Abstract
Dairy products, especially cheeses have a great nutritional value and a high consumption level around the world. Considering a widespread consumption of cheeses, there is a growing concern regarding safety and microbiological quality. The current study was designed to conduct a recent evaluation of cheeses microbiological quality. Sixty cheese samples from retailing Egyptian markets were analyzed on different selective microbiological media and 64 bacteria, 35 yeasts and 8 molds were isolated. Out of 60 samples; 26.6% were contaminated with Escherichia coli, 73.3% with Staphylococcus scuiri, 3.33% with Bacillus cereus, 1.66% with Salmonella enterica, and 1.66% with Pseudomonas aeruginosa. The presence of such microorganisms in cheeses referred to the wrong management in cheese manufacturing. These organisms are significant from public health view as they have been associated with the base of human food poisoning. Promising antagonistic behavior was observed using the tested lactic acid bacteria (LAB) either single or in combinations toward the undesired isolates. Lactobacillus helveticus CNRZ 32 (Lb. helveticus) was the most potent culture; recording ≥95% reduction in undesired microbial counts.
Keywords: Food science, Microbiology, Food technology, Biotechnology, Food safety
1. Introduction
Milk and dairy products represent excellent growth media for many opportunistic food spoilage and disease-causing microorganisms. In industrial countries, Staphylococcus aureus (Staph. aureus), Salmonella spp. (Sal. spp.), Listeria monocytogenes and Escherichia coli (E. coli) are the most commonly detected pathogens associated with milk and dairy products. Also, these organisms resemble the main microbiological risks linked to raw milk, post-processing contaminated cheese or improperly treated milk (Cancino-Padilla et al., 2017).
Cheese is one of the most common foods over the world, and according to the International Dairy Foods Association report (IDFA, 2010), cheese is the major manufactured dairy product, with the developing importance of the dairy industry.
Outbreaks due to consumption of cheese contaminated with pathogenic bacteria and/or their toxins have the most importance of public health and economic consequences. Losses due to outbreaks include medications, charges, increased production wastes, loss of business, recall and damage of products, and investigation of the outbreaks (Röhr et al., 2005).
Salmonellosis; illness caused by Salmonella spp. is one of the most important disease problems for human and animal health, that causes worldwide human and animal sickness and sometimes death (Pal et al., 2015).
In addition, several strains of Bacillus cereus (B. cereus) are well known as toxin producers causing food poisoning (Tewari and Abdullah, 2015).
A variety of psychrotrophic bacterial species that primarily represented by Pseudomonas spp. (Ps. spp.) in the cold chain of raw milk can produce heat-stable proteases. These enzymes may spoil pasteurized milk (72–75 °C/15–20 s) even Ultra High Temperature-treated milk (130–150 °C/2–4 s) and dairy products (Marchand et al., 2009). Besides exhibiting spoilage features, some psychrotrophs possess an opportunistic pathogenicity. Also, these species are inherently toxin producers and/or resistant to antibiotics. Regarding quality, this bacterial group has become a key problem for today's dairy industry as causing of spoilage and significant economic sufferers (Samaržija et al., 2012).
Molds and yeasts are important contaminants of dairy product as a favorable niche for their growth. They are causes of visible or non-visible defects, such as off-odor and off-flavor, leading to cheeses waste and loss (Garnier et al., 2017).
Recently, consumers have an increasing awareness of their health risks caused by the utilization of chemical preservatives. So, this is a growing need in the dairy industry to extend product shelf-life and prevent spoilage by natural preservatives and/or new methods of conservation (Silva et al., 2018).
The group of lactic acid bacteria has a long history of safe application in dairy and other industries. Since they possess no health risks, bacteriocins and other bioactive compounds, that excreted by LAB strains, are a great substitute for chemical preservatives in dairy products. Among the most protective and health promoting LAB cultures; are Lb. helveticus, Lb. plantarum, Lb. reuteri, Lb. rhamnosus, Lactococcus lactis, Streptococcus thermophilus (Ranadheera et al., 2017) and Enterococcus faecium (Cavallini et al., 2011).
The main objective of this study was to recently evaluate and estimate the incidence of different foodborne pathogens and/or food spoilage microorganisms in some Egyptian dairy markets. Secondly, identification of a model set of the total isolates by sequencing of PCR-amplified rDNA gene fragments. Screening of different LAB for availability as protective cultures to inhibit the growth of foodborne isolates; bacteria, yeasts, and fungi was the last objective.
2. Materials and methods
2.1. Materials
Nutrient agar was provided from Panreac Quimica, Spain; Pseudomonas agar and MacConkey broth were provided from LAB m, UK; MRS agar was purchased from SRL, India; M17 agar was supplemented by CONDA, Spain; Pseudomonas C-N supplement, Baird-Parker agar, Bacillus cereus agar, Reinforced Clostridial agar and their supplements were purchased from Oxoid, England; EMB agar was obtained from HIMEDIA, India; and Malt extract agar was imported from Biolife, Italy.
2.2. Sample collection
Sixty cheese samples representing four different types; Talaga, Ras, Domiati, and Feta cheeses were collected from retailing markets in three different Egyptian Governorates; Cairo, Giza, and Menofia.
2.3. Lactic acid bacteria strains
Seven LAB cultures were obtained from the collection of dairy Microbiological Lab., National Research Centre, Dokki, Giza, Egypt. Table 1 shows sources and references of these strains.
Table 1.
Protective cultures of lactic acid bacteria included in the study.
| MRS agar-cultivated cultures | Origin (Sources or reference) |
|---|---|
| Lactobacillus plantarum DSA 20174 | Collection of dairy Microbiological Lab., (supplemented with Cairo MIRCEN, Faculty of Agriculture, Ain Shams University. Egypt) |
| Lactobacillus helveticus CNRZ 32 | Collection of dairy Microbiological Lab., (supplemented with Centre National de Recherche Zootechnique, Jouy-en-Josas, France) |
| Lactobacillus reuteri NRRL B-14171 | Collection of dairy Microbiological Lab., (obtained from the Northern Regional Research Laboratory, Illinois, USA) |
| Lactobacillus rhamnosus NRRL B-442 | Collection of dairy Microbiological Lab., (obtained from the Northern Regional Research Laboratory, Illinois, USA) |
| M17 agar-cultivated cultures | |
| Streptococcus thermophilus CH-1 | Collection of dairy Microbiological Lab., (obtained from Chr. Hansen Lab., Denmark) |
| Enterococcus faecium FSD | Provided by Dr. Mohammed G. Shehata, City of Scientific Research and Technological Application, Egypt. He isolated it from a homemade Egyptian karish cheese (Deraz et al., 2015) |
| Lactococcus lactis subsp. lactis | Provided by Dr. Mohammed G. Shehata, City of Scientific Research and Technological Application, Egypt. He isolated from an Egyptian Rayeb Milk (Shehata et al., 2016) |
2.4. Microbiological examination
2.4.1. Recent evaluation of the microbiological quality
Isolation of pathogenic bacteria, molds and yeasts from the four cheese types were preceded under full aseptic conditions.
Ten grams of each sample were homogenized in 90 ml of 2% (w/v) sodium citrate solution. And serial 10 fold dilutions were prepared from this homogenate using 0.89 % (w/v) physiological saline solution.
As described by El-Hadedy and Abu El-Nour (2012), Bromo-Cresol Purple MaCconkey broth tubes (LABm) were used for the total Coliforms counting. Fecal Coliforms were detected on EMB Agar.
According to Ollis et al. (1995), Baird-Parker agar medium was used for Staphylococci counting. The presence of Staphylococcus aureus was confirmed by the coagulase test (Baird-Parker, 1962).
For detection of Salmonella and Shigella, 25 g of sample were added to 225 ml of sterile buffered peptone water for pre enrichment. After incubation at 37 °C/24 h, 10 ml of growth suspension, were transferred to 90 ml boiling-sterilized Selenite broth. After incubation, XLD plates were streaked from Selenite broth (Taylor and Harris, 1965).
Molds and yeasts were counted on malt extract agar. Plates were incubated for 72 h at 30 °C, and then analyzed for fungal population (log CFU/gram).
2.5. Identification of the foodborne isolates
2.5.1. Morphological and biochemical characterization
For detection of Shigella and Salmonella; pink colonies with negative or positive H2S were respectively detected on XLD agar after 24 h/37 °C. Salmonella; the positive H2S (black centre) colonies were then streaked on urease slants to confirm the negative reaction.
E. coli was distinguished by purple colonies with a green metallic sheen on EMB agar. Pseudomonas aeruginosa developed bluish green pigmented colonies on Pseudomonas agar.
For the last 4 organisms, colonies showed positive in the catalase test (Staphylex, Oxoid) and negative gram reaction, had been confirmed.
Bacillus cereus developed peacock blue colonies with a precipitate on a Bacillus cereus agar plate. Characteristic colonies showing a positive gram reaction, positive catalase test, and spore forming rod-shaped cells were confirmed as Bacillus cereus.
Baird–Parker agar was used to isolate Staphylococci where representative gram-positive clustered cocci, typical black appearance colonies and surrounded by clear zone were picked up, and tested for catalase. Colonies showed egg yolk lysis and positive catalase were confirmed as Staph. aureus.
2.5.2. Molecular identification
2.5.2.1. DNA extraction
A single microbial colony from each selective agar plate was aseptically sent to the GIS Research Centre (6th October City, Egypt) for DNA extraction, rRNA PCR amplification and sequencing. DNA was extracted, amplified and purified using Quick-DNA Miniprep Plus Kits, ZYMO RESEARCH CORP., Irvine, California, United States, according to manufacturer protocol for solid tissues.
2.5.2.2. PCR conditions and primers
Reactions were performed using Delbes et al. (2007) method with slight modifications as follows: in a final volume of 50 μl. The reaction mixture contained 8 μl template, 25 μl MyTaq Red Mix (2x), and 1 μl of each primer (conc. 20 μmol/l). The amplification program was denaturation; 1–5 min, 95 °C (1 cycle), then 30–36 cycles of denaturation (40–120 Sec, 95 °C), annealing (90–120 Sec, 55 °C), while annealing temperature was 60 °C in case of bacteria; and extension (5–7 min, 72 °C). Ten μl of PCR-amplified product was analyzed by electrophoresis on 0.9 % ethidium bromide-stained agarose gel where the DNA ladder (0.3 mg/L) or bacterial suspension (2 μl) was used as a template. Table 2 contains specific primers with expected amplicon sizes for bacteria, fungi and yeast:
Table 2.
Primers used for PCR, with the expected amplicon size.
| Isolate type | Primers | Amplicon size |
|---|---|---|
| Bacteria (16S rDNA) | F 5′-CAGGCCTAACACATGCAAGTC--3′ | 1500 bp |
| R 5′-CGGCGGWGTGTACAAGGC -3′ | ||
| Mold (18S rDNA) | F 5′ CTT GGT CAT TTA GAG GAA GTA A 3′ | 750 bp |
| R 5′ TCC TCC GCT TAT TGA TAT GC 3′ | ||
| Yeast (26S rDNA) | F 5′-GCATATCAATAAGCGGAGGA AAAG-3′ | 700 bp |
| R 5′- GGTCCGTGTTTCAAGACGG-3′ |
Then the resulted sequences were aligned with the National Center for Biotechnology Information (NCIB) database using Basic Local Alignment Search Tool (BLAST) (https://blast.ncbi.nlm.nih.gov/Blast.cgi), to choose the highest matching (Brindha and Mathew, 2012).
2.6. Antimicrobial assays
2.6.1. Qualitative screening of LAB against spoilage and pathogenic isolates
The assays were performed using the agar overlay technique as described by Shokryazdan et al. (2014) with slight modifications. Overnight cultures of lactic acid bacterial strains that listed in Table 1a were separately mixed with MRS or M17 agar (≈105 CFU/100 μl). One hundred μl from each strain were pipetted to fill wells that previously made in agar plates, using a micropipette, and the inoculated plates were incubated at 37 °C for 48 h.
The agar plates containing the growth of lactic acid bacteria in spot forms (5 mm diameter) were then overlaid with soft nutrient agar (0.8% agar) pre-mixed with 108 CFU of the indicator stains, and incubated, after the solidification of the overlaid agar layer, at 37 °C for 24 h. The zone diameter of inhibition values obtained were measured and interpreted. The antifungal activity of LAB was investigated with an overlay assay using the modified method of (Magnusson and Schnürer, 2001; Lind et al., 2005). LAB cultures on agar were poured in 2 cm × 1 cm grooves on MRS agar plates. Ten milliliter of soft (0.8%) malt extract agar containing 1 ml of inoculum of mold spore or yeast suspension was then poured onto the agar plates and incubated at 30 °C. After 48 h, the zone of inhibition was measured. The degree of inhibition was calculated as the area of inhibited growth.
2.6.2. Co-culturing of foodborne isolates with LAB (quantitative interaction)
As described by Ammor et al. (2006), the most potent antimicrobial LAB was co-cultivated with the previously isolated foodborne microbial cultures separately in dual species planktonic cultures. Log CFU was recorded for the foodborne isolates after 24 h in comparison with the control.
2.7. Statistical analysis
Statistical significance was determined using Statistica Version 9 (State Soft, Tulsa, Okla., USA). The means were determined by analysis of variance test (ANOVA, two way analysis) (p < 0.05). Fisher's LSD (Least Significant Difference) Method (α = 0.05) was applied to compare significant differences between treatments (Williams and Abdi, 2010).
3. Results and discussion
3.1. Microbiological quality evaluation of cheese
The results of the recent microbiological evaluation conducted on 60 cheese samples that categorized under main four cheese types were collected from different markets from three different Egyptian governorates; Cairo, Giza and Menofia. The results are presented in Tables 3, 4, 5, 6, and 7.
Table 3.
Microbiological analyses of different cheese types.
| Tested microorganisms | Microbial counts (Log CFU/g) |
|||
|---|---|---|---|---|
| Talaga (n = 15) | Ras (n = 15) | Domiati (n = 15) | Feta (n = 15) | |
| Coliform group | 2.5 ± 0.80 | 2.1 ± 1.41 | 0.3 ± 0.47 | Nil |
| Staphylococci | 5.8 ± 1.77 | 6.5 ± 1.07 | 3.6 ± 1.86 | 1.6 ± 1.06 |
| Aerobic spores | 3.4 ± 0.72 | 3.7 ± 1.06 | 2.4 ± 0.83 | Nil |
| Anaerobic spores | Not tested | Not tested | Not tested | Nil |
| Psychrotrophic bacteria | 1.8 ± 0.49 | Nil | Nil | 3.6 ± 2.92 |
| Molds | 1.1 ± 0.73 | 2.4 ± 1.69 | 2 ± 1.43 | Nil |
| Yeasts | 2.05 ± 1.44 | 4.5 ± 1.96 | 4.2 ± 2.48 | Nil |
| Total viable count | 6.8 ± 2.06 | 7.3 ± 0.85 | 6.1 ± 1.59 | 7 ± 4.28 |
Data expressed as mean ± standard error, Nil: No growth detected.
Table 4.
Incidence of coliforms in the examined cheese samples.
| Cheese type | Positive samples | % |
|---|---|---|
| Talaga | 13/15 | 87 |
| Domiati | 12/15 | 80 |
| Ras | 2/15 | 13.3 |
| Feta | 0/15 | 0 |
| All | 27/60 | 45 |
Table 5.
Distribution of the bacterial contaminants in different cheeses types.
| Pathogen | Positive samples by Cheese type |
Total positive samples | Positive samples % | |||
|---|---|---|---|---|---|---|
| Talaga | Domiati | Ras | Feta | |||
| E. coli | 4/15 | 2/15 | 10/15 | 0/15 | 16/60 | 26.6 |
| Sal. enterica | 0/15 | 1/15 | 0/15 | 0/15 | 1/60 | 1.66 |
| Ps. aeruginosa | 0/15 | 1/15 | 0/15 | 0/15 | 1/60 | 1.66 |
| Staph. scuiri | 13/15 | 15/15 | 13/15 | 3/15 | 44/60 | 73.3 |
| Bacillus cereus | 0/15 | 2/15 | 0/15 | 0/15 | 2/60 | 3.33 |
Table 6.
Morphological and biochemical grouping of bacterial isolates.
| Isolation media | Isolates number | Microscope | Gram staining | Catalase | Sporulation | others |
|---|---|---|---|---|---|---|
| EMB agar | 16 | Short rods | Negative | Positive | Negative | H2S (−) |
| XLD agar | 1 | Short rods | Negative | Positive | Negative | H2S (+) |
| Urease (−) | ||||||
| Pseudomonas agar | 1 | Rods | Negative | Positive | Negative | Blue green pigment (+) |
| Bacillus cereus agar | 2 | Rods | Positive | Positive | Positive | Peacock blue colonies |
| Baird Parker agar | 44 | Cocci in clusters | Positive | Positive | Negative | Coagulase (−) |
| Total isolates | 64 | |||||
−: negative, +: positive.
Table 7.
Homology percentages of isolates nucleotide sequences.
| Code | Identification | Isolation media | Accession number | Homology % |
|---|---|---|---|---|
| 11 | Escherichia coli | EMB agar | KY780346.1 | 98 % |
| 12 | Salmonella enterica | XLD agar | CP030207.1 | 96 % |
| 13 | Pseudomonas aeruginosa | Pseudomonas agar | KY549641.1 | 98 % |
| 14 | Bacillus cereus | Bacillus cereus agar | KY820914.1 | 99 % |
| 15 | Staphylococcus sciuri | Baird-Parker agar | MH491952.1 | 100 % |
| 16 | Penicillium chrysogenum | Malt extract agar | KF572447.1 | 99 % |
| 50 | Candida parapsilosis | Malt extract agar | KP852497.1 | 99 % |
It is well-known that the effectiveness of the pasteurization process and the resulting quality of cheese and other dairy products directly reflect the microbiological quality of the raw milk (Nörnberg et al., 2010). Table 3 illustrates the microbial counts (log CFU/g) of the dominant foodborne pathogens in the tested cheeses. Feta cheese contained the highest total viable count (7 ± 4.28 log CFU/g), and the predominant factor was Psychrotrophic bacteria whose count reached (3.6 log CFU/g), while the coliform group were not being detected. This may reflect the improper heat treatment of high microbial-loaded raw milk in some factories, the presence of biofilms in production lines and/or weak hygienic system as reported by Marchand et al. (2012). Lower than feta cheese, Talaga cheese whose total viable count reached 6.8 ± 2.06 log CFU/g, and Staphylococcal count achieved 5.8 ± 1.77 log CFU/g, Total coliforms count 2.5 ± 0.8 log CFU/g, molds and yeasts 1.1 ± 0.73 and 2.05 ± 1.44 log CFU/g respectively. Domiati and Ras types were distinguished by their high Staphylococcal contents (3.6 ± 1.86 and 6.5 ± 1.07 log CFU/g, respectively), while Ras cheese had the highest coliforms load that reached 4.3 log CFU/g in some samples. Domiati and Ras samples included the largest fungal loads (2 ± 1.43 and 2.4 ± 1.69 log CFU/g for fungi and 4.2 ± 2.48 and 4.5 ± 1.96 log CFU/g for yeasts, respectively). According to Kean et al. (2017), there was a reported relationship between the yeast count and Staphylococci existence in food samples. It was thought that some yeast can structurally and nutritionally support the existence (adherence and colonization) of Staphylococcus sp. if the both exist in the same environment. According to that reported by Johler et al. (2015), Staph. aureus was determined as a causative agent of a food poisoning outbreak On October 1, 2014. Within the first 7 hours after the meal, all persons consumed a soft cheese produced from raw cow milk fell ill. Investigations revealed that the soft cheese exhibited low levels of staphylococcal enterotoxin A and high levels of enterotoxin D. It was considered that temperature abuse above 10 °C and the reduced contribution of inactive starter culture during fermentation are key factors in staphylococcal-based dairy outbreaks (Cretenet et al., 2011).
Table 4 records the percentage of coliforms contamination of different cheeses. The total coliforms count was increasingly noted in the direction from Feta to Talaga cheeses. Coliforms presence indicates the insufficient processing and post-processing stages, and failure of the Good Manufacturing Practices (GMP) or hygienic control either during manufacture or distribution of these products. Focusing on cheese samples contaminated with coliforms, Trmčić et al. (2016) results represented 50% of these study findings. In addition, this suggested explanation of bad quality was supported by that mentioned by Hervert et al. (2017) and his team. They reported that the Enterobacteriaceae and total gram-negative groups more accurately reflect the hygienic status of the processed milk, processing, and storage environments.
The importance of these bacteria is that they have become components of the microbiological evaluating programs of both industry and guiding organizations. Principally, they may be considered as signs of safety or quality of dairy products. Presence of these signs in food products proposes the presence of conditions associated or suitable for other undetectable pathogens. In addition, they have a history of constant ecological association with the pathogen in the environment where contamination initiates (Price and Wildeboer, 2017). So these quality signs assess important conditions for product manufacturer or customer acceptability (Tortorello, 2003).
Table 5 represents the numbers and percentages of different contaminants in the examined cheese types. About 26.6% of the total samples were positive for E. coli; the most indicator organism of fecal contamination. Some serotypes of this organism have a high virulence e.g. E. coli O157:H7; a Shiga toxin-producing Escherichia coli (STEC) which can cause disease at a dose of 5–50 cells (Farrokh et al., 2013).
The main source of STEC is ruminants that contaminate milk over subclinical mastitis or fecal ways, and the bacteria can stay in milking apparatus.
3.2. Identification of the isolated pathogenic and spoilage microorganisms
Table 6 summarizes the morphological and biochemical grouping of bacterial isolates. Depending on the most important biochemical and morphological tests, out of the 64 bacterial isolates 16 were identified as E. coli, one Salmonella sp., one Pseudomonas aeruginosa, two Bacillus cereus and 44 were coagulase negative Staphylococcus sp.
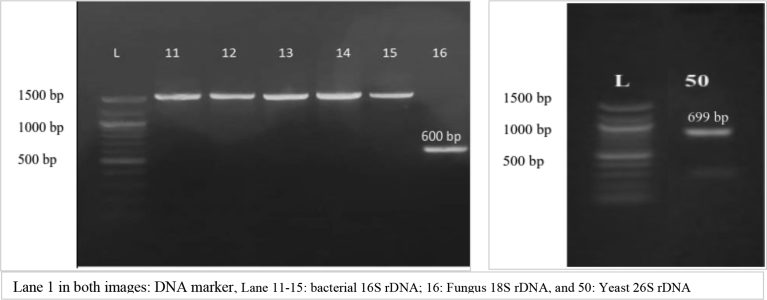
Fig. 1 shows gel electrophoresis of the purified PCR products of representative isolates. Bands from 11 to 15 represented the bacterial 16S rDNA at 1500 bp., while bands 16 and 50 represented the fungus 18S rDNA and the yeast 26S rDNA at 600 and 699 bp. respectively. These PCR products were next subjected to nucleotide sequencing and then compared with the available sequences in the NCBI GenBank database using BLAST; one of the most greatly used sequence analysis tools available in the public domain. Table 7 represents homology % of the highest matched strains with their accession numbers.
Fig. 1.
Gel images of rDNA PCR products for the representative cheese contaminants.
There is now a wide choice of BLAST systems that can be used to search much different sequence databases and finds regions of matching between biological sequences (McGinnis and Madden, 2004). The obtained sequence matching for the isolates were very reliable (≥96% similarity with the database).
According to Table 7, the alignment showed that the representative isolates had the highest identity with Escherichia coli strain E11 (accession number KY780346.1), Salmonella enterica strain SA19992307 (accession number CP030207.1), Pseudomonas aeruginosa strain Kasamber5 (accession number KY549641.1), Bacillus cereus strain 151007-R3-K09-40-27F (accession number KY820914.1), and Staphylococcus sciuri strain 2-6 (accession number MH491952.1). Ruaro et al. (2013) identified different coagulase-negative staphylococci from Italian raw milk and cheeses; Staphylococcus sciuri was among the recovered species and represented about 10% of the total community.
Mold and yeast representative isolates had the highest identity with Penicillium chrysogenum (P. chrysogenum) strain J127 (KF572447.1) and Candida parapsilosis (C. parapsilosis) strain F2-17 (KP852497.1) respectively.
3.3. Qualitative screening of LAB for antimicrobial activity against isolated pathogens
Table 8 presents antagonistic effect of different LAB cells against the representative microbial isolates. All the applied LAB strains showed inhibitory effects against all indictor food spoilage and pathogenic microorganisms, except Str. thermophilus and Ent. faecium in the case of Ps. aeruginosa. Also, Fig. 2 clearly showed model results of inhibition with different zone diameters confirming what presented in Table 8. As shown in Fig. 2, zones of inhibition are greater for bacterial isolates than fungus than yeast. The observed greatest inhibition in the case of Lb. plantarum and Lb. helveticus may be caused mainly by excessive acid production. Making this suggestion more acceptable is the weak response of yeast; Candida parapsilosis toward the inhibition. Many studies guaranteed these results explaining as those LAB strains can produce antimicrobial agents that – in different mechanisms-develop a strong inhibitory activity against many microorganisms, including pathogenic and spoilage ones. Metabolites such as organic acids (lactic and acetic acid), hydrogen peroxide, ethanol, diacetyl, acetaldehyde, acetoin, carbon dioxide, and bacteriocins (Šušković et al., 2010), are examples of antimicrobial agents produced by LAB. Organic acid produced by LAB resulted in low pH levels and activity of hydrogen peroxide (Ponce et al., 2008). Organic acids and other products exhibit antibacterial activity against various pathogenic Gram-positive and Gram-negative microorganisms (Maragkoudakis et al., 2009).
Table 8.
Antimicrobial efficacy of pure LAB cells against representative contaminants.
| LAB cultures | Pathogenic & spoilage strains |
||||||
|---|---|---|---|---|---|---|---|
| Staph. scuiri | B. cereus | Sal. enterica | E. coli | Ps. aeruginosa | P. chrysogenum | C. parapsilosis | |
| Lb. plantarum | +++ | ++ | +++ | +++ | + | +++ | ++ |
| Lb. helveticus | +++ | ++ | +++ | +++ | + | +++ | ++ |
| Lb. rhamnosus | ++ | ++ | + | ++ | + | + | + |
| Lb. reuteri | ++ | ++ | + | ++ | ++ | + | + |
| Str. thermophiles | ++ | + | + | + | ND | + | + |
| Ent. faecium | + | + | + | + | ND | + | + |
| Lactococcus lactis | ++ | ++ | + | ++ | ++ | + | + |
+: 5 mm, ++: 6–10 mm & +++: 10:15 mm and ND: non-detected inhibition.
Fig. 2.
Interaction between selected LAB and some cheese contaminants.
As data showed, most of LAB cultures possess antimicrobial activities in different degrees against all indicator isolates. The observed degree of inhibition seemed to be specific to the protective culture and the indicator stain. As mentioned above, it is clear that both Lb. helveticus and Lb. plantarum showed the strongest antagonism of 10–15 mm zone of inhibition against Staph. scuiri, Sal. enterica, E. coli and P. chrysogenum, while lower activity (6–10 mm) was shown against B. cereus, Ps. aeruginosa and C. parapsilosis. Mols et al. (2010) observed the same behavior in B. cereus stating that such behavior was due to the activation of some important enzymes that drive intracellular pH hemostasis. And Desriac et al. (2013) reported that the response of B. cereus to acid stress could be a general stress response, pH homeostasis, metabolic rearrangements, or secondary oxidative stress response. Also, Ps. aeruginosa was reported by Slabbert (2013) to show intermediate tolerance toward low pH.
In Tables 9 and 10, the shown data represented the effect of different LAB culture combinations in enhancing the antimicrobial activity against the indicator isolates.
Table 9.
Antimicrobial efficacy of LAB mixed cultures against representative contaminants.
| Combination | Pathogenic & spoilage strains |
|||||||
|---|---|---|---|---|---|---|---|---|
| Staph. scuiri | B. cereus | Sal. enterica | E. coli | Ps. aeruginos | P. chrysogenum | C. parapsilosis | ||
| Lb. p | Lactococcus lactis | + | ++ | ++ | +++ | ++ | ND | ND |
| Str. thermophilus | + | ++ | ++ | +++ | ND | ND | ND | |
| Ent. faecium | + | ++ | ++ | +++ | ND | ND | ND | |
| Lb. h | Lactococcus lactis | +++ | ++ | ++ | +++ | + | ND | ND |
| Str. thermophilus | +++ | ++ | +++ | +++ | ND | ND | ND | |
| Ent. faecium | +++ | ++ | +++ | +++ | ND | ND | ND | |
Lb.rh: Lb. rhamnosus, Lb.re: Lb. reuteri, +: 5 mm, ++: 6–10 mm & +++: 10:15 mm, and ND: non-detected inhibition.
Table 10.
Antimicrobial efficacy of LAB mixed cultures against representative contaminants.
| Combination | Pathogenic & spoilage strains |
|||||||
|---|---|---|---|---|---|---|---|---|
| Staph. scuiri | B. cereus | Sal. enterica | E. coli | Ps. aeruginosa | P. chrysogenum | C. parapsilosis | ||
| Lb. rh | Lactococcus lactis | ++ | + | + | ++ | ++ | ND | ND |
| Str. thermophilus | ++ | + | + | ++ | ND | ND | ND | |
| Ent. faecium | ++ | + | + | ++ | ND | ND | ND | |
| Lb. re | Lactococcus lactis | + | + | + | + | + | ND | ND |
| Str. thermophilus | + | + | + | + | ND | ND | ND | |
| Ent. faecium | + | + | + | + | ND | ND | ND | |
Lb.rh: Lb. rhamnosus, Lb.re: Lb. reuteri, +: 5 mm, ++: 6–10 mm & +++: 10:15 mm, and ND: non-detected inhibition.
It was noted that the antifungal activity disappeared in all cases, while the antibacterial activity was stimulated, especially in case of Lb. helveticus-based combinations. This pattern appeared as there is a synergistic relationship drives the antimicrobial activity by Lb. helveticus-based combinations. Considering the antibacterial activity, some researchers reported that the S-layer –upon combination with bacteriocins-can act synergistically to inhibit the growth of both gram positive and gram negative bacteria (Prado-Acosta et al., 2010). Also, they suggested that the surface layer of lactobacillus membrane enables bacteriocin to cross the pathogen cell wall, while bacteriocin provides requirements for S-layer murein hydrolase activity.
3.4. Co-culturing of foodborne isolates with protective culture
Table 11 shows the role of Lb. helveticus in the reduction of microbial count (Log CFU/ml). As presented, after co-culturing of Lb. helveticus with different indicator strains, bacterial count (log CFU/ml) was reduced by > 99.99% and fungal counts became undetected after 24 h, while the yeast count was reduced only by about 99.5% over the same incubation period. The observed behavior of C. parapsilosis confirmed its weak response to different LAB cultures which was observed in Tables 8, 9, and 10. This suggestion ensured that the major inhibitory effect of LAB is due to organic acids; Furthermore, Mawgoud et al. (2016) confirmed this by achieving the highest yield of lactic acid from Lactobacillus bulgaricus at 37 °C.
Table 11.
Microbial count reduction of cheese contaminants by Lb. helveticus.
| Cultures | Pathogenic & spoilage isolates count (log CFU/ml) after 24 h |
||||||
|---|---|---|---|---|---|---|---|
| Staph. scuiri | B. cereus | Salm. enterica | E. coli | Ps. aeruginosa | P. chrysogenum | C. parapsilosis | |
| Lb. helveticus | 2.8 ± 0.06A | 3.9 ± 0.06 AB | 4 ± 0.10 AB | 4.5 ± 0.06 AB | 4 ± 0.15 AB | 0A | 5.7 ± 0.12 AB |
| Control | 10.6 ± 0.15B | 10.4 ± 0.25B | 9.4 ± 0.15C | 10.7 ± 0.06E | 9.1 ± 015C | 5.1 ± 0.06D | 8.1 ± 0.15C |
Data expressed as mean ± standard error, A−E Different letters in a same column mean significant difference at P < 0.05.
The observed strong antagonism between Lb. helveticus and other undesired isolates is a clear evidence of its metabolic efficiency to synthesize different antimicrobial substances or a broad-spectrum antimicrobial compound (Oldak et al., 2017). Several studies used the live Lb. helveticus culture to attain a successful protection from enterohaemorrhagic E. coli (Jandu et al., 2009), and other in vivo studies recommended the utilization of Lb. helveticus culture to benefit its ability to prevent gastrointestinal infections, enhance protection against pathogens, modulate the host immune responses, and affect the composition of the intestinal microbiota (Taverniti and Guglielmetti, 2012).
4. Conclusion
The current study reported the microbiological status of different cheese types from local retailing markets of three Egyptian governorates. Sixty four isolates represented in five bacteria, one mold, and one yeast. The isolates were molecularly identified by rDNA gene fragments sequencing. Alignment with NCIB database sequences revealed that most of them had highest identities with potentially pathogenic and/or food spoilage strains. LAB cultures showed a promising antagonistic behavior toward these undesired isolates either as single or in combinations.
The most potent culture; Lb. helveticus was further applied in co-cultures against different isolates showing ≥95% reduction in undesired microbial counts. Further studies are recommended, for extraction, purification, identification and structure elucidation of the bioactive compounds that can contribute to the development of a novel bio preservation system.
Declarations
Author contribution statement
Mamdouh S. Al-Gamal, Osama M. Sharaf, Ahmed A. Radwan, Nadia M. Dabiza: Conceived and designed the experiments; Wrote the paper.
Gamal A. Ibrahim: Conceived and designed the experiments; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Ahmed M. Youssef: Contributed reagents, materials, analysis tools or data; Wrote the paper.
Mohamed F. El-ssayad: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Funding statement
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
References
- Ammor S., Tauveron G., Dufour E., Chevallier I. Antibacterial activity of lactic acid bacteria against spoilage and pathogenic bacteria isolated from the same meat small-scale facility: 1—screening and characterization of the antibacterial compounds. Food Contr. 2006;17(6):454–461. [Google Scholar]
- Baird-Parker A.C. An improved diagnostic and selective medium for isolating coagulase positive staphylococci. J. Appl. Bacteriol. 1962;25(1):12–19. [Google Scholar]
- Brindha V., Mathew A.A. Molecular characterization and identification of unknown bacteria from waste water. Indian J. Innov. Dev. 2012;1(2):87–91. [Google Scholar]
- Cancino-Padilla N., Fellenberg M.A., Franco W., Ibáñez R.A., Vargas-Bello-Pérez E. Foodborne bacteria in dairy products: detection by molecular techniques. Cienc. Invest. Agrar. 2017;44(3):215–229. [Google Scholar]
- Cavallini D.C., Suzuki J.Y., Abdalla D.S., Vendramini R.C., Pauly-Silveira N.D., Roselino M.N., Pinto R.A., Rossi E.A. Influence of a probiotic soy product on fecal microbiota and its association with cardiovascular risk factors in an animal model. Lipids Health Dis. 2011;10(1):126. doi: 10.1186/1476-511X-10-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cretenet M., Even S., Le Loir Y. Unveiling Staphylococcus aureus enterotoxin production in dairy products: a review of recent advances to face new challenges. Dairy Sci. Technol. 2011;91(2):127–150. [Google Scholar]
- Delbès C., Ali-Mandjee L., Montel M.C. Monitoring bacterial communities in raw milk and cheese by culture-dependent and -independent 16S rRNA gene-based analyses. Appl. Environ. Microbiol. 2007;73:1882–1891. doi: 10.1128/AEM.01716-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deraz S.F., Shehata M.G., Khalil A.A. Significant industrial properties of Enterococcus faecium SFD as a probiotic and bacteriocin-producing strain. Life Sci. J. 2015;12(3) [Google Scholar]
- Desriac N., Broussolle V., Postollec F., Mathot A.G., Sohier D., Coroller L., Leguerinel I. Bacillus cereus cell response upon exposure to acid environment: toward the identification of potential biomarkers. Front. Microbiol. 2013;4:284. doi: 10.3389/fmicb.2013.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Hadedy D., El-Nour S.A. Identification of Staphylococcus aureus and Escherichia coli isolated from Egyptian food by conventional and molecular methods. J. Genet. Eng. Biotechnol. 2012;10(1):129–135. [Google Scholar]
- Farrokh C., Jordan K., Auvray F., Glass K., Oppegaard H., Raynaud S., Thevenot D., Condron R., De Reu K., Govaris A., Heggum K. Review of Shiga-toxin-producing Escherichia coli (STEC) and their significance in dairy production. Int. J. Food Microbiol. 2013;162(2):190–212. doi: 10.1016/j.ijfoodmicro.2012.08.008. [DOI] [PubMed] [Google Scholar]
- Garnier L., Valence F., Mounier J. Diversity and control of spoilage fungi in dairy products: an update. Microorganisms. 2017;5(3):42. doi: 10.3390/microorganisms5030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hervert C.J., Martin N.H., Boor K.J., Wiedmann M. Survival and detection of coliforms, Enterobacteriaceae, and gram-negative bacteria in Greek yogurt. J. Dairy Sci. 2017;100(2):950–960. doi: 10.3168/jds.2016-11553. https://blast.ncbi.nlm.nih.gov/Blast.cgi [DOI] [PubMed] [Google Scholar]
- IDFA (International Dairy Foods Association) International Dairy Foods Association; Washington, DC, USA: 2010. Dairy Facts, 2010; pp. 66–76. [Google Scholar]
- Jandu N., Zeng Z.J., Johnson-Henry K.C., Sherman P.M. Probiotics prevent enterohaemorrhagic Escherichia coli O157: H7-mediated inhibition of interferon-γ-induced tyrosine phosphorylation of STAT-1. Microbiology. 2009;155(2):531–540. doi: 10.1099/mic.0.021931-0. [DOI] [PubMed] [Google Scholar]
- Johler S., Weder D., Bridy C., Huguenin M.C., Robert L., Hummerjohann J., Stephan R. Outbreak of staphylococcal food poisoning among children and staff at a Swiss boarding school due to soft cheese made from raw milk. J. Dairy Sci. 2015;98(5):2944–2948. doi: 10.3168/jds.2014-9123. [DOI] [PubMed] [Google Scholar]
- Kean R., Rajendran R., Haggarty J., Townsend E.M., Short B., Burgess K.E., Lang S., Millington O., Mackay W.G., Williams C., Ramage G. Candida albicans mycofilms support Staphylococcus aureus colonization and enhances miconazole resistance in dual-species interactions. Front. Microbiol. 2017;8:258. doi: 10.3389/fmicb.2017.00258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lind H., Jonsson H., Schnürer J. Antifungal effect of dairy propionibacteria—contribution of organic acids. Int. J. Food Microbiol. 2005;98(2):157–165. doi: 10.1016/j.ijfoodmicro.2004.05.020. [DOI] [PubMed] [Google Scholar]
- Magnusson J., Schnürer J. Lactobacillus coryniformis subsp. coryniformis strain Si3 produces a broad-spectrum proteinaceous antifungal compound. Appl. Environ. Microbiol. 2001;67(1):1–5. doi: 10.1128/AEM.67.1.1-5.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragkoudakis P.A., Mountzouris K.C., Psyrras D., Cremonese S., Fischer J., Cantor M.D., Tsakalidou E. Functional properties of novel protective lactic acid bacteria and application in raw chicken meat against Listeria monocytogenes and Salmonella enteritidis. Int. J. Food Microbiol. 2009;130(3):219–226. doi: 10.1016/j.ijfoodmicro.2009.01.027. [DOI] [PubMed] [Google Scholar]
- Marchand S., Vandriesche G., Coorevits A., Coudijzer K., De Jonghe V., Dewettinck K., De Vos P., Devreese B., Heyndrickx M., De Block J. Heterogeneity of heat-resistant proteases from milk Pseudomonas species. Int. J. Food Microbiol. 2009;133(1–2):68–77. doi: 10.1016/j.ijfoodmicro.2009.04.027. [DOI] [PubMed] [Google Scholar]
- Marchand S., De Block J., De Jonghe V., Coorevits A., Heyndrickx M., Herman L. Biofilm formation in milk production and processing environments; influence on milk quality and safety. Compr. Rev. Food Sci. Food Saf. 2012;11(2):133–147. [Google Scholar]
- Mawgoud Y.A., Ibrahim G.A., El-Ssayad M.F. Studying the influence of nitrogen source on lactic acid production from whey permeate by immobilized Lactobacillus bulgaricus Lb-12. Res. J. Pharmaceut. Biol. Chem. Sci. 2016;7(1):693–705. [Google Scholar]
- McGinnis S., Madden T.L. BLAST: at the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004;32(Suppl. 2):W20–W25. doi: 10.1093/nar/gkh435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mols M., Van Kranenburg R., Van Melis C.C., Moezelaar R., Abee T. Analysis of acid-stressed Bacillus cereus reveals a major oxidative response and inactivation-associated radical formation. Environ. Microbiol. 2010;12(4):873–885. doi: 10.1111/j.1462-2920.2009.02132.x. [DOI] [PubMed] [Google Scholar]
- Nörnberg M.F., Friedrich R.S., Weiss R.D., Tondo E.C., Brandelli A. Proteolytic activity among psychrotrophic bacteria isolated from refrigerated raw milk. Int. J. Dairy Technol. 2010;63(1):41–46. [Google Scholar]
- Oldak A., Zielińska D., Rzepkowska A., Kołożyn-Krajewska D. Comparison of antibacterial activity of Lactobacillus plantarum strains isolated from two different kinds of regional cheeses from Poland: Oscypek and Korycinski Cheese. BioMed Res. Int. 2017 doi: 10.1155/2017/6820369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollis G.W., Rawluk S.A., Schoonderwoerd M., Schipper C. Detection of Staphylococcus aureus in bulk tank milk using modified Baird-Parker culture media. Can. Vet. J. 1995;36(10):619–623. PMID: 8640634. [PMC free article] [PubMed] [Google Scholar]
- Pal M., Merera O., Abera F., Rahman M.T., Hazarika R.A. Salmonellosis: a major foodborne disease of global significance. Beverage Food World. 2015;42(12):21–24. [Google Scholar]
- Ponce A.G., Moreira M.R., Del Valle C.E., Roura S.I. Preliminary characterization of bacteriocin-like substances from lactic acid bacteria isolated from organic leafy vegetables. LWT Food Sci. Technol. 2008;41(3):432–444. [Google Scholar]
- Prado-Acosta M., Ruzal S.M., Allievi M.C., Palomino M.M., Rivas C.S. Synergistic effects of the Lactobacillus acidophilus surface layer and nisin on bacterial growth. Appl. Environ. Microbiol. 2010;76(3):974–977. doi: 10.1128/AEM.01427-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price R.G., Wildeboer D. Escherichia coli-Recent Advances on Physiology, Pathogenesis and Biotechnological Applications. InTech; 2017. E. coli as an indicator of contamination and health risk in environmental waters. [Google Scholar]
- Ranadheera C., Vidanarachchi J., Rocha R., Cruz A., Ajlouni S. Probiotic delivery through fermentation: dairy vs. non-dairy beverages. Fermentation. 2017;3(4):67. [Google Scholar]
- Röhr A., Lüddecke K., Drusch S., Müller M.J., Alvensleben R.V. Food quality and safety––consumer perception and public health concern. Food Contr. 2005;16(8):649–655. [Google Scholar]
- Ruaro A., Andrighetto C., Torriani S., Lombardi A. Biodiversity and characterization of indigenous coagulase-negative staphylococci isolated from raw milk and cheese of North Italy. Food Microbiol. 2013;34(1):106–111. doi: 10.1016/j.fm.2012.11.013. [DOI] [PubMed] [Google Scholar]
- Samaržija D., Zamberlin Š., Pogačić T. Psychrotrophic bacteria and milk and dairy products quality. Mljekarstvo. 2012;62(2):77–95. [Google Scholar]
- Shehata M.G., El Sohaimy S.A., El-Sahn M.A., Youssef M.M. Screening of isolated potential probiotic lactic acid bacteria for cholesterol lowering property and bile salt hydrolase activity. Ann. Agric. Sci. 2016;61(1):65–75. [Google Scholar]
- Shokryazdan P., Sieo C.C., Kalavathy R., Liang J.B., Alitheen N.B., Faseleh Jahromi M., Ho Y.W. Probiotic potential of Lactobacillus strains with antimicrobial activity against some human pathogenic strains. BioMed Res. Int. 2014;2:1–16. doi: 10.1155/2014/927268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slabbert R.S. Acid tolerance and organic acid susceptibility of selected food-borne pathogens. Interim Interdiscip. J. 2013;12(1):42–50. http://hdl.handle.net/11462/296 [Google Scholar]
- Silva C.C., Silva S.P., Ribeiro S.C. Application of bacteriocins and protective cultures in dairy food preservation. Front. Microbiol. 2018;9:594. doi: 10.3389/fmicb.2018.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šušković J., Kos B., Beganović J., Pavunc A.L., Habjanič K., Matošić S. Antimicrobial activity – the most important property of probiotic and starter lactic acid bacteria. Food Technol. Biotechnol. 2010;48(3):296–307. [Google Scholar]
- Taverniti V., Guglielmetti S. Health-promoting properties of Lactobacillus helveticus. Front. Microbiol. 2012;3:392. doi: 10.3389/fmicb.2012.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor W.I., Harris B. Isolation of shigellae. II. Comparison of plating media and enrichment broths. Am. J. Clin. Pathol. 1965;44(4):476–479. [PubMed] [Google Scholar]
- Tewari A., Abdullah S. Bacillus cereus food poisoning: international and Indian perspective. J. Food Sci. Technol. 2015;52(5):2500–2511. doi: 10.1007/s13197-014-1344-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tortorello M.L. Indicator organisms for safety and quality—uses and methods for detection: minireview. J. AOAC Int. 2003;86(6):1208–1217. [PubMed] [Google Scholar]
- Trmčić A., Chauhan K., Kent D.J., Ralyea R.D., Martin N.H., Boor K.J., Wiedmann M. Coliform detection in cheese is associated with specific cheese characteristics, but no association was found with pathogen detection. J. Dairy Sci. 2016;99(8):6105–6120. doi: 10.3168/jds.2016-11112. [DOI] [PubMed] [Google Scholar]
- Williams L., Abdi H. Fisher's least significant difference test. Encycl. Res. Des. 2010:492–495. [Google Scholar]