Abstract
Background
Klebsiella pneumoniae has been the leading causative pathogen for adult bacterial meningitis in several Asian countries. The clinical and microbiological characteristics of K. pneumoniae meningitis in mainland China are still unknown.
Materials and methods
The clinical data of patients with K. pneumoniae meningitis from January 2011 to July 2017 in a tertiary hospital were retrospectively evaluated. The isolates were tested for antibiotic-resistance genes, virulence-associated genes, and molecular subtypes. Hyper-virulent K. pneumoniae (hvKP) was defined as the presence of pLVPK-like virulence plasmid.
Results
During the study period, a total of 48 patients with meningitis caused by K. pneumoniae were identified, accounting for 21.2% (48/226) of Gram-negative bacilli meningitis. Of the 44 available isolates, 65.9% (29/44) were carbapenem resistant, and all except one har-bored blaKPC-2. K64 was the most common serotype (n=13), followed by K47 (n=11) and K1 (n=5). The pLVPK-related genetic loci were found in about half of isolates (iutA: 56.8%, iucA: 56.8%, rmpA2:50.0%, rmpA: 43.2%, and iroN: 40.9%). Twenty-two strains carrying pLVPK-derived virulence plasmid were defined as hvKP. Notably, the coexistence of blaKPC-2-encoding plasmid and the pLVPK-derived virulence plasmid was detected in 15 strains (34.1%, 15/44), suggesting K. pneumoniae carbapenemase-2 (KPC-2)-producing hvKP. The proportion of KPC-2-producing hvKP by year increased remarkably from 0% (2011) to 71.4% (2017). Of the 15 KPC-2-producing hvKP strains, 80.0% (12/15) were assigned to sequence type 11 and 2 strains (13.3%) belonged to clonal complex 23. Most of the patients infected with KPC-2-producing hvKP had preceding postneurosurgical state (93.3%, 14/15) and severe pneumonia (73.3%, 11/15). All the cases (100%, 15/15) had fatal outcome.
Conclusion
The high prevalence and mortality of K. pneumoniae, especially KPC-2-producing hvKP meningitis, in China should be of concern. The implementation of epidemiological surveillance and identification of an effective clinical treatment are paramount.
Keywords: meningitis, hypervirulent K. pneumonia, blaKPC-2, rmpA2, pLVPK-like virulence plasmid
Introduction
Bacterial meningitis is one of the most devastating infectious diseases. It is often associated with substantial mortality and long-term neurologic complications. Gram-negative bacilli, such as Acinetobacter baumannii and Enterobacteriaceae, are prominent causative pathogens of meningitis in adult patients, especially postoperative meningitis.1,2 Among the implicated Gram-negative pathogens, Klebsiella pneumoniae is the most common in Taiwan and several Southeast Asian countries.3–5 In these regions, K. pneumoniae meningitis was associated with a new variant, designated as hypervirulent K. pneumoniae (hvKP) during the past few decades, with reported mortality ranging from 33.3% to 48.5%.6,7
Described as hvKP, this new variant commonly exhibited hypermucoviscosity (HM) phenotype and frequently belonged to specific sequence types (STs), mainly including ST23, ST65, and ST86.8 Since first described in 1986, hvKP increasingly caused pyogenic liver abscesses (PLA) complicated by devastating metastatic infections including endophthalmitis, necrotizing fasciitis, and meningitis in young and healthy individuals.6,9,10 However, the pathogenesis of hvKP causing metastatic infections is still not well elucidated hitherto. Over the past years, HM has been regarded as an important in vitro parameter for hvKP identification, but several controversies regarding the association of HM phenotype and virulence have been raised.11,12 The large virulence plasmid pLVPK carrying capsular polysaccharides regulator genes (rmpA and rmpA2) and several siderophore gene clusters were recognized as essential contributors to the virulence of hvKP.13 A correlation between carriage of pLVPK-derived virulence plasmid and pyogenic infection has been confirmed.14 Most recently, Russo et al15 demonstrated several pLVPK-derived locus could accurately differentiate hvKP from non-hvKP, and they might serve as potential biomarkers for hvKP.
So far, most hvKP were susceptible to commonly used antibiotics except ampicillin. In contrast, carbapenem-resistant K. pneumoniae (CRKP) were usually non-hvKP and resistant to almost all currently available antibiotics. At present, ~70%–90% of clinical carbapenem-resistant Enterobacteriaceae infections were attributed to CRKP in Europe and China.16,17 Most clinical CRKP were K. pneumoniae carbapenemase (KPC)-producing strains18 and belonged to clonal group 258, with ST11 the most predominant clone in Asia, especially in China.17 As CRKP are highly transmissible and antibiotic resistant, they constitute a great threat to public health.
In most cases, CRKP and hvKP were largely nonover-lapping. Nevertheless, along with the spread of plasmids, carbapenem-resistant hvKP have been increasingly reported since first described by Zhang et al in 2015 in China.11 For its capability of causing severe and difficult-to-treat infection, carbapenem-resistant hvKP has posed a considerable concern to the public health and will be the next “superbug” if a successful epidemic clone emerges. Up to now, most carbapenem-resistant hvKP infections generally occurred as isolated or sporadic cases,19–21 with only two outbreaks of a small scale.22,23 More recently, a nationwide screen revealed only 3% (11/387) of ST11 CRKP isolated from blood or sputum harbored the pLVPK-like virulence plasmid in mainland China,22 while Ku et al24 discovered none of the 22 hvKP strains that caused meningitis was resistant to carbapenems in Taiwan. However, information regarding the antimicrobial resistance and virulence characteristics of K. pneumoniae isolated from cerebrospinal fluid (CSF) in mainland China is still lacking. In the present study, we sought to investigate the epidemiology, antibiotic profiles, and representative virulence factors including pLVPK-derived genetic loci of meningitis-causing K. pneumoniae in a tertiary hospital over a 6-year period in China.
Materials and methods
Identification of patients
This retrospective cohort study was conducted in The First Affiliated Hospital, College of Medicine, Zhejiang University, a 2500-bed tertiary hospital havinĝ131,000 admissions each year in Hangzhou, East China, from January 2011 to July 2017. The hospital has three intensive care units with 73 beds, and three neurosurgery departments with 126 beds. Patients who satisfied the following three criteria were included in the analysis: 1) aged >18 years; 2) CSF culture yielded K. pneumoniae; 3) present with abnormal CSF examination and symptoms of meningitis. The classification of community-acquired or nosocomial acquired meningitis was according to the report by Chang et al.4 The relevant clinical and microbiological data were extracted from electronic or paper medical records and microbiological database.
This study was conducted in accordance with the Declaration of Helsinki and approved by the hospital’s Ethics Committees. Written informed consents were obtained from all patients.
Bacterial isolates and antimicrobial susceptibility testing
The VITEK-2 compact system (bioMérieux, Craponne, France) was used to establish strain identification and antimicrobial susceptibility testing. The results were interpreted according to the guideline document established by Clinical and Laboratory Standards Institute (CLSI, 2018). For tigecycline and colistin, the minimum inhibitory concentrations were determined by using broth microdilution method, and the results were categorized in accordance with the breakpoints defined by the European Committee on Antimicrobial Susceptibility Testing criteria (version 7.1, http://www.eucast.org/clinical_breakpoints/) and CLSI, respectively. The species identification of all isolates was verified by matrix-assisted laser desorption/ionization mass spectrometry (Bruker Daltonics Inc., Fremont, CA, USA). K. pneumoniae ATCC 700603 and Escherichia coli ATCC 25922 were used as the quality control strain for species identification and antimicrobial susceptibility testing, respectively.
Determination of HM phenotype
The HM phenotype was determined by “string test” as described previously.10 Briefly, when using a bacteriology loop to stretch bacterial colony cultured on an agar plate overnight at 37°C, a formation of viscous string with >5 mm in length is considered to be positive.
Capsular serotyping and detection of resistance and virulence-associated genes
The capsular type of K. pneumoniae was determined by PCR and sequencing of wzi loci as previously described.25 The sequences of products were compared to the wzi sequences deposited in the database of Institut Pasteur to identify the corresponding capsular types using BLAST program (https://bigsdb.pasteur.fr/klebsiella/klebsiella. html). The extended-spectrum β-lactamase genes (blaCTX-M and blaSHV), carbapenemase genes (blaKPC, blaNDM, blaVIM, blaIMP, and blaOXA-48) and 14 representative virulence genes including the pLVPK-related genetic loci (magA, kfu, allS, fimH, wabG, ybtS, mrkD, uge, entB, iutA, rmpA, rmpA2, iucA, and iroN) were amplified by PCR.22,26 The sequences of primers are listed in Table S1. The amplicons of β-lactamase genes were purified and sequenced in an ABI 3730 DNA sequencer (Applied Biosystems, Foster City, CA, USA), and the sequences obtained compared to those in the NCBI database using BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Multilocus sequence typing (MLST)
MLST for all 44 isolates was done with seven housekeeping genes (gapA, infB, mdh, phoE, pgi, rpoB, and tonB) according to the protocol on the MLST website (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html). STs that had not been described previously were submitted to the database.
Pulsed-field gel electrophoresis (PFGE)
PFGE was performed on all of the isolates. In brief, genomic DNA was digested with XbaI (Bio-Rad Laboratories, Hercules, CA, USA), and the restriction fragments were separated in a CHEF Mapper XA System (Bio-Rad Laboratories). Cluster analysis was performed with Bionumerics software (Applied Maths NV, Sint-Martens-Latem, East Flanders, Belgium) using the Dice similarity coefficients and unweighted-pair group matching algorithm. The strains sharing >75% similarity were defined as the same PFGE pattern.
S1-PFGE and southern blot hybridization
To detect the pLVPK-like virulence plasmid and carbapenemase gene-encoding plasmid, whole chromosomal DNA of 15 strains positive for both rmpA2 and blaKPC-2 were subjected to S1 nuclease (Takara, Shiga, Japan) digestion. Digested fragments were subjected to PFGE. Then, the gels were blotted onto nylon membranes (Millipore, Burlington, MA, USA) according to standard technique. The membranes were hybridized with digoxigenin-labeled rmpA2 probe and blaKPC-2 probe, respectively. Consistent with the findings from Russo et al,15 K. pneumoniae strains that were confirmed to contain pLVPK-like virulence plasmid by S1 nuclease PFGE and southern blot hybridization were defined as hvKP in the present study.22
Results
Prevalence and clinical characteristics of hvKP meningitis
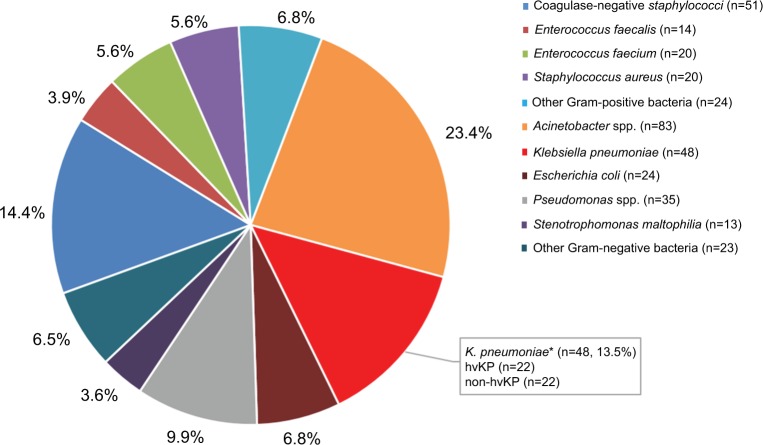
From January 2011 to July 2017, a total of 355 cases of bacterial meningitis were identified in our hospital. The detailed distribution of all meningitis causing species is shown in Figure 1. Among the 48 K. pneumoniae causing meningitis, 44 isolates were available for further experiments (4 isolates died). Twenty-two isolates carrying the pLVPK-like plasmid were identified as hvKP. The clinical characteristics of patients with hvKP and non-hvKP meningitis are shown in Table 1. Overall, there were no significant differences between the two groups. Co-occurrence of blaKPC-2-encoding plasmid and pLVPK-like virulence plasmid was detected in 68.2% (15/22) of hvKP, suggesting KPC-2-producing hvKP (Figure 2). The clinical features of 15 patients with KPC-2-producing hvKP meningitis are summarized in Table 2. In particular, most of the patients (73.3%, 11/15) suffered from severe pneumonia, and all except one patient (93.3%, 14/15) had a preceding neurosurgical state. All the cases (100%, 15/15) had fatal outcome.
Figure 1.
The distribution of all meningitis-causing species from January 2011 to July 2017.
Note: *Including four dead isolates.
Abbreviations: hvKP, hypervirulent K. pneumonia; non-hvKP, non-hypervirulent K. pneumonia.
Table 1.
Demographic profiles and clinical features of 48 patients with Klebsiella pneumoniae meningitis
| Characteristics | Available isolates (n=44) | P-value (hvKP vs non-hvKP) | ||
|---|---|---|---|---|
| Total (n=48) | hvKP (n=22) | Non-hvKP (n=22) | ||
| Demographic data | ||||
| Age (years), mean (± SD) | 50.3±16.0 | 55.2±13.4 | 48.3±17.6 | 0.15 |
| Male, n (%) | 30 (62.5) | 13 (59.1) | 16 (72.2) | 0.34 |
| Acquired infection model, n (%) | ||||
| Community-acquired infection | 3 (6.3) | 1 (4.5) | 1 (4.5) | 1.00 |
| Hospital-acquired infection | 45 (93.8) | 21 (95.5) | 21 (95.5) | 1.00 |
| Types of infection, n (%) | ||||
| Spontaneous meningitis | 3 (6.3) | 1 (4.5) | 1 (4.5) | 1.00 |
| Postneurosurgical/post-trauma meningitis | 45 (93.8) | 21 (95.5) | 21 (95.5) | 1.00 |
| Comorbidity, n (%) | ||||
| Diabetes mellitus | 10 (20.8) | 5 (22.7) | 5 (22.7) | 1.00 |
| Hypertension | 14 (29.2) | 6 (27.3) | 8 (36.4) | 0.52 |
| Brain tumor | 12 (25.0) | 4 (18.2) | 6 (27.3) | 0.47 |
| Head trauma | 18 (37.5) | 9 (40.9) | 9 (40.9) | 1.00 |
| Intracerebral hemorrhage | 15 (31.3) | 7 (31.8) | 8 (36.4) | 0.75 |
| Extrameningeal infection, n (%) | ||||
| Bacteremia | 17 (35.4) | 9 (40.9) | 7 (31.8) | 0.53 |
| Pneumonia | 18 (37.5) | 11 (50.0) | 7 (31.8) | 0.22 |
| Liver abscess | 1 (2.1) | 1 (4.5) | 0 (0.0) | 1.00 |
| Brain abscess | 2 (4.2) | 1 (4.5) | 0 (0.0) | 1.00 |
| Chronic otitis media | 1 (2.1) | 0 (0.0) | 0 (0.0) | – |
| CSF profiles, median (IQR) | ||||
| RBC count (per µL) | 150 (20.0–862.5) | 50 (155.0–1,500.0) | 10 (95.0–962.5) | 0.46 |
| WBC count (per µL) | 900 (72.5–6,265.0) | 60 (210–3,050.0) | 140 (1,040.0–9,100.0) | 0.41 |
| Sugar (mmol/L) | 0.6 (0.1–2.9) | 0.1 (0.4–2.9) | 0.1 (0.5–2.7) | 0.88 |
| Chlorine (mmol/L) | 112 (105.5–119.8) | 106.5 (112.0–120.8) | 106.5 (111.5–118.5) | 0.66 |
| Protein (g/L) | 2.7 (1.7–6.1) | 1.6 (2.7–7.2) | 1.6 (3.0–5.7) | 0.98 |
| 30-day mortality, n (%) | 30 (62.5) | 17 (77.3) | 12 (54.5) | 0.11 |
Abbreviations: CSF, cerebrospinal fluid; hvKP, hypervirulent K. pneumoniae; RBC, red blood cell; WBC, while blood cell.
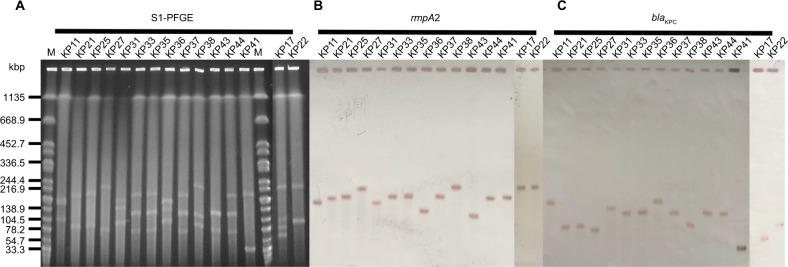
Figure 2.
The S1-PFGE and Southern hybridization analysis of 15 Klebsiella pneumoniae strains co-harboring blaKPC-2 and rmpA2.
Notes: (A) S1 nuclease digestion of genomic DNA of K. pneumoniae strains was followed by PFGE. Plasmid bands are shown as linearized fragment on the gel. (B) Southern blot hybridization of the marker gene (rmpA2) of the virulence plasmid. (C) Southern blot hybridization of blaKPC-2. Lane M, reference standard strain Salmonella serotype Braenderup H9812 restricted with Xbal.
Abbreviations: M, marker; S1-PFGE, S1 nuclease pulsed-field gel electrophoresis.
Table 2.
Clinical features of 15 patients with KPC-2-producing hvKP meningitis
| Patient no. | Strain ID | Age (years) | Sex | Unit | Comorbidity | Neurosurgical procedures | Treatment | Outcome |
|---|---|---|---|---|---|---|---|---|
| 1 | KP11 | 68 | F | ICU | Severe pneumonia, bacteremia | Yes | MRP | Died |
| 2 | KP17 | 27 | F | Surgery | ICH, pneumonia, bacteremia | Yes | MRP, MXF | Died |
| 3 | KP21 | 41 | M | ICU | Head trauma, pneumonia, bacteremia | Yes | MRP, TGC | Died |
| 4 | KP22 | 63 | F | Surgery | SAH, hypertension, pneumonia | Yes | MRP, LEV, AMI | Died |
| 5 | KP25 | 38 | M | Surgery | Head trauma, pneumonia | Yes | MRP, C/S, PXE | Died |
| 6 | KP27 | 67 | M | ICU | Meningioma, DM, heart disease | Yes | MRP | Died |
| 7 | KP31 | 67 | M | ICU | Cerebral infarction, DM, COPD | No | C/S, TGC | Died |
| 8 | KP33 | 53 | M | Surgery | AVM rupture, pneumonia | Yes | MRP, TGC | Died |
| 9 | KP35 | 58 | M | Surgery | Head trauma, SDH, bacteremia | Yes | MRP, TGC, C/S | Died |
| 10 | KP36 | 67 | F | ICU | SAH, hypertension, pneumonia | Yes | TGC, C/S | Died |
| 11 | KP37 | 63 | M | ICU | ICH, Alzheimer disease, hypertension, pneumonia, bacteremia | Yes | MRP, TGC | Died |
| 12 | KP38 | 46 | M | Surgery | Head trauma, renal calculus | Yes | MRP, TGC | Died |
| 13 | KP41 | 80 | F | ICU | SAH, DM, heart disease, pneumonia | Yes | C/S | Died |
| 14 | KP43 | 72 | M | ICU | Head trauma, hypertension, pneumonia, bacteremia | Yes | C/S, TGC, PXE | Died |
| 15 | KP44 | 52 | F | Surgery | Hydrocephalus, pneumonia | Yes | MRP, TGC, C/S | Died |
Abbreviations: AMI, amikacin; AVM, arteriovenous malformation; C/S, cefoperazone/sulbactam; DM, diabetes mellitus; F, female; hvKP, hypervirulent Klebsiella pneumoniae; ICH, intracerebral hemorrhage; ICU, intensive care unit; KPC-2, K. pneumoniae carbapenemase-2; LEV, levofloxacin; M, male; MRP, meropenem; MXF, moxifloxacin; PXE, polymyxin E; SAH, subarachnoid hemorrhage related to a brain aneurysm; SDH, subdural hemorrhage; TGC, tigecycline.
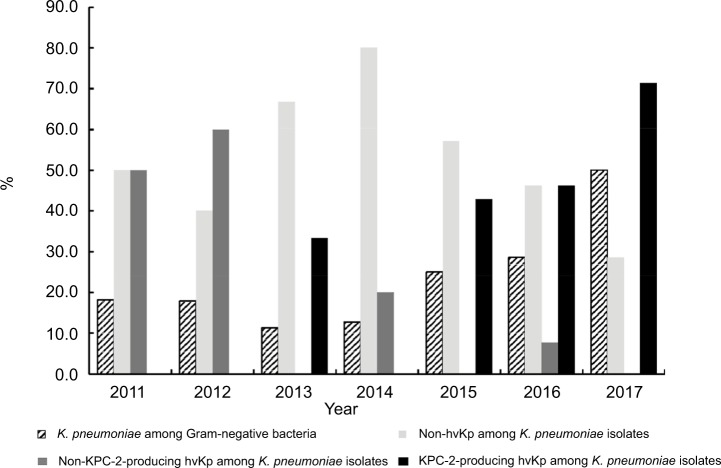
As shown in Figure 3, the proportion of K. pneumoniae meningitis among Gram-negative bacteria causing meningitis increased continuously and obviously during the last 5 years (11.4% in 2013, 12.8% in 2014, 25.0% in 2015, 28.6% in 2016, and 50.0% in 2017, respectively), and the proportion of hvKP among K. pneumonia isolates increased from 2014 to 2017 (20.0% to 71.4%) as well. It was noted that among the 15 cases of KPC-2-producing hvKP meningitis, the earliest case emerged in 2013, and an increase in the proportion of these strains within K. pneumoniae was observed, 42.9% in 2015 (from January to December), 46.2% in 2016 (from January to December), and 71.4% in 2017 (from January to July).
Figure 3.
The prevalence of four types of Klebsiella pneumoniae strains from January 2011 to July 2017.
Abbreviations: KPC-2, K. pneumoniae carbapenemase-2; hvKP, hypervirulent K. pneumoniae.
Antimicrobial susceptibility and resistance mechanism among hvKP and non-hvKP isolates
The detailed antimicrobial resistance profiles of the 16 drugs are listed in Table 3. 68.2% (15/22) of hvKP strains were resistant to carbapenem. Meanwhile, resistance to all tested antimicrobials, except tigecycline and colistin, was observed in a high proportion of hvKP strains. However, the overall resistance rates of hvKP were similar to those of non-hvKP.
Table 3.
The antimicrobial resistance profile of 44 Klebsiella pneumoniae strains from cerebrospinal fluid
| Antimicrobials | No. (%) of resistant isolates | P-value (hvKP vs non-hvKP) | ||
|---|---|---|---|---|
| Total (n=44) | hvKP (n=22) | Non-hvKP (n=22) | ||
| Ampicillin–sulbactam | 35 (79.5) | 17 (77.3) | 18 (81.8) | 1.00 |
| Piperacillin–tazobactam | 29 (65.9) | 15 (68.2) | 14 (63.6) | 0.75 |
| Ceftazidime | 29 (65.9) | 13 (59.1) | 16 (72.7) | 0.34 |
| Ceftriaxone | 34 (77.3) | 17 (77.3) | 17 (77.3) | 1.00 |
| Cefepime | 27 (61.4) | 13 (59.1) | 14 (63.6) | 0.76 |
| Aztreonam | 32 (72.7) | 16 (72.7) | 16 (72.7) | 1.00 |
| Ertapenem | 29 (65.9) | 15 (68.2) | 14 (63.6) | 0.75 |
| Imipenem | 28 (63.6) | 15 (68.2) | 13 (59.1) | 0.53 |
| Amikacin | 15 (34.1) | 10 (45.5) | 5 (22.7) | 0.11 |
| Gentamicin | 21 (47.7) | 10 (45.5) | 11 (50.0) | 0.76 |
| Tobramycin | 17 (38.6) | 10 (45.5) | 7 (31.8) | 0.35 |
| Ciprofloxacin | 26 (59.1) | 13 (59.1) | 13 (59.1) | 1.00 |
| Levofloxacin | 24 (54.5) | 13 (59.1) | 11 (50.0) | 0.55 |
| Trimethoprim–sulfamethoxazole | 17 (38.6) | 9 (40.9) | 8 (36.4) | 0.76 |
| Tigecycline | 0 (0.0) | 0 (0.0) | 0 (0.0) | – |
| Colistin | 0 (0.0) | 0 (0.0) | 0 (0.0) | – |
Abbreviations: hvKP, hypervirulent K. pneumonia; non-hvKP, non-hypervirulent K. pneumonia.
Twenty-eight K. pneumoniae strains were positive for blaKPC including 15 hvKP and 13 non-hvKP. The CTX-M-9 group was detected in 13 hvKP strains (blaCTX-M-14, n=4; blaCTX-M-65, n=9) and 10 non-hvKP strains (blaCTX-M-14, n=2; blaCTX-M-65, n=8), respectively. Interestingly, the CTX-M-1 group β-lactamases blaCTX-M-3 was exclusively detected in hvKP (13.6%, 3/22), while blaCTX-M-15 was found only in three non-hvKP strains. Various SHV genes were detected in 20 strains (hvKP, n=10; non-hvKP, n=10). None of the strains was positive for blaNDM, blaVIM, blaIMP, and blaOXA-48.
Capsular serotyping and virulence-associated genes among hvKP and non-hvKP isolates
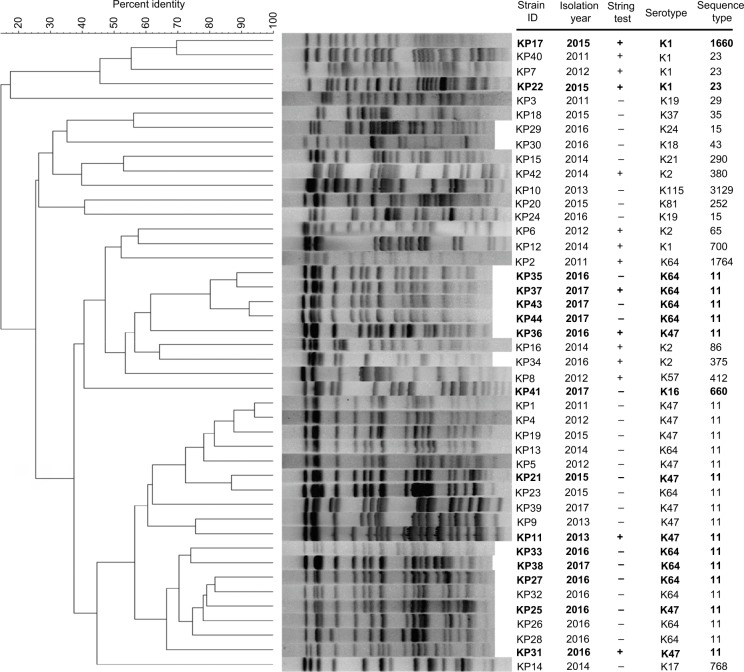
According to the results of the wzi typing, a total of 14 different capsular serotypes were identified (Figure 4). Overall, K64 was the most common serotype (n=13), followed by K47 (n=11), K1 (n=5), and K2 (n=4). In the hvKP group, K64 was the predominant serotype (36.4%) and K47 in non-hvKP (27.3%) group. Among the 15 KPC-2-producing hvKP strains, 7 (46.7%), 5 (33.3%), 2 (13.3), and 1 (6.7%) belonged to K64, K47, K1, and K16, respectively.
Figure 4.
PFGE dendrogram of 44 Klebsiella pneumoniae strains.
Notes: The 15 KPC-2-producing hvKP strains are indicated by boldface type. The similarity percentage of profiles was calculated by the Dice coefficient.
Abbreviations: hvKP, hypervirulent K. pneumoniae; KPC-2, K. pneumoniae carbapenemase-2; PFGE, pulsed-field gel electrophoresis.
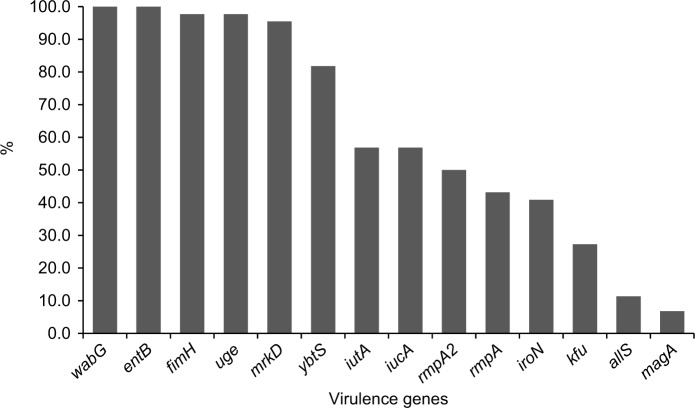
The positive rates of the 14 virulence genes in hvKP and non-hvKP isolates are shown in Figure 5. The virulence-associated genes with >50% of positive rates in hvKP and non-hvKP isolates included wabG (100% and 100%), entB (100% and 100%), fimH (100% and 95.5%), uge (95.5% and 100%), mrkD (90.9% and 100%), and ybtS (86.4% and 77.3%). The positive rates of all the five pLVPK-derived locus iutA, iucA, rmpA, rmpA2, iroN were significantly higher in hvKP isolates than non-hvKP isolates (P<0.001). Additionally, allS was exclusively detected in hvKP isolates (22.7%, 5/22). The HM phenotype was detected more frequently in hvKP isolates than in non-hvKP (59.1% vs 9.1%, P<0.001) as well.
Figure 5.
The distributions of virulence-associated genes among hvKP and non-hvKP strains.
Abbreviations: hvKP, hypervirulent Klebsiella pneumonia; non-hvKP, non-hypervirulent K. pneumonia.
Molecular characteristics of the K. pneumoniae isolates
MLST analysis revealed a total of 19 STs including one novel ST (ST3129). ST11 was the most predominant ST (52.3%, 23/44) followed by ST23 (6.8%, 3/44) and ST15 (4.5%, 2/44), while the remaining STs were found only in one strain each. In addition, the five strains with K1 serotype belonged to three STs (ST23, n=3; ST1660, n=1; and ST700, n=1) and the four strains with K2 serotype belonged to four STs (ST65, ST86, ST375, and ST380). Most strains belonged to ST11 both in hvKP and non-hvKP groups (54.5% and 50.0%, respectively). The PFGE results showed that the homology of 44 isolates was diverse (Figure 3). Only three clusters with >75% similarity were identified, and each cluster accounted for four strains.
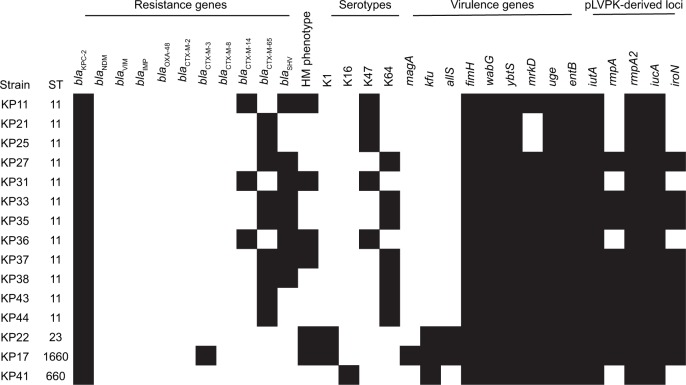
Among the 15 KPC-2-producing hvKP, four distinct STs were identified. ST11 was the predominant ST, accounting for 80% (12/15), and ST23, ST1660, and ST660 for each strain (Figure 6). Of the 12 ST11 KPC-2-producing hvKP, 7 strains belonged to serotype K64 and 5 strains belonged to K47. In addition to blaKPC-2, all of the 12 ST11 KPC-2-producing hvKP carried blaCTX-M-9. Four strains exhibited HM phenotype. The strains of ST23 and ST1660 belonged to K1 serotype and displayed HM phenotype. In general, ST11 KPC-2-producing hvKP strains harbored more antibiotic-resistant genes and less virulence factors when compared with the non-ST11 KPC-2-producing hvKP (Figure 5). Among the 15 KPC-2-producing hvKP strains, 4 strains (KP35, KP37, KP43, and KP44) shared >75% similarity and classified into one cluster (Figure 3), while the remaining 11 strains exhibited distinct genetic relationship.
Figure 6.
Antimicrobial resistance and virulence profiles of 15 KPC-2-producing hvKP strains.
Note: The black color indicated the presence of a gene in the corresponding strain.
Abbreviations: KPC-2, Klebsiella pneumoniae carbapenemase-2; hvKP, hypervirulent K. pneumoniae.
Discussion
Up to now, although some cases of K. pneumoniae meningitis have been reported in mainland China,27,28 the clinical and microbiological features of K. pneumoniae causing meningitis from our area remain unclear. Our study highlights a high prevalence and mortality of K. pneumoniae, especially KPC-2-producing hvKP causing meningitis, in a tertiary hospital in Eastern China, which posed a serious threat to public health.
Consistent with the epidemiology trend of K. pneumoniae meningitis in some Asian countries especially in Taiwan,3,4 where K. pneumoniae is found to be the leading pathogen for meningitis, our present study shows that the proportion of K. pneumoniae among Gram-negative bacilli causing meningitis increased consistently from 2013 to 2017 and K. pneumoniae gradually becomes to be the predominant Gram-negative pathogen responsible for meningitis. However, in contrast to the reports from Taiwan, describing that most of the K. pneumoniae meningitis belonged to community-acquired cases,3,4,29 only a few cases of community-acquired K. pneumoniae meningitis (n=3, 6.3%) were observed in our study. Sought to the possible explanations, besides the proved genetic predisposition unique to people being in different geographical locations, a higher incidence of postneurosurgical conditions as the preceding event than that reported in Taiwan (85.4% vs 56.9%) was of note. However, an inadequate infectious control program in our hospital might also contribute to the development of K. pneumoniae meningitis in postneurosurgical patients.
Previous studies have consistently confirmed that K. pneumoniae was capable of causing meningitis commonly complicated by PLA, and diabetes mellitus and liver diseases seemed to be the well-established predisposing factors for K. pneumoniae meningitis.3,6,29–31 Nevertheless, diabetes was noted in 20.8% of patients and liver diseases including PLA occurred in four patients in our study. These disparities can be attributed to the fact that the previous results were obtained based on the epidemiology investigations mainly involving community-acquired cases.3,29,31 Further exploring the detailed clinical features of the three cases of community-acquired meningitis in our study, we found that all of the patients had liver diseases with liver abscess, liver cirrhosis, and chronic hepatitis for each case, respectively, and two patients had diabetes. However, no definitive conclusions could be drawn due to the small size of the enrolled community-acquired cases.
The overall 30-day mortality was 62.5%, which was higher than that reported in Korea5 and Taiwan.29,30 It was believed that the interaction between host and bacterial variables contributed to the outcomes during bacterial infections. Adult patients with K. pneumoniae meningitis commonly suffered from severe pneumoniae and developed to bacteremia.29 These conditions were also found in the current study. But, in contrast to the previous studies mainly from Taiwan,3,24,29 a remarkable high prevalence of CRKP causing meningitis was observed, which implied that most of our patients did not benefit from the use of carbapenem as the initial empiric therapy. Therefore, it should be highly considered that carbapenem is indicated for meningitis empirically. hvKP was identified in 50% isolates, which was lower than that reported from Taiwan (66.7%) most recently.24 This difference might partly be attributed to the distinct indexes for hvKP. Nine isolates designated as hvKP were determined as string negative in our study. Hence, it seemed that the prevalence of hvKP might be overestimated due to the lack of definitive diagnostic methods.
KPC has been proved to be one of the most important genetic mechanisms of carbapenem resistance for K. pneu-moniae.18 Since KPC-2-producing K. pneumoniae firstly reported in China in 2007, it has spread widely and rapidly in our region.32 63.6% of the strains in the present study were positive for KPC-2. This percentage is much higher than that reported in bloodstream infection samples (33.3%) by our previous study.33 In accordance with the report by Qi et al,34 which described ST11 was the dominant clone of KPC-2-producing K. pneumoniae in China, 78.6% of the KPC-2-positive isolates belonged to ST11 in this study. These results suggested the nosocomial clonal dissemination of KPC-2-producing ST11 K. pneumoniae played a significant role in the high detection rate of blaKPC-2.
It had been common sense that the coexistence of plasmids encoding resistance genes and virulence factors in bacteria was a rare event. However, most recently, Gu et al22 reported that there was a fatal outbreak of ST11 carbapenem-resistant hvKP in a Chinese hospital in 2016. They also demonstrated the size of the acquired pLVPK-like virulence plasmid and its intrinsic blaKPC-carrying plasmid was nearly the same. In our study, both the blaKPC-2 and rmpA2 probes hybridized to the ~170 kb plasmid band in KP 11 strain; so, we speculated that the genetic background of KP11 was similar to the strains described by Gu et al. However, further whole genome sequencing or plasmid curing experiment might prove the speculation. ST23 was the most common ST among hvKP, which was frequently associated with PLA. Though hvKP belonging to ST23 always displayed a wide antibiotics susceptibility profile, several cases of ST23 KPC-2-producing hvKP infection had been described in China.35 In the present study, two out of four ST23 (including one ST23-like variant) isolates were found to harbor the blaKPC-2-bearing plasmid. Therefore, our results suggested that both ST11 and ST23 K. pneumoniae strains could evolve to KPC-2-producing hvKP causing meningitis, and patients with KPC-2-producing hvKP meningitis were always associated with extremely high mortality.
There were some limitations to our study. The study was a single-center study with retrospectively enrolled relatively small size of cases, which might limit its epidemiologic scope. In addition, because of the lack of a “reference standard” genotypic/phenotypic marker for hvKP up to now, K. pneumoniae strains harboring the pLVPK-like virulence plasmid were defined as hvKP as described by Gu et al.22 Studies combining with cell survival tests and whole-genome framework are needed to further illustrate the virulence characteristics and key genomic elements of these isolates.
Conclusion
In summary, K. pneumoniae meningitis has been increasing during the last 5 years, in which a high proportion of KPC-2-producing hvKP causing meningitis was observed. Such strains were likely to cause serious infections, particularly in patients with postneurosurgical meningitis. Our study highlights the urgent need to enhance clinical awareness and management of K. pneumoniae, especially KPC-2-producing hvKP-causing meningitis. Effective surveillance and strict infection control strategies are of prime importance to prevent such strains from further dissemination in China.
Ethics approval and informed consent
Ethical approval was granted from the ethics committee and review board of the First Affiliated Hospital, College of Medicine, Zhejiang University, and written informed consent for publication of individual details was obtained from all patients.
Supplementary material
Table S1.
Primers used to detect the resistance and virulence-associated genes in this study
| Target genes | Primer name | Primer sequence (5′–3′) | Product size (bp) |
|---|---|---|---|
| blaKPC | KPC-F | ATGTCACTGTATCGCCGTCT | 893 |
| KPC-R | TTTTCAGAGCCTTACTGCCC | ||
| blaNDM | NDM-F | GGTTTGGCGATCTGGTTTTC | 624 |
| NDM-R | CGGAATGGCTCATCACGATC | ||
| blaVIM | VIM-F | GATGGTGTTTGGTCGCATA | 390 |
| VIM-R | CGAATGCGCAGCACCAG | ||
| blaIMP | IMP-F | GGAATAGAGTGGCTTAAYTCTC | 188 |
| IMP-R | GGTTTAAYAAAACAACCACC | ||
| blaOXA-48 | OXA48-F | GCGTGGTTAAGGATGAACAC | 437 |
| OXA48-R | CATCAAGTTCAACCCAACCG | ||
| blaSHV | SHV-F | GCCTTTATCGGCCTTCACTCAAG | 898 |
| SHV-R | TTAGCGTTGCCAGTGCTCGATCA | ||
| blaCTX-M-1 group | CTX-M-1-F | CAGCGCTTTTGCCGTCTAAG | 944 |
| CTX-M-1-R | GGCCCATGGTTAAAAAATCACTGC | ||
| blaCTX-M-2 group | CTX-M-2-F | GCATTCGCCGCTCAATGTTA | 942 |
| CTX-M-2-R | GGTTCGTTGCAAGACAAGAC | ||
| blaCTX-M-8 group | CTX-M-8-F | ACTTCAGCCACACGGATTCA | 977 |
| CTX-M-8-R | CGAGTACGTCACGACGACTT | ||
| blaCTX-M-9 group | CTX-M-9-F | GTTACAGCCCTTCGGCGATGATTC | 877 |
| CTX-M-9-R | GCGCATGGTGACAAAGAGAATGCAA | ||
| magA | magA-F | GGTGCTCTTTACATCATTGC | 1,283 |
| magA-R | GCAATGGCCATTTGCGTTAG | ||
| Kfu | kfu-F | ATAGTAGGCGAGCACCGAGA | 530 |
| kfu-R | AGAACCTTCCTCGCTGAACA | ||
| allS | allS-F | CCGAAACATTACGCACCTTT | 508 |
| allS-R | ATCACGAAGAGCCAGGTCAC | ||
| fimH | fimH-F | TGCTGCTGGGCTGGTCGATG | 909 |
| fimH-F | GGGAGGGTGACGGTGACATC | ||
| wabG | wabG-F | ACCATCGGCCATTTGATAGA | 683 |
| wabG-R | CGGACTGGCAGATCCATATC | ||
| ybtS | ybtS-F | GACGGAAACAGCACGGTAAA | 782 |
| ybtS-R | GAGCATAATAAGGCGAAAGA | ||
| mrkD | mrkD-F | AAGCTATCGCTGTACTTCCGGCA | 340 |
| mrkD-R | GGCGTTGGCGCTCAGATAGG | ||
| uge | uge-F | TCTTCACGCCTTCCTTCACT | 534 |
| uge-R | GATCATCCGGTCTCCCTGTA | ||
| entB | entB-F | GTCAACTGGGCCTTTGAGCCGTC | 400 |
| entB-R | TATGGGCGTAAACGCCGGTGAT | ||
| iutA | iutA-F | GGGAAAGGCTTCTCTGCCAT | 300 |
| iutA-R | TTATTCGCCACCACGCTCTT | ||
| rmpA | rmpA-F | TACATATGAAGGAGTAGTTAAT | 505 |
| rmpA-R | GAGCCATCTTTCATCAAC | ||
| rmpA2 | rmpA2-F | TGTGCAATAAGGATGTTACATTAGT | 609 |
| rmpA2-R | TTTGATGTGCACCATTTTTCA | ||
| iucA | iucA-F | ATAAGGCAGGCAATCCAG | 2,927 |
| iucA-R | TAACGGCGATAAACCTCG | ||
| iroN | iroN-F | GTCCGGCGGTAACTTCAGCC | 829 |
| iroN-R | TCAGAATGAAACTACCGCCC |
Acknowledgments
We thank the entire staff at the Department of Microbiology, The First Affiliated Hospital, College of Medicine, Zhejiang University, for their daily contributions to this study. We also thank the team of curators from the Institut Pasteur MLST system (Paris, France) for importing novel alleles and profiles at http://bigsdb.pasteur.fr/klebsiella/klebsiella.html. This work was supported by the National Natural Science Foundation (Grant Number 81301459), Natural Science Foundation of Zhejiang Province (Grant Number LQ13H190001), Medical Science and Technology Project of Zhejiang Province (Grant Number 2014KYB096), and National Key Program for Infectious Diseases of China (Grant Number 2017ZX10103008).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Chang JB, Wu H, Wang H, Ma BT, Wang RZ, Wei JJ. Prevalence and antibiotic resistance of bacteria isolated from the cerebrospinal fluid of neurosurgical patients at Peking Union Medical College Hospital. Antimicrob Resist Infect Control. 2018;7:41. doi: 10.1186/s13756-018-0323-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bodilsen J, Brouwer MC, Kjærgaard N, et al. Community-acquired meningitis in adults caused by Escherichia coli in Denmark and The Netherlands. J Infect. 2018;77(1):25–29. doi: 10.1016/j.jinf.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Chang WN, Huang CR, Lu CH, Chien CC. Adult Klebsiella pneumoniae meningitis in Taiwan: an overview. Acta Neurol Taiwan. 2012;21(2):87–96. [PubMed] [Google Scholar]
- 4.Chang WN, Lu CH, Huang CR, et al. Changing epidemiology of adult bacterial meningitis in southern Taiwan: a hospital-based study. Infection. 2008;36(1):15–22. doi: 10.1007/s15010-007-7009-8. [DOI] [PubMed] [Google Scholar]
- 5.Moon SY, Chung DR, Kim SW, et al. Changing etiology of community-acquired bacterial meningitis in adults: a nationwide multicenter study in Korea. Eur J Clin Microbiol Infect Dis. 2010;29(7):793–800. doi: 10.1007/s10096-010-0929-8. [DOI] [PubMed] [Google Scholar]
- 6.Fang CT, Lai SY, Yi WC, Hsueh PR, Liu KL, Chang SC. Klebsiella pneumoniae genotype K1: an emerging pathogen that causes septic ocular or central nervous system complications from pyogenic liver abscess. Clin Infect Dis. 2007;45(3):284–293. doi: 10.1086/519262. [DOI] [PubMed] [Google Scholar]
- 7.Lu CH, Chang WN, Chang HW, Ch L, Chang HW. Adult bacterial meningitis in southern Taiwan: epidemiologic trend and prognostic factors. J Neurol Sci. 2000;182(1):36–44. doi: 10.1016/s0022-510x(00)00445-7. [DOI] [PubMed] [Google Scholar]
- 8.Bialek-Davenet S, Criscuolo A, Ailloud F, et al. Genomic definition of hypervirulent and multidrug-resistant Klebsiella pneumoniae clonal groups. Emerg Infect Dis. 2014;20(11):1812–1820. doi: 10.3201/eid2011.140206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fazili T, Sharngoe C, Endy T, Kiska D, Javaid W, Polhemus M. Klebsiella pneumoniae liver abscess: an emerging disease. Am J Med Sci. 2016;351(3):297–304. doi: 10.1016/j.amjms.2015.12.018. [DOI] [PubMed] [Google Scholar]
- 10.Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4(2):107–118. doi: 10.4161/viru.22718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang Y, Zeng J, Liu W, et al. Emergence of a hypervirulent carbapenem-resistant Klebsiella pneumoniae isolate from clinical infections in China. J Infect. 2015;71(5):553–560. doi: 10.1016/j.jinf.2015.07.010. [DOI] [PubMed] [Google Scholar]
- 12.Lin YC, Lu MC, Tang HL, et al. Assessment of hypermucoviscosity as a virulence factor for experimental Klebsiella pneumoniae infections: comparative virulence analysis with hypermucoviscosity-negative strain. BMC Microbiol. 2011;11:50. doi: 10.1186/1471-2180-11-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen YT, Chang HY, Lai YC, Pan CC, Tsai SF, Peng HL. Sequencing and analysis of the large virulence plasmid pLVPK of Klebsiella pneumoniae CG43. Gene. 2004;337:189–198. doi: 10.1016/j.gene.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 14.Tang HL, Chiang MK, Liou WJ, et al. Correlation between Klebsiella pneumoniae carrying pLVPK-derived loci and abscess formation. Eur J Clin Microbiol Infect Dis. 2010;29(6):689–698. doi: 10.1007/s10096-010-0915-1. [DOI] [PubMed] [Google Scholar]
- 15.Russo TA, Olson R, Fang CT, et al. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol. 2018;56(9) doi: 10.1128/JCM.00776-18. pii: e00776-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grundmann H, Glasner C, Albiger B, et al. Occurrence of carbapen-emase-producing Klebsiella pneumoniae and Escherichia coli in the European survey of carbapenemase-producing Enterobacteriaceae (EuSCAPE): a prospective, multinational study. Lancet Infect Dis. 2017;17(2):153–163. doi: 10.1016/S1473-3099(16)30257-2. [DOI] [PubMed] [Google Scholar]
- 17.Zhang R, Liu L, Zhou H, et al. Nationwide surveillance of clinical carbapenem-resistant Enterobacteriaceae (CRE) strains in China. EBioMedicine. 2017;19:98–106. doi: 10.1016/j.ebiom.2017.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Munoz-Price LS, Poirel L, Bonomo RA, et al. Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect Dis. 2013;13(9):785–796. doi: 10.1016/S1473-3099(13)70190-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arena F, Henrici de Angelis L, D’Andrea MM, et al. Infections caused by carbapenem-resistant Klebsiella pneumoniae with hypermuco-viscous phenotype: a case report and literature review. Virulence. 2017;8(8):1900–1908. doi: 10.1080/21505594.2017.1286439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang L, Li Y, Shen W, Wang SM, Wang G, Zhou Y. Whole-genome sequence of a carbapenem-resistant hypermucoviscous Klebsiella pneumoniae isolate SWU01 with capsular serotype K47 belonging to ST11 from a patient in China. J Glob Antimicrob Resist. 2017;11:87–89. doi: 10.1016/j.jgar.2017.09.001. [DOI] [PubMed] [Google Scholar]
- 21.Mei YF, Liu PP, Wan LG, et al. Virulence and genomic feature of a virulent Klebsiella pneumoniae sequence type 14 strain of serotype K2 harboring blaNDM-5 in China. Front Microbiol. 2017;8:335. doi: 10.3389/fmicb.2017.00335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu D, Dong N, Zheng Z, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18(1):37–46. doi: 10.1016/S1473-3099(17)30489-9. [DOI] [PubMed] [Google Scholar]
- 23.Zhan L, Wang S, Guo Y, et al. Outbreak by hypermucoviscous Klebsiella pneumoniae ST11 isolates with carbapenem resistance in a Tertiary Hospital in China. Front Cell Infect Microbiol. 2017;7:182. doi: 10.3389/fcimb.2017.00182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ku YH, Chuang YC, Chen CC, et al. Klebsiella pneumoniae isolates from meningitis: epidemiology, virulence and antibiotic resistance. Sci Rep. 2017;7(1):6634. doi: 10.1038/s41598-017-06878-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brisse S, Passet V, Haugaard AB, et al. wzi Gene sequencing, a rapid method for determination of capsular type for Klebsiella strains. J Clin Microbiol. 2013;51(12):4073–4078. doi: 10.1128/JCM.01924-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Compain F, Babosan A, Brisse S, et al. Multiplex PCR for detection of seven virulence factors and K1/K2 capsular serotypes of Klebsiella pneumoniae. J Clin Microbiol. 2014;52(12):4377–4380. doi: 10.1128/JCM.02316-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qian Y, Wong CC, Lai SC, et al. Klebsiella pneumoniae invasive liver abscess syndrome with purulent meningitis and septic shock: a case from mainland China. World J Gastroenterol. 2016;22(9):2861–2866. doi: 10.3748/wjg.v22.i9.2861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khaertynov KS, Anokhin VA, Davidyuk YN, et al. Case of meningitis in a neonate caused by an extended-spectrum-beta-lactamase-producing strain of hypervirulent Klebsiella pneumoniae. Front Microbiol. 2017;8:1576. doi: 10.3389/fmicb.2017.01576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee PY, Chang WN, Lu CH, et al. Clinical features and in vitro antimicrobial susceptibilities of community-acquired Klebsiella pneumoniae meningitis in Taiwan. J Antimicrob Chemother. 2003;51(4):957–962. doi: 10.1093/jac/dkg158. [DOI] [PubMed] [Google Scholar]
- 30.Lee SS, Chen YS, Tsai HC, et al. Predictors of septic metastatic infection and mortality among patients with Klebsiella pneumoniae liver abscess. Clin Infect Dis. 2008;47(5):642–650. doi: 10.1086/590932. [DOI] [PubMed] [Google Scholar]
- 31.Huang CR, Lu CH, Chang HW, Lee PY, Lin MW, Chang WN. Community-acquired spontaneous bacterial meningitis in adult diabetic patients: an analysis of clinical characteristics and prognostic factors. Infection. 2002;30(6):346–350. doi: 10.1007/s15010-002-3010-4. [DOI] [PubMed] [Google Scholar]
- 32.Wei ZQ, Du XX, Yu YS, Shen P, Chen YG, Li LJ. Plasmid-mediated KPC-2 in a Klebsiella pneumoniae isolate from China. Antimicrob Agents Chemother. 2007;51(2):763–765. doi: 10.1128/AAC.01053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xu M, Fu Y, Kong H, et al. Bloodstream infections caused by Klebsiella pneumoniae: prevalence of blaKPC, virulence factors and their impacts on clinical outcome. BMC Infect Dis. 2018;18(1):358. doi: 10.1186/s12879-018-3263-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 2011;66(2):307–312. doi: 10.1093/jac/dkq431. [DOI] [PubMed] [Google Scholar]
- 35.Zhang R, Lin D, Chan EW, Gu D, Chen GX, Chen S. Emergence of carbapenem-resistant serotype K1 hypervirulent Klebsiella pneumoniae strains in China. Antimicrob Agents Chemother. 2016;60(1):709–711. doi: 10.1128/AAC.02173-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1.
Primers used to detect the resistance and virulence-associated genes in this study
| Target genes | Primer name | Primer sequence (5′–3′) | Product size (bp) |
|---|---|---|---|
| blaKPC | KPC-F | ATGTCACTGTATCGCCGTCT | 893 |
| KPC-R | TTTTCAGAGCCTTACTGCCC | ||
| blaNDM | NDM-F | GGTTTGGCGATCTGGTTTTC | 624 |
| NDM-R | CGGAATGGCTCATCACGATC | ||
| blaVIM | VIM-F | GATGGTGTTTGGTCGCATA | 390 |
| VIM-R | CGAATGCGCAGCACCAG | ||
| blaIMP | IMP-F | GGAATAGAGTGGCTTAAYTCTC | 188 |
| IMP-R | GGTTTAAYAAAACAACCACC | ||
| blaOXA-48 | OXA48-F | GCGTGGTTAAGGATGAACAC | 437 |
| OXA48-R | CATCAAGTTCAACCCAACCG | ||
| blaSHV | SHV-F | GCCTTTATCGGCCTTCACTCAAG | 898 |
| SHV-R | TTAGCGTTGCCAGTGCTCGATCA | ||
| blaCTX-M-1 group | CTX-M-1-F | CAGCGCTTTTGCCGTCTAAG | 944 |
| CTX-M-1-R | GGCCCATGGTTAAAAAATCACTGC | ||
| blaCTX-M-2 group | CTX-M-2-F | GCATTCGCCGCTCAATGTTA | 942 |
| CTX-M-2-R | GGTTCGTTGCAAGACAAGAC | ||
| blaCTX-M-8 group | CTX-M-8-F | ACTTCAGCCACACGGATTCA | 977 |
| CTX-M-8-R | CGAGTACGTCACGACGACTT | ||
| blaCTX-M-9 group | CTX-M-9-F | GTTACAGCCCTTCGGCGATGATTC | 877 |
| CTX-M-9-R | GCGCATGGTGACAAAGAGAATGCAA | ||
| magA | magA-F | GGTGCTCTTTACATCATTGC | 1,283 |
| magA-R | GCAATGGCCATTTGCGTTAG | ||
| Kfu | kfu-F | ATAGTAGGCGAGCACCGAGA | 530 |
| kfu-R | AGAACCTTCCTCGCTGAACA | ||
| allS | allS-F | CCGAAACATTACGCACCTTT | 508 |
| allS-R | ATCACGAAGAGCCAGGTCAC | ||
| fimH | fimH-F | TGCTGCTGGGCTGGTCGATG | 909 |
| fimH-F | GGGAGGGTGACGGTGACATC | ||
| wabG | wabG-F | ACCATCGGCCATTTGATAGA | 683 |
| wabG-R | CGGACTGGCAGATCCATATC | ||
| ybtS | ybtS-F | GACGGAAACAGCACGGTAAA | 782 |
| ybtS-R | GAGCATAATAAGGCGAAAGA | ||
| mrkD | mrkD-F | AAGCTATCGCTGTACTTCCGGCA | 340 |
| mrkD-R | GGCGTTGGCGCTCAGATAGG | ||
| uge | uge-F | TCTTCACGCCTTCCTTCACT | 534 |
| uge-R | GATCATCCGGTCTCCCTGTA | ||
| entB | entB-F | GTCAACTGGGCCTTTGAGCCGTC | 400 |
| entB-R | TATGGGCGTAAACGCCGGTGAT | ||
| iutA | iutA-F | GGGAAAGGCTTCTCTGCCAT | 300 |
| iutA-R | TTATTCGCCACCACGCTCTT | ||
| rmpA | rmpA-F | TACATATGAAGGAGTAGTTAAT | 505 |
| rmpA-R | GAGCCATCTTTCATCAAC | ||
| rmpA2 | rmpA2-F | TGTGCAATAAGGATGTTACATTAGT | 609 |
| rmpA2-R | TTTGATGTGCACCATTTTTCA | ||
| iucA | iucA-F | ATAAGGCAGGCAATCCAG | 2,927 |
| iucA-R | TAACGGCGATAAACCTCG | ||
| iroN | iroN-F | GTCCGGCGGTAACTTCAGCC | 829 |
| iroN-R | TCAGAATGAAACTACCGCCC |