Abstract
Background
We assessed the utility of a multi-target, real-time PCR assay for Bordetella pertussis detection and diagnosis in patients with severe respiratory illness (SRI), influenza-like illness (ILI), and asymptomatic controls.
Methods
Real-time PCR detection of IS481, pIS1001, hIS1001 and ptxS1 was performed on nasopharyngeal specimens (SRI, ILI and controls) and induced sputum (SRI) collected from June 2012 to May 2016 through respiratory illness surveillance. Using PCR cycle threshold (Ct) value cut-offs, IS481 positive cases were classified as confirmed (Ct < 35) or possible (Ct 35–39) pertussis disease.
Results
Among 12,922 samples, 146 (1.1%) were IS481 positive of which 62% (90/146) were classified as confirmed. The attributable fraction (AF) was 92.2% (95% CI, 65.6 to 98.2%) and 90.5% (95% CI, 57.5 to 97.9%) amongst SRI and ILI PCR-confirmed pertussis cases, respectively. Amongst possible pertussis cases, AF was 36.9% (95% CI, − 142.3 to 83.6%) and 67.5% (95% CI, − 30.6 to 91.9%) in the SRI and ILI groups, respectively.
Conclusion
All IS481 positive specimens could be considered as B. pertussis infection, and potentially pertussis disease with supportive clinical information.
Electronic supplementary material
The online version of this article (10.1186/s12879-019-3869-7) contains supplementary material, which is available to authorized users.
Keywords: Bordetella pertussis, South Africa, Severe respiratory illness, Influenza-like illness (ILI), Surveillance, Real-time PCR, Confirmed-pertussis, Possible-pertussis, Attributable fraction
Background
Pertussis (whooping cough) is caused by the vaccine-preventable, bacterial pathogen Bordetella pertussis [1, 2]. Despite high vaccine coverage with the whole-cell or acellular vaccines and an initial substantial decrease following vaccine introduction, the incidence of pertussis has increased globally during the last two decades [3–7]. This has been attributed to increased awareness by clinicians, more sensitive molecular techniques for diagnosis [4, 7], serological markers for identification of infection in adolescents and adults who usually present atypically with B. pertussis [3], pathogen adaptation and/or waning vaccine immunity [6, 8].
The changing epidemiology of pertussis disease requires new control strategies but establishing the burden of pertussis disease in low-resource settings is challenging due to the absence of effective surveillance systems. As such, pertussis surveillance can potentially be linked to surveillance for other respiratory illnesses, such as influenza or pneumonia surveillance [9]. Differences in case definitions, however, may result in reduced sensitivity for detecting pertussis cases using these platforms.
Different laboratory methods such as bacterial culture, molecular testing and serology may be used to confirm a clinical diagnosis of pertussis, however, the sensitivity of each method is dependent on the stage of disease and the age of the individual. Culture and molecular testing are recommended for diagnosis during the early (catarrhal and paroxysmal) phases of disease, whereas serology is recommended during the convalescent phase [10, 11]. The sensitivity of both culture and PCR appear to be highest in young infants and decline in adolescents and adults [12].
Real-time PCR detection of B. pertussis is limited by a lack of validated and specific gene targets. The most commonly used PCR target is the multi-copy insertion sequence gene IS481 (present in 50–250 copies per genome); however, this gene is also found in B. holmesii and B. bronchiseptica [13, 14]. Use of a second confirmatory target such as the pertussis toxin subunit gene (ptxS1) improves specificity of B. pertussis detection [7, 15, 16].
The high copy number of IS481 enables highly sensitive detection of B. pertussis, however, this results in uncertainty regarding the interpretation of results with high PCR cycle threshold (Ct) values that cannot always be confirmed by detection of a less sensitive, single copy ptxS1 assay. Multiple copies of IS481 increase the risk of laboratory contamination and detection of false positives [17, 18]. Bacterial load in the host and resulting Ct values may be influenced by time between symptom onset and specimen collection, immune status of the host, prior use of antimicrobial therapy or quality of the sample collected [19–21]. The interpretation and clinical relevance of high Ct values, in a setting where stringent laboratory control measures are available to prevent and detect IS481 contamination, is not clearly understood.
We aimed to evaluate a multi-target real-time PCR assay and the use of Ct value cut-offs for B. pertussis detection and diagnosis in patients with mild or severe respiratory illness at two sentinel sites in South Africa.
Methods
Surveillance population
We enrolled patients and asymptomatic controls through two surveillance systems (one for severe respiratory illness (SRI) and one for influenza-like illness (ILI)) from June 2012 through May 2016. Patients of all ages, hospitalised with lower respiratory tract infection, irrespective of symptom duration, were enrolled in prospective, active surveillance as part of severe respiratory illness (SRI) surveillance at Edendale Hospital and Klerksdorp-Tshepong Hospital Complex (KTHC) [22]. From August 2014, case investigation forms used for patient enrolment were amended to capture clinical symptoms consistent with pertussis disease, namely, paroxysms of cough, inspiratory whoop, apnea (with or without cyanosis) and posttussive vomiting. Patients with influenza-like illness (ILI), and asymptomatic controls were enrolled at the respective outpatient clinics affiliated to each hospital [Edendale Gateway and Jouberton clinics [23]]. A case of ILI was defined as an outpatient presenting with acute fever (> 38 °C) and/or self-reported fever and cough or sore throat within the last 10 days. A control was defined as a person presenting at the same outpatient clinic with no history of fever, respiratory or gastrointestinal symptoms during the 14 days preceding the visit. Controls were not followed-up for development of respiratory symptoms after enrolment. We aimed to enrol one HIV-infected and one HIV-uninfected control every week in each clinic within the following age categories: 0–1, 2–4, 5–14, 15–54 and ≥ 55 years (controls were enrolled by HIV status so that the study was powered sufficiently for HIV-stratified analysis). Demographic and clinical data were collected by surveillance officers through interview and hospital record review. For children aged < 5 years, immunization history (including pertussis immunization) was documented and confirmed from the vaccination card if available.
Specimen collection and processing
Nasopharyngeal (NP) specimens were collected from patients and controls as follows: NP aspirates were collected from children (< 5 years), whereas both NP and oropharyngeal (OP) swabs were collected from older individuals (≥5 years) and stored together in a single vial of a 3 ml solution of Universal Transport Medium (UTM) (Copan, Italia, Brescia, Italy). Induced sputum was collected from hospitalised (SRI) patients only. NPs were stored at 4 °C and transported on ice packs and induced sputum was stored at − 20 °C and transported on dry ice to the National Institute for Communicable Diseases (NICD) in Johannesburg for testing. From August 2014, all SRI- and ILI-enrolled individuals with pertussis symptoms had a second NP collected and inoculated into Regan-Lowe medium (Media Mage, Johannesburg, South Africa) for culture at the NICD. Induced sputum and Regan-Lowe swabs were cultured on charcoal agar (Diagnostic Media Products, Johannesburg, South Africa). Plates were incubated at 37 °C for seven days and inspected for growth on days three and seven. Suspected B. pertussis colonies were confirmed using matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF, Bruker, Massachusetts, United States) and by real-time PCR detection of IS481 and ptxS1 genes [16].
Detection of Bordetella species by real-time PCR
Total nucleic acids were extracted from 200 μl of UTM or digested induced sputum using a Roche MagNa Pure 96 instrument (Roche Diagnostics, Mannheim, Germany) and MagNa Pure 96 DNA and Viral NA SV Kit (Roche Diagnostics) using the Pathogen Universal protocol. Real-time PCR was performed using an Applied Biosystems 7500 Fast instrument (Applied Biosystems, Foster City, United States) targeting IS481, pIS1001, hIS1001 and ptxS1 as previously described [16]. The targets differentiate B. pertussis, B. parapertussis, B. holmesii and B. bronchiseptica. PCR assays were performed in 96-well plates including 16 negative extraction controls and 16 no-template controls distributed throughout the plate to detect IS481 contamination. An internal validation was performed for the assay and 100% sensitivity and specificity was obtained for the detection of B. parapertussis, B. bronchiseptica and B. holmesii; and 95% sensitivity and 100% specificity was obtained for the detection of B. pertussis. All testing was conducted at the NICD, at an accredited reference laboratory using standardised procedures and testing algorithms.
Real-time PCR result interpretation and PCR quality control
A specimen was considered positive for B. pertussis DNA if both IS481 and ptxS1 were detected with Ct values ≤39. Additionally, samples that were positive for IS481 only (with Ct values 35–39 and without confirmation of ptxS1) and negative for pIS1001 (B. parapertussis) or hIS1001 (B. holmesii) were recorded as B. pertussis positive. Samples with positive PCR results (IS481 and/or ptxS1) were further categorised as confirmed or possible B. pertussis based on IS481 Ct value cut-offs of < 35 and 35–39, respectively. The testing algorithm was as follows: specimens that tested positive for IS481 on initial screening underwent repeat testing, namely, new extraction, and PCR in duplicate. Results were recorded as positive if IS481 was detected in at least two replicates, taking into account the result from the primary screen.
Detection of the human ribonuclease P gene (RNase P) served as a control to exclude the presence of PCR inhibitors and confirm specimen quality [24]. To exclude spurious amplification, a 1014-bp fragment of the IS481 gene from a selection of confirmed (n = 13) and possible (n = 14) B. pertussis PCR-positive samples was amplified and sequenced as described previously [16]. Sequences were analysed using DNAStar Lasergene SeqMan Pro software (DNAStar, Wisconsin, United States) and the Basic Local Alignment Search Tool (BLAST).
Higher than expected PCR detection rates of IS481 (based on baseline data) at any point during the study period were investigated to exclude laboratory and/or environmental contamination: (i) all laboratory processes were audited, (ii) environmental samples were collected from appropriate areas at surveillance sites and laboratory surfaces were swabbed and tested by real-time PCR for IS481 and (iii) individuals that tested positive for B. pertussis during this time were retrospectively interviewed using a standardised questionnaire to collect data on pertussis-related symptoms.
Data analysis
The prevalence of B. pertussis by patient group (SRI, ILI and controls) and 95% confidence intervals (CI) was calculated. To determine the optimal specimen type for molecular detection of B. pertussis, the detection rates in NP and induced sputum were compared using the chi-squared test among individuals that had both specimen types collected and tested. We estimated the fraction of B. pertussis detection (IS481-positive PCR results) attributable (attributable fraction [AF]) to severe (SRI) (including only patients that tested positive on NP specimens) or mild (ILI) illness using unconditional logistic regression by comparing the B. pertussis detection rate among SRI or ILI cases to those of controls. All models were adjusted for HIV status and age. The AF was estimated from the odds ratio (OR) obtained from the regression models using the following equation: AF = (OR-1)/(OR) × 100 as previously described [25]. An OR was used for these calculations as the data were analysed considering the case-control design of this study. The AF in this study provides an estimate of the proportion of pertussis-positive patients who have symptomatic illness (either SRI or ILI as defined in this study) resulting from pertussis infection. This analysis was implemented overall as well as among confirmed (NP IS481 Ct < 35) and possible (NP IS481 Ct 35–39) cases. In addition, we compared the demographic and clinical characteristics of possible vs. confirmed IS481-positive patients with SRI or ILI using univariate unconditional conventional or penalised (for categorical variables with 0 outcome in one or more categories) logistic regression. For patients with SRI, confirmed and possible cases were compared for (i) patients that tested positive for IS481 on either one or both specimen types (where both specimen types were available and tested positive for B. pertussis, the lower of the two Ct values was recorded) and (ii) patients that tested positive for IS481 on NP specimens only. Statistical analyses were performed using Stata® version 14 (Statacorp, Texas, United States). Statistical significance was determined at p < 0.05 for all analyses.
Ethics approval
Ethics for SRI (M081042) and ILI (M120133) surveillance and B. pertussis testing were approved by the University of the Witwatersrand (M130260) and the University of KwaZulu Natal (BF081/12), South Africa.
Results
Study population
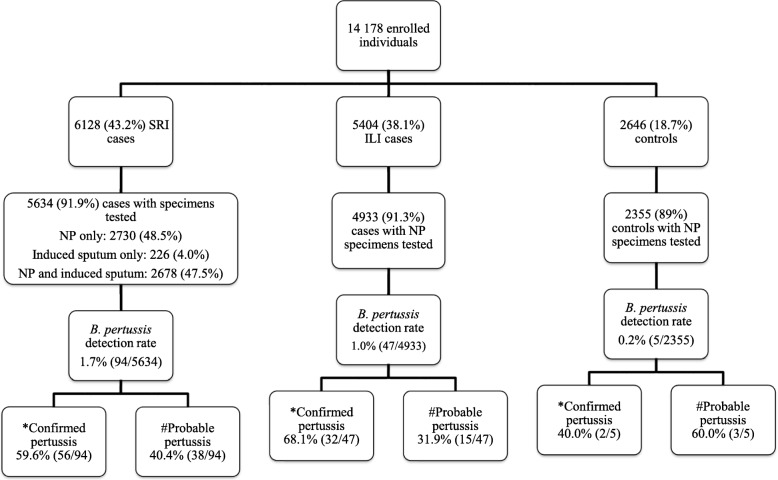
From June 2012 through May 2016, 14,178 individuals were enrolled. Testing was conducted on 12,922 (91%) specimens: 5634 (44%) from SRI patients, 4933 (38%) from ILI patients and 2355 (18%) from control subjects (Fig. 1). Specimens were not tested or available for 9% of enrolled individuals as participants were either too sick to have specimens taken or did not give consent for testing or were discharged before specimens could be collected. Children aged < 5 years accounted for 37% (2071/5618), 33% (1618/4921) and 33% (787/2352) of SRI cases, ILI cases, and controls, respectively (Table 1). The HIV prevalence was 51% (2731/5308) among SRI cases, 27% (1198/4451) among ILI cases and 42% (949/2263) among controls (enrolment criteria for controls was based on HIV status).
Fig. 1.
Flow diagram depicting breakdown of individuals enrolled, specimens collected and tested, and B. pertussis prevalence for severe respiratory (SRI) and influenza-like illness (ILI) surveillance, and controls, South Africa, June 2012 – May 2016. NP: nasopharyngeal aspirates from children < 5 years of age and combined nasopharyngeal and oropharyngeal swabs from individuals aged ≥5 years. *Confirmed = positive for B. pertussis with IS481 Ct < 35; Possible = positive for B. pertussis with IS481 35 ≥ Ct ≤ 39
Table 1.
Demographic and clinical characteristics of patients enrolled for severe respiratory and influenza-like illness surveillance and asymptomatic controls, South Africa, June 2012 – May 2016 (N = 12,922)
| Characteristic | Surveillance population | ||
|---|---|---|---|
| SRI n/N (%) N = 5634 | ILI n/N (%) N = 4933 | Controls n/N (%) N = 2355 | |
| Gender | |||
| Male | 2861/5626 (51) | 1757/4733 (37) | 774/2291 (34) |
| Female | 2765/5626 (49) | 2976/4733 (63) | 1517/2291 (66) |
| Race | |||
| Black | 5473/5604 (98) | 4723/4728 (100) | 2287/2292 (100) |
| Non-black | 131/5604 (2) | 5/4728 (0) | 5/2292 (0) |
| Age group (years) | |||
| < 1 | 1307/5618 (23) | 599/4921 (12) | 323/2352 (14) |
| 1–4 | 764/5618 (14) | 1019/4921 (21) | 464/2352 (20) |
| 5–14 | 168/5618 (3) | 727/4921 (15) | 520/2352 (22) |
| 15–24 | 276/5618 (5) | 649/4921 (13) | 191/2352 (8) |
| 25–44 | 1756/5618 (31) | 1388/4921 (28) | 371/2352 (16) |
| 45–64 | 1051/5618 (19) | 460/4921 (9) | 337/2352 (14) |
| ≥ 65 | 296/5618 (5) | 79/4921 (2) | 146/2352 (6) |
| Underlying illness* | |||
| No | 5023/5627 (89) | 4484/4723 (95) | 2172/2290 (95) |
| Yes | 604/5627 (11) | 239/4723 (5) | 118/2290 (5) |
| HIV status | |||
| Uninfected | 2577/5308 (49) | 3253/4451 (73) | 1314/2263 (58) |
| Infected | 2731/5308 (51) | 1198/4451 (27) | 949/2263 (42) |
| Hospital/Clinic | |||
| Edendale | 2507/5634 (45) | N/A | N/A |
| KTHC | 3127/5634 (56) | N/A | N/A |
| Edendale Gateway | N/A | 3655/4933 (74) | 1041/2355 (44) |
| Jouberton Clinic | N/A | 1278/4933 (26) | 1314/2355 (56) |
SRI = Severe respiratory illness; ILI = Influenza-like illness; N/A = Not applicable; KTHC = Klerksdorp Tshepong hospital complex
*Patients with previously diagnosed chronic conditions including asthma, chronic lung diseases, cirrhosis/liver failure, chronic renal failure, heart failure, valvular heart disease, coronary heart disease, immunosuppressive therapy, splenectomy, diabetes, burns, kwashiorkor/marasmus, nephrotic syndrome, spinal cord injury, seizure disorder, emphysema, or cancer. All percentages are rounded off
PCR detection rate of B. pertussis
Overall, B. pertussis was detected in 1.1% (146/12,922, 95% CI 1.4 to 1.8) of individuals with detection rates of 1.7% (94/5634, 95% CI 2.0 to 2.8), 1.0% (47/4933, 95% CI 0.9 to 1.5) and 0.2% (5/2355, 95% CI 0.1 to 0.6) in the SRI, ILI and control groups, respectively (SRI vs. ILI p < 0.0001) (Fig. 1). B. parapertussis was detected in 0.8% (46/5634), 0.4% (22/4933) and 0.2% (4/2355) of SRI, ILI and control individuals, respectively. We did not detect B. pertussis and B. parapertussis co-infections. In addition, neither B. holmesii nor B. bronchiseptica was detected.
Among SRI cases with both specimen types collected, the detection rate was 0.2% (6/2678) for NP specimens (using NP specimens only) and 1.2% (31/2678) for induced sputum (using induced sputum only) (p < 0.0001). In 0.9% (25/2678) of patients, both specimen types were positive for B. pertussis. Where both specimen types tested positive for B. pertussis, induced sputum yielded lower Ct values [mean Ct value (±standard deviation) 24 ± 7] compared to NP specimens (mean Ct value 31 ± 7) (p = 0.002).
Of 3104 induced sputum samples, 3 (0.1%) were culture positive for B. pertussis. From August 2014 to May 2016, 0.1% (4/4583) of NPs were culture positive for B. pertussis. All isolates were from IS481-positive samples with Ct values < 35.
Confirmed and possible pertussis cases
Overall, 146 PCR-positive B. pertussis individuals (patients positive on either NP or induced sputum) were identified in all three surveillance groups, of which 62% (90/146) were categorised as confirmed (Fig. 1). The proportion of confirmed cases did not differ significantly by study population (SRI: 60%, 56/94; ILI: 68%, 32/47; controls: 40%, 2/5 (40%); p = 0.37).
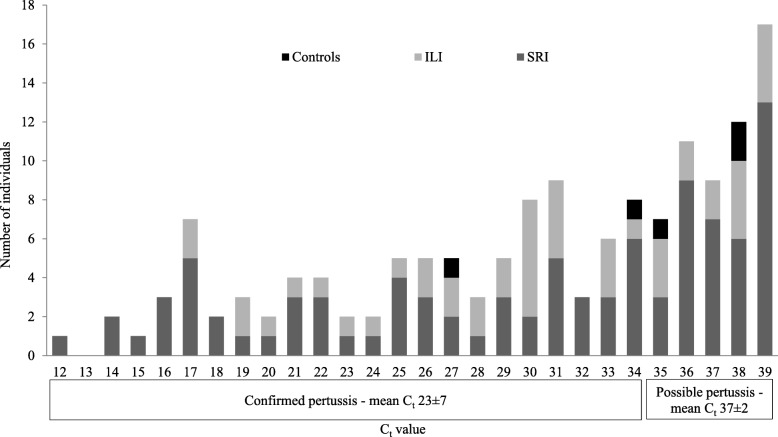
The mean Ct value was 23 ± 7 for confirmed cases and 37 ± 2 for possible cases (Fig. 2). Of the confirmed and possible cases, 92% (83/90) and 7% (4/56) were also positive for the ptxS1 gene, respectively. The sequences of IS481 amplicons from 12/13 (92%) samples from confirmed and 6/14 (43%) samples from possible cases were confirmed through BLAST analysis. IS481 amplicons from one confirmed (Ct = 34) and eight possible (Ct range 35–39) cases could not be confirmed by sequencing although PCR amplicons of the expected size were confirmed using agarose gel electrophoresis.
Fig. 2.
Distribution of real-time PCR IS481 Ct values for confirmed (n = 90) and possible (n = 56) pertussis cases, by surveillance population, South Africa, June 2012 – May 2016. SRI = severe respiratory illness; ILI = influenza-like illness
B. pertussis attributable fraction (AF)
For SRI patients compared to controls, the overall AF of B. pertussis detection to respiratory disease was 72.4% (95% CI, 41.0 to 87.1%); with 92.2% (95% CI, 65.6 to 98.2%) and 36.9% (95% CI, − 142.3 to 83.6%) for confirmed and possible cases, respectively. For ILI cases compared to controls, the overall AF was 77.3% (95% CI, 50.8 to 89.5%); with 90.5% (95% CI, 57.5 to 97.9%) and 67.5% (95% CI, − 30.6 to 91.9%) for confirmed and possible cases, respectively.
Patient characteristics among pertussis-confirmed and -possible cases
Among SRI patients, 1.7% (94/5634) were PCR positive for B. pertussis, of which 60% (56/94) were categorised as confirmed B. pertussis cases. Possible cases were more likely to occur among patients aged 45–64 years than among patients aged < 1 year (OR 6.3, 95% CI 1.7 to 23.8) (Table 2). Restricting the analysis to include cases that were positive on NP specimens only, there were no differences between the confirmed and possible cases (Additional file 1). There were no differences in the demographic and clinical characteristics between the confirmed and possible cases that tested positive on either NP and/or sputum specimens or on NP specimens only.
Table 2.
Comparison of confirmed (N = 56) and possible (N = 38) pertussis cases (real-time PCR positive for IS481 in nasopharyngeal or induced sputum specimens) in hospitalised patients with severe respiratory illness, South Africa, June 2012 – May 2016 (N = 94)
| Characteristic | Confirmed pertussis* n/N# (%) |
Possible pertussis* n/N# (%) |
OR† | P value |
|---|---|---|---|---|
| (95% CI) | ||||
| Year | ||||
| 2012 | 7/56 (12.5) | 4/38 (10.5) | Reference | |
| 2013 | 9/56 (16) | 9/38 (24) | 1.8 (0.4–8.1) | 0.48 |
| 2014 | 16/56 (29) | 13/38 (34) | 1.4 (0.3–5.9) | 0.63 |
| 2015 | 22/56 (39) | 10/38 (26) | 0.8 (0.1–3.4) | 0.76 |
| 2016 | 2/56 (4) | 2/38 (5) | 1.8 (0.2–17.7) | 0.64 |
| Gender | ||||
| Male | 24/56 (43) | 21/38 (55) | Reference | |
| Female | 32/56 (57) | 17/38 (45) | 0.6 (0.3–1.4) | 0.24 |
| Age group (years)¶ | ||||
| < 1 | 23/56 (41) | 9/38 (24) | Reference | |
| 1–4 | 6/56 (11) | 5/38 (13) | 2.1 (0.5–8.2) | 0.29 |
| 5–14 | 5/56 (9) | 2/38 (5) | 1.1 (0.2–6.0) | 0.89 |
| 15–24 | 1/56 (2) | 0/38 (0) | 0.8 (0.03–22.1) | 0.91 |
| 25–44 | 15/56 (27) | 11/38 (29) | 1.8 (0.6–5.4) | 0.27 |
| 45–64 | 4/56 (7) | 11/38 (29) | 6.3 (1.7–23.8) | 0.006 |
| ≥ 65 | 2/56 (4) | 0/38 (0) | 0.5 (0.02–11.3) | 0.66 |
| Fever history | ||||
| No | 28/55 (51) | 21/38 (55) | Reference | |
| Yes | 27/55 (49) | 17/38 (45) | 0.8 (0.4–1.9) | 0.68 |
| HIV status | ||||
| Uninfected | 27/53 (51) | 15/37 (41) | Reference | |
| Infected | 26/53 (49) | 22/37 (59) | 1.5 (0.7–3.6) | 0.33 |
| HIV treatment | ||||
| No | 7/22 (32) | 9/19 (47) | Reference | |
| Yes | 15/22 (68) | 10/19 (53) | 0.5 (0.1–1.8) | 0.31 |
| Symptom duration | ||||
| < 7 days | 35/56 (62.5) | 23/38 (60.5) | Reference | |
| 7–20 days | 10/56 (18) | 5/38 (13) | 0.8 (0.2–2.5) | 0.65 |
| ≥ 21 days | 11/56 (20) | 10/38 (26) | 1.4 (0.5–3.8) | 0.53 |
| Underlying illness‡ | ||||
| No | 50/56 (89) | 30/38 (79) | Reference | |
| Yes | 6/56 (11) | 8/38 (21) | 2.2 (0.7–7.0) | 0.17 |
| ICU | ||||
| No | 53/55 (96) | 37/38 (97) | Reference | |
| Yes | 2/55 (4) | 1/38 (3) | 0.7 (0.06–8.2) | 0.79 |
| Antibiotic treatment (24 h) | ||||
| No | 51/56 (91) | 36/38 (95) | Reference | |
| Yes | 5/56 (9) | 2/38 (5) | 0.6 (0.1–3.1) | 0.10 |
| Hospital duration | ||||
| < 2 days | 5/52 (10) | 5/38 (13) | Reference | |
| 2–4 days | 18/52 (35) | 17/38 (45) | 0.9 (0.2–3.9) | 0.94 |
| 5–7 days | 10/52 (19) | 5/38 (13) | 0.5 (0.1–2.6) | 0.41 |
| ≥ 8 days | 19/52 (36.5) | 11/38 (29) | 0.6 (0.1–2.5) | 0.56 |
| Viral co-infection | ||||
| Viral co-infectionNo | 32/56 (57) | 24/38 (63) | Reference | |
| Viral co-infectionYes | 24/56 (43) | 14/38 (37) | 0.8 (0.3–1.8) | 0.56 |
| Outcome | ||||
| Survived | 50/54 (93) | 37/38 (97) | Reference | |
| Died | 4/54 (7) | 1/38 (3) | 0.3 (0.04–3.1) | 0.34 |
| Vaccination for age§ | ||||
| Full coverage | 16/22 (73) | 10/14 (71) | Reference | |
| Incomplete | 6/22 (27) | 4/14 (29) | 0.9 (0.2–4.2) | 0.93 |
| Facility | ||||
| Edendale | 26/56 (46) | 12/38 (32) | Reference | |
| KTHC | 30/56 (54) | 26/38 (68) | 1.9 (0.8–4.4) | 0.15 |
OR = Odds ratio; CI = confidence interval; KTHC = Klerksdorp Tshepong Hospital complex. *Confirmed case = positive for B. pertussis with IS481 Ct < 35; Possible case = positive for B. pertussis with IS481 35 ≥ Ct ≤ 39. #Data unknown/missing for some cases accounting for the different denominators. †Odds ratio calculated for confirmed versus possible pertussis cases using univariate logistic regression. Bold font indicates statistical significance
‡Patients with previously diagnosed chronic conditions including asthma, chronic lung diseases, cirrhosis/liver failure, chronic renal failure, heart failure, valvular heart disease, coronary heart disease, immunosuppressive therapy, splenectomy, diabetes, burns, kwashiorkor/marasmus, nephrotic syndrome, spinal cord injury, seizure disorder, emphysema, or cancer. §For children ≤5 years of age where vaccine history was available and documented on vaccination card. All percentages are rounded off. ¶ Estimated using penalised logistic regression
Among ILI patients, 1% (47/4933) were PCR positive for B. pertussis, of which 68% (32/47) were confirmed cases. There was no difference in the demographic and clinical characteristics between confirmed and possible cases in this group (Additional file 2).
Laboratory and site investigation
In August 2014, there was a higher than usual PCR detection rate for B. pertussis (4.3%, 16/368) compared to previous months. The increase occurred in samples collected at one geographical site (KTHC and Jouberton Clinic). An evaluation of all laboratory systems and testing of 32 environmental samples collected from the affected hospital and clinic excluded facility and laboratory contamination, indicating a true increase in B. pertussis infection. In addition, 17 PCR-positive patients [9/17 (53%) SRI and 8/17 (47%) ILI], enrolled during the period when the increase in B. pertussis detection was observed, were retrospectively interviewed (Table 3). Eleven patients (65%) met the clinical case definition for pertussis disease, namely, cough for > 2 weeks (any duration in infants < 1 year) + one of the following: paroxysmal cough, inspiratory whoop, posttussive vomiting, apnea among infants < 1 year. Of these, 78% (7/9) and 50% (4/8) were confirmed and possible cases, respectively. No epidemiological links were identified between these individuals.
Table 3.
Characteristics of individuals with B. pertussis PCR-positive results that were interviewed following the unusual increase in B. pertussis PCR detection rate in August 2014 (N = 17)
| Case | Age group | HIV status | Confirmed/possible pertussis* | Cough ≥2 weeks | Paroxysmal cough | Inspiratory whoop | Posttussive vomiting | Apnea | Meets clinical case definition† |
|---|---|---|---|---|---|---|---|---|---|
| 1 | < 1y | Negative | Possible | No | No | No | No | No | No |
| 2 | 1-4y | Positive | Confirmed | Yes | Yes | No | No | N/A | Yes |
| 3 | 25-44y | Positive | Confirmed | No | No | No | No | No | No |
| 4 | 25-44y | Negative | Confirmed | Yes | Yes | No | No | N/A | Yes |
| 5 | 25-44y | Positive | Possible | Yes | No | No | No | N/A | No |
| 6 | 45-64y | Negative | Confirmed | Yes | No | No | No | N/A | No |
| 7 | 45-64y | Negative | Possible | Yes | No | No | No | N/A | No |
| 8 | 45-64y | Positive | Possible | Yes | Yes | Yes | No | N/A | Yes |
| 9 | 1y | Negative | Confirmed | Yes | Yes | No | No | Yes | Yes |
| 10 | 25-44y | Positive | Confirmed | Yes | Yes | Yes | No | N/A | Yes |
| 11 | < 1y | Unknown | Confirmed | Yes | Yes | Yes | Yes | Yes | Yes |
| 12 | 1-4y | Unknown | Possible | Yes | No | No | No | N/A | No |
| 13 | 1-4yy | Positive | Possible | Yes | No | No | Yes | N/A | Yes |
| 14 | 5-14y | Unknown | Possible | Yes | Yes | No | No | N/A | Yes |
| 15 | 25-44y | Negative | Possible | Yes | No | No | Yes | N/A | Yes |
| 16 | 25-44y | Negative | Confirmed | Yes | Yes | Yes | No | N/A | Yes |
| 17 | 5-14y | Unknown | Confirmed | Yes | Yes | Yes | Yes | N/A | Yes |
y = age in years. *Confirmed case = positive for B. pertussis with IS481 Ct < 35; Possible case = positive for B. pertussis with IS481 35 ≥ Ct ≤ 39. †Clinical case definition = cough > 2 weeks (any duration in infants <1 year) + one of the following: paroxysmal cough, inspiratory whoop, posttussive vomiting, apnea in infants < 1 year
Discussion
In the absence of pertussis-specific surveillance in South Africa, we evaluated the utility of real-time PCR detection of B. pertussis using an existing SRI and ILI surveillance platform. In our setting, laboratory methods were standardised and B. pertussis laboratory contamination could be excluded. As such, all individuals in whom IS481 was detected and B. holmesii, B. parapertussis and B. bronchiseptica were excluded, were considered to have B. pertussis DNA in their respiratory tract at the time of sampling and were therefore considered positive for B. pertussis infection. B. bronchiseptica has been associated with HIV-infected individuals [26–29]; however, this pathogen was not detected in our study. In confirmed cases, the attributable fraction of B. pertussis detection to respiratory disease was significant for both hospitalised (SRI) and mild (ILI) cases. However, for PCR-possible cases (both SRI and ILI) there was no significant association with B. pertussis disease.
In individuals with high Ct values (> 35), low bacterial loads may represent asymptomatic colonisation, or residual DNA post disease, or pertussis disease [19, 30]. Therefore, the use of stringent Ct value cut-offs may still result in the misdiagnosis of some true pertussis cases given that adults and adolescents have been shown to harbor lower bacterial loads and may present with milder disease or atypical symptoms [19, 30, 31]. In our study, just over one-third of B. pertussis PCR-positive cases had low bacterial loads (Ct > 35) and were defined as possible pertussis cases. This is potentially due to the case definition that was not specific for pertussis and the enrollment of individuals of all ages. We also showed that in hospitalised patients, pertussis-possible individuals were more likely to be adults (45–64 years of age) than infants (< 1 year of age) compared with pertussis-confirmed individuals.
In our study, a subset of 17 patients that were PCR positive for B. pertussis (confirmed and possible cases) from 2014 were retrospectively interviewed. The majority of confirmed cases reported clinical symptoms consistent with pertussis; however, 50% (4/8) of possible cases also had clinical pertussis. Since this was only a small group of individuals, we are prospectively conducting active case follow-up to understand the association between PCR Ct values and pertussis symptoms. In Canada, Public Health Ontario Laboratories determined the association between PCR Ct with pertussis severity and symptoms, and showed that the proportion of patients with pertussis symptoms did not differ between individuals with low (Ct < 36) and high IS481 Ct values [19].
IS481 is a multi-copy target with up to 250 copies per B. pertussis genome which increases assay sensitivity, but also increases the risk of false positives due to environmental or laboratory contamination [17, 18, 32]. As such, different Ct value cut-offs have been recommended when interpreting PCR data and these vary between studies. In a Tunisian study, B. pertussis cases were defined as PCR positive for IS481 and ptxS1 with a Ct < 45, or as Bordetella spp. if they were positive for IS481 only with a Ct < 45 [7]. The assay currently used by our laboratory detects one genomic equivalent of B. pertussis at an average Ct of 33.3 [16]. This, together with variable PCR testing and interpretation algorithms in some settings, supports the recommendation by the authors of using a more stringent IS481 Ct cut-off (< 35) [16]. However, the 95% confidence intervals (28.5–38.1) imply that higher Ct values could still indicate positivity and, in a setting where testing procedures are standardised, should not be ignored. Also, Ct values in clinical specimens may vary due to specimen composition or may be affected during storage or transit, and thus may not directly emulate experimental sensitivity in a controlled laboratory environment.
Sequencing confirmed the presence of the IS481 gene from a subset of PCR-positive samples, ruling out non-specific amplification, primer dimer formation and probe degradation, however, one B. pertussis confirmed, and eight possible samples could not be confirmed by sequencing likely due to insufficient DNA concentration for Sanger sequencing. Although only 7% of possible B. pertussis cases tested positive for the ptxS1 gene due to the lower sensitivity of the assay compared to IS481, we excluded B. holmesii from all IS481 PCR-positive samples, suggesting that these were positive for B. pertussis.
Earlier studies have indicated that NP is the preferred specimen type for laboratory detection of B. pertussis [10, 33, 34]. The use of sputum for B. pertussis detection has been suggested by WHO [34], however, supporting data are lacking. In our study, testing of induced sputum in addition to NP samples increased the diagnostic yield of B. pertussis 5-fold among hospitalised SRI patients. This is similar to another South African study where the B. pertussis detection rate was 3.7% when testing NP specimens and increased to 7% when induced sputum specimens were additionally included [35].
The World Health Organization has recommended that all countries perform pertussis surveillance and have suggested that for resource-limited settings it can be linked to surveillance for other respiratory illnesses. Due to the lack of pertussis data in South Africa, our aim was to establish pertussis surveillance by linking it to an already established syndromic SRI and ILI surveillance platform. While not pertussis-specific surveillance, this enabled us to obtain pertussis data for the country at a low cost by leveraging systems and staff already in place. An additional benefit of this surveillance is that children and adults with mild and severe respiratory illness that do not necessarily present with typical pertussis symptoms were included, adding to our understanding of pertussis disease in individuals that are not clinically suspected to have pertussis. However, use of syndromic surveillance platforms for pertussis surveillance also has several drawbacks, which need to be acknowledged. As the case definition for ILI cases and SRI adult cases included fever, it is likely that some pertussis cases were missed, and the data likely represents an underestimation of disease prevalence. In addition, the interpretation and clinical relevance of PCR-positive individuals that do not present with typical pertussis symptoms is not well understood. However, despite these limitations, we detected B. pertussis by PCR in 1.1% of patients with either mild or severe respiratory illness, which correlates with another South African study that enrolled an infant cohort from 2012 to 2014 as part of the Drakenstein Child Health Study in the Western Cape and found an overall pertussis prevalence of 2% among infants presenting with pneumonia [36].
The study has limitations that need to be considered. The surveillance case definition was not specific for pertussis therefore the time to swabbing following disease onset could not be determined. Overall, there was a small number of pertussis cases, which limited our statistical power for analyses, and as a result, differences between the characteristics of confirmed and possible cases which may exist, may not have been detected. Pertussis vaccination history was not available for adults, and for children < 5 years of age the collected data were poor, so the impact of vaccination on pertussis disease and Ct value could not be determined.
In a controlled setting with standardised procedures and when contamination can conclusively be excluded, we suggest that IS481-positive specimens with Ct values < 40 should be considered indicative of B. pertussis infection. Low IS481 Ct values (< 35) are more likely to represent true cases of pertussis disease, whereas specimens with high Ct values may represent B. pertussis carriage (as B. pertussis was detected in controls with higher Ct values), residual B. pertussis DNA following disease, or active disease especially in older individuals who may present atypically. As such, in a respiratory surveillance setting and for public health reporting, low bacterial loads may not reflect true cases of disease. However, for individual patient management, possible cases should be interpreted in the context of other clinical and epidemiological factors including patient symptoms, age, immune status and epidemiological links with laboratory-confirmed pertussis cases. B. pertussis carriage is not well understood, and further research is required to better understand the significance of PCR detection of B. pertussis in asymptomatic individuals and individuals lacking typical pertussis symptoms.
Conclusion
In conclusion, based on the results presented in this manuscript, we believe that in the absence of a pertussis-specific surveillance, leveraging of an already existing respiratory illness surveillance system as recommended by the WHO will allow countries to monitor the trend of infection over time, detect outbreaks and allow further research into cases that present with atypical pertussis symptoms.
Additional files
Comparison of confirmed (N = 38) and possible (N = 22) pertussis cases (real-time PCR positive for IS481 in nasopharyngeal specimens) in hospitalized patients with severe respiratory illness, South Africa, June 2012 – May 2016 (N = 60). (DOCX 18 kb)
Comparison of confirmed (N = 32) and probable (N = 15) pertussis cases (real-time PCR positive for IS481 in nasopharyngeal specimens) in patients with influenza-like illness, South Africa, June 2012 – May 2016 (N = 47). (DOCX 17 kb)
Acknowledgements
We thank the patients who consented to be enrolled in our surveillance, and all field and laboratory staff involved in pneumonia surveillance, and the data management team for data quality.
Funding
This work was funded by the National Institute for Communicable Diseases of the National Health Laboratory Service [grant number:94343], and the US Centers for Disease Control and Prevention [grant number: 5U51IP000155]. The funding bodies played no role in the design of the study; collection, analysis and interpretation of data or writing of the manuscript.
Availability of data and materials
The datasets generated and/or analysed during the current study are not publicly available due to reasons of patient confidentiality but are available from the corresponding author on reasonable request.
Disclaimer
The findings and conclusions in this report are those of the authors and do not necessarily represent the official position of the US Centers for Disease Control and Prevention, USA or the National Institute for Communicable Diseases, South Africa.
Abbreviations
- AF
Attributable Fraction
- BLAST
Basic Local Alignment Search Tool
- CI
Confidence Interval
- Ct
Cycle threshold
- ILI
Influenza-Like Illness
- KTHC
Klerksdorp-Tshepong Hospital Complex
- MALDI-TOF
Matrix-Assisted Laser Desorption/Ionization Time of Flight mass spectrometry
- NICD
National Institute for Communicable Diseases
- NP
Nasopharyngeal
- OP
Oropharyngeal
- PCR
Polymerase Chain Reaction
- RNaseP
Human ribonuclease P
- SRI
Severe Respiratory Illness
- UTM
Universal Transport Media
- WHO
World Health Organisation
Author’s contributions
The study was conceived by FM, MdP, NW, SW, CC and AvG. The analysis of the data was carried out by FM. The statistical analysis of the data was carried out by CvM and ST. The manuscript was drafted by FM and critically revised by MdP, NW, MC, CC, SW, CvM, ST, HD, EV and AvG. All authors have read and approved the final version of the manuscript.
Ethics approval and consent to participate
Ethics for SRI (M081042) and ILI (M120133) surveillance and B. pertussis testing were approved by the University of the Witwatersrand (M130260) and the University of KwaZulu Natal (BF081/12), South Africa. Written informed consent was obtained from all individuals enrolled into the study by trained surveillance officers upon interview. Surveillance officers approached adult patients directly to obtain consent. Parents, legal guardians or primary care giver were approached for cases who were minors; in addition, where a minor was old enough to understand, assent was obtained.
Consent for publication
Not applicable.
Competing interests
Halima Dawood has received honoraria/travel grants from Pfizer-South Africa, MSD329 South Africa and Mylan-South Africa. Anne von Gottberg has previously received grants from the US Centers for Disease Control and Prevention and Pfizer-South Africa. All other authors declare that they have no commercial or other association that may pose a conflict of interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Fahima Moosa, Email: fahimam@nicd.ac.za.
Mignon du Plessis, Email: mignond@nicd.ac.za.
Nicole Wolter, Email: nicolew@nicd.ac.za.
Maimuna Carrim, Email: maimunac@nicd.ac.za.
Cheryl Cohen, Email: cherylc@nicd.ac.za.
Claire von Mollendorf, Email: claire.vonmollendorf@mcri.edu.au.
Sibongile Walaza, Email: sibongilew@nicd.ac.za.
Stefano Tempia, Email: stefanot@nicd.ac.za.
Halima Dawood, Email: halimadawood@gmail.com.
Ebrahim Variava, Email: evariava@nwpg.gov.za.
Anne von Gottberg, Email: annev@nicd.ac.za.
References
- 1.Pertussis vaccines WHO position paper. Wkly Epidemiol Rec. 2010;85(40):385–400. [PubMed] [Google Scholar]
- 2.Pertussis epidemic--Washington, 2012. MMWR Morb Mortal Wkly Rep 2012 Jul;61(28):517–522. [PubMed]
- 3.Dalby T, Harboe ZB, Krogfelt KA. Seroprevalence of pertussis among Danish patients with cough of unknown etiology. Clin Vaccine Immunol. 2010;17(12):2016–2023. doi: 10.1128/CVI.00270-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fisman DN, Tang P, Hauck T, Richardson S, Drews SJ, Low DE, et al. Pertussis resurgence in Toronto, Canada: a population-based study including test-incidence feedback modeling. BMC Public Health. 2011;11:694. doi: 10.1186/1471-2458-11-694.:694-11.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Klein NP, Bartlett J, Fireman B, Aukes L, Buck PO, Krishnarajah G, et al. Waning protection following 5 doses of a 3-component diphtheria, tetanus, and acellular pertussis vaccine. Vaccine. 2017;35(26):3395–3400. doi: 10.1016/j.vaccine.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Witt MA, Katz PH, Witt DJ. Unexpectedly limited durability of immunity following acellular pertussis vaccination in preadolescents in a north American outbreak. Clin Infect Dis. 2012;54(12):1730–1735. doi: 10.1093/cid/cis287. [DOI] [PubMed] [Google Scholar]
- 7.Zouari A, Smaoui H, Brun D, Njamkepo E, Sghaier S, Zouari E, et al. Prevalence of Bordetella pertussis and Bordetella parapertussis infections in Tunisian hospitalized infants: results of a 4-year prospective study. Diagn Microbiol Infect Dis. 2012 Feb 6. [DOI] [PubMed]
- 8.Baxter R, Bartlett J, Rowhani-Rahbar A, Fireman B, Klein NP. Effectiveness of pertussis vaccines for adolescents and adults: case-control study. BMJ 2013 Jul 17;347:f4249.:f4249. [DOI] [PubMed]
- 9.Makokha C, Mott J, Njuguna HN, Khagayi S, Verani JR, Nyawanda B, et al. Comparison of severe acute respiratory illness (SARI) and clinical pneumonia case definitions for the detection of influenza virus infections among hospitalized patients, western Kenya, 2009-2013. Influenza Other Respir Viruses. 2016;10(4):333–339. doi: 10.1111/irv.12382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crowcroft NS, Pebody RG. Recent developments in pertussis. Lancet. 2006;367(9526):1926–1936. doi: 10.1016/S0140-6736(06)68848-X. [DOI] [PubMed] [Google Scholar]
- 11.Riffelmann M, Wirsing von Konig CH, Caro V, Guiso N. Nucleic acid amplification tests for diagnosis of Bordetella infections. J Clin Microbiol. 2005;43(10):4925–4929. doi: 10.1128/JCM.43.10.4925-4929.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fry NK, Tzivra O, Li YT, McNiff A, Doshi N, Maple PA, et al. Laboratory diagnosis of pertussis infections: the role of PCR and serology. J Med Microbiol. 2004;53(Pt 6):519–525. doi: 10.1099/jmm.0.45624-0. [DOI] [PubMed] [Google Scholar]
- 13.Preston A, Parkhill J, Maskell DJ. The bordetellae: lessons from genomics. Nat Rev Microbiol. 2004;2(5):379–390. doi: 10.1038/nrmicro886. [DOI] [PubMed] [Google Scholar]
- 14.Preston A. Bordetella pertussis: the intersection of genomics and pathobiology. CMAJ. 2005;173(1):55–62. doi: 10.1503/cmaj.050105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Farrell DJ, McKeon M, Daggard G, Loeffelholz MJ, Thompson CJ, Mukkur TK. Rapid-cycle PCR method to detect Bordetella pertussis that fulfills all consensus recommendations for use of PCR in diagnosis of pertussis. J Clin Microbiol. 2000;38(12):4499–4502. doi: 10.1128/jcm.38.12.4499-4502.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tatti KM, Sparks KN, Boney KO, Tondella ML. Novel multitarget real-time PCR assay for rapid detection of Bordetella species in clinical specimens. J Clin Microbiol. 2011;49(12):4059–4066. doi: 10.1128/JCM.00601-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lievano FA, Reynolds MA, Waring AL, Ackelsberg J, Bisgard KM, Sanden GN, et al. Issues associated with and recommendations for using PCR to detect outbreaks of pertussis. J Clin Microbiol. 2002;40(8):2801–2805. doi: 10.1128/JCM.40.8.2801-2805.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mandal S, Tatti KM, Woods-Stout D, Cassiday PK, Faulkner AE, Griffith MM, et al. Pertussis pseudo-outbreak linked to specimens contaminated by Bordetella pertussis DNA from clinic surfaces. Pediatrics. 2012;129(2):e424–e430. doi: 10.1542/peds.2011-1710. [DOI] [PubMed] [Google Scholar]
- 19.Bolotin S, Deeks SL, Marchand-Austin A, Rilkoff H, Dang V, Walton R, et al. Correlation of real-time PCR cycle threshold cut-off with Bordetella pertussis clinical severity. PLoS One. 2015;10(7):e0133209. doi: 10.1371/journal.pone.0133209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brotons P, de Paz HD, Toledo D, Villanova M, Plans P, Jordan I, et al. Differences in Bordetella pertussis DNA load according to clinical and epidemiological characteristics of patients with whooping cough. J Inf Secur. 2016;72(4):460–467. doi: 10.1016/j.jinf.2016.01.013. [DOI] [PubMed] [Google Scholar]
- 21.Mattoo S, Cherry JD. Molecular pathogenesis, epidemiology, and clinical manifestations of respiratory infections due to Bordetella pertussis and other Bordetella subspecies. Clin Microbiol Rev. 2005;18(2):326–382. doi: 10.1128/CMR.18.2.326-382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen C, Moyes J, Tempia S, Groom M, Walaza S, Pretorius M, et al. Severe influenza-associated respiratory infection in high HIV prevalence setting, South Africa, 2009-2011. Emerg Infect Dis. 2013;19(11):1766–1774. doi: 10.3201/eid1911.130546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pretorius MA, Tempia S, Treurnicht FK, Walaza S, Cohen AL, Moyes J, et al. Genetic diversity and molecular epidemiology of human rhinoviruses in South Africa. Influenza Other Respir Viruses. 2014;8(5):567–573. doi: 10.1111/irv.12264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Thurman KA, Warner AK, Cowart KC, Benitez AJ, Winchell JM. Detection of Mycoplasma pneumoniae, Chlamydia pneumoniae, and Legionella spp. in clinical specimens using a single-tube multiplex real-time PCR assay. Diagn Microbiol Infect Dis. 2011;70(1):1–9. doi: 10.1016/j.diagmicrobio.2010.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tempia S, Walaza S, Moyes J, Cohen AL, von MC, McMorrow ML, et al. Attributable fraction of influenza virus detection to mild and severe respiratory illnesses in HIV-infected and HIV-uninfected patients, South Africa, 2012-2016. Emerg Infect Dis. 2017;23(7):1124–1132. doi: 10.3201/eid2307.161959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de la Fuente J, Albo C, Rodriguez A, Sopena B, Martinez C. Bordetella bronchiseptica pneumonia in a patient with AIDS. Thorax. 1994;49(7):719–720. doi: 10.1136/thx.49.7.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dworkin MS, Sullivan PS, Buskin SE, Harrington RD, Olliffe J, MacArthur RD, et al. Bordetella bronchiseptica infection in human immunodeficiency virus-infected patients. Clin Infect Dis. 1999;28(5):1095–1099. doi: 10.1086/514761. [DOI] [PubMed] [Google Scholar]
- 28.Byrd LH, Anama L, Gutkin M, Chmel H. Bordetella bronchiseptica peritonitis associated with continuous ambulatory peritoneal dialysis. J Clin Microbiol. 1981;14(2):232–233. doi: 10.1128/jcm.14.2.232-233.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Woolfrey BF, Moody JA. Human infections associated with Bordetella bronchiseptica. Clin Microbiol Rev. 1991;4(3):243–255. doi: 10.1128/CMR.4.3.243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nakamura Y, Kamachi K, Toyoizumi-Ajisaka H, Otsuka N, Saito R, Tsuruoka J, et al. Marked difference between adults and children in Bordetella pertussis DNA load in nasopharyngeal swabs. Clin Microbiol Infect. 2011;17(3):365–370. doi: 10.1111/j.1469-0691.2010.03255.x. [DOI] [PubMed] [Google Scholar]
- 31.Paisley RD, Blaylock J, Hartzell JD. Whooping cough in adults: an update on a reemerging infection. Am J Med. 2012;125(2):141–143. doi: 10.1016/j.amjmed.2011.05.008. [DOI] [PubMed] [Google Scholar]
- 32.Muyldermans G, Soetens O, Antoine M, Bruisten S, Vincart B, Doucet-Populaire F, et al. External quality assessment for molecular detection of Bordetella pertussis in European laboratories. J Clin Microbiol. 2005;43(1):30–35. doi: 10.1128/JCM.43.1.30-35.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gregory DS. Pertussis: a disease affecting all ages. Am Fam Physician. 2006;74(3):420–426. [PubMed] [Google Scholar]
- 34.Guillot S, Guiso N. Laboratory manual for the diagnosis of whooping cough caused by Bordetella pertussis/Bordetella parapertussis. 2014. pp. 1–41. [Google Scholar]
- 35.Muloiwa R, Dube FS, Nicol MP, Zar HJ, Hussey GD. Incidence and diagnosis of pertussis in south African children hospitalized with lower respiratory tract infection. Pediatr Infect Dis J. 2016;35(6):611–616. doi: 10.1097/INF.0000000000001132. [DOI] [PubMed] [Google Scholar]
- 36.Zar HJ, Barnett W, Stadler A, Gardner-Lubbe S, Myer L, Nicol MP. Aetiology of childhood pneumonia in a well vaccinated South African birth cohort: a nested case-control study of the Drakenstein Child Health Study. Lancet Respir Med. 2016 Apr;21(16):10–2600. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Comparison of confirmed (N = 38) and possible (N = 22) pertussis cases (real-time PCR positive for IS481 in nasopharyngeal specimens) in hospitalized patients with severe respiratory illness, South Africa, June 2012 – May 2016 (N = 60). (DOCX 18 kb)
Comparison of confirmed (N = 32) and probable (N = 15) pertussis cases (real-time PCR positive for IS481 in nasopharyngeal specimens) in patients with influenza-like illness, South Africa, June 2012 – May 2016 (N = 47). (DOCX 17 kb)
Data Availability Statement
The datasets generated and/or analysed during the current study are not publicly available due to reasons of patient confidentiality but are available from the corresponding author on reasonable request.