Abstract
Early detection improves hepatocellular carcinoma (HCC) outcomes, but better noninvasive surveillance tools are needed. We aimed to identify and validate methylated DNA markers (MDMs) for HCC detection. Reduced representation bisulfite sequencing was performed on DNA extracted from 18 HCC and 35 control tissues. Candidate MDMs were confirmed by quantitative methylation specific PCR in DNA from independent tissues (74 HCC, 29 controls). A phase I plasma pilot incorporated quantitative allele-specific real time target and signal amplification assays on independent plasma-extracted DNA from 21 HCC cases and 30 cirrhotic controls. A phase II plasma study was then performed in 95 HCC cases, 51 cirrhosis controls, and 98 healthy controls using target enrichment long-probe quantitative amplified signal (TELQAS) assays. Recursive partitioning identified best MDM combinations. The entire MDM panel was statistically cross-validated by randomly splitting the data 2:1 for training and testing. Random forest regression models performed on the training set predicted disease status in the testing set; the median AUC (and 95% CI) were reported after 500 iterations. In phase II, a 6-marker MDM panel (HOXA1, EMX1, AK055957, ECE1, PFKP and CLEC11A, normalized by B3GALT6 level yielded a best fit AUC of 0.96 (95% CI, 0.93–0.99) with HCC sensitivity of 95% (88–98%) at specificity of 92% (86–96%). The panel detected 3/4 (75%) stage 0, 39/42 (93%) stage A, 13/14 (93%) ge B, 28/28 (100%) stage C and 7/7 (100%) stage D HCC. The AUC value for AFP was 0.80 (0.74–0.87) compared to 0.94 (0.9–0.97) for the cross-validated MDM panel, P<0.0001.
Conclusion:
Novel MDMs identified in this study proved to accurately detect HCC via plasma testing. Further optimization and clinical testing of this promising approach are indicated.
Keywords: Liver neoplasms, Early Detection of Cancer, Liver Cirrhosis/complications, DNA methylation, Biomarkers/analysis
INTRODUCTORY STATEMENT
Liver cancer is currently the 2nd leading cause of cancer death worldwide(1) and 6th leading cause in the United States (US), with hepatocellular carcinoma (HCC) being the most common form. (2) Incidence and mortality of HCC in the US has been steadily rising over the last 20 years (3) primarily due to epidemic rises in hepatitis C (HCV) infection and non-alcoholic fatty liver disease (NAFLD).(4) As a consequence, liver cancer is projected to become the 3rd leading cause of cancer deaths in the US by 2030.(5)
High-risk groups for HCC include chronic carriers of hepatitis B virus (HBV) and cirrhotic stage infectious, metabolic or alcoholic liver disease, in whom the annual incidence of HCC is 2–4% per year.(6) Among HBV carriers in China, a randomized controlled trial showed a near 40% reduction in mortality among patients offered surveillance for HCC.(7) There are also compelling observational data which show that HCC surveillance is associated with earlier stage detection, receipt of curative therapy, and improved overall survival.(8) For those at risk, the American Association for the Study of Liver Diseases recommends semiannual HCC surveillance by ultrasound imaging with or without alpha-fetoprotein (AFP).(9)
There are several limitations to this practice. First, despite reported moderate to high specificities (85–98%), ultrasound has low sensitivity for curable-stage disease, particularly those meeting Milan criteria for liver transplantation (63% pooled sensitivity, range 23–91%).(10) Second, surveillance is under-utilized, with up to 50% patient non-compliance.(11) Consequently, there is a strong rationale for the application of accurate biomarkers for HCC surveillance and potential for greater accessibility, lower cost and greater reporting objectivity. Currently available biomarkers include serum AFP, which remains the most commonly used surveillance tool world-wide. While AFP sensitivity for early stage HCC appears similar to ultrasound (66%), specificity may be lower (82%),(12) likely due to fluctuation of AFP levels in association with inflammation, sex and liver disease type.(13) Newer multi-protein, multi-variate models, notably GALAD, which incorporates gender, age, lectin-bound AFP, AFP and des-y carboxyprothromobin; appear to improve specificity; however, GALAD still misses about 30% of potentially curable HCC.(14)
Non-protein biomarkers for HCC include so-called “liquid biopsy” of circulating tumor cells and circulating DNA or exosome products.(15) Of these, methylated DNA of liver(16) or HCC tumor origin(17) appears to reliably distinguish HCC patients from healthy controls. Aberrantly methylated DNA sequences represent broadly informative potential markers of neoplasia(18). Methylated DNA markers were critical components of a multi-target stool DNA test, FDA-approved for average risk colorectal cancer screening.(19)
Our group and others have applied next-generation sequencing techniques to expand the list of candidate methylated DNA markers (MDMs) of gastro-intestinal and hepatobiliary cancers and have demonstrated feasibility of this approach in phase I clinical applications.(20, 21) Additionally, novel normalization methods and assay chemistry appear to achieve very high analytical sensitivity for circulating DNA in plasma,(21, 22) yet this approach has not yet been applied to HCC.
We hypothesized that: 1) next-generation DNA sequencing would identify novel and highly discriminant MDMs for HCC; 2) MDMs would be confirmed in independent tissues; 3.) MDMs would show strong clinical feasibility for detection of HCC when assayed from plasma-extracted DNA; and 4.) novel assay and normalization techniques could be applied to validate clinical feasibility of best performing MDMs in a larger, phase II case-control study.
EXPERIMENTAL PROCEDURES
Overview
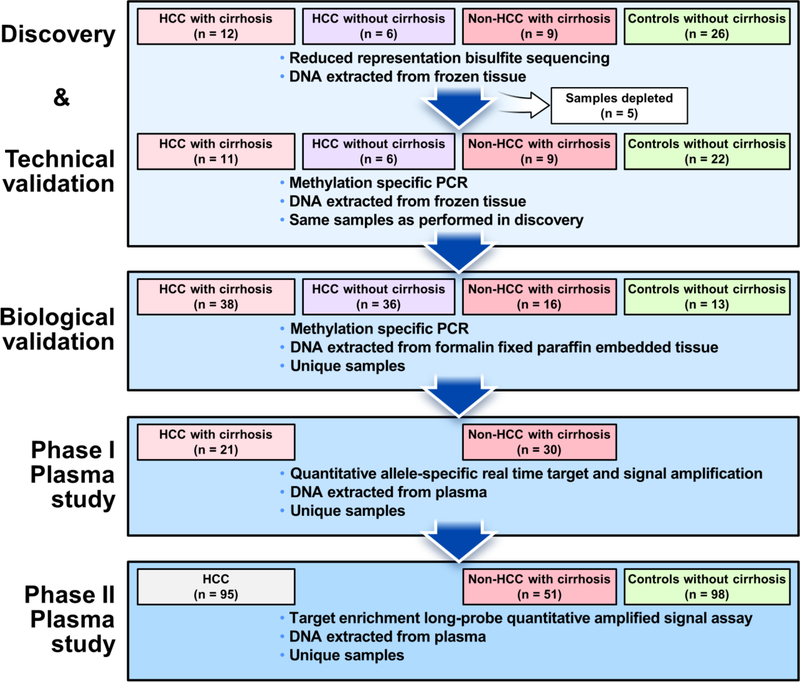
The study was conducted in 4 sequential case-control experiments (Figure 1). Briefly, we first performed discovery using reduced representation bisulfite sequencing (RRBS) & technically validated sequencing-derived candidate MDMs using methylation specific PCR (MSP) on DNA extracted from frozen primary tumor and control samples, in addition to normal buffy coat control samples. Candidate regions were selected where there was differential hypermethylation between cases referent to liver controls and between cases and buffy coat controls; to select candidates for a ctDNA assay, we sumed that background plasma DNA would primarily originate from leukocytes. Technically validated MDM candidates were then biologically validated on independent formalin-fixed paraffin-embedded (FFPE) tissue samples. Additional MDMs from other GI cancers were also tested on the basis of 1) hypermethylation in at least two other GI cancer types 2) AUC values in excess of 0.9 and 3) minimal cross-reactivity (<2% methylation) in leukocyte DNA. Selected MDMs were then tested using quantitative allele-specific target and signal amplification assays (QuARTS) on DNA extracted from archival plasma samples of HCC case and control patients (phase I plasma study). Lastly, a phase II plasma study was performed in which MDMs were assayed on DNA extracted from independent archival plasma samples using the Target Enrichment Long-probe Quantitative Amplified Signal (TELQAS™, Exact Sciences, Madison WI) say. TELQAS products were normalized by plasma volume and a target sequence of B3GALT6, which is methylated in normal liver, HCC primary tumor tissues and leukocytes.
Figure 1.
Study flow diagram
All experiments were performed by blinded personnel. For the phase II plasma study, all clinical data were reviewed and confirmed by a single clinician (JBK) prior to unblinding of laboratory data by the lead statistician (DWM).
Human Subjects
All study procedures were conducted after approval from the Mayo Clinic Institutional Review Board. No prisoners or institutionalized persons were asked to participate. Tissues used in RRBS & technical validation were obtained from the International Hepatobiliary Neoplasia Registry and Biorepository (IHNB, PI LRR) which has enrolled patients with HCC and cholangiocarcinoma, under informed consent, since January 2002. HCC tumor tissue was sampled at the time of segmental surgical resection from patients free from exposure to local/regional therapy or systemic emotherapy. Control tissues were non-adjacent, matched cirrhotic or non-cirrhotic liver parenchyma from HCC-affected individuals or tissues from individuals without HCC, balanced on age and sex to the HCC cases. Additional de-identified waste buffy coat samples were also sequenced to control for background leukocyte DNA. FFPE sues used for biological validation were obtained from the Mayo Clinic Tissue Registry, an archive of waste clinical tissue specimens maintained by the Mayo Clinic Department of Anatomic Pathology. All frozen and FFPE tissues underwent research histopathology review by one of two expert gastrointestinal pathologists (TCS or JTL) prior to macro-dissection and DNA extraction.
Plasma samples (≥1 mL, EDTA preserved) from HCC patients and controls with cirrhosis were obtained from the IHNB. HCC diagnosis was made by radiographic criteria. Referent to the time of plasma collection, patients were free from HCC treatment. All stages of HCC were included with a bias towards Barcelona Clinic Liver Cancer stage B or lower.(23) Cirrhosis controls were required to have at least two consecutive imaging studies free from HCC or indeterminate liver nodules. Sample selection was biased toward compensated liver disease (Child-Pugh A or B) to reflect the intended surveillance population.(9) Healthy control plasma, age- and sex-balanced to the HCC cases was obtained from a separate archive of patients without cancer (PI DAA) which has used informed consent to enroll from a 7-county regional population since September 2015. All patients were verified by a medical record review to be free from other cancers for ≥5 years. All plasma samples were processed and stored according to standardized institutional protocols in the central repository of the Mayo Clinic Biospecimens Accession and Processing laboratory.
Marker Identification
Discovery & Technical validation
RRBS discovery and qMSP technical validation were performed as previously described (Supplemental Methods).(20) Briefly, DNA from frozen liver tissue samples and buffy coat was isolated and quantitated. After MSPI digestion, DNA fragments were repaired and ligated to an indexing label. The product was bisulfite converted and size selected before sequencing on the HiSeq 2000 (Illumina, San Diego CA) in the Mayo Sequencing Core facility. Sequencing reads were called by standard Illumina pipeline software and aligned via SAAP-RRBS.(24) From this point forward short-sequence or fragmented DNA sequences were targeted by methylation-specific assays. We therefore used the term MDM for these candidate biomarkers rather than DMR which refers to genomic regional methylation differences.
Technical validation of these MDMs was performed using quantitative methylation specific PCR (MSP) assays on the discovery sample set, as previously described.(25)
Biological tissue validation
The same MSP assays were run on DNA extracted from independent FFPE case and control tissues. DNA was extracted using the Qiagen kit and bisulfite treated with the Zymo kit as above.
Phase 1 plasma study
Top MDM candidates from MSP testing were re-assessed using higher sensitivity QuARTs triplex assays, as previously described.(18, 26) In this study, 12 cycles of multiplex PCR reactions were first performed on bisulfite converted DNA followed by triplex QuARTS assays Triplexes were assayed on the LightCycler 480 (Roche) and all results were normalized to the β-actin product amplified from the same sample.
Phase II plasma study
For the MDMs with results that met the pre-specified criteria, below, additional MDM assay designs were developed using TELQAS chemistry, to allow for greater analytical sensitivity. The TELQAS chemistry is a modification to the QuARTS assay that utilizes probes that are longer and run at a higher temperature. MDMs interrogated in the phase I plasma study were assayed from independent plasma samples in phase II, following DNA extraction and bisulfite conversion (as above), using the TELQAS assay. TELQAS assays were normalized by products of a DNA region, B3GALT6 as previously described,(22) after verification from the RRBS library that this sequence was methylated in HCC, control tissues, and buffy coat.
For TELQAS, a limited number of cycles (12 cycles) multiplex PCR amplification of the MDMs as well as B3GALT6 was performed on the bisulfite converted DNA. The PCR products were then diluted 10-fold with a 10 mM Tris-HCl, 0.1 mM EDTA solution; 10 μL of the diluted amplicons were used in triplex LQAS assays in which two MDMs plus the B3GALT6 reference gene were quantified. TELQAS reactions were performed on ABI 7500DX equipment (Applied Biosystems, Foster City CA).
Statistical Analysis
Discovery & technical validation
From the RRBS data set, the difference in methylation percentage was mpared between HCC cases, tissue controls and buffy coat controls; a tiled reading frame within 100 base pairs of each mapped CpG was used to identify DMRs where ntrol methylation was <5%; DMRs were only analyzed if the total depth of coverage was 10 reads per subject on average and the variance across subgroups was >0. The sample size requirements were estimated as previously reported.(20) Assuming a biologically relevant increase in the odds ratio of >3x and a coverage depth of 10 reads, 8 samples per group were required to achieve 80% power with a two-sided test at a significance level of 5% and assuming binomial variance inflation factor of 1.
Following regression, DMRs were ranked by p-value, area under the receiver operating characteristic curve (AUC) and fold-change difference between cases and all controls. No adjustments for false discovery were made during this phase as independent validation was planned a priori. From these results, DMRs on the in silico genomic map were used to define the sequences targeted as MDMs in subsequent experiments.
Biological tissue validation
For confirmation of each MDM in FFPE samples, we aimed to achieve a 95% confidence interval ±10% around a specificity estimate of 95%; a minimum of 30 controls were required. To ensure that the 95% confidence interval was within ±10% for a sensitivity of 90%, a minimum of 70 cases were required (split equally between HCC cases with and without cirrhosis). To examine combinations of markers, a methylation intensity map was created by selecting a single MDM with the highest sensitivity at the % percentile value for each MDM in controls. Secondary MDMs were considered additive when additional cases were positive at the 100th percentile value in the controls (thus preserving an overall specificity for the panel of 95%). MDM combinations were limited to 5 predictors to minimize potential overfitting.
Phase I plasma study
It was estimated that a minimum of 20 patients in the case group would provide 80% power to distinguish an AUC of ≥70% from a null value of 0.5 with a one-sided one-sample proportion test at the 5% level. Marker combinations were studied using recursive partitioning trees (rPart) which first selected a single MDM that provided the greatest separation between cases and controls (branch split). Then, rPart searched for additional MDMs that provided the greatest separation between cases and controls under each branch. This process continued iteratively until a pre-specified cross-validated stopping rule was reached to avoid overfitting.
Phase II plasma study
With 100 HCC patients, 50 cirrhosis controls and 100 healthy controls, there was >80% power to distinguish an AUC of ≥70% from a null value of 0.5 with a one-sided Bonferroni corrected significance level of 5%/15 for each MDM. For combinations of markers, two techniques were used. First, the rPart technique was applied to the entire MDM set and limited to combinations of 10 MDMs, upon which an rPart predicted probability of cancer was calculated. The second approach used random forest regression (rForest) which generated 500 individual rPart models that were fit to boot strap samples of the original data (roughly 2/3 of the data for training) and used to estimate the cross-validation error (1/3 of the data for testing) of the entire MDM panel and was repeated 500 times to avoid spurious splits that either under- or overestimate the true cross-validation metrics. Results were then averaged across the 500 iterations. Separate models were performed for data standardized to plasma sample volume, and to sample B3GALT6 product level.
Serum AFP values were available for HCC cases and cirrhosis controls only. To mpare MDMs to AFP, two methods were used. First an AUC curve from the rPart model of TELQAS-assayed MDMs was assembled from a data set restricted HCC ses and cirrhosis controls; the AUC value estimated from AFP in the same patients. The second approach accounted for the unmeasured AFP (27) in the controls without cirrhosis using 500 iterations with imputation of AFP values from a log normal distribution with mean 3 and standard deviation of 1.9.(28) In the imputed data model, AUC values of the MDM panel were compared to that of the imputed AFP and to the combination of MDMs and AFP using rForest.
Estimates were reported with 95% confidence intervals (95% CI). The method of DeLong, DeLong and Clarke-Pearson was used to estimate the 95% CI of the area under the receiver operating characteristics curve and compare AUC values between MDMs.(29) Spearman’s correlation or Wilcoxon rank sum tests were used to assess the relationship of the rPart score with BCLC stage. The effect of group imbalance of underlying liver disease, age, and sex on the diagnostic accuracy of the rPart score was investigated by comparing stratified AUC values.
To further assess the functional relevance of the markers assayed in the phase II plasma study, the RNA-Seq by Expectation Maximization values were obtained from publically available data sets of RNA expression from HCC primary tumor samples (The Cancer Genome Atlas) and normal liver tissues (Genotype Tissue Expression Project) using the XENA browser (30); within XENA, the DESeq2 analysis was used to compare RNA expression between cases and controls(31).
RESULTS
Discovery & technical validation
Frozen liver tissue was available from 18 HCC patients and 35 controls. Additionally, buffy coat samples from 16 patients were included in the RRBS experiment.
Sequencing yielded 5.4 million CpGs mapped per sample with 2.8 million CpGs having 10X or greater coverage. From these, 1163 DMRs were mapped; 302 with AUCs greater than 0.75 with fold changes against normal liver ranging from 20–623 and against normal buffy coat from 3–120. Additionally, we identified 89 regions which contained hypermethylated CpGs in cancer samples as compared to buffy coat (leukocyte) derived DNA samples, irrespective of the methylation status of control tissues. We selected 30 non-overlapping DMRs with the highest AUCs and fold changes to undergo qMSP testing in the technical validation phase. Following technical validation, 14 DMRs had either lower AUCs or lower fold changes (or both) than earlier ults and were eliminated. The remaining 16 DMR sequences were chosen as candidate MDMs to carry forward to biological validation in independent tissues (Supplemental Table 1).
Biological tissue validation
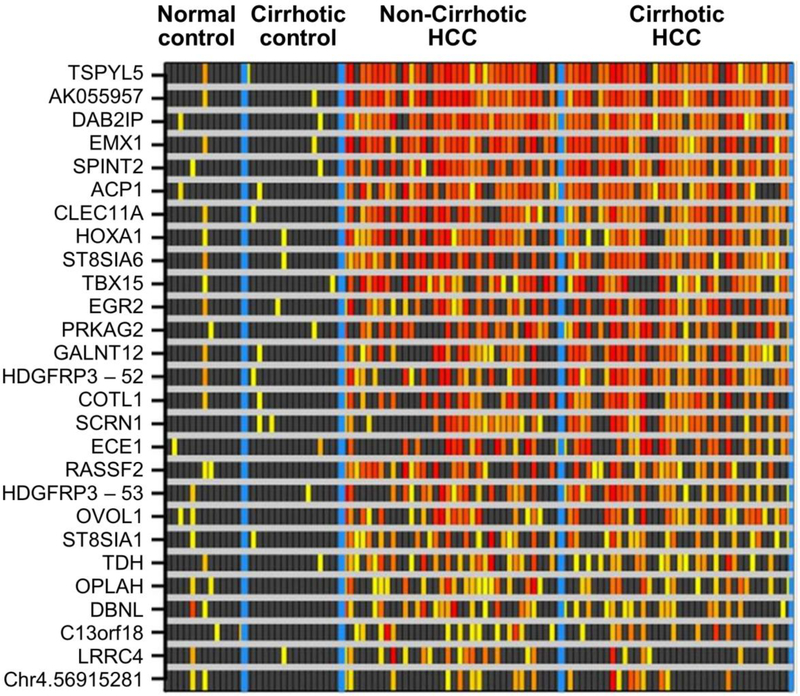
The 16 candidate MDMs were tested by qMSP on DNA extracted from 103 FFPE liver tissues including 74 HCC cases (38 from cirrhotic livers, 36 from non-cirrhotic livers) and 29 controls (16 cirrhotic livers without HCC, 13 normal livers). We also tested 11 MDMs identified in our previous RRBS discovery and tissue validations studies in other GI cancers, including colorectal, esophageal, pancreatic, biliary and gastric. In the independent FFPE sample set, AUC values of the 27 MDMs ranged from 0.64 or 0.94 with fold changes of 1.5–90. At specificity cut-off values of 95% set from normal and cirrhotic liver controls, a combination of 5 MDMs was positive in 70/74 (95% (95% CI, 87–99%)) of the HCC case tissues; these included AK055957, DAB2IP, EMX1, TBX15 and TSPYL5 (Figure 3).
Figure 3.
Methylation intensity of candidates confirmed in independent tissue samples: Increasing intensity of yellow-red color spectrum indicates methylation strand counts in decile values above the 95th percentile values for the control groups (normal liver and cirrhotic liver combined) of each candidate (rows) in each tissue sample (columns). Black boxes indicate values falling below the 95th percentile in controls.
Phase I plasma pilot
The 12 MDMs we selected for further testing in plasma included 10 from the biological validation (ACP1, AK055957, CLEC11A, DAB2IP, DBNL, EMX1, HOXA1, LRRC4, SPINT2 and TSPYL5). AK055957, DAB2IP, EMX1, and TSPYL5 all had tissue AUCs > 0.90 and low cross-reactivity in leukocyte DNA. HOXA1 had a lower AUC (0.80) but complemented other MDMs and had the lowest leukocyte signal in the panel. LRRC4 and DBNL had smaller fold changes with respect to normal liver DNA, but showed hypermethylation in the tissue samples (Supplemental Figure 1). Two additional markers, BDH1 and EFNB2, were identified in comparison of the HCC sequencing data to other RRBS data sets and selected for high specificity for liver cancer.
These candidate MDMs also annotate to genes which are known to play causal roles in tumorigenesis, (32) specifically transcriptional regulation, growth modulation and cell signaling (Table 1).
Table 1.
Biological roles of candidates confirmed in independent tissue samples
| Methylated DNA Marker | Biological Role | Genomic Coordinates* |
|---|---|---|
| ACP1 | Tyrosine phosphatase activity/cell growth modulation | chr2:264087–264151 |
| AK055957 | Uncharacterized | chr12:133484978–133485739 |
| CLEC11A | Growth factor | chr19:51228217–51228732 |
| DAB2IP | Cell growth modulation | chr9:124461305–124461420 |
| DBNL | Receptor-mediated endocytosis/immune cell activation | chr7:44080227–44080310 |
| EMX1 | Transcriptional regulation | chr2:73147710–73147772 |
| HOXA1 | Transcriptional regulation | chr7:27136145–27136425 |
| LRRC4 | Protein kinase inhibitor | chr7:127671993–127672310 |
| SPINT2 | Serine protease inhibitor | chr19:38755130–38755164 |
| TSPYL5 | Cell growth modulation | chr8:98289858–98290220 |
GRCh37/hg19 assembly
From the candidate MDMs selected from MSP tissue data, additional assays using the higher sensitivity QuARTs method were performed on unique plasma samples in a small pilot comprising 21 HCC cases and 30 cirrhosis controls. After standardizing all MDMs to the β-actin internal control, individual MDMs had AUCs of 0.65–0.90. The best performing single MDM in plasma, EMX1, had an AUC of 0.90 (95% CI, 0.80–0.99) with 76% (95% CI, 53–92%) sensitivity at 100% (95% CI, 88–100%) specificity. The combination of EMX1 and CLEC11A had an AUC of 0.91 (95% CI, 0.83–1.00); this corresponded to 86% (95% CI, 64–97%) sensitivity at 87% (95% CI, 69–96%) specificity.
Underlying liver diseases among HCC cases in the Discovery, Biological validation and Phase I plasma study are listed in Supplemental Table 2.
Phase II plasma study
Nine candidates were carried forward from phase I to phase II (ACP1, AK055957, CLEC11A, DAB2IP, EMX1, EFNB2, HOXA1, SPINT2, and TSPYL5). However, results of the phase I study also suggested a low contribution to HCC detection from BDH1, DBNL and LRRC4. A further interrogation of the original RRBS data against other GI cancer methylation sequencing libraries suggested that CCNJ_3124, CCNJ_3707, ECE1, PKFP and SCRN1 were found in strong association with HCC. These markers were then biologically validated in the FFPE tissue samples and moved forward to plasma testing at phase II (Supplemental Table 3) because DNA from phase I samples had been exhausted. Thus, we selected 14 candidate MDMs for the larger phase II study (ACP1, AK055957, CCNJ_3707, CCNJ_3124, CLEC11A, DAB2IP, ECE1, EFNB2, EMX1, HOXA1, PFKP, SPINT2, SCRN1 and TSPYL5) and the normalizer B3GALT6. The panel was assayed on each eligible patient sample with at least 1 mL plasma.
We studied 244 eligible patients including 95 HCC cases and 149 controls (51 with cirrhosis free from HCC and 98 healthy volunteers (Table 2). Age and sex were similar between HCC patients and healthy controls; however, cirrhotic control patients were younger and more likely to be women. HCC patients were more likely to be current smokers and healthy controls were more likely to be actively consuming alcohol. Only 7% of HCC and 2% of cirrhosis controls had decompensated liver disease (ChildPugh C). The majority of HCC patients had BCLC stage B cancer or earlier disease; 44% were stage A.
Table 2.
Patient characteristics of phase II clinical study
| HCC (n=95) | Cirrhosis controls (n=51) | Healthy controls (n=98) | p-value | |
|---|---|---|---|---|
| Median Age (IQR), years | 64 (57–70) | 58 (54–63) | 62 (56–68) | 0.09 |
| Men (%) | 75 (79) | 30 (59) | 67 (68) | 0.03 |
| Tobacco* | ||||
| Current (%) | 21 (22) | 4 (8) | 15 (15) | 0.05 |
| Former (%) | 38 (40) | 20 (39) | 43 (44) | |
| Never (%) | 31 (33) | 26 (51) | 40 (41) | |
| Alcohol intake* | ||||
| Current (%) | 35 (37) | 20 (40) | 84 (86) | <0.001 |
| Former (%) | 38 (40) | 25 (49) | 0 (0) | |
| Never (%) | 9 (9) | 5 (9) | 14 (14) | |
| Child-Pugh class | ||||
| A (%) | 68 (72) | 23 (45) | - | <0.001 |
| B (%) | 18 (19) | 27 (53) | - | |
| C (%) | 7 (7) | 1 (2) | - | |
| Median AFP ng/mL (IQR) | 17 (4.3–207) | 4.3 (2.5–6.5) | - | 0.153 |
| Underlying liver disease, n (%) Hepatitis C | 33 (35) | 8 (16) | 0.025 | |
| Alcohol | 23 (24) | 16 (31) | 0.462 | |
| NAFLD | 21 (22) | 17 (34) | 0.189 | |
| Other | 24 (25) | 11 (22) | 0.867 | |
| BCLC stage | ||||
| 0 (%) | 4 (4) | - | - | |
| A (%) | 42 (44) | - | - | |
| B (%) | 14 (15) | - | - | |
| C (%) | 28 (30) | - | - | |
| D (%) | 7 (7) | - | - | |
| Milan transplantation criteria† (% within) | 44 (47) | - | - |
Tobacco and alcohol exposure was not reported by all patients
Single tumor ≤5cm in diameter or ≤ 3 or fewer tumors ≤3 cm in diameter
AFP, alpha-fetorprotein
BCLC, Barcelona Clinic Liver Cancer
HCC, hepatocellular carcinoma
IQR, inter-quartile range
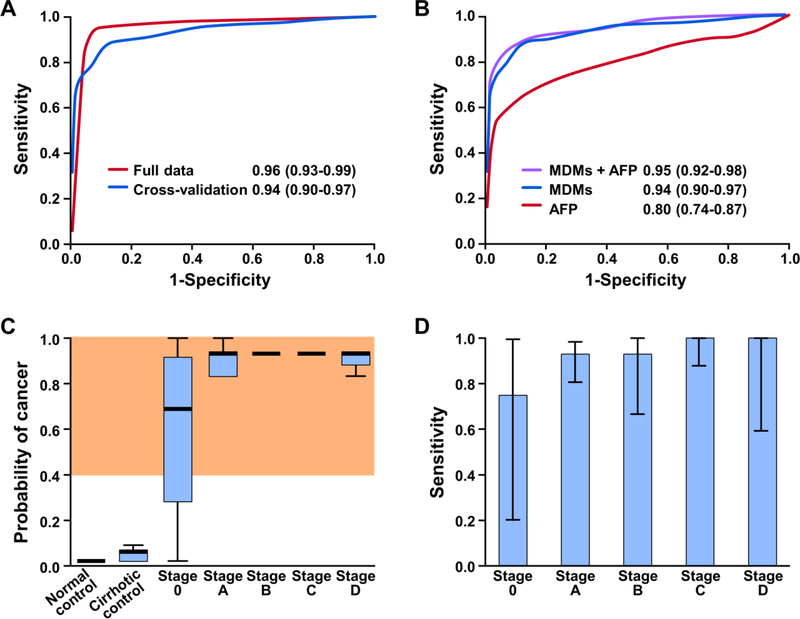
Distribution plots of candidate MDMs illustrate individual discrimination for HCC (Supplemental Figure 2). By rPart analysis, a combination of 6 MDMs achieved an AUC of 0.96 (95% CI, 0.93–0.99) (Figure 4A); this corresponded to a sensitivity for HCC of 95% (95% CI, 88–98%) at an overall specificity of 92% (95% CI, 86–96%) in all controls, 95% (88–98%) in healthy controls, 86% (73–94%) in cirrhotic controls). Cross validation of the panel performance averaged across the 500 iterations of training and test sets yielded an AUC of 0.94 (0.90–0.97) (Figure 4A); sensitivity for HCC was 85% (81–89%) at a specificity of 91% (90–91%).
Figure 4.
Results of phase II plasma pilot include: A) areas under the curve (AUC) and 95% confidence intervals of B3GALT6-corrected logistic fit model and boot strapped aggregating statistical cross-validation; B) comparison of methylated DNA markers (MDMs) with and without inclusion of alpha-fetoprotein (AFP) and AFP alone; C) RPart fit score estimated the probability of cancer for the given methylation distributions across normal controls, cirrhotic controls and HCC cases by BCLC stage, where gray shaded area depicts values above 92% specificity threshold; and, D) sensitivity and 95% confidence intervals of logistic fit score by BCLC stage at 92% specificity.
Limiting the data to that from HCC cases and cirrhosis controls, the MDM panel had an AUC of 0.93 (0.89–0.98); the AUC of AFP alone in the same two groups was 0.74 (0.66–0.82). Using the imputed data model for missing AFP values in non-cirrhosis controls, AFP was compared against the MDM panel (Figure 4B). The AUC value for AFP was 0.80 (95% CI, 0.74–0.87) compared to 0.94 (0.9–0.97) for the MDM panel, P<0.0001. In this group, AFP yielded a sensitivity for HCC of 60% (50–71%) at 91% specificity (80–98%) using a 10 ng/mL cut-off. Addition of AFP did not statistically improve the overall accuracy of the MDM panel (Figure 4B). None of the 5 HCCs missed by the MDM panel were AFP positive.
Based on plots of composite scores for the 6 MDM panel, marker levels increased with BCLC stage of HCC, p=0.02 (Figure 4C). Using the rPart model to assess HCC detection accuracy by stage, the panel detected 3/4 (75%) stage 0, 39/42 (93%) stage A, 13/14 (93%) stage B, 28/28 (100%) stage C and 7/7 (100%) stage D HCC at 92% specificity (Figure 4D).
The overall accuracies of the panel of markers for both assay normalization techniques (including plasma volume and B3GALT6 to correct MDM TELQAS products) were compared across the 500 iterations of the training and test set modeling. The average classification error for HCC with each approach was 13% and 11% respectively (p=0.4).
Due to significant differences in baseline variables, the rPart model score was plotted against healthy control and cirrhotic control distributions for age, sex, and baseline imbalance in the proportion of patients with hepatitis C infection. Importantly, there were no significant differences in the accuracy of the best fit rPart model after these stratifications (Table 3).
Table 3.
RPart fit area under the curve and 95% confidence intervals stratified by clinical co-variates with baseline differences
| Yes | No | p-value | |
|---|---|---|---|
| Age >60 | 0.96 (0.92–0.99) | 0.97 (0.94–1.00) | 0.99 |
| Male sex | 0.95 (0.91–0.98) | 0.99 (0.97–1.00) | 0.98 |
| Current smoking | 0.97 (0.94–1.00) | 0.95 (0.92–0.99) | 0.99 |
| Current alcohol intake | 0.97 (0.93–1.00) | 0.93 (0.87–0.98) | 0.97 |
| Child-Pugh B or C | 0.97 (0.93–1.00) | 0.89 (0.81–0.97) | 0.95 |
| Hepatitis C | 0.87 (0.71–1.00) | 0.98 (0.96–0.99) | 0.93 |
Publically available RNA expression data was obtained for 12 of the 14 MDMs which could be annotated to unique coding regions; AK055957 is non-coding and CCNJ_3707 & CCNJ_3124 annotate to the same region. The XENA browser could cess RNA expression data from 396 primary HCC tumors and 108 normal liver tissues. Significant differences in RNA levels were seen for 11/12 regions when comparing cases to controls. CLEC11A and TSPYL5 were significantly down regulated in tumors, whereas ACP1, DAB2IP, ECE1, EFNB2, EMX1, HOXA1, PFKP, SPINT2 and SCRN1 were up-regulated (Supplemental Table 4).
DISCUSSION
We report a panel of novel MDMs that, when assayed from plasma, highly discriminate HCC cases from both healthy and cirrhotic controls. Following a whole methylome sequencing discovery and biological confirmation on independent tissues, candidate MDMs were then tested on plasma in phase I and phase II case-control studies to substantiate the clinical feasibility of MDMs for early detection of HCC. The panel was superior to AFP, despite potential cross-sectional design biases, including patients who may have been referred on the basis of elevated AFP. The phase II plasma study was biased towards early-stage HCC and compensated cirrhosis to best reflect the intended surveillance population.
There are formidable challenges in developing a clinically useful early cancer detection test from circulating tumor DNA (ctDNA). Because plasma levels of ctDNA are correlated with increasing cancer stage.(33) the clinical applications utilizing ctDNA have focused on monitoring for relapse in patients with known cancers.(34) Advances in assay technology, including BEAMing,(34) digital droplet PCR,(35) and QuARTS(22) have progressively increased the ability to detect an intended mutant or methylated DNA target among the total DNA in a clinical sample. This is the first report of a clinical feasibility study in human cancers using TELQAS assay chemistry, a novel modification of the QuARTS assay that has a theoretical analytical sensitivity threshold of just 2–4 strands per mL of plasma. Based on the observations of the present study, it is anticipated that TELQAS will be well-suited to applications using ctDNA for early cancer detection. While the FDA-approved QuARTS stool DNA assay uses β-actin as the internal reference, this may be sub-optimal in ctDNA assays as non-methylated sequences will sustain greater damage during bisulfite conversion. It was for this reason that we previously identified, applied and reported B3GALT6 as a novel internal reference gene for ctDNA assays (22) and adapted this to HCC detection by the TELQAS method.
To detect early HCC, an unbiased marker selection approach was used to account for the potential heterogeneity in epigenetic profiles among primary liver tumors and underlying liver disease.(36) We have previously shown that DNA methylation is more broadly informative than DNA mutation in colorectal cancer.(18) With this in mind, our discovery strategy was directed to identify MDMs with the greatest representation in HCC tumors, while controlling for epigenetic differences due to underlying cirrhosis or non-cirrhotic liver disease.
We also controlled for the epigenetic signatures of white blood cells, which contribute the largest proportion of circulating DNA in plasma.(16) We also studied a sub-set of MDMs that were methylated in both HCC and control liver tissue but not in white blood cells. These could potentially indicate HCC vascular invasion, or hepatocyte cell death.(16)
In addition to using an analytically sensitive platform, the detection of ctDNA in plasma from curable stage HCC appears feasible for several strong biological reasons. The liver receives roughly 25% of cardiac output.(37) The highly vascular architecture of the liver may result in multiple foci for entry of apoptotic and necrotic DNA. Unlike the other visceral and glandular GI organs which might exfoliate DNA into portal circulation, nucleic acids of hepatic origin can enter systemic circulation via hepatic venous outflow. As a result, a recent analysis using a methylation deconvolution process estimated that 10–13% of circulating cfDNA in plasma originates from the normal liver and rises to 1944% when cancer is present.(16) These phenomena may explain why primary liver tumors were detected at a higher rate than other tumor types in a recent non-invasive multi-analyte multi-cancer detection study.(38) Our findings build on these by reporting high sensitivity (93%) for stage A and B HCC, high specificity even among cirrhotic ntrols and superiority of methylated DNA analytes over AFP.
Though the MDM markers were identified without consideration of biological function, these were subsequently annotated to genes with clear roles in the tumorigenic cascade. Of the 27 MDMs in our tissue validation, at least 17 are involved in mechanistic pathways which regulate cellular growth and division. Referent to the UCSC browser, almost all MDMs lie within CpG islands in association with known regulatory signposts such as ChIP-seq transcription factors, altered histone modifications, and ENCODE RRBS-derived methylation sites.(32) Additionally, public database interrogation of RNA expression levels in HCC and control liver tissues shows that genes to which the MDMs annotate are up- or down-regulated in cancers. These findings are similar to those reported in other solid tumors (Supplemental Table 4), strengthening the functional importance of DNA methylation at these sites. While causality in tumorigeneses is best ascertained through functional studies, these strong mechanistic associations with the MDMs in our panel provide further contextual validity, strengthen the biologic plausibility of the findings and argue against over-fitting during this large genome-wide biomarker discovery effort.
Despite these encouraging early results, there are several potential limitations to the present study. First, patients were enrolled from a single referral center. Sample sizes within each study phase were not sufficiently large enough to study associations between individual candidate MDMs and specific etiologies of cirrhosis and HCC. However, the most common diseases which predispose to HCC were represented at each stage. In Discovery and Biological validation steps, there were a relatively large proportion of patients who did not have cirrhosis or were not found to have an underlying liver disease at the time that they presented for surgical resection of their HCC. However confirmation of the MDMs in multiple downstream validation steps, the large case sample size in the phase II study and the inclusion of common underlying liver diseases help to generalize the findings. Second, definitive exclusion of HCC in the rhosis controls is clinically difficult since surveillance ultrasound may be insensitive for early-stage HCC.(10) We also anticipate that overall sensitivity for early stage disease will be improved with optimized methods of sample collection; these include obtaining larger blood volumes, utilization of collection tubes specifically designed for ctDNA recovery, and standardization of blood processing.
In conclusion, a novel MDM panel was highly accurate for the detection of HCC, including early stage lesions. The panel was significantly more accurate than AFP. Specificity was high among both cirrhotic controls free from HCC and healthy controls free from known underlying liver disease. This promising approach will require further refinement and confirmation in a larger phase II design using optimized sample collection and processing methods to establish firm MDM cut-off values prior to clinical validation in a phase III prospective cohort.
Supplementary Material
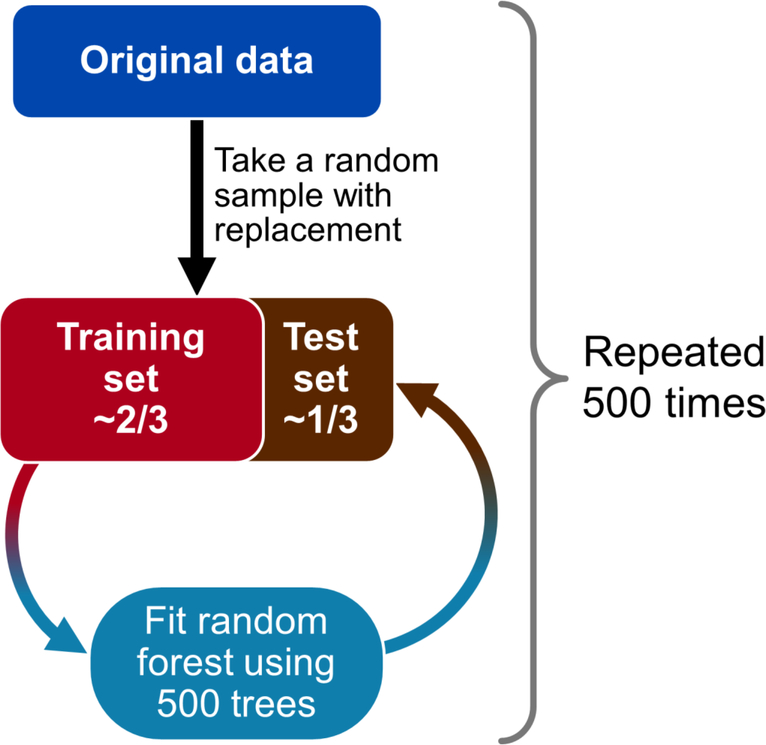
Figure 2.
Statistical methods for phase II plasma study cross-validation. The original data set was randomly sampled so training and test sets were created in 2:1 proportions. Random forest fits of 500 trees each were modeled on the training set data and applied to the test set data; this process was repeated 500 times.
Acknowledgments
Financial Support:
This work was supported by the Maxine and Jack Zarrow Family Foundation of Tulsa Oklahoma (to JBK), the Paul Calabresi Program in Clinical-Translational Research (NCI CA90628) (to JBK), R37 CA214679 (to JBK), the Dana and Edmund Gong Foundation (to DAA), the Carol M. Gatton endowment for Digestive Diseases Research (to DAA). RRBS sequencing costs, QuARTS assays and TELQAS assays were provided by Exact Sciences (Madison WI)
List of Abbreviations:
- 95% CI
95% confidence interval
- AFP
alpha-fetoprotein
- AUC
area under the receiver operating characteristics curve
- BCLC
Barcelona Clinic Liver Cancer staging
- ctDNA
circulating tumor DNA
- DMR
differentially methylated regions
- DNA
deoxyribonucleic acid
- FFPE
formalin-fixed paraffin-embedded
- GALAD
gender, age, lectin-bound AFP, AFP and des-y carboxyprothromobin
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- IHNB
International Hepatobiliary Neoplasia Registry and Biorepository
- IQR
interquartile range
- LQAS
long-probe quantitative amplified signal
- MDM
methylated DNA marker
- MSP
methylation specific polymerase chain reaction
- NAFLD
non-alcoholic fatty liver disease
- QuARTS
quantitative allele-specific realtime target and signal amplification
- ROC
receiver operating characteristics curve
- rForest
random forest modeling
- rPart
recursive partitioning decision analysis
- RRBS
reduced representation bisulfite sequencing
- TELQAS
target enrichment long-probe quantitative amplified signal
Footnotes
Publisher's Disclaimer: This article has been accepted for publication and undergone full peer review but has not been through the copyediting, typesetting, pagination and proofreading process, which may lead to differences between this version and the Version of Record. Please cite this article as doi: 10.1002/hep.30244
REFERENCES
- 1.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin 2015;65:87–108. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Jemal A. Cancer Statistics, 2017. CA: A Cancer Journal for Clinicians 2016:n/a–n/a. [DOI] [PubMed] [Google Scholar]
- 3.Petrick JL, Braunlin M, Laversanne M, Valery PC, Bray F, McGlynn KA. International trends in liver cancer incidence, overall and by histologic subtype, 1978–2007. Int J Cancer 2016;139:1534–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Makarova-Rusher OV, Altekruse SF, McNeel TS, Ulahannan S, Duffy AG, Graubard BI, Greten TF, et al. Population attributable fractions of risk factors for hepatocellular carcinoma in the United States. Cancer 2016;122:1757–1765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Petrick JL, Kelly SP, Altekruse SF, McGlynn KA, Rosenberg PS. Future of Hepatocellular Carcinoma Incidence in the United States Forecast Through 2030. J Clin Oncol 2016;34:1787–1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Serag HB. Hepatocellular carcinoma. N Engl J Med 2011;365:1118–1127. [DOI] [PubMed] [Google Scholar]
- 7.Zhang BH, Yang BH, Tang ZY. Randomized controlled trial of screening for hepatocellular carcinoma. J Cancer Res Clin Oncol 2004;130:417–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Singal AG, Pillai A, Tiro J. Early detection, curative treatment, and survival rates for hepatocellular carcinoma surveillance in patients with cirrhosis: a meta-analysis. PLoS Med 2014;11:e1001624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heimbach JK, Kulik LM, Finn R, Sirlin CB, Abecassis M, Roberts LR, Zhu A, et al. Aasld guidelines for the treatment of hepatocellular carcinoma. Hepatology 2017. [DOI] [PubMed] [Google Scholar]
- 10.Singal A, Volk ML, Waljee A, Salgia R, Higgins P, Rogers MA, Marrero JA. Meta-analysis: surveillance with ultrasound for early-stage hepatocellular carcinoma in patients with cirrhosis. Aliment Pharmacol Ther 2009;30:37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singal AG, Yopp A, C SS, Packer M, Lee WM, Tiro JA. Utilization of hepatocellular carcinoma surveillance among American patients: a systematic review. J Gen Intern Med 2012;27:861–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marrero JA, Feng Z, Wang Y, Nguyen MH, Befeler AS, Roberts LR, Reddy KR, et al. Alpha-fetoprotein, des-gamma carboxyprothrombin, and lectin-bound alpha-fetoprotein in early hepatocellular carcinoma. Gastroenterology 2009;137:110–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Richardson P, Duan Z, Kramer J, Davila JA, Tyson GL, El-Serag HB. Determinants of serum alpha-fetoprotein levels in hepatitis C-infected patients. Clin Gastroenterol Hepatol 2012;10:428–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berhane S, Toyoda H, Tada T, Kumada T, Kagebayashi C, Satomura S, Schweitzer N, et al. Role of the GALAD and BALAD-2 Serologic Models in Diagnosis of Hepatocellular Carcinoma and Prediction of Survival in Patients. Clin Gastroenterol Hepatol 2016;14:875–886e876. [DOI] [PubMed] [Google Scholar]
- 15.Labgaa I Liquit Biopsy in Liver Cancer. Discovery Medicine 2015;19:263–273. [PubMed] [Google Scholar]
- 16.Sun K, Jiang P, Chan KC, Wong J, Cheng YK, Liang RH, Chan WK, et al. Plasma DNA tissue mapping by genome-wide methylation sequencing for noninvasive prenatal, cancer, and transplantation assessments. Proc Natl Acad Sci U S A 2015;112:E5503–5512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu R-h, Wei W, Krawczyk M, Wang W, Luo H, Flagg K, Yi S, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. 2017. [DOI] [PubMed]
- 18.Zou H, Allawi H, Cao X, Domanico M, Harrington J, Taylor WR, Yab T, et al. Quantification of methylated markers with a multiplex methylation-specific technology. Clin Chem 2012;58:375–383. [DOI] [PubMed] [Google Scholar]
- 19.Imperiale TF, Ransohoff DF, Itzkowitz SH, Levin TR, Lavin P, Lidgard GP, Ahlquist DA, et al. Multitarget stool DNA testing for colorectal-cancer screening. N Engl J Med 2014;370:1287–1297. [DOI] [PubMed] [Google Scholar]
- 20.Kisiel JB, Raimondo M, Taylor W, Yab TC, Mahoney DW, Sun Z, Middha S, et al. New DNA methylation markers for pancreatic cancer: discovery, tissue validation, and pilot testing in pancreatic juice. Clin Cancer Res 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang JD, Yab TC, Taylor WR, Foote PH, Ali HA, Lavu S, Simonson JA, et al. Detection of Cholangiocarcinoma by Assay of Methylated DNA Markers in Plasma. Gastroenterology 2017;152:S1041–S1042. [Google Scholar]
- 22.Allawi HT, Giakoumopoulos M, Flietner E, Oliphant A, Volkman C, Aizenstein B, Sander T, et al. Detection of lung cancer by assay of novel methylated DNA markers in plasma. Cancer Research 2017;77:A. 712. [Google Scholar]
- 23.Llovet JM, Bru C, Bruix J. Prognosis of hepatocellular carcinoma: the BCLC staging classification. Semin Liver Dis 1999;19:329–338. [DOI] [PubMed] [Google Scholar]
- 24.Sun Z, Baheti S, Middha S, Kanwar R, Zhang Y, Li X, Beutler AS, et al. SAAP-RRBS: streamlined analysis and annotation pipeline for reduced representation bisulfite sequencing. Bioinformatics 2012;28:2180–2181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kisiel JB, Yab TC, Taylor WR, Chari ST, Petersen GM, Mahoney DW, Ahlquist DA. Stool DNA testing for the detection of pancreatic cancer: assessment of methylation marker candidates. Cancer 2012;118:2623–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lidgard GP, Domanico MJ, Bruinsma JJ, Light J, Gagrat ZD, Oldham-Haltom RL, Fourrier KD, et al. Clinical performance of an automated stool DNA assay for detection of colorectal neoplasia. Clin Gastroenterol Hepatol 2013;11:1313–1318. [DOI] [PubMed] [Google Scholar]
- 27.Little RJA, Rubin DB. Statistical Analysis with Missing Data. 2 ed: John Wiley & Sons, 2002. [Google Scholar]
- 28.Ball D, Rose E, Alpert E. Alpha-fetoprotein levels in normal adults. Am J Med Sci 1992;303:157–159. [DOI] [PubMed] [Google Scholar]
- 29.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 1988;44:837–845. [PubMed] [Google Scholar]
- 30.California RotUo. In.
- 31.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cruz GB GoUS In. Santa Cruz: The Regents of the University of California. [Google Scholar]
- 33.Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, Bartlett BR, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Science translational medicine 2014;6:224ra224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dawson SJ, Tsui DW, Murtaza M, Biggs H, Rueda OM, Chin SF, Dunning MJ, et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N Engl J Med 2013;368:1199–1209. [DOI] [PubMed] [Google Scholar]
- 35.Huang A, Zhang X, Zhou SL, Cao Y, Huang XW, Fan J, Yang XR, et al. Detecting Circulating Tumor DNA in Hepatocellular Carcinoma Patients Using Droplet Digital PCR Is Feasible and Reflects Intratumoral Heterogeneity. J Cancer 2016;7:1907–1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hlady RA, Tiedemann RL, Puszyk W, Zendejas I, Roberts LR, Choi JH, Liu C, et al. Epigenetic signatures of alcohol abuse and hepatitis infection during human hepatocarcinogenesis. Oncotarget 2014;5:9425–9443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lautt WW, Greenway CV. Conceptual review of the hepatic vascular bed. Hepatology 1987;7:952–963. [DOI] [PubMed] [Google Scholar]
- 38.Cohen JD, Li L, Wang Y, Thoburn C, Afsari B, Danilova L, Douville C, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science 2018;359:926–930. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.