Abstract
Stem cells have been widely used in tendon tissue engineering. The lack of refined and controlled differentiation strategy hampers the tendon repair and regeneration. This study aimed to find new effective differentiation factors for stepwise tenogenic differentiation. By microarray screening, the transcript factor Fos was found to be expressed in significantly higher amounts in postnatal Achilles tendon tissue derived from 1 day as compared with 7‐days‐old rats. It was further confirmed that expression of Fos decreased with time in postnatal rat Achilles tendon, which was accompanied with the decreased expression of multiply tendon markers. The expression of Fos also declined during regular in vitro cell culture, which corresponded to the loss of tendon phenotype. In a cell‐sheet and a three‐dimensional cell culture model, the expression of Fos was upregulated as compared with in regular cell culture, together with the recovery of tendon phenotype. In addition, significant higher expression of tendon markers was found in Fos‐overexpressed tendon stem/progenitor cells (TSPCs), and Fos knock‐down gave opposite results. In situ rat tendon repair experiments found more normal tendon‐like tissue formed and higher tendon markers expression at 4 weeks postimplantation of Fos‐overexpressed TSPCs derived nonscaffold engineering tendon (cell‐sheet), as compared with the control group. This study identifies Fos as a new marker and functional driver in the early stage teno‐lineage differentiation of tendon, which paves the way for effective stepwise tendon differentiation and future tendon regeneration. Stem Cells Translational Medicine 2017;6:2009–2019
Keywords: Differentiation, Stem/progenitor cell, Transgene expression, Transcription factor
Significance Statement.
This study identifies a new factor Fos for tendon early‐stage differentiation. It paves the way for the stepwise differentiation from stem cells to mature tenocytes, which is beneficial for stem cells‐based tendon regeneration.
Introduction
Tendon tissue engineering is promising for tendon repair and regeneration, which combines stem cells, scaffolds, and growth factors. However, current models are still far from ideal when it comes to tendon regeneration. A repaired tendon after injury is usually comprised of smaller‐sized collagen fibrils, which accounts for the poor mechanical strength 1.
Stem cells have been widely used in tendon tissue engineering, including embryonic stem cells (ESCs), mesenchymal stem cells (MSCs), tendon stem/progenitor cells (TSPCs), and induced pluripotent stem cells (iPSCs). The properties that stem cells harbor make them potentially ideal for tendon regeneration. However, controlled teno‐lineage differentiation is crucial for successful tendon regeneration and since stem cells have multi‐differentiation ability, this renders an uncertainty of cell fate. We stand by our firm belief that stem cells cannot fully differentiate into tenocytes, which causes the unsatisfactory repair effect in current tendon tissue engineering 2, 3, 4. Thus, new effective differentiation factors need to be found.
The normal in vivo tendon development process is the ultimate environment to find new important differentiation factors. The cell types during tendon development transit from ESCs to MSCs to TSPCs and eventually to mature tenocytes. The cell fate is gradually defined toward teno‐lineage during development, and this indicates that currently used stem cells may require different stimulation at different stages in order to achieve an effective and successful tendon differentiation. Actually, many known important genes have been found by studying the development process of tendons, such as Scleraxis (Scx) 5, 6, GDF‐5 7, and GDF‐6 8, most of which were discovered from the embryonic development stage. There are also some recent studies that have tried to elucidate the postnatal tendon development process 9, 10, 11, 12. They evaluated the structural alteration and the roles of some previously known genes during development. However, inherent molecular basis of tendon development and differentiation has not been clearly elucidated. Therefore, new genes crucial for tendon differentiation need to be discovered for successful tendon regeneration.
We hypothesize that there are new markers for the direction of early teno‐lineage differentiation to be found. Therefore, the aim of this study was to screen for potential candidate genes by microarray, and to confirm their role by doing gene overexpression and gene silence experiments. By comparing the tendon gene expression and in vivo function, we provide the first evidence that identifies the Fos gene as a tendon early stage differentiation factor.
Materials and Methods
Microarray Analyses
Achilles tendons at different development stages (postnatal 1 day and 7 days, n = 3, each sample contains at least two individuals) were harvested for microarray analyses. Total RNA was extracted using Trizol reagent and was further purified using Qiagen RNeasy Mini Kit according to the manufactures’ instructions. The RNA quality was assessed by formaldehyde agarose gel electrophoresis. An aliquot of 200 ng of total RNA was used to synthesize double‐stranded cDNA, and then produce biotin‐tagged aRNA using MessageAmp Premier RNA Amplification Kit (Life Technologies, Grand Island, NY, http://www.thermofisher.com/). The resulting bio‐tagged aRNA was fragmented to strands of 35–200 bases in length according to the protocols from Affymetrix. The fragmented aRNA was hybridized to Rat Genome 230 2.0 Array (Affymetrix, http://www.thermofisher.com/) containing 30,000 transcripts. Hybridization was performed at 45°C with rotation for 16 hours at hybridization oven 640. The GeneChip arrays were washed and then stained automatically on an Affymetrix Fluidics Station 450 followed by scanning on an Affymetrix GeneChip Scanner 3000 7G.
The scanned images were first assessed by visual inspection then analyzed to generate raw data files saved as CEL files using the default setting of Affymetrix GeneChip Command Console3.2 (AGCC) Software. Then, the raw data were normalized and summarized with the Affymetrix Microarray Suite 5.0 (MAS5) and with the Robust Multi‐array Average (RMA) algorithm 13. In a comparison analysis, we applied a two‐class unpaired method in the Significant Analysis of Microarray software (SAM, version 3.02) to identify significantly differentially expressed genes between two groups. Genes were determined to be significantly differentially expressed using fold change of 2.0 and q value < 5% as cutoffs in the SAM output result. Hierarchical clustering with the average linkage method was performed with Cluster3.0 software, and the cluster result was visualized through with the Treeview program. The array has been submitted to the GEO repository with accession number GSE70459.
Quantitative Polymerase Chain Reaction
RNA isolation, reverse transcription, and quantitative polymerase chain reaction (qPCR) were carried out as previously described 14. All primers (Invitrogen, http://www.thermofisher.com/) were designed using primer 5.0 software. Representative results are displayed as target genes expression normalized to house‐keeping gene.
Lentiviral Production and Infection
A third‐generation self‐inactivating lentivirus vector containing a CMV promoter upstream of the multiple cloning sites (MCS) was used. The Coding DNA Sequence sequences of rat Fos gene and Igfbp2 gene were inserted into MCS. Additionally, green fluorescence protein (GFP) was used as the control to discount any change in gene expression profile that may result from the delivery method. The constructed lentiviral vector and another three package vectors were cotransfected into 293FT cells (Invitrogen) with lipofectamine (Invitrogen) according to the manufacturer's instructions. The medium was replaced 16 hours after transfection. Forty‐eight hours later, the virus‐containing medium was pooled and passed through a 0.45 μm filter to remove cell debris and was immediately used to infect cells in the presence of 10 ng/ml polybrene (Sigma, St. Louis, MO, https://www.sigmaaldrich.com/). The infected cells before passage 4 were used for further study.
Immunocytochemistry
Monolayer cultures of cells were fixed in 4% (vol/vol) paraformaldehyde and subjected to immunostaining with the primary antibody, rabbit anti‐human MKX monoclonal antibody (LifeSpan BioSciences, Seattle, WA, https://www.lsbio.com/), followed with a goat anti‐rabbit secondary antibody (Invitrogen). 4′,6‐diamidino‐2‐phenylindole (DAPI) staining was used to reveal the nuclei of the cells.
Gene Expression of Fos in Postnatal Rat Achilles Tendon
Postnatal rat Achilles tendons were collected at 1, 3, 7, 14, 28, and 56 days (n = 3 for each time point). RNA isolation and reverse transcription were carried out to obtain cDNA. Gene expression of Fos and multiply tendon markers were compared by qPCR between different developmental time points.
Cell‐Sheet Model
As described previously 4, upon reaching confluence, cells were cultured in Dulbecco's Modified Eagle Medium (DMEM) supplemented with 10% (vol/vol) fetal bovine serum and 50 ug/ml ascorbic acid (Sigma). A multi‐layered cell‐sheet detached from the substratum and formed dense sphere‐like structure automatically within 2 weeks in culture. To evaluate the extracellular matrix (ECM) deposition speed, the percentage of samples forming sphere‐like structures was calculated. The cell aggregates were also harvested for transmission electron microscopy, RNA isolation, and animal experiments.
Three‐Dimensional Cell Culture Model
Human TSPCs were cultured in nonadherent culture dishes, with DMEM supplemented with 10% (vol/vol) fetal bovine serum. Cells aggregated into sphere‐like structures by themselves. RNA was isolated to evaluate the tendon‐related gene expression.
siRNA Transfection
siRNA transfection in TSPCs to knock‐down Fos was performed according to the manufacturer's protocol (RiboBio, Guangzhou, China, http://www.ribobio.com). Three Fos siRNA duplexes were used (siRfo1, siRfo2, and siRfo3). A scrambled siRNA was used as a control (siRNTC). All siRNA oligos were designed and synthesized by RiboBio Company. Forty‐eight hours post‐transfection, cells were harvested to assess the gene expression.
In Situ Rat Tendon Repair Model
Eight mature female Sprague Dawley rats weighing 200–220 g were used for this experiment. The Zhejiang University Institutional Animal Care and Use Committee approved the study protocol (ethical No. ZJU20170413). Under general anesthesia, a gap wound was created and the Patella tendon was removed to create a defect of 4 mm in length. Nonscaffold engineering tendons (cell sheet of Fos overexpressed TSPCs or control group transfected with GFP) were sutured to the remaining Patella tendon using a nonresorbable suture (6‐0 nylon). The wound was then irrigated and the skin was closed. The animals were allowed free cage activity after surgery. At 4 weeks postimplantation, eight samples from each group were harvested for the evaluation of histology and gene expression.
Histological Examination
Histological specimens treatment was performed as previously reported 15. The collected specimens were fixed in 10% (vol/vol) neutral buffered formalin, dehydrated through an alcohol gradient, cleared, and embedded in paraffin blocks. Histological sections (7 μm) were prepared by using a microtome and were stained with hematoxylin and eosin. A blinded semiquantitative scoring system was used to evaluate the repair of tendon tissue 16, which was based on six parameters (fiber structure, fiber arrangement, nuclear roundness, vascularity, inflammation, number of cells). For the evaluation of each parameter, 0 was allotted to normal tendon and 3 was allotted to maximally abnormal tissue.
Statistical Analysis
Student's t test (except for the microarray analysis) was performed to assess statistically significant differences in the results of different groups. Values of p < .05 were considered to be significantly different.
Results
Screen of Tendon Early Stage Marker Genes
Postnatal Achilles tendon tissues at 1 day (1d) and 7 days (7d) were harvested for microarray experiment. A total of 571 transcripts out of 30,000 transcripts were found to be more than twofold different when comparing 1d with 7d tendons (Supporting Information Table S1), in which 370 transcripts were upregulated (65%), and 201 transcripts were downregulated (35%) in tendon tissues at 1d as compared with tendon tissues at 7d. These transcripts were hierarchically clustered (Fig. 1A). The expression of six randomly chosen genes (Adipoq, Comp, Cpxm2, Cpz, Mpz, and Fos) were confirmed by qPCR (Fig. 1B). Gene Ontology (GO) analysis showed the top ten GO terms to be related to biological process (Fig. 1C) and pathway (Fig. 1D) between 1d and 7d tendon tissues. In biological process, the most significant GO term was “cell adhesion,” followed by “spermine biosynthesis” and “proteolysis.” The most significant GO term in pathway was “PPAR signaling pathway.” Other GO terms were related to cell adhesion molecules and graft‐versus‐host disease.
Figure 1.
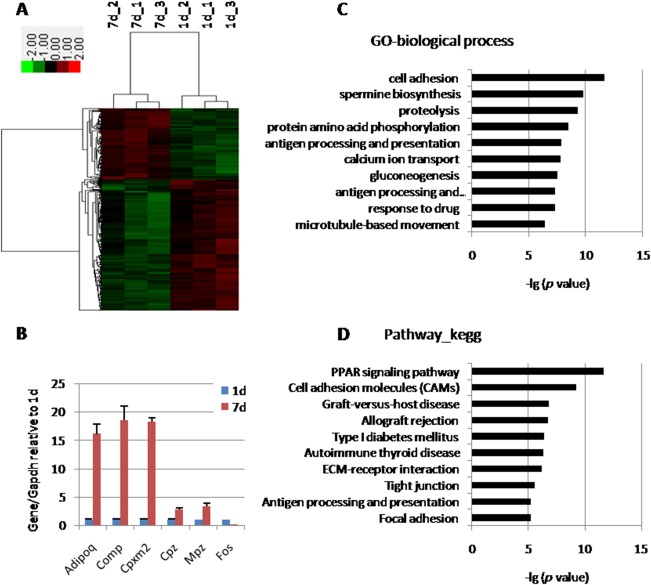
Microarray analysis of tendon tissues at postnatal 1d and 7d. (A): Gene cluster analysis of 571 transcripts, which showed more than twofold differences between 1d and 7d tendons. (B): qPCR results confirming six differently expressed genes from microarray. (C): GO related to biological process ordered according to –lg (p value). (D): GO related to pathway ordered according to –lg (p value). Abbreviation: GO, gene ontology.
To find out the early stage tendon marker genes for development and differentiation, the top 25 genes were ordered, which were enriched in postnatal 1d tendon tissue as compared with 7d tendon tissue (Supporting Information Table S2). These genes were upregulated in tendon tissues at 1d, as compared with 7d, with at least 3.7‐fold. Transcript factors usually play crucial roles in cell fate determination. Transcript factor Igfbp2 is the most abundant binding protein in tendon, and sensitively responsive to tendon injury 17. Transcript factor Fos plays an important role in mesoderm derived tissue differentiation and maintenance 18, 19, 20. Thus, Igfbp2and Fos were chosen as candidates in the current study.
Cluster analysis indicated Igfbp2 and Fos to be upregulated in postnatal tendon tissues at 1d as compared with 7d (Fig. 2A). qPCR results were consistent with microarray data. It was also found by qPCR that the expressions of Igfbp2 and Fos in postnatal 1d tissues were higher than 56d tissues (Fig. 2B). Overexpression vectors of these two candidate genes were constructed. The transfection efficiency came to nearly 100% (Fig. 2C, 2D). qPCR results showed that the expression of Igfbp2 and Fos in human ESC derived MSCs (hESC‐MSCs) were upregulated by 400‐fold and 30,000‐fold, respectively, (Fig. 2E).
Figure 2.
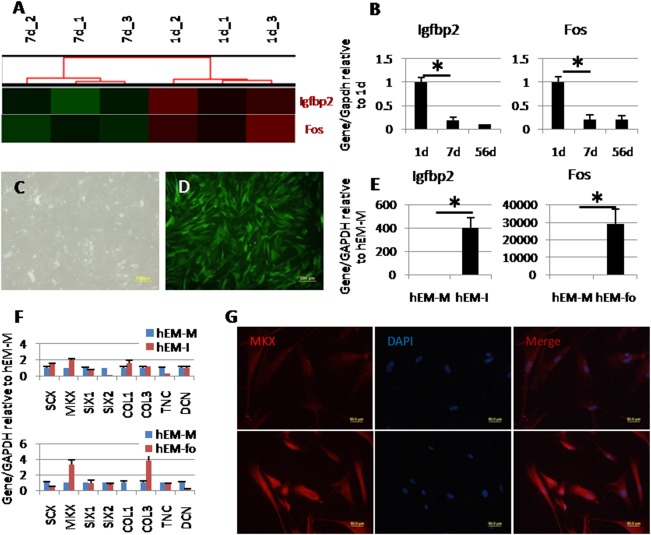
Screen of early stage differentiation factors. (A): Gene cluster analysis of Igfbp2 and Fos. (B): qPCR results showing higher expression of Igfbp2 and Fos in 1d tendon tissue compared with 7d and 56d. (C, D): Overexpression in human embryonic stem cells derived mesenchymal stem cells (hESC‐MSCs). Green fluorescence indicating the transfection efficiency of green fluorescence protein (GFP) (control) came to nearly 100%. (E): Gene expression of Igfbp2 or Fos in overexpressed hESC‐MSCs. (F): Expression of tendon related genes in Igfbp2 or Fos overexpressed hESC‐MSCs, compared with GFP overexpressed hESC‐MSCs. (G): MKX immunofluorescence in control group (upper) and Fos overexpressed hESC‐MSCs (lower). Scale bars = 200 μm (C, D) and 50 μm (G). *Significant difference between two groups at p < .05. Abbreviations: hEM‐fo, Fos overexpressed in hESC‐MSCs; hEM‐M, control group with GFP overexpressed; hEM‐I, Igfbp2 overexpressed in hESC‐MSCs.
The expression of tendon related transcript factors and ECM genes were evaluated after overexpression of Igfbp2 or Fos in human ESC‐MSCs (Fig. 2F). There were no obvious upregulations in the group in which Igfbp2 was overexpressed. However, the expression of MKX and COL3 increased by overexpression of Fos (3.37‐fold and 3.84‐fold, respectively). Immunofluorescence detected the increased expression of MKX protein in the Fos overexpressed group. MKX and COL3 are early stage related genes in tendon, thus Fos may play a role in maintaining early stage tendon phenotype.
Relevance Between Fos and Tendon Phenotype
The expression of Fos in postnatal rat Achilles tendon was compared at 1d, 3d, 7d, 14d, 28d, and 56d (Fig. 3). Continuous decrease in Fos gene expression was found during the development, in which significance was found since postnatal 7d, as compared with 1d. Similar decreases during development were found in expression levels of tendon markers, including Mkx, Egr1, Col14, Tnc, Gdf5, and Gdf6. As for Scx, Tnmd, Dcn, and Bgn, there was a peak expression at around 7d or 14d, which came to a low level when the tendon tissue matured. No obvious change was found for other tested tendon‐related genes, such as Hoxa11, Six1, Fmod, Col1, and Col3.
Figure 3.
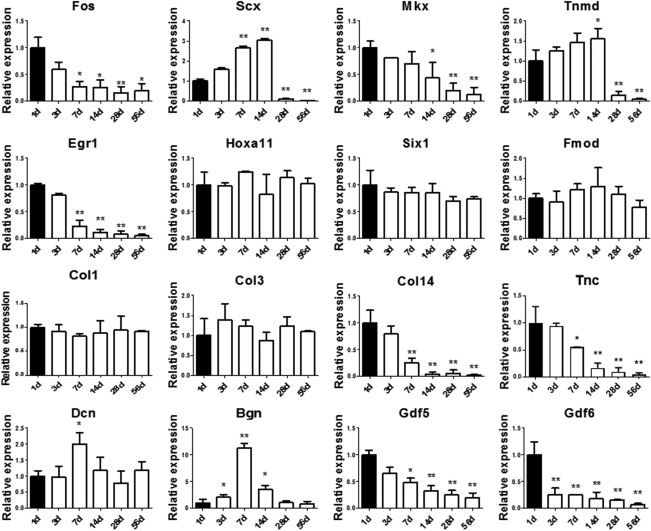
Expression of Fos and multiply tendon markers in postnatal rat Achilles tendon. The mRNA levels of Fos and multiply tendon markers in postnatal rat Achilles tendons were compared by qPCR at 1, 3, 7, 14, 28, and 56 days. The expression level of each gene at 1d was set as 1. *Significant difference between specific time point and 1d at p < .05. **Significant difference between specific time point and 1d at p < .01.
Cells gradually lose their tendon phenotype during regular cell culture in vitro. The expression of the tendon related genes or proteins were downregulated. It was noticed that TSPCs at passage 1 expressed more than 3.5‐fold higher Scx as compared with normal tendon tissues (Fig. 4A). Nevertheless, the expression of Scx decreased to 24% of normal tendon tissues in TSPCs at passage 4 in cell culture. The expression of Scx in in vitro cultured tenocytes was 63% of normal tendon tissues. Tnmd could only be detected in tendon tissue, but not in in vitro cultured TSPCs or tenocytes (Fig. 4A). Mkx and Col14, early tendon stage related genes, also gradually lost their expression during in vitro cell culture (Fig. 4A). Green fluorescence was shown in normal tendon tissue of Scx‐GFP mice. However, the fluorescence could not be detected in cells that were cultured in vitro (Fig. 4C–4E). Notably, the expression of Fos was downregulated in accordance with the loss of tendon phenotype in vitro (Fig. 4B).
Figure 4.
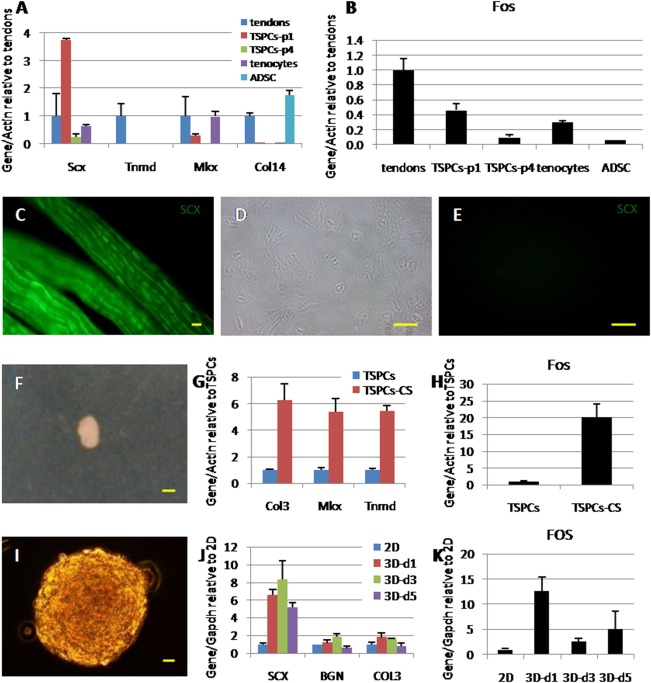
Relevance between Fos and tendon phenotype. (A): The expression of tendon related genes decreased during in vitro cell culture. (B): The expression of Fos in tendon tissues and cells cultured in vitro. (C): Green fluorescence could be observed in tendon tissue of Scx‐GFP mice. (D, E): Cells isolated from tendons of Scx‐GFP mice lost fluorescence when cultured in vitro. (F): Cell‐sheet model: cells self‐crimped into a sphere when cultured with ascorbic acid within 14 days. (G): Tendon related gene expressions compared between regular cell culture and the cell‐sheet model. (H): The expression of Fos in the cell‐sheet model. (I): Three‐dimensional (3D) model: cells cultured with a 3D sphere structure. (J): Tendon related gene expressions compared between two‐dimensional regular cell culture and 3D model at 1 day, 3 day, and 5 day. (K): The expression of FOS in 3D model. Scale bars = 50 μm (C), 100 μm (D, E), 1 mm (F), and 20 μm (I). Abbreviations: ADSC, adipose derived stem cells; TSPCs, tendon stem/progenitor cells; TSPCs‐CS, TSPCs formed cell‐sheet; TSPCs‐p1, TSPCs at passage 1.
When TSPCs were cultured in vitro using a cell‐sheet or a three‐dimensional (3D) cell culture model (Fig. 4F, 4I). The expression of tendon related genes was upregulated in the two different models as compared with that in regular cell culture (Fig. 4G, 4J), accompanied by a great upregulation of Fos expression (Fig. 4H, 4K).
Therefore, there is potential relevance between Fos and tendon phenotype.
Overexpression of Fos in Postnatal 7d Derived TSPCs
Fos was overexpressed in 7d derived TSPCs (TSPCs‐7d). The expression of Fos increased with about 7‐fold (Fig. 5A). The Fos overexpressed TSPCs had significant higher expression of tendon markers as compared with the control group (Fig. 5B, 5C), including Scx (2.70‐fold), Mkx (2.17‐fold), Tnmd (2.16‐fold), Egr1 (1.92‐fold), Col1 (3.17‐fold), Col3 (7.56‐fold), Dcn (5.31‐fold), Bgn (2.38‐fold), Gdf5 (3.57‐fold), and Gdf6 (4.75‐fold). When cells were cultured in a cell‐sheet model, qPCR results showed that Fos overexpression reversed the gene expression of Mmp3, Col14, and Col3 from being 7d postnatal to be more like 1d postnatal (Supporting Information Fig. S1).
Figure 5.
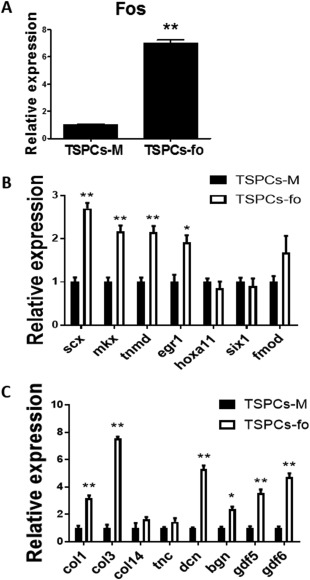
Overexpression of Fos in TSPCs caused upregulated gene expression of tendon markers. (A): Gene expression of Fos in overexpressed TSPCs. (B, C): Gene expression of multiply tendon markers in TSPCs‐fo versus TSPCs‐M. The expression level of each gene in TSPCs‐M group was set as 1. *Significant difference between two groups at p < .05. **Significant difference between two groups at p < .01. Abbreviations: TSPCs, tendon stem/progenitor cells; TSPCs‐fo, TSPCs transfected with Fos, TSPCs‐M, control group which is transfected with GFP.
Besides, the Fos overexpressed TSPCs had advantages in colony‐forming ability (Supporting Information Fig. 2), cell proliferation ability, and ECM deposition ability (Supporting Information Fig. S3), as compared with the control group. There was no significant difference of osteo‐lineage and adipo‐lineage differentiation potential between Fos overexpressed TSPCs and control group (Supporting Information Fig. S2).
siRNA Knock‐Down of Fos in TSPCs
The expression of Fos in TSPCs was knocked down by siRNA. qPCR experiments confirmed the knock‐down effect of Fos expression by the use of three siRNA sequences (69, 31, and 39%, Fig. 6A). The siRNA sequence siRfo1 showed highest knock‐down efficiency as compared with the other two siRNA sequence and was therefore used to knock down the expression of Fos in the following experiments. The Fos knock‐down TSPCs had significant lower expression of tendon markers as compared with the control group (Fig. 6B, 6C), such as Mkx (0.32‐fold), Tnmd (0.20‐fold), Egr1 (0.56‐fold), Fmod (0.60‐fold), Col14 (0.44‐fold), Tnc (0.52‐fold), Gdf5 (0.24‐fold), and Gdf6 (0.34‐fold). When cells were cultured in a cell‐sheet model, the expressions of Mmp3, Col14, and Col3 were the contrary when Fos was over‐expressed (Supporting Information Fig. S4). Besides, the Fos knocked‐down TSPCs had inferior ECM deposition ability as compared with the control group (Supporting Information Fig. S4).
Figure 6.
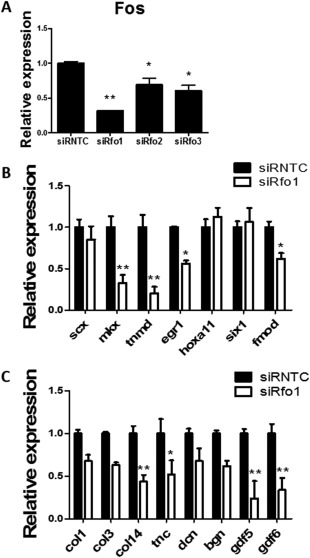
Fos knock‐down by siRNA in tendon stem/progenitor cells caused downregulated gene expression of tendon markers. (A): qPCR showed the downregulation of Fos by three different siRNA sequences (siRfo1, siRfo2, and siRfo3), compared with siRNTC. (B, C): Gene expression of multiply tendon markers in siRfo1 versus siRNTC. The expression level of each gene in siRNTC group was set as 1. *Significant difference between specific group and siRNTC group at p < .05. **Significant difference between specific group and siRNTC group at p < .01. Abbreviation: siRNTC, control siRNA.
Fos‐Overexpressed TSPCs Improved the Healing of Injured Rat Tendon
Nonscaffold engineering tendons (cell‐sheet) were constructed in vitro with Fos overexpressed TSPCs (FOS group) or control group transfected with GFP (Ctrl group). Implantation was performed in an in situ tendon repair model. After 4 weeks post‐surgery, the cells in the repaired tissue of FOS group exhibited a more spindle‐shaped morphology, as compared with the control group (Fig. 7A). More alignment of collagen fibers was also observed in the FOS group (Fig. 7A). Histology score evaluation found that FOS group had significantly better score than the control group (5.94 ± 1.17 vs. 8.13 ± 1.08, Fig. 7B). The mRNA transcript levels of multiply tendon markers were compared (Fig. 7C), in which significant upregulated expression was found in FOS group on Scx (5.7‐fold), Tnmd (14.9‐fold), Fmod (3.1‐fold), Col1 (3.6‐fold), Col3 (9.9‐fold), and Col14 (3.1‐fold), as compared with the control group. Increased expression without significance was found in FOS group on other tendon‐related genes.
Figure 7.
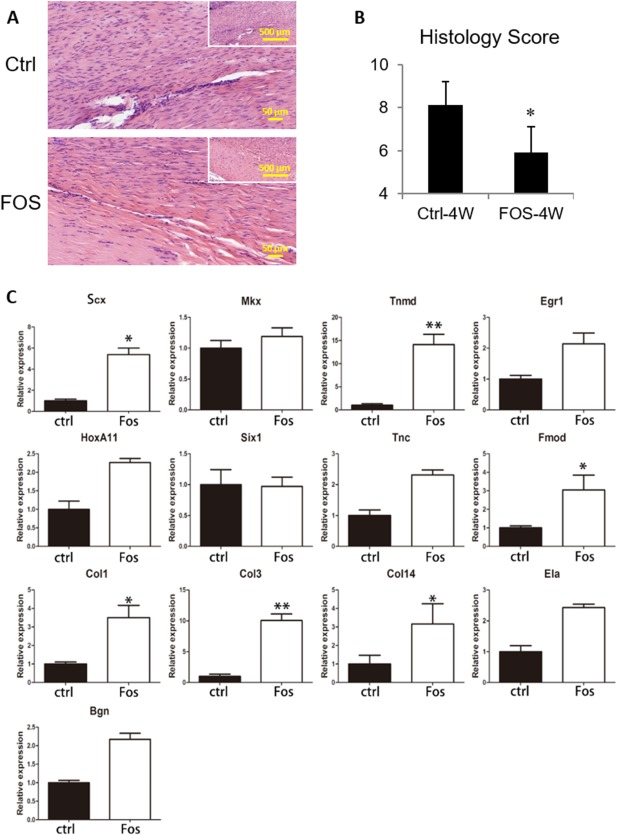
Fos‐overexpressed tendon stem/progenitor cells (TSPCs) improved the healing of injured rat tendon. Cell‐sheet constructed with Fos overexpressed TSPCs (FOS group) or control group transfected with GFP (Ctrl group) was compared in in situ rat Patella tendon repair model. (A): Hematoxylin and eosin staining after 4 weeks post‐surgery. (B): Histology score evaluation was performed to compare between FOS group and Ctrl group. (C): Gene expression of multiply tendon markers after 4 weeks post‐surgery. The expression level of each gene in Ctrl group was set as 1. *Significant difference between specific group and siRNTC group at p < .05. **Significant difference between specific group and siRNTC group at p < .01. Abbreviation: Ctrl, control.
Discussion
This study identifies the Fos gene as a tendon early‐stage differentiation marker. It was obvious that reduced expression of Fos corresponded to tendon phenotype loss during development and regular in vitro cell culture, and increased expression of Fos corresponded to upregulation of tendon related genes expression in cell‐sheet and 3D cell culture model. Overexpression of Fos in TSPCs caused upregulated gene expression of tendon markers, the opposite was seen in Fos knocked‐down cells. Finally, accelerated healing of injured tendon was found by using Fos overexpressed TSPCs. The results of this study strongly suggest that Fos is a candidate marker for early teno‐lineage differentiation and maintain.
The concept of stepwise differentiation is important for tissue engineering and tissue regeneration when using stem cells 2. It has been confirmed in other tissues that stepwise differentiation of stem cells into defined cell types brings success and new hope for tissue repair, such as myoblasts 21, 22, 23, 24, chondrocytes 25, liver 26, heart 27, insulin‐secreting beta cells 28, neurons 29, and oligodendrocytes 30. Goudenege and collaborators generated skeletal myoblasts from hESCs through mesodermal transition by two steps 24. The mesenchymal differentiation was achieved by culturing hESCs in myogenic medium. After that, the myogenic factor MyoD was overexpressed in cells to cause the final conversion. This stepwise differentiation not only overcame the difficulties of differentiating ESCs into myoblasts, but also lowered the risks of teratomas formation. Oldershaw and collaborators reported a refined protocol for differentiation of hESCs to chondrocytes using chemicals by three steps 25. Different chemicals were combined in a specific order based on the knowledge from tissue development. There is not much reported about the stepwise differentiation in tendon, and is far away from elaborate and controllable, which hinders work on tendon repair and regeneration. To apply hESCs in tendon regeneration, our group differentiated hESCs into ESC‐MSCs, which has shown good potential for tendon differentiation and regeneration when using scaffolds, mechanical stimulation, or overexpression of SCX in hESC‐MSCs 1, 3, 4, 14. It was found by Alberton and collaborators that overexpression of SCX in human BMSCs could convert the cells into tenogenic progenitor cells, which is an important step for fully differentiation of MSCs toward mature tenocytes 31. Our recent work 32 showed that TSPCs isolated at different postnatal time‐points possessed different self‐renew, multi‐potent differentiation potential, cell proliferation, and ECM deposition ability. The transition of TSPCs during development indicates the importance of stepwise differentiation from MSCs/TSPCs to mature tenocytes. Based on this, the current study analyzed the highly expressed genes in postnatal 1d tendon tissue, by microarray, to screen for important differentiation factors for stepwise tenogenic differentiation.
By comparison of the gene expression profiles, 370 transcripts were upregulated in tendon tissues at 1d as compared with 7 days. The gene Fos was picked out due to the following reasons: (a) The Fos gene was more highly expressed in postnatal 1d tendon tissue (more than fourfold) as compared with in 7d tendon, (b) Fos is a transcript factor, and it is well known that transcript factors are crucial for cell fate determination during development and differentiation. When Fos was overexpressed in hESC‐MSCs, it promoted the expression of MKX and COL3 by more than threefold as compared with the control group. Mkx is an important transcript factor for tendon differentiation 33, 34, 35. It was reported that dense Mkx mRNA could be detected in mice at E13.5 and E14.5 but decreased a lot at postnatal 0d and 14day 33. Col3 regulates the initial fibril assembly 36. During development, the amount of Col3 decreased with the increase of collagen fiber diameter 37, 38, 39. Thus, the elevation of MKX and COL3 expression after Fos overexpression indicates the conversion of cells into an early stage tendon. Besides, in regular in vitro cell culture or in vivo postnatal tendon development, the expression of tendon related genes or proteins were decreased. This is also seen in the expression of Fos. In vitro cell‐sheet and 3D models maintained the tendon phenotype, causing an increase of the tendon related genes and Fos. Overexpression of Fos in TSPCs caused upregulated gene expression of tendon markers, while siRNA knock‐down of Fos showed opposite results.
The expression of Fos has been detected in fetal liver, growing bone and developing central nervous system 40. It is a quickly responsive gene to various stimulations and is of importance for signal transduction, cell proliferation and differentiation 41, 42. Fos is the first transcription factor identified for the regulation of cell fate during development and maintenance of skeleton. It also plays an important role in the early osteogenic differentiation 19 and in the chondrocyte gene transcription 20. Tendon, bone, and cartilage are all derived from mesenchyme, thus Fos may play a potential role in the regulation of tenogenic differentiation. A recent report by Eliasson and collaborators showed that Fos is one of the four genes significantly upregulated (almost sevenfold) in mechanically induced rat tendon repair, which was screened by microarray containing 27,342 genes 43. However, to our knowledge, our research is the first that tries to find the relationship between Fos and tendon differentiation.
Previous studies have shown that Fos is sensitive to mechanical stimulation both in vitro and in vivo, and in many different cell types, such as tenocytes, ligament cells, osteoblasts, chondrocytes, and so forth 18, 20, 41, 42, 44, 45. The natural exposure to mechanical stimulation at birth might be the reason for the high expression of Fos in postnatal 1d tendon tissue. Besides, to some extent, the cell‐sheet and 3D models used in our study caused cell aggregation in vitro, and enhanced the mechanical strength during cell–cell interaction, which may induce Fos expression and tendon phenotype maintain. The promoter region of Mkx contains the binding site of Fos. Mkx plays important roles in tenogenesis and tendon regeneration 33, 34, 35 and thus may be the downstream effector of Fos. The inherent mechanism of Fos‐regulating tenogenic differentiation, and whether mechanics regulate Fos during this process, should be studied in our future work.
Conclusion
This study is the first to identify the Fos gene as a tendon early‐stage differentiation factor. The expression change of Fos is related to the loss and regain of tendon phenotype during in vivo tendon development, regular in vitro cell culture, and in cell‐sheet and 3D cell culture model. Overexpression of Fos in TSPCs induced upregulated gene expression of tendon markers. In the case of Fos knock‐down, the opposite results were seen. Cell‐sheet derived from Fos overexpressed TSPCs promoted the healing of injured tendon. This study paves the way for the stepwise differentiation from MSCs/TSPCs to mature tenocytes, which is beneficial for tendon regeneration.
Author Contributions
J.C. and E.Z.: conception and design, collection and assembly of data, data analysis and interpretation, manuscript writing; W.Z.: collection and assembly of data, data analysis and interpretation, manuscript writing; Z.L., P.L., T.Z., Z.Y., and H.L.: collection data; X.C.: conception and design, data analysis and interpretation, provision of study material, final approval of manuscript; L.J.B.: manuscript writing; H.O.: conception and design, financial support, final approval of manuscript.
Disclosure of Potential Conflicts of Interest
The authors indicated no potential conflicts of interest.
Supporting information
Supplemental Figure 1
Supplemental Figure 2
Supplemental Figure 3
Supplemental Figure 4
Supplemental Table 1
Supplemental Table 2
Supporting Information
Acknowledgments
We thank Wang Li for assistance with TEM imaging. This work was supported by National Key R&D Program of China (2017YFA0104902), NSFC grants (81330041, 81522029, 81772418, 31570987, 81401781, 81572157), Key scientific and Technological Innovation Team of Zhejiang Province (2013TD11), and Zhejiang Provincial Natural Science Foundation of China (LR14H060001).
Contributor Information
Xiao Chen, Email: chenxiao-610@zju.edu.cn.
Hongwei Ouyang, Email: hwoy@zju.edu.cn.
References
- 1. Chen X, Yin Z, Chen JL et al. Scleraxis‐overexpressed human embryonic stem cell‐derived mesenchymal stem cells for tendon tissue engineering with knitted silk‐collagen scaffold. Tissue Eng Part A 2014;20:1583–1592. [DOI] [PubMed] [Google Scholar]
- 2. Chen JL, Zhang W, Liu ZY et al. Physical regulation of stem cells differentiation into teno‐lineage: Current strategies and future direction. Cell Tissue Res 2015;360:195–207. [DOI] [PubMed] [Google Scholar]
- 3. Chen X, Yin Z, Chen JL et al. Force and Scleraxis synergistically promote the commitment of human ES cells derived MSCs to tenocytes. Sci Rep 2012;2:977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen X, Song XH, Yin Z et al. Stepwise differentiation of human embryonic stem cells promotes tendon regeneration by secreting fetal tendon matrix and differentiation factors. Stem Cells 2009;27:1276–1287. [DOI] [PubMed] [Google Scholar]
- 5. Schweitzer R, Chyung JH, Murtaugh LC et al. Analysis of the tendon cell fate using Scleraxis, a specific marker for tendons and ligaments. Development 2001;128:3855–3866. [DOI] [PubMed] [Google Scholar]
- 6. Brent AE, Schweitzer R, Tabin CJ. A somitic compartment of tendon progenitors. Cell 2003;113:235–248. [DOI] [PubMed] [Google Scholar]
- 7. Mikic B. Multiple effects of GDF‐5 deficiency on skeletal tissues: Implications for therapeutic bioengineering. Ann Biomed Eng 2004;32:466–476. [DOI] [PubMed] [Google Scholar]
- 8. Mikic B, Rossmeier K, Bierwert L. Identification of a tendon phenotype in GDF6 deficient mice. Anat Rec (Hoboken) 2009;292:396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang G, Ezura Y, Chervoneva I et al. Decorin regulates assembly of collagen fibrils and acquisition of biomechanical properties during tendon development. J Cell Biochem 2006;98:1436–1449. [DOI] [PubMed] [Google Scholar]
- 10. Ezura Y, Chakravarti S, Oldberg A et al. Differential expression of lumican and fibromodulin regulate collagen fibrillogenesis in developing mouse tendons. J Cell Biol 2000;151:779–788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ansorge HL, Adams S, Birk DE et al. Mechanical, compositional, and structural properties of the post‐natal mouse Achilles tendon. Ann Biomed Eng 2011;39:1904–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Liu CF, Aschbacher‐Smith L, Barthelery NJ et al. Spatial and temporal expression of molecular markers and cell signals during normal development of the mouse patellar tendon. Tissue Eng Part A 2012;18:598–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Irizarry RA, Bolstad BM, Collin F et al. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res 2003;31:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen JL, Yin Z, Shen WL et al. Efficacy of hESC‐MSCs in knitted silk‐collagen scaffold for tendon tissue engineering and their roles. Biomaterials 2010;31:9438–9451. [DOI] [PubMed] [Google Scholar]
- 15. Yin Z, Chen X, Chen JL et al. The regulation of tendon stem cell differentiation by the alignment of nanofibers. Biomaterials 2010;31:2163–2175. [DOI] [PubMed] [Google Scholar]
- 16. Shen W, Chen J, Yin Z et al. Allogenous tendon stem/progenitor cells in silk scaffold for functional shoulder repair. Cell Transplant 2012;21:943–958. [DOI] [PubMed] [Google Scholar]
- 17. Dahlgren LA, Mohammed HO, Nixon AJ. Expression of insulin‐like growth factor binding proteins in healing tendon lesions. J Orthop Res 2006;24:183–192. [DOI] [PubMed] [Google Scholar]
- 18. Rangaswami H, Marathe N, Zhuang S et al. Type II cGMP‐dependent protein kinase mediates osteoblast mechanotransduction. J Biol Chem 2009;284:14796–14808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Friedl G, Schmidt H, Rehak I et al. Undifferentiated human mesenchymal stem cells (hMSCs) are highly sensitive to mechanical strain: Transcriptionally controlled early osteo‐chondrogenic response in vitro. Osteoarthritis Cartilage 2007;15:1293–1300. [DOI] [PubMed] [Google Scholar]
- 20. Fitzgerald JB, Jin M, Chai DH et al. Shear‐ and compression‐induced chondrocyte transcription requires MAPK activation in cartilage explants. J Biol Chem 2008;283:6735–6743. [DOI] [PubMed] [Google Scholar]
- 21. Barberi T, Bradbury M, Dincer Z et al. Derivation of engraftable skeletal myoblasts from human embryonic stem cells. Nat Med 2007;13:642–648. [DOI] [PubMed] [Google Scholar]
- 22. Darabi R, Gehlbach K, Bachoo RM et al. Functional skeletal muscle regeneration from differentiating embryonic stem cells. Nat Med 2008;14:134–143. [DOI] [PubMed] [Google Scholar]
- 23. Darabi R, Arpke RW, Irion S et al. Human ES‐ and iPS‐derived myogenic progenitors restore DYSTROPHIN and improve contractility upon transplantation in dystrophic mice. Cell Stem Cell 2012;10:610–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Goudenege S, Lebel C, Huot NB et al. Myoblasts derived from normal hESCs and dystrophic hiPSCs efficiently fuse with existing muscle fibers following transplantation. Mol Ther 2012;20:2153–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Oldershaw RA, Baxter MA, Lowe ET et al. Directed differentiation of human embryonic stem cells toward chondrocytes. Nat Biotechnol 2010;28:1187–1194. [DOI] [PubMed] [Google Scholar]
- 26. Hay DC, Zhao D, Fletcher J et al. Efficient differentiation of hepatocytes from human embryonic stem cells exhibiting markers recapitulating liver development in vivo. Stem Cells 2008;26:894–902. [DOI] [PubMed] [Google Scholar]
- 27. Laflamme MA, Chen KY, Naumova AV et al. Cardiomyocytes derived from human embryonic stem cells in pro‐survival factors enhance function of infarcted rat hearts. Nat Biotechnol 2007;25:1015–1024. [DOI] [PubMed] [Google Scholar]
- 28. D'Amour KA, Bang AG, Eliazer S et al. Production of pancreatic hormone‐expressing endocrine cells from human embryonic stem cells. Nat Biotechnol 2006;24:1392–1401. [DOI] [PubMed] [Google Scholar]
- 29. Yan Y, Yang D, Zarnowska ED et al. Directed differentiation of dopaminergic neuronal subtypes from human embryonic stem cells. Stem Cells 2005;23:781–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Nistor GI, Totoiu MO, Haque N et al. Human embryonic stem cells differentiate into oligodendrocytes in high purity and myelinate after spinal cord transplantation. Glia 2005;49:385–396. [DOI] [PubMed] [Google Scholar]
- 31. Alberton P, Popov C, Pragert M et al. Conversion of human bone marrow‐derived mesenchymal stem cells into tendon progenitor cells by ectopic expression of scleraxis. Stem Cells Dev 2012;21:846–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen J, Zhang W, Liu Z et al. Characterization and comparison of post‐natal rat Achilles tendon‐derived stem cells at different development stages. Sci Rep 2016;6:22946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Liu W, Watson SS, Lan Y et al. The atypical homeodomain transcription factor Mohawk controls tendon morphogenesis. Mol Cell Biol 2010;30:4797–4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ito Y, Toriuchi N, Yoshitaka T et al. The Mohawk homeobox gene is a critical regulator of tendon differentiation. Proc Natl Acad Sci USA 2010;107:10538–10542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Liu H, Zhang C, Zhu S et al. Mohawk promotes the tenogenesis of mesenchymal stem cells through activation of the TGFbeta signaling pathway. Stem Cells 2015;33:443–455. [DOI] [PubMed] [Google Scholar]
- 36. Tozer S, Duprez D. Tendon and ligament: Development, repair and disease. Birth Defects Res C Embryo Today 2005;75:226–236. [DOI] [PubMed] [Google Scholar]
- 37. Birk DE, Mayne R. Localization of collagen types I, III and V during tendon development. Changes in collagen types I and III are correlated with changes in fibril diameter. Eur J Cell Biol 1997;72:352–361. [PubMed] [Google Scholar]
- 38. Zhang G, Young BB, Ezura Y et al. Development of tendon structure and function: Regulation of collagen fibrillogenesis. J Musculoskelet Neuronal Interact 2005;5:5–21. [PubMed] [Google Scholar]
- 39. Bland YS, Ashhurst DE. Fetal and postnatal development of the patella patellar tendon and suprapatella in the rabbit; changes in the distribution of the fibrillar collagens. J Anat 1997;190 (Pt 3):327–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Muller R, Tremblay JM, Adamson ED et al. Tissue and cell type‐specific expression of two human c‐onc genes. Nature 1983;304:454–456. [DOI] [PubMed] [Google Scholar]
- 41. Liu J, Liu T, Zheng Y et al. Early responses of osteoblast‐like cells to different mechanical signals through various signaling pathways. Biochem Biophys Res Commun 2006;348:1167–1173. [DOI] [PubMed] [Google Scholar]
- 42. Ott CE, Bauer S, Manke T et al. Promiscuous and depolarization‐induced immediate‐early response genes are induced by mechanical strain of osteoblasts. J Bone Miner Res 2009;24:1247–1262. [DOI] [PubMed] [Google Scholar]
- 43. Eliasson P, Andersson T, Hammerman M et al. Primary gene response to mechanical loading in healing rat Achilles tendons. J Appl Physiol (1985) 2013;114:1519–1526. [DOI] [PubMed] [Google Scholar]
- 44. Li KW, Lindsey DP, Wagner DR et al. Gene regulation ex vivo within a wrap‐around tendon. Tissue Eng 2006;12:2611–2618. [DOI] [PubMed] [Google Scholar]
- 45. Ziegler N, Alonso A, Steinberg T et al. Mechano‐transduction in periodontal ligament cells identifies activated states of MAP‐kinases p42/44 and p38‐stress kinase as a mechanism for MMP‐13 expression. BMC Cell Biol 2010;11:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1
Supplemental Figure 2
Supplemental Figure 3
Supplemental Figure 4
Supplemental Table 1
Supplemental Table 2
Supporting Information


