Abstract
p53 gain-of-function mutations are similar to driver mutations in cancer genes, with both promoting tumorigenesis. Most previous studies focused on residues lost by mutations, providing information related to a dominantly-negative effect. However, to understand gain-of-function mutations, it is also important to investigate what are the distributions of residues gained by mutations. We compile available p53/p63/p73 protein sequences and construct a non-redundant dataset. We analyze the amino acid and dipeptide composition of p53/p63/p73 proteins across evolution and compare them with the gain/loss of amino acids and dipeptides in human p53 following cancer-related somatic mutations. We find that the ratios of amino acids gained via somatic mutations during evolution to those lost through p53 cancer mutations correlate with the ratios found in single nucleotide polymorphisms in the human proteome. The dipeptide mutational gain/loss ratios are inversely correlated with those observed over p53 evolution but tend to follow the increasing p63/p73-like dipeptide propensities. We successfully simulated the p53 cancer mutation spectrum using the dipeptide composition across the p53 family accounting for the likelihood of mutations in p53 codons. The results revealed that the p53 mutation spectrum is dominated not only by p53 evolution but also by reversal of evolution to a certain degree. This article is part of a Special Issue entitled: Computational Proteomics, Systems Biology & Clinical Implications. Guest Editor: Yudong Cai.
Keywords: p53/p63/p73, Gene regulation, Phosphorylation, Cancer, Mutation, Evolution
1. Introduction
The sequencing of all genes related to cancer (the ‘cancer genome’) [1,2] has reinforced the concept that oncogenesis relates to evolution [3]. Among the mutations that accumulate in cancer genes, ‘driver’ mutations are positively selected during cancer development, unlike the accompanying ‘passenger’ mutations [1–3]. The landscape of driver mutations provides the mutational basis for p53 pathway deregulation in melanoma [4]. TP53 has the highest cancer mutation prevalence [1], accumulating thousands of somatic and inherited mutations. The p53 protein plays a central role in the cell by integrating pathways related to apoptosis, cell cycle arrest, and DNA repair, in response to various types of stress [5,6]. Apart from its critical role as a tumor suppressor, it regulates hundreds of genes and is a guardian of genome stability [5,7]. The multiple functions of p53 also correlate between cancer and neurodegeneration [8]. Two other p53 family proteins, p63 and p73, are structurally and functionally highly similar to p53, particularly in transactivation of similar genes and in maintaining similar interaction network [9–11]. Still, the p53/p63/p73 differs after a billion years of evolution [10,12], with p63/p73 acquiring new functions related to organism development while p53 evolved to be a tumor suppressor and the guardian of the somatic genome [12–14].
p53 gain-of-function mutations are similar to driver mutations in cancer genes because p53 gain-of-function mutants are also positively selected and tumorigenic [15,16]. p53 mutations have been implicated in more than 50% of all human cancers. Several p53 hot spot mutations appear with very high frequencies. While many can be explained by their destabilization of p53, often affecting p53-DNA interactions [17,18], others are still not entirely understood. For example, it is unclear why p53 has many cancer-related mutations, while mutations in p63/p73 are rare and the roles of p63/p73 in cancer are also less clear [19]. For p63/p73, altered expression rather than mutation could be more important in cancer [20,21].
A correlation between conservation and positive selection of p53 mutational hot spots has been implied [16,22]. Further, recently, using the cancer-specific high-throughput annotation of somatic mutations several of these hot spots were predicted to be driver mutations [23]. These studies indicated that destroying highly conserved functional sites is insufficient for inactivation and tumorigenesis, but the origin of the mutant gain-of-(tumorigenic)-function is still unclear. Most previous studies focused on residues lost in mutations, providing information related to a dominant-negative effect. To understand gain-of-function mutations, it is also important to investigate what are the distributions of residues gained from mutations.
In this study, we use dipeptide patterns in p53/p63/p73 proteins across evolution to investigate human cancer mutations, focusing on both loss and gain of dipeptides from somatic mutations. By comparing p53 mutations with single nucleotide polymorphisms (SNPs) in the human proteome, we show that while p53 somatic mutations tend to follow the human proteome trend, the amino acids whose frequencies are increased (or decreased) in p53 across evolution tend to differ from these trends. For all p53 somatic mutations, the most gained dipeptide pairs are those eliminated from p53 during evolution.
2. Methods
2.1. Gain/loss of amino acids and dipeptides in the p53 somatic mutation dataset
TP53 somatic mutations were downloaded from the IARC p53 mutation database Release R13, which contains 24,785 somatic mutations in sporadic cancers reported up to the end of 2007 [24] (http://www-p53.iarc.fr/). 18,266 non-synonymous mutations resulted from a single nucleotide change. These mutations are grouped according to the codon position and the amino acid substitutions and the corresponding frequencies are calculated. The frequencies of gain-and-loss mutation-containing dipeptides are obtained by replacing wild type dipeptide pairs by those with the corresponding mutations. We list the frequencies in Sup-Table 1. The changes are normalized by gain/loss ratios R = (Gain − Loss) / (Gain + Loss). We also compared the results using the current release R16, which contains 29,575 mutations. All observations reported in this paper have been confirmed.
2.2. p53/p63/p73 sequences used in the analysis
Recently, several studies investigated the homology and evolution of the p53/p63/p73 family by searching known p53/p63/p73 family sequences [25,26]. Nedelcu and Tan found that a p53/p63/p73 type sequence is present in the unicellular choanoflagellate, Monosiga brevicollis [25], which is also the species with the least homology with human p53 and diverged early from p63/p73 (Sup-Table 1). Even though the classification of Caenorhabditis elegans Cep-1 as a putative p53 homologue is sometimes questioned [26], the functions [27] and structures [28,29] of Cep-1 indeed show similarities to p53. Increasing evidence reveals additional similarities between Cep-1 and p53, for example, CBP-1, the worm ortholog of human p300/CBP, functions as a cofactor of CEP-1 [30].
We merged the datasets of Nedelcu and Tan [25] and Fernandes and Atchley [26]. We also searched the NCBI Entrez protein database (www.ncbi.nlm.nih.gov), adding more than twenty entries of p63/p73 related sequences. Our final compilation contains 46 p53 and 36 p63/p73 unique sequences. We follow Nedelcu and Tan’s suggestion to distinguish p53- and p63/p73 sequences based on the presence of a SAM domain in p63/p73. After removing highly similar sequences, the final non-redundant dataset has 36 p53 sequences with homology less than 90%. For p63/p73, we retained 22 sequences with homology less than 95% in the non-redundant dataset. The sequence dataset is listed in Supporting Table 2.
2.3. Dipeptide pair correlation and propensities
For each sequence in the non-redundant dataset (Sup-Table 1) of p53 and p63/p73 families, we count the number of dipeptides XY. We calculate the propensity of the dipeptides by normalizing the frequencies by the counts of individual amino acids:
| (1) |
where PXiYj is the propensity of the dipeptide. NXiYj is the total number dipeptides. The NX and NY are the total number of amino acids of type X and Y. A dipeptide has a high propensity if it is frequent in each sequence and across sequences. The above PXiYj is similar to that used by Vonderviszt et al. [31], who used NXiYj / (NXY * PX * PY) to measure the dipeptide propensity (NXY is the total number of all dipeptides, PX and PY are the relative abundances of amino acid types X and Y). PXiYj in Eq. (1) changes more smoothly with the variation of NX and NY in the calculation of the dipeptide propensities across evolution.
We calculate the degenerate dipeptide pair correlation propensity DPXiYj by adding PXiYj and PYiXj for non-diagonal elements in the correlation matrix.
| (2) |
Thus we do not distinguish between XY and YX and label such amino acid combinations X/Y, and the propensity X/Y is obtained from the combined XY and YX counts. Since we have 210 DPXiYj dipeptide pairs, the top 20 ranked pairs represent the top 10% of the pair correlation.
2.4. Evolution in the p53/p63/p73 family
We use ClustalX 2.0 [32] to align the sequences. We use sequence identities with human p53 as the measure of evolutionary distances from human p53. For the p63/p73 family, we use the averaged sequence identities with human p63 and p73. These sequence identities with human p53 are listed in Sup-Table 2.
The gain and loss of amino acids and dipeptide pairs in p53 during evolution are obtained by comparing the amino acid composition and dipeptide pair propensities in human p53 with the average values from species with less than 28% identity to human p53 (Sup-Table 2). The gain and loss of amino acids and dipeptide pairs in p63 and p73 are derived from comparisons of human p63 and p73 with the average values of (p63/p73-brafl1, p63/p73-XP-001184464, p63-DAA06085, p63-XP-001518439, P63-EEC11931). All the changes are normalized with gain/loss ratios R = (Gain − Loss) / (Gain + Loss).
2.5. Random mutation control
Random mutations were generated by randomly selecting a missense mutation from 9 possible single nucleotide mutations at each codon. A total of 18,266 random mutations were used to compare with the 18,266 cancer mutations. In the correction analysis, the random mutations were subtracted from the observed mutation counts at each codon. For a given mutation, if the number of random mutations was more than observed, the observed mutation count was set to zero.
3. Results
3.1. p53 and p63/p73 have different dipeptide propensities
In Table 1, we compare the top dipeptide correlations in p53 with the p63/p73 entries. As expected, the p53/p63/p73 family shares dipeptide pair similarities: among the top 20 ranked dipeptide pairs for p53, 9 are among the top 20 for p63/p73 (bold and italic, Table 1). Examination of the underlying dipeptide motifs corresponding to the common pairs reveals that they have high frequencies in the DNA binding region, oligomerization domain, and phosphorylation motifs. However, both student T-test and Wilcoxon signed-rank test indicated that there are significant differences between the top ranked amino acid pairs for p53 and p63/p73 (Table 1), while the overall amino acid pair propensities for p53 and p63/p73 are statistically similar (T-test, p = 0.413 for all 210 amino acid pairs) and correlated (Spearman rank-order correlation, rs = 0.68, p < 0.000001).
Table 1.
Top ranked amino acid pair propensitiesa for p53 and the corresponding p63/p73 values.
| Dipeptide pairb | Propensity p53 | Propensity p63/p73 |
|---|---|---|
| D/S | 11.425 | 4.648 |
| A/P | 10.716 | 6.603 |
| V/R | 10.482 | 7.759 |
| K/R | 8.905 | 6.42 |
| K/K | 8.482 | 5.009 |
| P/P | 8.355 | 7.264 |
| L/E | 8.248 | 8.065 |
| S/T | 8.232 | 10.443 |
| R/G | 7.962 | 8.472 |
| S/Q | 7.95 | 9.898 |
| L/N | 7.925 | 6.667 |
| P/S | 7.783 | 11.639 |
| S/G | 7.685 | 5.71 |
| M/C | 7.662 | 3.292 |
| V/E | 7.594 | 7.518 |
| L/P | 7.567 | 5.882 |
| V/P | 7.548 | 6.712 |
| L/S | 7.541 | 7.481 |
| S/S | 7.525 | 7.21 |
| A/L | 7.513 | 4.885 |
| Statistical differences | Wilcoxon test: P = 0.0116 T-test: P = 0.0095 |
Based on the non-redundant dataset in Sup-Table 2.
The bold and italic X/Y pairs are top 20 ranked for both p53 and p63/p73 subfamilies.
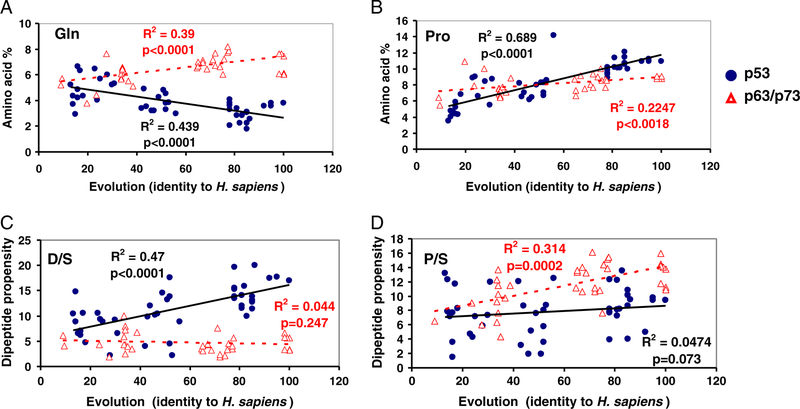
The differences in the dipeptide propensities for p53 and p63/p73 come from their amino acid composition, but could also relate to their functions. Dipeptide composition has been shown to be related to the nuclear receptors’ function [33] and to the subcellular locations of apoptosis-related proteins [34,35]. Evolution appears to have followed different pathways in the change of amino acid composition: for example, while the p53 subfamily has systematically reduced the Gln content, p63/p73 steadily increased it (Fig. 1A). As a result, the Gln content in p63/p73 is significantly higher than in p53 (Sup-Table 3). p53 and p63/p73 have relatively similar composition at the amino acid level, but different dipeptide preferences, as observed for Asp and Ser (D/S), and Pro and Ser (P/S). D/S dipeptides have the highest propensity in p53 sequences, while p63/p73 prefers Pro and Ser (P/S) dipeptides (Table 1). At the amino acid level, the content of Pro, Ser, and Asp is similar between p53 and p63/p73 (Sup-Table 2). During evolution, both p53 and p63/p73 have increased the Pro content, with p53 at a rate surpassing that of p63/p73 (Fig. 1B); however, they have different evolution trajectories for the D/S and P/S dipeptides. p53 gradually increased the D/S dipeptide propensity, which is fairly constant during the p63/p73 evolution (Fig. 1C). While the dipeptide propensity of P/S fluctuated during p53 evolution, it steadily increased for p63/p73 (Fig. 1D). We further found that D/S and P/S dipeptides have different likelihoods to be associated with cancer mutations near phosphorylation motifs in p53 (data not shown).
Fig. 1.
Evolution of amino acid and dipeptide composition in the p53/p63/p73 families. Some of the changes correlate with sequence identity to Homo sapiens. (A) and (B). While Gln composition decreased (A), Pro composition increased significantly (B); (C): The change of the D/S dipeptide propensity during evolution indicates that the D/S pair is positively selected in p53 (but not in p63/p73); (D). While dipeptide propensity of P/S in p53 did not correlate with p53 evolution, that in p63/p73 increased. Thus, both the most preferred dipeptide pairs in p53 (D/S) and in p63/p73 (P/S) were positively selected during evolution, for p53 and p63/p73 respectively.
3.2. Gain and loss of dipeptide pairs in p53 somatic cancer mutations
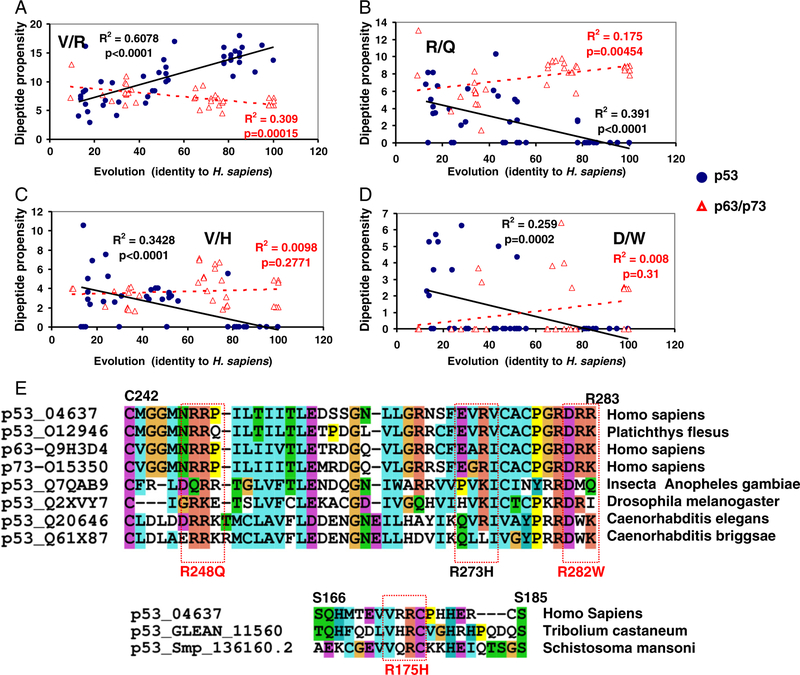
When a missense mutation occurs at a position i, two dipeptide pairs (i − 1, i; i, i + 1) change. The frequencies of all dipeptide pairs gained and lost upon mutations are in the Supporting material, Excel file Sup-Table 1. As expected, dipeptides related to six hot spot mutations (R175, G245, R248, R249, R273, and R282) contribute the most to the high counts for dipeptide lost (Table 2). The V/R pair is the most mutated dipeptide partly due to the high frequencies of the R273 mutation; the other six highly mutated pairs (R/R, R/N, R/C, D/R, M/G, and G/G) derive from hot spot mutations in R174RCP177, M243GGMNRR249, and R280DRR283. Even without the hot spots, the V/R pair still is the highest mutated dipeptide pair. On average, the dipeptide pairs lost most frequently have higher dipeptide propensity in p53 than in p63/p73 (Table 2). In Fig. 2A, we can see that V/R has increased during p53 evolution, but decreased in p63/p73, suggesting that V/R mutations may reverse the above evolution trend.
Table 2.
Top 20 dipeptide pairs lost by p53 mutations.
| Top 20 | Number | Number | Number lost | Propensityc | |
|---|---|---|---|---|---|
| lost pairs | losta | gained | (no hotspot)b | p53 | p63/p73 |
| V/R | 4502 | 188 | 1400 | 10.482 | 7.759 |
| R/R | 4110 | 176 | 122 | 5.866 | 4.955 |
| R/N | 1763 | 267 | 142 | 5.622 | 5.057 |
| R/C | 1581 | 121 | 429 | 5.585 | 4.915 |
| D/R | 1222 | 89 | 580 | 6.28 | 7.235 |
| M/G | 1126 | 31 | 381 | 5.914 | 3.765 |
| G/G | 958 | 59 | 213 | 3.567 | 4.457 |
| P/C | 929 | 412 | 929 | 7.29 | 6.391 |
| P/R | 680 | 248 | 107 | 3.338 | 2.897 |
| P/P | 662 | 36 | 662 | 8.355 | 7.264 |
| M/C | 639 | 222 | 639 | 7.662 | 3.292 |
| E/Y | 553 | 186 | 553 | 6.558 | 4.884 |
| R/G | 548 | 356 | 548 | 7.962 | 8.472 |
| P/G | 492 | 162 | 492 | 7.477 | 7.197 |
| H/H | 461 | 59 | 461 | 4.082 | 0.616 |
| E/E | 455 | 50 | 455 | 5.017 | 3.101 |
| V/E | 441 | 99 | 441 | 7.594 | 7.518 |
| E/H | 423 | 69 | 423 | 4.336 | 7.358 |
| V/V | 404 | 15 | 404 | 5.849 | 3.015 |
| S/C | 401 | 278 | 401 | 4.396 | 2.182 |
| Average | 1117.5 | 156.15 | 489.1 | 6.1616 | 5.1165 |
| Statistical differencesd between p53 and p63/p73 P = 0.0066 | |||||
All missense mutations counted.
Gain/loss due to hot spot mutations are excluded.
Propensity for wild type p53 and p63/p73.
Student test of the significance between the dipeptide propensities for p53 and p63/p73.
Fig. 2.
The evolution of dipeptide composition in the p53 is reversed by hot spot mutations. (A) Dipeptide evolution for V/R pairs. The V/R dipeptide decreases the most in p53 mutations; however it has the highest increase rate in p53 evolution. (B) R/Q is among the highly gained dipeptides following p53 mutations. It has been eliminated during p53 evolution while increasing in p63/p73. (C) The dipeptide V/H is the most gained in p53 cancer mutations; on the other hand, it was eliminated during evolution. (D) The dipeptide pair D/W, which relates to R282W mutation, was eliminated during p53 evolution. (E) Several hot spot mutants gained dipeptide patterns which were eliminated during p53 evolution; for example, R175H mutant in human compared to Tribolium castaneum; R248Q in human compared to Platichthys flesus and Insecta Anopheles gambiae, and R282W in human compared to C. elegans and C. briggsae.
It is important to note in Table 3 that the most gained dipeptide pairs are those not present in wild type p53 sequences. Among the top ten amino-acid pairs in the mutation database seven are not observed in the human p53 sequence. Only a few amino-acid pairs which are generated by mutations have relatively high propensity in the p53 family (PS, SG, LP and PC). The average dipeptide propensity in the top 20 gained pairs is only 3.92, significantly lower than the average propensity for the dipeptide lost (6.16, Table 2, p = 0.0066). A clear trend is observed: the less preferred the dipeptide is in the p53 family, the higher the chance that it will be gained in a cancer-related mutation (Sup-Figure 1A, Sup-Table 1).
Table 3.
The top 20 most gained dipeptides from p53 mutations.
| Dipeptide pairs | Number lost | Number gained | Dipeptide propensity | |
|---|---|---|---|---|
| p53 | p63/p73 | |||
| V/H | 0 | 16222 | 2.469 | 3.843 |
| R/H | 170 | 1424 | 3.984 | 3.451 |
| V/C | 231 | 1321 | 3.788 | 3.255 |
| R/W | 0 | 1308 | 0.098 | 1.302 |
| H/C | 0 | 1262 | 1.491 | 2.367 |
| R/Q | 0 | 966 | 2.649 | 7.407 |
| N/Q | 0 | 834 | 3.231 | 5.94 |
| N/W | 0 | 714 | 2.043 | 1.314 |
| P/S | 99 | 674 | 7.783 | 11.639 |
| V/L | 2 | 654 | 4.767 | 7.052 |
| S/G | 119 | 652 | 7.685 | 5.71 |
| F/R | 195 | 638 | 5.11 | 3.835 |
| R/S | 3 | 635 | 4.71 | 3.226 |
| M/S | 0 | 626 | 1.417 | 5.092 |
| L/R | 184 | 584 | 4.198 | 3.102 |
| D/W | 0 | 552 | 1.147 | 1.029 |
| E/C | 0 | 497 | 1.225 | 1.415 |
| E/R | 149 | 429 | 5.706 | 2.61 |
| L/P | 54 | 415 | 7.567 | 5.882 |
| P/C | 929 | 412 | 7.29 | 6.391 |
| Average | 106.75 | 810.95 | 3.9179 | 4.2931 |
| Statistical differencesa of dipeptide propensity between the lost and gain groups P = 0.04 | Statistical differencesa between p53 and p63/p73 P = 0.2225 | |||
Student test of the significance of the difference.
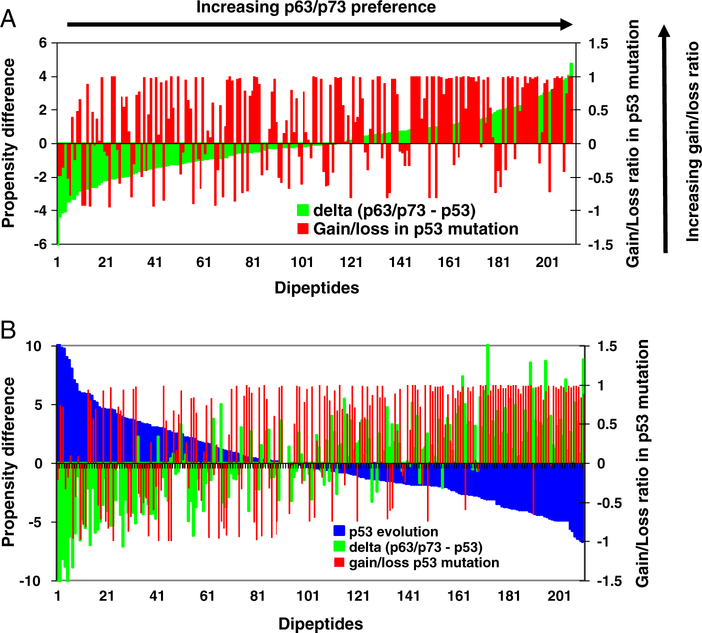
It can be seen from Table 3 that the propensities for the top 20 most gained dipeptides from p53 mutations are higher for p63/p73 than for p53. The statistical differences for these 20 dipeptides are not significant (p = 0.22). However, when we examine all 210 dipeptide pairs, we find that there is a correlation between the dipeptide propensity in p63/p73 and the likelihood that the dipeptide will be gained by p53 mutations. The correlation is obtained by calculating the normalized gain/loss ratio for each dipeptide pair observed in p53 cancer mutations, and then comparing the ratio with the dipeptide propensity difference in p63/p73 and in p53 (Δ(p63 / p73 − p53)). The two measures are correlated (Spearman rank-order correlation, p = 0.055 for all dipeptides with positive Δ(p63 / p73 − p53)). This observation can also be seen in Fig. 3A, where the green lines represent the differences between dipeptide propensities in p63/p73 and in p53, and the red lines are gain/loss ratios for the corresponding dipeptides. The dipeptides are ordered by Δ(p63 / p73 − p53); dipeptides with larger Δ(p63 / p73 − p53) are gained more frequently by p53 mutations.
Fig. 3.
The correlations of p53 dipeptide gain/loss ratios with evolution and with the gap of dipeptide propensities in p63/p73 and in p53. (A). p53 dipeptide gain/loss ratios increase with increasing gap of peptide propensities in p63/p73 and in p53. The green lines represent the differences between dipeptide propensities in p63/p73 and in p53, and the red lines are gain/loss ratios for the corresponding dipeptides. The dipeptides are ordered with the value of Δ(p63/p73−p53). (B). p53 dipeptide gain/loss ratios increase with the decreasing dipeptide propensities in evolution. The green lines represent the differences between dipeptide propensities in human p63/p73 and p53, and the red lines are gain/loss ratios for the corresponding dipeptides. The blue lines are the changing of dipeptide propensities in evolution. The dipeptides are ordered with the decreasing value of dipeptide propensities in evolution.
3.3. p53 somatic mutations correlate with gain and loss of amino acids in human SNPs
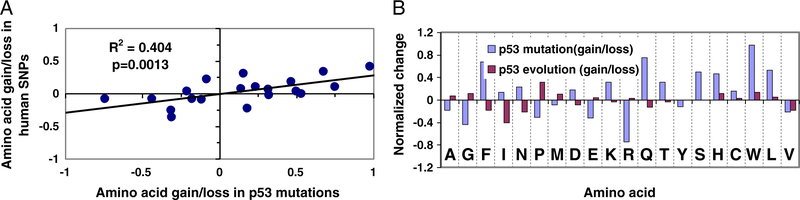
Surprisingly, when we examined the gain/loss of amino acids during p53 mutation/evolution against a previous study of the human protein evolution [36], we found that the single amino acids that gained/lost during the p53 evolution tend to follow a trend opposite to that of the human proteins (Sup-Figure 1B, p = 0.0013). Comparing Jordan’s amino acid data [36] with the p53 frequencies provides insights into the p53 mutations/evolution. As shown in Sup-Figure 1B, the normalized amino acid gain/loss ratio during human p53 evolution decreases while it increases in human proteins. In comparison, p63 follows the trend (Sup-Figure 1C), and p73 fluctuates (Sup-Figure 1D). The differences among p53, p63, and p73 are consistent with the notion that p53 evolved from p63. It seems that p53 deviated while p63 followed the mainstream evolution path suggesting that p53 has been under evolutionary pressure.
However, the gain/loss of amino acids in p53 by cancer-related mutations follows the trend observed in human SNPs (Fig. 4A). Jordan et al. [36] compared proteins encoded by triplets of closely related genomes and provided the trend of the gain/loss of amino acids in human non-synonymous SNPs since the human–chimpanzee divergence. This can be readily compared with the gain/loss of amino acids in the p53 mutations. As shown in Fig. 3B, the mutations in p53 illustrate a gain/loss of amino acids essentially reflecting the substitutions in human SNPs, with R2 = 0.4, indicating that p53-related cancer cell evolution may mimic the human proteome evolution (p = 0.0013). The randomly generated p53 mutations do not correlate with the substitutions in human SNPs. Following the random corrections, the observed p53 mutations still present a similar correlation with the human SNPs.
Fig. 4.
(A) Comparison between amino acid gained/lost in p53 mutations with those gained/lost in human protein evolution, which are taken from reference [36]. (B) The trend of amino acid gain/loss in p53 mutations differs from that of p53 evolution.
3.4. Reversal of p53 evolution may relate to p53 gain-of-function mutants and p63/p73 interference
Comparing the amino acid composition during evolution with amino acid gain/loss ratio in the p53 mutation database (Fig. 4B) we observe that residues lost (or gained) in evolution are those gained (or lost) in cancer-related mutations. These residues include A, G, F, I, N, P, M, D, E, K, R, Q, and T. Two (S and Y) show no trend, and only five amino acids (V, C, H, L and W) follow the same trend in evolution and gain/loss ratio.
This counter-evolution pattern is more prominent in the dipeptide gain/loss. As shown in Fig. 3B, the dipeptides gained the most through mutations are strong losers in p53 evolution. To exclude random mutations, we subtract random mutation counts from observed counts. If the number of random mutations was more than observed, the observed mutation count was set to zero. Still the Spearman rank-order correlation is very high (rs = 0.56, p1 < 0.00001). We also noted that the dipeptide gain/loss ratios in p53 mutations are similarly correlated with the differences of dipeptide propensities in human p63/p73 and p53 (rs = 0.35, p1 < 0.00001).
The similarities of p53 mutant dipeptides to those in homologue sequences could be rationalized by two scenarios: one is that mutant dipeptides appeared in ancient p53 species; another is that mutant dipeptides are also more frequent in p63/p73. Our analysis cannot distinguish between these, and the gain/loss of p53 cancer mutations correlates with both p53 evolution and p63/p73 sequences. Intuitively, given a wild type p53 sequence, it may not be surprising to see that the gained dipeptides in mutants are those found in homologues. The question is if these similarities have functional correlations.
There are indications that the reversal of p53 evolution may relate to p53 gain-of-function mutants. The two top mutation gainers (R/Q and V/H, Fig. 2B and 2C) have been completely eliminated during evolution. R175H and R248Q are two hot spot mutants related to the R/Q and V/H dipeptide pairs. Both R175H and R248Q are gain-of-function mutants, known to induce GEF-H1 gene (Guanine exchange factor-H1) [37], and R175H also regulates the expression of other genes [38]. R273H was suggested to be a driver mutation [23], and it also accompanies the mutational switch from the evolutionary favored V/R pair to evolutionary eliminated V/H pair (Fig. 2). The percentages of Gln and His have increased during the evolution of human proteins. Consequently, R248Q, R175H, and R273H may have been ‘passenger’ mutations during the evolution of human proteins, but ‘driver’ mutations in p53. R282W was also suggested to be a driver mutation [23]. Similarly, the R282W hot spot mutation generated the R/W and D/W dipeptides which are rare in p53, and evolution eliminated D/W (Fig. 2D).
The R175 and R248 motifs are highly conserved in most p53/p63/p73 members; however, in several earlier p53 species there are R/Q and R/H dipeptides at these locations (Fig. 2E). The motif corresponding to VVRR175C in Red flour beetle (p53 GLEAN_11560, Sup-Table 2) is LVHRC, comparable to VVRH175C with an R175H substitution in human mutants. For the human p53 MNR248RPI motif, the corresponding sequences in European flounder (p53_O12946) and Insecta Anopheles gambiae (p53_Q7QAB9, Sup-Table 2) are respectively MNRRQI, and LDQRRT (Fig. 2E), which are comparable to MNQ248RPI in the R248Q mutant. Thus, these mutations could relate to some p53 gained functions that have been eliminated during its evolution. Indeed, sequence alignment also revealed that the R282W hot spot mutation left a trace in C. elegans and Caenorhabditis briggsae (Fig. 2E). The worm Cep-1 transactivates the consensus human p53 binding sites [39] with similar p53 functions [27]. To examine a relationship between p53 mutants’ functions and the differences of dipeptide propensities between human p63/p73 and p53, we compare three groups of p53 mutants: dominant negative mutants (DNMs), general gain-of-function-1 (GOF-1), and a group known to interfere with p63/p73 functions (GOF-2). We follow the classification by Waldman et al. [40] (Sup-Table 4). For each group of mutants, we calculate the average change of dipeptide propensities of Δ(p63 / p73 − p53) caused by mutations. We found that on average, the GOF-2 group has the largest increase of p63/p73 dipeptide propensity (0.78), followed by the GOF-1 group (0.4); the DNM group has no p63/p73 dipeptide propensity gain (−0.17) as compared to p53. Thus it seems that dipeptide propensity changes could relate to the functional outcomes.
When examining p53 and p63/p73 proteins during evolution, we used the entire protein sequence. Since both the N-terminal and C-terminal regions have diverged more than the core domain (Sup-Table 2), it is interesting to analyze only the DNA binding core domain. Furthermore, the majority of the oncogenic p53 mutations fall in the core domain, hence their impact on amino acid gain or loss and dipeptide propensity could be more relevant to p53 functions. We also investigated the p53 evolution in the core domain (200–300). As can be seen in Sup-Table 5, the correlations of dipeptide propensities with the core domain evolution are close to those obtained using the entire protein sequence.
3.5. p53 mutation spectrum can be simulated using p53 evolution and reversal of evolution
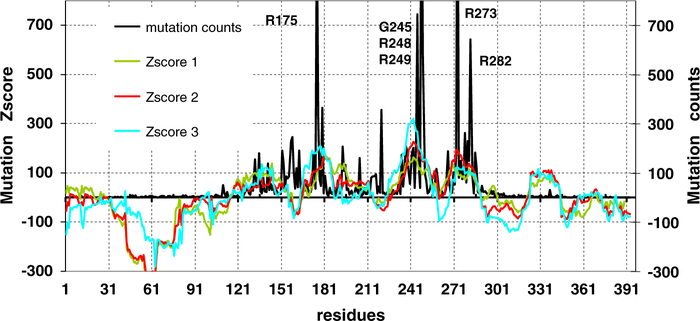
To further study the relationship between p53 evolution and its cancer-related mutations, we simulate the spectrum of human p53 mutations considering only the evolution of p53. We assign scores to dipeptides: higher scores are generated for low propensities, reflecting the observation that the most gained dipeptide pairs are those with low propensity in p53 sequences (see Sup material method). In Fig. 5, we compare the Z-score of the mutation index with the actual counts in the mutations database. Surprisingly, the simple concept captures most of the mutation spectrum. We further tested additional factors. First, we include the likelihood of a mutation to deviate from the human protein evolution. The second modification considers the ratio of non-synonymous/synonymous mutations reflecting the ‘standard’ driver mutation definition. The Zscore-2 and Zscore-3 with the additional factors are plotted in Fig. 5 to compare with the observed mutations as well. As can be seen in Fig. 5, accounting for p63/p73 dipeptide propensities increased the coverage of two hot spot regions, R248 and R273–R282 (Zscore 2, red line). The explicit term of synonymous mutations (Zscore-3) improves the fitting of the R175 and R248 regions, but decreases that of R273.
Fig. 5.
The simulated p53 mutation spectrum and the actual mutation frequencies counted from the IARC p53 mutation database Release R13. The number of hot spot positions was truncated from 750 for easy comparison with prediction (the actual counts are: 1152 for R175, 745 for G245, 1621 for R248, 573 for R249, 1551 for R273, 642 for R282). Several evolutionary factors may lower the mutation rate in the N- and C-termini (NC regions). In our simulation, on average the propensities of dipeptides gained from mutations are higher in the NC regions as compared to the core domain, indicating lower likelihood for mutations in the NC regions. The right Y-axis is the actual frequencies from IARC p53 mutation database Release R13.
The 1–100 N-terminal region is predicted to have a low mutation probability. Most of the C-terminal region is also predicted to have low probability; on the other hand, as Fig. 5 indicates, the 323–343 region in the tetramerization domain was overestimated. The overestimation could suggest that even though the tetramerization domain is highly conserved and the mutations could disrupt the tetramerization, the mutants are less prone to have gain-of-function effects [41]. Most core domain regions are correctly predicted to have high mutation probability, except that the short segments near residues 161 and 221 were underestimated. In our simulation, several factors are still not included, like tandem mutations (with hundreds observed). Nevertheless, accounting for the p53 and p63/p73 evolution, the likelihood of synonymous mutations almost reproduces the observed p53 mutations. Thus, we propose that the large number of mutations in p53 is due to p53 divergence from the human protein evolution, while its mutation probability tends to follow the mainstream.
Using the SIFT websever (http://sift.jcvi.org) [42], we also tested other evolutionary measures to predict deleterious mutations. The SIFT is a sequence homology based method to separate tolerant and intolerant mutations. Using wt p53 sequence as input, the SFIT predicts that most core domain residues are intolerant to mutations (Sup-Figure 2), while both N- and C- terminal regions may allow non-deleterious mutations. However, the SIFT cannot reproduce the observed p53 mutation spectrum.
4. Discussion
Current computational tools designed to predict the functional outcome of amino acid mutations often generate inconsistent and thus difficult to interpret results [43]. It is still challenging to distinguish driver from passenger mutations. The extensive network related to the functions of p53 complicates the delineation of the functions gained by the mutants, making the understanding of p53 mutants’ gain of function a difficult task. Many p53 mutants, hot spots and non-hot spots, promote a broad spectrum of cancer phenotypes [38]. The different cancer types have very different mutation spectra, and the mutant’s role as “driver” and “passenger” may change with the microenvironment of the cancer cell. Our study does not provide details of p53 mutations in different cancer types. Rather, our goal is to illuminate p53 evolution and somatic mutations.
The large number of p53 missense mutations collectively reveals mutational trends. The two major findings from this study are that the gain/loss ratio of amino acids following mutations resembles the average trend observed in the evolution of human proteins (Fig. 4A), and that the dipeptide gain/loss is counter to that in p53 evolution but tends to increase p63/p73-like dipeptide propensities (Fig. 3). These findings may help us look at p53 mutants’ gain-of-function and at cancer evolution from novel angles.
(1) The adaptation of p53 in cancer cells to selection pressure could involve a ‘reverse evolution’ toward to the p53 ancestral form. Such gain-of-function mutations can reverse the evolution [44] either leading to cryptic innovation (the formation of a new structure that mimics the old structure by gain-of-function mutations) or regaining lost functions [45] by recapturing a conserved network [45]. Therefore, p53 gain-of-function mutants can be related to ancestral p53 functions. We have shown that several hotspot mutants (R175H, R248Q, and R282W) resemble other species like C. elegans and C. briggsae. The function of p53 as a tumor suppressor is unlikely to be an ancestral function of the p53 gene [46]. A related p53 function in aging appears a more likely candidate for the ancestral function. However, it remains to be revealed what kinds of functions have been lost during p53 evolution.
(2) The correlation of p53 somatic mutations with the gain and loss of amino acids in human SNPs may indicate that cancer cell evolution in the human body is extremely rapid, mimicking the evolution of the human proteome. Alternatively, it may suggest that these mutations are selected because the mutants are more similar to p63. We have shown that the amino acid gain/loss ratio during human p53 evolution decreases whereas it increases in human proteins, and p63 follows the trend of human proteins (Sup-Figure 1B and 1C). In Table 2, we see that the mutations significantly decrease the p53 (most lost) dipeptide propensities, more than in p63/p73 thus decreasing the p53–p63/p73 gap (Table 3, Fig. 3). This trend is con-firmed by comparing the p53 gain-of-function mutants known to interfere with p63/p73 with other type of GOF and negative function groups (Sup-Table 4). The reversal of dipeptide frequencies in p53 vs. p63/p73 can be best illustrated by three dipeptide pairs (V/R, V/H, and R/Q) related to hotspot mutations: (a) The dipeptide pair V/R is the most frequently lost pair (Table 2). The dipeptide propensity of V/R in p53 has greatly increased during evolution, while the corresponding propensity in p63/p73 has been steadily decreasing (Fig. 2A). (b) The dipeptide V/H is the most frequently gained pair (among p53 somatic mutations), which has been eliminated during the evolution of p53 (but not of p63/p73, Fig. 2C). (c) The dipeptide R/Q is also a highly frequently gained pair (Table 3), and the occurrence of the R/Q has been eliminated during the evolution of p53 but increased during the p63/p73 evolution (Fig. 2B). Thus, our results support the hypothesis that the p53 gain-of-function mutants may interfere with p63/p73 transactivation either directly by forming complexes with p63 or p73, or by changing the p63/p73 networks [12] because p53 mutants are more similar to p63/p73 than to wild type p53.
It is easy to understand the dominant-negative effect of a p53 mutation since a point mutation can lead to p53 core domain unfolding or to disruption of p53-DNA interaction. However, it is still unclear how a point mutation can render a new oncogenic function of p53. Mutations can change the properties of both p53 mRNA and protein. It is known that p53 mRNA is also actively involved in the p53 regulation network [47], and it has been shown that p53 cancerous mutations exhibit selection for translational efficiency [40]. From the protein structural point of view, the notion that a cancer mutant gains ancestral functions (or p63/p73) because this substitution is found in an ancestral protein that shows otherwise relatively little sequence conservation is unlikely. For example, C. elegans has an equivalent R282W mutation but the structure in the vicinity of the mutation site, the L1 loop, is significantly different, which results in a completely different packing environment and structural constraints [28]. However, Cep-1 is still able to transactivate the consensus human p53 binding sites [39] with similar p53 functions [27], probably because the loop is highly flexibly fluctuating between different substates. Even though we do not know the evolutionary pressure on p53 to increase certain dipeptides (like V/R) and decrease others (like V/H), functional changes would accompany the sequence and structural evolution. Beyond the static crystal structures, function is exerted by protein conformational dynamics and conformational selection in transactivation [48–50]. Even if a point mutation induces a minor change in the static structure, it could lead to differences in conformational dynamics and affect the subsequent assembly of the transcriptional machinery via allosteric effects [51], where dynamics has been shown to play a key role [52]. A point mutation can also lead to re-distribution of the possible binding modes of p53 with the DNA [53]. These changes not only relate to ancestral p53 functions, but can also facilitate p53’s interference with for example p63/p73 [54].
Mutations shift the energy landscape of proteins, redistributing the relative populations of the conformational ensemble [55–57], including of disordered states [58], as in p53 monomers. These effects on protein conformations, i.e. their shapes and dynamics may help in the understanding of the apparent paradox of p53 gain-of-function mutants. The most frequent cancer-related mutations either result in lost DNA contacts that are strictly conserved in p63/p73 or conformationally destabilize the protein to an extent where it denatures at physiological temperature. p63 is more stable than wild type p53 [59], and particularly more than most of the p53 mutants. However, interestingly it has been found that p53 mutants are still able to interfere with the p63/p73 network and contribute to the gain of function in hepatocellular carcinoma [60]. Both R175H and R273H have V/R → V/H dipeptide change. While the R175H mutant has 98% native fold at 37 °C, the R273H is globally denatured [61]. Nonetheless, R174H and R273H similarly interact with p63/p73, interfering with the p63/p73 transcriptional activity [60], indicating that gained function (like function in general) is a property of the ensemble, and should not be viewed in terms of the static protein structure.
Evolutionary conservation has often been employed to predict deleterious mutations [42]. Our approach is based on mRNA sequences and is limited to single nucleotide mutations in each position. Our mutation indices are based on evolution–mutation pattern for the p53 family only, since other proteins may not necessarily share the p53 mutational trend: the frequently generated dipeptides by somatic mutations are more frequently lost in p53 evolution. In this sense, our mutation indices are not only a useful index of protein mutability; they can also be used to verify the dominant factor in p53 somatic mutation spectrum.
5. Conclusions
In conclusion, amino acid substitutions over evolutionary time and spontaneous mutations are different processes, in particular for key hub-encoding genes. Here we show that the p53 mutational spectrum can be defined by factors such as dipeptide propensities which relate to p53 evolution and mutational reversion of p53 evolution. The p53 gain-of-function mutants may relate to p53 ancestral function which was lost during evolution. Interfering with p63/p73 appears among the many mechanisms of p53 gain-of-function mutants. For example, gain of function of mutant p53 is partly mediated by its ability to form a complex with p63/p73 [62,63]. One possible explanation of the p53 mutant gain-of-function mechanism could be that such mutants shift closer to p63/p73 in terms of dipeptide propensities.
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbapap.2013.04.002.
Supplementary Material
Acknowledgements
This project has been funded in whole or in part with Federal funds from the National Cancer Institute, National Institutes of Health, under contract number HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. Huang and Yu were supported by grants from the National Natural Science Foundation of China (30570406, 30024001), the HI-tech Research and Development Program of China (2008AA02Z311), and the National Key Sci-Tech Special Projects of China (2008ZX10002–020). A. J. Levine thanks grants from the NCI and the Verto Institute.
Footnotes
This article is part of a Special Issue entitled: Computational Proteomics, Systems Biology & Clinical Implications. Guest Editor: Yudong Cai.
References
- [1].Sjoblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, Szabo S, Buckhaults P, Farrell C, Meeh P, Markowitz SD, Willis J, Dawson D, Willson JK, Gazdar AF, Hartigan J, Wu L, Liu C, Parmigiani G, Park BH, Bachman KE, Papadopoulos N, Vogelstein B, Kinzler KW, Velculescu VE, The consensus coding sequences of human breast and colorectal cancers, Science 314 (2006) 268–274. [DOI] [PubMed] [Google Scholar]
- [2].Greenman C, Stephens P, Smith R, Dalgliesh GL, Hunter C, Bignell G, Davies H, Teague J, Butler A, Stevens C, Edkins S, O’Meara S, Vastrik I, Schmidt EE, Avis T, Barthorpe S, Bhamra G, Buck G, Choudhury B, Clements J, Cole J, Dicks E, Forbes S, Gray K, Halliday K, Harrison R, Hills K, Hinton J, Jenkinson A, Jones D, Menzies A, Mironenko T, Perry J, Raine K, Richardson D, Shepherd R, Small A, Tofts C, Varian J, Webb T, West S, Widaa S, Yates A, Cahill DP, Louis DN, Goldstraw P, Nicholson AG, Brasseur F, Looijenga L, Weber BL, Chiew YE, DeFazio A, Greaves MF, Green AR, Campbell P, Birney E, Easton DF, Chenevix-Trench G, Tan MH, Khoo SK, Teh BT, Yuen ST, Leung SY, Wooster R, Futreal PA, Stratton MR, Patterns of somatic mutation in human cancer genomes, Nature 446 (2007) 153–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Stratton MR, Campbell PJ, Futreal PA, The cancer genome, Nature 458 (2009) 719–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hodis E, Watson IR, Kryukov GV, Arold ST, Imielinski M, Theurillat JP, Nickerson E, Auclair D, Li L, Place C, Dicara D, Ramos AH, Lawrence MS, Cibulskis K, Sivachenko A, Voet D, Saksena G, Stransky N, Onofrio RC, Winckler W, Ardlie K, Wagle N, Wargo J, Chong K, Morton DL, Stemke-Hale K, Chen G, Noble M, Meyerson M, Ladbury JE, Davies MA, Gershenwald JE, Wagner SN, Hoon DS, Schadendorf D, Lander ES, Gabriel SB, Getz G, Garraway LA, Chin L, A landscape of driver mutations in melanoma, Cell 150 (2012) 251–263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Vogelstein B, Lane D, Levine AJ, Surfing the p53 network, Nature 408 (2000) 307–310. [DOI] [PubMed] [Google Scholar]
- [6].Vousden KH, Prives C, Blinded by the light: the growing complexity of p53, Cell 137 (2009) 413–431. [DOI] [PubMed] [Google Scholar]
- [7].Riley T, Sontag E, Chen P, Levine A, Transcriptional control of human p53-regulated genes, Nat. Rev. Mol. Cell Biol 9 (2008) 402–412. [DOI] [PubMed] [Google Scholar]
- [8].Lanni C, Racchi M, Memo M, Govoni S, Uberti D, p53 at the crossroads between cancer and neurodegeneration, Free Radic. Biol. Med 52 (2012) 1727–1733. [DOI] [PubMed] [Google Scholar]
- [9].Tozluoglu M, Karaca E, Haliloglu T, Nussinov R, Cataloging and organizing p73 interactions in cell cycle arrest and apoptosis, Nucleic Acids Res. 36 (2008) 5033–5049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Marcel V, Dichtel-Danjoy ML, Sagne C, Hafsi H, Ma D, Ortiz-Cuaran S, Olivier M, Hall J, Mollereau B, Hainaut P, Bourdon JC, Biological functions of p53 isoforms through evolution: lessons from animal and cellular models, Cell Death Differ. 18 (2011) 1815–1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Tsai CJ, Ma B, Nussinov R, Protein–protein interaction networks: how can a hub protein bind so many different partners? Trends Biochem. Sci 34 (2009) 594–600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Belyi VA, Levine AJ, One billion years of p53/p63/p73 evolution, Proc. Natl. Acad. Sci. U. S. A 106 (2009) 17609–17610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Yang A, Kaghad M, Caput D, McKeon F, On the shoulders of giants: p63, p73 and the rise of p53, Trends Genet. 18 (2002) 90–95. [DOI] [PubMed] [Google Scholar]
- [14].Blandino G, Dobbelstein M, p73 and p63: why do we still need them? Cell Cycle 3 (2004) 886–894. [PubMed] [Google Scholar]
- [15].Suad O, Rozenberg H, Brosh R, Diskin-Posner Y, Kessler N, Shimon LJ, Frolow F, Liran A, Rotter V, Shakked Z, Structural basis of restoring sequence-specific DNA binding and transactivation to mutant p53 by suppressor mutations, J. Mol. Biol 385 (2009) 249–265. [DOI] [PubMed] [Google Scholar]
- [16].Koonin EV, Rogozin IB, Glazko GV, p53 gain-of-function: tumor biology and bioinformatics come together, Cell Cycle 4 (2005) 686–688. [DOI] [PubMed] [Google Scholar]
- [17].Cho Y, Gorina S, Jeffrey PD, Pavletich NP, Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations, Science 265 (1994) 346–355. [DOI] [PubMed] [Google Scholar]
- [18].Joerger AC, Fersht AR, Structural biology of the tumor suppressor p53, Annu. Rev. Biochem 77 (2008) 557–582. [DOI] [PubMed] [Google Scholar]
- [19].Murray-Zmijewski F, Lane DP, Bourdon JC, p53/p63/p73 isoforms: an orchestra of isoforms to harmonise cell differentiation and response to stress, Cell Death Differ. 13 (2006) 962–972. [DOI] [PubMed] [Google Scholar]
- [20].Graziano V, De Laurenzi V, Role of p63 in cancer development, Biochim. Biophys. Acta 1816 (2011) 57–66. [DOI] [PubMed] [Google Scholar]
- [21].Kadakia MP, de-Fromentel CC, Sabapathy K, The 5th International p63/p73 Workshop: much more than just tumour suppression, Cell Death Differ. 19 (2012) 549–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Glazko GV, Koonin EV, Rogozin IB, Mutation hotspots in the p53 gene in tumors of different origin: correlation with evolutionary conservation and signs of positive selection, Biochim. Biophys. Acta 1679 (2004) 95–106. [DOI] [PubMed] [Google Scholar]
- [23].Carter H, Chen S, Isik L, Tyekucheva S, Velculescu VE, Kinzler KW, Vogelstein B, Karchin R, Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations, Cancer Res. 69 (2009) 6660–6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P, The IARC TP53 database: new online mutation analysis and recommendations to users, Hum. Mutat 19 (2002) 607–614. [DOI] [PubMed] [Google Scholar]
- [25].Nedelcu AM, Tan C, Early diversification and complex evolutionary history of the p53 tumor suppressor gene family, Dev. Genes Evol 217 (2007) 801–806. [DOI] [PubMed] [Google Scholar]
- [26].Fernandes AD, Atchley WR, Biochemical and functional evidence of p53 homology is inconsistent with molecular phylogenetics for distant sequences, J. Mol. Evol 67 (2008) 51–67. [DOI] [PubMed] [Google Scholar]
- [27].Derry WB, Putzke AP, Rothman JH, Caenorhabditis elegans p53: role in apoptosis, meiosis, and stress resistance, Science 294 (2001) 591–595. [DOI] [PubMed] [Google Scholar]
- [28].Huyen Y, Jeffrey PD, Derry WB, Rothman JH, Pavletich NP, Stavridi ES, Halazonetis TD, Structural differences in the DNA binding domains of human p53 and its C. elegans ortholog Cep-1, Structure (Camb) 12 (2004) 1237–1243. [DOI] [PubMed] [Google Scholar]
- [29].Ou HD, Lohr F, Vogel V, Mantele W, Dotsch V, Structural evolution of C-terminal domains in the p53 family, EMBO J. 26 (2007) 3463–3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yang M, Sun J, Sun X, Shen Q, Gao Z, Yang C, Caenorhabditis elegans protein arginine methyltransferase PRMT-5 negatively regulates DNA damage-induced apoptosis, PLoS Genet. 5 (2009) e1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Vonderviszt F, Matrai G, Simon I, Characteristic sequential residue environment of amino acids in proteins, Int. J. Pept. Protein Res 27 (1986) 483–492. [DOI] [PubMed] [Google Scholar]
- [32].Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG, Clustal W and Clustal X version 2.0, Bioinformatics 23 (2007) 2947–2948. [DOI] [PubMed] [Google Scholar]
- [33].Bhasin M, Raghava GP, Classification of nuclear receptors based on amino acid composition and dipeptide composition, J. Biol. Chem 279 (2004) 23262–23266. [DOI] [PubMed] [Google Scholar]
- [34].Gu Q, Ding YS, Jiang XY, Zhang TL, Prediction of subcellular location apoptosis proteins with ensemble classifier and feature selection, Amino Acids 38 (2008) 975–983. [DOI] [PubMed] [Google Scholar]
- [35].Habib T, Zhang C, Yang JY, Yang MQ, Deng Y, Supervised learning method for the prediction of subcellular localization of proteins using amino acid and amino acid pair composition, BMC Genomics 9 (Suppl. 1) (2008) S16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Jordan IK, Kondrashov FA, Adzhubei IA, Wolf YI, Koonin EV, Kondrashov AS, Sunyaev S, A universal trend of amino acid gain and loss in protein evolution, Nature 433 (2005) 633–638. [DOI] [PubMed] [Google Scholar]
- [37].Mizuarai S, Yamanaka K, Kotani H, Mutant p53 induces the GEF-H1 oncogene, a guanine nucleotide exchange factor-H1 for RhoA, resulting in accelerated cell proliferation in tumor cells, Cancer Res. 66 (2006) 6319–6326. [DOI] [PubMed] [Google Scholar]
- [38].Weisz L, Oren M, Rotter V, Transcription regulation by mutant p53, Oncogene 26 (2007) 2202–2211. [DOI] [PubMed] [Google Scholar]
- [39].Schumacher B, Hofmann K, Boulton S, Gartner A, The C elegans homolog of the p53 tumor suppressor is required for DNA damage-induced apoptosis, Curr. Biol 11 (2001) 1722–1727. [DOI] [PubMed] [Google Scholar]
- [40].Waldman YY, Tuller T, Sharan R, Ruppin E, TP53 cancerous mutations exhibit selection for translation efficiency, Cancer Res. 69 (2009) 8807–8813. [DOI] [PubMed] [Google Scholar]
- [41].Chene P, The role of tetramerization in p53 function, Oncogene 20 (2001) 2611–2617. [DOI] [PubMed] [Google Scholar]
- [42].Kumar P, Henikoff S, Ng PC, Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm, Nat. Protoc 4 (2009) 1073–1081. [DOI] [PubMed] [Google Scholar]
- [43].Lee W, Yue P, Zhang Z, Analytical methods for inferring functional effects of single base pair substitutions in human cancers, Hum. Genet 126 (2009) 481–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Teotonio H, Rose MR, Perspective: reverse evolution, Evolution 55 (2001) 653–660. [DOI] [PubMed] [Google Scholar]
- [45].Cronk QC, Evolution in reverse gear: the molecular basis of loss and reversal, Cold Spring Harb. Symp. Quant. Biol (2009). [DOI] [PubMed] [Google Scholar]
- [46].Lu WJ, Amatruda JF, Abrams JM, p53 ancestry: gazing through an evolutionary lens, Nat. Rev. Cancer 9 (2009) 758–762. [DOI] [PubMed] [Google Scholar]
- [47].Ma B, Nussinov R, Regulating highly dynamic unstructured proteins and their coding mRNAs, Genome Biol. 10 (2009) 204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Pan Y, Tsai CJ, Ma B, Nussinov R, How do transcription factors select specific binding sites in the genome? Nat. Struct. Mol. Biol 16 (2009) 1118–1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Pan Y, Tsai CJ, Ma B, Nussinov R, Mechanisms of transcription factor selectivity, Trends Genet. 26 (2010) 75–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Ma B, Tsai CJ, Pan Y, Nussinov R, Why does binding of proteins to DNA or proteins to proteins not necessarily spell function? ACS Chem. Biol 5 (2010) 265–272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Sinha N, Nussinov R, Point mutations and sequence variability in proteins: redistributions of preexisting populations, Proc. Natl. Acad. Sci. U. S. A 98 (2001) 3139–3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Gunasekaran K, Ma B, Nussinov R, Is allostery an intrinsic property of all dynamic proteins? Proteins 57 (2004) 433–443. [DOI] [PubMed] [Google Scholar]
- [53].Ma B, Levine AJ, Probing potential binding modes of the p53 tetramer to DNA based on the symmetries encoded in p53 response elements, Nucleic Acids Res. 35 (2007) 7733–7747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Menendez D, Inga A, Resnick MA, The expanding universe of p53 targets, Nat. Rev. Cancer 9 (2009) 724–737. [DOI] [PubMed] [Google Scholar]
- [55].Kumar S, Ma B, Tsai CJ, Sinha N, Nussinov R, Folding and binding cascades: dynamic landscapes and population shifts, Protein Sci. 9 (2000) 10–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Tsai CJ, Kumar S, Ma B, Nussinov R, Folding funnels, binding funnels, and protein function, Protein Sci. 8 (1999) 1181–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Ma B, Kumar S, Tsai CJ, Nussinov R, Folding funnels and binding mechanisms, Protein Eng. 12 (1999) 713–720. [DOI] [PubMed] [Google Scholar]
- [58].Tsai CJ, Ma B, Sham YY, Kumar S, Nussinov R, Structured disorder and conformational selection, Proteins 44 (2001) 418–427. [DOI] [PubMed] [Google Scholar]
- [59].Klein C, Georges G, Kunkele KP, Huber R, Engh RA, Hansen S, High thermostability and lack of cooperative DNA binding distinguish the p63 core domain from the homologous tumor suppressor p53, J. Biol. Chem 276 (2001) 37390–37401. [DOI] [PubMed] [Google Scholar]
- [60].Schilling T, Kairat A, Melino G, Krammer PH, Stremmel W, Oren M, Muller M, Interference with the p53 family network contributes to the gain of oncogenic function of mutant p53 in hepatocellular carcinoma, Biochem. Biophys. Res. Commun 394 (2010) 817–823. [DOI] [PubMed] [Google Scholar]
- [61].Bullock AN, Fersht AR, Rescuing the function of mutant p53, Nat. Rev. Cancer 1 (2001) 68–76. [DOI] [PubMed] [Google Scholar]
- [62].Liu K, Ling S, Lin WC, TopBP1 mediates mutant p53 gain of function through NF-Y and p63/p73, Mol. Cell. Biol 31 (2011) 4464–4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Xu J, Reumers J, Couceiro JR, De Smet F, Gallardo R, Rudyak S, Cornelis A, Rozenski J, Zwolinska A, Marine JC, Lambrechts D, Suh YA, Rousseau F, Schymkowitz J, Gain of function of mutant p53 by coaggregation with multiple tumor suppressors, Nat. Chem. Biol 7 (2011) 285–295. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.