of 0.3 mM. Installation of a tetrahydrofuran ring at the terpene scaffold (i.e. FC-THF), achieved through a previous semi-synthesis,28 reduced the cooperativity factor to 0.1 × 103, potentially via a steric clash with the TASK3 peptide. Small modifications to the glycone (e.g. FC-A ′3 deAc and FC-A (′3,19) dideAc) seem to have either a very modest or no effect on the α-factor and KDII of FC-A, whereas complete removal of the glycone unit from FC-A substantially lowers the KDII value towards the mM regime, with a concomitant modest reduction in α-factor. The FC-aglycone derivatives (FC-A aglycone, FC-J aglycone), and FC-THF all measured a similar moderate α-factor. This finding is interesting because in classical 2D dose-response assays, the two aglycones are detectable only at high concentrations compared to FC-A (ESI Fig. 3†), whereas FC-THF produces a robust response. This difference here can be explained by the different KDII values. The previously published FC-NAc stabilizer35 has an α-factor identical to FC-A but an increased intrinsic affinity towards 14-3-3, making FC-NAc a 30-fold more potent stabilizer, thus revealing a structural entry for further potency enhancement or hit identification.
Fig. 5. (A) Structure–activity relationship of fusicoccin (FC) analogues as stabilizers for the 14-3-3/TASK3 interaction determined through 2D titrations (Fig. 5B). Variations in overall potency can now be expressed in terms of differences in either the cooperativity factor α or the intrinsic affinity KDII or both. (B) 2D-FP titrations with all fusicoccin analogs from this study and their corresponding cooperativity factor α and intrinsic affinity KDII for the 14-3-3/TASK3 PPI.
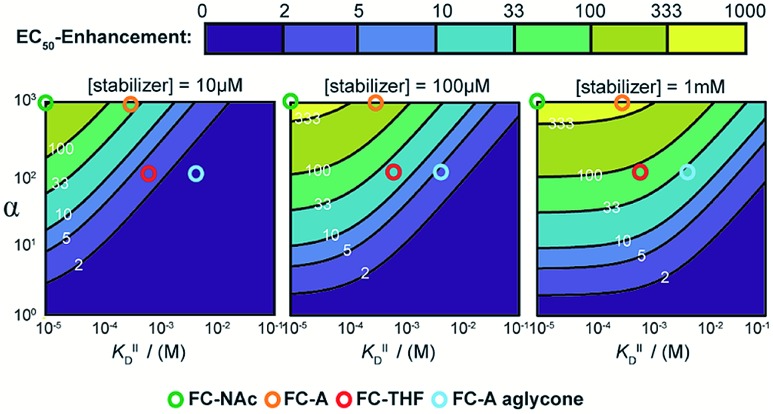
To guide future development of PPI stabilizers, we used our model to determine the boundaries at which stabilization could be observed in terms of both cooperativity factor α and intrinsic affinity (KDII). We simulated protein titration at three different constant stabilizer concentrations (i.e. 10 μM, 100 μM, and 1 mM) for different α′s and KDII and mapped the expected enhancement in EC50 value (Fig. 6). The corresponding position of four FC analogues are annotated as well. At a 10 μM dose of stabilizer, larger portions of the simulated space gave a less than two-fold enhancement in EC50 value (area indicated in purple), and are therefore close to limit of detection. In this case, higher doses of stabilizer would be required to observe an effect. Indeed, an effect is observed for FC-A aglycone at 1 mM. The cooperativity paradigm thus provides an additional benefit in that it avoids false negatives; i.e. it allows the detection of weak binding stabilizers, which would otherwise be missed in conventional dose-response assays.
Fig. 6. Expected EC50-enhancement, defined as the ratio of non-stabilized and stabilized EC50, for protein titrations with different stabilizers concentration. The EC50's are determined via simulated protein titrations depending on α and KDII at 10 μM, 100 μM and 1 mM stabilizer. The corresponding position of FC-derivatives are annotated as circles.
Conclusions
This study provides a systematic framework to interpret PPI stabilization using a cooperativity model. The model describes stabilizer efficacy in terms of its cooperativity factor (α) as well as its intrinsic affinity towards one of the proteins (KDII). Both parameters are obtained in a straightforward manner through 2D-FP titrations, by varying stabilizer compound and receptor protein, in combination with a numerical model. Since absolute KD′s and cooperativity factors are fundamental thermodynamic parameters, they allow for an objective comparison of different stabilizers and the establishment of structure–activity relationships between analogue stabilizer compounds. In our examples, the affinities of the stabilizers for the 14-3-3 binding partners are negligibly low, but for PPI systems where such interactions are relevant this could be added to the model. Here, for the first time, we performed an objective structure–activity analysis for stabilizers of PPIs, tracing variation in potency back to differences in cooperativity or intrinsic affinity. Traditional dose-response assays of (such) ternary stabilizer-protein systems are biased towards stabilizers with both high affinity and large α-factors. However, in our experience these assays tend to miss stabilizers with weaker binding affinity (false negatives), even when their cooperativity factor is high, such is the case for the FC-aglycone in this present study. It is notable that the intrinsic affinities for the apo-form of 14-3-3 toward the analogue fusicoccin stabilizers are relativity weak (in the high micromolar range). These compounds only act as stabilizers when both partners are present, which prevents the occurrence of bell-shaped curves or the so-called “hook-effect” at increased stabilizer dosages.17,36 The cooperativity factor also provides an attractive entry to address the challenge of achieving selectivity in PPI stabilization.
The cooperativity framework presented here allows to understand PPI stabilization on a fundamental level and is not only important for the screening of 14-3-3 PPI stabilizers, but should also be of similarly high value for other PPIs, as well as for PPI complexes based on other cooperativity mechanisms.13 We are currently working on dedicated methods for more efficient screening of such PPI stabilizing molecules for diverse PPIs.
Conflicts of interest
There are no conflicts to declare.
Supplementary Material
Acknowledgments
This research was funded by the Netherlands Organization for Scientific Research (NWO) through Gravity program 024.001.035, VICI grant 016.150.366, and ECHO grant 717.014.001 and by JSPS KAKENHI Grant Number JP 16 K21138. We thank Minke Nijenhuis and Tom de Greef for support with the MATLAB script.
Footnotes
†Electronic supplementary information (ESI) available: Supporting Figures, exact sequences, synthesis, extensive clarification of the thermodynamic model. See DOI: 10.1039/c8sc05242e
References
- Milroy L.-G., Grossmann T. N., Hennig S., Brunsveld L., Ottmann C. Chem. Rev. 2014;114:4695–4748. doi: 10.1021/cr400698c. [DOI] [PubMed] [Google Scholar]
- Arkin M. R., Tang Y., Wells J. A. Chem. Biol. 2014;21:1102–1114. doi: 10.1016/j.chembiol.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrei S. A., Sijbesma E., Hann M., Davis J., O'Mahony G., Perry M. W. D., Karawajczyk A., Eickhoff J., Brunsveld L., Doveston R. G., Milroy L.-G., Ottmann C. Expert Opin. Drug Discovery. 2017;12:925–940. doi: 10.1080/17460441.2017.1346608. [DOI] [PubMed] [Google Scholar]
- Bondeson D. P., Smith B. E., Burslem G. M., Buhimschi A. D., Hines J., Jaime-Figueroa S., Wang J., Hamman B. D., Ishchenko A., Crews C. M. Cell Chem. Biol. 2018;25:78–87.e5. doi: 10.1016/j.chembiol.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bier D., Mittal S., Bravo-Rodriguez K., Sowislok A., Guillory X., Briels J., Heid C., Bartel M., Wettig B., Brunsveld L., Sanchez-Garcia E., Schrader T., Ottmann C. J. Am. Chem. Soc. 2017;139:16256–16263. doi: 10.1021/jacs.7b07939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petzold G., Fischer E. S., Thomä N. H. Nature. 2016;532:127–130. doi: 10.1038/nature16979. [DOI] [PubMed] [Google Scholar]
- Matyskiela M. E., Lu G., Ito T., Pagarigan B., Lu C.-C., Miller K., Fang W., Wang N.-Y., Nguyen D., Houston J., Carmel G., Tran T., Riley M., Nosaka L., Lander G. C., Gaidarova S., Xu S., Ruchelman A. L., Handa H., Carmichael J., Daniel T. O., Cathers B. E., Lopez-Girona A., Chamberlain P. P. Nature. 2016;535:252–257. doi: 10.1038/nature18611. [DOI] [PubMed] [Google Scholar]
- Huang H., Ceccarelli D. F., Orlicky S., St-Cyr D. J., Ziemba A., Garg P., Plamondon S., Auer M., Sidhu S., Marinier A., Kleiger G., Tyers M., Sicheri F. Nat. Chem. Biol. 2014;10:156–163. doi: 10.1038/nchembio.1412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petta I., Lievens S., Libert C., Tavernier J., De Bosscher K. Mol. Ther. 2015;24:707–718. doi: 10.1038/mt.2015.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löwe J., Li H., Downing K. H., Nogales E. J. Mol. Biol. 2001;313:1045–1057. doi: 10.1006/jmbi.2001.5077. [DOI] [PubMed] [Google Scholar]
- Choi J., Chen J., Schreiber S. L., Clardy J. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- Ottmann C., Marco S., Jaspert N., Marcon C., Schauer N., Weyand M., Vandermeeren C., Duby G., Boutry M., Wittinghofer A., Rigaud J.-L., Oecking C. Mol. Cell. 2007;25:427–440. doi: 10.1016/j.molcel.2006.12.017. [DOI] [PubMed] [Google Scholar]
- Whitty A. Nat. Chem. Biol. 2008;4:435–439. doi: 10.1038/nchembio0808-435. [DOI] [PubMed] [Google Scholar]
- von Krbek L. K. S., Schalley C. A., Thordarson P. Chem. Soc. Rev. 2017;46:2622–2637. doi: 10.1039/c7cs00063d. [DOI] [PubMed] [Google Scholar]
- Kenakin T. Chem. Rev. 2017;117:4–20. doi: 10.1021/acs.chemrev.5b00561. [DOI] [PubMed] [Google Scholar]
- Johnstone S., Albert J. S. Bioorg. Med. Chem. Lett. 2017;27:2239–2258. doi: 10.1016/j.bmcl.2017.03.084. [DOI] [PubMed] [Google Scholar]
- Gadd M. S., Testa A., Lucas X., Chan K.-H., Chen W., Lamont D. J., Zengerle M., Ciulli A. Nat. Chem. Biol. 2017;13:514–521. doi: 10.1038/nchembio.2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevers L. M., Sijbesma E., Botta M., MacKintosh C., Obsil T., Landrieu I., Cau Y., Wilson A. J., Karawajczyk A., Eickhoff J., Davis J., Hann M., O'Mahony G., Doveston R. G., Brunsveld L., Ottmann C. J. Med. Chem. 2018;61:3755–3778. doi: 10.1021/acs.jmedchem.7b00574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aghazadeh Y., Papadopoulos V. Drug Discovery Today. 2016;21:278–287. doi: 10.1016/j.drudis.2015.09.012. [DOI] [PubMed] [Google Scholar]
- de Vink P. J., Briels J. M., Schrader T., Milroy L.-G., Brunsveld L., Ottmann C. Angew. Chem., Int. Ed. 2017;56:8998–9002. doi: 10.1002/anie.201701807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevers L. M., de Vink P. J., Ottmann C., Huskens J., Brunsveld L. J. Am. Chem. Soc. 2018;140:14498–14510. doi: 10.1021/jacs.8b09618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevers L. M., Lam C. V., Leysen S. F. R., Meijer F. A., van Scheppingen D. S., de Vries R. M. J. M., Carlile G. W., Milroy L. G., Thomas D. Y., Brunsveld L., Ottmann C. Proc. Natl. Acad. Sci. U.S.A. 2016;113:E1152–E1161. doi: 10.1073/pnas.1516631113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gwinn D. M., Shackelford D. B., Egan D. F., Mihaylova M. M., Mery A., Vasquez D. S., Turk B. E., Shaw R. J. Mol. Cell. 2008;30:214–226. doi: 10.1016/j.molcel.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masters S. C., Pederson K. J., Zhang L., Barbieri J. T., Fu H. Biochemistry. 1999;38:5216–5221. doi: 10.1021/bi982492m. [DOI] [PubMed] [Google Scholar]
- Molzan M., Kasper S., Röglin L., Skwarczynska M., Sassa T., Inoue T., Breitenbuecher F., Ohkanda J., Kato N., Schuler M., Ottmann C. ACS Chem. Biol. 2013;8:1869–1875. doi: 10.1021/cb4003464. [DOI] [PubMed] [Google Scholar]
- De Vries-van Leeuwen I. J., da Costa Pereira D., Flach K. D., Piersma S. R., Haase C., Bier D., Yalcin Z., Michalides R., Feenstra K. A., Jiménez C. R., de Greef T. F. A., Brunsveld L., Ottmann C., Zwart W., de Boer A. H. Proc. Natl. Acad. Sci. U.S.A. 2013;110:8894–8899. doi: 10.1073/pnas.1220809110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doveston R. G., Kuusk A., Andrei S. A., Leysen S., Cao Q., Castaldi M. P., Hendricks A., Brunsveld L., Chen H., Boyd H., Ottmann C. FEBS Lett. 2017;591:2449–2457. doi: 10.1002/1873-3468.12723. [DOI] [PubMed] [Google Scholar]
- Anders C., Higuchi Y., Koschinsky K., Bartel M., Schumacher B., Thiel P., Nitta H., Preisig-Müller R., Schlichthörl G., Renigunta V., Ohkanda J., Daut J., Kato N., Ottmann C. Chem. Biol. 2013;20:583–593. doi: 10.1016/j.chembiol.2013.03.015. [DOI] [PubMed] [Google Scholar]
- Ehlert F. J. Mol. Pharmacol. 1988;33:187. [PubMed] [Google Scholar]
- Douglass E. F., Miller C. J., Sparer G., Shapiro H., Spiegel D. A. J. Am. Chem. Soc. 2013;135:6092–6099. doi: 10.1021/ja311795d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Hamer A., Lemmens L. J. M., Nijenhuis M. A. D., Ottmann C., Merkx M., de Greef T. F. A., Brunsveld L. ChemBioChem. 2017;18:331–335. doi: 10.1002/cbic.201600631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Würtele M., Jelich-Ottmann C., Wittinghofer A., Oecking C. EMBO J. 2003;22:987–994. doi: 10.1093/emboj/cdg104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milroy L.-G., Brunsveld L., Ottmann C. ACS Chem. Biol. 2013;8:27–35. doi: 10.1021/cb300599t. [DOI] [PubMed] [Google Scholar]
- Maki T., Kawamura A., Kato N., Ohkanda J. Mol. BioSyst. 2013;9:940–943. doi: 10.1039/c2mb25388g. [DOI] [PubMed] [Google Scholar]
- Andrei S. A., de Vink P., Sijbesma E., Han L., Brunsveld L., Kato N., Ottmann C., Higuchi Y. Angew. Chem., Int. Ed. 2018;57:13470–13474. doi: 10.1002/anie.201806584. [DOI] [PubMed] [Google Scholar]
- Ercolani G., Schiaffino L. Angew. Chem., Int. Ed. 2011;50:1762–1768. doi: 10.1002/anie.201004201. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.