Abstract
Immunotherapies have revolutionized cancer treatment. Immunotherapy is effective for the treatment of a wide range of cancer types and can mediate complete and durable tumor regression. Nonetheless, the field still faces many significant challenges, such as the need for personalized therapeutic strategies and better biomarkers, the difficulty of selecting the right combination therapy, and resistance to currently available immunotherapies. Both cancer and host immunity comprise significantly diverse and complex ecosystems, making immunogenomics an ideal field for functional genomics analysis. In this review, we describe the cancer–immunity cycle, how cancer cells manage to evade immune attack and the current hurdles in the path of cancer immunotherapy. Then, we discuss how functional genomics approaches can pave the way for more successful cancer immunotherapies.
Keywords: cancer, functional genomics, immunotherapy, checkpoints, immunity
The concept of immunotherapy has revolutionized cancer treatment. Immunotherapeutics are designed to stimulate efficient, diverse and highly specific antitumor immune responses to fight cancer. The goal of immunotherapy is to enhance the immune system to search for any cancer cell, regardless of its cellular origin or location in the body, and to eliminate it effectively. Ideally, this immune-mediated tumor attack leaves neighboring normal cells intact and protects the body from relapse. This unique mechanism of action may explain why some forms of immunotherapy are effective for the treatment of a wide range of cancer types. It may also explain why some immunotherapeutic modalities can mediate complete durable tumor regression beyond 5 years.
Although immunotherapies are effective cancer treatments, not all treated individuals show favorable responses, and many responders develop acquired resistance. Not surprisingly, great effort has been made to identify biomarkers that can select the right immunotherapeutic strategy for the right patient. However, no perfect biomarkers have been identified yet and no clear understanding of how immunotherapy resistance emerges. To improve the effectiveness of immunotherapy, we need to gain a comprehensive understanding of the cancer−immune system interaction. Nonetheless, cancers and host immunity are both best characterized as complex cellular ecosystems. Typically, cancers contain multiple heterogeneous clones and subclones. Likewise, the immune system consists of many different cell types and subtypes, each of which varies in its effector functions and maturation status. Additionally, all tumor cells continually experience highly sophisticated and dynamic interactions with cells and molecules in the surrounding microenvironment, including components of the immune system. Moreover, although host immunity can recognize and destroy cancer cells, it can also enhance the selection of immune-resistant populations. To address this complexity, functional genomics, which evaluates a wide spectrum of genes and their products, might be more informative than studying isolated genes and their products.
In this review, we briefly discuss how the immune system interacts with cancer, how cancer immunotherapeutics work and the current challenges in the field. Finally, we explain how functional genomics analysis may allow us to better understand the cancer–host immunity interactions and to design more effective immunotherapeutic strategies. Computational tools and mathematical algorithms have been extensively reviewed elsewhere. Therefore, detailed discussion of this area is beyond the scope of this article.
The cancer–immunity cycle
During tumorigenesis, cancer undergoes strong selection pressure. For cancer cells to grow and evolve, they need to compete with neighboring cells for nutrients and space [1]. They also need to escape immune recognition and attack. Although some cells can successfully adapt and survive, many others die. The death of cancer cells, which are genetically and epigenetically modified as compared with normal cells, can result in the release of mutant proteins, which may be recognized as non-self (i.e. antigens) [2], and thus stimulate host immunity (Figure 1). There are two main types of cancer-related antigens: neoantigens, also called tumor-specific antigens (TSA) that are specifically and exclusively expressed by cancer cells, and tumor-associated antigens, which are self-proteins [3] that include differentiation antigens, overexpressed cellular antigens and cancer testis antigens [4].
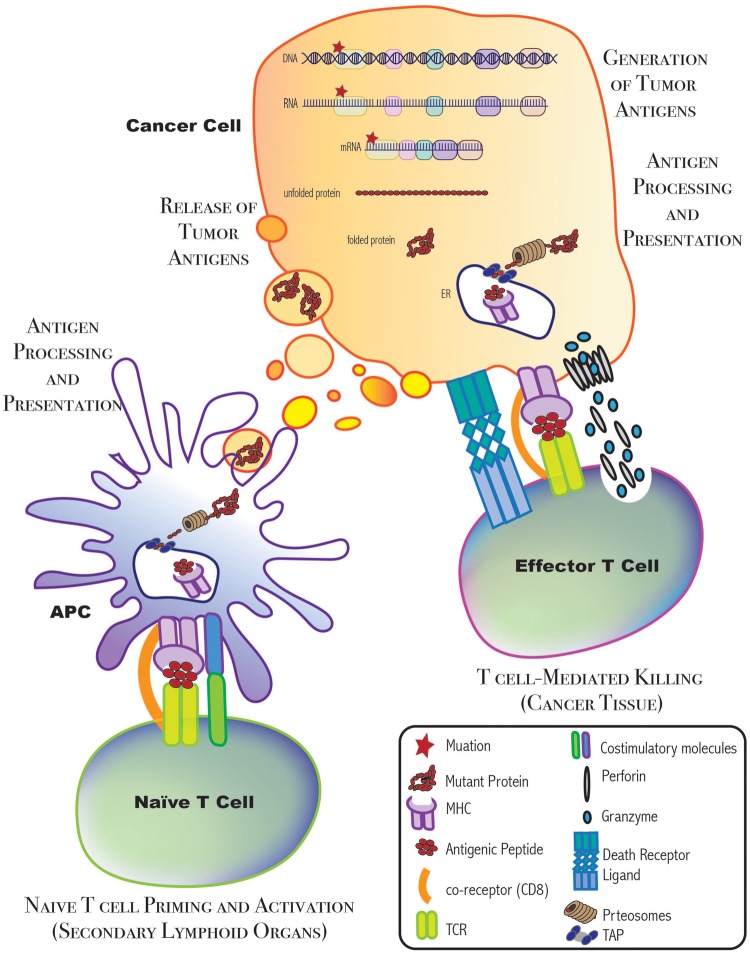
Figure 1:
Immune and cancer cell interactions. When a mutation arises in the genetic sequence of a cancer cell, it is transcribed, and eventually translated, until the mutation is carried in the complete, functional protein product. This mutant protein may eventually be processed and presented on an MHC molecule on the surface of cancer cells or APCs. In the secondary lymphoid organs, APCs prime and activate naïve T cells. Then, these activated T cells recognize the antigen and mediate T cell killing.
The initiation of an immune response begins when tumor-infiltrating antigen-presenting cells (APCs), exemplified by dendritic cells (DCs), recognize dying cancer cells and/or their related secreted particles. This can lead to DC activation. Active DCs then capture these antigens, process them and present them on their cell surface loaded onto the major histocompatibility complex (MHC) molecules. Thereafter, the DCs migrate to T-cell-rich areas of secondary lymphoid tissues, such as lymph nodes. There, they interact with antigen-specific-naïve T cells that have not yet been exposed to this antigen. DC−T cell interactions result in T cell priming and activation. Next, through a coordinated cascade of cytokines and chemokines, the now active T cells migrate toward the cancer tissue. Once they arrive and infiltrate the tumor, cytotoxic T cells (CTLs; CD8+) can directly kill the cancer cells expressing the given antigen by either activating the death receptor pathway or the granule exocytosis pathway. In contrast, helper T cells (Th; CD4+) mainly stimulate other downstream effector immune cells that are capable of eliminating the cancer cells [5]. Importantly, this immune attack can result in the generation of more immunogenic antigens, and thus continue to feed progression through this cycle [2] (Figure 2).
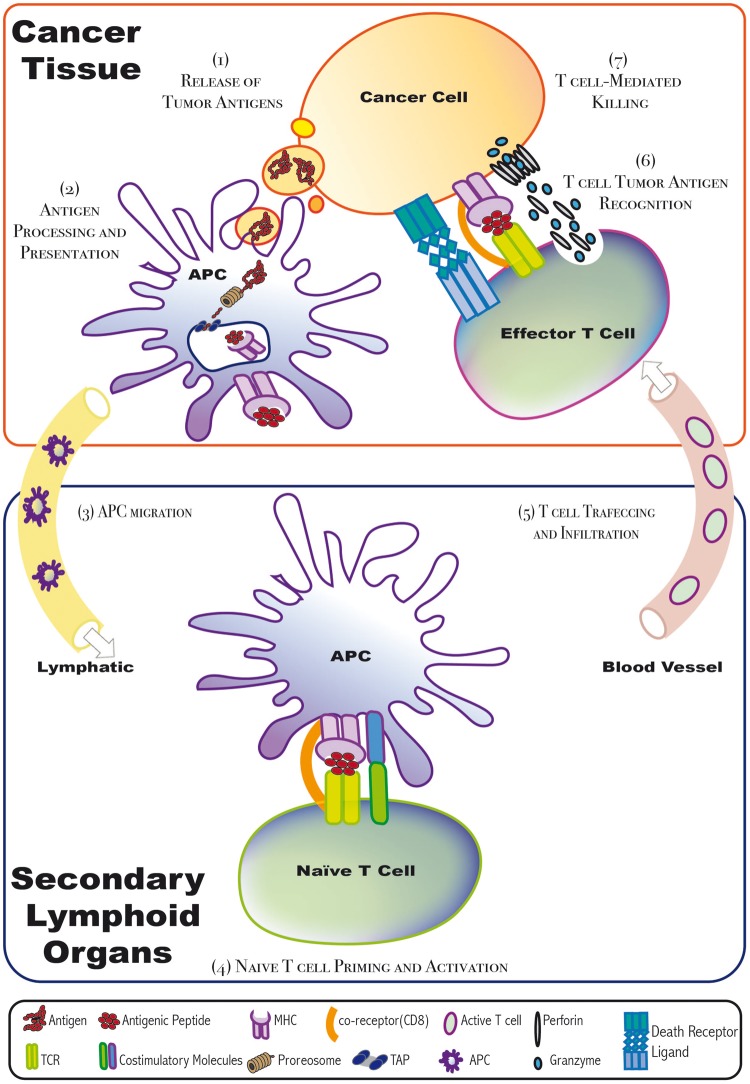
Figure 2:
Cancer–immunity cycle. When tumor-infiltrating APCs recognize tumor antigens, they get activated and thus migrate to secondary lymphoid organs. There, the APC interacts and promotes the priming and activation of antigen-specific-naïve T cells. Then, these active T cells, when infiltrated to the tumors through blood vessels, can recognize tumor antigens and thus kill the antigen-expressing cancer cells. Cytotoxic T cells kill their targets via either the death receptor pathway or the granule exocytosis pathway. ER = endoplasmic reticulum; APC = antigen presenting cell; MHC = major histocompatibility complex molecule; TCR = T cell receptor; TAP = transporter associated with antigen processing.
Cancer cells block progression through the cancer–immunity cycle to escape immune attack
The immune system is well adapted to differentiating between ‘self’ and ‘non-self’ tissue when determining whether or not to mount an effector response. However, based on murine tumor models and clinical data, it is now clear that cancer cells can use many escape routes to subdue or circumvent an antitumor immune response [2].
The first step in the cancer–immunity cycle is the release of immunogenic antigens from dying cancer cells. Every day, millions of cells die as a result of normal tissue turnover, tissue injury, infection and other causes. Interestingly, host immunity is educated to react against certain dying cells (i.e. immunogenic cell death) but not others (i.e. tolerogenic cell death). The goal of this distinction is to mount an immune response against dangerous cells without inducing an unwanted immune response against self. However, over the course of cancer progression, malignant cells tend to accumulate genetic and epigenetic changes. Some of these changes can enable cancer cells to shift the balance toward immune tolerance. Supporting evidence comes from the finding that the endoplasmic reticulum (ER) chaperone calreticulin (CALR), along with a few other proteins that act as ‘eat-me’ signals (i.e. phagocytosis signals) are downregulated in cancer. In contrast, the CALR antagonist CD47, which acts as a ‘don’t eat-me’ signal (i.e. an inhibitory signal that suppresses phagocytosis) is upregulated in many malignancies [6]. As a result, CD47 blockage is currently being tested in clinical trials as a therapeutic strategy [7].
Another crucial step is the priming and activation of naïve T cells. These cells require two independent signals to become activated. The first, the ‘recognition signal’, is antigen-dependent and involves the interaction between a T-cell receptor (TCR) and an antigen/MHC complex. The second signal, the ‘co-stimulatory signal’, is antigen independent and involves the binding of co-stimulatory molecules on T cells to their ligands and counter-receptors on APCs [2]. Although T-cell immunity is needed to fight infections and cancer, it is also critical to deactivate T cells once they have done their jobs and before the development of chronic inflammation and tissue damage [8]. The immune system is capable of achieving this balance by stimulating immunosuppressive cells, such as Regulatory T (Treg) cells, and cytokines, as well as by inducing the expression of co-inhibitory signals (checkpoints) [5]. For illustration, one of the most well-studied co-stimulatory molecules expressed by T cells is CD28, which binds its receptors B7.1 or B7.2 on APCs. This binding results in naïve T-cell priming and activation. On their activation, T cells express the co-inhibitory molecule cytotoxic T-lymphocyte-associated antigen 4 (CTLA-4) on their surfaces. CTLA-4 binds B7.1 or B7.2 with a high affinity, outcompeting CD28 binding, and thus inhibiting T-cell activity. Interestingly, cancer cells can also use these subversive tactics, including the expression of immune checkpoints and the stimulation of immunosuppressive cells and cytokines, to defeat antitumor immunity [9]. In 1995, Allison et al. first showed that in vivo administration of CTLA-4-targeted antibodies restores inhibited anti-tumor T-cell immunity [9–11]. This observation was further supported in subsequent clinical trials, resulting in the U.S. Food & Drug Administration (FDA)-approval of the first immune checkpoint inhibitor, ipilimumab, in 2011 for the treatment of melanoma [12–16].
If naïve T cells become successfully primed and activated, they next travel through the vasculature and extravasate toward the tumor tissue. Cytokines and chemokines play a central role in both attracting, as is the case for C-X-C motif chemokine ligand 9 (CXCL9) and CXCL10, and diverting, as is the case for CXCL12, T cells. Interestingly, it has been observed that cancer cells exhibit changes in cytokines’ and chemokines’ gene expression and epigenetic profiles [17]. Not surprisingly, changes that favor cancer cell growth and evasion are selected. Among these are the genetic and epigenetic alterations that enable cancer cells to prevent T-cell infiltration into the tumor site (i.e. ignored cold tumors), or allow the T cells to be recruited but prevent them from penetrating deeply into the tumor core (i.e. cold excluded tumors). Furthermore, malignant cells can promote angiogenesis, which provides them the nutrients and oxygen supply needed for growth. T cells may also travel through these newly formed blood vessels toward the tumor tissue. As an additional escape mechanism, cancer cells tend to downregulate adhesion molecules, including E-selectin, intercellular adhesion molecule 1/2, vascular cell adhesion molecule 1 and CD34, on the surface of endothelial cells. As a result, cancer may disallow T cells from adhering to blood vessels’ wall, and thus prevent them from infiltrating tumors [2, 18].
The following step in this cycle is antigen recognition by T cells. For T cells to specifically elicit an immune response against their targets, they need to recognize the antigens that were processed into small peptides with a specific length and configuration, and were presented to them on an MHC molecule. Not surprisingly, malignant cells can impair antigen processing and reduce antigen presentation. These modifications make T cells blind to the presence of tumor antigens, thus, allowing cancer cells to evade immune recognition and attack [2]. This is supported by several lines of evidence, including the loss of up to 90% of normal MHC I expression in many types of cancer [19], and the loss of members of the antigen-processing machinery, including the transporter associated with antigen processing (TAP) [20]. Another important escape mechanism is the selection of less immunogenic cancer cell clones through the process of cancer immunoediting [21–23]. Besides hiding their antigens, malignant cells can also hide themselves physically within the dense collagenous stroma [24], or in immune privileged sites [25].
Because T cells have a decidedly crucial role in the cancer–immunity cycle, immunologists and oncologists have translated this information to develop cancer therapeutics. The two main types of T-cell-based immunotherapies are the adoptive transfer of autologous tumor-infiltrating lymphocyte (TIL) populations and biologically engineered T cells. Autologous T-cell therapy can result in about 50% overall response rates and about 20% complete response rates for melanoma patients with metastatic disease [26, 27]. Despite these encouraging therapeutic outcomes, the development of autologous T cells does not consider the direct identification of tumor antigens [28]. In an attempt to improve the effectiveness of T cell-based immunotherapies, engineered T cells have been created to efficiently recognize and kill cancer cells when administered to patients. These engineered T cells entail the cloning of TCRs [28–30], components of the activated TCR complex, or a chimeric antigen receptor (CAR), which extraordinarily combines the effects of antibody recognition with T-cell cytotoxicity [31–33]. In 2017, the FDA announced the approval for CTL019 (tisagenlecleucel), a CD19-directed CAR-T-cell therapy, for relapsed/refractory pediatric and young adult patients with B-cell acute lymphocytic leukemia. Strikingly, 83% of patients involved in the clinical trial had complete remission within 3 months of infusion [31–33]. A few weeks later, another CD-19-directed CAR-T therapy, axicabtagene ciloleucel (YESCARTA™), which is manufactured by Kite Pharma, Incorporated, was also approved by FDA for the treatment of adult patients with relapsed or refractory large B-cell lymphoma. In the clinical trial that led to this approval, 82% of the patients, who previously had treatment-resistant or relapsed aggressive non-Hodgkin lymphoma, showed favorable responses, and 54% experienced complete responses. More interestingly, this therapy can cause durable complete remissions [34]. Of notice, most solid tumors do not share the same TSAs. Therefore, identifying such targets for each individual is more challenging than targeting CD19-positive lymphoma and leukemia.
Furthermore, even if T cells were properly activated, infiltrated the tumor and recognized their antigens, cancer cells often harbor the ability to express many immune checkpoints. These inhibitory molecules inactivate T cells and prevent direct T-cell-mediated killing [8]. Notably, although this concept is similar to the CTLA-4−B7.1/B7.2 interaction that occurs in the secondary lymphoid organs, CTLA-4 mainly regulates T cells at the initial stage of naive T-cell activation. In contrast, many other immune checkpoints regulate the latter part of this cycle [35]. One of the well-studied checkpoint receptor –ligand interactions is that of programmed death-ligand 1 (PD-L1), which interacts with PD-1 expressed by activated T cells [36]. Several monoclonal antibodies that target PD-L1, including atezolizumab, durvalumab and avelumab, or target PD-1, including pembrolizumab and nivolumab, have received FDA approval for cancer treatment. These PD1/PD-L1-targeted antibodies vary based on immunoglobulin (Ig) isotype, but they all act by blocking PD-L1−PD-1 interactions, and thus effectively release anti-tumor T-cell responses [37]. In fact, these therapies have resulted in significantly longer median overall survival, with minimal toxicity compared to ipilimumab [38, 39]. Also, PD1/PD-L1-targeted therapies are approved for the treatment of multiple malignancies, including adult and pediatric melanoma, non-small cell lung cancer, renal cell carcinoma (RCC), head and neck cancers, Hodgkin lymphoma, Merkel cell carcinoma, gastric/esophageal cancer and hepatocellular carcinoma [37, 40–41]. This list is continuously expanding.
Emerging challenges in cancer immunotherapy
Despite the considerable progress that has been made in the field of cancer immunotherapy, significant challenges remain.
Cancer immunotherapy requires personalization
During tumorigenesis, cancer cells acquire different numbers and types of mutations. Also, as a result of the immunoediting process, mutations that stimulate an antitumor immunity might be lost, whereas less immunogenic mutations might be selectively maintained. Consequently, neoantigens are rarely shared between patients [42], and thus neoantigen-based cancer immunotherapies require personalization.
In addition, although some immune escape strategies, such as the upregulation of PD-L1 expression, are more commonly used than others [43], this observation may not justify the use of a single immunotherapy approach to treat all cancer patients. Indeed, for more successful therapeutic outcomes, it might be necessary to have a complete understanding of how a given individual tumor managed to escape antitumor immunity. In support of this hypothesis is the fact that only a small percentage of patients may benefit from immune checkpoint blockades [44]. In fact, nivolumab or ipilimumab induce durable responses in only 10–30% of melanoma patients when either agent is used alone [45, 46].
The need for better biomarkers in cancer immunotherapy
In an attempt to increase the proportion of responders to immunotherapies, a great amount of recent effort has been devoted to identifying predictive biomarkers. To date, many biomarkers have been evaluated in preclinical and clinical studies. These biomarkers include PD-1/PD-L1 expression, tumor-infiltrating immune cells, absolute lymphocyte counts, TCR clonality, tumor mutational load, immune-related gene expression profiles, MHC class I epitope frequency/specificity and tumor mismatch–repair status [47]. In addition, serum biomarkers have received much attention recently. This approach relies on the expectation that it might be possible to identify molecules that can predict the response to immunotherapies with ease and reliability [48]. Nonetheless, biomarker studies have illustrated that predicting which patients are likely to respond using a single biomarker is quite challenging. As an example, melanoma patients who have high PD-L1 expression show higher response rates compared with patients with low PD-L1 expression. Nevertheless, not all patients with high PD-L1 expression respond to the treatment, and, at the same time, PD1/PD-L1-targeted therapies are effective in some PD-L1 negative patients [49]. More strikingly, PD-L1 expression is not significantly associated with anti-PD-1/PD-L1 treatment responses in some other malignancies, such as RCC [50]. Collectively, these findings illustrate that there is still much work to be done and that integrative analysis of multiple biomarkers might be necessary to improve the prediction of therapeutic response. In line with that, the stratification of tumors based on both the presence/absence of tumor-infiltrating T cell in addition to PD-L1 expression has been suggested as a better predictive method to design and identify ideal immunotherapies, rather than evaluating these two factors individually [51].
The difficulty of selecting the right combination therapy
The combination of PD1/PD-L1- and CTLA-4-targeted immunotherapy is a major breakthrough in cancer treatment. Immune checkpoint inhibitor combinations have improved the median overall survival of melanoma patients with advanced disease from <1 year [52] to 37.6 months using nivolumab, 19.9 months using ipilimumab and >3 years with nivolumab-plus-ipilimumab. This dramatic improvement was seen in more than half of nivolumab- and nivolumab-plus-ipilimumab-treated patients, and in about one-third of the ipilimumab-receiving group [53]. Likewise, the inhibition of the enzyme indoleamine 2, 3-dioxygenase (IDO), a T-cell-response suppressor, in conjunction with other immune checkpoint inhibitors, resulted in a significant increase in patients’ response rates and overall survival rates. This observation leads to the development of several IDO inhibitors that have been/are currently being tested in Phase 1, 2 and 3 clinical trials [54]. Although these are encouraging findings, not all of the treated patients had complete responses. Also, these numbers are less dramatic in the case of other malignancies [55]. Therefore, there remains a need to identify combination treatment options.
Furthermore, besides combining different modalities of immunotherapy, conventional cancer treatments may act synergistically with immunity-based treatments. For example, some conventional cancer treatments trigger immunogenic cancer cell death. This effect results in the release of more tumor antigens, and thus it stimulates host antitumor immunity [56]. Supporting evidence comes from the observation that the combination of ipilimumab with whole-brain radiation therapy or stereotactic radiosurgery increased average overall survival by 13 months, compared with those only receiving radiation, in patients with melanoma brain metastases. The risk of death was also significantly reduced [57]. However, there are thousands of possible drug combinations, making it challenging to know where to begin in solidifying the optimal combination of treatments.
Resistance to cancer immunotherapy
One of the major obstacles to effective response to cancer immunotherapy is resistance. For example, cancer patients undergoing transplantation, HIV positive individuals, and elderly people have preexisting systemic intrinsic resistance to immunity-based treatments [58]. Moreover, many individuals have an intact immune system but may lack antitumor immune activity only at the site of the cancer. In fact, immunological factors, including the density and geography of tumor-infiltrating CD8 T cells (immunoscore) [59–61], and CD4:CD8 T cells, can play an important role in immunotherapy resistance [62]. Furthermore, over the course of cancer progression, tumor cells consistently acquire changes in their genetic, epigenetic, transcriptional and metabolic profiles, as well as alterations in their oncogenic signaling [63]. Similarly, stromal cells continually change the expression of their cell surface molecules, the activity of their intracellular signaling pathways and their cellular metabolism [64–68]. As might be expected, some of these modifications can confer immune resistance and thus attenuate the effectiveness of cancer treatments [64]. Notably, immunotherapies may also promote these cellular changes [58]. As a result, to overcome emerged resistance to immunotherapy, it would be necessary to perform a comprehensive integrative analysis to consider all of these factors.
The immune network in cancer is large and complex
To overcome these challenges, it is important to both step back and recognize the complexity of cancer tissue, and to dive in and focus on the details. Considering both approaches would allow us to create a comprehensive picture of cancer with the highest resolution.
Cancer contains many heterogeneous cell populations. Each of them has different morphological features, cellular activities and proliferative and metastatic capabilities. Also, each malignant tumor clone harbors distinct mutational, transcriptional, phenotypic and metabolic profiles [69]. Moreover, cancer cells live in a diverse environment. They are surrounded by many different cellular and non-cellular structural components, including cells of the innate and adaptive immune systems, cytokines and chemokines, blood and lymphatic vessels, adipocytes, fibroblasts and collagen fibers. Furthermore, each of these components has a wide range of specialized types and subtypes [70]. Focusing only on the immune system, it consists of >200 cell types with >300 immune cell state transitions [71, 72]. Even among cells of the same subpopulation, such as T cells, each clone expresses a unique receptor. Thus, there are roughly 1020 TCRs in each human being [71], and each TCR can respond to more than a million different peptides [73]. Moreover, each subtype of cells secretes various cytokines and chemokines, resulting in different biological functions [74]. Beyond this, the immune cells vary in maturation stages ranging from immature and mature but naïve to mature effector or memory cells. Additionally, all the components of the microenvironment communicate with each other either directly or indirectly, in a bidirectional or multidimensional, and synergistic or opposing manner. For example, tumor cells, tumor-infiltrating immune cells and other stromal cells can secrete different cytokines and chemokines [70]. Although many cytokines exert tumor suppressor effects, such as interferon-α (IFN-α) and interleukin-2 (IL-2), many others, such as IL-10, IL-4 and IL-13, promote tumor initiation and progression. In addition to this redundancy of cytokine signaling, some cytokines, such as transforming growth factor-β (TGF-β), act on various cell types and have diverse and multiple functional properties with pleiotropic roles. Indeed, many cytokines were tested in clinical trials for the treatment of cancer, including IL-10, TGF-β, granulocyte-macrophage colony-stimulating factor (GM-CSF), IL-7, IL-12, IL-15, IL-18 and IL-21, but most failed to show significant therapeutic effects, except IFN-α and IL-2 [74]. These observations suggest that the complex cytokine signaling relationships are still poorly understood and remain to be elucidated.
Another example that illustrates the complexity of cancer−immune system interactions is the expression of immune checkpoints and their signaling networks. To date, many co-stimulatory and co-inhibitory molecules have been identified on the surface of T cells. Many of these molecules can bind more than one ligand or counter-receptor. Additionally, they can be expressed on multiple cell types, including cancer cells, DCs, macrophages and/or endothelial cells. Therefore, the presence of only one immune checkpoint does not necessarily mean complete T-cell suppression. In fact, the level of T-cell activity depends on the overall outcome of the balance between these signals [8]. As a result, we need a deeper comprehension of each of these signaling pathways, as well as how they integrate. With this in mind, it becomes clear that cancer immunology is an ideal field for the application of functional genomics analysis.
Functional genomics
As the name implies, functional genomics involves large-scale evaluation of the relationship between genes and their functions. This field studies the wide spectrum of transcriptional, translational and epigenetic properties, as well as DNA−RNA, RNA−protein, and protein−protein interactions, and it fits together all of these pieces [75] (Figure 3). Indeed, conducting multi-omic studies, such as genomics, transcriptomics, proteomics or metabolomics, by processing bulk tumor tissue is useful. Nonetheless, they only explain what the entire population of cancer cells does to survive and escape immune attack, and what the overall effect of host immunity is. To determine what each tumor cell does to evade antitumor immunity, and how each immune cell clone responds, either loss and gain of function screens or single-cell approaches are used. Examples of loss of function screens include but are not limited to RNA interference (RNAi) and clustered regularly interspaced short palindromic repeats-associated nucleus 9 (CRISPR/Cas9) whole-genome libraries. Notably, the field of cancer immunogenomics applies the same genomics techniques that have been used in the past decade to study cancer biology. However, cancer immunogenomics focuses not only on cancer cells but also on immune cells and their products [71].
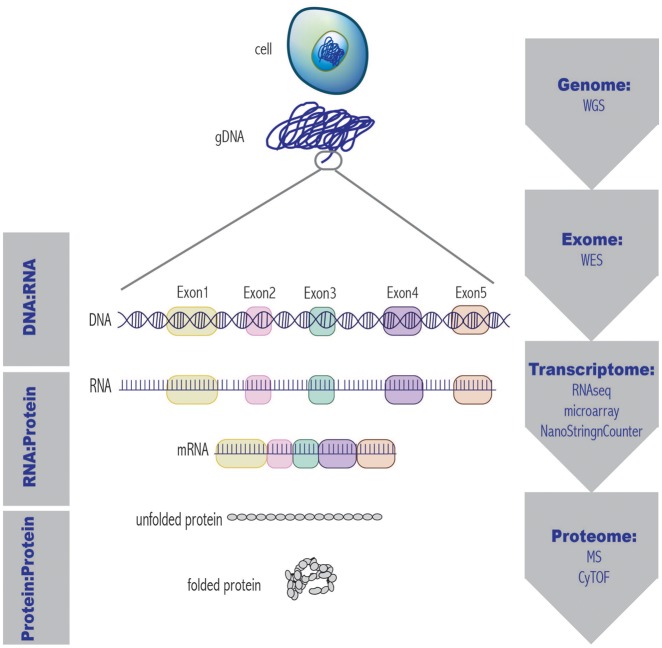
Figure 3:
The central dogma of molecular biology and examples of related functional genomics approaches. At each tier of genetic organization, from the whole genome to the complete protein, there exists a high-throughput technique to analyze a given molecule. WGS= whole genome sequencing; WES= whole exome sequencing; RNAseq= RNA sequencing; MS= mass spectrometry; CyTOF= cytometry time-of-flight.
Functional genomics can pave the way for more effective cancer immunotherapy
The use of functional genomics to characterize tumor immune infiltrates
Each tumor has a unique collection of infiltrated immune cells. Although some have antitumor immunostimulatory properties, others are immunosuppressive. Therefore, in-depth characterization of immune cells is necessary to distinguish between enemies and allies. The main contemporary strategies for cellular characterization are imaging, proteomic profiling, flow cytometry or cytometry time-of-flight (CyTOF) and gene expression profiling, using microarray, RNA sequencing (RNAseq) or NanoString nCounter technologies [76]. Imaging technologies allow for the assessment of specific markers and their cellular co-localization in tissues. Flow cytometry, on the other hand, can determine the identity and quantity of cellular proteins. However, even recently developed multispectral microscopy and flow cytometry instruments are limited in the number of markers they are able to simultaneously detect [71]. Therefore, they do not clearly distinguish between some immune cell subtypes, nor do they detect rare immune cell populations [76].
To develop a more comprehensive picture of immune infiltrates, techniques such as gene expression profiling have been used. This approach identifies immunophenotypes based on predefined gene expression signatures of immune cell types and subtypes, activation and exhaustion markers and maturation stages [76]. Moreover, transcriptomic profiles can be used to determine the proportion of each cell population and to calculate differential gene expressions [71]. To illustrate this concept, Charoentong et al. used publically available data from The Cancer Genome Atlas to characterize the intratumoral cellular genotypes and phenotypes of 20 different solid malignancies. Interestingly, by conducting integrative functional genomics analysis, this group was able to identify 28 subpopulations of TILs and to estimate tumor antigens and cancer heterogeneity. They were also able to predict response to immune checkpoint inhibitors. Finally, they transformed this powerful analysis into a web-accessible resource [77]. A similar scheme focused specifically on samples obtained from colorectal cancer patients. In this study, Angelova et al. [42] characterized the phenotypes of infiltrated immune cells, identified molecular determinants of immunogenicity and detected B-cell maturation antigen as a potential therapeutic target.
Despite the importance of conventional gene expression profiling, such techniques are not capable of generating high-resolution phenotypic and functional profiles of every single immune cell. To address heterogeneity at the cellular level, single-cell RNAseq and CyTOF have emerged as useful options. CyTOF labels cells with stable heavy metal isotopes and thus enables the analysis of >40 markers using 135 detection channels [78]. Recently, in-depth proteomic profiling of immune infiltrates using this technique has been used extensively to understand the immune network in cancer [79] and to reveal how immunotherapies work [80]. For example, Chevrier et al. [79] were able to deduce seventeen phenotypes of tumor-associated macrophages (TAMs) and 22 phenotypes of T cells in clear cell RCC. Moreover, they were able to identify populations of TAMs that correlates with exhausted T cells, and an immune composition that correlates with progression-free survival in this group of patients. However, unlike RNAseq, CyTOF does not evaluate the whole transcriptome or proteome. In a study that illustrates the power of using single-cell RNAseq, Chung et al. evaluated intratumoral heterogeneity of tumor cells, as well as cells of the surrounding microenvironment within different types of breast cancer tissues. Additionally, they identified the cellular phenotypes of infiltrated immune cells and showed that many of them are in fact immunosuppressive [81].
Furthermore, in an attempt to improve current immunogenomic methodologies, Stoeckius et al. have developed a novel technique referred to as CITE-seq. This approach is a bimodal methodology that integrates cellular protein markers and transcriptome detection using an oligonucleotide-labeled antibody. Thus, Stoeckius et al. suggested that CITE-seq is more efficient compared with either procedure on its own. To support their hypothesis, they demonstrated the ability of this technology to thoroughly characterize immune cell populations by both expression and lineage marker [82].
The use of functional genomics to identify tumor-specific immunogenic antigens
The identification of neoantigens is clinically useful for at least two reasons. First, it provides essential information for the development of tumor antigen-based immunotherapies. Second, it estimates the tumor mutational load, which might correlate with responsiveness to cancer immunotherapies, such as immune checkpoint-targeted antibodies [83–85]. However, the process of tumor-specific antigen identification requires a combination of genomics, bioinformatics and immunological approaches, making it quite challenging [23].
The basic workflow consists of four main steps. First, mutant proteins are identified in the resected tumor sample, as compared with matched normal tissue. Then, the human leukocyte antigen (HLA) haplotypes of the patient are also identified. Next, among all detected mutant proteins, neoantigens that bind the patient’s HLA molecules with high affinity are selected. Finally, neoantigens that stimulate efficient antitumor immune responses are determined. This information can then be used to generate neoantigen-specific autologous TILs and/or to synthesize personalized therapeutic cancer vaccines [23, 86].
Several methods can be applied to identify mutated proteins, including whole-exome sequencing (WES), RNAseq and mass spectrometry (MS), which seek to identify genetic mutations at the DNA level, mutated transcripts and mutant proteins, respectively. WES allows for the identification of the vast majority of somatic mutations in all exons of the genomic DNA, including mutations in genes with low levels of expression. Although WES is certainly informative to detect a multitude of variants, many of these mutations may not be expressed as proteins. In contrast, RNAseq evaluates mutated transcripts. Thus, it focuses on mutations that are more likely to be expressed in the majority of the tumor cells. It also estimates the relevant mutant alleles’ expression levels, which indirectly predicts mutant peptide abundance. Additionally, in contrast to WES, RNAseq data can be used to detect splice variants and structural rearrangements, such as fusion transcripts [23, 86, 87]. Finally, the most direct evidence of the presence and relative abundance of mutant proteins lies in the identification of their related peptides using MS [88]. However, because of the low sensitivity of currently available instruments and the requirement for large tumor samples, this approach is also imperfect. Another major challenge of MS is that it uses public proteomic databases that lack information regarding patient-specific mutations [87]. As each of these high-throughput techniques has its own strengths and weaknesses, there remains controversy in the field concerning which method is most reliable and practical in the identification of cancer neoantigens. There is also disagreement with regard to the necessity of combining multiple approaches. Although it might be effective to integrate sequencing and MS strategies, it would be expensive, labor intensive, time consuming and technically challenging. Thus, integrative analysis may not be suitable for potential clinical applications given technological and practical concerns.
The next steps of the neoantigen identification process are HLA typing, followed by epitope prediction and prioritization. These steps rely mainly on the use of mathematical algorithms and computational tools, which are not covered in this review [23, 71]. However, it is worth noting that although there are several currently available bioinformatics algorithms and tools, there is still much room for improvement. To date, it remains exceedingly difficult to predict all potential neoantigens, including mutant proteins that arise from mistakes in translation, and peptides that interact with MHC II molecules [23].
It is necessary to determine which of the computationally predicted neoantigens can trigger the activity of autologous TILs. Such approaches include MHC multimer-based screens and cytokine induction assays [23]. In addition, TCR sequencing can also estimate T-cell clonality, which is clinically useful [89–92]. In fact, the number of TCR clones could be associated with mutational burden, and thus may contribute to differential response to immunotherapy [93]. Notably, the step of determining which tumor antigen can stimulate T-cell immunity is singularly crucial because not all mutated proteins can be processed successfully into small peptides and carried on MHC molecules. Further, not all processed peptides can be recognized by TCRs (Figure 4). Although neoantigen-based immunotherapies have not yet been approved for cancer treatment, such techniques are currently being explored in clinical trials [26, 94–98].
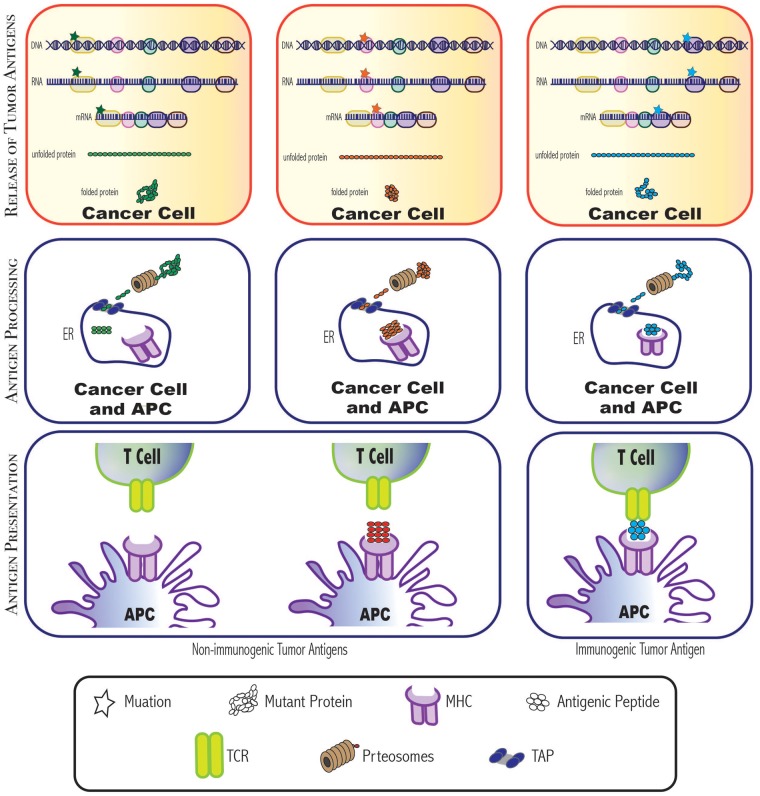
Figure 4:
Not all mutilations result in immunogenic tumor antigens. For a mutant protein to become an immunogenic tumor antigen, it has to be cleaved into small peptides with a specific length and configuration. Then, one of these peptides, which binds to an MHC molecule on an APC with high affinity, is recognized by a TCR, and elicits a T-cell response. Those immunogenic antigens provide promising options for therapeutic development. ER = endoplasmic reticulum; APC = antigen presenting cell; MHC = major histocompatibility complex molecule; TCR = T cell receptor; TAP = transporter associated with antigen processing.
Finally, it must be noted that current immunotherapies have focused only on αβ T-cell clones. Although αβ T cells are more common and exhibit very high specificity than γδ T cells, γδ TCRs have greater variability than αβ TCRs (i.e. about 1018 combinations versus 1016 combinations, respectively, in human), suggesting that they may recognize not only proteins but also smaller molecules. Moreover, the majority of γδ TCRs is not HLA restricted. Thus, it is claimed that they can be used for off-the-shelf non-personalized cancer immunotherapies. However, because of the diversity of γδ TCRs, it might be more challenging to perform TCR profiling [99, 100].
The use of functional genomics to identify better biomarkers for immunotherapy
As mentioned earlier, there is still uncertainty regarding the patient-specific correlation between current biomarkers and the clinical response to immunotherapies. In an attempt to overcome this challenge, Noguchi et al. used both a CRISPR loss of function screen and gain of function screen to more accurately monitor the expressions of PD-1 and PD-L1 in murine tumor models. Interestingly, this study revealed that the induction of PD-L1 expression in tumor cells is transient, whereas its expression by host immune cells is more stable. This observation suggests that PD-L1 expression on the surface of the cells of the microenvironment could become a more reliable prognostic biomarker [64]. Therefore, functional genomics approaches may give us a deeper insight into the biomarkers’ mechanisms of action, allowing for identifying better and more reliable biomarkers.
The use of functional genomics for determining cancer immunotherapy resistance mechanisms
Because of the complexity of cancer tissue and its surrounding microenvironment, a comprehensive view is necessary to deeply understand immunotherapy resistance mechanisms. In a clinical trial that demonstrated the power of functional genomics analysis, Zaretsky et al. reported that, in this particular study, about 25% of melanoma patients who had an objective response to pembrolizumab had acquired resistance at a median follow-up of 21 months. Genetic profiling of tumors from patients who had late cancer relapse was performed before and after treatment. Interestingly, WES analysis revealed that these patients developed mutations in genes important for regulating interferon–receptor signaling and antigen presentation pathways. Interestingly, this observation suggests that, in response to immune selection pressure, acquired mutations that confer immune resistance were selected over time, resulting in immunotherapy resistance after a strong initial response [66]. Similarly, using both whole-exome and transcriptome sequencing approaches, the Rosenberg group was able to uncover the resistance mechanism of a progressing tumor lesion in a patient treated with adoptive cell therapy against the tumor neoantigen expressed by the KRAS oncogene. Chromosomal copy number analysis revealed that although this particular metastatic lesion harbored the same targeted KRAS mutation, it also harbored genetic loss of the HLA allele required for its recognition by T cells [98]. The significance of these two clinical studies lies not only in their ability to identify resistance mechanisms for immunotherapies, but also in their ability to illustrate the effectiveness and applicability of precision cancer immunotherapy.
Nonetheless, evaluating the impact of changes in tumors’ characteristics on clinical response is not always straightforward, and it is not always associated with acquired mutations. Indeed, several groups have indicated that not only tumors of different patients but even distinct tumors within a single patient can be heterogeneous. In fact, they can differ in their genetic profiles, transcriptional profiles, the expression of immunotherapy targets and/or the number and activity of tumor-infiltrating immune cells. Importantly, any of these factors can lead to treatment resistance. More strikingly, tumor heterogeneity has been identified even between different cells within an individual tumor in an individual patient. Although advances in functional genomics analysis may determine the mechanism of resistance of tumor clones and subclones, tumor heterogeneity is clinically challenging [62, 93, 101, 102].
In addition to clinical studies, functional genomics strategies can be used in preclinical settings to identify unknown mediators of immune evasion and immunotherapy resistance. For example, Manguso et al. [103] used an in vivo whole genome-wide CRISPR screen to identify the genes associated with responsiveness to immunotherapy in a B16 murine melanoma model. Furthermore, in a separate study, Patel et al. [104] used an in vitro CRISPR library to identify the essential genes for the effector function of T cells. Remarkably, these screens were able to identify promising targets for cancer immunotherapies.
The use of functional genomics to understand cancer–immune system interactions
Recently, our group used an unbiased in vivo genome-wide RNAi screening platform to understand how immune selection pressure can manipulate cancer epithelial cells at a single-cell level. In this study, Shuptrine et al. inoculated whole-genome RNAi library-transduced EO771 breast cancer cells into syngeneic immune-competent and immunodeficient C57Bl/6 mice. By comparing the enriched and deleted short hairpin RNAs (shRNAs) between the groups of mice, we were able to identify 709 genes whose knockdown led to significantly different representation of the targeted cells in the two groups. Some of which selectively regulate adaptive antitumor immunity. This hypothesis was supported by functional validation experiments of several prioritized hits, including CD47 and TGFβ1, which are potential targets for single or combination immunotherapy [105].
Conclusion
Cancer immunotherapy has changed the paradigm for cancer treatment. It is possible to overcome current challenges by applying functional genomics approaches and performing integrative analysis. Today, there are many technologies and bioinformatics tools available, and many more will be developed in the next few years to fill current gaps, such as considering γδ TCRs in the analysis. Future efforts will need to balance the utilization of multiple functional genomics approaches, which require the analysis of massive amounts of data, to make them relevant to cancer care. Also, to improve the applicability of functional genomics technologies for routine clinical use, it is important to consider the time and money consumption.
Key Points
Cancer immunotherapy is a promising and effective cancer treatment modality that reinvigorates the natural defenses of a host against its own tumors. However, significant challenges remain.
Functional genomics approaches are equipped to survey the tremendous diversity and complexity of the dynamic ecosystems: cancer and host immunity.
Functional genomics approaches can be used to deeply understand host immunity by characterizing tumor-infiltrating immune cells and their products
Functional genomics technologies provide needed information for the generation of neoantigen-based cancer treatments, and may identify biomarkers and resistance mechanisms against already available immunotherapies.
There is a pressing need to improve currently available technologies and computational analysis to improve their applicability for routine cancer care.
Funding
This research was supported by NCI R01 CA50633 and NCI CA51008 (LMW).
Reham Ajina is a current PhD candidate in Tumor Biology at Georgetown University. Her primary research interest is cancer immunotherapy.
Danielle Zamalin is an undergraduate student at Georgetown University, receiving a BS in Human Science degree in 2018. Her research interests focus on clinical and translational projects, which she plans to continue to work on as she enters medical school.
Louis M Weiner is the director of Georgetown Lombardi Comprehensive Cancer Center (LCCC) at Georgetown University. He also serves as the chair of the Department of Oncology and director of the MedStar Georgetown Cancer Institute. His laboratory focuses on the application of whole-genome screens to identify cancer resistance mechanisms to immune selection pressure and cancer immunotherapies.
References
- 1. Ferreira SC, Martins ML, Vilela MJ.. Reaction-diffusion model for the growth of avascular tumor. Phys Rev E Stat Nonlin Soft Matter Phys 2002;65(2):021907. [DOI] [PubMed] [Google Scholar]
- 2. Chen DS, Mellman I.. Oncology meets immunology: the cancer-immunity cycle. Immunity 2013;39(1):1–10. [DOI] [PubMed] [Google Scholar]
- 3. Zilberberg J, Feinman R, Korngold R.. Strategies for the identification of T cell-recognized tumor antigens in hematological malignancies for improved graft-versus-tumor responses after allogeneic blood and marrow transplantation. Biol Blood Marrow Transplant 2015;21(6):1000–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Criscitiello C. Tumor-associated antigens in breast cancer. Breast Care 2012;7(4):262–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Weiner LM. Cancer immunology for the clinician. Clin Adv Hematol Oncol 2015;13(5):299–306. [PubMed] [Google Scholar]
- 6. Galluzzi L, Buqué A, Kepp O.. Immunogenic cell death in cancer and infectious disease. Nat Rev Immunol 2017;17(2):97–111. [DOI] [PubMed] [Google Scholar]
- 7. Huang Y, Ma Y, Gao P, Targeting CD47: the achievements and concerns of current studies on cancer immunotherapy. J Thorac Dis 2017;9(2):E168–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012;12(4):252–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chambers CA, Kuhns MS, Egen JG, CTLA-4-mediated inhibition in regulation of T cell responses: mechanisms and manipulation in tumor immunotherapy. Annu Rev Immunol 2001;19(1):565–94. [DOI] [PubMed] [Google Scholar]
- 10. Leach DR, Krummel MF, Allison JP.. Enhancement of antitumor immunity by CTLA-4 blockade. Science 1996;271(5256):1734–6. [DOI] [PubMed] [Google Scholar]
- 11. Hurst JH. Cancer immunotherapy innovator James Allison receives the 2015 Lasker∼DeBakey Clinical Medical Research Award. Am Soc Clin Investig 2015;125(10):3732–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wolchok JD, Hodi FS, Weber JS, Development of ipilimumab: a novel immunotherapeutic approach for the treatment of advanced melanoma. Ann N Y Acad Sci 2013;1291(1):1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hodi FS, O'Day SJ, McDermott DF, Improved survival with ipilimumab in patients with metastatic melanoma. N Engl J Med 2010;363:711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hoos A, Eggermont AMM, Janetzki S, Improved endpoints for cancer immunotherapy trials. J Natl Cancer Inst 2010;102(18):1388–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Robert C, Thomas L, Bondarenko I, Ipilimumab plus dacarbazine for previously untreated metastatic melanoma. N Engl J Med 2011;364(26):2517–26. [DOI] [PubMed] [Google Scholar]
- 16. Ribas A, Kefford R, Marshall MA, Phase III randomized clinical trial comparing tremelimumab with standard-of-care chemotherapy in patients with advanced melanoma. J Clin Oncol 2013;31(5):616–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lunardi S, Jamieson NB, Lim SY, IP-10/CXCL10 induction in human pancreatic cancer stroma influences lymphocytes recruitment and correlates with poor survival. Oncotarget 2014;5(22):11064–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. van der Woude LL, Gorris MAJ, Halilovic A, Migrating into the tumor: a roadmap for T cells. Trends Cancer 2017;3(11):797–808. [DOI] [PubMed] [Google Scholar]
- 19. Aptsiauri N, García-Lora AM, Garrido F.. ‘Hard’ and ‘soft’ loss of MHC class I expression in cancer cells In: Rees RC. (ed), Tumor Immunology and Immunotherapy. Oxford University Press; 2014, pp. 63–78. [Google Scholar]
- 20. Fruci D, Benevolo M, Cifaldi L, Major histocompatibility complex class i and tumour immuno-evasion: how to fool T cells and natural killer cells at one time. Curr Oncol 2012;19(1):39–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Shankaran V, Ikeda H, Bruce AT, IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature 2001;410(6832):1107–11. [DOI] [PubMed] [Google Scholar]
- 22. Dunn GP, Bruce AT, Ikeda H, Cancer immunoediting: from immunosurveillance to tumor escape. Nat Immunol 2002;3(11):991–8. [DOI] [PubMed] [Google Scholar]
- 23. Gubin MM, Artyomov MN, Mardis ER, Tumor neoantigens: building a framework for personalized cancer immunotherapy. J Clin Invest 2015;125(9):3413–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bartholin L. Pancreatic cancer and the tumor microenvironment: mesenchyme’s role in pancreatic carcinogenesis In: Grippo PJ, Munshi HG (eds). Pancreatic Cancer and Tumor Microenvironment. Trivandrum, India: Transworld Research Network, 2012. [PubMed] [Google Scholar]
- 25. Igney FH, Krammer PH.. Immune escape of tumors: apoptosis resistance and tumor counterattack. J Leukoc Biol 2002;71(6):907–20. [PubMed] [Google Scholar]
- 26. Rosenberg SA, Yang JC, Sherry RM, Durable complete responses in heavily pretreated patients with metastatic melanoma using T-cell transfer immunotherapy. Clin Cancer Res 2011;17(13):4550–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hinrichs CS, Rosenberg SA.. Exploiting the curative potential of adoptive T-cell therapy for cancer. Immunol Rev 2014;257(1):56–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Lu Y-, Zheng CZ, Robbins PF, An efficient single-cell RNA-seq approach to identify neoantigen-specific T cell receptors. Mol Ther 2018;26:379–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Morgan RA, Dudley ME, Wunderlich JR, Cancer regression in patients after transfer of genetically engineered lymphocytes. Science 2006;314(5796):126–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Morgan RA, Dudley ME, Yu YY, High efficiency TCR gene transfer into primary human lymphocytes affords avid recognition of melanoma tumor antigen glycoprotein 100 and does not alter the recognition of autologous melanoma antigens. J Immunol 2003;171(6):3287–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Kalos M, Levine BL, Porter DL, T cells with chimeric antigen receptors have potent antitumor effects and can establish memory in patients with advanced leukemia. Sci Transl Med 2011;3(95):95ra73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pettitt D, Arshad Z, Smith J, CAR-T cells: a systematic review and mixed methods analysis of the clinical trial landscape. Mol Ther 2017;26:342–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kochenderfer JN, Wilson WH, Janik JE, Eradication of B-lineage cells and regression of lymphoma in a patient treated with autologous T cells genetically engineered to recognize CD19. Blood 2010;116(20):4099–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Neelapu SS, Locke FL, Bartlett NL, Axicabtagene ciloleucel CAR T-cell therapy in refractory large B-cell lymphoma. N Engl J Med 2017;377(26):2531–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fife BT, Bluestone JA.. Control of peripheral T-cell tolerance and autoimmunity via the CTLA-4 and PD-1 pathways. Immunol Rev 2008;224(1):166–82. [DOI] [PubMed] [Google Scholar]
- 36. Okazaki T, Honjo T.. PD-1 and PD-1 ligands: from discovery to clinical application. Int Immunol 2007;19(7):813–24. [DOI] [PubMed] [Google Scholar]
- 37. Lee HT, Lee JY, Lim H, Molecular mechanism of PD-1/PD-L1 blockade via anti-PD-L1 antibodies atezolizumab and durvalumab. Sci Rep 2017;7(1):5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Robert C, Schachter J, Long GV, Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med 2015;372(26):2521–32. [DOI] [PubMed] [Google Scholar]
- 39. Schachter J, Ribas A, Long GV, Pembrolizumab versus ipilimumab for advanced melanoma: final overall survival results of a multicentre, randomised, open-label phase 3 study (KEYNOTE-006). Lancet 2017;390(10105):1853–62. [DOI] [PubMed] [Google Scholar]
- 40. Kaufman HL, Russell J, Hamid O, Avelumab in patients with chemotherapy-refractory metastatic Merkel cell carcinoma: a multicentre, single-group, open-label, phase 2 trial. Lancet Oncol 2016;17(10):1374–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chen R, Zinzani PL, Fanale MA, Phase II study of the efficacy and safety of pembrolizumab for relapsed/refractory classic hodgkin lymphoma. J Clin Oncol 2017;35(19):2125–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Angelova M, Charoentong P, Hackl H, Characterization of the immunophenotypes and antigenomes of colorectal cancers reveals distinct tumor escape mechanisms and novel targets for immunotherapy. Genome Biol 2015;16(1):64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang X, Teng F, Kong L, PD-L1 expression in human cancers and its association with clinical outcomes. Onco Targets Ther 2016;9:5023–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Bu X, Mahoney KM, Freeman GJ.. Learning from PD-1 resistance: new combination strategies. Trends Mol Med 2016;22(6):448–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wolchok JD, Kluger H, Callahan MK, Nivolumab plus ipilimumab in advanced melanoma. N Engl J Med 2013;369(2):122–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sathyanarayanan V, Neelapu SS.. Cancer immunotherapy: strategies for personalization and combinatorial approaches. Mol Oncol 2015;9(10):2043–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Masucci GV, Cesano A, Hawtin R, Validation of biomarkers to predict response to immunotherapy in cancer: volume I - pre-analytical and analytical validation. J Immunother Cancer 2016;4(1):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wu X, Giobbie-Hurder A, Liao X, Angiopoietin-2 as a biomarker and target for immune checkpoint therapy. Cancer Immunol Res 2017;5(1):17–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Gnjatic S, Bronte V, Brunet LR, Identifying baseline immune-related biomarkers to predict clinical outcome of immunotherapy. J Immunother Cancer 2017;5(1):44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Qu H-X, Zhao L-P, Zhan S-H, Clinicopathological and prognostic value of programmed death ligand-1 (PD-L1) in renal cell carcinoma: a meta-analysis. Int J Clin Exp Med 2016;8(11):14595–603. [PMC free article] [PubMed] [Google Scholar]
- 51. Teng MWL, Ngiow SF, Ribas A, Classifying cancers based on T-cell infiltration and PD-L1. Cancer Res 2015;75(11):2139–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. SandruVoinea A, Panaitescu SE, Survival rates of patients with metastatic malignant melanoma. J Med Life 2014;7(4):572–76. [PMC free article] [PubMed] [Google Scholar]
- 53. Wolchok JD, Chiarion-Sileni V, Gonzalez R, Overall survival with combined nivolumab and ipilimumab in advanced melanoma. N Engl J Med 2017;377(14):1345–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Brochez L, Chevolet I, Kruse V.. The rationale of indoleamine 2, 3-dioxygenase inhibition for cancer therapy. Eur J Cancer 2017;76:167–82. [DOI] [PubMed] [Google Scholar]
- 55. Ott PA, Hodi FS, Kaufman HL, Combination immunotherapy: a road map. J Immunother Cancer 2017;5(1):16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Gebremeskel S, Johnston B.. Concepts and mechanisms underlying chemotherapy induced immunogenic cell death: impact on clinical studies and considerations for combined therapies. Oncotarget 2015;6(39):41600–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Silk AW, Bassetti MF, West BT, Ipilimumab and radiation therapy for melanoma brain metastases. Cancer Med 2013;2(6):899–906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Kelderman S, Schumacher TN, Haanen JB.. Acquired and intrinsic resistance in cancer immunotherapy. Mol Oncol 2014;8(6):1132–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Galon J, Pagès F, Marincola FM, The immune score as a new possible approach for the classification of cancer. J Transl Med 2012;10(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Galon J, Lugli A, Bifulco C, World-wide immunoscore task force: meeting report from the ‘Melanoma Bridge’, Napoli, November 30th-December 3rd, 2016. J Transl Med 2017;15(1):212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Galon J, Pagès F, Marincola FM, Cancer classification using the immunoscore: a worldwide task force. J Transl Med 2012;10(1):205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ascierto ML, Makohon-Moore A, Lipson EJ, Transcriptional mechanisms of resistance to anti-PD-1 therapy. Clin Cancer Res 2017;23(12):3168–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Jenkins RW, Barbie DA, Flaherty KT.. Mechanisms of resistance to immune checkpoint inhibitors. Br J Cancer 2018;118(1):9–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Noguchi T, Ward JP, Gubin MM, Temporally distinct PD-L1 expression by tumor and host cells contributes to immune escape. Cancer Immunol Res 2017;5(2):106–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schelker M, Feau S, Du J, Estimation of immune cell content in tumour tissue using single-cell RNA-seq data. Nat Commun 2017;8(1):2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Zaretsky JM, Garcia-Diaz A, Shin DS, Mutations associated with acquired resistance to PD-1 blockade in Melanoma. N Engl J Med 2016;375(9):819–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kong B, Bruns P, Behler NA, Dynamic landscape of pancreatic carcinogenesis reveals early molecular networks of malignancy. Gut 2018;67(1):146–56. [DOI] [PubMed] [Google Scholar]
- 68. Sukumar M, Kishton RJ, Restifo NP.. Metabolic reprograming of anti-tumor immunity. Curr Opin Immunol 2017;46:14–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Gay L, Baker A-M, Graham TA.. Tumour cell heterogeneity. F1000Res 2016;5:238.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wörmann SM, Diakopoulos KN, Lesina M, The immune network in pancreatic cancer development and progression. Oncogene 2014;33(23):2956–67. [DOI] [PubMed] [Google Scholar]
- 71. Hackl H, Charoentong P, Finotello F, Computational genomics tools for dissecting tumour-immune cell interactions. Nat Rev Genet 2016;17(8):441–58. [DOI] [PubMed] [Google Scholar]
- 72. Heng TSP, Painter MW; Immunological Genome Project Consortium. The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 2008;9(10):1091–4. [DOI] [PubMed] [Google Scholar]
- 73. Wooldridge L, Ekeruche-Makinde J, van den Berg HA, A single autoimmune T cell receptor recognizes more than a million different peptides. J Biol Chem 2012;287(2):1168–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Lee S, Margolin K.. Cytokines in cancer immunotherapy. Cancers 2011;3(4):3856–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bunnik EM, Le Roch KG.. An introduction to functional genomics and systems biology. Adv Wound Care 2013;2(9):490–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Lyons YA, Wu SY, Overwijk WW, Immune cell profiling in cancer: molecular approaches to cell-specific identification. NPJ Precis Oncol 2017;1(1):26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Charoentong P, Finotello F, Angelova M, Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep 2017;18(1):248–62. [DOI] [PubMed] [Google Scholar]
- 78. Matos TR, Liu H, Ritz J.. Research techniques made simple: experimental methodology for single-cell mass cytometry. J Invest Dermatol 2017;137(4):e31–8. [DOI] [PubMed] [Google Scholar]
- 79. Chevrier S, Levine JH, Zanotelli VRT, An immune atlas of clear cell renal cell carcinoma. Cell 2017;169(4):736–49.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Wei SC, Levine JH, Cogdill AP, Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. Cell 2017;170(6):1120–33.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Chung W, Eum HH, Lee H-O, Single-cell RNA-seq enables comprehensive tumour and immune cell profiling in primary breast cancer. Nat Commun 2017;8:15081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Stoeckius M, Hafemeister C, Stephenson W, Simultaneous epitope and transcriptome measurement in single cells. Nat Methods 2017;14(9):865–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Rizvi NA, Hellmann M, Snyder DA, Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015;348(6230):124–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yarchoan M, Hopkins A, Jaffee EM.. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med 2017;377(25):2500–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Chan TA, Wolchok JD, Snyder A.. Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 2015;373(20):1984–4. [DOI] [PubMed] [Google Scholar]
- 86. Liu XS, Mardis ER.. Applications of immunogenomics to cancer. Cell 2017;168(4):600–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Yadav M, Jhunjhunwala S, Phung QT, Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 2014;515(7528):572–6. [DOI] [PubMed] [Google Scholar]
- 88. Bassani-Sternberg M, Bräunlein E, Klar R, Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat Commun 2016;7:13404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Kirsch I, Vignali M, Robins H.. T-cell receptor profiling in cancer. Mol Oncol 2015;9(10):2063–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Park J-H, Jang M, Tarhan YE, Clonal expansion of antitumor T cells in breast cancer correlates with response to neoadjuvant chemotherapy. Int J Oncol 2016;49(2):471–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Ikeda Y, Kiyotani K, Yew PY, Clinical significance of T cell clonality and expression levels of immune-related genes in endometrial cancer. Oncol Rep 2017;37(5):2603–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Subudhi SK, Aparicio A, Gao J, Clonal expansion of CD8 T cells in the systemic circulation precedes development of ipilimumab-induced toxicities. Proc Natl Acad Sci USA 2016;113(42):11919–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Reuben A, Spencer CN, Prieto PA, Genomic and immune heterogeneity are associated with differential responses to therapy in melanoma. NPJ Genom Med 2017;2(1):2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hu Z, Ott PA, Wu CJ.. Towards personalized, tumour-specific, therapeutic vaccines for cancer. Nat Rev Immunol 2018;18:168–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Ott P, Hu AZ, Keskin DB, An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 2017;547(7662):217–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Sahin U, Derhovanessian E, Miller M, Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 2017;547(7662):222–6. [DOI] [PubMed] [Google Scholar]
- 97. Carreno BM, Magrini V, Becker-Hapak M, Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 2015;348(6236):803–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Tran E, Robbins PF, Lu Y-C, T-Cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med 2016;375(23):2255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Guo X-ZJ, Dash P, Calverley M, Rapid cloning, expression, and functional characterization of paired αβ and γδ T-cell receptor chains from single-cell analysis. Mol Ther Methods Clin Dev 2016;3:15054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Legut M, Cole DK, Sewell AK.. The promise of γδ T cells and the γδ T cell receptor for cancer immunotherapy. Cell Mol Immunol 2015;12(6):656–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Jiménez-Sánchez A, Memon D, Pourpe S, Heterogeneous tumor-immune microenvironments among differentially growing metastases in an ovarian cancer patient. Cell 2017;170(5):927–38.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Fox EJ, Loeb LA.. Cancer one cell at a time. Nature 2014;512(7513):143–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Manguso RT, Pope HW, Zimmer MD, In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 2017;547(7664):413–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Patel SJ, Sanjana NE, Kishton RJ, Identification of essential genes for cancer immunotherapy. Nature 2017;548(7669):537–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Shuptrine CW, Ajina R, Fertig EJ, An unbiased in vivo functional genomics screening approach in mice identifies novel tumor cell-based regulators of immune rejection. Cancer Immunol Immunother 2017;66(12):1529–44. [DOI] [PMC free article] [PubMed] [Google Scholar]