Abstract
Background
Despite the advances in early detection and treatment methods, breast cancer still has a high mortality rate, even in those patients predicted to have a good prognosis. The purpose of this study is to identify a microRNA signature that could better predict prognosis in breast cancer and add new insights to the current classification criteria.
Materials and methods
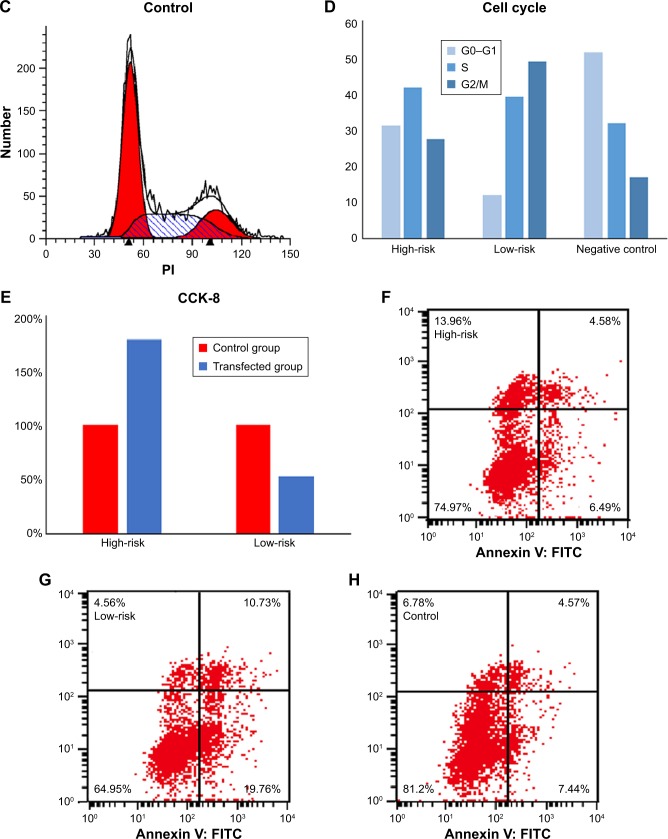
We downloaded microRNA sequencing data along with corresponding clinicopathological data from The Cancer Genome Atlas (TCGA). Of 1,098 breast cancer patients identified, 253 patients with fully characterized microRNA profiles were selected for analysis. A three-microRNA signature was generated in the training set. Subsequently, the performance of the signature was confirmed in a validation set. After construction of the signature, we conducted additional experiments, including flow cytometry and the Cell Counting Kit-8 assay, to illustrate the correlation of this microRNA signature with breast cancer cell cycle, apoptosis, and proliferation.
Results
Three microRNAs (hsa-mir-31, hsa-mir-16-2, and hsa-mir-484) were identified to be significantly and independently correlated with patient prognosis, and performed with good stability. Our results suggest that higher expression of hsa-mir-484 indicated worse prognosis, while higher expression of hsa-mir-31 and hsa-mir-16-2 indicated better prognosis. Moreover, additional experiments confirmed that this microRNA signature was related to breast cancer cell cycle and proliferation.
Conclusion
Our results indicate a three-microRNA signature that can accurately predict the prognosis of breast cancer, especially in basal-like and hormone receptor-positive breast cancer subtypes. We recommend more aggressive therapy and more frequent follow-up for high-risk groups.
Keywords: microRNA, breast cancer, TCGA, prognosis
Introduction
Breast cancer is one of the most common malignancies among women, and despite the discovery of early detection methods and effective treatment therapies, it is still the second leading cause of cancer-related death in females.1 Breast cancer is a group of molecularly distinct neoplasms classified into four main subgroups based on their expression of estrogen receptor (ER),2 progesterone receptor (PR), and human epidermal growth factor receptor 2 (Her2). These subgroups require different treatment therapies and experience different clinical outcomes. However, even within the subgroups, there are different subsets of genetic and epigenetic abnormalities leading to different patient prognoses;3 thus, more research is needed to understand the mechanisms related to the prognosis within different breast cancer subgroups.
MicroRNAs are a class of endogenously expressed small, single-stranded, non-coding RNAs. Over the past decade, the aberrant expression of microRNAs has been increasingly reported in human cancers and has often been associated with diagnosis,4 prognosis, and response to clinical therapies.5 They are involved in the post-transcriptional regulation of gene expression via base pairing with target mRNAs (usually in the 3′ untranslated region), causing degradation and translation repression of mRNAs.6 MicroRNAs are now widely regarded as the most powerful regulators of gene expression in complex cellular processes including cancer cell proliferation, metastasis, migration, and apoptosis.7 Of particular importance is the association with cancer cell proliferation and metastasis, as these are two hallmarks of malignancy and the leading causes of cancer-related death.5 In addition, many studies have shed light on tumor-targeting therapies using microRNAs as novel diagnostic and therapeutic tools.8,9
The Cancer Genome Atlas (TCGA) project provides researchers with a set of comprehensive tools that can be used to analyze clinical and genetic signatures of a variety of cancers including breast carcinoma. In this study, we retrieved breast carcinoma data from TCGA to construct a three-microRNA signature that can be used to predict the prognosis of breast cancer, and we verified the signature using both statistical and experimental methods.
Materials and methods
TCGA breast invasive carcinoma data set
The clinical information and expression levels from 1,158 microRNAs of 1,098 patients with breast as the primary cancer site were downloaded from TCGA (https://cancergenome.nih.gov/) on May 4, 2017. Patients were screened by the following criteria for inclusion: 1) the patients were female; 2) the patients had no preoperative treatment; 3) the patients’ sample types were primary tumor; 4) the patients had fully characterized microRNA profiles; and 5) the percentage of necrosis in samples was <40% on both the top and bottom slides. Patients who were alive but missing the date of last contact were excluded. A total of 253 breast invasive carcinoma patients were identified for further study according to the selection criteria. The total set was randomly separated into a training set (153 patients) and a validation set (100 patients).
Construction and validation of the integrated microRNA signature
The microRNA signature was constructed in the training set. A total of 1,158 microRNA expression levels were presented as reads per million (RPM) microRNA mapped data. Any microRNA expression level reads where microRNAs equaled 0 RPM in >40% observations were excluded. After transformation into binary variables according to the median expression level, univariate Cox models were generated for preliminary screening of microRNAs that were significantly correlated with overall survival (OS). A cut-off P-value of <0.05 was used to filter out significant parameters. Clinical characteristics that were previously reported to be associated with prognosis, including age at diagnosis, N stage, T stage, metastasis, ER, PR, and Her2, were also similarly evaluated in the univariate Cox models. We then generated general multivariate stepwise Cox regression models to determine which of the significant microRNA identified by univariate proportional hazards regression was an independent predictor of prognosis. OS time was calculated from the date of the initial pathological diagnosis to the date of death.
The permutation test was used to evaluate the performance and randomness of the final multivariate model. Using the combination of patient OS time and vital status as a label, each patient was assigned a label and risk score under the microRNA scoring system. A random system was constructed by assigning labels while the risk score was kept consistent within each individual. The random system was tested for significance in predicting survival. If the model performed well, the random system was not a predictor of prognosis, and the area under the curve (AUC) of the receiver operating characteristics (ROC) curve would approach 0.5. We generated 1,000 random systems. A cut-off P-value of <0.05 was used to indicate a significant association between AUCs of the random system and the label system. We would conclude that the label system had no effect on outcome unless the calculated P-value was smaller than 0.05. A validation set containing 100 patients was used to test the prognostic value of the microRNA signature. These analyses were performed using R software (version 3.3.2, https://www.r-project.org/).
Bioinformatics analysis
Targetscan7.1 (http://www.targetscan.org/vert_71/), DIANA-microT,10 miRWalk,11 miRanda (http://www.microrna.org/microrna/home.do), PicTar (http://www.pictar.org/), and miRDB12 were used to identify the target genes of three microRNAs. To increase accuracy, only target genes predicted by a minimum of three programs were retained for further analysis. Lists of target genes were submitted to DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov/) to annotate the biological functions of the candidate microRNAs. Subsequently, Gene Ontology (GO) function, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis,13 and PANTHER™ Version 11 analyses were conducted. Pathways with fold enrichment >1.5 and P<0.05 were considered to be of interest.14
Cell lines and culturing method
After evaluating qRT-PCR (data not shown) for the expression of the three microRNAs together with our statistical analysis results, we ultimately chose the cell line MDA-MB-231 to continue further study. MDA-MB-231 was obtained from the American Type Culture Collection (Manassas, VA, USA), cultured according to the instructions, and used within 6 months after recovery from liquid nitrogen.
Transfection, cell proliferation assay, and flow cytometry
Cells were plated in six-well plates, transfected with microRNA mimic, microRNA inhibitor, and their corresponding negative controls using Lipofectamine™ 3000 Transfection Reagent (Thermo Fisher Scientific, Waltham, MA, USA) following established protocols (transfection efficiency was at least 60% as confirmed by qRT-PCR; data not shown). All microRNA oligonucleotides were synthesized by RiboBio (Guangzhou, China) and quantification was performed with a stem-loop real-time PCR microRNA kit (RiboBio, Guangzhou, China). Transfected MDA-MB-231 was seeded at a density of 5×103 cells per well into 96-well plates and incubated at 37°C for 72 hours. Cell viability was assessed using the Cell-Counting Kit-8 (CCK-8) assay (Dojindo, Kumamoto, Japan); absorbance values were determined at 450 nm using a microplate spectrophotometer. Flow cytometry was performed using propidium iodide (PI) staining solution (Chinese Academy of Sciences, Shanghai, China) and Annexin V: fluorescein isothiocyanate (FITC) Apoptosis Detection Kit I (BD Bioscience) following the instructions provided.
Statistical analyses
Apart from the above methods, other statistical analyses were performed using IBM SPSS Statistics version 22.0 (IBM Corp., Armonk, NY, USA). Survival analysis was conducted using the Kaplan–Meier method with the log-rank test. Means ± SDs of continuous variables were calculated from at least three independent experiments. Student’s t-test was used to compare groups and Pearson’s chi-squared test to assess the correlation between variables. All statistical tests were two-sided and a P-value <0.05 was considered statistically significant.
Results
Construction of microRNA prognostic signature
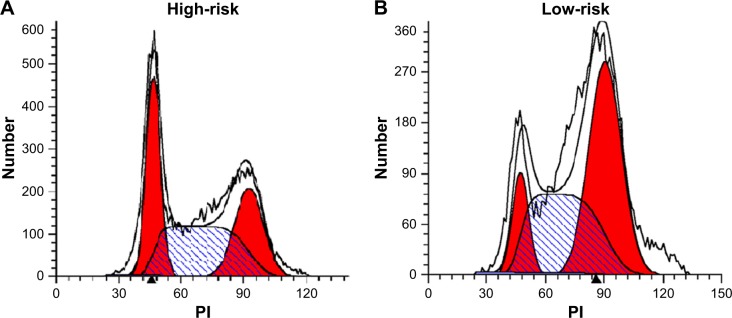
Six microRNAs were identified as prognostic markers after univariate Cox model screening (Table 1). Three microRNAs (hsa-mir-31, hsa-mir-16-2, and hsa-mir-484) were identified to be independently correlated with patient prognosis in multivariate Cox regression (Table 2); higher expression of hsa-mir-484 indicated worse prognosis, while higher expression of hsa-mir-31 and hsa-mir-16-2 indicated improved prognosis. The β-coefficients (microRNA weight on OS) and status of every selected microRNA were used to calculate the risk score, as follows: risk score = (0.494 * Status of hsa-mir-484) − (0.786 * Status of hsa-mir-16-2) − (0.620 * Status of hsa-mir-31). The patients were assigned to the high-risk group if their risk score was greater than the median; otherwise, they were assigned to the low-risk group.
Table 1.
Univariate Cox analysis of 1,158 microRNAs
| MicroRNA | P-value | Coefficient | Type |
|---|---|---|---|
| hsa-mir-31 | 0.008361862 | −0.625612446 | Protective |
| hsa-mir-16-2 | 0.007335068 | −0.629745321 | Protective |
| hsa-mir-484 | 0.007238498 | 0.636249043 | Increased risk |
| hsa-mir-877 | 0.00619359 | 0.652427525 | Increased risk |
| hsa-let-7b | 0.00126726 | −0.781058038 | Protective |
| hsa-mir-937 | 0.001580468 | 0.777204799 | Increased risk |
Table 2.
Multivariate Cox analysis of 1,158 microRNAs
| MicroRNA | P-value | Coefficient | Type |
|---|---|---|---|
| hsa-mir-31 | 0.011486 | −0.62045 | Protective |
| hsa-mir-16-2 | 0.001398 | −0.78621 | Protective |
| hsa-mir-484 | 0.042246 | 0.493782 | Increased risk |
Performance of microRNA signature
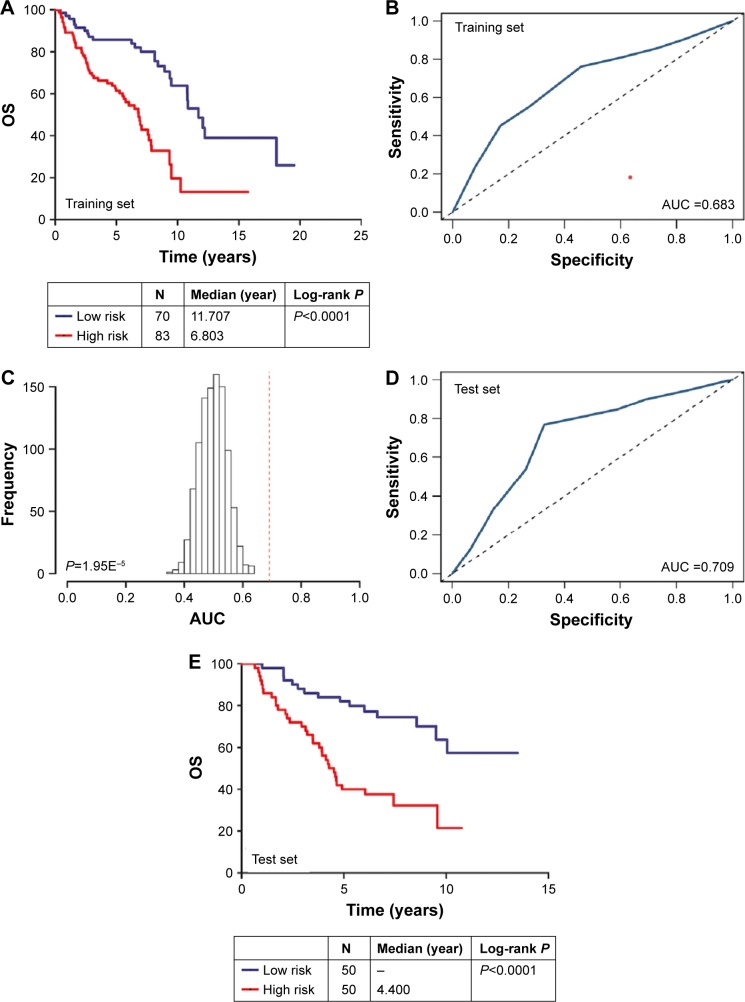
The Kaplan–Meier and ROC analyses were applied to test the performance of the three-microRNA signature in the training set. The patients in the high-risk group had significantly worse OS than those in the low-risk group (P<0.0001) (Figure 1A). The AUC of the signature was 0.683 (Figure 1B). These results confirmed that the three-microRNA signature was powerful enough to divide breast cancer patients into high-risk and low-risk groups.
Figure 1.
(A) Kaplan–Meier analysis of OS in the training set: OS rates between the high-risk group and low-risk group showed statistically significant differences using the log-rank test (P<0.0001); (B) ROC curve of the training set. (C) The permutation test found that the AUC of the random system showed great significance with high-risk and low-risk groups (P=1.95E-05); (D) ROC curves of the validation set, AUC =0.709. (E) Kaplan–Meier analysis of OS in the test set: OS rates between the high-risk group and low-risk group showed statistically significant differences using the log-rank test (P<0.0001). All of these results suggest that our three-microRNA signature can be used as a better diagnostic marker to distinguish breast cancer patients into high-risk and low-risk groups.
Abbreviations: AUC, area under the curve; OS, overall survival; ROC, receiver operating characteristics.
Next, we conducted a permutation test and leave-one-out cross-validation (LOO-CV) to test whether the three-microRNA signature was applicable to other breast cancer patients in the test set.15 The permutation test found that the AUC of the random system showed great significance with high-risk and low-risk groups (P=1.95E-05) (Figure 1C). In addition, the LOO-CV AUC was 0.709 (Figure 1D) and the Kaplan–Meier curve indicated that the high-risk patients had significantly worse OS (P<0.0001) (Figure 1E), which together validated the performance of the three-microRNA signature.
Subgroup analysis
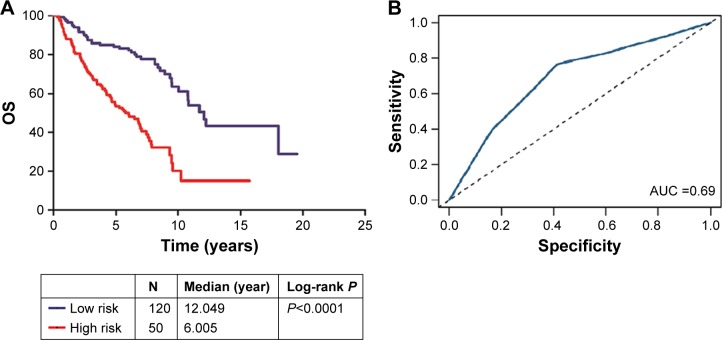
After the construction and validation of the three-microRNA signature, we constructed Kaplan–Meier and ROC curves of OS in the total set (Figure 2). We then divided these patients into different subgroups according to their clinicopathological features to assess the performance of the three-microRNA signature in different groups.
Figure 2.
(A) Kaplan–Meier analysis of OS in the total set; (B) the ROC curve of the total set AUC was 0.69.
Abbreviations: AUC, area under the curve; OS, overall survival; ROC, receiver operating characteristics.
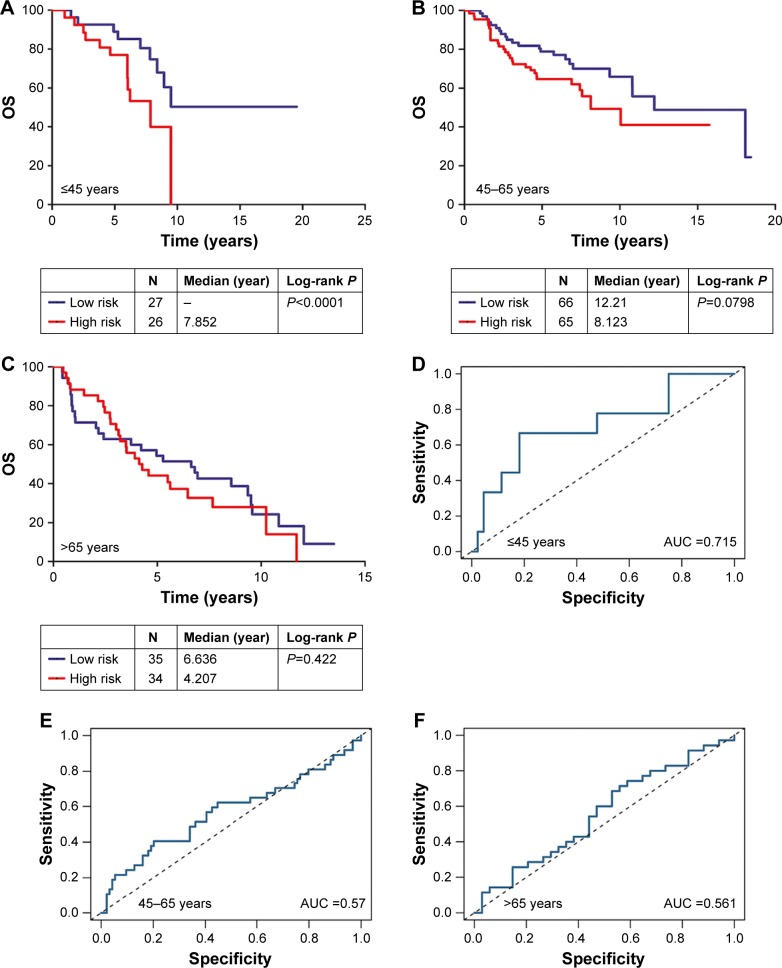
First, the patients were separated into three groups based on their age at diagnosis (≤45 years, 46–65 years, and >65 years). In the ≤45-year-old group, the AUC of the signature was 0.715 with a Kaplan–Meier curve P-value <0.0001 (Figure 3A and D). However, in the 46–65-year-old and >65-year-old groups, the AUCs were 0.57 and 0.561, respectively, and the Kaplan–Meier curve P-values were 0.0798 and 0.422, respectively (Figure 3B, C, E, and F).
Figure 3.
(A) Kaplan–Meier analysis of OS in the ≤45-year age group: OS rates between the high-risk group and low-risk group showed statistically significant differences using the log-rank test (P<0.0001); (B) the ROC curve AUC was 0.715. (C) Kaplan–Meier analysis of OS in the 46–65-year age group: OS rates between the high-risk group and low-risk group showed no significant differences (P=0.0798); (D) the ROC curve AUC was 0.57. (E) Kaplan–Meier analysis of OS in the >65-year age group: OS rates between the high-risk group and low-risk group showed no significant differences (P=0.561); (F) the ROC curve AUC was 0.422. This signature performs better in younger patients (≤45 years) than older patients (>65 years).
Abbreviations: AUC, area under the curve; OS, overall survival; ROC, receiver operating characteristics.
Next, we grouped the patients based on their molecular subtype. For basal-like carcinoma patients, the AUC and P-value were 0.755 and 0.003, respectively (Figure 4A and B). For luminal carcinoma patients, the AUC and P-value were 0.688 and <0.0001, respectively (Figure 4C and D). However, in the Her2-enriched subgroup, the AUC and P-value were 0.545 and 0.5532, respectively (Figure 4E and F).
Figure 4.
(A) Kaplan–Meier analysis of OS in the basal-like carcinoma group: OS rates between the high-risk group and low-risk group showed statistically significant differences using the log-rank test (P=0.003); (B) the ROC curve AUC was 0.755. (C) Kaplan–Meier analysis of OS in the luminal carcinoma group: OS rates between the high-risk group and low-risk group showed statistically significant differences using the log-rank test (P<0.0001); (D) the ROC curve AUC was 0.688. (E, F) Kaplan–Meier analysis of OS in the Her2-enriched subgroup showed no significant differences between the high-risk group and low-risk group; the AUC and P-value were 0.545 and 0.5532, respectively. This signature showed better performance in basal-like and luminal patients than in Her2-enriched patients.
Abbreviations: AUC, area under the curve; OS, overall survival; ROC, receiver operating characteristics.
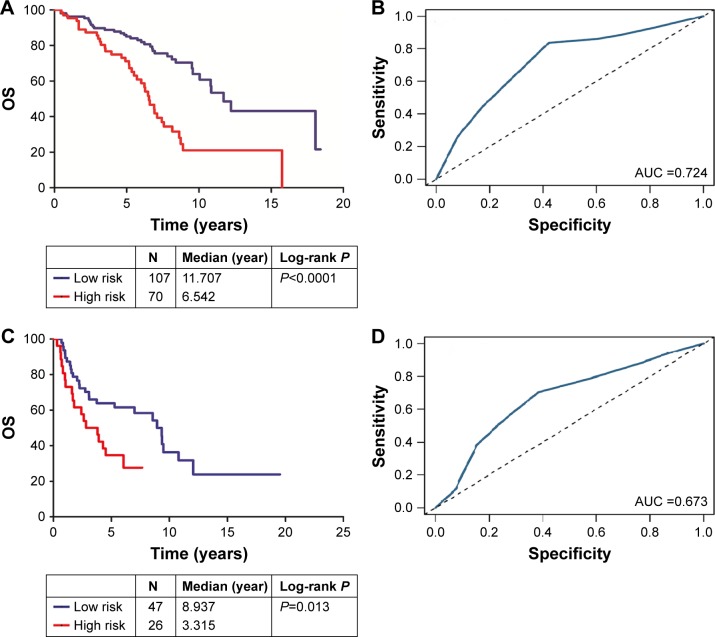
Finally, we analyzed the relationship between tumor stage and the microRNA signature. In the American Joint Committee on Cancer (AJCC) stage I and II group, the AUC and P-value were 0.724 and <0.0001, respectively (Figure 5A and B); in the stage III and IV group, the AUC and P-value were 0.673 and <0.013, respectively (Figure 5C and D). There was no significant difference between these two groups.
Figure 5.
(A) Kaplan–Meier analysis of OS in stage I and II groups: OS rates between the high-risk group and low-risk group showed statistically significant differences (P<0.0001); (B) the ROC curve AUC was 0.724. (C) Kaplan–Meier analysis of OS in stage III and IV groups: OS rates between the high-risk group and low-risk group showed statistically significant differences (P=0.0013); (D) the ROC curve AUC was 0.673. The performance of the signature was not associated with the AJCC stage of the patients.
Abbreviations: AJCC, American Joint Committee on Cancer; AUC, area under the curve; OS, overall survival; ROC, receiver operating characteristics.
Clinical and pathological features and microRNA signature
The clinical characteristics that were utilized to fit the univariate Cox model are shown in Table 3. In our study, age at diagnosis, ER status, PR status, Her2 status, and T stage were not associated with prognosis. N stage and metastasis had significant prognostic value, with P-values of 0.000 and 0.000, respectively. After adjustment for N stage and metastasis, hsa-mir-31, hsa-mir-16-2, and hsa-mir-484 were all still independent prognostic factors (Table 4).
Table 3.
Univariate Cox analysis of clinicopathological parameters
| Variables | n | HR | 95% CI | P-value |
|---|---|---|---|---|
| Age | 253 | 1.448 | 0.905–2.317 | 0.112 |
| ER | 243 | 0.719 | 0.481–1.076 | 0.108 |
| PR | 243 | 0.715 | 0.491–1.041 | 0.080 |
| Her2 | 232 | 1.165 | 0.693–1.958 | 0.565 |
| T stage | 250 | 1.106 | 0.892–1.371 | 0.361 |
| N stage | 251 | 1.471 | 1.205–1.795 | 0.000 |
| Metastasis | 238 | 3.260 | 1.787–5.948 | 0.000 |
Abbreviations: ER, estrogen receptor; Her2, human epidermal growth factor receptor 2; PR, progesterone receptor.
Table 4.
Multivariate Cox analysis of clinicopathological parameters and microRNAs
| Variables | n | HR | 95% CI | P-value |
|---|---|---|---|---|
| N stage | 237 | 1.355 | 1.080–1.702 | 0.009 |
| Metastasis | 237 | 1.845 | 0.870–3.914 | 0.110 |
| hsa-mir-16-2 | 237 | 0.556 | 0.379–0.817 | 0.003 |
| hsa-mir-484 | 237 | 1.560 | 1.043–2.332 | 0.030 |
| hsa-mir-31 | 237 | 0.486 | 0.333–0.711 | 0.000 |
| hsa-mir-877 | 237 | 1.476 | 0.968–2.251 | 0.071 |
| hsa-mir-937 | 237 | 1.223 | 0.815–1.837 | 0.331 |
| hsa-let-7b | 237 | 0.670 | 0.437–1.027 | 0.066 |
The correlation between patient clinicopathological characteristics and the microRNA signature is presented in Table 5. The microRNA signature was not associated with age at diagnosis, ER status, PR status, Her2 status, T stage, N stage or metastasis.
Table 5.
Correlation between microRNA expression level and clinical pathological parameters in breast cancer patients
| Parameters | Total (n) | MicroRNA score | P-value | |
|---|---|---|---|---|
| Low (n=120) | High (n=133) | |||
| Age, years | 0.791 | |||
| ≤45 | 53 | 26 (21.7) | 27 (20.3) | |
| >45 | 200 | 94 (78.3) | 106 (79.7) | |
| Missing (%) | 0 | |||
| ER (%) | 0.723 | |||
| Negative | 66 | 30 (45.5) | 36 (54.5) | |
| Positive | 177 | 85 (48.0) | 92 (52.0) | |
| Missing | 10 | |||
| PR (%) | 0.778 | |||
| Negative | 91 | 42 (46.2) | 49 (53.8) | |
| Positive | 152 | 73 (48.0) | 79 (52.0) | |
| Missing | 10 | |||
| Her2 (FISH) (%) | 0.343 | |||
| Negative | 197 | 96 (48.7) | 101 (51.3) | |
| Positive | 35 | 14 (40.0) | 21 (60.0) | |
| Missing | 21 | |||
| T stage (%) | 0.177 | |||
| T1 | 65 | 29 (44.6) | 36 (55.4) | |
| T2 | 133 | 59 (44.4) | 74 (55.6) | |
| T3 | 38 | 22 (57.9) | 16 (42.1) | |
| T4 | 14 | 8 (57.1) | 6 (42.9) | |
| Missing | 3 | |||
| Nodal stage (%) | 0.564 | |||
| N0 | 114 | 53 (46.5) | 61 (53.5) | |
| N1 | 91 | 49 (53.8) | 42 (46.2) | |
| N2 | 32 | 13 (40.6) | 19 (59.4) | |
| N3 | 14 | 5 (35.7) | 9 (64.3) | |
| Missing | 2 | |||
| Metastasis (%) | 0.947 | |||
| M0 | 225 | 106 (47.1) | 119 (52.9) | |
| M1 | 13 | 6 (46.2) | 7 (53.8) | |
| Missing | 15 | |||
Abbreviations: ER, estrogen receptor; FISH, fluorescence in situ hybridization; Her2, human epidermal growth factor receptor 2; PR, progesterone receptor.
GO annotation and KEGG pathway analysis of hsa-mir-31, hsa-mir-16-2, and hsa-mir-484
Target genes of hsa-mir-16-2, hsa-mir-31, and hsa-mir-484, as predicted by five programs, are listed in Table 6. There were 254, 149, and 336 target genes predicted by at least three programs for hsa-mir-16-2, hsa-mir-31, and hsa-mir-484, respectively. GO annotation analysis included biological processes, cellular components, and molecular function, as shown in Table 7 (fold enrichment >1.5, P<0.05). These results indicate that these candidate targets are significantly related to biosynthesis, metabolic processes, DNA binding, and system development. Furthermore, they could be protein complex or transcription factor complex components. KEGG and PANTHER analyses indicate that the candidate targets were significantly enriched in several oncogenic signaling pathways, including Hippo (P=0.0025), Wnt (P=0.000852), epidermal growth factor (EGF) receptor (P=0.00712), fibroblast growth factor (FGF) (P=0.000458), angiogenesis (P=0.003092), adherens junction (P=0.003865), and cytokine–cytokine receptor interaction (P=0.001133), as shown in Table 8. The three microRNAs are related to breast cancer cell cycle, viability, and apoptosis in vitro.
Table 6.
Target genes of three microRNAs
| MicroRNA | Target gene | Annotation |
|---|---|---|
| hsa-mir-31 | NR2C2 | nuclear receptor subfamily 2 group C member 2 |
| hsa-mir-31 | MLXIP | MLX interacting protein |
| hsa-mir-31 | STAU2 | staufen double-stranded RNA binding protein 2 |
| hsa-mir-31 | ATF7IP | activating transcription factor 7 interacting protein |
| hsa-mir-31 | PRKAA2 | protein kinase AMP-activated catalytic subunit alpha 2 |
| hsa-mir-31 | ZNF16 | zinc finger protein 16 |
| hsa-mir-31 | RHBDL3 | rhomboid like 3 |
| hsa-mir-31 | GPRC5A | G protein-coupled receptor class C group 5 member A |
| hsa-mir-31 | ARID1A | AT-rich interaction domain 1A |
| hsa-mir-31 | KHDRBS3 | KH RNA binding domain containing, signal transduction associated 3 |
| hsa-mir-31 | UCN2 | urocortin 2 |
| hsa-mir-31 | CTNND2 | catenin delta 2 |
| hsa-mir-31 | KLF13 | Kruppel like factor 13 |
| hsa-mir-31 | IQSEC2 | IQ motif and Sec7 domain 2 |
| hsa-mir-31 | RAB6B | RAB6B, member RAS oncogene family |
| hsa-mir-31 | TFRC | transferrin receptor |
| hsa-mir-31 | SLC24A3 | solute carrier family 24 member 3 |
| hsa-mir-31 | KCNN3 | potassium calcium-activated channel subfamily N member 3 |
| hsa-mir-31 | APBB2 | amyloid beta precursor protein binding family B member 2 |
| hsa-mir-31 | TACC2 | transforming acidic coiled-coil containing protein 2 |
| hsa-mir-31 | NDRG3 | NDRG family member 3 |
| hsa-mir-31 | DICER1 | dicer 1, ribonuclease III |
| hsa-mir-31 | SPRED1 | sprouty related EVH1 domain containing 1 |
| hsa-mir-31 | NFAT5 | nuclear factor of activated T-cells 5 |
| hsa-mir-31 | BAHD1 | bromo adjacent homology domain containing 1 |
| hsa-mir-31 | RTL9 | retrotransposon Gag like 9 |
| hsa-mir-31 | KLF7 | Kruppel like factor 7 |
| hsa-mir-31 | PRSS8 | protease, serine 8 |
| hsa-mir-31 | PIK3C2A | phosphatidylinositol-4-phosphate 3-kinase catalytic subunit type 2 alpha |
| hsa-mir-31 | FNDC5 | fibronectin type III domain containing 5 |
| hsa-mir-31 | ZNHIT6 | zinc finger HIT-type containing 6 |
| hsa-mir-31 | BTBD11 | BTB domain containing 11 |
| hsa-mir-31 | PHF8 | PHD finger protein 8 |
| hsa-mir-31 | ZNF662 | zinc finger protein 662 |
| hsa-mir-31 | TMPRSS11F | transmembrane protease, serine 11F |
| hsa-mir-31 | CCNC | cyclin C |
| hsa-mir-31 | FZD4 | frizzled class receptor 4 |
| hsa-mir-31 | SATB2 | SATB homeobox 2 |
| hsa-mir-31 | SLC43A2 | solute carrier family 43 member 2 |
| hsa-mir-31 | RSF1 | remodeling and spacing factor 1 |
| hsa-mir-31 | RAP2B | RAP2B, member of RAS oncogene family |
| hsa-mir-31 | FMNL3 | formin like 3 |
| hsa-mir-31 | TM9SF4 | transmembrane 9 superfamily member 4 |
| hsa-mir-31 | PPP1R12B | protein phosphatase 1 regulatory subunit 12B |
| hsa-mir-31 | SLC39A14 | solute carrier family 39 member 14 |
| hsa-mir-31 | AKAP7 | A-kinase anchoring protein 7 |
| hsa-mir-31 | HOXC13 | homeobox C13 |
| hsa-mir-31 | RAB14 | RAB14, member RAS oncogene family |
| hsa-mir-31 | PPBP | pro-platelet basic protein |
| hsa-mir-31 | KIAA1429 | KIAA1429 |
| hsa-mir-31 | KRT6C | keratin 6C |
| hsa-mir-31 | FTMT | ferritin mitochondrial |
| hsa-mir-31 | IGSF11 | immunoglobulin superfamily member 11 |
| hsa-mir-31 | RSBN1 | round spermatid basic protein 1 |
| hsa-mir-31 | SEPHS1 | selenophosphate synthetase 1 |
| hsa-mir-31 | PDZD2 | PDZ domain containing 2 |
| hsa-mir-31 | TBXA2R | thromboxane A2 receptor |
| hsa-mir-31 | LBH | limb bud and heart development |
| hsa-mir-31 | PRKCE | protein kinase C epsilon |
| hsa-mir-31 | SH2D1A | SH2 domain containing 1A |
| hsa-mir-31 | GXYLT1 | glucoside xylosyltransferase 1 |
| hsa-mir-31 | LATS2 | large tumor suppressor kinase 2 |
| hsa-mir-31 | CAMK2D | calcium/calmodulin dependent protein kinase II delta |
| hsa-mir-31 | SYDE2 | synapse defective Rho GTPase homolog 2 |
| hsa-mir-31 | KIAA1024 | KIAA1024 |
| hsa-mir-31 | ELAVL1 | ELAV like RNA binding protein 1 |
| hsa-mir-31 | DCBLD2 | discoidin, CUB and LCCL domain containing 2 |
| hsa-mir-31 | MAP4K5 | mitogen-activated protein kinase kinase kinase kinase 5 |
| hsa-mir-31 | RGS4 | regulator of G protein signaling 4 |
| hsa-mir-31 | MAP1B | microtubule associated protein 1B |
| hsa-mir-31 | PPP1R9A | protein phosphatase 1 regulatory subunit 9A |
| hsa-mir-31 | PAX9 | paired box 9 |
| hsa-mir-31 | KANK1 | KN motif and ankyrin repeat domains 1 |
| hsa-mir-31 | WNK1 | WNK lysine deficient protein kinase 1 |
| hsa-mir-31 | WDR5 | WD repeat domain 5 |
| hsa-mir-31 | SLC1A2 | solute carrier family 1 member 2 |
| hsa-mir-31 | INSC | inscuteable homolog (Drosophila) |
| hsa-mir-31 | NUP153 | nucleoporin 153 |
| hsa-mir-31 | MBOAT2 | membrane bound O-acyltransferase domain containing 2 |
| hsa-mir-31 | RNF144A | ring finger protein 144A |
| hsa-mir-31 | MYO5A | myosin VA |
| hsa-mir-31 | VPS26B | VPS26, retromer complex component B |
| hsa-mir-31 | TNS1 | tensin 1 |
| hsa-mir-31 | NR5A2 | nuclear receptor subfamily 5 group A member 2 |
| hsa-mir-31 | SLC6A6 | solute carrier family 6 member 6 |
| hsa-mir-31 | PPP2R2A | protein phosphatase 2 regulatory subunit Balpha |
| hsa-mir-31 | MGAT1 | mannosyl (alpha-1,3-)-glycoprotein beta-1,2-N-acetylglucosaminyltransferase |
| hsa-mir-31 | RHOBTB1 | Rho related BTB domain containing 1 |
| hsa-mir-31 | IL34 | interleukin 34 |
| hsa-mir-31 | ZNF384 | zinc finger protein 384 |
| hsa-mir-31 | RASA1 | RAS p21 protein activator 1 |
| hsa-mir-31 | TMED10 | transmembrane p24 trafficking protein 10 |
| hsa-mir-31 | ZFP30 | ZFP30 zinc finger protein |
| hsa-mir-31 | PSMB11 | proteasome subunit beta 11 |
| hsa-mir-31 | VAV3 | vav guanine nucleotide exchange factor 3 |
| hsa-mir-31 | CRYBG3 | crystallin beta-gamma domain containing 3 |
| hsa-mir-31 | PEX5 | peroxisomal biogenesis factor 5 |
| hsa-mir-31 | RETREG1 | reticulophagy regulator 1 |
| hsa-mir-31 | PPP3CA | protein phosphatase 3 catalytic subunit alpha |
| hsa-mir-31 | NUMB | NUMB, endocytic adaptor protein |
| hsa-mir-31 | PC | pyruvate carboxylase |
| hsa-mir-31 | CEP85L | centrosomal protein 85 like |
| hsa-mir-31 | YWHAE | tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein epsilon |
| hsa-mir-31 | BACH2 | BTB domain and CNC homolog 2 |
| hsa-mir-31 | EIF5 | eukaryotic translation initiation factor 5 |
| hsa-mir-31 | VEZT | vezatin, adherens junctions transmembrane protein |
| hsa-mir-31 | TACC1 | transforming acidic coiled-coil containing protein 1 |
| hsa-mir-31 | UBE2K | ubiquitin conjugating enzyme E2 K |
| hsa-mir-31 | TM9SF3 | transmembrane 9 superfamily member 3 |
| hsa-mir-31 | SGMS1 | sphingomyelin synthase 1 |
| hsa-mir-31 | ARHGEF2 | Rho/Rac guanine nucleotide exchange factor 2 |
| hsa-mir-31 | COPS2 | COP9 signalosome subunit 2 |
| hsa-mir-31 | SPARC | secreted protein acidic and cysteine rich |
| hsa-mir-31 | CACNB2 | calcium voltage-gated channel auxiliary subunit beta 2 |
| hsa-mir-31 | ZSWIM6 | zinc finger SWIM-type containing 6 |
| hsa-mir-31 | CLCN3 | chloride voltage-gated channel 3 |
| hsa-mir-31 | AHCYL1 | adenosylhomocysteinase like 1 |
| hsa-mir-31 | JAZF1 | JAZF zinc finger 1 |
| hsa-mir-31 | RIMS3 | regulating synaptic membrane exocytosis 3 |
| hsa-mir-31 | TESK2 | testis-specific kinase 2 |
| hsa-mir-31 | HIF1AN | hypoxia inducible factor 1 alpha subunit inhibitor |
| hsa-mir-31 | KCTD20 | potassium channel tetramerization domain containing 20 |
| hsa-mir-31 | STX12 | syntaxin 12 |
| hsa-mir-31 | OXSR1 | oxidative stress responsive 1 |
| hsa-mir-31 | CLOCK | clock circadian regulator |
| hsa-mir-31 | EDNRB | endothelin receptor type B |
| hsa-mir-31 | ATF6 | activating transcription factor 6 |
| hsa-mir-31 | VAPB | VAMP associated protein B and C |
| hsa-mir-31 | BICRA | BRD4 interacting chromatin remodeling complex associated protein |
| hsa-mir-31 | VPS53 | VPS53, GARP complex subunit |
| hsa-mir-31 | MBNL3 | muscleblind like splicing regulator 3 |
| hsa-mir-31 | OSBP2 | oxysterol binding protein 2 |
| hsa-mir-31 | MFAP3 | microfibrillar associated protein 3 |
| hsa-mir-31 | CCNT1 | cyclin T1 |
| hsa-mir-31 | ATP8A1 | ATPase phospholipid transporting 8A1 |
| hsa-mir-31 | SIKE1 | suppressor of IKBKE 1 |
| hsa-mir-31 | HERPUD2 | HERPUD family member 2 |
| hsa-mir-31 | PTGFRN | prostaglandin F2 receptor inhibitor |
| hsa-mir-31 | EPC1 | enhancer of polycomb homolog 1 |
| hsa-mir-31 | GNA13 | G protein subunit alpha 13 |
| hsa-mir-31 | RPH3A | rabphilin 3A |
| hsa-mir-31 | MAP3K1 | mitogen-activated protein kinase kinase kinase 1 |
| hsa-mir-31 | CBL | Cbl proto-oncogene |
| hsa-mir-31 | JMJD8 | jumonji domain containing 8 |
| hsa-mir-31 | STK40 | serine/threonine kinase 40 |
| hsa-mir-31 | FZD3 | frizzled class receptor 3 |
| hsa-mir-31 | PPP6C | protein phosphatase 6 catalytic subunit |
| hsa-mir-31 | SUPT16H | SPT16 homolog, facilitates chromatin remodeling subunit |
| hsa-mir-31 | EBF3 | early B-cell factor 3 |
| hsa-mir-484 | PRR14L | proline rich 14 like |
| hsa-mir-484 | NFATC2 | nuclear factor of activated T-cells 2 |
| hsa-mir-484 | PTPRF | protein tyrosine phosphatase, receptor type F |
| hsa-mir-484 | HSPG2 | heparan sulfate proteoglycan 2 |
| hsa-mir-484 | RSPO4 | R-spondin 4 |
| hsa-mir-484 | PLCXD2 | phosphatidylinositol specific phospholipase C X domain containing 2 |
| hsa-mir-484 | AGAP2 | ArfGAP with GTPase domain, ankyrin repeat and PH domain 2 |
| hsa-mir-484 | DOLPP1 | dolichyldiphosphatase 1 |
| hsa-mir-484 | M6PR | mannose-6-phosphate receptor, cation dependent |
| hsa-mir-484 | CMPK1 | cytidine/uridine monophosphate kinase 1 |
| hsa-mir-484 | SLC46A3 | solute carrier family 46 member 3 |
| hsa-mir-484 | AP1G1 | adaptor related protein complex 1 gamma 1 subunit |
| hsa-mir-484 | TBC1D16 | TBC1 domain family member 16 |
| hsa-mir-484 | THUMPD2 | THUMP domain containing 2 |
| hsa-mir-484 | LDLRAD3 | low density lipoprotein receptor class A domain containing 3 |
| hsa-mir-484 | FARP1 | FERM, ARH/RhoGEF and pleckstrin domain protein 1 |
| hsa-mir-484 | PREB | prolactin regulatory element binding |
| hsa-mir-484 | DND1 | DND microRNA-mediated repression inhibitor 1 |
| hsa-mir-484 | ANAPC11 | anaphase promoting complex subunit 11 |
| hsa-mir-484 | SEC24C | SEC24 homolog C, COPII coat complex component |
| hsa-mir-484 | SLC1A4 | solute carrier family 1 member 4 |
| hsa-mir-484 | UPF3A | UPF3 regulator of nonsense transcripts homolog A (yeast) |
| hsa-mir-484 | TBL1X | transducin beta like 1X-linked |
| hsa-mir-484 | CDS1 | CDP-diacylglycerol synthase 1 |
| hsa-mir-484 | TAGLN2 | transgelin 2 |
| hsa-mir-484 | CD4 | CD4 molecule |
| hsa-mir-484 | HR | HR, lysine demethylase and nuclear receptor corepressor |
| hsa-mir-484 | RPL26 | ribosomal protein L26 |
| hsa-mir-484 | TNNI1 | troponin I1, slow skeletal type |
| hsa-mir-484 | IPO9 | importin 9 |
| hsa-mir-484 | COG2 | component of oligomeric golgi complex 2 |
| hsa-mir-484 | MAP10 | microtubule associated protein 10 |
| hsa-mir-484 | SPOCD1 | SPOC domain containing 1 |
| hsa-mir-484 | HIC2 | HIC ZBTB transcriptional repressor 2 |
| hsa-mir-484 | GUCD1 | guanylyl cyclase domain containing 1 |
| hsa-mir-484 | SGMS2 | sphingomyelin synthase 2 |
| hsa-mir-484 | MCTP1 | multiple C2 and transmembrane domain containing 1 |
| hsa-mir-484 | ST6GAL1 | ST6 beta-galactoside alpha-2,6-sialyltransferase 1 |
| hsa-mir-484 | UBR2 | ubiquitin protein ligase E3 component n-recognin 2 |
| hsa-mir-484 | NFIB | nuclear factor I B |
| hsa-mir-484 | YTHDF3 | YTH N6-methyladenosine RNA binding protein 3 |
| hsa-mir-484 | USP2 | ubiquitin specific peptidase 2 |
| hsa-mir-484 | SEC31B | SEC31 homolog B, COPII coat complex component |
| hsa-mir-484 | SH3PXD2A | SH3 and PX domains 2A |
| hsa-mir-484 | SPTLC2 | serine palmitoyltransferase long chain base subunit 2 |
| hsa-mir-484 | GLG1 | golgi glycoprotein 1 |
| hsa-mir-484 | DCTN5 | dynactin subunit 5 |
| hsa-mir-484 | SHANK1 | SH3 and multiple ankyrin repeat domains 1 |
| hsa-mir-484 | S100PBP | S100P binding protein |
| hsa-mir-484 | AMPD2 | adenosine monophosphate deaminase 2 |
| hsa-mir-484 | NBPF14 | NBPF member 14 |
| hsa-mir-484 | DACH2 | dachshund family transcription factor 2 |
| hsa-mir-484 | ZNF341 | zinc finger protein 341 |
| hsa-mir-484 | VAPB | VAMP associated protein B and C |
| hsa-mir-484 | TRIOBP | TRIO and F-actin binding protein |
| hsa-mir-484 | CCR9 | C-C motif chemokine receptor 9 |
| hsa-mir-484 | TACR1 | tachykinin receptor 1 |
| hsa-mir-484 | DCBLD2 | discoidin, CUB and LCCL domain containing 2 |
| hsa-mir-484 | KALRN | kalirin, RhoGEF kinase |
| hsa-mir-484 | OGDH | oxoglutarate dehydrogenase |
| hsa-mir-484 | CYFIP2 | cytoplasmic FMR1 interacting protein 2 |
| hsa-mir-484 | CYP3A43 | cytochrome P450 family 3 subfamily A member 43 |
| hsa-mir-484 | TRPS1 | transcriptional repressor GATA binding 1 |
| hsa-mir-484 | DCHS1 | dachsous cadherin-related 1 |
| hsa-mir-484 | TARBP2 | TARBP2, RISC loading complex RNA binding subunit |
| hsa-mir-484 | NCAN | neurocan |
| hsa-mir-484 | SERPINF2 | serpin family F member 2 |
| hsa-mir-484 | EMC6 | ER membrane protein complex subunit 6 |
| hsa-mir-484 | THPO | thrombopoietin |
| hsa-mir-484 | TMEM184A | transmembrane protein 184A |
| hsa-mir-484 | TRMT10B | tRNA methyltransferase 10B |
| hsa-mir-484 | MLLT6 | MLLT6, PHD finger domain containing |
| hsa-mir-484 | ZBTB47 | zinc finger and BTB domain containing 47 |
| hsa-mir-484 | TCEANC2 | transcription elongation factor A N-terminal and central domain containing 2 |
| hsa-mir-484 | TEX261 | testis expressed 261 |
| hsa-mir-484 | CLOCK | clock circadian regulator |
| hsa-mir-484 | NR6A1 | nuclear receptor subfamily 6 group A member 1 |
| hsa-mir-484 | MPRIP | myosin phosphatase Rho interacting protein |
| hsa-mir-484 | TRIM66 | tripartite motif containing 66 |
| hsa-mir-484 | MLXIP | MLX interacting protein |
| hsa-mir-484 | EIF4G2 | eukaryotic translation initiation factor 4 gamma 2 |
| hsa-mir-484 | SERTAD1 | SERTA domain containing 1 |
| hsa-mir-484 | MBNL3 | muscleblind like splicing regulator 3 |
| hsa-mir-484 | NEUROD4 | neuronal differentiation 4 |
| hsa-mir-484 | DBNDD2 | dysbindin domain containing 2 |
| hsa-mir-484 | PAX5 | paired box 5 |
| hsa-mir-484 | IPO11 | importin 11 |
| hsa-mir-484 | RFC5 | replication factor C subunit 5 |
| hsa-mir-484 | GRB10 | growth factor receptor bound protein 10 |
| hsa-mir-484 | RNF40 | ring finger protein 40 |
| hsa-mir-484 | SORBS2 | sorbin and SH3 domain containing 2 |
| hsa-mir-484 | CYB561D1 | cytochrome b561 family member D1 |
| hsa-mir-484 | GAPVD1 | GTPase activating protein and VPS9 domains 1 |
| hsa-mir-484 | SLC41A3 | solute carrier family 41 member 3 |
| hsa-mir-484 | MAP2 | microtubule associated protein 2 |
| hsa-mir-484 | POU2AF1 | POU class 2 associating factor 1 |
| hsa-mir-484 | CREM | cAMP responsive element modulator |
| hsa-mir-484 | HHIPL2 | HHIP like 2 |
| hsa-mir-484 | NAGA | alpha-N-acetylgalactosaminidase |
| hsa-mir-484 | RTN3 | reticulon 3 |
| hsa-mir-484 | NPNT | nephronectin |
| hsa-mir-484 | IL6R | interleukin 6 receptor |
| hsa-mir-484 | RFFL | ring finger and FYVE like domain containing E3 ubiquitin protein ligase |
| hsa-mir-484 | SLC25A45 | solute carrier family 25 member 45 |
| hsa-mir-484 | WASF3 | WAS protein family member 3 |
| hsa-mir-484 | OPN4 | opsin 4 |
| hsa-mir-484 | FAM46B | family with sequence similarity 46 member B |
| hsa-mir-484 | DBNL | drebrin like |
| hsa-mir-484 | ADD2 | adducin 2 |
| hsa-mir-484 | DPYSL3 | dihydropyrimidinase like 3 |
| hsa-mir-484 | VTI1A | vesicle transport through interaction with t-SNAREs 1A |
| hsa-mir-484 | CENPB | centromere protein B |
| hsa-mir-484 | LRRC32 | leucine rich repeat containing 32 |
| hsa-mir-484 | TOX4 | TOX high mobility group box family member 4 |
| hsa-mir-484 | SNRNP200 | small nuclear ribonucleoprotein U5 subunit 200 |
| hsa-mir-484 | PHF19 | PHD finger protein 19 |
| hsa-mir-484 | FBXO31 | F-box protein 31 |
| hsa-mir-484 | IL18BP | interleukin 18 binding protein |
| hsa-mir-484 | SEMA4F | ssemaphorin 4F |
| hsa-mir-484 | GTDC1 | glycosyltransferase like domain containing 1 |
| hsa-mir-484 | COLQ | collagen like tail subunit of asymmetric acetylcholinesterase |
| hsa-mir-484 | PRM1 | protamine 1 |
| hsa-mir-484 | LMAN2L | lectin, mannose binding 2 like |
| hsa-mir-484 | LPL | lipoprotein lipase |
| hsa-mir-484 | WWC1 | WW and C2 domain containing 1 |
| hsa-mir-484 | MAP3K11 | mitogen-activated protein kinase kinase kinase 11 |
| hsa-mir-484 | ANGPT1 | angiopoietin 1 |
| hsa-mir-484 | ZNF37A | zinc finger protein 37A |
| hsa-mir-484 | SGSM2 | small G protein signaling modulator 2 |
| hsa-mir-484 | EMX1 | empty spiracles homeobox 1 |
| hsa-mir-484 | LENG9 | leukocyte receptor cluster member 9 |
| hsa-mir-484 | FBXO11 | F-box protein 11 |
| hsa-mir-484 | HNF1A | HNF1 homeobox A |
| hsa-mir-484 | SPATA2L | spermatogenesis associated 2 like |
| hsa-mir-484 | TXNRD3 | thioredoxin reductase 3 |
| hsa-mir-484 | CPSF4 | cleavage and polyadenylation specific factor 4 |
| hsa-mir-484 | NEO1 | neogenin 1 |
| hsa-mir-484 | TCF7 | transcription factor 7 (T-cell specific, HMG-box) |
| hsa-mir-484 | HOXA5 | homeobox A5 |
| hsa-mir-484 | MTF2 | metal response element binding transcription factor 2 |
| hsa-mir-484 | PIK3CD | phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit delta |
| hsa-mir-484 | NCOA2 | nuclear receptor coactivator 2 |
| hsa-mir-484 | RIN1 | Ras and Rab interactor 1 |
| hsa-mir-484 | TRIM71 | tripartite motif containing 71 |
| hsa-mir-484 | DDX31 | DEAD-box helicase 31 |
| hsa-mir-484 | ACBD5 | acyl-CoA binding domain containing 5 |
| hsa-mir-484 | ABR | active BCR-related |
| hsa-mir-484 | GPR63 | G protein-coupled receptor 63 |
| hsa-mir-484 | RARG | retinoic acid receptor gamma |
| hsa-mir-484 | YAP1 | Yes associated protein 1 |
| hsa-mir-484 | RANBP17 | RAN binding protein 17 |
| hsa-mir-484 | POLD4 | DNA polymerase delta 4, accessory subunit |
| hsa-mir-484 | FAM160B2 | family with sequence similarity 160 member B2 |
| hsa-mir-484 | LYSMD1 | LysM domain containing 1 |
| hsa-mir-484 | PPARD | peroxisome proliferator activated receptor delta |
| hsa-mir-484 | COL20A1 | collagen type XX alpha 1 chain |
| hsa-mir-484 | SCP2 | sterol carrier protein 2 |
| hsa-mir-484 | IL20RB | interleukin 20 receptor subunit beta |
| hsa-mir-484 | TMC8 | transmembrane channel like 8 |
| hsa-mir-484 | SOX5 | SRY-box 5 |
| hsa-mir-484 | MAPKAPK3 | mitogen-activated protein kinase-activated protein kinase 3 |
| hsa-mir-484 | ZNF667 | zinc finger protein 667 |
| hsa-mir-484 | GRAMD1C | GRAM domain containing 1C |
| hsa-mir-484 | CRTC2 | CREB regulated transcription coactivator 2 |
| hsa-mir-484 | SERF1B | small EDRK-rich factor 1B |
| hsa-mir-484 | FLVCR2 | feline leukemia virus subgroup C cellular receptor family member 2 |
| hsa-mir-484 | TRIM74 | tripartite motif containing 74 |
| hsa-mir-484 | STAG3L3 | stromal antigen 3-like 3 (pseudogene) |
| hsa-mir-484 | PKNOX1 | PBX/knotted 1 homeobox 1 |
| hsa-mir-484 | SOX21 | SRY-box 21 |
| hsa-mir-484 | GLDN | gliomedin |
| hsa-mir-484 | HOXC8 | homeobox C8 |
| hsa-mir-484 | FFAR2 | free fatty acid receptor 2 |
| hsa-mir-484 | SH2D1B | SH2 domain containing 1B |
| hsa-mir-484 | KDM4A | lysine demethylase 4A |
| hsa-mir-484 | BCL7B | BCL tumor suppressor 7B |
| hsa-mir-484 | PCDH19 | protocadherin 19 |
| hsa-mir-484 | SERF1A | small EDRK-rich factor 1A |
| hsa-mir-484 | EIF3J | eukaryotic translation initiation factor 3 subunit J |
| hsa-mir-484 | NGRN | neugrin, neurite outgrowth associated |
| hsa-mir-484 | C3ORF62 | chromosome 3 open reading frame 62 |
| hsa-mir-484 | MYCBP2 | MYC binding protein 2, E3 ubiquitin protein ligase |
| hsa-mir-484 | PDE11A | phosphodiesterase 11A |
| hsa-mir-484 | AXIN2 | axin 2 |
| hsa-mir-484 | BRD9 | bromodomain containing 9 |
| hsa-mir-484 | CLCN4 | chloride voltage-gated channel 4 |
| hsa-mir-484 | FCF1 | FCF1 rRNA-processing protein |
| hsa-mir-484 | SUSD5 | sushi domain containing 5 |
| hsa-mir-484 | SP6 | Sp6 transcription factor |
| hsa-mir-484 | LAMB3 | laminin subunit beta 3 |
| hsa-mir-484 | MFRP | membrane frizzled-related protein |
| hsa-mir-484 | THRSP | thyroid hormone responsive |
| hsa-mir-484 | MED8 | mediator complex subunit 8 |
| hsa-mir-484 | CCDC142 | coiled-coil domain containing 142 |
| hsa-mir-484 | FOXH1 | forkhead box H1 |
| hsa-mir-484 | LGI4 | leucine rich repeat LGI family member 4 |
| hsa-mir-484 | CHD8 | chromodomain helicase DNA binding protein 8 |
| hsa-mir-484 | VLDLR | very low density lipoprotein receptor |
| hsa-mir-484 | PGGHG | protein-glucosylgalactosylhydroxylysine glucosidase |
| hsa-mir-484 | CSRNP1 | cysteine and serine rich nuclear protein 1 |
| hsa-mir-484 | N4BP2L2 | NEDD4 binding protein 2 like 2 |
| hsa-mir-484 | CYB5B | cytochrome b5 type B |
| hsa-mir-484 | PROM2 | prominin 2 |
| hsa-mir-484 | CNTFR | ciliary neurotrophic factor receptor |
| hsa-mir-484 | SEMA4D | semaphorin 4D |
| hsa-mir-484 | DOK4 | docking protein 4 |
| hsa-mir-484 | TOMM5 | translocase of outer mitochondrial membrane 5 |
| hsa-mir-484 | DKK2 | dickkopf WNT signaling pathway inhibitor 2 |
| hsa-mir-484 | DACH1 | dachshund family transcription factor 1 |
| hsa-mir-484 | CLEC6A | C-type lectin domain containing 6A |
| hsa-mir-484 | TTC39A | tetratricopeptide repeat domain 39A |
| hsa-mir-484 | TGFBRAP1 | transforming growth factor beta receptor associated protein 1 |
| hsa-mir-484 | VCP | valosin containing protein |
| hsa-mir-484 | F2RL3 | F2R like thrombin/trypsin receptor 3 |
| hsa-mir-484 | SNN | stannin |
| hsa-mir-484 | ARL15 | ADP ribosylation factor like GTPase 15 |
| hsa-mir-484 | CNKSR3 | CNKSR family member 3 |
| hsa-mir-484 | IGBP1 | immunoglobulin binding protein 1 |
| hsa-mir-484 | TINF2 | TERF1 interacting nuclear factor 2 |
| hsa-mir-484 | SMYD4 | SET and MYND domain containing 4 |
| hsa-mir-484 | ACVR1B | activin A receptor type 1B |
| hsa-mir-484 | IL21R | interleukin 21 receptor |
| hsa-mir-484 | DACT3 | disheveled binding antagonist of beta catenin 3 |
| hsa-mir-484 | PDGFA | platelet derived growth factor subunit A |
| hsa-mir-484 | NUP62 | nucleoporin 62 |
| hsa-mir-484 | TAF1L | TATA-box binding protein associated factor 1 like |
| hsa-mir-484 | CDH1 | cadherin 1 |
| hsa-mir-484 | MRFAP1L1 | Morf4 family associated protein 1 like 1 |
| hsa-mir-484 | NDUFA2 | NADH:ubiquinone oxidoreductase subunit A2 |
| hsa-mir-484 | CCNL1 | cyclin L1 |
| hsa-mir-484 | COL25A1 | collagen type XXV alpha 1 chain |
| hsa-mir-484 | HERC3 | HECT and RLD domain containing E3 ubiquitin protein ligase 3 |
| hsa-mir-484 | TRIM73 | tripartite motif containing 73 |
| hsa-mir-484 | C9ORF62 | chromosome 9 open reading frame 62 |
| hsa-mir-484 | SMUG1 | single-strand-selective monofunctional uracil-DNA glycosylase 1 |
| hsa-mir-484 | PYGO2 | pygopus family PHD finger 2 |
| hsa-mir-484 | PEX6 | peroxisomal biogenesis factor 6 |
| hsa-mir-484 | CTAGE1 | cutaneous T-cell lymphoma-associated antigen 1 |
| hsa-mir-484 | IGLON5 | IgLON family member 5 |
| hsa-mir-484 | ESR2 | estrogen receptor 2 |
| hsa-mir-484 | LIN28B | lin-28 homolog B |
| hsa-mir-484 | CTTNBP2NL | CTTNBP2 N-terminal like |
| hsa-mir-484 | GJD4 | gap junction protein delta 4 |
| hsa-mir-484 | SREBF2 | sterol regulatory element binding transcription factor 2 |
| hsa-mir-484 | TSTD2 | thiosulfate sulfurtransferase like domain containing 2 |
| hsa-mir-484 | GIGYF1 | GRB10 interacting GYF protein 1 |
| hsa-mir-484 | RETREG1 | reticulophagy regulator 1 |
| hsa-mir-484 | SLC6A1 | solute carrier family 6 member 1 |
| hsa-mir-484 | GTF3C4 | general transcription factor IIIC subunit 4 |
| hsa-mir-484 | TMIE | transmembrane inner ear |
| hsa-mir-484 | HIPK1 | homeodomain interacting protein kinase 1 |
| hsa-mir-484 | HIVEP2 | human immunodeficiency virus type I enhancer binding protein 2 |
| hsa-mir-484 | ANAPC7 | anaphase promoting complex subunit 7 |
| hsa-mir-484 | THBD | thrombomodulin |
| hsa-mir-484 | PTGER4 | prostaglandin E receptor 4 |
| hsa-mir-484 | HOXA11 | homeobox A11 |
| hsa-mir-484 | RHOBTB1 | Rho related BTB domain containing 1 |
| hsa-mir-484 | IFNAR1 | interferon alpha and beta receptor subunit 1 |
| hsa-mir-484 | JPT1 | Jupiter microtubule associated homolog 1 |
| hsa-mir-484 | FGF1 | fibroblast growth factor 1 |
| hsa-mir-484 | PTPRE | protein tyrosine phosphatase, receptor type E |
| hsa-mir-484 | DPYSL2 | dihydropyrimidinase like 2 |
| hsa-mir-484 | SORBS1 | sorbin and SH3 domain containing 1 |
| hsa-mir-484 | ZSWIM6 | zinc finger SWIM-type containing 6 |
| hsa-mir-484 | NUP54 | nucleoporin 54 |
| hsa-mir-484 | RIMS2 | regulating synaptic membrane exocytosis 2 |
| hsa-mir-484 | STEAP3 | STEAP3 metalloreductase |
| hsa-mir-484 | ABLIM2 | actin binding LIM protein family member 2 |
| hsa-mir-484 | TNRC6C | trinucleotide repeat containing 6C |
| hsa-mir-484 | TNFSF9 | TNF superfamily member 9 |
| hsa-mir-484 | PIKFYVE | phosphoinositide kinase, FYVE-type zinc finger containing |
| hsa-mir-484 | CPLX3 | complexin 3 |
| hsa-mir-484 | PEA15 | phosphoprotein enriched in astrocytes 15 |
| hsa-mir-484 | KIAA1549 | KIAA1549 |
| hsa-mir-484 | SLC20A2 | solute carrier family 20 member 2 |
| hsa-mir-484 | CDK9 | cyclin dependent kinase 9 |
| hsa-mir-484 | MAPKAPK2 | mitogen-activated protein kinase-activated protein kinase 2 |
| hsa-mir-484 | CSF1 | colony stimulating factor 1 |
| hsa-mir-484 | PITPNA | phosphatidylinositol transfer protein alpha |
| hsa-mir-484 | CSRNP2 | cysteine and serine rich nuclear protein 2 |
| hsa-mir-484 | NFATC4 | nuclear factor of activated T-cells 4 |
| hsa-mir-484 | AVL9 | AVL9 cell migration associated |
| hsa-mir-484 | POT1 | protection of telomeres 1 |
| hsa-mir-484 | HLA-DOB | major histocompatibility complex, class II, DO beta |
| hsa-mir-484 | DAG1 | dystroglycan 1 |
| hsa-mir-484 | STX5 | syntaxin 5 |
| hsa-mir-484 | PRPF4B | pre-mRNA processing factor 4B |
| hsa-mir-484 | STRN | striatin |
| hsa-mir-484 | CRTC3 | CREB regulated transcription coactivator 3 |
| hsa-mir-484 | B3GNT9 | UDP-GlcNAc:betaGal beta-1,3-N-acetylglucosaminyltransferase 9 |
| hsa-mir-484 | WFS1 | wolframin ER transmembrane glycoprotein |
| hsa-mir-484 | SLC17A9 | solute carrier family 17 member 9 |
| hsa-mir-484 | TRIM33 | tripartite motif containing 33 |
| hsa-mir-484 | KCNJ14 | potassium voltage-gated channel subfamily J member 14 |
| hsa-mir-484 | TSPAN17 | tetraspanin 17 |
| hsa-mir-484 | ELMO2 | engulfment and cell motility 2 |
| hsa-mir-484 | RAPGEF3 | Rap guanine nucleotide exchange factor 3 |
| hsa-mir-484 | GTPBP10 | GTP binding protein 10 |
| hsa-mir-484 | TSGA10 | testis specific 10 |
| hsa-mir-484 | ZFYVE1 | zinc finger FYVE-type containing 1 |
| hsa-mir-484 | ADAM33 | ADAM metallopeptidase domain 33 |
| hsa-mir-484 | MINK1 | misshapen like kinase 1 |
| hsa-mir-484 | NAF1 | nuclear assembly factor 1 ribonucleoprotein |
| hsa-mir-484 | VKORC1 | vitamin K epoxide reductase complex subunit 1 |
| hsa-mir-484 | TNR | tenascin R |
| hsa-mir-484 | PNRC1 | proline rich nuclear receptor coactivator 1 |
| hsa-mir-484 | PRRT2 | proline rich transmembrane protein 2 |
| hsa-mir-484 | SAMD4B | sterile alpha motif domain containing 4B |
| hsa-mir-484 | GOSR2 | golgi SNAP receptor complex member 2 |
| hsa-mir-484 | TMEM130 | transmembrane protein 130 |
| hsa-mir-484 | FAM71E2 | family with sequence similarity 71 member E2 |
| hsa-mir-484 | DCLK3 | doublecortin like kinase 3 |
| hsa-mir-484 | TMEM56 | transmembrane protein 56 |
| hsa-mir-484 | TRAT1 | T-cell receptor associated transmembrane adaptor 1 |
| hsa-mir-484 | ALPK3 | alpha kinase 3 |
| hsa-mir-484 | GRPEL2 | GrpE like 2, mitochondrial |
| hsa-mir-484 | RIPOR2 | RHO family interacting cell polarization regulator 2 |
| hsa-mir-484 | MAN1A2 | mannosidase alpha class 1A member 2 |
| hsa-mir-484 | STC1 | stanniocalcin 1 |
| hsa-mir-484 | ZMIZ1 | zinc finger MIZ-type containing 1 |
| hsa-mir-484 | TCHP | trichoplein keratin filament binding |
| hsa-mir-484 | BSDC1 | BSD domain containing 1 |
| hsa-mir-484 | TOX2 | TOX high mobility group box family member 2 |
| hsa-mir-484 | FLOT1 | flotillin 1 |
| hsa-mir-484 | GRM1 | glutamate metabotropic receptor 1 |
| hsa-mir-484 | BMP1 | bone morphogenetic protein 1 |
| hsa-mir-484 | WDR3 | WD repeat domain 3 |
| hsa-mir-484 | HK2 | hexokinase 2 |
| hsa-mir-484 | PCDHA9 | protocadherin alpha 9 |
| hsa-mir-484 | XKR9 | XK related 9 |
| hsa-mir-484 | CYB5RL | cytochrome b5 reductase like |
| hsa-mir-484 | SUSD2 | sushi domain containing 2 |
| hsa-mir-484 | RBM24 | RNA binding motif protein 24 |
| hsa-mir-484 | DLG2 | discs large MAGUK scaffold protein 2 |
| hsa-mir-484 | DENND5A | DENN domain containing 5A |
| hsa-mir-484 | SAP130 | Sin3A associated protein 130 |
| hsa-mir-16-2 | CCNB2 | cyclin B2 |
| hsa-mir-16-2 | C22ORF29 | chromosome 22 open reading frame 29 |
| hsa-mir-16-2 | CMTM7 | CKLF like MARVEL transmembrane domain containing 7 |
| hsa-mir-16-2 | PRKG1 | protein kinase, cGMP-dependent, type I |
| hsa-mir-16-2 | PTER | phosphotriesterase related |
| hsa-mir-16-2 | FAM49B | family with sequence similarity 49 member B |
| hsa-mir-16-2 | TSHZ1 | teashirt zinc finger homeobox 1 |
| hsa-mir-16-2 | KIAA2022 | KIAA2022 |
| hsa-mir-16-2 | PRDM15 | PR/SET domain 15 |
| hsa-mir-16-2 | KAT6A | lysine acetyltransferase 6A |
| hsa-mir-16-2 | KCTD15 | potassium channel tetramerization domain containing 15 |
| hsa-mir-16-2 | DIP2B | disco interacting protein 2 homolog B |
| hsa-mir-16-2 | NEGR1 | neuronal growth regulator 1 |
| hsa-mir-16-2 | ACTN1 | actinin alpha 1 |
| hsa-mir-16-2 | ZBTB44 | zinc finger and BTB domain containing 44 |
| hsa-mir-16-2 | ABTB2 | ankyrin repeat and BTB domain containing 2 |
| hsa-mir-16-2 | CNR1 | cannabinoid receptor 1 |
| hsa-mir-16-2 | PCDH11Y | protocadherin 11 Y-linked |
| hsa-mir-16-2 | RAB1A | RAB1A, member RAS oncogene family |
| hsa-mir-16-2 | RAB6B | RAB6B, member RAS oncogene family |
| hsa-mir-16-2 | FAM135A | family with sequence similarity 135 member A |
| hsa-mir-16-2 | ANKRD44 | ankyrin repeat domain 44 |
| hsa-mir-16-2 | CFL2 | cofilin 2 |
| hsa-mir-16-2 | PHLPP1 | PH domain and leucine rich repeat protein phosphatase 1 |
| hsa-mir-16-2 | STAG2 | stromal antigen 2 |
| hsa-mir-16-2 | LMNB1 | lamin B1 |
| hsa-mir-16-2 | SHANK2 | SH3 and multiple ankyrin repeat domains 2 |
| hsa-mir-16-2 | TANC2 | tetratricopeptide repeat, ankyrin repeat and coiled-coil containing 2 |
| hsa-mir-16-2 | MAP3K5 | mitogen-activated protein kinase kinase kinase 5 |
| hsa-mir-16-2 | ELOA | elongin A |
| hsa-mir-16-2 | SNRK | SNF related kinase |
| hsa-mir-16-2 | CLIC4 | chloride intracellular channel 4 |
| hsa-mir-16-2 | DGKB | diacylglycerol kinase beta |
| hsa-mir-16-2 | TENM1 | teneurin transmembrane protein 1 |
| hsa-mir-16-2 | AMOTL2 | angiomotin like 2 |
| hsa-mir-16-2 | PBRM1 | polybromo 1 |
| hsa-mir-16-2 | ANKRD12 | ankyrin repeat domain 12 |
| hsa-mir-16-2 | ZNF260 | zinc finger protein 260 |
| hsa-mir-16-2 | GLS | glutaminase |
| hsa-mir-16-2 | GRHL2 | grainyhead like transcription factor 2 |
| hsa-mir-16-2 | KDM2A | lysine demethylase 2A |
| hsa-mir-16-2 | GDPD1 | glycerophosphodiester phosphodiesterase domain containing 1 |
| hsa-mir-16-2 | PTPN12 | protein tyrosine phosphatase, non-receptor type 12 |
| hsa-mir-16-2 | SBNO1 | strawberry notch homolog 1 |
| hsa-mir-16-2 | MPPED2 | metallophosphoesterase domain containing 2 |
| hsa-mir-16-2 | IL13RA1 | interleukin 13 receptor subunit alpha 1 |
| hsa-mir-16-2 | CASP3 | caspase 3 |
| hsa-mir-16-2 | SYVN1 | synoviolin 1 |
| hsa-mir-16-2 | USP16 | ubiquitin specific peptidase 16 |
| hsa-mir-16-2 | FAM120C | family with sequence similarity 120C |
| hsa-mir-16-2 | TMBIM4 | transmembrane BAX inhibitor motif containing 4 |
| hsa-mir-16-2 | INTU | inturned planar cell polarity protein |
| hsa-mir-16-2 | RAB6A | RAB6A, member RAS oncogene family |
| hsa-mir-16-2 | PABPC4L | poly(A) binding protein cytoplasmic 4 like |
| hsa-mir-16-2 | CPEB2 | cytoplasmic polyadenylation element binding protein 2 |
| hsa-mir-16-2 | FAM126B | family with sequence similarity 126 member B |
| hsa-mir-16-2 | CNTN4 | contactin 4 |
| hsa-mir-16-2 | SEC24A | SEC24 homolog A, COPII coat complex component |
| hsa-mir-16-2 | TLK1 | tousled like kinase 1 |
| hsa-mir-16-2 | RNF6 | ring finger protein 6 |
| hsa-mir-16-2 | SPOPL | speckle type BTB/POZ protein like |
| hsa-mir-16-2 | RAD21 | RAD21 cohesin complex component |
| hsa-mir-16-2 | AMOTL1 | angiomotin like 1 |
| hsa-mir-16-2 | CHML | CHM like, Rab escort protein 2 |
| hsa-mir-16-2 | RAP1A | RAP1A, member of RAS oncogene family |
| hsa-mir-16-2 | CADM2 | cell adhesion molecule 2 |
| hsa-mir-16-2 | CDK17 | cyclin dependent kinase 17 |
| hsa-mir-16-2 | SGIP1 | SH3 domain GRB2 like endophilin interacting protein 1 |
| hsa-mir-16-2 | FRS2 | fibroblast growth factor receptor substrate 2 |
| hsa-mir-16-2 | HSPA5 | heat shock protein family A (Hsp70) member 5 |
| hsa-mir-16-2 | PAPD7 | poly(A) RNA polymerase D7, non-canonical |
| hsa-mir-16-2 | TSHZ3 | teashirt zinc finger homeobox 3 |
| hsa-mir-16-2 | PLAGL1 | PLAG1 like zinc finger 1 |
| hsa-mir-16-2 | ACER3 | alkaline ceramidase 3 |
| hsa-mir-16-2 | RCN2 | reticulocalbin 2 |
| hsa-mir-16-2 | CYP26B1 | cytochrome P450 family 26 subfamily B member 1 |
| hsa-mir-16-2 | BTG3 | BTG anti-proliferation factor 3 |
| hsa-mir-16-2 | ZNF770 | zinc finger protein 770 |
| hsa-mir-16-2 | AEBP2 | AE binding protein 2 |
| hsa-mir-16-2 | HNRNPLL | heterogeneous nuclear ribonucleoprotein L like |
| hsa-mir-16-2 | FMNL2 | formin like 2 |
| hsa-mir-16-2 | SP3 | Sp3 transcription factor |
| hsa-mir-16-2 | FGL2 | fibrinogen like 2 |
| hsa-mir-16-2 | PTPN13 | protein tyrosine phosphatase, non-receptor type 13 |
| hsa-mir-16-2 | BCL11B | B-cell CLL/lymphoma 11B |
| hsa-mir-16-2 | LLGL1 | LLGL1, scribble cell polarity complex component |
| hsa-mir-16-2 | DPP10 | dipeptidyl peptidase like 10 |
| hsa-mir-16-2 | ZSWIM6 | zinc finger SWIM-type containing 6 |
| hsa-mir-16-2 | GRIA2 | glutamate ionotropic receptor AMPA type subunit 2 |
| hsa-mir-16-2 | GALNT1 | polypeptide N-acetylgalactosaminyltransferase 1 |
| hsa-mir-16-2 | PDE10A | phosphodiesterase 10A |
| hsa-mir-16-2 | HIF1A | hypoxia inducible factor 1 alpha subunit |
| hsa-mir-16-2 | PRRX1 | paired related homeobox 1 |
| hsa-mir-16-2 | DSTYK | dual serine/threonine and tyrosine protein kinase |
| hsa-mir-16-2 | KAT6B | lysine acetyltransferase 6B |
| hsa-mir-16-2 | PCGF3 | polycomb group ring finger 3 |
| hsa-mir-16-2 | EMB | embigin |
| hsa-mir-16-2 | TMLHE | trimethyllysine hydroxylase, epsilon |
| hsa-mir-16-2 | TMEM161B | transmembrane protein 161B |
| hsa-mir-16-2 | EIF1AX | eukaryotic translation initiation factor 1A, X-linked |
| hsa-mir-16-2 | ADCYAP1 | adenylate cyclase activating polypeptide 1 |
| hsa-mir-16-2 | NAT2 | N-acetyltransferase 2 |
| hsa-mir-16-2 | PEX5L | peroxisomal biogenesis factor 5 like |
| hsa-mir-16-2 | AGL | amylo-alpha-1, 6-glucosidase, 4-alpha-glucanotransferase |
| hsa-mir-16-2 | COL11A1 | collagen type XI alpha 1 chain |
| hsa-mir-16-2 | RBFOX1 | RNA binding protein, fox-1 homolog 1 |
| hsa-mir-16-2 | CAV2 | caveolin 2 |
| hsa-mir-16-2 | TDG | thymine DNA glycosylase |
| hsa-mir-16-2 | IYD | iodotyrosine deiodinase |
| hsa-mir-16-2 | FRK | fyn related Src family tyrosine kinase |
| hsa-mir-16-2 | CLOCK | clock circadian regulator |
| hsa-mir-16-2 | MEX3B | mex-3 RNA binding family member B |
| hsa-mir-16-2 | SATB1 | SATB homeobox 1 |
| hsa-mir-16-2 | DPY19L4 | dpy-19 like 4 (C. elegans) |
| hsa-mir-16-2 | ZNF254 | zinc finger protein 254 |
| hsa-mir-16-2 | CREB1 | cAMP responsive element binding protein 1 |
| hsa-mir-16-2 | ANKRD26 | ankyrin repeat domain 26 |
| hsa-mir-16-2 | VDAC1 | voltage dependent anion channel 1 |
| hsa-mir-16-2 | LRIG1 | leucine rich repeats and immunoglobulin like domains 1 |
| hsa-mir-16-2 | INPP1 | inositol polyphosphate-1-phosphatase |
| hsa-mir-16-2 | ZFP36 | ZFP36 ring finger protein |
| hsa-mir-16-2 | HORMAD1 | HORMA domain containing 1 |
| hsa-mir-16-2 | TBC1D12 | TBC1 domain family member 12 |
| hsa-mir-16-2 | C1ORF21 | chromosome 1 open reading frame 21 |
| hsa-mir-16-2 | PAIP2 | poly(A) binding protein interacting protein 2 |
| hsa-mir-16-2 | HNRNPUL2 | heterogeneous nuclear ribonucleoprotein U like 2 |
| hsa-mir-16-2 | STX12 | syntaxin 12 |
| hsa-mir-16-2 | RORA | RAR related orphan receptor A |
| hsa-mir-16-2 | TTC39B | tetratricopeptide repeat domain 39B |
| hsa-mir-16-2 | ARAP2 | ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 2 |
| hsa-mir-16-2 | IGSF11 | immunoglobulin superfamily member 11 |
| hsa-mir-16-2 | MTF2 | metal response element binding transcription factor 2 |
| hsa-mir-16-2 | CPEB3 | cytoplasmic polyadenylation element binding protein 3 |
| hsa-mir-16-2 | ZNF615 | zinc finger protein 615 |
| hsa-mir-16-2 | MIER3 | MIER family member 3 |
| hsa-mir-16-2 | AHCTF1 | AT-hook containing transcription factor 1 |
| hsa-mir-16-2 | ZNF280D | zinc finger protein 280D |
| hsa-mir-16-2 | UBE2V2 | ubiquitin conjugating enzyme E2 V2 |
| hsa-mir-16-2 | SCN2A | sodium voltage-gated channel alpha subunit 2 |
| hsa-mir-16-2 | PTAR1 | protein prenyltransferase alpha subunit repeat containing 1 |
| hsa-mir-16-2 | EYA4 | EYA transcriptional coactivator and phosphatase 4 |
| hsa-mir-16-2 | KRTAP4-5 | keratin associated protein 4–5 |
| hsa-mir-16-2 | LPAR1 | lysophosphatidic acid receptor 1 |
| hsa-mir-16-2 | TAOK3 | TAO kinase 3 |
| hsa-mir-16-2 | AFF2 | AF4/FMR2 family member 2 |
| hsa-mir-16-2 | NYAP2 | neuronal tyrosine-phosphorylated phosphoinositide-3-kinase adaptor 2 |
| hsa-mir-16-2 | DLL1 | delta like canonical Notch ligand 1 |
| hsa-mir-16-2 | RNF44 | ring finger protein 44 |
| hsa-mir-16-2 | SEPSECS | Sep (O-phosphoserine) tRNA:Sec (selenocysteine) tRNA synthase |
| hsa-mir-16-2 | CD226 | CD226 molecule |
| hsa-mir-16-2 | HAND2 | heart and neural crest derivatives expressed 2 |
| hsa-mir-16-2 | ST13 | ST13, Hsp70 interacting protein |
| hsa-mir-16-2 | ICK | intestinal cell kinase |
| hsa-mir-16-2 | ZNF117 | zinc finger protein 117 |
| hsa-mir-16-2 | OAZ1 | ornithine decarboxylase antizyme 1 |
| hsa-mir-16-2 | ATP11B | ATPase phospholipid transporting 11B (putative) |
| hsa-mir-16-2 | HSDL1 | hydroxysteroid dehydrogenase like 1 |
| hsa-mir-16-2 | MME | membrane metalloendopeptidase |
| hsa-mir-16-2 | PURA | purine rich element binding protein A |
| hsa-mir-16-2 | RGS4 | regulator of G protein signaling 4 |
| hsa-mir-16-2 | AUH | AU RNA binding methylglutaconyl-CoA hydratase |
| hsa-mir-16-2 | SOAT1 | sterol O-acyltransferase 1 |
| hsa-mir-16-2 | TBX18 | T-box 18 |
| hsa-mir-16-2 | HS6ST2 | heparan sulfate 6-O-sulfotransferase 2 |
| hsa-mir-16-2 | ZNF569 | zinc finger protein 569 |
| hsa-mir-16-2 | AZIN1 | antizyme inhibitor 1 |
| hsa-mir-16-2 | IRF6 | interferon regulatory factor 6 |
| hsa-mir-16-2 | RGS5 | regulator of G-protein signaling 5 |
| hsa-mir-16-2 | ANKIB1 | ankyrin repeat and IBR domain containing 1 |
| hsa-mir-16-2 | TPP2 | tripeptidyl peptidase 2 |
| hsa-mir-16-2 | SCARB2 | scavenger receptor class B member 2 |
| hsa-mir-16-2 | KIAA1107 | KIAA1107 |
| hsa-mir-16-2 | ZNF624 | zinc finger protein 624 |
| hsa-mir-16-2 | BLOC1S2 | biogenesis of lysosomal organelles complex 1 subunit 2 |
| hsa-mir-16-2 | CHIC1 | cysteine rich hydrophobic domain 1 |
| hsa-mir-16-2 | TUBB2B | tubulin beta 2B class IIb |
| hsa-mir-16-2 | ZNF681 | zinc finger protein 681 |
| hsa-mir-16-2 | ZNF236 | zinc finger protein 236 |
| hsa-mir-16-2 | B2M | beta-2-microglobulin |
| hsa-mir-16-2 | PRKAA1 | protein kinase AMP-activated catalytic subunit alpha 1 |
| hsa-mir-16-2 | CUL2 | cullin 2 |
| hsa-mir-16-2 | NAB1 | NGFI-A binding protein 1 |
| hsa-mir-16-2 | CAMK1D | calcium/calmodulin dependent protein kinase ID |
| hsa-mir-16-2 | SLC2A13 | solute carrier family 2 member 13 |
| hsa-mir-16-2 | FGF14 | fibroblast growth factor 14 |
| hsa-mir-16-2 | KL | klotho |
| hsa-mir-16-2 | HS2ST1 | heparan sulfate 2-O-sulfotransferase 1 |
| hsa-mir-16-2 | ARID2 | AT-rich interaction domain 2 |
| hsa-mir-16-2 | KIAA0408 | KIAA0408 |
| hsa-mir-16-2 | STRBP | spermatid perinuclear RNA binding protein |
| hsa-mir-16-2 | CLIP4 | CAP-Gly domain containing linker protein family member 4 |
| hsa-mir-16-2 | DSC3 | desmocollin 3 |
| hsa-mir-16-2 | SLC9C2 | solute carrier family 9 member C2 (putative) |
| hsa-mir-16-2 | RC3H1 | ring finger and CCCH-type domains 1 |
| hsa-mir-16-2 | ATF3 | activating transcription factor 3 |
| hsa-mir-16-2 | TAF5L | TATA-box binding protein associated factor 5 like |
| hsa-mir-16-2 | HNRNPR | heterogeneous nuclear ribonucleoprotein R |
| hsa-mir-16-2 | SSX2IP | SSX family member 2 interacting protein |
| hsa-mir-16-2 | RAI2 | retinoic acid induced 2 |
| hsa-mir-16-2 | RPS6KA3 | ribosomal protein S6 kinase A3 |
| hsa-mir-16-2 | CYBB | cytochrome b-245 beta chain |
| hsa-mir-16-2 | NKRF | NFKB repressing factor |
| hsa-mir-16-2 | ARHGEF6 | Rac/Cdc42 guanine nucleotide exchange factor 6 |
| hsa-mir-16-2 | ARFGEF2 | ADP ribosylation factor guanine nucleotide exchange factor 2 |
| hsa-mir-16-2 | USP25 | ubiquitin specific peptidase 25 |
| hsa-mir-16-2 | UBE2E2 | ubiquitin conjugating enzyme E2 E2 |
| hsa-mir-16-2 | UBP1 | upstream binding protein 1 (LBP-1a) |
| hsa-mir-16-2 | ZNF512 | zinc finger protein 512 |
| hsa-mir-16-2 | STRN | striatin |
| hsa-mir-16-2 | BCL11A | B-cell CLL/lymphoma 11A |
| hsa-mir-16-2 | MAP3K2 | mitogen-activated protein kinase kinase kinase 2 |
| hsa-mir-16-2 | GSTCD | glutathione S-transferase C-terminal domain containing |
| hsa-mir-16-2 | TRPC3 | transient receptor potential cation channel subfamily C member 3 |
| hsa-mir-16-2 | RAPGEF2 | Rap guanine nucleotide exchange factor 2 |
| hsa-mir-16-2 | CLCN3 | chloride voltage-gated channel 3 |
| hsa-mir-16-2 | CDH12 | cadherin 12 |
| hsa-mir-16-2 | DNAJC21 | DnaJ heat shock protein family (Hsp40) member C21 |
| hsa-mir-16-2 | SNX18 | sorting nexin 18 |
| hsa-mir-16-2 | ZBTB38 | zinc finger and BTB domain containing 38 |
| hsa-mir-16-2 | CCDC50 | coiled-coil domain containing 50 |
| hsa-mir-16-2 | RBPJ | recombination signal binding protein for immunoglobulin kappa J region |
| hsa-mir-16-2 | USP46 | ubiquitin specific peptidase 46 |
| hsa-mir-16-2 | MOB1B | MOB kinase activator 1B |
| hsa-mir-16-2 | PARM1 | prostate androgen-regulated mucin-like protein 1 |
| hsa-mir-16-2 | CNKSR3 | CNKSR family member 3 |
| hsa-mir-16-2 | CDK13 | cyclin dependent kinase 13 |
| hsa-mir-16-2 | PCDHA6 | protocadherin alpha 6 |
| hsa-mir-16-2 | PCDHAC1 | protocadherin alpha subfamily C, 1 |
| hsa-mir-16-2 | RBM27 | RNA binding motif protein 27 |
| hsa-mir-16-2 | USP49 | ubiquitin specific peptidase 49 |
| hsa-mir-16-2 | SAMD9L | sterile alpha motif domain containing 9 like |
| hsa-mir-16-2 | PEG10 | paternally expressed 10 |
| hsa-mir-16-2 | SMARCA2 | SWI/SNF related, matrix associated, actin dependent regulator of chromatin, subfamily a, member 2 |
| hsa-mir-16-2 | ZNF483 | zinc finger protein 483 |
| hsa-mir-16-2 | ASTN2 | astrotactin 2 |
| hsa-mir-16-2 | FOXP2 | forkhead box P2 |
| hsa-mir-16-2 | CALU | calumenin |
| hsa-mir-16-2 | NUP205 | nucleoporin 205 |
| hsa-mir-16-2 | TMEM178B | transmembrane protein 178B |
| hsa-mir-16-2 | DCAF4L2 | DDB1 and CUL4 associated factor 4 like 2 |
| hsa-mir-16-2 | FBXO32 | F-box protein 32 |
| hsa-mir-16-2 | KBTBD3 | kelch repeat and BTB domain containing 3 |
| hsa-mir-16-2 | MAB21L1 | mab-21 like 1 |
| hsa-mir-16-2 | RGCC | regulator of cell cycle |
| hsa-mir-16-2 | NALCN | sodium leak channel, non-selective |
| hsa-mir-16-2 | TEX30 | testis expressed 30 |
| hsa-mir-16-2 | RASSF8 | Ras association domain family member 8 |
| hsa-mir-16-2 | C12ORF66 | chromosome 12 open reading frame 66 |
| hsa-mir-16-2 | DYRK2 | dual specificity tyrosine phosphorylation regulated kinase 2 |
| hsa-mir-16-2 | TRPM7 | transient receptor potential cation channel subfamily M member 7 |
| hsa-mir-16-2 | WDR72 | WD repeat domain 72 |
| hsa-mir-16-2 | IREB2 | iron responsive element binding protein 2 |
| hsa-mir-16-2 | ZNF790 | zinc finger protein 790 |
| hsa-mir-16-2 | ZNF558 | zinc finger protein 558 |
Table 7.
Gene Ontology annotation analysis
| MicroRNA | Category | ID | Term | P-value | Fold enrichment |
|---|---|---|---|---|---|
| hsa-mir-16-2 | Biological process | GO:0032774 | RNA biosynthetic process | 3.31E-02 | 1.82 |
| GO:0010556 | Regulation of macromolecule biosynthetic process | 4.31E-05 | 1.77 | ||
| GO:2000112 | Regulation of cellular macromolecule biosynthetic process | 2.76E-04 | 1.74 | ||
| GO:0051252 | Regulation of RNA metabolic process | 8.69E-04 | 1.73 | ||
| GO:1903506 | Regulation of nucleic acid-templated transcription | 3.70E-03 | 1.72 | ||
| GO:2001141 | Regulation of RNA biosynthetic process | 4.09E-03 | 1.71 | ||
| GO:0006355 | Regulation of transcription, DNA-templated | 5.83E-03 | 1.71 | ||
| GO:0009889 | Regulation of biosynthetic process | 2.13E-04 | 1.7 | ||
| GO:0019219 | Regulation of nucleobase-containing compound metabolic process | 7.37E-04 | 1.69 | ||
| GO:0031326 | Regulation of cellular biosynthetic process | 4.32E-04 | 1.69 | ||
| GO:0048523 | Negative regulation of cellular process | 6.20E-04 | 1.68 | ||
| GO:0048519 | Negative regulation of biological process | 1.52E-03 | 1.61 | ||
| GO:0051171 | Regulation of nitrogen compound metabolic process | 1.02E-04 | 1.58 | ||
| GO:0060255 | Regulation of macromolecule metabolic process | 7.20E-05 | 1.57 | ||
| GO:0010468 | Regulation of gene expression | 2.39E-02 | 1.57 | ||
| GO:0080090 | Regulation of primary metabolic process | 1.22E-04 | 1.57 | ||
| GO:0031323 | Regulation of cellular metabolic process | 2.27E-04 | 1.55 | ||
| GO:0048856 | Anatomical structure development | 3.73E-02 | 1.51 | ||
| GO:0019222 | Regulation of metabolic process | 7.13E-04 | 1.5 | ||
| Cellular component | GO:0005634 | Nucleus | 4.54E-06 | 1.52 | |
| Molecular function | GO:0003700 | Transcription factor activity, sequence-specific DNA binding | 1.11E-02 | 2.29 | |
| GO:0001071 | Nucleic acid binding transcription factor activity | 1.13E-02 | 2.29 | ||
| GO:0003677 | DNA binding | 1.48E-02 | 1.83 | ||
| GO:0046872 | Metal ion binding | 1.88E-05 | 1.77 | ||
| GO:0043169 | Cation binding | 5.41E-05 | 1.73 | ||
| GO:0043167 | Ion binding | 5.96E-04 | 1.51 | ||
| hsa-mir-31 | Biological process | GO:0042325 | Regulation of phosphorylation | 4.93E-02 | 2.57 |
| GO:0031325 | Positive regulation of cellular metabolic process | 7.67E-03 | 2.07 | ||
| GO:0051173 | Positive regulation of nitrogen compound metabolic process | 4.79E-02 | 2.01 | ||
| GO:0009893 | Positive regulation of metabolic process | 2.07E-02 | 1.98 | ||
| GO:0048522 | Positive regulation of cellular process | 3.44E-05 | 1.93 | ||
| GO:0048518 | Positive regulation of biological process | 2.12E-06 | 1.92 | ||
| GO:0051171 | Regulation of nitrogen compound metabolic process | 5.65E-03 | 1.67 | ||
| GO:0060255 | Regulation of macromolecule metabolic process | 3.60E-03 | 1.67 | ||
| GO:0031323 | Regulation of cellular metabolic process | 4.80E-03 | 1.66 | ||
| GO:0080090 | Regulation of primary metabolic process | 7.25E-03 | 1.65 | ||
| GO:0019222 | Regulation of metabolic process | 4.29E-03 | 1.62 | ||
| hsa-mir-484 | Biological process | GO:0048666 | Neuron development | 3.68E-02 | 2.48 |
| GO:0010557 | Positive regulation of macromolecule biosynthetic process | 1.00E-03 | 2.08 | ||
| GO:0031328 | Positive regulation of cellular biosynthetic process | 2.42E-03 | 2 | ||
| GO:0010628 | Positive regulation of gene expression | 3.84E-03 | 1.99 | ||
| GO:0051254 | Positive regulation of RNA metabolic process | 4.90E-02 | 1.98 | ||
| GO:0009891 | Positive regulation of biosynthetic process | 4.15E-03 | 1.97 | ||
| GO:0045935 | Positive regulation of nucleobase-containing compound metabolic process | 9.25E-03 | 1.96 | ||
| GO:0010604 | Positive regulation of macromolecule metabolic process | 4.02E-05 | 1.84 | ||
| GO:0051173 | Positive regulation of nitrogen compound metabolic process | 4.58E-04 | 1.79 | ||
| GO:0031325 | Positive regulation of cellular metabolic process | 2.55E-04 | 1.79 | ||
| GO:0009893 | Positive regulation of metabolic process | 8.18E-05 | 1.78 | ||
| GO:0009892 | Negative regulation of metabolic process | 1.73E-03 | 1.77 | ||
| GO:0031324 | Negative regulation of cellular metabolic process | 8.96E-03 | 1.77 | ||
| GO:0010605 | Negative regulation of macromolecule metabolic process | 4.78E-02 | 1.71 | ||
| GO:0048869 | Cellular developmental process | 3.51E-02 | 1.57 | ||
| GO:0048731 | System development | 8.96E-03 | 1.55 | ||
| GO:0048523 | Negative regulation of cellular process | 5.89E-03 | 1.54 | ||
| GO:0048522 | Positive regulation of cellular process | 1.07E-03 | 1.54 | ||
| GO:0007275 | Multicellular organism development | 2.81E-03 | 1.53 | ||
| Cellular component | GO:0005667 | Transcription factor complex | 8.69E-03 | 3.49 | |
| GO:0043234 | Protein complex | 6.53E-04 | 1.68 | ||
| GO:0032991 | Macromolecular complex | 3.60E-05 | 1.56 | ||
| Molecular function | GO:0043565 | Sequence-specific DNA binding | 1.34E-02 | 2.23 |
Table 8.
KEGG and PANTHER analyses
| MicroRNA | Term | Database | ID | Input number | Background number | P-value |
|---|---|---|---|---|---|---|
| hsa-mir-16-2 | Circadian rhythm | KEGG pathway | hsa04710 | 4 | 30 | 0.000365555 |
| MAPK signaling pathway | KEGG pathway | hsa04010 | 8 | 257 | 0.005579127 | |
| Gap junction | KEGG pathway | hsa04540 | 4 | 88 | 0.014007203 | |
| ALS | KEGG pathway | hsa05014 | 3 | 51 | 0.017107832 | |
| Progesterone-mediated oocyte maturation | KEGG pathway | hsa04914 | 4 | 97 | 0.019095593 | |
| Glycosaminoglycan biosynthesis – heparan sulfate/heparin | KEGG pathway | hsa00534 | 2 | 25 | 0.029980769 | |
| Long-term potentiation | KEGG pathway | hsa04720 | 3 | 66 | 0.032404978 | |
| Renal cell carcinoma | KEGG pathway | hsa05211 | 3 | 69 | 0.036094866 | |
| Dorsoventral axis formation | KEGG pathway | hsa04320 | 2 | 28 | 0.036434645 | |
| Oocyte meiosis | KEGG pathway | hsa04114 | 4 | 120 | 0.036784673 | |
| Neurotrophin signaling pathway | KEGG pathway | hsa04722 | 4 | 122 | 0.038652503 | |
| Thyroid hormone synthesis | KEGG pathway | hsa04918 | 3 | 71 | 0.03866969 | |
| Antigen processing and presentation | KEGG pathway | hsa04612 | 3 | 71 | 0.03866969 | |
| RNA degradation | KEGG pathway | hsa03018 | 3 | 77 | 0.046937327 | |
| FAS signaling pathway | PANTHER | P00020 | 3 | 31 | 0.004779418 | |
| Integrin signaling pathway | PANTHER | P00034 | 6 | 166 | 0.008045548 | |
| Cadherin signaling pathway | PANTHER | P00012 | 5 | 154 | 0.022562161 | |
| FGF signaling pathway | PANTHER | P00021 | 4 | 103 | 0.023047641 | |
| Heterotrimeric G-protein signaling pathway – Gi alpha and Gs alpha-mediated pathway | PANTHER | P00026 | 5 | 157 | 0.02421771 | |
| Apoptosis signaling pathway | PANTHER | P00006 | 4 | 108 | 0.026693923 | |
| CCKR signaling map | PANTHER | P06959 | 5 | 176 | 0.036514088 | |
| hsa-mir-31 | Heterotrimeric G-protein signaling pathway-Gq alpha and Go alpha mediated pathway | PANTHER | P00027 | 4 | 121 | 0.037711913 |
| Hippo signaling pathway | KEGG pathway | hsa04390 | 5 | 153 | 0.00249984 | |
| Oxytocin signaling pathway | KEGG pathway | hsa04921 | 5 | 160 | 0.003013067 | |
| Melanogenesis | KEGG pathway | hsa04916 | 4 | 100 | 0.003392532 | |
| Sphingolipid signaling pathway | KEGG pathway | hsa04071 | 4 | 123 | 0.006872453 | |
| AMPK signaling pathway | KEGG pathway | hsa04152 | 4 | 125 | 0.007255462 | |
| Dopaminergic synapse | KEGG pathway | hsa04728 | 4 | 129 | 0.008063183 | |
| Proteoglycans in cancer | KEGG pathway | hsa05205 | 5 | 208 | 0.008756401 | |
| Ubiquitin-mediated proteolysis | KEGG pathway | hsa04120 | 4 | 137 | 0.009850899 | |
| Wnt signaling pathway | KEGG pathway | hsa04310 | 4 | 142 | 0.011089117 | |
| Circadian rhythm | KEGG pathway | hsa04710 | 2 | 30 | 0.015281252 | |
| cGMP-PKG signaling pathway | KEGG pathway | hsa04022 | 4 | 173 | 0.021002596 | |
| Axon guidance | KEGG pathway | hsa04360 | 4 | 178 | 0.022982044 | |
| Calcium signaling pathway | KEGG pathway | hsa04020 | 4 | 179 | 0.023391098 | |
| Glucagon signaling pathway | KEGG pathway | hsa04922 | 3 | 102 | 0.024384407 | |
| T-cell receptor signaling pathway | KEGG pathway | hsa04660 | 3 | 107 | 0.02748699 | |
| Insulin resistance | KEGG pathway | hsa04931 | 3 | 111 | 0.030113491 | |
| Oocyte meiosis | KEGG pathway | hsa04114 | 3 | 120 | 0.036489064 | |
| Neurotrophin signaling pathway | KEGG pathway | hsa04722 | 3 | 122 | 0.037992724 | |
| Vascular smooth muscle contraction | KEGG pathway | hsa04270 | 3 | 123 | 0.038756301 | |
| ALS | KEGG pathway | hsa05014 | 2 | 51 | 0.039184796 | |
| Natural killer cell-mediated cytotoxicity | KEGG pathway | hsa04650 | 3 | 130 | 0.044318798 | |
| Basal cell carcinoma | KEGG pathway | hsa05217 | 2 | 55 | 0.044700392 | |
| FGF signaling pathway | PANTHER | P00021 | 5 | 103 | 0.000457823 | |
| EGF receptor signaling pathway | PANTHER | P00018 | 5 | 114 | 0.000712265 | |
| Angiogenesis | PANTHER | P00005 | 5 | 161 | 0.003092145 | |
| Endothelin signaling pathway | PANTHER | P00019 | 3 | 79 | 0.012699626 | |
| T-cell activation | PANTHER | P00053 | 3 | 79 | 0.012699626 | |
| CCKR signaling map | PANTHER | P06959 | 4 | 176 | 0.022177133 | |
| Apoptosis signaling pathway | PANTHER | P00006 | 3 | 108 | 0.028131602 | |
| Alzheimer disease – presenilin pathway | PANTHER | P00004 | 3 | 112 | 0.030790107 | |
| Lonotropic glutamate receptor pathway | PANTHER | P00037 | 2 | 46 | 0.03269137 | |
| Wnt signaling pathway | PANTHER | P00057 | 5 | 295 | 0.032927779 | |
| Inflammation mediated by chemokine and cytokine signaling pathway | PANTHER | P00031 | 4 | 202 | 0.034033451 | |
| Oxytocin receptor mediated signaling pathway | PANTHER | P04391 | 2 | 55 | 0.044700392 | |
| Thyrotropin-releasing hormone receptor signaling pathway | PANTHER | P04394 | 2 | 57 | 0.047559619 | |
| hsa-mir-484 | Wnt signaling pathway | KEGG pathway | hsa04310 | 8 | 142 | 0.000852343 |
| HTLV-I infection | KEGG pathway | hsa05166 | 11 | 259 | 0.000948874 | |
| Cytokine–cytokine receptor interaction | KEGG pathway | hsa04060 | 11 | 265 | 0.001132924 | |
| Adherens junction | KEGG pathway | hsa04520 | 5 | 74 | 0.003864995 | |
| Hippo signaling pathway | KEGG pathway | hsa04390 | 7 | 153 | 0.005339916 | |
| Jak-STAT signaling pathway | KEGG pathway | hsa04630 | 7 | 160 | 0.006710057 | |
| Endometrial cancer | KEGG pathway | hsa05213 | 4 | 54 | 0.00710104 | |
| Hippo signaling pathway – multiple species | KEGG pathway | hsa04392 | 3 | 28 | 0.007685262 | |
| Axon guidance | KEGG pathway | hsa04360 | 7 | 178 | 0.011418848 | |
| VEGF signaling pathway | KEGG pathway | hsa04370 | 4 | 64 | 0.012312511 | |
| CAMs | KEGG pathway | hsa04514 | 6 | 143 | 0.014087791 | |
| SNARE interactions in vesicular transport | KEGG pathway | hsa04130 | 3 | 36 | 0.014469204 | |
| PPAR signaling pathway | KEGG pathway | hsa03320 | 4 | 73 | 0.018678443 | |
| Melanoma | KEGG pathway | hsa05218 | 4 | 73 | 0.018678443 | |
| PI3K-Akt signaling pathway | KEGG pathway | hsa04151 | 10 | 343 | 0.018921484 | |
| Protein processing in endoplasmic reticulum | KEGG pathway | hsa04141 | 6 | 167 | 0.027083189 | |
| ECM-receptor interaction | KEGG pathway | hsa04512 | 4 | 83 | 0.027778622 | |
| RNA transport | KEGG pathway | hsa03013 | 6 | 171 | 0.029828233 | |
| N-glycan biosynthesis | KEGG pathway | hsa00510 | 3 | 49 | 0.030909326 | |
| Hematopoietic cell lineage | KEGG pathway | hsa04640 | 4 | 86 | 0.030942 | |
| Mismatch repair | KEGG pathway | hsa03430 | 2 | 23 | 0.042292382 | |
| Insulin signaling pathway | KEGG pathway | hsa04910 | 5 | 141 | 0.043874311 | |
| Pathways in cancer | KEGG pathway | hsa05200 | 10 | 399 | 0.044847988 | |
| Acute myeloid leukemia | KEGG pathway | hsa05221 | 3 | 59 | 0.048121632 | |
| Angiogenesis | PANTHER | P00005 | 7 | 161 | 0.006925275 | |
| Wnt signaling pathway | PANTHER | P00057 | 10 | 295 | 0.007391165 | |
| Pyrimidine metabolism | PANTHER | P02771 | 2 | 10 | 0.01040345 | |
| Axon guidance mediated by netrin | PANTHER | P00009 | 3 | 32 | 0.010767727 | |
| Blood coagulation | PANTHER | P00011 | 3 | 38 | 0.016557181 | |
| Axon guidance mediated by semaphorins | PANTHER | P00007 | 2 | 19 | 0.030634121 |
Abbreviations: ALS, amyotrophic lateral sclerosis; AMPK, AMP-activated protein kinase; CAM, cell adhesion molecule; CCKR, cholecystokinin receptor; cGMP-PKG, cyclic guanosine monophosphate-dependent protein kinase G; ECM, extracellular matrix; EGF, epidermal growth factor; FAS, fatty acid synthase; FGF, fibroblast growth factor; HTLV-I, human T-cell lymphotropic virus I; Jak-STAT, janus kinase–STAT; KEGG, Kyoto Encyclopedia of Genes and Genomes; MAPK, mitogen-activated protein kinase; PPAR, peroxisome-proliferator-activated receptor; VEGF, vascular endothelial growth factor.
MDA-MB-231 cells were transfected according to the β-coefficient. One group was transfected with hsa-mir-484 inhibitor, hsa-mir-16-2 mimic, and hsa-mir-31 mimic (low-risk group), a second group was transfected with hsa-mir-484 mimic, hsa-mir-16-2 inhibitor, and hsa-mir-31 inhibitor (high-risk group), and a final group was transfected with control sequences (negative control group). Cell cycle flow cytometry showed that the cell counts of S and G2/M phase were increased in both high-risk and low-risk groups compared to the negative control group (Figure 6A–D). The CCK-8 assay showed that cell viability of the high-risk group was significantly increased compared to the control group, while the viability of the low-risk group was decreased (Figure 6E). We then used an apoptosis assay to confirm whether cell apoptosis was increased in the experimental groups. Our results revealed that the apoptosis rate was 11.07% in the high-risk group (Figure 6F) and 30.49% in low-risk group (Figure 6G), while it was 12.01% in the control group (Figure 6H).
Figure 6.
(A–D) Flow cytometry analysis of the cell cycle revealed that low-risk group cells were arrested at S and G2/M phase, while the cell cycle was activated in the high-risk group compared to the control group. (E) The cell viability of the high-risk group was significantly increased compared to the control group, while the viability of the low-risk group was decreased. (F–H) Flow cytometry analysis of apoptosis revealed that the apoptosis rate was 11.07% in the high-risk group, 30.49% in the low-risk group, and 12.01% in the control group.
Abbreviations: CCK-8, Cell-Counting Kit-8; FITC, fluorescein isothiocyanate; PI, propidium iodide.
Discussion
Accumulating evidence has shown that microRNA deregulation plays a pivotal role in multiple cellular and biological processes, including cell proliferation and cell apoptosis,16–19 and targets a variety of pathways as oncogenes or tumor suppressors. Recently, microRNA-based anticancer therapies have been explored, either alone or in combination with other therapies.20,21 However, only a few articles have constructed a microRNA scoring system to predict the outcome of breast carcinoma.22,23 Here, we built a three-microRNA signature (hsa-mir-31, hsa-mir-484, and hsa-mir-16-2) that proved powerful enough to be an independent prognostic factor after rounds of statistical analysis.
According to our analysis, all three microRNAs target many cancer-related pathways, including the MAPK signaling pathway,24 Hippo signaling pathway,25 EGF receptor signaling pathway,26 and Wnt signaling pathway;27 some of these have been confirmed by previous studies.28 To be specific, hsa-mir-484 was found to be associated with poor prognosis in patients receiving gemcitabine treatment for breast cancer or sunitinib treatment for metastatic renal cell carcinoma and in ovarian cancer patients demonstrating chemosensitivity.28–30 In addition, we found that circulating hsa-mir-484 is significantly differentially expressed, with decreased expression in the tumor tissue and increased expression in plasma compared to healthy volunteers.28,31–33 The microRNA hsa-mir-16-2 plays a tumor suppressor role by inducing cell cycle arrest, DNA damage repair, and apoptosis.33–35 Of the three microRNAs, hsa-mir-31 is the most studied. Previous studies show that hsa-mir-31 is a major contributor to breast cancer progression and metastasis by regulating metastasis-related genes, including RhoA, Radexin,36 WAVE3,37 RDX, SATB2,38,39 FOXP3,40 GNA13,41 and several integrin subunits,42 all involved in key steps in the invasion–metastasis cascade. In addition, hsa-mir-31 expression level is high in early-stage breast cancer tissues, diminishes as the tumor progresses to more advanced stages, and is even sometimes undetectable in metastatic tumors.36,37 Loss of hsa-mir-31 expression is accompanied by increased expression of its target genes, allowing the tumor to become more invasive and ultimately metastasize.37 In summary, these three microRNAs are involved in chemoresistance, cell cycle arrest, and metastasis, and therefore, they can theoretically predict the prognosis of breast cancer.
Of note, our analysis indicates that our prognostic signature performed especially well in young patients (age ≤45 years) with basal-like breast carcinoma. To our knowledge, triple-negative breast cancer is characterized by the lack of hormone receptors (ER and PR) and HER2 expression, a common basal-like subtype, and a high propensity for distant site metastases.43 Furthermore, effective targeted therapies beyond chemotherapy and radiotherapy are absent for triple-negative breast cancer, leading to poor clinical outcomes and a high mortality rate.44,45 These features make our signature even more valuable. We propose that high-risk patients, as determined by the calculations derived from our model, should be treated more aggressively and have a shorter follow-up interval.
Moreover, our experimental results also verified our signature. In the low-risk group, cell proliferative ability was inhibited, and S and G2/M phase cell counts were significantly increased, indicating that the cell cycle was arrested at the G2/M phase. In the high-risk group, cell proliferative ability was significantly increased combined with low cell counts in S and G2/M phase, indicating that the cells were proliferating rapidly. We also conducted an apoptosis assay in which the cell apoptosis rate was significantly increased in the low-risk group compared to the control group. Meanwhile, there was no significant difference between the high-risk group and the control group. This was not consistent with our prediction, and we propose that perhaps this signature could not significantly affect the apoptosis of breast cancer cells. Combined together, these results suggest that our signature was associated with the viability and cell cycle of breast cancer cells.
Limitations
We must acknowledge some limitations of our study. Since we excluded patients with insufficient data for analysis (such as RNA sequencing data, histological data, and follow-up data), there could be an influence of selection bias on our final results. Despite this, our microRNA signature demonstrated performance stability. As it is well accepted that microRNAs can be secreted and/or released to the local microenvironment and into the circulation,46 it may be possible to use blood or tissue samples to detect the expression level of these three microRNAs as a reference to guide the treatment of breast cancer patients.
Conclusion
We recommend more aggressive therapy and appropriate shorter follow-up intervals for patients in the high-risk group.
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Acknowledgments
This work was supported by the National Natural Science Foundation of China: Jie Ming (grant no. 81672611) and Hui Guo (grant no. 81602350).
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Parker JS, Mullins M, Cheang MC, et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J Clin Oncol. 2009;27(8):1160–1167. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Polyak K. Heterogeneity in breast cancer. J Clin Invest. 2011;121(10):3786–3788. doi: 10.1172/JCI60534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer. Annu Rev Pathol. 2014;9:287–314. doi: 10.1146/annurev-pathol-012513-104715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Abba ML, Patil N, Leupold JH, et al. MicroRNAs as novel targets and tools in cancer therapy. Cancer Lett. 2017;387:84–94. doi: 10.1016/j.canlet.2016.03.043. [DOI] [PubMed] [Google Scholar]
- 6.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rupaimoole R, Slack FJ. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat Rev Drug Discov. 2017;16(3):203–222. doi: 10.1038/nrd.2016.246. [DOI] [PubMed] [Google Scholar]
- 8.Mlcochova J, Faltejskova-Vychytilova P, Ferracin M, et al. MicroRNA expression profiling identifies miR-31-5p/3p as associated with time to progression in wild-type RAS metastatic colorectal cancer treated with cetuximab. Oncotarget. 2015;6(36):38695–38704. doi: 10.18632/oncotarget.5735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gandellini P, Giovannetti E, Nicassio F. MicroRNAs in cancer management: big challenges for small molecules. Biomed Res Int. 2015;2015:1–2. doi: 10.1155/2015/982156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maragkakis M, Vergoulis T, Alexiou P, et al. DIANA-microT web server upgrade supports fly and worm miRNA target prediction and bibliographic miRNA to disease association. Nucleic Acids Res. 2011;39(Web Server issue):W145–W148. doi: 10.1093/nar/gkr294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dweep H, Gretz N. miRWalk2.0: a comprehensive atlas of microRNA-target interactions. Nat Methods. 2015;12(8):697. doi: 10.1038/nmeth.3485. [DOI] [PubMed] [Google Scholar]
- 12.Wong N, Wang X. miRDB: an online resource for microRNA target prediction and functional annotations. Nucleic Acids Res. 2015;43(Database issue):D146–D152. doi: 10.1093/nar/gku1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene Lists using David bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 14.Mi H, Huang X, Muruganujan A, et al. Panther version 11: expanded annotation data from gene ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017;45(D1):D183–D189. doi: 10.1093/nar/gkw1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng H, Garrick DJ, Fernando RL. Efficient strategies for leave-one-out cross validation for genomic best linear unbiased prediction. J Anim Sci Biotechnol. 2017;8:38. doi: 10.1186/s40104-017-0164-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pan YZ, Morris ME, Yu AM. MicroRNA-328 negatively regulates the expression of breast cancer resistance protein (BCRP/ABCG2) in human cancer cells. Mol Pharmacol. 2009;75(6):1374–1379. doi: 10.1124/mol.108.054163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller TE, Ghoshal K, Ramaswamy B, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283(44):29897–29903. doi: 10.1074/jbc.M804612200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun F, Fu H, Liu Q, et al. Downregulation of CCND1 and CDK6 by miR-34a induces cell cycle arrest. FEBS Lett. 2008;582(10):1564–1568. doi: 10.1016/j.febslet.2008.03.057. [DOI] [PubMed] [Google Scholar]
- 19.Mei M, Ren Y, Zhou X, et al. Downregulation of miR-21 enhances chemotherapeutic effect of taxol in breast carcinoma cells. Technol Cancer Res Treat. 2010;9(1):77–86. doi: 10.1177/153303461000900109. [DOI] [PubMed] [Google Scholar]
- 20.Jain CK, Gupta A, Dogra N, Kumar VS, Wadhwa G, Sharma SK. MicroRNA therapeutics: the emerging anticancer strategies. Recent Pat Anticancer Drug Discov. 2014;9(3):286–296. doi: 10.2174/1574892809666140307101519. [DOI] [PubMed] [Google Scholar]
- 21.Dai X, Tan C. Combination of microRNA therapeutics with small-molecule anticancer drugs: mechanism of action and co-delivery nanocarriers. Adv Drug Deliv Rev. 2015;81:184–197. doi: 10.1016/j.addr.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 22.Volinia S, Croce CM. Prognostic microRNA/mRNA signature from the integrated analysis of patients with invasive breast cancer. Proc Natl Acad Sci U S A. 2013;110(18):7413–7417. doi: 10.1073/pnas.1304977110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Buffa FM, Camps C, Winchester L, et al. microRNA-associated progression pathways and potential therapeutic targets identified by integrated mRNA and microRNA expression profiling in breast cancer. Cancer Res. 2011;71(17):5635–5645. doi: 10.1158/0008-5472.CAN-11-0489. [DOI] [PubMed] [Google Scholar]
- 24.Sebolt-Leopold JS, Herrera R. Targeting the mitogen-activated protein kinase cascade to treat cancer. Nat Rev Cancer. 2004;4(12):937–947. doi: 10.1038/nrc1503. [DOI] [PubMed] [Google Scholar]
- 25.Johnson R, Halder G. The two faces of Hippo: targeting the Hippo pathway for regenerative medicine and cancer treatment. Nat Rev Drug Discov. 2014;13(1):63–79. doi: 10.1038/nrd4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ciardiello F, Tortora G. EGFR antagonists in cancer treatment. N Engl J Med. 2008;358(11):1160–1174. doi: 10.1056/NEJMra0707704. [DOI] [PubMed] [Google Scholar]
- 27.Reya T, Clevers H. Wnt signalling in stem cells and cancer. Nature. 2005;434(7035):843–850. doi: 10.1038/nature03319. [DOI] [PubMed] [Google Scholar]
- 28.Vecchione A, Belletti B, Lovat F, et al. A microRNA signature defines chemoresistance in ovarian cancer through modulation of angiogenesis. Proc Natl Acad Sci U S A. 2013;110(24):9845–9850. doi: 10.1073/pnas.1305472110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ye FG, Song CG, Cao ZG, et al. Cytidine deaminase axis modulated by miR-484 differentially regulates cell proliferation and chemoresistance in breast cancer. Cancer Res. 2015;75(7):1504–1515. doi: 10.1158/0008-5472.CAN-14-2341. [DOI] [PubMed] [Google Scholar]
- 30.Prior C, Perez-Gracia JL, Garcia-Donas J, et al. Identification of tissue microRNAs predictive of sunitinib activity in patients with metastatic renal cell carcinoma. PLoS One. 2014;9(1):e86263. doi: 10.1371/journal.pone.0086263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kjersem JB, Ikdahl T, Lingjaerde OC, Guren T, Tveit KM, Kure EH. Plasma microRNAs predicting clinical outcome in metastatic colorectal cancer patients receiving first-line oxaliplatin-based treatment. Mol Oncol. 2014;8(1):59–67. doi: 10.1016/j.molonc.2013.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hu Z, Dong J, Wang LE, et al. Serum microRNA profiling and breast cancer risk: the use of miR-484/191 as endogenous controls. Carcinogenesis. 2012;33(4):828–834. doi: 10.1093/carcin/bgs030. [DOI] [PubMed] [Google Scholar]
- 33.Lovat F, Fassan M, Gasparini P, et al. miR-15b/16-2 deletion promotes B-cell malignancies. Proc Natl Acad Sci U S A. 2015;112(37):11636–11641. doi: 10.1073/pnas.1514954112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sherr CJ. The Pezcoller Lecture: cancer cell cycles revisited. Cancer Res. 2000;60(14):3689–3695. [PubMed] [Google Scholar]
- 35.Rahman M, Lovat F, Romano G, et al. miR-15b/16-2 regulates factors that promote p53 phosphorylation and augments the DNA damage response following radiation in the lung. J Biol Chem. 2014;289(38):26406–26416. doi: 10.1074/jbc.M114.573592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valastyan S, Reinhardt F, Benaich N, et al. RETRACTED: a pleiotropically acting microRNA, miR-31, inhibits breast cancer metastasis. Cell. 2009;137(6):1032–1046. doi: 10.1016/j.cell.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Sossey-Alaoui K, Downs-Kelly E, Das M, Izem L, Tubbs R, Plow EF. WAVE3, an actin remodeling protein, is regulated by the metastasis suppressor microRNA, miR-31, during the invasion-metastasis cascade. Int J Cancer. 2011;129(6):1331–1343. doi: 10.1002/ijc.25793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.O’Day E, Lal A. MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res. 2010;12(2):201. doi: 10.1186/bcr2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aprelikova O, Yu X, Palla J, et al. The role of miR-31 and its target gene SATB2 in cancer-associated fibroblasts. Cell Cycle. 2010;9(21):4387–4398. doi: 10.4161/cc.9.21.13674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rouas R, Fayyad-Kazan H, El Zein N, et al. Human natural Treg microRNA signature: role of microRNA-31 and microRNA-21 in FOXP3 expression. Eur J Immunol. 2009;39(6):1608–1618. doi: 10.1002/eji.200838509. [DOI] [PubMed] [Google Scholar]
- 41.Rasheed SAK, Teo CR, Beillard EJ, et al. MicroRNA-31 controls G protein alpha-13 (GNA13) expression and cell invasion in breast cancer cells. Mol Cancer. 2015;14(1):67. doi: 10.1186/s12943-015-0337-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Augoff K, Das M, Bialkowska K, McCue B, Plow EF, Sossey-Alaoui K. miR-31 is a broad regulator of β1-integrin expression and function in cancer cells. Mol Cancer Res. 2011;9(11):1500–1508. doi: 10.1158/1541-7786.MCR-11-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vaz-Luis I, Ottesen RA, Hughes ME, et al. Outcomes by tumor subtype and treatment pattern in women with small, node-negative breast cancer: a multi-institutional study. J Clin Oncol. 2014;32(20):2142–2150. doi: 10.1200/JCO.2013.53.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Anders C, Carey LA. Understanding and treating triple-negative breast cancer. Oncology. 2008;22(11):1233–1239. 1239–1240, 1243. [PMC free article] [PubMed] [Google Scholar]
- 45.Carey L, Winer E, Viale G, Cameron D, Gianni L. Triple-negative breast cancer: disease entity or title of convenience? Nat Rev Clin Oncol. 2010;7(12):683–692. doi: 10.1038/nrclinonc.2010.154. [DOI] [PubMed] [Google Scholar]
- 46.Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids – the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011;8(8):467–477. doi: 10.1038/nrclinonc.2011.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.