Abstract
Background
A previous study indicated that microRNA(miR)-328-3p upregulation might be critical for resveratrol-mediated suppression of metastatic ability in human osteosarcoma, implying its potential role in this malignancy. However, the clinical significance and the biological functions of miR-328-3p in osteosarcoma remain unclear.
Methods
miR-328-3p expression in 88 pairs of osteosarcoma and matched non-cancerous bone tissues were detected by quantitative real-time PCR. Then, the associations of miR-328-3p expression with various clinicopathological features of osteosarcoma patients were statistically analyzed. Cell Counting Kit-8 and scratch-wound healing were performed to evaluate roles of miR-328-3p in human osteosarcoma cells. After that, luciferase reporter assay, western blot analysis and rescue assay were applied to determine the underlying molecular mechanisms of miR-328-3p in osteosarcoma cells.
Results
miR-328-3p expression in osteosarcoma tissues was significantly lower than those in non-cancerous bone tissues (P<0.001). miR-328-3p downregulation was significantly associated with advanced surgical stage, positive metastasis and recurrence (all P<0.05). Functionally, enforced expression of miR-328-3p efficiently suppressed cell proliferation and migration in vitro. Moreover, matrix metalloprotease 16 (MMP16) was identified as a direct target of miR-328-3p in osteosarcoma cells. Notably, MMP16 overexpression partially reversed the miR-328-3p-inhibited cell proliferation and migration of osteosarcoma cells.
Conclusions
Our data indicated that the aberrant expression of miR-328-3p may play a crucial role in malignant progression of human osteosarcoma. More importantly, miR-328-3p may function as a tumor suppressor inhibiting osteosarcoma cell proliferation and migration partially mediated by regulating of MMP16.
Keywords: microRNA-328-3p, matrix metalloprotease 16, osteosarcoma, clinicopathological feature, cell proliferation, cell migration
Introduction
Osteosarcoma, the most prevalent primary malignant bone tumor, often happens in children and adolescents, as well as is characterized by fast growth, metastasis at early stage, and poor prognosis.1 Although significant advances in early diagnosis and treatment have been achieved in recent years, the clinical outcome of osteosarcoma patients with metastasis and recurrence remains to be unfavorable.2 A previous epidemiological survey revealed that the osteosarcoma patients without metastasis have a 5-year survival rate of 60%–70%, whereas patients with metastasis have a survival rate of <30%.3 Accumulating studies were performed to investigate the underlying mechanisms in the development and progression of osteosarcomas.4–6 However, it has not been fully elucidated. Therefore, it is of great clinical significance to find out the crucial metastatic-related markers and potential therapeutic targets for preventing early-stage metastases and improving the patients’ prognosis.
MicroRNAs (miRs), a group of abundant, short (with ~22–28 nucleotides in length), and ncRNAs play important roles in various aspects of cellular and molecular biology by negatively regulating posttranscriptional expression of target genes through the binding of the corresponding 3′-UTR of the miRs.7,8 Growing evidence shows that the expression of miRs may be dysregulated in various types of human cancers, and these miRs function either as oncogenes or tumor suppressors implying their important position in cancer development and progression.9–11 Especially, in human osteosarcoma, Chen et al12 observed the under-expression of miR-145 in metastatic osteosarcoma, and demonstrated that miR-145 might act as a tumor metastasis suppressor; Sun et al13 reported that miR-183 could suppress the vitality, invasion, and migration of human osteosarcoma cells; Yao et al14 showed the decreased expression of miR-96 in human osteosarcoma tissues compared to human paracancerous tissues, and also revealed that the upregulation of miR-96 could inhibit cell proliferation, migration, invasion, and tumor formation ability, as well as increase the percentage of cell apoptosis in osteosarcoma cells. These previous findings may contribute to the development of novel clinical therapeutic approaches for the treatment of osteosarcoma.
As a dysregulated miR in several types of human cancers, miR-328-3p upregulation was demonstrated to be critical for the resveratrol-mediated suppression of metastatic ability in human osteosarcoma, implying its potential role in this malignancy.15 However, the clinical significance and the biological functions of miR-328-3p in osteosarcoma remain unclear. In the current study, we aimed to investigate the expression patterns of miR-328-3p in osteosarcoma tissues, and also clarify its associations with patients’ tumor progression and cellular malignant phenotypes.
Patients and methods
Patients and tissue samples
The current study was approved by the Research Ethics Committee of Huai’an Second Hospital (No HEYLL201503). Written informed consent was obtained from all patients. All specimens were handled and made anonymous in accordance with the Declaration of Helsinki.
In total, 88 pairs of tumor tissues and self-matched noncancerous bone tissues were collected from 88 osteosarcoma patients who were retrospectively enrolled according to the surgical pathology records of Huai’an Second Hospital from 2008 to 2015. All the tumors were confirmed pathologically according to the surgically resected specimens. All patients received the uniform preoperative multi-agent chemotherapy following the initial biopsy. The cytotoxic drugs used as preoperative chemotherapy were cis-diamminedichloroplatinum, adriamycin, vincristine, ifosfamide, and high-dose methotrexate. Table 1 summarizes the clinicopathological characteristics of 88 osteosarcoma patients.
Table 1.
Associations of miR-328-3p expression with various clinicopathological characteristics of primary osteosarcoma
| Clinicopathological characteristics | No. of patients (n, %) | miR-328-3p low (%) | P-value |
|---|---|---|---|
| Age (years) | |||
| <20 | 38 (43.18) | 20 (52.64) | NS |
| ≥20 | 50 (56.82) | 25 (50.00) | |
| Gender | |||
| Male | 52 (59.09) | 29 (55.77) | NS |
| Female | 36 (40.91) | 16 (44.44) | |
| Tumor size (cm) | |||
| ≤5 | 40 (45.45) | 20 (50.00) | NS |
| >5 | 48 (54.55) | 25 (52.08) | |
| Tumor location | |||
| Femur | 46 (52.27) | 21 (45.65) | NS |
| Tibia | 20 (22.73) | 12 (60.00) | |
| Humerus | 18 (20.45) | 10 (55.56) | |
| Fibula | 4 (4.55) | 2 (50.00) | |
| Surgical stage | |||
| IIA | 33 (37.50) | 10 (30.30) | 0.01 |
| IIB/III | 55 (62.50) | 35 (63.64) | |
| Metastasis | |||
| No | 53 (60.23) | 13 (24.53) | 0.008 |
| Yes | 35 (39.77) | 32 (91.43) | |
| Recurrence | |||
| No | 54 (61.36) | 20 (37.04) | 0.01 |
| Yes | 34 (38.64) | 25 (73.53) | |
| Histological subtype | |||
| Osteoblastic | 54 (61.36) | 35 (64.81) | NS |
| Chondroblastic | 18 (20.45) | 6 (33.33) | |
| Fibroblastic | 11 (12.50) | 2 (18.18) | |
| Mixed type | 3 (3.41) | 2 (66.67) | |
| Small type | 2 (2.27) | 0 (0.00) | |
Abbreviations: miR, microRNA; NS, Not significant.
Cell culture and transfection
Two human osteosarcoma cell lines (HOS and U2OS) and one human osteoblast cell line (hFOB1.19) were purchased from Cell Bank, Shanghai Institutes for Biological Sciences, Chinese Academy of Sciences, Shanghai, People’s Republic of China. All cell lines were cultured in DMEM (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% FBS at 37°C in a humidified 5% CO2 incubator.
miR-328-3p mimics (miR-328-mimics), negative control mimics (NC-mimics), matrix metalloprotease 16 (MMP16) expression plasmid (MMP16-ex), and negative control expression plasmid (NC-ex) were all purchased from GenePharma (Shanghai, People’s Republic of China). Transfection of miR mimics or plasmids was transfected into the cells using Lipofectamine 2000 (Thermo Fisher Scientific). Transfection efficiencies were evaluated in every experiment by quantitative real-time (qRT)-PCR.
Quantita tive real-time-PCR
qRT-PCR was performed to detect the expression levels of miR-328-3p in human tissues and cell lines according to our previous study.16 Total RNA was extracted from fresh tissues and cells using TRIzol reagent (Thermo Fisher Scientific). TaqMan miR Assays (Thermo Fisher Scientific) with primers specific to miR-328-3p were used. Reverse transcription was performed using One Step PrimeScript miR cDNA Synthesis Kit (Thermo Fisher Scientific), and qRT-PCR was performed using SYBR Premix Ex Taq™ II (Thermo Fisher Scientific). RNU6B was used as an internal control. qRT-PCR primer sequences were as follows: miR-328-3p, forward 5′-CTG GCC CTC TCT GCC C-3′, and reverse 5′-GTG CAG GGT CCG AGG T-3′; RNU6B, forward 5′-ACA GUA GUC UGC ACA UUG GUU A-3′, and reverse 5′-ACG CAA ATT CGT GAA GCG TT-3′. Quantitative PCR was performed using an ABI 7500 Real-Time PCR Detection System (Thermo Fisher Scientific). The threshold cycle (Ct) was defined as the fractional cycle number at which the fluorescence passed the fixed threshold. Each sample was detected thrice, and the relative expression level of miR-328-3p to RNU6 was calculated using the equation 2−ΔCt, where ΔCT = (CTmiR-328-3p/CTRNU6). The median value of miR-328-3p expression in osteosarcoma tissues was used as a cutoff point for dividing miR-328-3p-low/high groups.
Western blot analysis
MMP16 protein expression was detected by Western blot analysis. Proteins were isolated from the transfected cell lines using a RIPA lysis buffer (Sigma-Aldrich Co., St Louis, MO, USA), and the protein concentration was determined by a bicinchoninic acid protein assay kit (Beyotime Institute of Biotechnology, Jiangsu, People’s Republic of China) based on the manufacturers’ instruction. Equal protein was separated by 12% SDS-PAGE and then transferred to polyvinylidene fluoride membranes (EMD Millipore, Billerica, MA, USA). After being blocked with 5% skimmed milk in PBS for 1 hour at room temperature, the membrane was incubated with primary antibodies against MMP16 and GAPDH at 4°C overnight. After that, the membranes were washed with PBS thrice, and then incubated with horseradish peroxidase-conjugated secondary antibodies for 2 hours at room temperature. ECL detection systems (SuperSignal™ West Femto, Pierce™; Thermo Fisher Scientific) were used for the detection of protein expression.
Luciferase reporter assay
The 3′-UTR of MMP16 containing the putative binding site of miR-328-3p and the mutated target seed sequence were amplified and subcloned into pGL3 luciferase promoter vector (Promega Corporation, Fitchburg, WI, USA) to construct the report vectors pGL3-MMP16 and pGL3-MUT MMP16, respectively. These vectors were cotransfected with the miR-328-3p mimics into U2OS cells for 48 hours. After collecting the cells, we detected the relative luciferase activity using a Dual-Luciferase Reporter Assay Kit (Promega Corporation) based on the manufacturer’s instructions. All experiments were performed at least three times.
Cell proliferation assay
Cell proliferation ability was evaluated by Cell Counting Kit-8 ([CCK-8]; Dojindo Laboratories, Kumamoto, Japan). Cells in different groups were cultured in 96-well plates about 1.0×104 cells/mL for 24, 48, 72, 96, and 120 hours. The CCK-8 solution (10 µL) was added into each well, incubated for 4 hours, and then the optical density at 450 nm was measured to calculate the number of proliferative cells. A total of six parallel wells were analyzed for each group.
Cell migration assay
Cell migrative ability was evaluated by wound healing assay. A scratch was produced in the cell monolayer using a micro-pipette tip when the cells in different groups had grown to 90% confluence. Images were captured from six randomly selected fields in each sample, and the migration distance between the leading edge of the migrating cells and the edge of the wound was calculated by NIH ImageJ software.
Statistical analyses
All statistical analyses in the current study were performed by SPSS software for Windows (version 11.0; SPSS Inc., Chicago, IL, USA). All data represent the mean ± SD. Fisher’s exact test was used for any 2×2 tables, Pearson chi-squared test for non- 2×2 tables, and chi-square trend test for ordinal datum. Comparison among different groups was performed using a one-way ANOVA test followed by Tukey’s multiple comparison post hoc analysis. Differences were considered statistically significant when P<0.05.
Results
Downregulation of miR-328-3p associates with aggressive progression of osteosarcomas
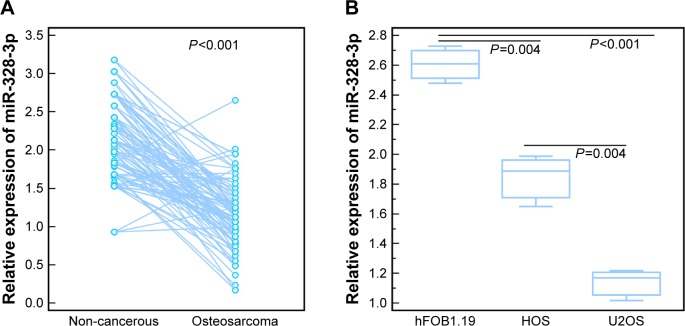
As shown in Figure 1A, the expression levels of miR-328-3p in osteosarcoma tissues were significantly lower than those in noncancerous bone tissues (tumor vs normal: 1.16±0.44 vs 2.14±0.52, P<0.001, Figure 1A). Of 88 osteosarcoma patients, 45 (51.14%) and 43 (48.86%) displayed low and high miR-328-3p expressions, respectively.
Figure 1.
Expression patterns of miR-328-3p in human osteosarcoma tissues and cells.
Notes: (A) Expression levels of miR-328-3p in osteosarcoma tissues and noncancerous bone tissues. (B) Expression levels of miR-328-3p in osteosarcoma cells (HOS and U2OS) and osteoblast cells (hFOB1.19). Each assay was conducted in triplicate and was repeated thrice. The data show the mean of triplicate repeats ± SD.
Abbreviation: miR, microRNA.
Table 1 summarized the associations of miR-328-3p expression with various clinicopathological characteristics of osteosarcoma patients. The statistical analyses revealed that miR-328-3p downregulation was significantly associated with advanced surgical stage (P=0.01), positive metastasis (P<0.001), and recurrence (P=0.01). However, there was no significant associations with age, gender, tumor size, tumor location, or histological subtype (all P>0.05, Table 1).
Elevated expression of miR-328-3p suppresses cell proliferation and migration in osteosarcoma cells
To determine the biological function of miR-328-3p in osteosarcoma, we first detected its expression levels in two human osteosarcoma cell lines (HOS and U2OS) and one human osteoblast cell line (hFOB1.19). As shown in Figure 1B, miR-328-3p expression levels in HOS and U2OS cells were both significantly decreased compared to hFOB1.19 cells (both P<0.05). Since the expression levels of miR-328-3p in U2OS cells were distinctively lower than HOS cells (P<0.05), we chose U2OS cells to perform the following functional experiments.
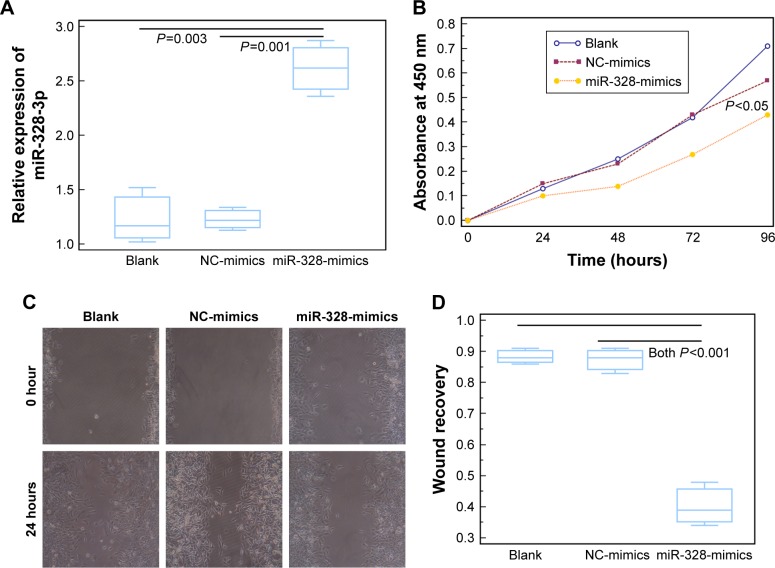
miR-328-3p expression in U2OS cells was significantly elevated following the transfection with miR-328-3p mimic (P<0.05, Figure 2A). CCK-8 assay showed that U2OS cells transfected with miR-328-3p mimics displayed a significant suppression in the proliferation at five consecutive time points (0, 24, 48, 72, and 96 hours) compared to the NC and blank control groups (P<0.05, Figure 2B). Wound healing assays demonstrated the inhibition of migration ability of osteosarcoma cells by miR-328-3p mimics (P<0.05, Figure 2C and D).
Figure 2.
Elevated expression of miR-328-3p suppresses cell proliferation and migration in osteosarcoma cells.
Notes: (A) Expression levels of miR-328-3p in U2OS cells transfected with miR-328-3p mimics and NC-mimics, or in blank group. (B) Absorbance of U2OS cells transfected with miR-328-3p mimics and NC-mimics, or in blank group at five consecutive time points (0, 24, 48, 72, and 96 hours) detected by CCK-8 assay. (C and D) The number of migrating U2OS cells transfected with miR-328-3p mimics and NC-mimics, or in blank group detected by wound healing assay. Each assay was conducted in triplicate and repeated thrice. The data show the mean of triplicate repeats ± SD.
Abbreviations: CCK-8, Cell Counting Kit-8; miR, microRNA; NC-mimics, negative control mimics.
MMP16 is a potential target of miR-328-3p in osteosarcoma cells
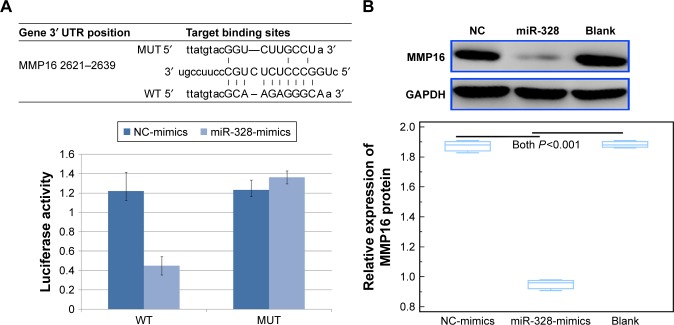
To further investigate the molecular mechanisms underlying the tumor suppressive role of miR-328-3p in osteosarcoma, we collected its potential targets from miRTarBase, the experimentally validated miR-target interactions database (http://mirtarbase.mbc.nctu.edu.tw/php/index.php, Release 7.0). As a result, MMP16 might be a potential target of miR-328-3p. Then, luciferase reporter assay was performed to verify this hypothesis. As shown in Figure 3A, the relative luciferase activity of miR-328-3p overexpressed U2OS cells cotransfected with the construct containing the wild type 3′-UTR of MMP16 was significantly lower than those of the mutant MMP16 3′-UTR. Further Western blot analysis showed that the enforced expression of miR-328-3p suppressed the expression levels of MMP16 protein in osteosarcoma cells (P<0.01, Figure 3B).
Figure 3.
MMP16 is a potential target of miR-328-3p in osteosarcoma cells.
Notes: (A) The binding sites of miR-328-3p to MMP16 mRNA and the relative luciferase activity of miR-328-3p overexpressed U2OS cells cotransfected with the constructs containing the WT or the MUT 3′-UTR of MMP16. (B) Western blot analysis of the relative expression levels of MMP16 protein in U2OS cells with the enforced expression of miR-328-3p. Each assay was conducted in triplicate and repeated thrice. The data show the mean of triplicate repeats ± SD.
Abbreviations: GAPDH, glyceraldehyde-3-phosphate dehydrogenase; miR, microRNA; MMP16, matrix metalloprotease 16; MUT, mutant; NC-mimics, negative control mimics; WT, wild type.
MMP16 overexpression partially reverses the miR-328-3p-inhibited cell proliferation and migration of osteosarcoma cell
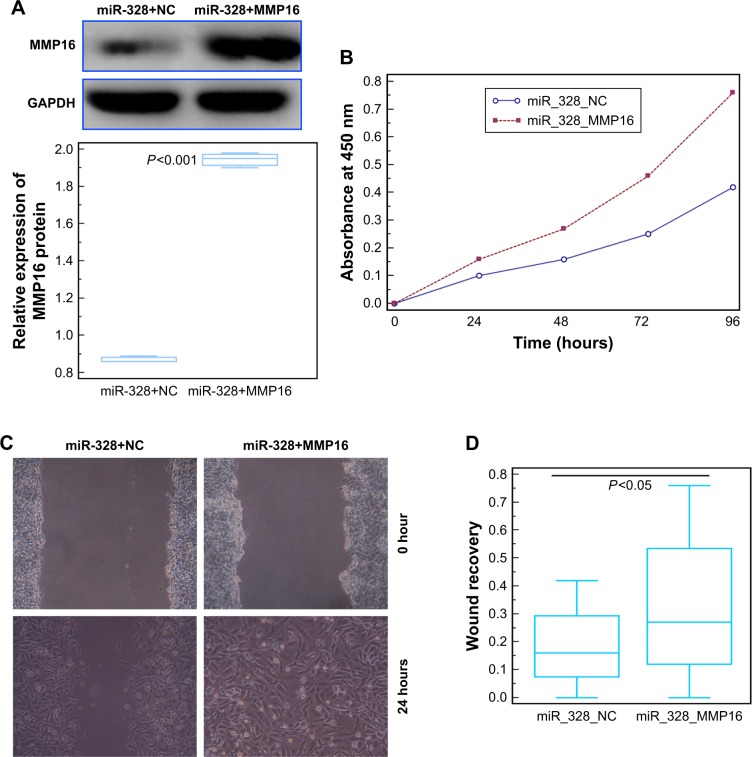
To confirm the role of MMP16 in the tumor suppressive function of miR-328-3p in osteosarcoma, we performed rescue experiments by transfecting MMP16-ex into miR-328-3p-overexpressed U2OS cells. As shown in Figure 4A, the expression levels of MMP16 protein in U2OS cells cotransfected with MMP16-ex and miR-328-3p-mimic were significantly higher in those only transfected with miR-328-3p-mimic (P<0.05, Figure 4A). Functionally, both CCK-8 and wound healing assays indicated that MMP16 overexpression partially rescued the impaired cell proliferation and migration abilities of U2OS cells induced by miR-328-3p mimics (both P<0.05, Figure 4B–D).
Figure 4.
MMP16 overexpression partially reverses the miR-328-3p-inhibited cell proliferation and migration of osteosarcoma cell.
Notes: (A) The expression levels of MMP16 protein in U2OS cells cotransfected with MMP16-ex and miR-328-3p-mimic, or with NC-ex and miR-328-3p-mimic. (B) Absorbance of U2OS cells cotransfected with MMP16-ex and miR-328-3p-mimic, or with NC-ex and miR-328-3p-mimic at five consecutive time points (0, 24, 48, 72, and 96 hours) detected by CCK-8 assay. (C and D) The representative images and the number of migrating U2OS cells cotransfected with MMP16-ex and miR-328-3p-mimic, or with NC-ex and miR-328-3p-mimic detected by Wound healing assay. Each assay was conducted in triplicate and repeated thrice. The data show the mean of triplicate repeats ± SD.
Abbreviations: CCK-8, Cell Counting Kit-8; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; miR, microRNA; MMP16, matrix metalloprotease 16; MMP16-ex, MMP16 expression plasmid; NC, negative control; NC-ex, negative control expression plasmid.
Discussion
Growing evidence shows that it is compelling to seek out crucial metastatic-related molecules and to elucidate the underlying mechanisms in osteosarcoma. In the current study, we demonstrated that miR-328-3p expression in osteosarcoma tissues and cells were both markedly lower than those in noncancerous bone tissues and cells. Clinically, miR-328-3p downregulation was significantly associated with advanced surgical stage, positive metastasis, and recurrence. In addition, the enforced expression of miR-328-3p efficiently suppressed cell proliferation and migration in vitro. Mechanically, MMP16 was identified as a direct target of miR-328-3p in osteosarcoma cells, and MMP16 overexpression partially reversed the miR-328-3p-inhibited cell proliferation and migration of osteosarcoma cells. These findings indicated for the first time that miR-328-3p may function as a tumor suppressor by negatively regulating MMP16 in human osteosarcoma.
An increasing number of studies have indicated that miR-328-3p may play either oncogenic or tumor suppressive role in variety of human cancers via regulating different targets.17–20 For example, Xue et al17 found that miR-328 negatively interfered with the viability of early gastric cancer cells, and it was also recognized as a potential biomarker to predict recurrence after endoscopic submucosal dissection (ESD) in early gastric cancer patients via analysis of the recurrence-free rate among different groups of the patients. Luo et al18 indicated that miR-328 could enhance cellular motility through posttranscriptional regulation of protein tyrosine phosphatase receptor type J in human hepatocellular carcinoma. Lin et al19 observed that miR-328 overexpression might inhibit migration and epithelial-mesenchymal transition by targeting CD44 in nasopharyngeal carcinoma cells. Arora et al20 also confirmed that miR-328 was associated with the brain metastasis of non-small-cell lung cancer and mediated cancer migration. In contrast, the results of Han et al21 showed that overexpression of miR-328 markedly inhibited esophageal cancer cell proliferation and invasion, and enhanced apoptosis; whereas, inhibition of miR-328 significantly promoted cell proliferation and invasion, and suppressed apoptosis in esophageal cancer cells. Wang and Xia22 provided the first evidence for the growth suppressive activity of miR-328 in cervical cancer, which was largely ascribed to downregulation of TCF7L2. Santasusagna et al23 also revealed that miR-328 may be involved in the orchestration of the Warburg effect in colon cancer cells, and its reduced expression in colon cancer patients was inversely correlated with the classically reported upregulated SLC2A1/GLUT1 expression in tumors. These previous findings suggest a complex role for miR-328 in tumor biology. However, the expression and functions of miR-328 in osteosarcoma remain unclear. In the current study, our data pinpointed to the decreased expression of miR-328-3p in both human osteosarcoma tissues and cells, and confirmed miR-328-3p downregulation was dramatically associated with tumor aggressive progression of osteosarcoma patients. Then, it is noteworthy that miR-328-3p may play a tumor suppressive role and can inhibit tumor cell proliferation and migration.
Having characterized the suppressive role of miR-328 in cervical cancer, we next integrated bioinformatics prediction, luciferase report assay, and functional validation to identify MMP16 as one of the potential target genes mediating its function in osteosarcoma. MMP16, also known as membrane type (MT)-3 MMP, belongs to the type-1 MT MMP family, together with MMP14, MMP15, and MMP24.24 Under the physiological conditions, MMP16 mainly localizes on microglial cells, melanocytes, and endothelial cells. Similar to other members of the MMPs family, MMP16 can exert proteolytic activity on components of the extracellular matrix.25 Accumulating studies have observed the overexpression of MMP16 in various human cancer tissues, and also revealed that its aberrant expression may help facilitate cancer metastasis and progression.26–29 Chen et al12 identified MMP16 as a target gene of miR-145 in OS using a dual luciferase reporter system, and also determined that miR-145 might function as a tumor metastasis suppressor gene by downregulating MMP16 in OS. Moreover, Xu et al30 indicated that MMP16 might be a direct target of miR-328 in cancer stem cell-like cells in colorectal cancer. Consistently, our data here also validated that miR-328-3p directly targeted MMP16 in osteosarcoma cells, and it also was associated with the tumor suppressive role of miR-328-3p in this malignancy.
Conclusion
Our data indicated that the aberrant expression of miR-328-3p may play a crucial role in malignant progression of human osteosarcomas. More importantly, miR-328-3p may function as a tumor suppressor inhibiting osteosarcoma cell proliferation and migration partially mediated by regulation of MMP16.
Acknowledgments
This research was supported by grants from Jiangsu Natural Science Foundation (BK20171265) and Huai’an Key Laboratory funded projects (HAP201608).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Bielack SS, Hecker-Nolting S, Blattmann C, Kager L. Advances in the management of osteosarcoma. F1000Res. 2016;5:2767. doi: 10.12688/f1000research.9465.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang Z, Li X, Yang Y, He Z, Qu X, Zhang Y. Long noncoding RNAs in the progression, metastasis, and prognosis of osteosarcoma. Cell Death Dis. 2016;7(9):e2389. doi: 10.1038/cddis.2016.272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Adamopoulos C, Gargalionis AN, Basdra EK, Papavassiliou AG. Deciphering signaling networks in osteosarcoma pathobiology. Exp Biol Med (Maywood) 2016;241(12):1296–1305. doi: 10.1177/1535370216648806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meazza C, Scanagatta P. Metastatic osteosarcoma: a challenging multidisciplinary treatment. Expert Rev Anticancer Ther. 2016;16(5):543–556. doi: 10.1586/14737140.2016.1168697. [DOI] [PubMed] [Google Scholar]
- 5.Varshney J, Scott M, Largaespada D, Subramanian S. Understanding the osteosarcoma pathobiology: a comparative oncology approach. Vet Sci. 2016;3(1):3. doi: 10.3390/vetsci3010003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Anderson ME. Update on survival in osteosarcoma. Orthop Clin North Am. 2016;47(1):283–292. doi: 10.1016/j.ocl.2015.08.022. [DOI] [PubMed] [Google Scholar]
- 7.Lekka E, Hall J. Noncoding RNAs in disease. FEBS Lett. 2018;592(17):2884–2900. doi: 10.1002/1873-3468.13182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dragomir M, Mafra A, Dias S, Vasilescu C, Calin G. Using microRNA networks to understand cancer. Int J Mol Sci. 2018;19(7):1871. doi: 10.3390/ijms19071871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 10.Sethi S, Sethi S, Bluth MH. Clinical implication of microRNAs in molecular pathology: an update for 2018. Clin Lab Med. 2018;38(2):237–251. doi: 10.1016/j.cll.2018.02.003. [DOI] [PubMed] [Google Scholar]
- 11.Lei K, Bai H, Wei Z, et al. The mechanism and function of circular RNAs in human diseases. Exp Cell Res. 2018;368(2):147–158. doi: 10.1016/j.yexcr.2018.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Chen B, Huang Z, Zhang Y, Chen Y, Li Z. MicroRNA-145 suppresses osteosarcoma metastasis via targeting MMP16. Cell Physiol Biochem. 2015;37(6):2183–2193. doi: 10.1159/000438575. [DOI] [PubMed] [Google Scholar]
- 13.Sun X, Xu Y, Zhang S, et al. MicroRNA-183 suppresses the vitality, invasion and migration of human osteosarcoma cells by targeting metastasis-associated protein 1. Exp Ther Med. 2018;15(6):5058–5064. doi: 10.3892/etm.2018.6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yao Q, Pei Y, Zhang X, Xie B. microRNA-96 acts as a tumor suppressor gene in human osteosarcoma via target regulation of ezrin. Life Sci. 2018;203:1–11. doi: 10.1016/j.lfs.2018.04.012. [DOI] [PubMed] [Google Scholar]
- 15.Yang SF, Lee WJ, Tan P, et al. Upregulation of miR-328 and inhibition of CREB-DNA-binding activity are critical for resveratrol-mediated suppression of matrix metalloproteinase-2 and subsequent metastatic ability in human osteosarcomas. Oncotarget. 2015;6(5):2736–2753. doi: 10.18632/oncotarget.3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao J, Chen F, Zhou Q, et al. Aberrant expression of microRNA-328-3p and its target gene mTOR associated with malignant progression and poor prognosis in patients with osteosarcoma. Onco Targets Ther. 2016;9:1589–1597. doi: 10.2147/OTT.S102421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xue HG, Yang AH, Sun XG, Lu YY, Tian ZB. Expression of microRNA-328 functions as a biomarker for recurrence of early gastric cancer (EGC) after endoscopic submucosal dissection (ESD) by modulating CD44. Med Sci Monit. 2016;22:4779–4785. doi: 10.12659/MSM.896225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Luo X, Yang S, Zhou C, Pan F, Li Q, Ma S. MicroRNA-328 enhances cellular motility through posttranscriptional regulation of PTPRJ in human hepatocellular carcinoma. Onco Targets Ther. 2015;8:3159–3167. doi: 10.2147/OTT.S93056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin CH, Chiang MC, Chen YJ. MicroRNA-328 inhibits migration and epithelial-mesenchymal transition by targeting CD44 in nasopharyngeal carcinoma cells. Onco Targets Ther. 2018;11:2375–2385. doi: 10.2147/OTT.S151665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arora S, Ranade AR, Tran NL, et al. MicroRNA-328 is associated with (non-small) cell lung cancer (NSCLC) brain metastasis and mediates NSCLC migration. Int J Cancer. 2011;129(11):2621–2631. doi: 10.1002/ijc.25939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han N, Zhao W, Zhang Z, Zheng P. MiR-328 suppresses the survival of esophageal cancer cells by targeting PLCE1. Biochem Biophys Res Commun. 2016;470(1):175–180. doi: 10.1016/j.bbrc.2016.01.020. [DOI] [PubMed] [Google Scholar]
- 22.Wang X, Xia Y. microRNA-328 inhibits cervical cancer cell proliferation and tumorigenesis by targeting TCF7L2. Biochem Biophys Res Commun. 2016;475(2):169–175. doi: 10.1016/j.bbrc.2016.05.066. [DOI] [PubMed] [Google Scholar]
- 23.Santasusagna S, Moreno I, Navarro A, et al. miR-328 mediates a metabolic shift in colon cancer cells by targeting SLC2A1/GLUT1. Clin Transl Oncol. 2018;20(9):1161–1167. doi: 10.1007/s12094-018-1836-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Itoh Y. Membrane-type matrix metalloproteinases: their functions and regulations. Matrix Biol. 2015:44–46. 207–223. doi: 10.1016/j.matbio.2015.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Ueno H, Nakamura H, Inoue M, et al. Expression and tissue localization of membrane-types 1, 2, and 3 matrix metalloproteinases in human invasive breast carcinomas. Cancer Res. 1997;57(10):2055–2060. [PubMed] [Google Scholar]
- 26.Tatti O, Arjama M, Ranki A, Weiss SJ, Keski-Oja J, Lehti K. Membrane-type-3 matrix metalloproteinase (MT3-MMP) functions as a matrix composition-dependent effector of melanoma cell invasion. PLoS One. 2011;6(12):e28325. doi: 10.1371/journal.pone.0028325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Plaisier M, Kapiteijn K, Koolwijk P, et al. Involvement of membrane-type matrix metalloproteinases (MT-MMPs) in capillary tube formation by human endometrial microvascular endothelial cells: role of MT3-MMP. J Clin Endocrinol Metab. 2004;89(11):5828–5836. doi: 10.1210/jc.2004-0860. [DOI] [PubMed] [Google Scholar]
- 28.Tatti O, Gucciardo E, Pekkonen P, et al. MMP16 mediates a proteolytic switch to promote cell-cell adhesion, collagen alignment, and lymphatic invasion in melanoma. Cancer Res. 2015;75(10):2083–2094. doi: 10.1158/0008-5472.CAN-14-1923. [DOI] [PubMed] [Google Scholar]
- 29.Jiang C, Wang J, Dong C, Wei W, Li J, Li X. Membranous type matrix metalloproteinase 16 induces human prostate cancer metastasis. Oncol Lett. 2017;14(3):3096–3102. doi: 10.3892/ol.2017.6536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xu XT, Xu Q, Tong JL, et al. MicroRNA expression profiling identifies miR-328 regulates cancer stem cell-like SP cells in colorectal cancer. Br J Cancer. 2012;106(7):1320–1330. doi: 10.1038/bjc.2012.88. [DOI] [PMC free article] [PubMed] [Google Scholar]