Abstract
Traumatic brain and spinal cord injuries cause permanent disability. Although progress has been made in understanding the cellular and molecular mechanisms underlying the pathophysiological changes that affect both structure and function after injury to the brain or spinal cord, there are currently no cures for either condition. This may change with the development and application of multi-layer omics, new sophisticated bioinformatics tools, and cutting-edge imaging techniques. Already, these technical advances, when combined, are revealing an unprecedented number of novel cellular and molecular targets that could be manipulated alone or in combination to repair the injured central nervous system with precision. In this review, we highlight recent advances in applying these new technologies to the study of axon regeneration and rebuilding of injured neural circuitry. We then discuss the challenges ahead to translate results produced by these technologies into clinical application to help improve the lives of individuals who have a brain or spinal cord injury.
Keywords: axon regeneration, brain injury, spinal cord injury, epigenomics, transcriptomics, kinomics, phosphoproteomics, metagenomics
Introduction
Central nervous system (CNS) trauma causes permanent disability, imposing huge economic and emotional burdens on affected family and society. No therapies exist that will effectively restore function to individuals who have an injury to the brain or spinal cord. Recovery of brain and spinal cord functions in adults might be achieved by promoting axon sprouting and regeneration, either alone or in combination with other promising approaches such as neuroprotection 1– 3, cell reprogramming and transplantation 4– 8, brain–computer interface and epidural stimulation 9– 14. In the injured adult mammalian CNS, however, axon sprouting and regeneration are limited and this is due in part to both the poor intrinsic regenerative potential of adult CNS neurons 15, 16 and the hostile cellular and molecular environment that develops at the site of injury 17– 21. These are major obstacles that must be overcome to effectively promote axon regeneration, sprouting, and functional recovery after CNS trauma 22– 24. Recent data indicate that it is possible to overcome such barriers. For example, it is feasible to reprogram adult mammalian neurons into a growth-competent state and remove extracellular growth inhibitors to promote regrowth of axons that project to the brain and spinal cord 16, 25– 32.
Here, we review the most recent data, emphasizing how omics technologies are improving our insight into novel mechanisms that regulate axon regeneration and also the feasibility of rebuilding functional neuronal circuits after CNS injury. We also discuss the challenges to applying these new discoveries in the clinic to maximize recovery of function.
Omics approaches to study spinal cord injury
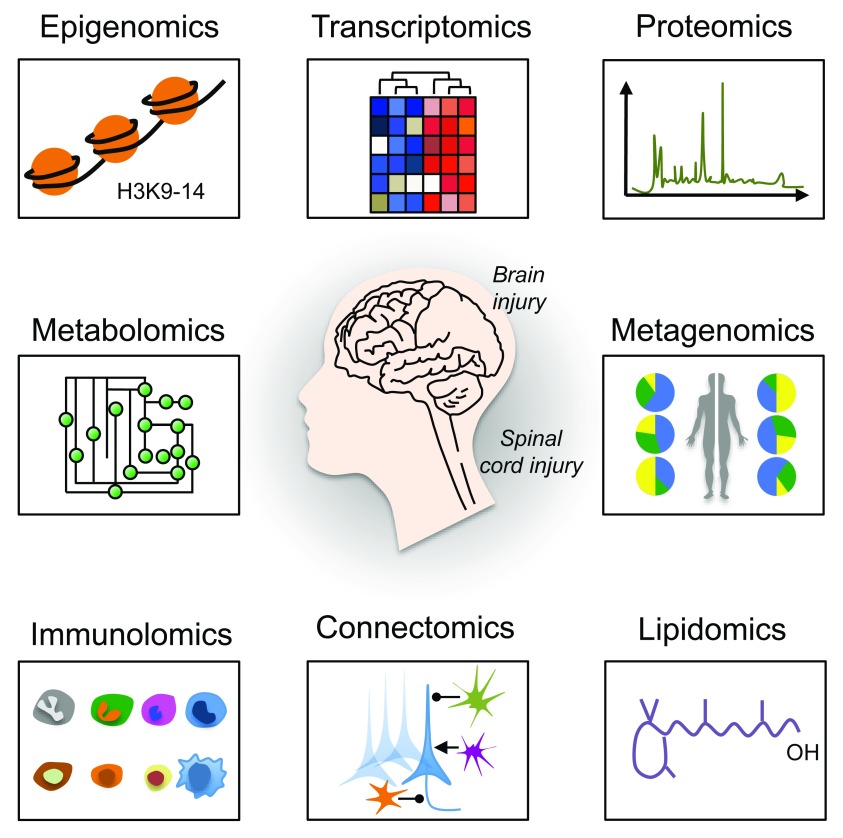
High-throughput omics technologies, including epigenomics, transcriptomics, proteomics, metabolomics, metagenomics, immunolomics, connectomics, and lipidomics have revolutionized the way we study brain and spinal cord injury (SCI) 27, 33– 41 ( Figure 1). When combined with high-content screening 42– 44, data from these technologies are providing unprecedented insight into how an injury to the brain or spinal cord affects the roles and interrelationships of various genes, molecules, cells, and body systems. As a result, novel targets and pathways are emerging as critical regulators of effective axon growth as well as regeneration and remodeling of both injured and spared neural circuits. In this review, we will focus on a subset of new omics data, using examples from epigenomics, transcriptomics, kinomics, phosphoproteomics, and metagenomics studies. We will then discuss how these technologies have helped identify novel biological mechanisms, such as neuronal metabolism and mitochondria transport, that contribute to axon regeneration failure.
Figure 1. A multi-layer omics approach to study axon regeneration.
A schematic representation of omics approaches to study axon regeneration is shown. Epigenomics studies epigenetic modifications on DNA or histone proteins that alter gene expression. DNA methylation and histone modifications are the most well-characterized epigenetic modifications. Transcriptomics examines the transcriptome that comprises all RNA transcripts (for example, mRNA and non-coding RNAs) in a given cell population. New technologies, including RNA sequencing at single-cell resolution, have been developed that allow the identification of genes and transcript variants that are actively expressed, co-expressed, or repressed. After epigenomics and transcriptomics, proteomics represents the next step in the study of any given biological system. Indeed, proteomics is the large-scale study of the proteome (for example, the set of proteins produced in an organism, system, or biological context). Protein activity is also regulated by many different factors in health and disease other than the gene expression level. Modern high-throughput technologies allow the investigation of protein location, turnover, post-translational modification, activity, and interactions in depth. Phosphoproteomics represents a branch of proteomics that focuses entirely on the identification and characterization of phosphorylated proteins. Metabolomics is the study of substrates and products (also called metabolites) of cellular metabolism and their interactions within a biological system. Each cellular activity is reflected by the presence of specific metabolites. Therefore, metabolomics represents a powerful approach to study the state and phenotype of any given biological system. Metagenomics is the genomic analysis of microbial communities from environmental and biological samples, such as the gut microbiota. Indeed, metagenomics allows the study of intestinal microbiome diversity and dysbiosis as well as its relationship between human health and disease. Immunolomics profiles cells of the immune system, antibodies, and cytokine responses in a comprehensive manner. With the advent of powerful imaging methods and molecular and genetic tools, it is now possible to create comprehensive maps of connections within the nervous system. Connectomics refers to the production and study of such connections and the molecular interactions that pair cells. One of the emerging fields of biomedical research is certainly lipidomics. Lipidomics is the large-scale study of cellular lipids at both the structural and functional levels.
Epigenomics
Recent data indicate that the induction of regenerative gene expression, a prerequisite for activating axon growth programs, relies partly on creating a more permissive chromatin environment in the nucleus of injured neurons 45– 51. Epigenomic screens of adult dorsal root ganglia (DRG) neurons injured by a peripheral nerve lesion (PNL), an experimental condition that switches DRG neurons into a regenerative-competent state, identified Tet methylcytosine dioxygenase 3 (Tet3) as a critical regulator of axon growth and regeneration. After PNL, Tet3 is upregulated along with the epigenetic mark 5-hydroxymethylcytosine (5hmC) in DRG neurons 52. By oxidizing 5hmC, Tet3 reverses DNA methylation. Interestingly, epigenomic mapping in DRG neurons after injury to the peripheral (regenerative) or central (no regenerative effect) projecting axons triggered differential 5hmC changes that were associated with distinct signaling pathways 52. Nearly half of the genes that were differentially regulated after peripheral lesion contain 5hmC alterations 52, suggesting that 5hmC is a previously unrecognized mechanism that controls the regenerative potential of injured neurons.
Although transcriptional events that turn on the expression of regeneration-associated genes are recognized as important steps in the activation of cell-autonomous regeneration programs 53, far less is known about how gene inactivation affects these programs. A recent study identified ubiquitin-like containing PHD ring finger 1 (UHRF1)-dependent DNA methylation as a critical epigenetic mechanism responsible for silencing expression of genes that are required to promote axon regeneration in DRG neurons 54. After PNL, a decrease in miR-9 causes a transient increase in the expression of the RE1-silencing transcription factor (REST) and UHRF1 54. During embryogenesis, REST acts as master regulator by inhibiting the expression of many neuronal genes 55. While a transient increase in REST primes injured DRG neurons for enhanced axon regeneration, UHRF1 interacts with DNA methyltransferases and methyl groups on histone H3, creating epigenetic marks that silence promoter elements of tumor suppressor genes such as the phosphatase and tensin homolog ( PTEN) and REST 54. Since sustained expression of REST in neurons is known to cause axon guidance defects 56, UHRF1-dependent epigenetic silencing may be required to fine-tune REST activity and thus axon regeneration programs. Together, these data support the idea that neurons may need to revert to an immature or intermediate state to successfully unlock developmental programs for axon regeneration 57– 62.
Transcriptomics
Advanced transcriptomics analyses have identified several genes and gene networks that regulate axon regeneration success and failure 27, 34, 63, 64. However, the efficiency and success of translation of these genes have received less attention. By ribosome pull-down and metabolic isotopic labeling, a recent study analyzed gene translation and protein synthesis within the regeneration-associated program in DRG neurons. Of the proteins that undergo de novo synthesis in regenerating DRG neurons, apolipoprotein E (ApoE), which has been previously implicated in axon growth and regeneration 65– 68, is one of the most robustly synthesized proteins 69. DRG neurons cultured in the presence of an ApoE receptor inhibitor extend shorter neurites, providing evidence that neuronal ApoE is an autocrine regulator of axon growth 69. It is likely, though speculative, that ApoE facilitates recycling of cholesterol from degenerating axons for integration into new membranes during the process of axon regeneration. Alternatively, cholesterol may be synthesized in the cell body and then efficiently delivered to the axonal compartment via anterograde transport of lipid-containing vesicles.
An unbiased genome-wide loss-of-function screen in cerebral cortical projection neurons in vitro identified Rab27b, a member of the Rab subfamily of GTPases, as a cell-autonomous factor that restricts axon regeneration 39. Adult worms lacking Rab27 exhibit greater regeneration of GABA neurons. Moreover, optic nerve regeneration, raphespinal sprouting, and locomotor recovery all are enhanced in mice lacking Rab27 39. Interestingly, Caenorhabditis elegans Rab27 mutants have defects in synaptic transmission 70. Given that Rab27 localizes in synaptic-rich regions and participates in the transport of synaptic vesicles 71, removing or blocking Rab27 in adult neurons may promote axon regeneration by shifting the trafficking of new cell membrane from synapses to the axolemma. Indeed, new membrane insertion is necessary for axon elongation 72.
Interestingly, data from an independent study show that selective exclusion of Rab11 vesicles, which are needed for axon elongation, contributes to axon regeneration failure. Rab GTPases coordinate vesicle trafficking 73, thereby allowing growth-promoting cargoes to be delivered to the axon. In cultured rat cortical neurons, overexpressing Rab11 decreases axon retraction and augments new growth cone formation and enhanced axon regeneration occurs in an integrin-dependent manner 74. It is likely that changes in spatiotemporal interaction between Rab GTPases and specific guanine nucleotide exchange factors contribute to diversify the role of Rab GTPases in axon growth and regeneration.
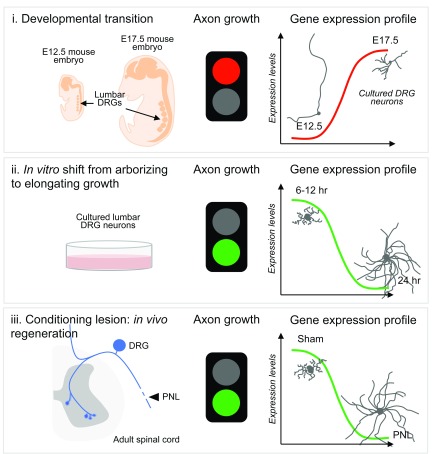
When the transcriptional landscape of mouse DRG neurons was explored in both growth-competent and -incompetent states at different developmental stages, Cacna2d2, the gene encoding the α2δ2 subunit of voltage-gated calcium channels 75, was identified as a developmental switch that limits axon growth and regeneration 27 ( Figure 2). Interestingly, in these neurons, the developmental transition from a growth-competent (electrically dormant) to a transmitting (electrically active) phase is associated with a marked increase in the expression of genes that control synapse formation and synaptic transmission. Deletion or silencing of Cacna2d2 in adult DRG neurons promotes axon growth in vitro. Pharmacological blockade of α2δ2 via systemic injection of gabapentinoids promotes regeneration of sensory axons after SCI in adult mice 27. Precisely how gabapentinoids enhance axon regeneration is unknown, but a mechanistic understanding is important, especially since these drugs are often used in humans to treat various neurological disorders, including neuropathic pain. Moreover, a multi-center cohort study found that motor recovery is improved in SCI individuals receiving gabapentinoids 76. Together, these data highlight the need to consider repurposing gabapentinoids as a novel treatment for CNS repair.
Figure 2. The transcriptional landscape of mouse dorsal root ganglia (DRG) neurons in both growth-competent and -incompetent states.
DRG neurons have been instrumental in dissecting key molecular mechanisms of axon growth and regeneration failure. Whole transcriptome sequencing of DRG neurons from different stages of axon growth, including a developmental transition from axon growth to synapse formation, a shift from arborizing to elongating growth, and axon regeneration after a peripheral nerve lesion (PNL), has identified novel negative regulators of axon growth and regeneration, thus expanding the number of targets that could be manipulated for therapeutic gain. E, embryonic day.
During development, a discrete number of transcription factors act as master regulators of gene expression. Among others, Sox11 is highly expressed in many developing organs and its expression is turned off in adults. Whereas Sox11 expression is not changed after CNS injury, its expression increases after peripheral injury, facilitating regeneration of injured peripheral nerves 34, 77. In normally non-regenerative cortical motor neurons, forcing Sox11 expression enables sprouting and regeneration of corticospinal tract (CST) axons after unilateral pyramidotomy and cervical SCI, respectively 28. However, forced expression of Sox11 in CST neurons impairs, rather than improves, skilled forelimb functions 28. Thus, improved axon regeneration does not necessarily predict that functional recovery also will improve. Intuitively, this makes sense since functionally significant axon regeneration is a multi-step repair process in which regenerating axons must re-establish proper synaptic connectivity in order to effectively integrate into existing or regrowing neuronal circuitry. Another study tested whether overexpression of Sox11 or other master regulators of gene transcription can enhance regeneration of retinal ganglion cell (RGC) axons after optic nerve crush injury in adult mice. Only overexpression of Sox11, among the seven candidates tested, robustly increased RGC regeneration 64. Interestingly, a gene ontology analysis of transcriptomics data derived from RGC neurons overexpressing Sox11 revealed that most genes that are suppressed by Sox11 are associated with synaptic transmission 64, highlighting similarities with the α2δ2 findings described above. Together, these data suggest that genetic gain-of-function manipulations can rejuvenate adult neurons, enhancing their growth potential; however, these same manipulations may inadvertently impair synaptic function in the neural circuitry.
In a search for mechanisms underlying neural plasticity, a recent study profiled the transcriptome of sprouting intact neurons isolated from mice lacking Nogo receptor 1 (for example, a receptor for myelin-associated axon growth inhibitors) after incomplete SCI. In these mice, structural plasticity and regeneration of CST axons are enhanced in various CNS injury models 78, 79. The authors found that the lysophosphatidic acid (LPA) signaling modulators, LPPR1 and LPAR1, are intrinsic regulators of axon growth in corticospinal neurons. More specifically, LPPR1 overexpression or LPAR1 inhibition promoted collateral sprouting of intact CST axons and enhanced functional recovery after unilateral pyramitomy in adult wild-type mice. LPA is a bioactive lipid species derived from membrane phospholipids, and among the many cellular mechanisms that LPA signaling is known to affect (including oligodendrocyte maturation, myelination, astrocyte proliferation, and inflammation), synaptic transmission is also affected by LPA 80.
Single-cell RNA sequencing has emerged as a powerful technology that enables researchers to identify expression changes of thousands of genes in heterogeneous cell populations 81, 82. A recent study applied this technology to reveal DRG neuron heterogeneity and molecular dynamics after sciatic nerve transection 40. DRG neurons can be classified into several functionally distinct subtypes with very different gene expression patterns 83– 85. Such heretogeneity is reflected by the fact that injury to the peripheral branch of DRG neurons is often associated with mixed responses such as pain, cell death, plasticity, and axon regeneration. After segregating DRG neurons into different subtypes, weighted gene co-expression network analysis revealed injury-responsive gene modules with distinct expression patterns among the different subtypes. Interestingly, the cell death genes—programmed cell death-2 and neuron survival-like ISL LIM homeobox—were upregulated and downregulated, respectively, in a subset of non-peptidergic nociceptor neurons 3 days after injury 40. The fact that caspase-3 was upregulated in all injured subtypes suggests that these neurons may be more susceptible to cell death and therefore not able to regenerate. Indeed, a prerequisite for axon regeneration is that injured neurons survive. Dynamic changes in gene transcription in DRG neuronal subtypes were identified by completing an analysis of the transcriptome at 3 and 7 days after axotomy. Genes related to nervous system development, axonogenesis, regulation of metabolic process, and actin cytoskeleton reorganization were gradually upregulated in large myelinated neurons 40. In contrast, many genes related to learning or memory or nucleus organization were downregulated in these neurons 40, further implicating gene inactivation as an important regulator of axon growth programs (see above).
Kinomics
A phenotypic screen of kinase inhibitors (that is, kinomics) combined with machine learning identified the ribosomal S6 kinase 1 (S6K1) as a negative regulator of axon regeneration in rodents 42. In C. elegans, ribosomal S6 kinase loss of function elicits new axon growth cone formation after injury and accelerates axon elongation 86. In vitro, S6K1 inhibition enhances growth of primary mouse hippocampal neurons. In vivo administration of a selective S6K1 inhibitor (for example, PF-4708671) promotes regeneration of CST axons into and beyond the lesion site in a model of cervical SCI. Functional recovery also is achieved in SCI animals treated with PF-4708671. The benefits of inhibiting S6K1, a known effector of the mammalian target of rapamycin (mTOR), conflict with data showing that mTOR is a positive regulator of axon regeneration in mammals 16, 87– 89. Hence, new kinomics data have enriched our understanding of molecular mechanisms of axon regeneration by showing that PI3K/mTOR signaling is negatively regulated by S6K1.
Phosphoproteomics
Growth cones are specialized structures that are required during axon growth and regeneration 90. A better understanding of the signaling pathways that control growth cone activity may be necessary to gain control of axon growth, guidance, and regeneration as well as formation of neural circuits. Reversible protein phosphorylation is one of the most studied post-translational modifications. Phosphorylation of proline, serine, threonine, and tyrosine residues plays a crucial role in function, subcellular localization, and degradation of proteins, thus participating in various cellular processes, including signal transduction. A phosphoproteomic study of growth cone membranes isolated from postnatal day 1 rat forebrain identified 4596 phosphorylation sites from 1223 phosphoproteins 41. Of these phosphorylation sites, proline phosphorylation was the most represented. Analysis of the identified phosphoproteins suggested that cytoskeletal components and signaling proteins were the most abundant 41. Using a kinase-specific phosphorylation site prediction tool, the authors of this study revealed that proline phosphorylation was due to activation of the mitogen-activated protein kinase (MAPK) pathway 41. Of note, coordinated activation of highly conserved MAPK pathways is required for axon growth and regeneration 68, 91– 93. Strikingly, the most abundant phosphorylation site was an uncharacterized serine 96 of the growth-associated protein 43 (GAP-43), which is highly expressed during development and regeneration 94– 97. Of the different kinases involved in signal transduction, c-Jun N-terminus kinase (JNK) was responsible for numerous phosphorylated sites in the phosphoproteomic data set 41.
Another interesting study applied quantitative phosphoproteomics to study changes in protein phosphorylation in primary cerebellar granule neurons plated on growth-inhibitory chondroitin sulfate proteoglycans (CSPGs). Of the differentially phosphorylated proteins, phosphorylation increased on 41 peptides and decreased on 77 in neurons exposed to CSPGs 98. Cytoskeletal proteins were the top annotated category, representing 25 of the 118 phosphopeptides identified. Among these cytoskeletal proteins, 14 were of the actin family of cytoskeleton proteins. Cofilin, an actin depolymerization factor regulated by phosphorylation, plays an important role in growth cone behavior and neurite outgrowth 99. In neurons exposed to myelin-associated inhibitor, phosphorylation and inactivation of cofilin have been shown to be regulated via LIM kinase and slingshot phosphatase 100, contributing to axon growth inhibition and regeneration failure. Similarly, overexpression of the transcription factor serum response factor enhances axon regeneration through cytoplasmic localization and cofilin-mediated reactivation of actin dynamics in growth-inert retraction bulbs 101. The top three signaling pathways representing the 118 phosphopeptides were pyrimidine metabolism, p38MAPK pathway, and synaptic vesicle trafficking. Together, these results suggest that phosphoproteomics can serve as a powerful approach to unmask promising targets and signaling pathways to overcome regeneration failure in the adult CNS.
Metagenomics
Successful axon regeneration may require a detailed understanding of genetic, proteomic, metabolic, and immunologic functions that occur in the body outside the nervous system. Metagenomics is a collection of high-throughput genetic analyses of transorganismal behaviors and the biosphere. Currently, most metagenomics studies focus on non-eukaryotic microbes, especially those found in the gastrointestinal tract (that is, the “gut”), to learn how these microbes affect organs and cells throughout the body, in both health and disease. Compelling data indicate that microbial metabolites, derived from gut microbes, directly affect the function of neurons and glia in the CNS. How or whether these metabolites will affect regeneration in the injured CNS has not been explored; however, robust and lasting changes in gut microbial communities do occur after a brain injury or SCI 102– 105. Injury-induced change in microbial populations is a potentially novel target for regulating the structure and function of injured neurons. Indeed, the magnitude and diversity of the microbial “payload” are remarkable—unique microbial genes outnumber mammalian genes by about 150 to 1 106, and an impressive number of microbial enzymes and metabolites are already known to affect the metabolism and function of mammalian cells, including those in the immune and nervous systems 107– 109. Thus, it is easy to speculate that metagenomic and metabolomic techniques, when applied in the context of axon regeneration models, will reveal novel roles for microbes in affecting axon regeneration.
Although recent progress in the field of axon regeneration clearly illustrates the power of using omics-based approaches to reveal novel molecular mechanisms to target for therapeutic enhancement of axon growth, we believe that a better understanding of the mechanisms controlling presynaptic biogenesis, synaptic alignment, and connectivity will be necessary to rebuild injured neural circuits in a functionally meaningful way.
Neuronal metabolism and mitochondrial transport
Promoting successful axon regeneration will likely require that we understand more than those genes and proteins that directly affect the physical structure of axons and synapses. Indeed, axon regeneration is a metabolically active, multi-step process. Omics technologies have revealed that optimal axon regeneration also depends on efficient mitochondrial transport and energy production in injured axons.
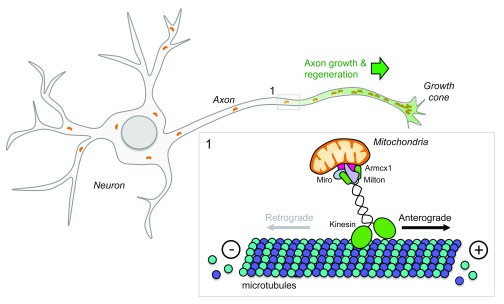
Recent evidence indicates that enhancing mitochondrial transport promotes neuron survival and axon regeneration in experimental models of axotomy in worms 110 and mice 25, 111 ( Figure 3). Anterograde mitochondrial transport requires kinesin-1 motors, whereas dynein motors control retrograde transport back to the soma 112– 114. Live imaging of laser axotomized GABA motor neurons in mutant worms with an enhanced regenerative capacity has shown that mitochondrial density increases in regenerating axons and that regeneration can be enhanced further by experimentally boosting mitochondrial transport. Conversely, axon regeneration is poor in mutant worms with deficient mitochondrial transport. Mitochondrial localization to the axon is regulated in part by dual-leucine zipper kinase 1 (DLK-1) 110, an evolutionarily conserved intrinsic regulator of axon growth and regeneration in worms, flies, and mice 92, 115– 118.
Figure 3. Mitochondria participate in axon regeneration.
Mitochondria are actively transported to axons via axonal microtubules with their plus ends pointing toward the distal part and their minus ends facing the cell soma. Members of the kinesin family are responsible for mitochondria anterograde transport. Kinesin motors generate force by hydrolyzing adenosine triphosphate. Given that mitochondria provide energy for axonal functions (including active transport and membrane fusion), alteration of mitochondrial distribution along the axon leads to defects during development, maintenance, and regeneration of the nervous system.
Again, using the optic nerve injury model, high-throughput analysis of gene expression in RGC neurons has revealed that the armadillo repeat containing x-linked 1 (Armcx1) is a critical regulator of mitochondrial transport and plays a key role in promoting axon regeneration after optic nerve crush injury. Armcx1 localizes to mitochondria and interacts with components of the mitochondrial transport machinery, such as Miro 1 25. Whereas Armcx1 overexpression enhances mitochondrial transport in mouse retinal explants and promotes RGC neuron survival and regeneration after optic nerve crush injury 25, its downregulation negatively impacts axon regeneration.
Adenosine triphosphate (ATP) is the major source of cellular energy produced by mitochondria. In injured CNS axons, mitochondria acutely depolarize, causing energy deficits along the injured axons 111. Recent data indicate that it is possible to reverse injury-induced energy loss and restore regenerative capacity in cultured mouse neurons. Indeed, overexpressing Miro 1 or knocking down the mitochondria-anchoring protein syntaphilin enhances mitochondrial transport and restores the energy balance in injured axons, leading to enhanced axon regeneration 111.
Using proteomics and bioinformatics techniques to analyze the injury response in axotomized RGCs, Belin et al. identified 12 signaling hubs, including several neuronal intrinsic regulators of axon growth and regeneration 33. The top three connected nodes are the tumor suppressor p53 119, c-Myc 120, and Rictor. The authors focused on c-myc because of its role as master transcriptional regulator of several target genes that coordinate the de novo synthesis of new lipids and proteins that are needed for axon elongation. Indeed, forced expression of c-Myc in RGCs promotes neuron survival and regeneration after optic nerve crush injury 33. Although it is possible to manipulate oncogenes to achieve regenerative growth in CNS neurons in animal models, whether it is safe or prudent to do so in humans is questionable.
A recent study in C. elegans has shed some light on metabolic regulation controlling neuron repair after axotomy. O-linked β-N-acetylglucosamine (O-GlcNAc), a post-translational modification of serine and threonine residues of nuclear and cytoplasmic proteins, functions as a nutrient sensor and metabolic mediator by linking glucose metabolism to the hexosamine biosynthetic pathway. Twenty-four hours following laser axotomy in vivo, a decrease in O-GlcNAc levels promotes axon regeneration of either the anterior or posterior lateral microtubule neurons via ARK-1/AKT-1 signaling, using glycolysis as the primary source of energy 121. Blocking glucose transport or inhibiting glycolysis leads to axon regeneration failure in mutant worms with decreased O-GlcNAc levels 121. By contrast, increasing O-GlcNAc levels acts on mitochondrial function and enhances axon regeneration in C. elegans through FOXO/DAF-16–dependent mechanisms 121. These seemingly contradictory results may be explained by the fact that O-GlcNAc levels drive distinct branches of the insulin pathway to promote regeneration in worms.
The liver kinase B1 (LKB1) links cellular metabolism and energy homeostasis to cell polarity and growth 122, 123. LKB1 phosphorylates the central metabolic sensor AMPK, whose activation regulates cholesterol, lipid, and glucose metabolism 123. LKB1 overexpression in corticospinal neurons of adult mice was recently shown to promote long-distance regeneration of CST axons in experimental models of SCI 31. Also, systemic overexpression of LKB1 in mice causes descending serotonergic and tyrosine hydroxylase-positive axons to regrow into caudal segments of the injured spinal cord. Mechanistically, the AMP-activated protein kinase alpha, NUAK family SNF1-like kinase 1, and extracellular signal–regulated kinase act as effectors of LKB1 to promote axon growth and regeneration 31. Importantly, enhanced axon growth and regeneration in LKB1-overexpressing mice correlated with improved recovery of locomotor function 31.
Together, the above examples highlight the importance of achieving efficient mitochondrial transport and energy production in injured axons to fuel axon regeneration. Whether boosting neuronal metabolism and other metabolic pathways are sufficient to repair the injured CNS requires further investigation.
Conclusions
During the last three years, novel candidates and combinatorial approaches that can promote structural plasticity, regeneration, and some degree of functional recovery have been identified 27, 28, 31, 39, 124– 128. To maximize chances to achieve functional recovery, however, axon regeneration, neuronal metabolism, synapse formation, and functional connectivity need to be spatially and temporally controlled to allow the establishment, refinement, and consolidation of essential neural circuitry 23. Thus far, data suggest that prolonged activation of neuron-intrinsic pathways causes defects in target innervation in several experimental injury models 33, 129, 130. Failure to re-innervate target neurons negatively impacts functional recovery and can cause neurobehavioral abnormalities that impair normal daily activities and thus quality of life. In addition, cardiovascular disease and autonomic dysfunction have become a growing concern for individuals with SCI 131, 132. Thus, turning off or reducing intrinsic axon growth ability together with cardiovascular rehabilitation, activity-based training, or other facilitators may indeed facilitate synapse formation, refinement, and consolidation of functional connectivity in the injured CNS. Several lines of evidence also suggest that adult regenerating axons can be guided toward specific target areas by providing chemoattraction 133, 134. Modulating astrocyte behavior to control synapse formation and elimination represents another intriguing direction for future studies 135, 136.
Several signaling pathways described above are conserved across different species. This will likely facilitate the translation of data obtained in smaller organisms and animal models to larger and more complex mammalian systems, including humans. Still, genetic variation exists within each model organism 36, 137, 138, so exploring the robustness of treatment strategies across different genetic backgrounds, within and between species, will be prudent before embarking on randomized clinical trials in humans. Lately, despite the generation of large omics data sets, a significant amount of information remains hidden. In our opinion, validation of omics approaches with stringent criteria and additional assays is an essential step to facilitate translation of breakthrough discoveries from the laboratory into clinical practice. Although there is no optimal strategy for integrating multi-omics data sets, more integration is likely to provide the most realistic picture about true biology. It is now possible to integrate data from transcriptomics, phosphoproteomics, and metabolomics. When combined with multi-layer omics 139– 141, the recent development of powerful computational methods 142– 144, machine learning and artificial intelligence 145, 146 will allow data mining and extracting principles and key biological information on a broad range of normal and disease conditions. Hence, automated inference methods should allow the rapid development and testing of new hypotheses and establish potential causal relationships in large data sets. As we enter a new era of regenerative medicine, we will be able to select combinations of treatment strategies for a personalized medicine approach to aid CNS repair.
Editorial Note on the Review Process
F1000 Faculty Reviews are commissioned from members of the prestigious F1000 Faculty and are edited as a service to readers. In order to make these reviews as comprehensive and accessible as possible, the referees provide input before publication and only the final, revised version is published. The referees who approved the final version are listed with their names and affiliations but without their reports on earlier versions (any comments will already have been addressed in the published version).
The referees who approved this article are:
Simone Di Giovanni, Department of Medicine, Imperial College London, London, UK
Shuxin Li, Shriners Hospitals Pediatric Research Center and Department of Anatomy and Cell Biology, Lewis Katz School of Medicine at Temple University, Philadelphia, PA, USA
Melissa R Andrews, Biological Sciences, University of Southampton, Southampton, UK
Funding Statement
AT is supported by the Craig H. Neilsen Foundation, the Marina Romoli Onlus Association, the Discovery Themes Initiative on Chronic Brain Injury, and The Ohio State University. PGP is supported by the National Institute of Neurological Disorders and Stroke and the Ray W. Poppleton Endowment.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
[version 1; peer review: 3 approved]
References
- 1. Fehlings MG, Vaccaro A, Wilson JR, et al. : Early versus delayed decompression for traumatic cervical spinal cord injury: Results of the Surgical Timing in Acute Spinal Cord Injury Study (STASCIS). PLoS One. 2012;7(2):e32037. 10.1371/journal.pone.0032037 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 2. Fehlings MG, Wilson JR, Frankowski RF, et al. : Riluzole for the treatment of acute traumatic spinal cord injury: Rationale for and design of the NACTN Phase I clinical trial. J Neurosurg Spine. 2012;17(1 Suppl):151–6. 10.3171/2012.4.AOSPINE1259 [DOI] [PubMed] [Google Scholar]
- 3. Festoff BW, Ameenuddin S, Arnold PM, et al. : Minocycline neuroprotects, reduces microgliosis, and inhibits caspase protease expression early after spinal cord injury. J Neurochem. 2006;97(5):1314–26. 10.1111/j.1471-4159.2006.03799.x [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 4. Su Z, Niu W, Liu ML, et al. : In vivo conversion of astrocytes to neurons in the injured adult spinal cord. Nat Commun. 2014;5:3338. 10.1038/ncomms4338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kadoya K, Lu P, Nguyen K, et al. : Spinal cord reconstitution with homologous neural grafts enables robust corticospinal regeneration. Nat Med. 2016;22(5):479–87. 10.1038/nm.4066 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 6. Lu P, Woodruff G, Wang Y, et al. : Long-distance axonal growth from human induced pluripotent stem cells after spinal cord injury. Neuron. 2014;83(4):789–96. 10.1016/j.neuron.2014.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 7. Rosenzweig ES, Brock JH, Lu P, et al. : Restorative effects of human neural stem cell grafts on the primate spinal cord. Nat Med. 2018;24(4):484–90. 10.1038/nm.4502 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 8. Assinck P, Duncan GJ, Hilton BJ, et al. : Cell transplantation therapy for spinal cord injury. Nat Neurosci. 2017;20(5):637–47. 10.1038/nn.4541 [DOI] [PubMed] [Google Scholar]
- 9. Capogrosso M, Milekovic T, Borton D, et al. : A brain-spine interface alleviating gait deficits after spinal cord injury in primates. Nature. 2016;539(7628):284–8. 10.1038/nature20118 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 10. Harkema S, Gerasimenko Y, Hodes J, et al. : Effect of epidural stimulation of the lumbosacral spinal cord on voluntary movement, standing, and assisted stepping after motor complete paraplegia: a case study. Lancet. 2011;377(9781):1938–47. 10.1016/S0140-6736(11)60547-3 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 11. Minev IR, Musienko P, Hirsch A, et al. : Biomaterials. Electronic dura mater for long-term multimodal neural interfaces. Science. 2015;347(6218):159–63. 10.1126/science.1260318 [DOI] [PubMed] [Google Scholar]
- 12. Angeli CA, Boakye M, Morton RA, et al. : Recovery of Over-Ground Walking after Chronic Motor Complete Spinal Cord Injury. N Engl J Med. 2018;379(13):1244–50. 10.1056/NEJMoa1803588 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 13. Formento E, Minassian K, Wagner F, et al. : Electrical spinal cord stimulation must preserve proprioception to enable locomotion in humans with spinal cord injury. Nat Neurosci. 2018;21(12):1728–41. 10.1038/s41593-018-0262-6 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 14. Gill ML, Grahn PJ, Calvert JS, et al. : Neuromodulation of lumbosacral spinal networks enables independent stepping after complete paraplegia. Nat Med. 2018;24(11):1677–82. 10.1038/s41591-018-0175-7 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 15. Curcio M, Bradke F: Axon Regeneration in the Central Nervous System: Facing the Challenges from the Inside. Annu Rev Cell Dev Biol. 2018;34:495–521. 10.1146/annurev-cellbio-100617-062508 [DOI] [PubMed] [Google Scholar]
- 16. Liu K, Lu Y, Lee JK, et al. : PTEN deletion enhances the regenerative ability of adult corticospinal neurons. Nat Neurosci. 2010;13(9):1075–81. 10.1038/nn.2603 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 17. Chen M, Geoffroy CG, Meves JM, et al. : Leucine Zipper-Bearing Kinase Is a Critical Regulator of Astrocyte Reactivity in the Adult Mammalian CNS. Cell Rep. 2018;22(13):3587–97. 10.1016/j.celrep.2018.02.102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dias DO, Kim H, Holl D, et al. : Reducing Pericyte-Derived Scarring Promotes Recovery after Spinal Cord Injury. Cell. 2018;173(1):153–165.e22. 10.1016/j.cell.2018.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 19. Geoffroy CG, Zheng B: Myelin-associated inhibitors in axonal growth after CNS injury. Curr Opin Neurobiol. 2014;27:31–8. 10.1016/j.conb.2014.02.012 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 20. Schwab ME, Strittmatter SM: Nogo limits neural plasticity and recovery from injury. Curr Opin Neurobiol. 2014;27:53–60. 10.1016/j.conb.2014.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Silver J, Schwab ME, Popovich PG: Central nervous system regenerative failure: Role of oligodendrocytes, astrocytes, and microglia. Cold Spring Harb Perspect Biol. 2014;7(3):a020602. 10.1101/cshperspect.a020602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. He Z, Jin Y: Intrinsic Control of Axon Regeneration. Neuron. 2016;90(3):437–51. 10.1016/j.neuron.2016.04.022 [DOI] [PubMed] [Google Scholar]
- 23. Tedeschi A, Bradke F: Spatial and temporal arrangement of neuronal intrinsic and extrinsic mechanisms controlling axon regeneration. Curr Opin Neurobiol. 2017;42:118–27. 10.1016/j.conb.2016.12.005 [DOI] [PubMed] [Google Scholar]
- 24. Tran AP, Warren PM, Silver J: The Biology of Regeneration Failure and Success After Spinal Cord Injury. Physiol Rev. 2018;98(2):881–917. 10.1152/physrev.00017.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cartoni R, Norsworthy MW, Bei F, et al. : The Mammalian-Specific Protein Armcx1 Regulates Mitochondrial Transport during Axon Regeneration. Neuron. 2016;92(6):1294–307. 10.1016/j.neuron.2016.10.060 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 26. Luo X, Ribeiro M, Bray ER, et al. : Enhanced Transcriptional Activity and Mitochondrial Localization of STAT3 Co-induce Axon Regrowth in the Adult Central Nervous System. Cell Rep. 2016;15(2):398–410. 10.1016/j.celrep.2016.03.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Tedeschi A, Dupraz S, Laskowski CJ, et al. : The Calcium Channel Subunit Alpha2delta2 Suppresses Axon Regeneration in the Adult CNS. Neuron. 2016;92(2):419–34. 10.1016/j.neuron.2016.09.026 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 28. Wang Z, Reynolds A, Kirry A, et al. : Overexpression of Sox11 promotes corticospinal tract regeneration after spinal injury while interfering with functional recovery. J Neurosci. 2015;35(7):3139–45. 10.1523/JNEUROSCI.2832-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Warren PM, Steiger SC, Dick TE, et al. : Rapid and robust restoration of breathing long after spinal cord injury. Nat Commun. 2018;9(1):4843. 10.1038/s41467-018-06937-0 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 30. Burnside ER, De Winter F, Didangelos A, et al. : Immune-evasive gene switch enables regulated delivery of chondroitinase after spinal cord injury. Brain. 2018;141(8):2362–2381. 10.1093/brain/awy158 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 31. Ohtake Y, Sami A, Jiang X, et al. : Promoting Axon Regeneration in Adult CNS by Targeting Liver Kinase B1. Mol Ther. 2019;27(1):102–17. 10.1016/j.ymthe.2018.10.019 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 32. O'Donovan KJ, Ma K, Guo H, et al. : B-RAF kinase drives developmental axon growth and promotes axon regeneration in the injured mature CNS. J Exp Med. 2014;211(5):801–14. 10.1084/jem.20131780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Belin S, Nawabi H, Wang C, et al. : Injury-induced decline of intrinsic regenerative ability revealed by quantitative proteomics. Neuron. 2015;86(4):1000–14. 10.1016/j.neuron.2015.03.060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Chandran V, Coppola G, Nawabi H, et al. : A Systems-Level Analysis of the Peripheral Nerve Intrinsic Axonal Growth Program. Neuron. 2016;89(5):956–70. 10.1016/j.neuron.2016.01.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Li S, Nie EH, Yin Y, et al. : GDF10 is a signal for axonal sprouting and functional recovery after stroke. Nat Neurosci. 2015;18(12):1737–45. 10.1038/nn.4146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Omura T, Omura K, Tedeschi A, et al. : Robust Axonal Regeneration Occurs in the Injured CAST/Ei Mouse CNS. Neuron. 2015;86(5):1215–27. 10.1016/j.neuron.2015.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Huebner EA, Budel S, Jiang Z, et al. : Diltiazem Promotes Regenerative Axon Growth. Mol Neurobiol. 2018. 10.1007/s12035-018-1349-5 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 38. Posti JP, Dickens AM, Orešic M, et al. : Metabolomics Profiling As a Diagnostic Tool in Severe Traumatic Brain Injury. Front Neurol. 2017;8:398. 10.3389/fneur.2017.00398 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 39. Sekine Y, Lin-Moore A, Chenette DM, et al. : Functional Genome-wide Screen Identifies Pathways Restricting Central Nervous System Axonal Regeneration. Cell Rep. 2018;23(2):415–28. 10.1016/j.celrep.2018.03.058 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 40. Hu G, Huang K, Hu Y, et al. : Single-cell RNA-seq reveals distinct injury responses in different types of DRG sensory neurons. Sci Rep. 2016;6: 31851. 10.1038/srep31851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kawasaki A, Okada M, Tamada A, et al. : Growth Cone Phosphoproteomics Reveals that GAP-43 Phosphorylated by JNK Is a Marker of Axon Growth and Regeneration. iScience. 2018;4:190–203. 10.1016/j.isci.2018.05.019 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 42. Al-Ali H, Ding Y, Slepak T, et al. : The mTOR Substrate S6 Kinase 1 (S6K1) Is a Negative Regulator of Axon Regeneration and a Potential Drug Target for Central Nervous System Injury. J Neurosci. 2017;37(30):7079–95. 10.1523/JNEUROSCI.0931-17.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 43. Al-Ali H, Lemmon VP, Bixby JL: Phenotypic Screening of Small-Molecule Inhibitors: Implications for Therapeutic Discovery and Drug Target Development in Traumatic Brain Injury. Methods Mol Biol. 2016;1462:677–88. 10.1007/978-1-4939-3816-2_37 [DOI] [PubMed] [Google Scholar]
- 44. Blackmore MG, Moore DL, Smith RP, et al. : High content screening of cortical neurons identifies novel regulators of axon growth. Mol Cell Neurosci. 2010;44(1):43–54. 10.1016/j.mcn.2010.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Cho Y, Sloutsky R, Naegle KM, et al. : Injury-induced HDAC5 nuclear export is essential for axon regeneration. Cell. 2013;155(4):894–908. 10.1016/j.cell.2013.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 46. Finelli MJ, Wong JK, Zou H: Epigenetic regulation of sensory axon regeneration after spinal cord injury. J Neurosci. 2013;33(50):19664–76. 10.1523/JNEUROSCI.0589-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mahar M, Cavalli V: Intrinsic mechanisms of neuronal axon regeneration. Nat Rev Neurosci. 2018;19(6):323–37. 10.1038/s41583-018-0001-8 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 48. Palmisano I, Di Giovanni S: Advances and Limitations of Current Epigenetic Studies Investigating Mammalian Axonal Regeneration. Neurotherapeutics. 2018;15(3):529–40. 10.1007/s13311-018-0636-1 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 49. Puttagunta R, Tedeschi A, Sória MG, et al. : PCAF-dependent epigenetic changes promote axonal regeneration in the central nervous system. Nat Commun. 2014;5: 3527. 10.1038/ncomms4527 [DOI] [PubMed] [Google Scholar]
- 50. Venkatesh I, Mehra V, Wang Z, et al. : Developmental Chromatin Restriction of Pro-Growth Gene Networks Acts as an Epigenetic Barrier to Axon Regeneration in Cortical Neurons. Dev Neurobiol. 2018;78(10):960–77. 10.1002/dneu.22605 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 51. Weng YL, An R, Cassin J, et al. : An Intrinsic Epigenetic Barrier for Functional Axon Regeneration. Neuron. 2017;94(2):337–346.e6. 10.1016/j.neuron.2017.03.034 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 52. Loh YE, Koemeter-Cox A, Finelli MJ, et al. : Comprehensive mapping of 5-hydroxymethylcytosine epigenetic dynamics in axon regeneration. Epigenetics. 2017;12(2):77–92. 10.1080/15592294.2016.1264560 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 53. Tedeschi A: Tuning the orchestra: transcriptional pathways controlling axon regeneration. Front Mol Neurosci. 2011;4:60. 10.3389/fnmol.2011.00060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Oh YM, Mahar M, Ewan EE, et al. : Epigenetic regulator UHRF1 inactivates REST and growth suppressor gene expression via DNA methylation to promote axon regeneration. Proc Natl Acad Sci U S A. 2018;115(52):E12417–E12426. 10.1073/pnas.1812518115 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 55. Chen ZF, Paquette AJ, Anderson DJ: NRSF/REST is required in vivo for repression of multiple neuronal target genes during embryogenesis. Nat Genet. 1998;20(2):136–42. 10.1038/2431 [DOI] [PubMed] [Google Scholar]
- 56. Paquette AJ, Perez SE, Anderson DJ: Constitutive expression of the neuron-restrictive silencer factor (NRSF)/REST in differentiating neurons disrupts neuronal gene expression and causes axon pathfinding errors in vivo. Proc Natl Acad Sci U S A. 2000;97(22):12318–23. 10.1073/pnas.97.22.12318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Enes J, Langwieser N, Ruschel J, et al. : Electrical activity suppresses axon growth through Ca v1.2 channels in adult primary sensory neurons. Curr Biol. 2010;20(13):1154–64. 10.1016/j.cub.2010.05.055 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 58. Filbin MT: Recapitulate development to promote axonal regeneration: Good or bad approach? Philos Trans R Soc Lond B Biol Sci. 2006;361(1473):1565–74. 10.1098/rstb.2006.1885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Hilton BJ, Bradke F: Can injured adult CNS axons regenerate by recapitulating development? Development. 2017;144(19):3417–29. 10.1242/dev.148312 [DOI] [PubMed] [Google Scholar]
- 60. Kaplan A, Bueno M, Hua L, et al. : Maximizing functional axon repair in the injured central nervous system: Lessons from neuronal development. Dev Dyn. 2018;247(1):18–23. 10.1002/dvdy.24536 [DOI] [PubMed] [Google Scholar]
- 61. Lorenzana AO, Lee JK, Mui M, et al. : A surviving intact branch stabilizes remaining axon architecture after injury as revealed by in vivo imaging in the mouse spinal cord. Neuron. 2015;86(4):947–54. 10.1016/j.neuron.2015.03.061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tedeschi A, He Z: Axon regeneration: electrical silencing is a condition for regrowth. Curr Biol. 2010;20(17):R713–4. 10.1016/j.cub.2010.07.006 [DOI] [PubMed] [Google Scholar]
- 63. Fink KL, López-Giráldez F, Kim IJ, et al. : Identification of Intrinsic Axon Growth Modulators for Intact CNS Neurons after Injury. Cell Rep. 2017;18(11):2687–701. 10.1016/j.celrep.2017.02.058 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 64. Norsworthy MW, Bei F, Kawaguchi R, et al. : Sox11 Expression Promotes Regeneration of Some Retinal Ganglion Cell Types but Kills Others. Neuron. 2017;94(6):1112–1120.e4. 10.1016/j.neuron.2017.05.035 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 65. Comley LH, Fuller HR, Wishart TM, et al. : ApoE isoform-specific regulation of regeneration in the peripheral nervous system. Hum Mol Genet. 2011;20(12):2406–21. 10.1093/hmg/ddr147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Jiménez CR, Stam FJ, Li KW, et al. : Proteomics of the injured rat sciatic nerve reveals protein expression dynamics during regeneration. Mol Cell Proteomics. 2005;4(2):120–32. 10.1074/mcp.M400076-MCP200 [DOI] [PubMed] [Google Scholar]
- 67. Li FQ, Fowler KA, Neil JE, et al. : An apolipoprotein E-mimetic stimulates axonal regeneration and remyelination after peripheral nerve injury. J Pharmacol Exp Ther. 2010;334(1):106–15. 10.1124/jpet.110.167882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yin C, Guo ZD, He ZZ, et al. : Apolipoprotein E Affects In Vitro Axonal Growth and Regeneration via the MAPK Signaling Pathway. Cell Transplant. 2018; 963689718808736. 10.1177/0963689718808736 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 69. Rozenbaum M, Rajman M, Rishal I, et al. : Translatome Regulation in Neuronal Injury and Axon Regrowth. eNeuro. 2018;5(2): pii: ENEURO.0276-17.2018. 10.1523/ENEURO.0276-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 70. Mahoney TR, Liu Q, Itoh T, et al. : Regulation of synaptic transmission by RAB-3 and RAB-27 in Caenorhabditis elegans. Mol Biol Cell. 2006;17(6):2617–25. 10.1091/mbc.e05-12-1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Pavlos NJ, Grønborg M, Riedel D, et al. : Quantitative analysis of synaptic vesicle Rabs uncovers distinct yet overlapping roles for Rab3a and Rab27b in Ca 2+-triggered exocytosis. J Neurosci. 2010;30(40):13441–53. 10.1523/JNEUROSCI.0907-10.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Quiroga S, Bisbal M, Cáceres A: Regulation of plasma membrane expansion during axon formation. Dev Neurobiol. 2018;78(3):170–80. 10.1002/dneu.22553 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 73. Stenmark H: Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 2009;10(8):513–25. 10.1038/nrm2728 [DOI] [PubMed] [Google Scholar]
- 74. Koseki H, Donegá M, Lam BY, et al. : Selective rab11 transport and the intrinsic regenerative ability of CNS axons. eLife. 2017;6: pii: e26956. 10.7554/eLife.26956 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 75. Dolphin AC: Calcium channel auxiliary α 2δ and β subunits: Trafficking and one step beyond. Nat Rev Neurosci. 2012;13(8):542–55. 10.1038/nrn3311 [DOI] [PubMed] [Google Scholar]
- 76. Warner FM, Cragg JJ, Jutzeler CR, et al. : Early Administration of Gabapentinoids Improves Motor Recovery after Human Spinal Cord Injury. Cell Rep. 2017;18(7):1614–8. 10.1016/j.celrep.2017.01.048 [DOI] [PubMed] [Google Scholar]
- 77. Jankowski MP, McIlwrath SL, Jing X, et al. : Sox11 transcription factor modulates peripheral nerve regeneration in adult mice. Brain Res. 2009;1256:43–54. 10.1016/j.brainres.2008.12.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Fink KL, Strittmatter SM, Cafferty WB: Comprehensive Corticospinal Labeling with mu-crystallin Transgene Reveals Axon Regeneration after Spinal Cord Trauma in ngr1 -/- Mice. J Neurosci. 2015;35(46):15403–18. 10.1523/JNEUROSCI.3165-15.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Meves JM, Geoffroy CG, Kim ND, et al. : Oligodendrocytic but not neuronal Nogo restricts corticospinal axon sprouting after CNS injury. Exp Neurol. 2018;309:32–43. 10.1016/j.expneurol.2018.07.013 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 80. Yung YC, Stoddard NC, Mirendil H, et al. : Lysophosphatidic Acid signaling in the nervous system. Neuron. 2015;85(4):669–82. 10.1016/j.neuron.2015.01.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Tabula Muris Consortium; Overall coordination; Logistical coordination, et al. : Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562(7727):367–72. 10.1038/s41586-018-0590-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Häring M, Zeisel A, Hochgerner H, et al. : Neuronal atlas of the dorsal horn defines its architecture and links sensory input to transcriptional cell types. Nat Neurosci. 2018;21(6):869–80. 10.1038/s41593-018-0141-1 [DOI] [PubMed] [Google Scholar]
- 83. Chiu IM, Barrett LB, Williams EK, et al. : Transcriptional profiling at whole population and single cell levels reveals somatosensory neuron molecular diversity. eLife. 2014;3:e04660. 10.7554/eLife.04660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Li CL, Li KC, Wu D, et al. : Somatosensory neuron types identified by high-coverage single-cell RNA-sequencing and functional heterogeneity. Cell Res. 2016;26(1):83–102. 10.1038/cr.2015.149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Usoskin D, Furlan A, Islam S, et al. : Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat Neurosci. 2015;18(1):145–53. 10.1038/nn.3881 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 86. Hubert T, Wu Z, Chisholm AD, et al. : S6 kinase inhibits intrinsic axon regeneration capacity via AMP kinase in Caenorhabditis elegans. J Neurosci. 2014;34(3):758–63. 10.1523/JNEUROSCI.2886-13.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Christie KJ, Webber CA, Martinez JA, et al. : PTEN inhibition to facilitate intrinsic regenerative outgrowth of adult peripheral axons. J Neurosci. 2010;30(27):9306–15. 10.1523/JNEUROSCI.6271-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Park KK, Liu K, Hu Y, et al. : Promoting axon regeneration in the adult CNS by modulation of the PTEN/mTOR pathway. Science. 2008;322(5903):963–6. 10.1126/science.1161566 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 89. Yang L, Miao L, Liang F, et al. : The mTORC1 effectors S6K1 and 4E-BP play different roles in CNS axon regeneration. Nat Commun. 2014;5: 5416. 10.1038/ncomms6416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Bradke F, Fawcett JW, Spira ME: Assembly of a new growth cone after axotomy: The precursor to axon regeneration. Nat Rev Neurosci. 2012;13(3):183–93. 10.1038/nrn3176 [DOI] [PubMed] [Google Scholar]
- 91. Widmann C, Gibson S, Jarpe MB, et al. : Mitogen-activated protein kinase: Conservation of a three-kinase module from yeast to human. Physiol Rev. 1999;79(1):143–80. 10.1152/physrev.1999.79.1.143 [DOI] [PubMed] [Google Scholar]
- 92. Hammarlund M, Nix P, Hauth L, et al. : Axon Regeneration Requires a Conserved MAP Kinase Pathway. Science. 2009;323(5915):802–6. 10.1126/science.1165527 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 93. Nix P, Hisamoto N, Matsumoto K, et al. : Axon regeneration requires coordinate activation of p38 and JNK MAPK pathways. Proc Natl Acad Sci U S A. 2011;108(26):10738–43. 10.1073/pnas.1104830108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Basi GS, Jacobson RD, Virág I, et al. : Primary structure and transcriptional regulation of GAP-43, a protein associated with nerve growth. Cell. 1987;49(6):785–91. 10.1016/0092-8674(87)90616-7 [DOI] [PubMed] [Google Scholar]
- 95. Benowitz LI, Perrone-Bizzozero NI, Finklestein SP, et al. : Localization of the growth-associated phosphoprotein GAP-43 (B-50, F1) in the human cerebral cortex. J Neurosci. 1989;9(3):990–5. 10.1523/JNEUROSCI.09-03-00990.1989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Nakamura F, Strittmatter P, Strittmatter SM: GAP-43 Augmentation of G Protein-Mediated Signal Transduction Is Regulated by Both Phosphorylation and Palmitoylation. J Neurochem. 1998;70(3):983–92. 10.1046/j.1471-4159.1998.70030983.x [DOI] [PubMed] [Google Scholar]
- 97. Strittmatter SM, Igarashi M, Fishman MC: GAP-43 amino terminal peptides modulate growth cone morphology and neurite outgrowth. J Neurosci. 1994;14(9):5503–13. 10.1523/JNEUROSCI.14-09-05503.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Yu P, Pisitkun T, Wang G, et al. : Global analysis of neuronal phosphoproteome regulation by chondroitin sulfate proteoglycans. PLoS One. 2013;8(3):e59285. 10.1371/journal.pone.0059285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Meberg PJ, Bamburg JR: Increase in Neurite Outgrowth Mediated by Overexpression of Actin Depolymerizing Factor. J Neurosci. 2000;20(7):2459–69. 10.1523/JNEUROSCI.20-07-02459.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Hsieh SHK, Ferraro GB, Fournier AE: Myelin-associated inhibitors regulate cofilin phosphorylation and neuronal inhibition through LIM kinase and Slingshot phosphatase. J Neurosci. 2006;26(3):1006–15. 10.1523/JNEUROSCI.2806-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Stern S, Haverkamp S, Sinske D, et al. : The transcription factor serum response factor stimulates axon regeneration through cytoplasmic localization and cofilin interaction. J Neurosci. 2013;33(48):18836–48. 10.1523/JNEUROSCI.3029-13.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Gungor B, Adiguzel E, Gursel I, et al. : Intestinal Microbiota in Patients with Spinal Cord Injury. PLoS One. 2016;11(1):e0145878. 10.1371/journal.pone.0145878 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 103. Kigerl KA, Hall JC, Wang L, et al. : Gut dysbiosis impairs recovery after spinal cord injury. J Exp Med. 2016;213(12):2603–20. 10.1084/jem.20151345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Ma EL, Smith AD, Desai N, et al. : Bidirectional brain-gut interactions and chronic pathological changes after traumatic brain injury in mice. Brain Behav Immun. 2017;66:56–69. 10.1016/j.bbi.2017.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 105. O'Connor G, Jeffrey E, Madorma D, et al. : Investigation of Microbiota Alterations and Intestinal Inflammation Post-Spinal Cord Injury in Rat Model. J Neurotrauma. 2018;35(18):2159–66. 10.1089/neu.2017.5349 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 106. Vázquez-Baeza Y, Callewaert C, Debelius J, et al. : Impacts of the Human Gut Microbiome on Therapeutics. Annu Rev Pharmacol Toxicol. 2018;58:253–70. 10.1146/annurev-pharmtox-042017-031849 [DOI] [PubMed] [Google Scholar]
- 107. El Aidy S, Dinan TG, Cryan JF: Gut Microbiota: The Conductor in the Orchestra of Immune-Neuroendocrine Communication. Clin Ther. 2015;37(5):954–67. 10.1016/j.clinthera.2015.03.002 [DOI] [PubMed] [Google Scholar]
- 108. Hill DA, Artis D: Intestinal bacteria and the regulation of immune cell homeostasis. Annu Rev Immunol. 2010;28:623–67. 10.1146/annurev-immunol-030409-101330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Ochoa-Repáraz J, Mielcarz DW, Begum-Haque S, et al. : Gut, bugs, and brain: role of commensal bacteria in the control of central nervous system disease. Ann Neurol. 2011;69(2):240–7. 10.1002/ana.22344 [DOI] [PubMed] [Google Scholar]
- 110. Han SM, Baig HS, Hammarlund M: Mitochondria Localize to Injured Axons to Support Regeneration. Neuron. 2016;92(6):1308–23. 10.1016/j.neuron.2016.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 111. Zhou B, Yu P, Lin MY, et al. : Facilitation of axon regeneration by enhancing mitochondrial transport and rescuing energy deficits. J Cell Biol. 2016;214(1):103–19. 10.1083/jcb.201605101 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 112. Glater EE, Megeath LJ, Stowers RS, et al. : Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J Cell Biol. 2006;173(4):545–57. 10.1083/jcb.200601067 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 113. Lin MY, Sheng ZH: Regulation of mitochondrial transport in neurons. Exp Cell Res. 2015;334(1):35–44. 10.1016/j.yexcr.2015.01.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Saxton WM, Hollenbeck PJ: The axonal transport of mitochondria. J Cell Sci. 2012;125(Pt 9):2095–104. 10.1242/jcs.053850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Hao Y, Frey E, Yoon C, et al. : An evolutionarily conserved mechanism for cAMP elicited axonal regeneration involves direct activation of the dual leucine zipper kinase DLK. eLife. 2016;5: pii: e14048. 10.7554/eLife.14048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Shin JE, Cho Y, Beirowski B, et al. : Dual leucine zipper kinase is required for retrograde injury signaling and axonal regeneration. Neuron. 2012;74(6):1015–22. 10.1016/j.neuron.2012.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Tedeschi A, Bradke F: The DLK signalling pathway--a double-edged sword in neural development and regeneration. EMBO Rep. 2013;14(7):605–14. 10.1038/embor.2013.64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Yan D, Wu Z, Chisholm AD, et al. : The DLK-1 kinase promotes mRNA stability and local translation in C. elegans synapses and axon regeneration. Cell. 2009;138(5):1005–18. 10.1016/j.cell.2009.06.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Tedeschi A, Di Giovanni S: The non-apoptotic role of p53 in neuronal biology: enlightening the dark side of the moon. EMBO Rep. 2009;10(6):576–83. 10.1038/embor.2009.89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Stine ZE, Walton ZE, Altman BJ, et al. : MYC, Metabolism, and Cancer. Cancer Discov. 2015;5(10):1024–39. 10.1158/2159-8290.CD-15-0507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Taub DG, Awal MR, Gabel CV: O-GlcNAc Signaling Orchestrates the Regenerative Response to Neuronal Injury in Caenorhabditis elegans. Cell Rep. 2018;24(8):1931–1938.e3. 10.1016/j.celrep.2018.07.078 [DOI] [PubMed] [Google Scholar]; F1000 Recommendation
- 122. Hardie DG: AMPK: positive and negative regulation, and its role in whole-body energy homeostasis. Curr Opin Cell Biol. 2015;33:1–7. 10.1016/j.ceb.2014.09.004 [DOI] [PubMed] [Google Scholar]
- 123. Shackelford DB, Shaw RJ: The LKB1-AMPK pathway: metabolism and growth control in tumour suppression. Nat Rev Cancer. 2009;9(8):563–75. 10.1038/nrc2676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Geoffroy CG, Lorenzana AO, Kwan JP, et al. : Effects of PTEN and Nogo Codeletion on Corticospinal Axon Sprouting and Regeneration in Mice. J Neurosci. 2015;35(16):6413–28. 10.1523/JNEUROSCI.4013-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Gobrecht P, Andreadaki A, Diekmann H, et al. : Promotion of Functional Nerve Regeneration by Inhibition of Microtubule Detyrosination. J Neurosci. 2016;36(14):3890–902. 10.1523/JNEUROSCI.4486-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Liu Y, Wang X, Li W, et al. : A Sensitized IGF1 Treatment Restores Corticospinal Axon-Dependent Functions. Neuron. 2017;95(4):817–833.e4. 10.1016/j.neuron.2017.07.037 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 127. Ruschel J, Hellal F, Flynn KC, et al. : Axonal regeneration. Systemic administration of epothilone B promotes axon regeneration after spinal cord injury. Science. 2015;348(6232):347–52. 10.1126/science.aaa2958 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 128. Wang XW, Li Q, Liu CM, et al. : Lin28 Signaling Supports Mammalian PNS and CNS Axon Regeneration. Cell Rep. 2018;24(10):2540–2552.e6. 10.1016/j.celrep.2018.07.105 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 129. Carlin D, Golden JP, Mogha A, et al. : Deletion of Tsc2 in Nociceptors Reduces Target Innervation, Ion Channel Expression, and Sensitivity to Heat. eNeuro. 2018;5(2): pii: ENEURO.0436-17.2018. 10.1523/ENEURO.0436-17.2018 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 130. Pernet V, Joly S, Dalkara D, et al. : Long-distance axonal regeneration induced by CNTF gene transfer is impaired by axonal misguidance in the injured adult optic nerve. Neurobiol Dis. 2013;51:202–13. 10.1016/j.nbd.2012.11.011 [DOI] [PubMed] [Google Scholar]
- 131. Myers J, Lee M, Kiratli J: Cardiovascular disease in spinal cord injury: an overview of prevalence, risk, evaluation, and management. Am J Phys Med Rehabil. 2007;86(2):142–52. 10.1097/PHM.0b013e31802f0247 [DOI] [PubMed] [Google Scholar]
- 132. Prüss H, Tedeschi A, Thiriot A, et al. : Spinal cord injury-induced immunodeficiency is mediated by a sympathetic-neuroendocrine adrenal reflex. Nat Neurosci. 2017;20(11):1549–59. 10.1038/nn.4643 [DOI] [PubMed] [Google Scholar]
- 133. Alto LT, Havton LA, Conner JM, et al. : Chemotropic guidance facilitates axonal regeneration and synapse formation after spinal cord injury. Nat Neurosci. 2009;12(9):1106–13. 10.1038/nn.2365 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 134. Anderson MA, O'Shea TM, Burda JE, et al. : Required growth facilitators propel axon regeneration across complete spinal cord injury. Nature. 2018;561(7723):396–400. 10.1038/s41586-018-0467-6 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 135. Chung WS, Allen NJ, Eroglu C: Astrocytes Control Synapse Formation, Function, and Elimination. Cold Spring Harb Perspect Biol. 2015;7(9):a020370. 10.1101/cshperspect.a020370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Faissner A, Pyka M, Geissler M, et al. : Contributions of astrocytes to synapse formation and maturation - Potential functions of the perisynaptic extracellular matrix. Brain Res Rev. 2010;63(1–2):26–38. 10.1016/j.brainresrev.2010.01.001 [DOI] [PubMed] [Google Scholar]
- 137. Tedeschi A, Omura T, Costigan M: CNS repair and axon regeneration: Using genetic variation to determine mechanisms. Exp Neurol. 2017;287(Pt 3):409–22. 10.1016/j.expneurol.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138. Lisi V, Singh B, Giroux M, et al. : Enhanced Neuronal Regeneration in the CAST/Ei Mouse Strain Is Linked to Expression of Differentiation Markers after Injury. Cell Rep. 2017;20(5):1136–47. 10.1016/j.celrep.2017.07.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139. Bakker OB, Aguirre-Gamboa R, Sanna S, et al. : Integration of multi-omics data and deep phenotyping enables prediction of cytokine responses. Nat Immunol. 2018;19(7):776–86. 10.1038/s41590-018-0121-3 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 140. Hasin Y, Seldin M, Lusis A: Multi-omics approaches to disease. Genome Biol. 2017;18(1):83. 10.1186/s13059-017-1215-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Kawata K, Hatano A, Yugi K, et al. : Trans-omic Analysis Reveals Selective Responses to Induced and Basal Insulin across Signaling, Transcriptional, and Metabolic Networks. iScience. 2018;7:212–29. 10.1016/j.isci.2018.07.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Huang S, Chaudhary K, Garmire LX: More Is Better: Recent Progress in Multi-Omics Data Integration Methods. Front Genet. 2017;8:84. 10.3389/fgene.2017.00084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Kohl M, Megger DA, Trippler M, et al. : A practical data processing workflow for multi-OMICS projects. Biochim Biophys Acta. 2014;1844(1 Pt A):52–62. 10.1016/j.bbapap.2013.02.029 [DOI] [PubMed] [Google Scholar]
- 144. Rohart F, Gautier B, Singh A, et al. : mixOmics: An R package for 'omics feature selection and multiple data integration. PLoS Comput Biol. 2017;13(11):e1005752. 10.1371/journal.pcbi.1005752 [DOI] [PMC free article] [PubMed] [Google Scholar]; F1000 Recommendation
- 145. Chaudhary K, Poirion OB, Lu L, et al. : Deep Learning-Based Multi-Omics Integration Robustly Predicts Survival in Liver Cancer. Clin Cancer Res. 2018;24(6):1248–59. 10.1158/1078-0432.CCR-17-0853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Costello Z, Martin HG: A machine learning approach to predict metabolic pathway dynamics from time-series multiomics data. NPJ Syst Biol Appl. 2018;4:19. 10.1038/s41540-018-0054-3 [DOI] [PMC free article] [PubMed] [Google Scholar]