Abstract
Adults older than 65 account for most of the deaths caused by respiratory influenza A virus (IAV) infections, but the underlying mechanisms for this susceptibility are poorly understood. IAV RNA is detected by the cytosolic sensor retinoic acid-inducible gene I (RIG-I), which induces the production of type I interferons (IFNs) that curtail the spread of the virus and promote the elimination of infected cells. We have previously identified a marked defect in the IAV-inducible secretion of type I IFNs, but not proinflammatory cytokines, in monocytes from healthy older (>65 years) human donors. Here, we found that monocytes from older adults exhibited decreased abundance of the adaptor protein TRAF3 because of its increased proteasomal degradation with age, thereby impairing the primary RIG-I signaling pathway for the induction of type I IFNs. We determined that monocytes from older adults also failed to effectively stimulate the production of the interferon regulatory transcription factor IRF8, which compromised IFN induction through secondary RIG-I signaling. IRF8 played a central role in IFN induction in monocytes, because knocking down IRF8 in monocytes from younger adults was sufficient to replicate the IFN defects observed in monocytes from older adults, whereas restoring IRF8 expression in older adult monocytes was sufficient to restore RIG-I-induced IFN responses. Aging thus compromises both the primary and secondary RIG-I signaling pathways that govern expression of type I IFN genes, thereby impairing antiviral resistance to IAV.
Introduction
Influenza A viruses (IAV) are human respiratory pathogens that cause half a million deaths worldwide annually (1, 2). Older adults over the age of 65 exhibit increased susceptibility to IAV, accounting for 90% of annual IAV-related fatalities (2, 3). The mechanisms that govern this increase in IAV-associated mortality with age are complex and are an important area of study. We previously identified a defect in the induction of type I interferon (IFN) and antiviral gene expression in response to IAV infection in older human monocytes (4). Upon IAV infection, monocytes are rapidly recruited to the respiratory tract, where they differentiate into dendritic cells (DCs) and macrophages (5-9), serving as a major source of inflammatory and antiviral cytokines and antigen-presenting cells (5, 10). Understanding the basis for the age-related defect in monocyte function will thus provide valuable insight into defects in innate and adaptive antiviral immunity that compromise IAV control in older individuals.
Within the infected target cell, IAV viral RNA bearing a 5’ triphosphate motif is detected by the cytosolic sensor retinoic acid-inducible gene I (RIG-I), an RNA helicase that is activated by recognition of this 5’-ppp RNA (11-13). RIG-I activation initiates a series of signaling events mediated by the adaptor protein mitochondrial antiviral-signaling (MAVS) that lead to the phosphorylation and both hetero- and homodimerization of the interferon regulatory transcription factors (IRFs) IRF3 and IRF7 (14, 15). These factors translocate to the nucleus where they drive the rapid expression and subsequent secretion of type I IFNs, thereby initiating an antiviral response. IFN induction upon RIG-I signaling occurs in three phases (16). The primary phase relies on the activation of constitutively-expressed RIG-I, IRF3, and IRF7, resulting in the phosphorylation of IRF3 and IRF7 by the kinase TBK1 that facilitates their dimerization, nuclear translocation, and activation of primary target genes including IFN-β (16). IRF3 and IRF7 are thus class I transcription factors. The secondary phase of RIG-I signaling utilizes de novo synthesized transcription factors (class II) that activate the transcription of secondary response genes. IRF7 serves as both a constitutively expressed (class I) and an inducible (class II) transcription factor mediating IFN induction. Feedback amplification of IFN signaling occurs when secreted type I IFNs bind to the interferon-α/β receptor (IFNAR) to stimulate the assembly of the interferon-stimulated gene factor 3 (ISGF3) transcription factor complex, which activates transcription of IRF7 to further amplify type I IFN induction (17). Of note, in addition to these transcription factors that function in most all nucleated cells, the myeloid-specific transcription factor IRF8 (class III) can participate in IFN induction in monocyte and dendritic cell lineages by serving as a scaffold that stabilizes transcription from the IFN promoter (18, 19).
Defects in any one of these phases of IFN induction may underlie age-linked defects in IFN production. Using primary peripheral blood monocytes from healthy younger (age 21-30) and older (age 65-89) adult donors, we sought to clarify which aspects of IFN induction are disrupted by the aging process and to elucidate the underlying molecular mechanisms. We determined that increased basal proteasomal degradation of the adaptor protein tumor necrosis factor receptor–associated factor 3 (TRAF3) in monocytes from older adults was associated with impaired primary IFN induction during the first phase of RIG-I signaling. We further observed defective induction of IRF8 in monocytes from older adults, which compromised the secondary phase of IFN induction downstream of RIG-I. In contrast, IFNAR-driven signaling for IRF7 induction remained intact in monocytes from older adults. Restoring the expression of TRAF3 or IRF8 was sufficient to enhance the ability of monocytes from older adults to produce IFN in response to RIG-I stimulation. These results contribute to the understanding of the multifaceted defects in innate antiviral signaling that arise with age and highlight potential targets for therapeutic intervention.
Results
Monocytes from Older Adults Exhibit Cell-Intrinsic Defects in IFN Induction
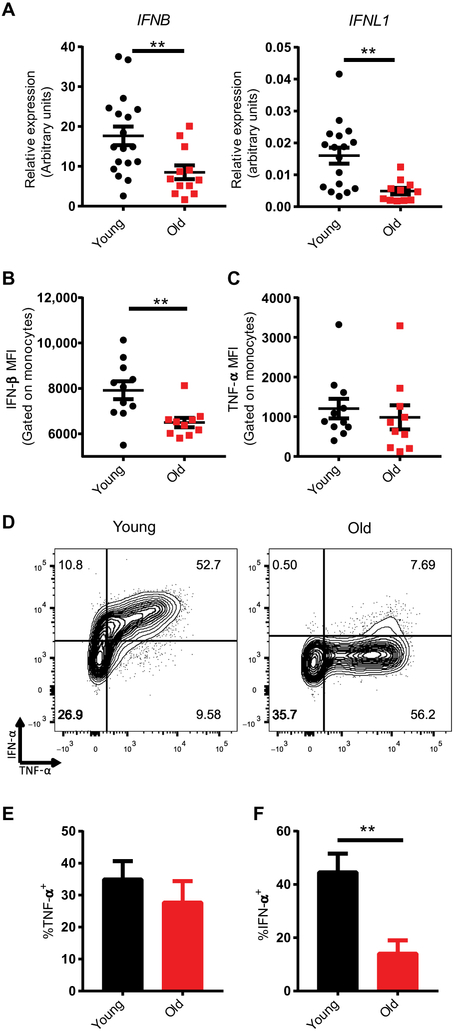
To analyze the defects in RIG-I signaling that arise with age, we quantified RIG-I signaling responses in peripheral blood monocytes from younger (age: 21-30 years) or older (age: 65-89 years) healthy human donors (table S1; fig. S1) following transfection with a 5’-ppp-double-stranded RNA (dsRNA) ligand specific to RIG-I (20). IFN induction in human monocytes depends on the 5’ triphosphate motif of this ligand, as illustrated by the failure of an identical dsRNA ligand bearing a 5’ hydroxyl group to induce an IFN response, consistent with the RIG-I specificity of this ligand (fig. S2). Both mRNA expression and subsequent production of IFN-β protein were reduced in monocytes from older adults as compared to those from younger adults, whereas the production of tumor necrosis factor α (TNF-α), which is an inflammatory cytokine induced by RIG-I stimulation, was comparable in the two monocyte populations (Fig. 1A-C). Expression of IFNL1, which encodes IFN-λ1, a type III IFN closely related to IFN-β, was also reduced in older monocytes, emphasizing the broad impairment of antiviral IFN induction with age (Fig. 1A). Basal expression of IFNB, which encodes IFN-β, in monocytes from older and younger donors was comparable, highlighting the inducible nature of this impairment (fig. S3). This defect in inducible IFN production could be a consequence of a cell-intrinsic defect in IFN production downstream of RIG-I signaling in monocytes from older adult donors, or it could be a consequence of the loss of a specialized subset of IFN-secreting, but not TNF-α–secreting, monocytes in older individuals. To differentiate between these two possibilities we performed intracellular cytokine staining analyses of RIG-I–stimulated monocytes to identify the sources of inflammatory and antiviral cytokines (Fig. 1D). This analysis revealed that the same population of monocytes in young donors produced both inflammatory TNF-α and antiviral IFN-α, and although monocytes from older donors retained the ability to induce TNF-α in response to RIG-I stimulation (Fig. 1D, E), their ability to induce IFN-α was markedly impaired (Fig. 1F). Pretreating monocytes with recombinant human IFN-β was insufficient to ablate age-related differences in RIG-I–induced IFN secretion, indicating that the phenotype we observed was not the consequence of increased IFN in younger monocytes’ supernatants at early time points, leading to exaggerated IFN secretion from these cells at later time points (fig. S4). Together these data indicate that an age-linked cell-intrinsic defect in IFN induction lies downstream of RIG-I activation, and we next investigated the possible causes of this defect in a sequential manner.
Fig. 1. Monocytes from older humans exhibit cell-intrinsic impairment of RIG-I–induced type I IFN expression.
Human monocytes enriched from the blood of younger (age 20-30; n=18) and older (age 65-89; n=12) healthy donors were transfected with a RIG-I–specific 5’-ppp 14-bp dsRNA ligand. (A) RNA was isolated from monocytes 12 hours after stimulation and analyzed by quantitative polymerase chain reaction (qPCR) to measure IFNB and IFNL1 expression. Expression values for each donor were normalized to HPRT. (B) Monocytes were treated with a protein transport inhibitor cocktail 3 hours after stimulation, and at 6 hours poststimulation cells were fixed, permeabilized, and labeled for intracellular IFN-β or (C) TNF-α protein, the median fluorescence intensity (MFI) of which was measured using flow cytometry. (D) Representative flow cytometry plots of fixed and permeabilized monocytes stained for both intracellular TNF-α and IFN-α. Using these plots, the frequency of (E) TNF-α+ monocytes and (F) IFN=α+ monocytes were calculated. Data are presented as means ± SEM. ** P<0.01; Student t-test.
Aging Impairs the Rapid Phosphorylation of IRF3 and IRF7 upon RIG-I Stimulation
We first investigated the stage at which the older monocytes fail to produce type I IFNs. Type I IFN production depends on RIG-I signaling through MAVS. Because TNF-α production was not perturbed with increasing age (Fig. 1C), we concluded that RIG-I expression and activation of MAVS remain intact in older adults. Thus, we reasoned that the defect in IFN production may arise due to defects in either the IFN-inducing arm of the primary RIG-I signaling pathway downstream of MAVS, the secondary RIG-I signaling pathway that requires de novo synthesis of a transcription factor, and/or the feedforward amplification of IFN production in response to IFNAR signaling.
We examined primary RIG-I signaling downstream of MAVS for defects compromising type I IFN induction. There were no age-dependent changes in baseline expression of the RIG-I sensor (encoded by DDX58) or of the key interferon regulatory transcription factors (IRFs) IRF3 or IRF7, which promote IFN gene expression (21), at either the RNA or protein level (fig. S5). In contrast, upon RIG-I stimulation, the phosphorylation of both IRF3 (Fig. 2A; fig. S6) and IRF7 (Fig. 2B) was impaired in monocytes from older adults. Phosphorylation of these transcription factors is necessary for their dimerization and subsequent nuclear translocation, suggesting that defects in primary RIG-I signaling through the phosphorylation of IRF3 and IRF7 contribute to the underlying impairments in IFN induction with age.
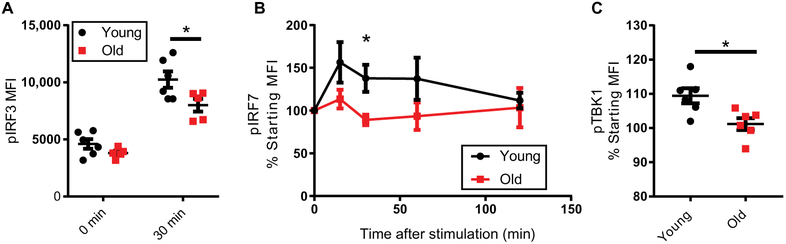
Fig. 2. TBK1, IRF3, and IRF7 phosphorylation in response to RIG-I stimulation is impaired in older monocytes.
(A) Monocytes from younger (n=6) and older (n=5) donors were transfected with a RIG-I–specific ligand and labeled for intracellular phosphorylated IRF3 (pIRF3) or (B) phosphorylated IRF7 (pIRF7) at the indicated timepoints (younger n=9, older n=7). (C) Phosphorylated TBK1 (pTBK1) levels in monocytes from older (n=6) or younger (n=6) donors was assessed 30 minutes after transfection with a RIG-I–specific ligand. Data are presented as means ± SEM. * P<0.05; Student T-test, Two-way ANOVA, or, for pIRF7, a linear mixed model (with a Toeplitz covariance structure).
Because TBK1 is a key kinase that directly phosphorylates IRF3 and IRF7 in response to RIG-I stimulation (22), we next assessed the activation status of TBK1 itself in our monocyte samples. We found that TBK1 phosphorylation was significantly decreased in monocytes from older donors 30 minutes after RIG-I stimulation (Fig 2C). This is consistent with the altered IRF phosphorylation dynamics that we observe with age, suggesting that the impaired activation of IRF3 and IRF7 in monocytes from older adults is a consequence in the impaired activation of TBK1 in these same cells.
Proteasomal Degradation of TRAF3 in Older Adult Monocytes Impairs IFN Induction
TRAF3 is an adaptor protein that has E3 ubiquitin ligase activity and acts downstream of RIG-I signaling to mediate activation of TBK1, which in turn phosphorylates IRF3 and IRF7 (23, 24). Notably, TRAF3 deficiency does not affect the induction of proinflammatory genes or the activation of nuclear factorκB (NF-κB) downstream of RIG-I (25). Given this key role for TRAF3 upstream of TBK1, IRF3, and IRF7 activation, we quantified the amount of TRAF3 protein in our monocyte samples. We identified a significant reduction in basal TRAF3 protein abundance in monocytes from older donors (Fig. 3A, B) that correlated with decreased IFN-β induction upon RIG-I stimulation (fig. S7). Although TRAF3 protein abundance decreased with age, TRAF3 mRNA expression was comparable between age groups (Fig. 3C) indicating that this decline in TRAF3 protein is post-transcriptionally regulated.
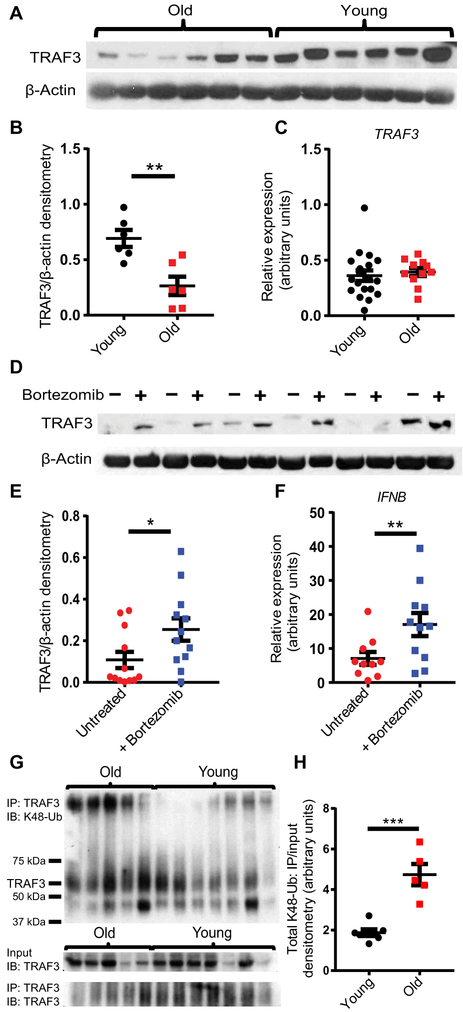
Fig. 3. Increased proteasomal degradation of TRAF3 in older monocytes contributes to impaired IFN Induction.
(A) TRAF3 and β-actin abundance in older (n=6) and younger (n=6) unstimulated monocytes were measured by Western blotting, and (B) the abundance of TRAF3 was quantified by densitometry and normalized to β-actin using ImageJ. (C) TRAF3 expression was measured by qPCR in unstimulated younger (n=19) and older (n=11) monocytes. Expression values for each donor were normalized to HPRT. (D) Older monocytes (n=12) were treated with bortezomib for 4 hours before TRAF3 and β-actin abundance were measured by Western blotting and (E) quantified by densitometry. (F) Older monocytes (n=12) were pretreated with bortezomib for 4 hours prior to transfection with a RIG-I–specific ligand for 6 hours, and IFNB expression was quantified by qPCR. (G) TRAF3 was immunoprecipitated from lysates of old (n=5) and young (n=7) human monocytes, and both input and TRAF3 immunoprecipitates (IP: TRAF3) were subjected to Western blotting using antibodies recognizing TRAF3 (IB: TRAF3) and K48-polyubiquitin (IB: K48-Ub). (H) Quantification of K48-polyubiquitin by densitometry. Data are presented as means ± SEM. * P<0.05,** P<0.01, *** P<0.001; Paired or Unpaired Student t-test.
Differential ubiquitination of TRAF3 is known to play a key role in TRAF3 signaling dynamics. Polyubiquitination of TRAF3 at Lys48 (K48-polyubiquitination) leads to its proteasomal degradation, facilitating alternative activation of NF-κB in response to CD40 signaling, whereas a distinct K63-polyubiquitintion is necessary for IRF activation in response to TLR or RIG-I signaling (24, 26). In order to determine whether proteolysis contributes to the reduced TRAF3 present in the monocytes from older adults, we treated these cells with the proteasome inhibitor bortezomib for 4 hours. This treatment was sufficient to increase the total amount of TRAF3 in monocytes from older donors, supporting a role for proteasomal degradation in the observed age-related basal decrease in TRAF3 protein abundance (Fig. 3D,E). Treatment of monocytes from older donors with bortezomib was sufficient to enhance the induction of IFNB expression in response to stimulation with a RIG-I–specific ligand (Fig. 3F). Bortezomib treatment also reduced viral gene expression after infection with the PR8 strain of IAV (fig. S8). To investigate whether TRAF3 degradation in older monocytes was associated with increased K48-polyubiquitination, we measured the abundance of K48-linked polyubiquitin (K48-Ub) in TRAF3. immunoprecipitated from old and young donor monocytes. We found that K48-Ub abundance was significantly higher in samples of TRAF3 immunoprecipitated from older donor monocytes, despite no apparent age-related increase in total K48-Ub (Fig. 3G-H). These results are consistent with a model wherein increased basal proteasomal degradation of TRAF3 due to increased K48-polyubiquitination in monocytes from older donors compromises the activation of TBK1 and subsequent phosphorylation of the transcription factors IRF3 and IRF7 upon RIG-I stimulation, leading to impaired IFN induction and a consequent reduction in viral control.
Decreased TRAF3 protein abundance in monocytes from older donors may compromise signaling downstream of multiple distinct cytosolic nucleic acid sensors. To test this possibility, we assessed IFN signaling responses to cytosolic DNA in monocytes from old and young donors (fig. S9). We did not detect a statistically significant difference in these responses, suggesting that further study into the role of TRAF3 in distinct signaling contexts with age is needed in order to clarify why decreased TRAF3 levels do not adversely affect IFN induction in response to all nucleic acid stimuli.
Secondary IFN Induction upon RIG-I signaling is Defective in Older Adult Monocytes
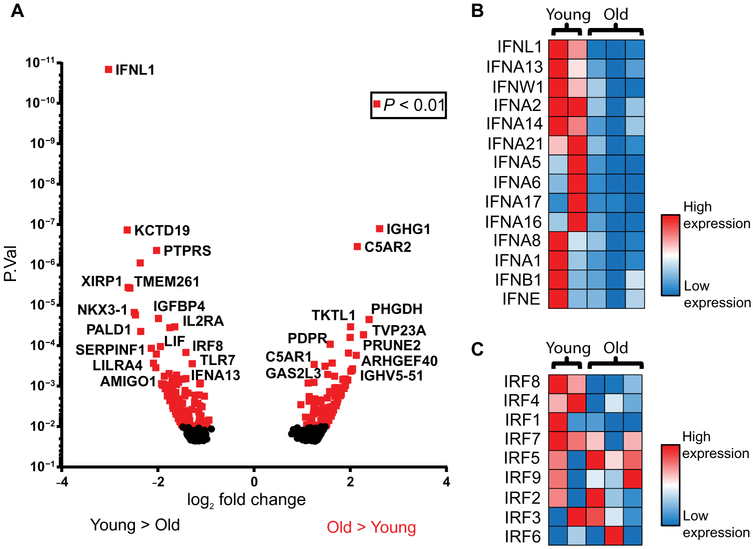
Defects in primary RIG-I signaling caused by TRAF3 degradation would be expected to lead to defects in secondary IFN induction upon RIG-I signaling. Because secondary RIG-I signaling requires the de novo synthesis of transcription factors, we performed RNA sequencing analysis of monocytes from older and young adults that had been stimulated with a RIG-I–specific ligand for 6 hours in order to screen for alterations in expression of class II transcription factors that could also contribute to age-dependent IFN induction defects (Fig. 4A). Pathway analyses revealed that IFNs, RIG-I, IRFs, and IAV-related signaling components were among the most differentially expressed transcripts with age (fig. S10). All sequenced type I and type III IFN genes exhibited significantly reduced expression or trended toward decreased expression in monocytes from older donors relative to younger donors (Fig. 4B, table S2), supporting the existence of a broad defect in the transcriptional activation of antiviral IFNs with age. We identified no significant differences in IRF3 or IRF7 induction at the mRNA level (Fig. 4C, table S3). However, examination of larger number of donors revealed a statistically significant decrease in the abundance of IRF7 protein (fig. S11).
Fig. 4. RNA-seq reveals impairment of the amplification phase of the IFN response to RIG-I stimulation in older monocytes.
Human monocytes from younger (n=2) and older (n=3) healthy donor blood were transfected with a RIG-I–specific 5’-ppp 14-bp dsRNA ligand. After 6 hours, RNA was isolated from these cells and was used for exploratory RNA sequencing. (A) Volcano plot of differentially regulated genes. Normalized values of transcripts encoding both (B) type I/III IFNs and (C) IRFs were compiled into heat maps using Microsoft Excel in order to highlight trends in differential induction of these genes.
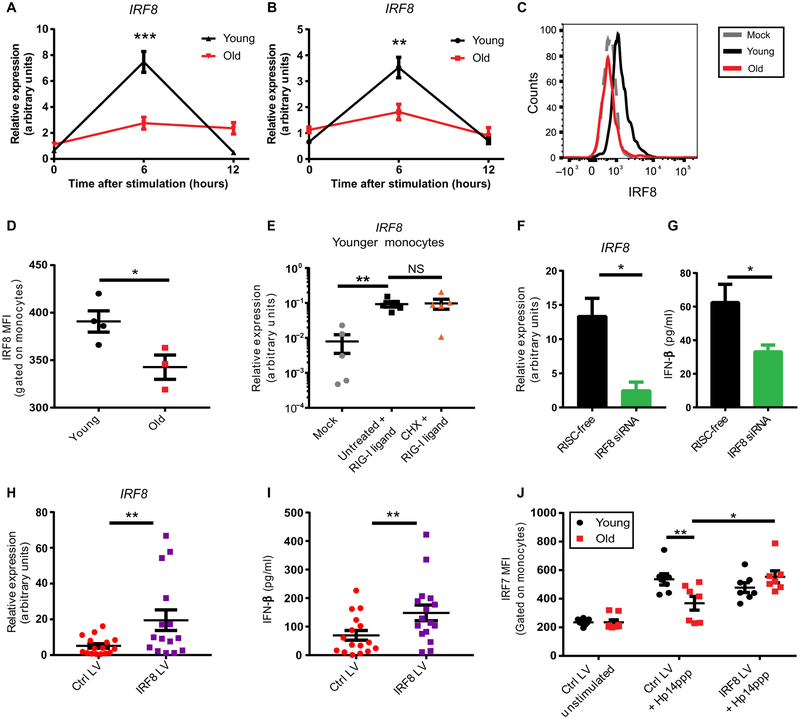
Notably, the most differentially expressed IRF member in our RNA-seq data was IRF8, the expression of which was decreased in older adult monocytes (Fig. 4C). Although IRF8 is most frequently studied in the context of monocyte and DC lineage development, it has also been shown to play a key role in the induction of IFNs in human monocytes, DCs, and plasmacytoid DCs (18, 19, 27). We observed no differences in basal IRF8 mRNA or protein abundance with age (fig. S12); however, induction of IRF8 was markedly impaired in monocytes from older donors in response to both RIG-I stimulation (Fig. 5A) and IAV infection (Fig. 5B). This induction depended on RIG-I (fig. S13), and RIG-I–induced IRF8 expression was transient, with maximal IRF8 expression 6 hours post-stimulation, consistent with previous reports of IRF8 induction kinetics indicating peak IRF8 induction 4 hours after LPS stimulation (28, 29). An age-linked difference in IRF8 induction was also evident at the protein level (Fig. 5C-D). IRF8 expression in stimulated monocytes was positively correlated with IFN-β secretion by monocytes from those same donors (fig. S14), supporting a role for differential IRF8 induction in IFN production. We found that IRF8 induction upon RIG-I stimulation was enhanced in older monocytes pretreated with bortezomib, suggesting that decreased IRF8 with age may be at least partially a consequence of increased proteasomal degradation of TRAF3 (fig. S15). To assess whether IRF8 is induced de novo as a primary transcript in response to RIG-I signaling, we assessed IRF8 mRNA expression in monocytes from younger adults treated with cyclohexamide (CHX) to inhibit protein translation. IRF8 induction remained intact upon CHX treatment (Fig. 5E), supporting its status as a primary RIG-I signaling transcriptional target that is activated in response to IRF3 or other class I transcription factors.
Fig. 5. Impaired IRF8 induction compromises amplification of the IFN response in older monocytes, and restoring IRF normalizes this response.
Monocytes from younger (n=19) and older (n=12) human donors were (A) transfected with a RIG-I–specific ligand or (B) infected with PR8 IAV, and IRF8 expression was measured by qPCR at the indicated timepoints. Expression values for each donor were normalized to HPRT. (C, D) Younger (n=4) and older (n=3) monocytes were transfected with a RIG-I–specific ligand for 6 hours, IRF8 protein abundance was measured by flow cytometry (C), and mean fluorescence intensity (MFI) values (D) were quantified. Data is representative of two independent experiments. (E) Monocytes from 5 younger donors were left untreated or were treated with cyclohexamide (CHX) for 4 hours prior to mock transfection or RIG-I stimulation for 6 hours, after which IRF8 expression was quantified by qPCR. (F, G) Monocytes from younger donors were treated with an IRF8-specific siRNA or RNA-induced silencing complex (RISC)-free control for 48 hours, then stimulated with a RIG-I–specific ligand. IRF8 expression was quantified by qPCR after 6 hours of RIG-I stimulation (F), and IFN-β secreted into the supernatant was quantified by ELISA after 12 hours of RIG-I stimulation. Data is representative of two independent experiments. (H, I) Monocytes from older donors (n=16) were transfected with an IRF8 or control lentiviral (LV) construct for 48 hours, then stimulated with a RIG-I–specific ligand. IRF8 expression was quantified by qPCR after 6 hours of RIG-I stimulation (H), and secreted IFN-β was measured by ELISA after 12 hours of RIG-I stimulation (I). (J) Monocytes from younger (n=7) and older (n=7) donors were transfected with a control or IRF8 lentiviral construct for 48 hours, stimulated with a RIG-I–specific ligand for 6 hours, and IRF7 MFI was quantified by flow cytometry. Data are means ± SEM. * P<0.05,** P<0.01, ***P<0.001; Unpaired or Paired Student t-test or Two-way Repeated Measures ANOVA.
Impaired IRF8 Induction Compromises IFN Feedback Amplification
To determine the role of IRF8 in IFN-β induction in primary human monocytes, we used an IRF8-specific siRNA to knock down IRF8 in monocytes from young donors (Fig. 5F). This knockdown was sufficient to reduce RIG-I–induced IFN-β secretion in monocytes from younger donors to levels similar in magnitude to those observed in monocytes from older donors (Fig. 5G) (4). Conversely, using a lentivirus to express IRF8 in older adult monocytes (Fig. 5H) was sufficient to bolster IFN-β secretion by these cells (Fig. 5I). Increasing IRF8 expression was also sufficient to restore IRF7 protein abundance in monocytes from older donors but did not alter IRF7 dynamics in younger controls (Fig. 5J), highlighting an important role for decreased IRF8 induction as a mediator of observed impairments to IFN induction in older adults. Collectively, these results show that monocytes from older adults have impaired RIG-I primary signaling due to lower TRAF3 protein abundance. This leads to impaired induction of IRF8, which mediates activation of IFN genes in secondary RIG-I signaling.
Induction of IRF8 downstream of IFNAR signaling is impaired with age
To test whether the IFNAR-mediated feedback loop for IFN synthesis may be defective in older adults, we stimulated monocytes from younger and older donors with recombinant IFN-β (rIFN-β). We observed no general age-related impairment in interferon-stimulated gene (ISG) induction in monocytes from older relative to younger donors (fig. S16A-B), indicating that the ability of rIFN-β to protect monocytes from IAV infection did not depend on the age of the monocyte donor (fig. S16C-D). In addition, induction of the IFN-stimulating transcription factor IRF7 remained intact in monocytes from older donors following rIFN-β stimulation (fig. S16E). These results indicate that general IFNAR signaling and consequent induction of ISGs and antiviral responses remain largely intact with increasing age.
To examine whether IFNAR-mediated amplification of RIG-I signaling may be impaired in monocytes from older adults, we examined whether rIFN-β induced IRF8 expression. We found that rIFN-β induced IRF8 expression in monocytes from both old and young donors (fig. S17A). However, when compared on a per-person basis, we found that IRF8 induction in response to recombinant IFN-β treatment decreased with age (fig. S17B).
Decreased IRF8 induction in response to both RIG-I stimulation and IFN treatment suggests that IRF8 may be differentially regulated at the epigenetic level with age such that its induction is impaired in multiple distinct contexts. Given previous reports that the IRF8 locus exhibits increased DNA methylation in hematopoietic stem cells from older human donors (30), we hypothesized that altered DNA methylation at the IRF8 locus constrains inducible IRF8 activity in older monocytes. To test this hypothesis, we performed whole-genome methylation profiling with a chip-based assay platform (fig. S18). We did not observe any differential methylation of the IRF8 locus in older monocytes, suggesting that altered epigenetic control of IRF8 induction in older monocytes is not regulated at the level of DNA methylation.
Although the underlying mechanisms governing age-related shifts in IRF8 induction remain to be fully clarified, these data suggest that primary defects in the RIG-I signaling response (Fig. 1-3) may be further amplified by a reduction in IFNAR-induced IRF8 expression in older monocytes. The decreased induction of IRF8 thereby compromises both secondary RIG-I signaling as well as IFNAR-dependent amplification of the IFN response, ultimately resulting in marked impairment of IFN production in older adults.
Discussion
It is well known that aging is associated with increased influenza morbidity and mortality in humans, and multiple factors contribute to this increase in susceptibility. Many studies have identified serious defects in the development of effective immunological memory in older adults (31), and these defects are associated with impaired vaccine responsiveness and a consequent impediment to preventative medical care (32, 33). In contrast to the extensive examination of aging and the adaptive immune response to IAV, how innate immunity is shaped by aging has remained incompletely understood (34). Given our previous demonstration that IFN production and antiviral gene induction in response to influenza infection are markedly impaired in monocytes from older human donors (4), characterizing the underlying nature of this defect is vital to better develop treatment strategies for this vulnerable population.
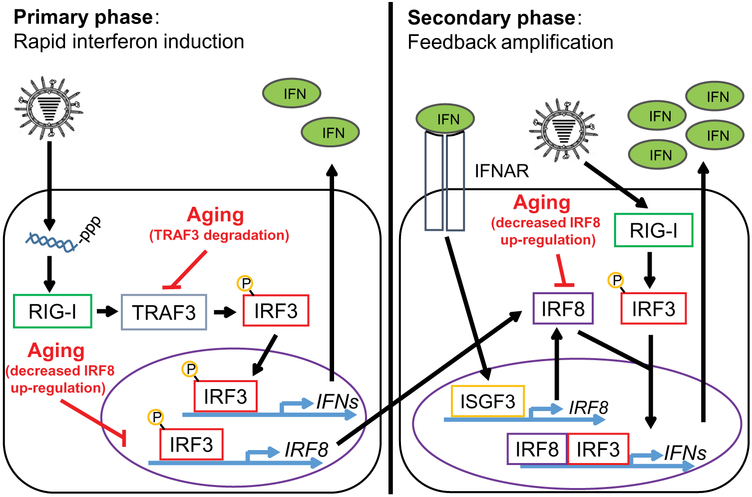
Using primary human monocytes from older and younger healthy donors, we demonstrated that monocytes from older adults exhibit a cell-intrinsic defect in IFN production while maintaining robust production of inflammatory cytokines. We identified three key factors that underlie this decrease in IFN production with age, each of which acts at a different stage in the IFN induction pathway (Fig. 6). Increased K48-polyubiquitin–mediated proteasomal degradation of the TRAF3 adaptor protein in older monocytes impairs the primary induction of IFN expression downstream of RIG-I signaling, limiting the ability of these cells to quickly activate antiviral defense mechanisms. This leads to impaired expression of the secondary transcription factor IRF8, further dampening the feed-forward amplification of IFN transcription. Finally, monocytes from older donors are impaired in their ability to induce IRF8 expression downstream of IFNAR signaling, further diminishing the positive feedback amplification of IFN responses. Taken together, our results demonstrate that the ability of older adults to induce IFNs is compromised at multiple levels. Our data also indicate that increasing expression of the secondary transcription factor IRF8 in older monocytes was sufficient to restore IFN induction in response to RIG-I stimulation, highlighting the possibility of targeting these defects individually to restore effective antiviral immunity to at-risk individuals.
Fig. 6. A model of aging-related impairment of IFN Induction in monocytes.
During the primary phase of the interferon response (minutes to hours after IAV infection or RIG-I stimulation), RIG-I mediates phosphorylation of the transcription factor IRF3 through a mechanism that depends on TRAF3. Once phosphorylated, IRF3 homodimerizes and mediates the rapid transcription of genes encoding type I IFNs, which are secreted from the cell, and promote the expression of other primary transcripts such as IRF8. Aging is associated with decreased TRAF3 protein abundance as a consequence of increased K48-polyubiquitin–mediated TRAF3 proteasomal degradation, which impairs IRF3 phosphorylation and consequent IFN secretion. During the secondary feedback phases of the interferon response, secreted IFNs and continued RIG-I signaling promote the further induction of IRF8 within the cell. IFNs produced during the primary response activate the interferon-α/β receptor (IFNAR) to stimulate the assembly of the interferon-stimulated gene factor 3 (ISGF3) complex, which further stimulates the production of IRF8. IRF8 cooperates with other IRF family members to amplify the production of IFNs, producing a robust antiviral interferon response. Aging impairs IRF8 induction, thereby further compromising interferon production.
Given its central role as an adaptor molecule in the RIG-I pathway, the lower abundance of TRAF3 is the most proximal cause of the decreased activation of the downstream kinase TBK1 in monocytes from older individuals, and this impairment in TBK1 activation consequently impairs IRF3 and IRF7 phosphorylation in these same cells. TRAF3 protein abundance is decreased with age despite preserved TRAF3 mRNA expression, consistent with a model wherein TRAF3 undergoes increased proteasomal degradation in older monocytes. This is supported by the fact that the proteasome inhibitor bortezomib increased TRAF3 abundance and restored IFN expression in monocytes from older donors. In further support of this model, TRAF3 from unstimulated monocytes from older individuals is associated with a greater amount of K48-polyubiquitin. K48-polyubiquitination is known to mediate TRAF3 degradation and is distinct from the K63-polyubiquitin modification necessary to facilitate TRAF3-mediated TBK1 activation upon RIG-I or TLR stimulation (24, 26, 35-37). In addition to leading to decreased TRAF3 protein abundance, whether the aging process may affect other aspects of TRAF3 signaling such as its E3 ubiquitin ligase activity remains to be determined, and as such a more extensive investigation of how aging shapes TRAF3 expression and function is an important area for future research.
TRAF3 has been found to facilitate IFN production downstream of many pattern recognition receptors (PRRs), including both Toll-like receptors (TLRs) and RIG-I–like receptors (RLRs) (24). In addition to our findings highlighting defective RLR signaling with age, multiple previous reports have identified defects in TLR signaling responses in myeloid cells from older individuals (34). IFN induction by plasmacytoid DCs in response to TLR7 stimulation was found to be decreased in older mice, correlating with reduced nuclear translocation of IRF7 (38), and similar defects in type I and type III IFN production induced by TLR7 and TLR9, IAV, or West Nile virus have previously been observed in dendritic cells from older humans (39-43). Our data indicate that TRAF3 is less abundant in monocytes from older adults. If a similar reduction in TRAF3 is present in other cells types with age, this may be a common thread that can explain the reduced IFN induction downstream of TLRs and RLRs.
IRF8 requires partner transcription factors to bind to DNA, utilizing the Ets family member PU.1 to mediate basal transcriptional control of lineage-specifying genes (44). IRF8 can also partner with other IRFs and members of the AP-1 family of transcription factors, allowing it to have broad regulatory roles both basally and in response to stimulation in myeloid cells (29, 45). IRF8 and PU.1 binding to the IFN promoter creates a scaffold that facilitates IRF3 recruitment and stabilizes its binding to the promoter (19). Our data suggest that in monocytes from older donors, RIG-I induces only suboptimal expression of IRF8 in part due to the upstream defect in TRAF3. This lower induction of IRF8 is correlated with reduced phosphorylation of TBK1, IRF3, and IRF7, and may be insufficient to serve as a scaffold to stabilize the binding of IRF3 to the IFN promoter, thereby diminishing IFN transcription. This defect in primary IRF8 expression in monocytes from older donors is compounded by the impaired induction of IRF8 in response to IFNAR signaling in these same cells. Given that IRF8 has been reported to additionally serve as a scaffold that stabilizes transcription from IFN promoters during the feedback phase of the IFN response by enhancing RNA Pol II binding to this region (18), the reduced expression of IRF8 as a primary transcript in older monocytes therefore contributes substantially to reduced IFN production in monocytes from older individuals, and both reduced IFN secretion and reduced IFNAR-driven IRF8 induction further dampen this feed-forward loop.
These observed dual impairments in basal TRAF3 protein abundance and both TRAF3- and IFNAR-mediated IRF8 induction ultimately result in a profoundly impaired IFN response to infection or RIG-I stimulation. The underlying cause of this diminished IRF8 induction in monocytes from older donors remains to be characterized and may be a consequence of differential epigenetic modification of the IRF8 locus with age. Indeed, others have found that hematopoietic stem cells (HSCs) from older individuals exhibit hypermethylation of the IRF8 locus (30), as well as decreased IRF8 expression (46). It is interesting to note that loss of IRF8 expression is associated with a skewing towards myeloid cell generation in mouse HSCs (47), a hallmark of human aging (34, 48). The loss of effective IRF8 induction in the context of the IFN response may thus be a secondary consequence of broader changes in IRF8 regulation that arise with age and which may be related to appropriate shaping of the hematopoietic compartment. Given the importance of IRF8 as a mediator of the feedback amplification of the IFN response, these potential epigenetic regulatory mechanisms may link the well-known myeloid skewing of HSCs with impaired IFN induction with age. Because we did not identify a difference in DNA methylation of the IRF8 locus in our monocyte samples, future examination of chromatin accessibility is warranted to better understand the epigenetic regulation of IRF8 with increasing age.
Given that TRAF3 is an adapter protein important in signaling downstream of other PRRs and that IRF8 is likely involved in IFN induction downstream of these same sensors, it is probable that the defects we have identified in this study contribute to previously reported age-related defects in TLR-induced IFN production, but further study will be needed to confirm this. How aging shapes intracellular cytosolic DNA sensing by cyclic GMP-AMP synthase (cGAS) is unknown, and thus establishing whether TRAF3 or IRF8 defects further compromise this pathway will be informative. To date we have conducted preliminary studies of cytosolic DNA sensing in our human monocytes (Fig. S9), but our analyses only indicate trends towards decreased IFN-β production with age and lack sufficient power to resolve a clear phenotype, highlighting the need for additional study.
In conclusion, our results demonstrate two key defects in innate immunity that together serve to compromise the induction of an IFN-mediated antiviral gene program in monocytes from older donors. First, we show that reduced abundance of the adaptor protein TRAF3 impairs IFN induction owing to defects in TBK1, IRF3, and IRF7 phosphorylation. We additionally show that impairment of IRF8 induction in cells from older donors is exaggerated by these initial defects in IFN secretion, resulting in markedly impaired positive feedback amplification of the IFN response in these cells. Finally, we show that restoring the expression of these intermediates is sufficient to restore the IFN response to RIG-I stimulation. Therapeutic strategies that seek to normalize TRAF3 and/or IRF8 dynamics in older adults may thus represent powerful strategies to bolster antiviral responses to influenza in this at-risk population, thereby reducing the seasonal morbidity and mortality associated with this disease.
Materials and Methods
Clinical Study Design and Recruitment of Participants
All subjects were healthy and were recruited as volunteers at influenza clinics organized by Yale Health Services during the 2013-2017 vaccine seasons. Blood was collected with written informed consent in accordance with the regulations of the Human Research Protection Program of Yale University. Samples were randomly chosen for experiments over 4 years for assays under study at the time of donor recruitment, thus it was not possible to test every sample for every assay. In this study, young individuals were defined as people 21 to 30 years of age, and old subjects were defined as individuals 65 years of age and older. Consenting adults were screened using a questionnaire determining their demographic information, usage of medication, and comorbid conditions. Participants were excluded from the study for primary or acquired immunodeficiency or immunomodulating medications (such as steroids or chemotherapy), pregnancy, history of cancer (other than localized skin and prostate cancer), a history of cirrhosis or renal failure requiring hemodialysis, or for acute illness or antibiotic use within two weeks prior to the day of recruitment.
Isolation and Stimulation of Human Monocytes
Monocytes were isolated as previously described (4) from human peripheral blood from donors who had received the influenza vaccine 28 days earlier. Briefly, peripheral blood mononuclear cells (PBMCs) were isolated from heparinized blood using Ficoll-Paque PLUS (GE Healthcare Sciences) gradient centrifugation. For experiments using purified human monocytes, cells were negatively enriched from total PBMCs using the EasySep™ Human Monocyte Enrichment Kit without CD16 Depletion (Stemcell Technologies), according to manufacturer’s instructions. Isolated monocytes were plated at a density of 2.0×105 cells/well in round 96-well plates in a volume of 100 μL complete RPMI media (10% FBS, Pen-Strep). For experiments involving IAV infection, monocytes were infected with A/PR8 influenza at an MOI of 10 in a total volume of 100 μL PBS, containing 0.1% BSA, for a period of 1 hour, after which cells were resuspended in 500 μL complete RPMI for 12 hours. To induce RIG-I stimulation, monocytes were stimulated with a RIG-I ligand, 5’-ppp 14 dsRNA, generously provided by Dr. Anna Pyle (20). dsRNA was allowed to form complexes with Lipofectamine 2000 (Life Technologies) for 20 minutes in Opti-MEM media (Life Technologies) prior to transfection of cells with 100 μL of complexes for 1 hour. Cells were resuspended in 250 μL of complete RPMI for experimentally indicated amounts of time. For treatment of cells with recombinant human IFNβ, 1 or 10 units of IFNβ (Sigma-Aldrich) were added to cell culture media for 4 hours prior to infection or RNA isolation. To prevent translation, monocytes were pretreated with cyclohexamide (500 μM) for 4 hours prior to stimulation, and after 1 hour of transfection cells were resuspended in growth media containing cyclohexamide for an additional 5 hours. To induce proteasome inhibition, cells were pretreated with bortezomib (EMD Millipore; 100 ng/mL) for 4 hours prior to stimulation or infection. Where indicated, supernatants were collected after stimulation and frozen at −80°C, and monocytes were lysed in 350 μL RLT buffer with β-mercaptoethanol (Qiagen) for RNA storage at −80°C.
IRF8 Knockdown and Overexpression
For IRF8 knockdown experiments, siRNA-mediated knockdown in human monocytes was conducted as previously described (49). Briefly, for each well of a 24-well plate to be treated, a mix was prepared of 3.75 μL of a 20 μM solution of control RISC-free or IRF8-specific siRNA constructs (GE Dharmacon) were complexed with 11 μL HiPerFect transfection reagent (Qiagen) and 110.25 μL unsupplemented RPMI. After 20 minutes, this solution was added to a 24 well plate containing 3×105 human monocytes in 250 μL complete RPMI and allowed to incubate for 4 hours at 37°C. Silencing was then terminated through addition of 500 μL complete RPMI containing 10 ng/mL human M-CSF (Affymetrix) and cells were incubated for an additional 48 hours prior to stimulation or infection. For IRF8 overexpression experiments, 2.5×105 monocytes were infected at an MOI = 1 with a control or human IRF8 lentiviral vector under the control of a CMV promoter (Applied Biological Materials Inc.). Infection was conducted in a total volume of 2 mL, containing 8μg/mL Polybrene (Santa Cruz Biotechnology, Inc.), while spinning in a 32°C centrifuge at 800 x g. Cell pellets were then resuspended in 1250 μL complete RPMI containing 10 ng/mL human M-CSF, and 500 μL of cells were plated per well of a 24 well plate. Cells were incubated for an additional 48 hours prior to stimulation or infection.
Quantitative reverse transcription polymerase chain reaction (qRT-PCR)
RNA was isolated from human monocytes using the RNeasy Mini Kit with on-column DNAse digestion (Qiagen). cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad). Quantitative PCR was performed on a CFX Connect instrument (Bio-Rad) using SYBR Green-based quantification (Bio-Rad). For all human gene qPCR assays, gene expression was normalized against HPRT. Primers were as follows:
IFNB (F: CTTTCGAAGCCTTTGCTCTG; R: GGAGAGCAATTTGGAGGAGAC),
IRF3 (F: TGGGCCCCCAGATCTGATTA; R: GCACAACCTTGACCATCACG),
DDX58 (F: CAGAGCACTTGTGGACGCTT; R: AGCAACTGAGGTGGCAATCA),
IRF7 (F: CGCGGCACTAACGACAGGCGA; R: GCTGCCGTGCCCGGAATTCCA),
IRF8 (F: CAGCTCCTTCCAGACTGGTG; R: GGAGAATGCTGAATGGTGCG),
MX1 (F: AGAGAAGGTGAGAAGCTGATCC; R: TTCTTCCAGCTCCTTCTCCTG),
IFITM2 (F: CTCCGTGAAGTCTAGGGACAG; R: CCTCCTGATCTATCGCTGGG),
IFIT2 (F: CTGCAACCATGAGTGAGAACAA; R: CTCCCTCCATCAAGTTCCAGG),
HPRT (F: TGTAGGATATGCCCTTGACTATA; R: CAATAGGACTCCAGATGTTTCCA),
PR8 NS (F: CAGGACATACTGATGAGGATG; R: GTTTCAGAGACTCGAACTGTG).
Enzyme-linked immunosorbent assays (ELISAs)
Cell supernatants were harvested from stimulated and/or infected human monocytes 12 hours after stimulation. Supernatants were centrifuged for 5 minutes at 3,000xg to remove cellular debris and were stored at −80°C prior to analysis. Measurement of human IFNβ in cell supernatants was conducted using the VeriKine Human Interferon Beta ELISA Kit (PBL Assay Science) according to manufacturer’s instructions.
Flow Cytometry
Monocytes used for intracellular cytokine staining had Protein Transport Inhibitor Cocktail (eBioscience) added for the final 3 hours of stimulation. Cells were then washed twice with FACS buffer (PBS containing 1% FBS), fixed/permeabilized using BD Cytofix/Cytoperm (BD Biosciences) according to manufacturer’s instructions, and stained using antibodies against human TNF-α (eBioscience), IFN-α, or IFN-β (Antigenix America) for 30 minutes in 50 μL Perm/Wash buffer (BD Biosciences). Cells were then washed twice and resuspended in a final volume of 200 μL of FACS buffer. For intracellular IRF7 and IRF8 staining, cells were treated as above without the addition of Protein Transport Inhibitor cocktail, and cells were stained with antibodies against human IRF7 or IRF8 (eBioscience). Fluorescence was detected using an LSR II flow cytometer (BD Biosciences) and analysis was conducted using FlowJo software (version 10). Monocytes used for intracellular phospho-protein staining were stimulated for the indicated amount of time in a total volume of 100 μL. BD Cytofix solution (BD Biosciences; 100 μL/well) was then added to stimulated cells and cells were incubated at 37°C for 20 minutes. Fixed cells were then pelleted and resuspended in 200 μL cold BD Cytoperm Solution III (BD Biosciences) for 30 minutes at 4°C to permeabilize cells. Cells were labeled with antibodies against intracellular phospho-IRF7 (pS477/pS479; BD Biosciences; (50-52)) or intracellular phospho-IRF3 (Ser396; EMD Millipore) for 20 minutes in 50 μL of Cytoperm Solution III at 4°C. For pIRF3 staining, a secondary antibody specific for rabbit IgG conjugated to AlexaFluor 647 (BioLegend) was used with identical staining conditions.
Western Blots
Stimulated or unstimulated monocyte samples (1×106 cells/sample) were washed once with cold PBS and lysed for 30 minutes at 4°C in 100 μL or RIPA buffer containing Complete protease inhibitor cocktail (Roche) and PhosStop phosphates inhibitor cocktail (Roche). Western blotting was conducted according to standard protocols using a Bio Rad western blotting system and a 10% SDS gel. Protein transfer to a PVDF membrane (Millipore) was conducted at 4°C for 1 hour at 100V. Membrane blocking was conducted using a solution of TBST containing 5% bovine serum albumin (Sigma). Primary antibodies against human RIG-I, IRF3, IRF7, IRF8, pIRF3, K48-polyubiquitin, and β-actin as well as a secondary antibody specific for rabbit IgG conjugated to HPRT were purchased from Cell Signaling Technology. Blots were developed using SuperSignal West Pico Chemiluminescent Substrate (Thermo Fisher Scientific) and film. Densitometry was calculated for individual protein bands using the ImageJ software package.
TRAF3 Immunoprecipitation
Unstimulated monocyte samples lysed in RIPA buffer were incubated overnight under constant agitation with 1μg of a mouse antibody recognizing TRAF3 (Santa Cruz Biotechnology). Samples were then incubated with 20μL of magnetic protein G–conjugated SureBeads (Bio-Rad) for 1 hour. A magnetic rack was used to separate out these beads, which were then washed three times with cold RIPA buffer and attached sample was eluted by heating to 95°C for 5 minutes in 2X sample buffer. Eluted protein was loaded into an SDS-PAGE gel and analyzed as above, using an HRP-conjugated conformation-specific secondary antibody recognizing only non-denatured rabbit IgG to detect TRAF3 or K48-polyubiquitin in resultant Western blots.
RNA Sequencing
RNA was isolated from cellular lysates in the same manner as for qPCR experiments. RNA sequencing was performed by the HudsonAlpha Institute for Biotechnology. RNA samples were sequenced using an Illumina HiSeq 2500 machine, with 50 base pair paired end reads. The raw reads of RNA-seq experiments were trimmed off sequencing adaptors and low quality regions by btrim (53). The trimmed reads were mapped to human genome (GRCh37) by tophat2 (54). The counts of reads for each gene were based on Ensembl annotation (release 70). After the counts are collected, the differential expression analysis was done by DEseq2 (55), which calculated the adjusted p-values.
Methylation Profiling
DNA was isolated from unstimulated monocytes using the Qiagen DNA Mini kit according to the manufacturer’s instructions. DNA methylation profiling was assessed using an Infinium Methylation EPIC 8 sample array kit (Illumina) according to manufacturer’s instructions. Methylation data were mapped and methylation frequencies for each donor assessed were calculated and arranged into a heat map using Microsoft Excel.
Statistical Analyses
Two-way ANOVA tests (with Sidak’s multiple comparisons test) were performed to compare groups at multiple time points. Student t tests were used to compare two samples at a single time point, with paired tests being used when samples were matched between groups. All statistical tests were calculated using GraphPad Prism.
Supplementary Material
Fig. S1. Monocyte purity and plasmacytoid DC numbers are comparable between age groups.
Fig. S2. IFN Induction in response to dsRNA transfection depends on a 5’-ppp motif in human monocytes.
Fig. S3. Basal IFNB expression is comparable in human monocytes from older and younger donors.
Fig. S4. IFN-β pretreatment is insufficient to ablate age-related differences in IFN-β secretion upon RIG-I stimulation.
Fig. S5. Basal levels of RIG-I, IRF3, and IRF7 protein are comparable in older and younger monocytes.
Fig. S6. IRF3 phosphorylation is impaired in monocytes from older donors.
Fig. S7. Basal TRAF3 protein levels are positively correlated with IFNβ secretion in response to RIG-I stimulation.
Fig. S8. Bortezomib improves antiviral responses in monocytes from older donors.
Fig. S9. Lack of age-related differences in IFN-β secretion in response to cGAMP stimulation of human monocytes.
Fig. S10. RNA-seq highlights differential regulation of RIG-I, IRF, and IAV-related signaling pathways in older RIG-I–stimulated monocytes.
Fig. S11. Amplification-stage increase in IRF7 protein is defective in older RIG-I–stimulated monocytes.
Fig. S12. Basal IRF8 protein and RNA levels are comparable in old and young monocytes.
Fig. S13. Inducible IRF8 expression is age-dependent.
Fig. S14. Inducible IRF8 levels are positively correlated with RIG-I–inducible IFNβ secretion.
Fig. S15. Bortezomib treatment of older monocytes enhances IRF8 induction upon RIG-I stimulation.
Fig. S16. IFNβ-stimulated gene induction and inducible protection from influenza is preserved with age in human monocytes.
Fig. S17. IRF8 is an interferon-inducible gene, and its induction is impaired in older monocytes.
Fig. S18. Lack of age-related difference in CpG DNA methylation of the IRF8 gene in human monocytes.
Table S1. Human Donor Characteristics.
Table S2. Differences in IFN Expression in RNA-Seq Data from RIG-I Stimulated Older and Younger Monocytes.
Table S3. Differences in IRF Expression in RNA-Seq Data from RIG-I Stimulated Older and Younger Monocytes.
Acknowledgments
Funding: This study was supported in part by the Howard Hughes Medical Institute, awards from the National Institutes of Health (U19 AI089992 (A.C.S., R.R.M., A.I.)), HHSN272201100019C (A.C.S. and R.R.M), K24 AG042489 (A.C.S.), T32 AI007019-38, T32 AI055403 (to R.D.M.) and the Claude D. Pepper Older Americans Independence Center at Yale P30 AG0212342 (A.C.S.)), and the Francis Trudeau Trainee Fellowship (to R.D.M.). A.I. is an investigator of the Howard Hughes Medical Institute.
Footnotes
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The data presented in this manuscript are tabulated in the main paper and in the supplementary materials. RNA sequencing data have been publicly released to the general scientific community and are available in the NCBI Gene Expression Omnibus (Accession Number: GSE100709).
References and Notes
- 1.Johnson NB, Hayes LD, Brown K, Hoo EC, Ethier KA, CDC National Health Report: leading causes of morbidity and mortality and associated behavioral risk and protective factors—United States, 2005–2013. MMWR Surveill Summ 63, 3–27 (2014). [PubMed] [Google Scholar]
- 2.Thompson WW, Shay DK, Weintraub E, Brammer L, Cox N, Anderson LJ, Fukuda K, Mortality associated with influenza and respiratory syncytial virus in the united states. JAMA 289, 179–186 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Simonsen L, Clarke MJ, Schonberger LB, Arden NH, Cox NJ, Fukuda K, Pandemic versus epidemic influenza mortality: a pattern of changing age distribution. Journal of Infectious Diseases 178, 53–60 (1998). [DOI] [PubMed] [Google Scholar]
- 4.Pillai PS, Molony RD, Martinod K, Dong H, Pang IK, Tal MC, Solis AG, Bielecki P, Mohanty S, Trentalange M, Homer RJ, Flavell RA, Wagner DD, Montgomery RR, Shaw AC, Staeheli P, Iwasaki A, Mx1 reveals innate pathways to antiviral resistance and lethal influenza disease. Science 352, 463–466 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lin KL, Suzuki Y, Nakano H, Ramsburg E, Gunn MD, CCR2+ monocyte-derived dendritic cells and exudate macrophages produce influenza-induced pulmonary immune pathology and mortality. The Journal of Immunology 180, 2562–2572 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Landsman L, Jung S, Lung macrophages serve as obligatory intermediate between blood monocytes and alveolar macrophages. The Journal of Immunology 179, 3488–3494 (2007). [DOI] [PubMed] [Google Scholar]
- 7.Gill MA, Long K, Kwon T, Muniz L, Mejias A, Connolly J, Roy L, Banchereau J, Ramilo O, Differential recruitment of dendritic cells and monocytes to respiratory mucosal sites in children with influenza virus or respiratory syncytial virus infection. Journal of Infectious Diseases 198, 1667–1676 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi C, Pamer EG, Monocyte recruitment during infection and inflammation. Nature Reviews Immunology 11, 762–774 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Iwasaki A, Foxman EF, Molony RD, Early local immune defences in the respiratory tract. Nature Reviews Immunology 17, 7–20 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kallfass C, Lienenklaus S, Weiss S, Staeheli P, Visualizing the beta interferon response in mice during infection with influenza A viruses expressing or lacking nonstructural protein 1. Journal of virology 87, 6925–6930 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pichlmair A, Schulz O, Tan CP, Näslund TI, Liljeström P, Weber F, e Sousa CR, RIG-I-mediated antiviral responses to single-stranded RNA bearing 5'-phosphates. Science 314, 997–1001 (2006). [DOI] [PubMed] [Google Scholar]
- 12.Hornung V, Ellegast J, Kim S, Brzózka K, Jung A, Kato H, Poeck H, Akira S, Conzelmann K-K, Schlee M, 5'-Triphosphate RNA is the ligand for RIG-I. Science 314, 994–997 (2006). [DOI] [PubMed] [Google Scholar]
- 13.Kato H, Takeuchi O, Sato S, Yoneyama M, Yamamoto M, Matsui K, Uematsu S, Jung A, Kawai T, Ishii KJ, Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441, 101–105 (2006). [DOI] [PubMed] [Google Scholar]
- 14.Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S, IPS-1, an adaptor triggering RIG-I-and Mda5-mediated type I interferon induction. Nature immunology 6, 981–988 (2005). [DOI] [PubMed] [Google Scholar]
- 15.Kato H, Sato S, Yoneyama M, Yamamoto M, Uematsu S, Matsui K, Tsujimura T, Takeda K, Fujita T, Takeuchi O, Akira S, Cell type-specific involvement of RIG-I in antiviral response. Immunity 23, 19–28 (2005). [DOI] [PubMed] [Google Scholar]
- 16.Medzhitov R, Horng T, Transcriptional control of the inflammatory response. Nature Reviews Immunology 9, 692–703 (2009). [DOI] [PubMed] [Google Scholar]
- 17.Honda K, Taniguchi T, IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nature Reviews Immunology 6, 644–658 (2006). [DOI] [PubMed] [Google Scholar]
- 18.Tailor P, Tamura T, Kong HJ, Kubota T, Kubota M, Borghi P, Gabriele L, Ozato K, The Feedback Phase of Type I Interferon Induction in Dendritic Cells Requires Interferon Regulatory Factor 8. Immunity 27, 228–239 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li P, Wong JJ-Y, Sum C, Sin W-X, Ng K-Q, Koh MB, Chin K-C, IRF8 and IRF3 cooperatively regulate rapid interferon-β induction in human blood monocytes. Blood 117, 2847–2854 (2011). [DOI] [PubMed] [Google Scholar]
- 20.Vela A, Fedorova O, Ding SC, Pyle AM, The thermodynamic basis for viral RNA detection by the RIG-I innate immune sensor. Journal of Biological Chemistry 287, 42564–42573 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honda K, Takaoka A, Taniguchi T, Type I inteferon gene induction by the interferon regulatory factor family of transcription factors. Immunity 25, 349–360 (2006). [DOI] [PubMed] [Google Scholar]
- 22.Fitzgerald KA, McWhirter SM, Faia KL, Rowe DC, Latz E, Golenbock DT, Coyle AJ, Liao S-M, Maniatis T, IKKε and TBK1 are essential components of the IRF3 signaling pathway. Nature immunology 4, 491–496 (2003). [DOI] [PubMed] [Google Scholar]
- 23.Paz S, Vilasco M, Werden SJ, Arguello M, Joseph-Pillai D, Zhao T, Nguyen TL-A, Sun Q, Meurs EF, Lin R, Hiscott J, A functional C-terminal TRAF3-binding site in MAVS participates in positive and negative regulation of the IFN antiviral response. Cell Res 21, 895–910 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Häcker H, Tseng P-H, Karin M, Expanding TRAF function: TRAF3 as a tri-faced immune regulator. Nat Rev Immunol 11, 457–468 (2011). [DOI] [PubMed] [Google Scholar]
- 25.Oganesyan G, Saha SK, Guo B, He JQ, Shahangian A, Zarnegar B, Perry A, Cheng G, Critical role of TRAF3 in the Toll-like receptor-dependent and-independent antiviral response. Nature 439, 208–211 (2006). [DOI] [PubMed] [Google Scholar]
- 26.Tseng P-H, Matsuzawa A, Zhang W, Mino T, Vignali DA, Karin M, Different modes of ubiquitination of the adaptor TRAF3 selectively activate the expression of type I interferons and proinflammatory cytokines. Nature immunology 11, 70–75 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sichien D, Scott CL, Martens L, Vanderkerken M, Van Gassen S, Plantinga M, Joeris T, De Prijck S, Vanhoutte L, Vanheerswynghels M, IRF8 Transcription Factor Controls Survival and Function of Terminally Differentiated Conventional and Plasmacytoid Dendritic Cells, Respectively. Immunity 45, 626–640 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Xu H, Zhu J, Smith S, Foldi J, Zhao B, Chung AY, Outtz H, Kitajewski J, Shi C, Weber S, Saftig P, Li Y, Ozato K, Blobel CP, Ivashkiv LB, Hu X, Notch-RBP-J signaling regulates the transcription factor IRF8 to promote inflammatory macrophage polarization. Nat Immunol 13, 642–650 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mancino A, Termanini A, Barozzi I, Ghisletti S, Ostuni R, Prosperini E, Ozato K, Natoli G, A dual cis-regulatory code links IRF8 to constitutive and inducible gene expression in macrophages. Genes & development, gad. 257592.257114 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beerman I, Bock C, Garrison Brian S., Smith Zachary D., Gu H, Meissner A, Rossi Derrick J., Proliferation-Dependent Alterations of the DNA Methylation Landscape Underlie Hematopoietic Stem Cell Aging. Cell Stem Cell 12, 413–425 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weng N.-p., Aging of the immune system: how much can the adaptive immune system adapt? Immunity 24, 495–499 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thakar J, Mohanty S, West AP, Joshi SR, Ueda I, Wilson J, Meng H, Blevins TP, Tsang S, Trentalange M, Aging-dependent alterations in gene expression and a mitochondrial signature of responsiveness to human influenza vaccination. Aging (Albany NY) 7, 38–52 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reber AJ, Chirkova T, Kim JH, Cao W, Biber R, Shay DK, Sambhara S, Immunosenescence and challenges of vaccination against influenza in the aging population. Aging and disease 3, 68–90 (2014). [PMC free article] [PubMed] [Google Scholar]
- 34.Shaw AC, Goldstein DR, Montgomery RR, Age-dependent dysregulation of innate immunity. Nat Rev Immunol 13, 875–887 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kayagaki N, Phung Q, Chan S, Chaudhari R, Quan C, O'Rourke KM, Eby M, Pietras E, Cheng G, Bazan JF, A deubiquitinase that regulates type I interferon production. Science 318, 1628–1632 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Mao A-P, Li S, Zhong B, Li Y, Yan J, Li Q, Teng C, Shu H-B, Virus-triggered ubiquitination of TRAF3/6 by cIAP1/2 is essential for induction of interferon-β (IFN-β) and cellular antiviral response. Journal of Biological Chemistry 285, 9470–9476 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nakhaei P, Mesplede T, Solis M, Sun Q, Zhao T, Yang L, Chuang T-H, Ware CF, Lin R, Hiscott J, The E3 Ubiquitin Ligase Triad3A Negatively Regulates the RIG-I/MAVS Signaling Pathway by Targeting TRAF3 for Degradation. PLOS Pathogens 5, e1000650 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stout-Delgado HW, Yang X, Walker WE, Tesar BM, Goldstein DR, Aging impairs IFN regulatory factor 7 up-regulation in plasmacytoid dendritic cells during TLR9 activation. J Immunol 181, 6747–6756 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Canaday DH, Amponsah NA, Jones L, Tisch DJ, Hornick TR, Ramachandra L, Influenza-induced production of interferon-alpha is defective in geriatric individuals. Journal of clinical immunology 30, 373–383 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Qian F, Wang X, Zhang L, Lin A, Zhao H, Fikrig E, Montgomery RR, Impaired interferon signaling in dendritic cells from older donors infected in vitro with West Nile virus. Journal of Infectious Diseases 203, 1415–1424 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Panda A, Qian F, Mohanty S, van Duin D, Newman FK, Zhang L, Chen S, Towle V, Belshe RB, Fikrig E, Allore HG, Montgomery RR, Shaw AC, Age-associated decrease in TLR function in primary human dendritic cells predicts influenza vaccine response. J Immunol 184, 2518–2527 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jing Y, Shaheen E, Drake RR, Chen N, Gravenstein S, Deng Y, Aging is associated with a numerical and functional decline in plasmacytoid dendritic cells, whereas myeloid dendritic cells are relatively unaltered in human peripheral blood. Human immunology 70, 777–784 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Prakash S, Agrawal S, Cao JN, Gupta S, Agrawal A, Impaired secretion of interferons by dendritic cells from aged subjects to influenza : role of histone modifications. Age (Dordr) 35, 1785–1797 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tamura T, Thotakura P, Tanaka TS, Ko MS, Ozato K, Identification of target genes and a unique cis element regulated by IRF-8 in developing macrophages. Blood 106, 1938–1947 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Glasmacher E, Agrawal S, Chang AB, Murphy TL, Zeng W, Vander Lugt B, Khan AA, Ciofani M, Spooner CJ, Rutz S, A genomic regulatory element that directs assembly and function of immune-specific AP-1–IRF complexes. Science 338, 975–980 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stirewalt DL, Choi YE, Sharpless NE, Pogosova-Agadjanyan EL, Cronk MR, Yukawa M, Larson EB, Wood BL, Appelbaum FR, Radich JP, Heimfeld S, Decreased IRF8 Expression in Aging Hematopoietic Progenitor/Stem Cells. Leukemia : official journal of the Leukemia Society of America, Leukemia Research Fund, U.K 23, 391–393 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wang H, Lee CH, Qi C, Tailor P, Feng J, Abbasi S, Atsumi T, Morse HC, IRF8 regulates B-cell lineage specification, commitment, and differentiation. Blood 112, 4028–4038 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Geiger H, de Haan G, Florian MC, The ageing haematopoietic stem cell compartment. Nature Reviews Immunology 13, 376–389 (2013). [DOI] [PubMed] [Google Scholar]
- 49.Troegeler A, Lastrucci C, Duval C, Tanne A, Cougoule C, Maridonneau-Parini I, Neyrolles O, Lugo-Villarino G, An efficient siRNA-mediated gene silencing in primary human monocytes, dendritic cells and macrophages. Immunology and cell biology 92, 699–708 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Chiang EY, Yu X, Grogan JL, Immune complex-mediated cell activation from systemic lupus erythematosus and rheumatoid arthritis patients elaborate different requirements for IRAK1/4 kinase activity across human cell types. The Journal of Immunology 186, 1279–1288 (2011). [DOI] [PubMed] [Google Scholar]
- 51.van de Laar L, van den Bosch A, Boonstra A, Binda RS, Buitenhuis M, Janssen HL, Coffer PJ, Woltman AM, PI3K-PKB hyperactivation augments human plasmacytoid dendritic cell development and function. Blood 120, 4982–4991 (2012). [DOI] [PubMed] [Google Scholar]
- 52.Wang L, Zhao J, Ren J, Hall KH, Moorman JP, Yao ZQ, Ning S, Protein phosphatase 1 abrogates IRF7-mediated type I IFN response in antiviral immunity. European journal of immunology 46, 2409–2419 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kong Y, Btrim: a fast, lightweight adapter and quality trimming program for next-generation sequencing technologies. Genomics 98, 152–153 (2011). [DOI] [PubMed] [Google Scholar]
- 54.Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL, TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome biology 14, 1 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Love MI, Huber W, Anders S, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome biology 15, 1 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. Monocyte purity and plasmacytoid DC numbers are comparable between age groups.
Fig. S2. IFN Induction in response to dsRNA transfection depends on a 5’-ppp motif in human monocytes.
Fig. S3. Basal IFNB expression is comparable in human monocytes from older and younger donors.
Fig. S4. IFN-β pretreatment is insufficient to ablate age-related differences in IFN-β secretion upon RIG-I stimulation.
Fig. S5. Basal levels of RIG-I, IRF3, and IRF7 protein are comparable in older and younger monocytes.
Fig. S6. IRF3 phosphorylation is impaired in monocytes from older donors.
Fig. S7. Basal TRAF3 protein levels are positively correlated with IFNβ secretion in response to RIG-I stimulation.
Fig. S8. Bortezomib improves antiviral responses in monocytes from older donors.
Fig. S9. Lack of age-related differences in IFN-β secretion in response to cGAMP stimulation of human monocytes.
Fig. S10. RNA-seq highlights differential regulation of RIG-I, IRF, and IAV-related signaling pathways in older RIG-I–stimulated monocytes.
Fig. S11. Amplification-stage increase in IRF7 protein is defective in older RIG-I–stimulated monocytes.
Fig. S12. Basal IRF8 protein and RNA levels are comparable in old and young monocytes.
Fig. S13. Inducible IRF8 expression is age-dependent.
Fig. S14. Inducible IRF8 levels are positively correlated with RIG-I–inducible IFNβ secretion.
Fig. S15. Bortezomib treatment of older monocytes enhances IRF8 induction upon RIG-I stimulation.
Fig. S16. IFNβ-stimulated gene induction and inducible protection from influenza is preserved with age in human monocytes.
Fig. S17. IRF8 is an interferon-inducible gene, and its induction is impaired in older monocytes.
Fig. S18. Lack of age-related difference in CpG DNA methylation of the IRF8 gene in human monocytes.
Table S1. Human Donor Characteristics.
Table S2. Differences in IFN Expression in RNA-Seq Data from RIG-I Stimulated Older and Younger Monocytes.
Table S3. Differences in IRF Expression in RNA-Seq Data from RIG-I Stimulated Older and Younger Monocytes.