Abstract
Our previous studies indicate that the mitochondrial redox state and its intratumor heterogeneity are associated with invasiveness and metastatic potential in human breast cancer cell models and mouse xenografts. To further study the molecular basis of redox heterogeneity, we obtained the fluorescence images of Fp (oxidized flavoproteins containing flavin adenine dinucleotide, i.e., FAD), NADH (reduced nicotinamide adenine dinucleotide), and the Fp redox ratio (FpR = Fp/(Fp + NADH)) of MDA-MB-231 xenografts by the Chance redox scanner, then isolated the intratumoral redox subpopulations by dissection according to the redox ratio image. A total of 12 subpopulations were isolated from 4 tumors (2–4 locations from each tumor). The 12 subpopulations were classified into 3 FpR groups: high FpR (HFpR, n = 4, FpR range 0.78–0.92, average 0.85), medium FpR (MFpR, n = 5, FpR range 0.39–0.68, average 0.52), and low FpR (LFpR, n = 3, FpR range 0.15–0.28, average 0.20). The RT-PCR (reverse transcription polymerase chain reaction) analysis on these redox subpopulations showed that PGC-1α is significantly upregulated in the HFpR redox group compared to the MFpR group (fold change 2.1, p = 0.008), but not significantly different between MFpR and LFpR groups, or between HFpR and LFpR groups. These results indicate that optical redox imaging (ORI)-based redox subpopulations exhibit differential expression of PGC1α gene and suggest that PGC1α might play a role in redox mediation of breast cancer progression.
1. Introduction
Intratumor heterogeneity has been studied at genetic, epigenetic, cellular and molecular levels [1–3] and is likely associated with metastasis [1, 4]. Employing the ORI technique, we previously identified intratumor redox heterogeneity in cancer mouse models and clinical breast tumor specimens based on fluorescence signals from NADH and Fp and the calculated redox ratio FpR [5]. NADH and Fp signals are contributed mainly by mitochondria. The redox ratio can be regarded as a surrogate indicator of the mitochondrial redox state and is shown to correlate linearly with NAD-coupled redox potential NAD+/NADH as determined biochemically [6, 7]. Our studies have shown that the heterogeneity-based mitochondrial redox indices (NADH, Fp and FpR) can differentiate between tumors with different levels of aggressiveness and between premalignant and normal tissues [8–10]. Higher mitochondrial redox indices were found in breast cancer clinical specimens compared to the adjacent normal tissues [11–13].
Redox imbalance in cancer associates with biochemical changes [14, 15] and altered gene expressions that may modulate cellular metabolic and signaling pathways leading to cancer progression and metastasis [16–19]. However, no studies have reported on the differential gene expression associated with ORI-based redox states. It is likely that abnormal redox states as detected by ORI are associated with genome wide transcriptome profiles mediating cancer invasion and metastasis. In this study, we examined PGC1α gene specifically since it is a master regulator in the mitochondrial biogenesis and function, energy production and homeostasis, and cancer progression [20–23].
2. Materials and Methods
MDA –MB-231 cell culture and xenograft
MDA-MB-231 cells were cultured in RPMI 1640 medium with 10% FBS and subcutaneously inoculated into hind thighs of athymic nude mice [24]. The induced tumors (diameter 12–14 mm) were harvested, snap-frozen, and stored in liquid nitrogen before redox scanning. All animal experiments were performed according to an Institutional Animal Care and Use Committee approved protocol.
Tissue embedding and redox scanning
The frozen mouse xenografts were embedded in a chilled mounting media (H2O: ethanol: glycerol = 60:30:10, freezing point −30 °C). NADH (100 μM) and FAD (100 μM) frozen solution standards were placed adjacent to the tumor and used as reference standards [24, 25]. The embedded tissue samples and standards were shaved flat to expose a surface plane, which was then scanned two-dimensionally under liquid nitrogen by the Chance redox scanner [26, 27]. The fluorescence signals of NADH and Fp were collected through optical filters for NADH (Excitation 365 ± 13 nm; Emission 455 ± 35 nm) and Fp channels (Excitation 440 ± 10 nm; Emission 515 ± 15 nm), respectively. The image resolution is 200 μm.
Imaging analysis
A customized MATLAB® program was used to reconstruct images of NADH and Fp and the redox ratios in tumors. The nominal concentrations of both Fp and NADH in the tissues were obtained by comparing their fluorescence intensities with that of the corresponding standards. The redox indices, including nominal concentrations of NADH, Fp, and FpR were extracted from each image.
Tumor tissue dissection
After imaging, subpopulations with different redox states were isolated by dissection using a surgical knife from the four embedded frozen mouse xenografts according to their redox images. A total of 12 subpopulations (each sample ~2 to 5 mm) for 12 regions were obtained. The dissected tissues were immediately frozen and stored in liquid nitrogen until use for RNA isolation.
RT-PCR
RNA was isolated from dissected tumor tissues with RNeasy Mini Kit (QIAGEN, Vencia, CA, USA). First strand cDNA was synthesized from the isolated RNA with SuperScript® First-Strand Synthesis System for RT-PCR (Invitrogen, Carlshad, USA). PGC1α gene expression was analyzed by PCR of cDNA in three repeated experiments for each dissected specimen. PCR was performed with the following denature steps: 95 °C for 4 min; followed by 30 cycles of 95 °C (each for 30 s), 58 °C for 1 min, and 72 °C for 30 s; then an extension step at 72 °C for 4 min. GAPDH was used as reference gene as in previous studies on cancers including breast cancer [28, 29]. PCR reaction volume (20 μl) contains various concentrations of cDNA and dH2O, 1.6 μl primer mix and 10 μl BlueTaq mix. The RT-PCR primers were forward 5′ tgaagacggattgccctcatt 3′ and reverse 5′ gctggtgccagtaagagctt 3′ for PGC1α; and forward 5′ ggagcgagatccctccaaaat 3′ and reverse 5′ ggctgttgtcatacttctcatgg 3′ for GAPDH. PCR products were separated by electrophoresis with 2.5% of agarose gel. Gel images were taken under UV light.
Quantitative analysis of gel images and statistical analysis of gel image data
The PCR gel images were analyzed using Bio-Rad Quantity One software (v4.5.0, Bio-Rad, Hercules, CA, USA). The trace quantity (intensity integrated over band area) of a specific gene band was normalized to the reference gene GAPDH from the same tissue sample to represent the level of gene expression. The data were expressed as mean ± standard deviation (SD) of replicates of at least three independent determinations. A p value from an unpaired Student’s t-test of <0.05 was considered statistically significant.
3. Results
3.1. Intratumor Redox Subpopulations Dissected from Mouse Xenografts
Redox scanning showed that all four xenografts are heterogeneous in terms of the ORI-based redox state. The redox heterogeneity and subpopulations in a representative tumor (T62) are shown in Fig. 1 as an example, where the left image is the FpR image and the right one is the corresponding white light photo (taken with a regular camera) with dissection marks.
Fig. 1.
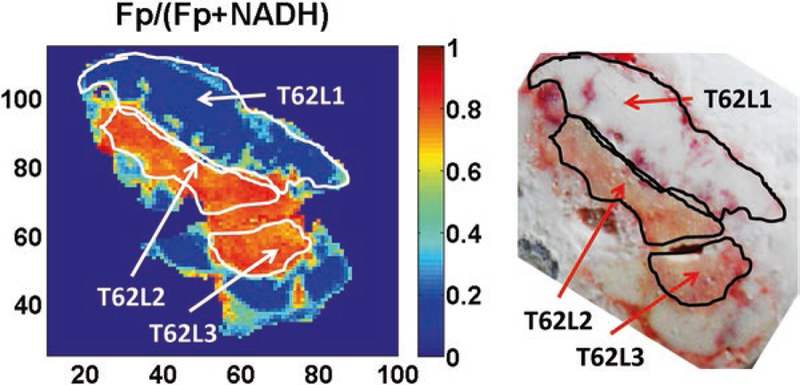
Dissected redox subpopulations from a representative MDA-MB-231 xenograft. (Left) An FpR image showing three subpopulations dissected from tumor T62 with dissection marks; (Right) The corresponding white light image taken with a regular photo camera
Based on the ranges of redox indices Fp, NADH, and redox ratio Fp/(Fp + NADH) of the 12 isolated subpopulations, these specimens were divided into high, medium, and low groups: HFpR (n = 4, FpR 0.78–0.92), MFpR (n = 5, FpR 0.39–0.68), and LFpR (n = 3, FpR 0.15–0.28); HFp (n = 4, Fp 110–341 μM), MFp (n = 3, Fp 84–90 μM), and LFp (n = 5, Fp 26–67 μM); and H-NADH (n = 5, NADH 120–162 μM), M-NADH (n = 2, NADH 48–101 μM), and L-NADH (n = 5, NADH 14–31 μM).
3.2. Differential Expression of PGC1α Among Redox Subpopulations
The RT-PCR results show that PGC1α is expressed differently among the 12 studied redox subpopulations. Figure 2a shows representative PGC1α expressions from low, medium, and high FpR groups. Statistical analysis of the RT-PCR results shows that PGC1α is significantly upregulated in the HFpR group compared with that of MFpR (fold change 2.10, p = 0.008), but shows no significant difference between HFpR and LFpR groups or between LFpR and MFpR groups (Fig. 2b). There is no significant difference in PGC1α expression among the three Fp groups or among the three NADH groups.
Fig. 2.
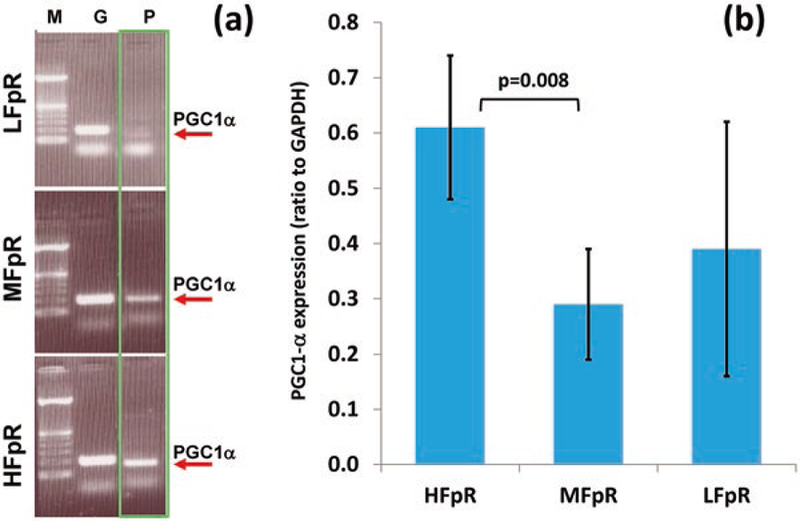
PGC1α gene expression in three FpR groups. (a) Representative PGC1α expression levels in three FpR groups, where lane M shows the molecular weight standards, lane G shows GAPDH level, and lane P shows PGC1α level; (b) PGC1α expression in three FpR groups (mean ± SD), with significant difference between HFpR and MFpR groups
4. Discussion and Conclusions
We previously observed distinct intratumor redox heterogeneity and identified the ORI-based redox state as potential diagnostic/prognostic biomarkers for breast tumor [5, 13]. This paper reports for the first time the differential gene expressions among the redox subpopulations based on ORI mapping. The gene expression level of PGC1α is upregulated in more oxidized subpopulations in breast tumor xenografts of MDA-MB-231. PGC1α, as a master regulator of mitochondrial function and bioenergetics, was shown to mediate metastasis of several types of cancer including human melanoma, prostate and breast cancer [21, 30, 31]. Thus, the results from this study that ORI-based redox heterogeneity is associated with differential gene expression of PGC1α support the ORI redox indices as prognostic biomarkers of breast cancer. However, the details of how the redox state and PGC1α may interact to regulate cancer progression remain unclear. Further studies are underway along this direction.
The tumor in Fig. 1 showed a similar pattern between the redox imaging and the white light photo. However, this is not always the case. Some small aggressive tumors such as MDA-MB-231 tumors (<5 mm) may not show distinct histological difference, but clearly show distinct redox difference. Although some histological differences were reported between cores and rims of human melanoma xenografts [32], we need to investigate more specifically the histological basis of breast cancer xenografts in the future.
Acknowledgments
This study is financially supported by the NIH Grants R01CA155348 and R01CA191207.
Contributor Information
Zhenwu Lin, Department of Radiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA.
He N. Xu, Department of Radiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA
Yunhua Wang, Department of Pediatrics, Pennsylvania State University, Hershey, PA, USA.
Joanna Floros, Department of Pediatrics, Pennsylvania State University, Hershey, PA, USA, Department of Obstetrics and Gynecology, College of Medicine, Pennsylvania State University, Hershey, PA, USA.
Lin Z. Li, Department of Radiology, Perelman School of Medicine, University of Pennsylvania, Philadelphia, PA, USA
References
- 1.Yates LR, Gerstung M, Knappskog S et al. (2015) Subclonal diversification of primary breast cancer revealed by multiregion sequencing. Nat Med 21:751–759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhong Q, Ruschoff JH, Guo T et al. (2016) Imagebased computational quantification and visualization of genetic alterations and tumour heterogeneity. Sci Rep 6:24146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Marusyk A, Almendro V, Polyak K (2012) Intratumour heterogeneity: a looking glass for cancer? Nature reviews. Cancer 12:323–334 [DOI] [PubMed] [Google Scholar]
- 4.Li LZ (2012) Imaging mitochondrial redox potential and its possible link to tumor metastatic potential. J Bioenerg Biomembr 44:645–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu HN, Li LZ (2014) Quantitative redox imaging biomarkers for studying tissue metabolic state and its heterogeneity. J Innov Opt Heal Sci 7:1430002–1-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Varone A, Xylas J, Quinn KP et al. (2014) Endogenous two-photon fluorescence imaging elucidates metabolic changes related to enhanced glycolysis and glutamine consumption in precancerous epithelial tissues. Cancer Res 74:3067–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ozawa K, Chance B, Tanaka A et al. (1992) Linear correlation between acetoacetate beta-hydroxybutyrate in arterial blood and oxidized flavoprotein reduced pyridine-nucleotide in freeze-trapped human livertissue. Biochim Biophys Acta 1138:350–352 [DOI] [PubMed] [Google Scholar]
- 8.Li LZ, Zhou R, Xu HN et al. (2009) Quantitative magnetic resonance and optical imaging biomarkers of melanoma metastatic potential. Proc Natl Acad Sci U S A 106:6608–6613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu HN, Nioka S, Li LZ (2013) Imaging heterogeneity in the mitochondrial redox state of premalignant pancreas in the pancreas-specific PTEN-null transgenic mouse model. Biomarker Res 1:6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu HN, Feng M, Moon L et al. (2013) Redox imaging of the p53-dependent mitochondrial redox state in colon cancer ex vivo. J Innov Opt Heal Sci 6:1350016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xu HN, Tchou J, Chance B et al. (2013) Imaging the redox states of human breast cancer core biopsies. Adv Exp Med Biol 765:343–349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu HN, Tchou J, Li LZ (2013) Redox imaging of human breast cancer core biopsies: a preliminary investigation. Acad Radiol 20:764–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu HN, Tchou J, Feng M et al. (2016) Optical redox imaging indices discriminate human breast cancer from normal tissues. J Biomed Opt 21:114003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jorgenson TC, Zhong W, Oberley TD (2013) Redox imbalance and biochemical changes in cancer. Cancer Res 73:6118–6123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Alberghina L, Gaglio D (2014) Redox control of glutamine utilization in cancer. Cell Death Dis 5:e1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerlinger M, Rowan AJ, Horswell S et al. (2012) Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 366:883–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amoedo ND, Rodrigues MF, Rumjanek FD (2014) Mitochondria: are mitochondria accessory to metastasis? Int J Biochem Cell Biol 51:53–57 [DOI] [PubMed] [Google Scholar]
- 18.Santidrian AF, Matsuno-Yagi A, Ritland M et al. (2013) Mitochondrial complex I activity and NAD+/NADH balance regulate breast cancer progression. J Clin Invest 123:1068–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cantor JR, Sabatini DM (2012) Cancer cell metabolism: one hallmark, many faces. Cancer Discov 2:881–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kelly DP, Scarpulla RC (2004) Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes & Dev 18:357–368 [DOI] [PubMed] [Google Scholar]
- 21.Luo C, Lim JH, Lee Y et al. (2016) A PGC1alpha- mediated transcriptional axis suppresses melanoma metastasis. Nature 537:422–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan Z, Luo X, Xiao L et al. (2016) The role of PGC1alpha in cancer metabolism and its therapeutic implications. Mol Cancer Ther 15:774–782 [DOI] [PubMed] [Google Scholar]
- 23.Zhang J, Wang C, Chen X et al. (2015) EglN2 associates with the NRF1-PGC1alpha complex and controls mitochondrial function in breast cancer. EMBO J 34:2953–2970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu HN, Nioka S, Glickson J et al. (2010) Quantitative mitochondrial redox imaging of breast cancer metastatic potential. J Biomed Opt 15:036010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Xu HN, Wu B, Nioka S et al. (2009) Quantitative redox scanning of tissue samples using a calibration procedure. J Innov Opt Heal Sci 2:375–385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Quistorff B, Haselgrove JC, Chance B (1985) High spatial resolution readout of 3-D metabolic organ structure: an automated, low-temperature redox ratio-scanning instrument. Anal Biochem 148:389–400 [DOI] [PubMed] [Google Scholar]
- 27.Li LZ, Xu HN, Ranji M et al. (2009) Mitochondrial redox imaging for cancer diagnostic and therapeutic studies. J Innov Opt Heal Sci 2:325–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kilic Y, Celebiler AC, Sakizli M (2014) Selecting housekeeping genes as references for the normalization of quantitative PCR data in breast cancer. Clin Transl Oncol 16:184–190 [DOI] [PubMed] [Google Scholar]
- 29.Liu LL, Zhao H, Ma TF et al. (2015) Identification of valid reference genes for the normalization of RT-qPCR expression studies in human breast cancer cell lines treated with and without transient transfection. PLoS One 10:e0117058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wallace M, Metallo CM (2016) PGC1alpha drives a metabolic block on prostate cancer progression. Nat Cell Biol 18:589–590 [DOI] [PubMed] [Google Scholar]
- 31.LeBleu VS, O’Connell JT, Gonzalez Herrera KN et al. (2014) PGC-1α mediates mitochondrial biogenesis and oxidative phosphorylation in cancer cells to promote metastasis. Nat Cell Biol 16:992–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xu HN, Zhou R, Nioka S et al. (2009) Histological basis of MR/optical imaging of human melanoma mouse xenografts spanning a range of metastatic potentials. Adv Exp Med Biol 645:247–253. [DOI] [PMC free article] [PubMed] [Google Scholar]


