Abstract
Redox-related plasma proteins are candidate reporters of protein signatures associated with endothelial structure/function. Thiol-proteins from protein disulfide isomerase (PDI) family are unexplored in this context. Here, we investigate the occurrence and physiological significance of a circulating pool of PDI in healthy humans. We validated an assay for detecting PDI in plasma of healthy individuals. Our results indicate high inter-individual (median = 330 pg/mL) but low intra-individual variability over time and repeated measurements. Remarkably, plasma PDI levels could discriminate between distinct plasma proteome signatures, with PDI-rich (>median) plasma differentially expressing proteins related to cell differentiation, protein processing, housekeeping functions and others, while PDI-poor plasma differentially displayed proteins associated with coagulation, inflammatory responses and immunoactivation. Platelet function was similar among individuals with PDI-rich vs. PDI-poor plasma. Remarkably, such protein signatures closely correlated with endothelial function and phenotype, since cultured endothelial cells incubated with PDI-poor or PDI-rich plasma recapitulated gene expression and secretome patterns in line with their corresponding plasma signatures. Furthermore, such signatures translated into functional responses, with PDI-poor plasma promoting impairment of endothelial adhesion to fibronectin and a disturbed pattern of wound-associated migration and recovery area. Patients with cardiovascular events had lower PDI levels vs. healthy individuals. This is the first study describing PDI levels as reporters of specific plasma proteome signatures directly promoting contrasting endothelial phenotypes and functional responses.
Keywords: Protein disulfide isomerase, Plasma proteome, Endothelial cells, Plasma protein signatures, Thiol proteins
Abbreviations: CM, conditioned medium; EC, endothelial cell; ELISA, enzyme-linked immunosorbent assay; ER, endoplasmic reticulum; ERp57, endoplasmic reticulum protein 57; ERp72, endoplasmic reticulum protein 72; FBS, fetal bovine serum; GSNO, S-nitrosoglutathione; GSSG, glutathione disulfide; HRG, histidine-rich glycoprotein; HUVEC, human umbilical vein endothelial cell line; MPB, 3-(N-maleimido-propionyl) biocytin; PDI, protein disulfide isomerase; pecPDI, peri/epicellular PDI; PRP, Platelet-rich plasma; TMB, tetramethylbenzidine
Graphical abstract
Highlights
-
•
A newly-validated plasma PDI assay shows high inter- but low intra-individual variability.
-
•
PDI-poor plasma carries protein signatures related to coagulation/immunoinflammation
-
•
PDI-rich plasma protein signatures relate to cell adhesion/housekeeping functions.
-
•
Endothelial exposure to such plasmas triggers corresponding gene and secretome patterns.
-
•
PDI-poor plasma impairs endothelial adhesion and migration vs. PDI-rich plasma.
1. Introduction
Redox processes play major integrative roles in plasma [1] and endothelial (patho)physiology. Redox-related plasma proteins are thus candidate reporters of vascular structure and function. In this context, thiol proteins from the protein disulfide isomerase (PDI) family remain unexplored [2]. The family prototype is PDIA1 (henceforth designated as PDI), a 55 kDa thiol oxidoreductase chaperone mainly from the endoplasmic reticulum (ER), which depicts a U-shaped structure comprising 4 thioredoxin tandem domains termed a-b-b'-a', plus a C-terminal domain. The a-type domains at each arm of the “U” contain catalytic active sites carrying Cys-Gly-His-Cys motifs, which enable PDI to undergo reduction-oxidation cycles [3,4]. The b-type domains at the bottom of the “U” are enriched in hydrophobic residues and involved in substrate binding. PDI also displays chaperone activity, per se thiol redox-independent, although enhanced upon oxidation [3,5]. Canonical PDI functions consist of introduction or isomerization of disulfide bonds in nascent ER proteins, as well as their retrograde cytoplasmic transport during ER-associated protein degradation [3]. Despite being primarily an ER protein, PDI has been reported at other intracellular locations and also at the cell surface and extracellular milieu (a pool we have termed peri/epicellular PDI = pecPDI) [6,7].
A pecPDI pool released from activated endothelial cells (EC) and platelets exerts critical roles in thrombus formation after vascular injury. PDI inhibition strongly inhibits both platelet accumulation and fibrin generation at the injury site [8]. The a′ rather than a-domain PDI cysteines are specifically involved in thrombosis and platelet activation [9]. In EC, pecPDI is estimated as <2% of cellular PDI and its externalization occurs without detectable plasma membrane damage via yet unclear secretion routes that are mostly Golgi-independent [10]. However, PDI extravasation substantially increases upon cell injury, given the very high intracellular expression of PDIs in general [10]. In EC, PDI is detectable in vesicles distinct from those containing Von Willebrand factor [[8], [11], [12]], while impairment of lysosome and dense granule biogenesis inhibits EC or platelet PDI secretion and enhances bleeding [12]. In platelets, PDI is located at the dense tubular system and likely exocytosed upon activation [13]. The roles of PDIs in thrombosis are at the forefront of translational advances in novel antithrombotic agents [8], with the list of new PDI inhibitors growing quite fast recently [[14], [15], [16]]. A multi-center efficacy clinical trial of isoquercetin as PDI inhibitor against cancer-associated thrombosis is currently under way [17].
Thrombosis-related extracellular PDI substrates have been actively investigated and include several integrins, which were well documented in platelets, EC and vessel wall [18]. The main effect of PDI (as well as ERp57 and ERp72) is to reduce integrin disulfide bonds to support transition from extended-moderate to extended-high affinity conformation [19,20], although our group reported pecPDI-mediated α5-integrin oxidation upon short-term EC mechanostimulation [10]. While PDI has no membrane-binding or transmembrane motifs, binding to integrins (e.g., β3) accounts for extracellular PDI retention [21]. In addition to integrins, kinetic trapping studies identified multiple PDI substrates in stimulated platelet-rich plasma, including vitronectin, complement factors 3 and 5, C4b-binding protein, histidine-rich glycoprotein (HRG), thrombospondin 1, coagulation factor V, serpin B6, glutaredoxin-1 [[22], [23], [24]]. Importantly, peculiar redox properties of PDI likely modulate the mode of PDI-substrate interaction. The slow reaction rate with hydrogen peroxide [25] indicates that PDI is unlikely to act as a mass redox sensor such as peroxiredoxins [7]. Also, the low redox potential of N-terminal PDI domain Cys may support PDI oxidative capacity over a larger range of ambient reducing potentials than, e.g., thioredoxin [[3], [26]]. Moreover, PDI supports protein complex organization, such as for NADPH oxidases [27]. These properties bring two implications. First, PDI likely promotes targeted rather than nonspecific redox modulation of its substrates [7]. Second, in the extracellular environment, in which PDI concentration is lower than that of its substrates - contrarily to the ER lumen - the amount of free PDI can be affected by type and binding capacity of substrates. Thus, PDI levels could potentially act as reporter of distinct proteome signatures in extracellular compartments such as plasma.
However, there is little information on circulating plasma levels of vascular thiol isomerases. One study reported a low-level pool of PDI in plasma from healthy humans [28] and some reports showed PDI release from platelets [29,30] or associated with EC and platelet microparticles [[31], [32], [33]]. In contrast, it has been recently suggested that extracellular PDI and other thiol isomerases are absent from the circulation as a way to suppress thrombus formation [22], while upon vascular wall injury PDI release from disrupted endothelium and bound platelets relieves such suppression and promotes thrombosis via disulfide modification of covalently-bound substrates. Here, we addressed the occurrence and physiological significance of a circulating pool of PDI in healthy humans, with a focus on its role as reporter of distinct plasma protein signatures involved in endothelial phenotype and functional responses potentially associated with disease risk.
2. Materials and methods
2.1. Study population and ethics approval
We selected 35 healthy volunteers with characteristics shown in Table 1. All participants had no prior history of chronic or acute illness including cardiovascular, digestive, metabolic or tumoral conditions. None of them made use of tobacco or other drugs, as well as chronic medications. The study protocol was approved by the Institutional Scientific/Ethics Committee (CAPPesq number 3334/09/085) and written informed consent was obtained from all participants prior to entering the study.
Table 1.
Baseline characteristics of healthy individual population.
| n | 35 |
|---|---|
| Age | 34.2 ± 1.6 |
| Males (%) | 40 |
| Females (%) | 60 |
| Total cholesterol, mg/dL | 186.5 ± 5.1 |
| HDL cholesterol, mg/dL | 58.3 ± 2.8 |
| LDL cholesterol, mg/dL | 111.3 ± 5.1 |
| Non-HDL cholesterol, mg/dL | 128.7 ± 5.6 |
| Triglycerides, mg/dL | 84.9 ± 8.2 |
| Whole blood platelet count (x109/L) | 263.6 ± 10.1 |
| Red blood cell count (x1012/L) | 4.7 ± 0.1 |
| White blood cell count (109/L) | 6.7 ± 0.3 |
| C-Reactive Protein, mg/L | 2.6 ± 0.8 |
| PDI levels, pg/mL | |
| Median | 330 |
| Mean | 539.4 |
| 25% percentile | 95 |
| 75% percentile | 918 |
Data represent mean ± SEM unless specified otherwise. HDL: High-density lipoprotein, LDL: Low-density lipoprotein.
2.2. Measurement of PDI by ELISA
Soluble PDI antigen was measured using the Human P4HB Pair Set enzyme-linked immunosorbent assay (ELISA) (SinoBiological–SEK10827, Lot numbers KW05NO2203 and KW09NO1102) according to the manufacturer's protocol with minor modifications. Briefly, 96-well microplates were coated with 100 μL/well of diluted capture antibody and incubated overnight at 4 °C, followed by addition of each sample (100 μL) for 2 h at room temperature (RT) with agitation. Thereafter, 100 μL of detection antibody conjugated to horseradish-peroxidase (HRP) was incubated for 2 h at room temperature (RT) with agitation. Plates were washed three times with phosphate buffered saline (PBS)-tween/20 (0.5%) after each incubation. Finally, 200 μL of tetramethylbenzidine (TMB) solution were added for 30 min with agitation. The optical density of each well was determined immediately using a microplate reader (SpectraMax-340, Molecular Devices) set to 450 nm. Values were determined according to a standard curve (log/log curve-fit). Platelet-poor plasma (no dilution), recombinant PDI solutions (reduced, oxidized, S-nitrosylated), human cell lysates, conditioned medium, and recombinant Erp57 (OriGene-TP305940) and ERp5 (OriGene-TP301710) were analyzed.
2.3. PDI purification
Histidine-tagged full length human PDI was cloned into the pET28a vector (Novagen) and overexpressed in E. coli strain BL21 (DE3) codon plus. PDI was first isolated and purified with an immobilized metal affinity resin as previously described [34].
2.4. Preparation of reduced, oxidized and S-nitrosylated PDI
PDI was reduced with 10 mM of GSH (Sigma) or oxidized with 10 mM of diamide (Sigma) overnight at 4 °C. For preparing the nitrosylated protein, PDI was incubated with 200-fold molar excess of S-nitrosoglutathione (GSNO) (Sigma) for 30 min at 37 °C. Samples were then run through a desalting column (Microcon YM-50, Amicon) to remove excess products. The concentrations of PDI in solutions were assessed at 280 nm (ɛ = 45,565 M−1 cm−1).
2.5. Western blot analysis
Equal amounts of protein from HUVEC, HCT116 and HKE3 lysates or conditioned medium (CM) were resolved by SDS-PAGE (12% gel density). The following primary antibodies used were anti-PDI (capture antibody from Human P4HB Pair Set ELISA kit or clone RL90, Thermo-Fisher), anti-ERp72 (ab82587, Abcam), anti-ERp57 (ADI-SPA-585, Enzo). Fluorescence-coupled secondary antibody was purchased from Odyssey and fluorescent immunoblottings were scanned with the Odyssey near-infrared imaging system (Li-cor).
2.6. Preparation of platelet-poor plasma
Blood for platelet-poor plasma samples were collected by venipuncture into EDTA tubes and centrifuged for 15 min at 2.500×g. After centrifugation the plasma phase was used for further analysis or for stored at −80 °C.
2.7. Immunoprecipitation
Fresh platelet-poor plasma samples pooled from 3 donors (10 mL) was diluted 1:1 in lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA and 1% Triton-X 100) supplemented with protease (1 mM PMSF, 1 μg/mL leupeptin and aprotinin) and phosphatase inhibitors (50 mM sodium fluoride, 2 mM sodium orthovanadate, 10 mM sodium pyrophosphate) and 10 mM MgCl2. The plasma sample was incubated overnight at 4 °C under agitation with anti-PDI antibody (10 μg, rabbit EnzoLife, SPA890) followed by incubation with 50 μL Protein G-coated magnetic beads (GE HealthCare) for 4 h at 4 °C. Beads were successively washed in lysis buffer to remove contaminating material. PDI was detected by reducing SDS-PAGE and immunoblotted with a mouse monoclonal antibody (clone RL90, Thermo-Fisher). Blots were scanned with the Odyssey near-infrared fluorescence imaging system (Li-cor). Results are representative from at least 3 independent experiments.
2.8. Measurement of PDI reductase activity
The probe di-eosin-glutathione disulfide (GSSG) was prepared through the incubation of 200 μM GSSG (Sigma) with 2 mM eosin-isothiocyanate (Thermo-Fisher) in 0.1 M potassium phosphate (PE) containing 2 mM EDTA (pH 8.5) at RT, overnight in the dark [35]. The mixture was run through a PD-G25 (GE HealthCare) column and different fractions were collected. A fold-change of fluorescence (Exc: 520 nm, Emis: 545 nm) was calculated using samples subjected to either buffer or 20 mM DTT. Any fraction with a fold-change >5 was kept. The concentration of di-eosin–GSSG was determined by using the molar absorption coefficient for eosin (ɛ = 56,000 M−1 cm−1) at 525 nm in PE buffer [36]. Reduction of plasma was carried out by incubation of 0.5 mL of fresh platelet-poor plasma in assay buffer (PE pH: 7.4, containing 2 mM EDTA), 5 μM DTT and 150 nM of di-eosin-GSSG in the presence or absence of 60 μM of rutin (quercetin-3-rutinoside) (Sigma). The increase in fluorescence was determined for 20 min by excitation at 520 nm and emission at 545 nm in a SpectraMax-M5. The reduction of 150 nM di-eosin-GSSG by 5 μM DTT served as a negative control. Five independent experiments were performed, and each sample was detected in duplicate. Data were normalized to the fluorescence of total plasma (control) and represented as percent reductase activity.
2.9. Separation of microparticle fraction
Protocol was performed as described previously by Ref. [37]. Briefly, platelet-poor plasma (5 mL) was diluted with an equal volume of PBS and centrifuged for 30 min at 2000×g, 4 °C. Supernatants were transferred to ultracentrifuge tubes and centrifuged for 45 min at 12,000×g, 4 °C. Subsequently, supernatants were submitted to centrifugation for 2 h at 110,000×g, 4 °C. After that, pellets were re-suspended in 1 mL PBS, filtered through a 0.22-μm filter and again centrifuged for 70 min at 110,000×g, 4 °C (twice). The final pellets were suspended in 100 μL PBS, lysed in lysis buffer (50 mM Tris-HCl, 1% Triton X-100, 150 mM NaCl) with protease inhibitors (1 mM PMSF, 1 μg/mL leupeptin and aprotinin) and stored at −80 °C.
2.10. EZ-Link Sulfo-NHS-biotin and MPB-labelled assay for plasma PDI redox state
To assess the redox state of plasma PDI, 200 μL of fresh platelet-poor plasma aliquots were separately labelled by a large excess of MPB [3-(N-maleimido-propionyl) biocytin] (4 mM, Molecular Probes) or EZ-Link Sulfo-NHS-(N-hydroxysuccinimido)-Biotin (4 mM, Thermo-Scientific) in the dark, under agitation at RT for 30 min. All reagents were prepared in 20 mM HEPES buffer containing 0.14 M NaCl, 1.5 mM CaCl2, 4 mM KCl, 0.5 mM Na2HPO4, at pH 7.4 (HBS) as described [38]. Thereafter, samples were diluted in the same buffer (1 mL) and incubated for additional 10 min, in the dark at RT. Unbound EZ-Link Sulfo-NHS-Biotin or MPB was then removed by acetone precipitation. The protein pellets were then re-suspended in 250 μL of PBS-Tween/20 (0.05%) and constitute the labelled plasma samples. Additionally, unlabelled plasma samples were also incubated only with HBS (without MPB or EZ-Link Sulfo-NHS-Biotin) as a control. As an internal control for variations in sample labelling with MPB and EZ-Link Sulfo-NHS-Biotin, a standard in-house pool of plasmas was used in each assay. All samples and controls were run concomitantly in parallel. To perform the ELISA for PDI determination, first 96-well plates were coated with anti-PDI capture antibody from Human P4HB Pair Set ELISA kit. Before adding labelled samples, PDI plates were pre-washed three times with PBS-tween/20 (0.5%) and blocked for 1 h with 2% BSA/PBS-tween. Labelled samples were then added to the 96-well plates (100 μL/well) in duplicate and incubated at RT for 2 h with agitation. After washing three times with PBS-tween, a mouse monoclonal anti-biotin (1:200, peroxidase-conjugate) (Clone BN-34, Sigma) diluted in 0.5% BSA/PBS-tween/20 was added (100 μL/well) and incubated at RT for 2 h with agitation. At the final step, wells were washed three times with PBS-tween, 200 μL of TMB solution was added for 30 min with agitation and samples were read at OD 450 nm (SpectraMax-340, Molecular Devices). Amine-labelled PDI sample was considered as total PDI, while MPB-labelled PDI sample was considered reduced PDI.
2.11. Cell culture
A selection-immortalized human umbilical vein endothelial cell line (HUVEC line, from ATCC) [10,39,40] was maintained in RPMI (GIBCO Cell Culture systems, Invitrogen) supplemented with 10% fetal bovine serum (FBS), 100 mg/mL streptomycin, 25 mg/mL penicillin and 10 mM HEPES at 37 °C in 95%O2/5%CO2. Human colon carcinoma cell lines HCT116 and HKE3 were a kind gift from Dr. Walter Kolch (University College Dublin, Belfield, Dublin 4, Ireland). HCT116 and HKE3 cells were maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS (GIBCO Cell Culture systems, Invitrogen) at 37 °C in 95%O2/5%CO2.
2.12. EC plasma incubation
Adhering confluent HUVEC cells were harvested and seeded at 2.5 × 106 cells in plastic petri dishes (100 mm) with a final volume of 10 mL culture medium (containing 10% FBS) per dish. After culturing for 22–24 h, cells were washed with PBS (pH: 7.4) and cultured for additional 24 h in medium in which FBS (5%) was replaced with 5% of either PDI-poor or PDI-rich plasma obtained from a pool of equal volumes of 3 individual samples. The 5% concentration was chosen from validation experiments; incubation with 10% human plasma associated with significant EC detachment, although they remained viable during the 24-h period.
2.13. Cell viability assessment
After 22–24 h in culture conditions, HUVEC were washed with PBS (pH 7.4) and cultured for 24 h in medium in which FBS was substituted for 5% of platelet-free plasma pools from 3 individuals with PDI-poor or PDI-rich plasma. After that, cells were mixed with Trypan Blue solution (0.4%). Colored (non-viable) and dye-excluding (viable) cells were counted on automated cell counter (Countess II FL, Thermo-Scientific).
2.14. Proteomic analysis
2.14.1. Sample preparation and UPLC-HDMSE analysis
2.14.1.1. Blood plasma sample preparation
Forty microliters of blood plasma from 6 healthy donors were diluted in 120 μL of buffer solution A, from the MARS Hu14 column manufacturer's protocol (Agilent) to remove the highest abundant proteins. The immunoaffinity chromatography was performed according to Ref. [41].
2.14.1.2. Conditioned medium (CM) from HUVEC and sample preparation
HUVEC were cultured and treated as describe above (‘EC plasma incubation’ section). To obtain the CM, cell culture media were removed, carefully washed with PBS and each CM sample was obtained following incubation with 6 mL of serum-free medium. After 8 h, supernatants were collected and depleted of cell debris by centrifugation (1400 rpm, 10 min, 4 °C), followed by medium centrifugation and 3-fold concentration, using SpeedVac. The concentrated medium was stored at −80 °C for additional analysis. Protein concentrations were determined by Bradford assay and protein samples (50 μg each sample) were transferred into SDS-loading buffer (156.25 mM Tris-HCl, pH 6.8, 2.3% (w/v) SDS, 4% (v/v) glycerol, 0.5% (v/v) 2-mercaptoethanol), boiled (5 min, 95 °C) and then subjected to SDS-PAGE (12% gel density). Each lane was divided into >20 fragments. The proteins within the gel fragments were in-gel-trypsinized at 37 °C overnight. To extract the peptides, 50 mM (NH4)CO3 (50 μL) and 2 × 50 μL 50% (v/v) acetonitrile, 5% (v/v) formic acid were added successively to the gel pieces. The volume of the extraction solution was reduced to <20 μL using SpeedVac.
2.14.1.3. Mass spectrometry acquisition
Peptides (0.5 μg) were automatically loaded into a high-throughput sample manager (SM) in a UPLC M-Class System (Waters Corporation, Milford, MA), coupled online to a Synapt G2-Si mass spectrometer (Waters Corporation, Milford, MA). Trapped peptides were eluted with an acetonitrile gradient from 7% to 40% (v/v) for 52 min at a flow rate of 0.5 μL/min and analyzed directly into a Synapt G2-Si. Peptides were ionized using a nano-electrospray source in positive mode. Results were obtained in a data-independent mode (DIA) combined with ion mobility acquisition mode (HDMSe). The mass spectrometer parameters were set according to Ref. [66].
2.14.2. Protein identification and quantitation
Proteins were identified and quantified by using dedicated algorithms of the Progenesis QI for Proteomics software (Waters Corporation). Data were searched against the UniProt human proteomic database: for blood plasma, we used the complete database (version: 2016_06) and for HUVEC cells we employed the reviewed database (version: 2017_10). The following parameters were considered to identify peptides in blood plasma samples: 1) Digestion by trypsin with at most one missed cleavage; 2) variable modifications by oxidation (M) and fixed modification by carbamidomethyl (C); 3) calculated false discovery rate (FDR) less than 4% across all conditions and replicates; 4) mass error less than 20 ppm. One or more ion fragments per peptide, three or more fragments per protein and one unique or more peptides per protein were required for ion matching. The following parameters were considered to identify peptides in HUVEC samples: 1) Digestion by trypsin with at most one missed cleavage; 2) variable modifications by oxidation (M) and fixed modification by carbamidomethyl (C); 3) calculated false discovery rate (FDR) less than 1% across all conditions and replicates; 4) mass error less than 20 ppm. Two or more ion fragments per peptide, five or more fragments per protein and one unique or more peptides per protein were required for ion matching. Identifications that did not satisfy these criteria were rejected. The final, confirmed list of proteins only considered proteins identified by at least 2 peptides. Also, differences between groups were analyzed using one-way ANOVA and only a p-value < 0.05 was considered significant Keratin proteins were excluded from our data.
2.14.3. Enrichment analysis
Metacore software (Thomson Reuters, MI, USA) was used to perform functional enrichment analysis and to create biological protein networks. Enriched pathways were considered significant at p value < 0.05.
2.15. Platelet aggregation
Platelet-rich plasma (PRP) samples were obtained from individuals who had not used antiplatelet medications for at least 10 days prior to venipuncture. Blood samples were anticoagulated with trisodium citrate 3.2% (blood/anticoagulant 9:1 v/v) and centrifuged at 190×g for 15 min to obtain PRP. PRP (2–3 × 108/mL−1) were incubated with ADP (10 μM), collagen (5 μg/mL) or calcium ionophore-A23187 (20.5 μM) for 5 min and then aggregation was measured using a whole blood/lumi-aggregometer (Chronolog, 560VS). Whole blood samples were anticoagulated with trisodium citrate 3.2% (blood/anticoagulant 9:1 v/v) and thereafter incubated with collagen (25 μM) for 10 min. Aggregation was measured as describe for PRP.
2.16. RT2 profiler PCR array
HUVEC were cultured and prepared as described in ‘EC plasma incubation’ section. After that, 2 × 106 cells were collected, centrifuged and homogenized in Trizol (Invitrogen) for RNA extraction. The quantity and quality of RNA samples were determined using Nanodrop ND2000 (Thermo-Fisher). RNA samples (1.5 μg) were reverse-transcribed to synthesize cDNA library using RT2 First Strand Kit (Qiagen). A cDNA library was used as the template for the subsequent PCR analysis in a StepOnePlus™ Real-Time PCR System. The expression levels of 84 genes (listed in Supplementary Table 3) were analyzed using RT2 Profile PCR array (PAHS-038, Qiagen). The PCR array experiment was performed from 3 independent experiments (each one performed in triplicate). Relative quantities of mRNA were calculated using ΔΔCt with β-2-microglobulin as the reference.
2.17. Cell adhesion assay
Flat bottom 96-well plates were coated with either fibronectin (Sigma) (5 μg/mL) or collagen type-I (Sigma) (10 μg/mL) overnight at 4 °C and then blocked for 2 h with 1% BSA/PBS. Cultured HUVEC prepared as described in ‘EC plasma incubation’ section were washed with PBS and detached with PBS-EDTA (500 μM, pH 7.4). After that, cells were re-suspended in RPMI containing 0.2% BSA (adhesion buffer), added to coated wells at a volume of 150 μL (4 × 104 cells/well) and allowed to adhere to the substrate for 1 h (fibronectin) or 4h30min (collagen) at 37 °C. Unattached cells were removed by gently washing three times with PBS. Residual attached cells were fixed by 2% glutaraldehyde for 30 min at RT, and then plates were washed three times with PBS and stained by 0.1% crystal violet (30 min at RT), followed by lysis with 1% SDS. Absorbance was then measured at 600 nm in a microplate reader (SpectraMax-340, Molecular Devices).
2.18. Wound healing assay
HUVEC migration was investigated by wound healing assay as described previously [42]. Cells were seeded (2 × 105) in flat bottom 24-well plates. After 48 h, confluent cell monolayers were incubated for 5 min with HBSS (Hanks’ Balanced Salt solution) without Ca2+ and Mg2+ (H6648, Sigma). Thereafter, a wound was created by scratching the cells once with a sterile tip (500 μm in diameter). The cultures were washed twice with HBSS to remove cell debris. HUVEC were then incubated with RPMI alone or supplemented with 5% of either FBS, PDI-poor plasma or PDI-rich plasma. Cells were then transferred to a microscope chamber at 37 °C with 95%O2/5%CO2 (Axiovert 200, Zeiss) and pictures were taken at 0 h, 8 h, 16 h and 24 h (3 separate fields/well) with 5× objective. Covered surface areas were measured using Image-J software.
2.19. Statistical analysis
Normally and non-normally distributed data were presented as mean ± standard error of the mean (SEM) and median with interquartile ranges, respectively. Differences between two groups were analyzed using paired Student's t-test to compare two groups or one-way ANOVA plus Student-Tukey's test or the Mann–Whitney test. Correlation coefficients were according to the method of Spearman. A p-value <0.05 was considered significant. Analyses were carried out using GraphPad Prism v.7. Linear regression was used to estimate the association between plasma PDI and traditional risk factors in patients from databank. All analyses were performed using at least three independent experiments repeated in distinct times, with 2 or 3 replicates for each measurement, as specified. Normality tests (Shapiro-Wilk and D'Agostino-Pearson Omnibus) were performed followed by frequency distribution of PDI levels using Prisma parameters.
3. Results
3.1. ELISA assay validation for plasma PDI
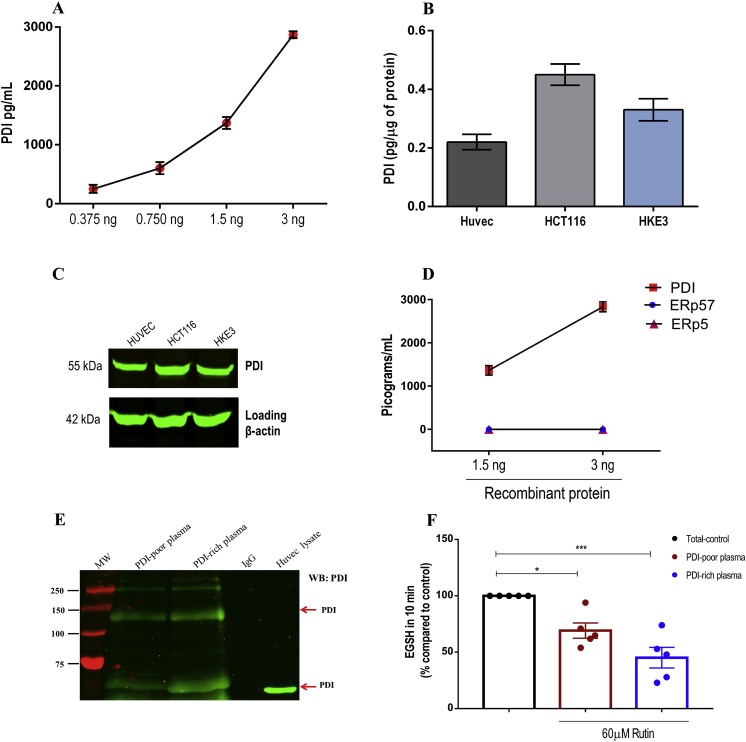
Circulating plasma PDI levels were assessed by enzyme-linked immunosorbent assay (ELISA). After testing distinct approaches, which proved to be insufficiently sensitive, we chose a commercially available kit not yet validated in the literature (Human P4HB Pair Set, SinoBiological), which was able to detect levels as low as 47 pg/mL (Suppl.Fig. S1-A and B). Assay curves with recombinant PDI confirmed its accurate detection (Fig. 1A), while the assay was equally able to detect reduced, oxidized or S-nitrosylated PDI – Suppl.Figs. S1–C). Additionally, we quantified intracellular PDI from human cell lysates (Fig. 1B) in parallel with western analysis using ELISA's capture antibody in the same samples, showing immunostaining of a single band consistent with PDI (Fig. 1C). We also tested possible cross-reactivity with other PDIs using recombinant PDI, ERp57 and ERp5 and showed that this assay recognized only PDI (Fig. 1D). Detection of circulating PDI was additionally confirmed by immunoprecipitation (Fig. 1E, Suppl.Figs. S1–D). Also, to assess whether circulating PDI is active, we measured the quercetin-3-rutinoside (rutin)-inhibitable fraction of total dieosin-GSSG reductase activity; the use of rutin, a well-reported PDI inhibitor [43], enhances the specificity of this otherwise nonspecific reductase assay. Rutin inhibited di-eosin-GSSG turnover by ca.70% and 45% in PDI-rich and PDI-poor plasma, respectively (Fig. 1F and Suppl.Figs. S1–E). A conceptually analogous approach allowed evidence of plasma PDI activity in sickle-cell disease transgenic mice [44]. These results support a plasma pool of active PDI in healthy individuals.
Fig. 1.
Validation of ELISA assay for specific detection of PDI. A. PDI concentration curves (0.375–3 ng/mL) were generated using purified PDI quantified by ELISA. Measurements were analyzed on 4 independent assays. Data represent mean ± SEM. B-C. Human cell lysates (HUVEC, HCT116 and HKE3) were (B) quantified by ELISA (n = 4; mean ± SEM) and (C) submitted to protein separation by reducing SDS-PAGE and immunoblotted with anti-PDI ELISA's capture antibody or anti-β actin (loading control) (n = 3). Uncropped western blots are shown in Supplementary Fig. S6. D. Cross-reactivity with other PDI family members was analyzed using recombinant PDI, ERp57 and ERp5 (1.5–3 ng/mL) quantified by ELISA. Measurements were analyzed on 4 independent assays. Data represent mean ± SEM. E. Plasma PDI immunoprecipitation. Platelet-poor plasma was diluted in lysis buffer and immunoprecipitated using rabbit anti-PDI antibody. Immunoblotting was performed using mouse anti-PDI (RL90) (n = 3 from independent experiments). F. Detection of plasma PDI reductase activity (di-eosin-GSSG assay) by formation of the reduced fluorogenic product eosin-5-isothiocyanate-coupled reduced glutathione (EGSH). It was measured in platelet-poor plasma exposed or not to 60 μM of rutin. Bar graphs represent the percentage cleavage of di-eosin-GSSG as compared to total reductase activity – Control (100%) in 10 min. The main result in this case is the rutin-inhibitable fraction of di-eosin-GSSG reductase activity. Data represent mean ± SEM from 5 independent experiments. *p < 0.05; ***p < 0.001 vs. control (One-way ANOVA followed by Tukey's post test).
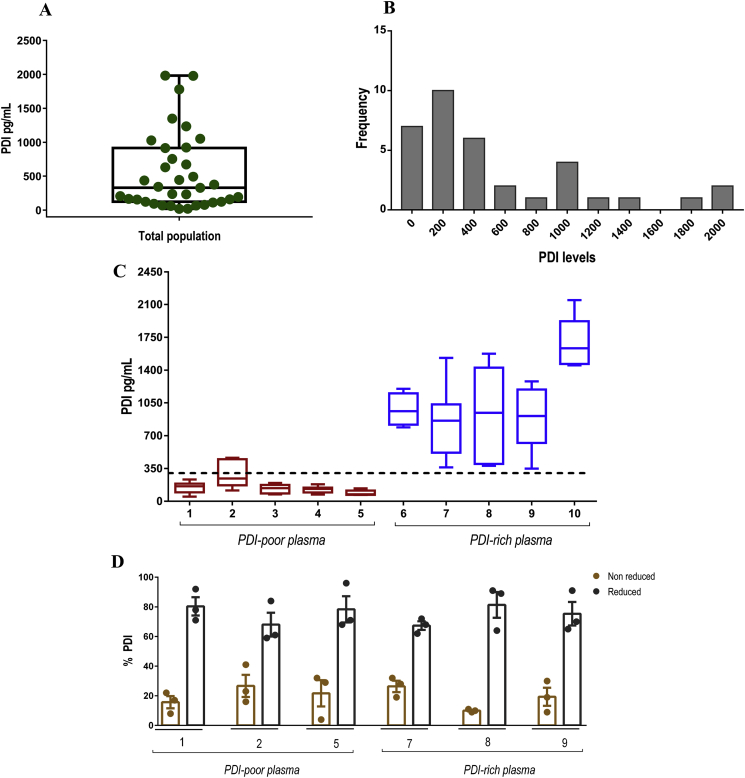
3.2. Plasma levels of PDI show high inter-individual but low intra-individual variability
The amount of PDI detected by our assay was low, at pM levels, and their distribution was not normal, as confirmed by specific tests (Fig. 2A and B). Microparticle-associated PDI accounted for a minor fraction of total PDI levels in our population (Suppl.Figs. S2–A). Plasma PDI concentrations depicted high inter-individual variability, with values running from undetectable/very low until >1000 pg/mL (∼18 pM). As the median value was 330 pg/mL (∼6 pM) (Fig. 2A), we designated values ≤ 330 pg/mL as PDI-poor plasma, as opposed to PDI-rich plasma (>330 pg/mL). Remarkably, opposite to inter-individual variability, the intra-individual variability was quite low, so that values assessed under distinct conditions over time were close and reproducible (Fig. 2C). For that, plasma was collected 10–15 times, spaced from days to weeks over a 10–15 month span and circulating PDI levels exhibited in most cases a small range of values (Fig. 2C). Also, in a cohort of 5 individuals, PDI levels were serially collected 3 times over a 9-h period; again, variability among results was negligible (Suppl.Figs. S2–D). Importantly, we assessed the possible influence of variables reflecting traditional cardiovascular and inflammation risk factors on PDI levels. There was no significant correlation between PDI plasma levels with age, triglyceride levels, high-density lipoprotein (HDL) cholesterol levels, C-reactive protein (CRP) values, white and red blood cell counts and platelet counts, while there was a non-robust direct correlation with total and LDL cholesterol (p < 0.05) (Suppl.Fig. S3).
Fig. 2.
Plasma PDI levels in healthy individuals. A. Distribution of plasma PDI levels are represented by box plot, indicating the median plus lower and upper quartiles (n = 35). B. Frequency distribution of plasma PDI levels shown in (A). C. Graphic representation of plasma PDI from selected individuals assessed over time. Each box plot represents one individual and their different plasma samples collected over time (n = 10–15 samples for each individual), as detailed in the text. The dashed line designates the median value (330 pg/mL) of the whole population shown in (A). Values ≤ 330 pg/mL were designated as PDI-poor plasma and >330 pg/mL as PDI-rich plasma. Data represent median plus lower and upper quartiles. D. Detection of PDI redox state in human plasma. Plasma samples labelled with either EZ-Link Sulfo-NHS-Biotin or MPB were acetone precipitated (to remove unbounded biotinylated probes) and protein pellets were re-suspended in PBS-tween. Samples were added to a plate coated with anti-PDI antibody and after this incubation probed with a monoclonal antibody to biotin (peroxidase conjugate). Bar graphics represent the percentage of reduced or non-reduced PDI comparing MPB vs. EZ-Link Sulpho-NHS-Biotin labelling. Numbers represent different individuals. Data represent mean ± SEM from 3 independent experiments.
3.3. Thiol redox state of plasma PDI pool
PDI exhibits a pool of distinct redox states: fully/partially reduced, fully/partially oxidized to intramolecular disulfides, oxidized to intermolecular disulfides (e.g., S-glutathionylated) or S-nitrosylated. We developed assays with biotinylated probes (see Materials and methods) using a pooled (3 individuals) in-house plasma sample as internal reference control for their labeling amount (Suppl.Figs. S2–C). Our results showed that the majority (60–80%) of plasma PDI was in the reduced state (Fig. 2D and Suppl.Figs. S2–B), with no difference in PDI-poor vs. PDI-rich plasma.
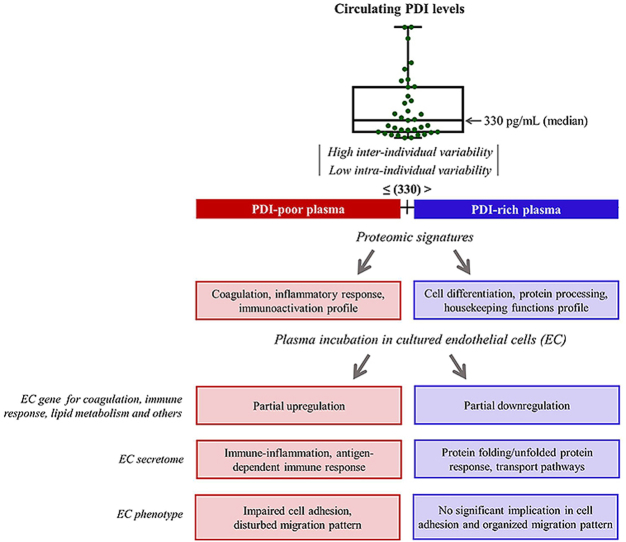
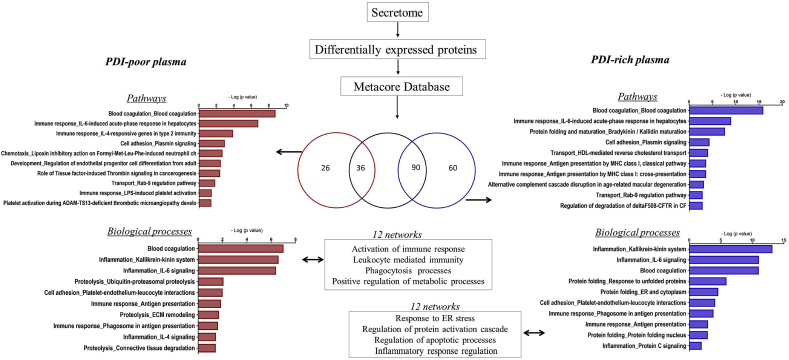
3.4. Distinct PDI concentrations uncover contrasting plasma protein signature patterns
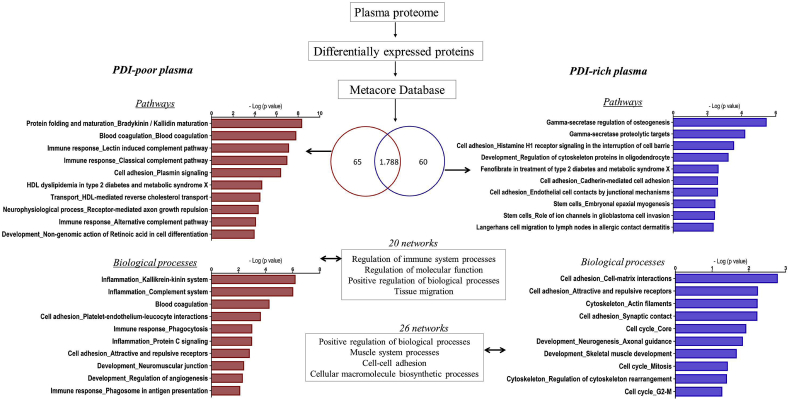
Given the low intra-individual variability despite widely distinct inter-individual plasma levels, we hypothesized that PDI concentrations could behave as a window to reveal individual plasma protein signatures. Thus, we performed quantitative proteome analyses comparing PDI-poor vs. PDI-rich plasma. Plasma samples were depleted of top 14 high-abundant proteins, as these can mask low-abundance protein detection given the huge concentration dynamic range. Low abundant proteins were analyzed by shotgun mass spectrometry-based proteomics, allowing identification of 1788 proteins. Quantitative analysis indicated 125 upregulated proteins differentially expressed between PDI-poor and PDI-rich plasma (65 and 60, respectively) (Suppl.Tables S1–2). To investigate whether these proteins compose signatures for specific processes, we used Metacore software database. Importantly, the protein subset upregulated in PDI-poor plasma was preferentially associated with immuno-inflammation, coagulation and platelet interactions, with 50% pathways and 70% biological processes including blood coagulation and immune response. Main examples included: coagulation factors XII and XI and kininogen 1, from the intrinsic coagulation pathway; proteins associated with plasminogen and fibrinogen interactions; proteins involved in platelet interactions, such as histidine-rich glycoprotein (HRG) [45,46]; immune-related proteins, such as leucine-rich alpha-2-glycoprotein [47] and CD14 [48]. In particular, several complement-related proteins were preferentially identified in PDI-poor plasma. Biological processes/pathways and main gene ontology processes identified in the biological networks are depicted in Fig. 3 (left diagram) and Suppl.Figs. S4–A. In contrast, the subset of proteins preferentially upregulated in PDI-rich plasma (vs. PDI-poor plasma) was related to cell adhesion (e.g. cadherin-5), cytoskeleton (e.g. myosin-8) and cell cycle (e.g. MAD2-like protein 1), as depicted in Fig. 3 (right diagram) and Suppl.Figs. S4–B. These results suggest that PDI levels can reflect distinct processes and gene ontology profiles of proteomic signatures in the plasma of healthy individuals with no other obvious differences in their conventional risk-associated indicators.
Fig. 3.
Total proteome and Metacore analysis of PDI-poor or PDI-rich plasma. Samples from individuals with PDI-poor plasma (n = 3) or PDI-rich plasma (n = 3) were submitted to shotgun mass spectrometry profiling analysis to identify proteins differentially expressed in each case. There were 65 proteins differentially upregulated in PDI-poor plasma samples and 60 proteins differentially upregulated in PDI-rich plasma samples. Further analysis using Metacore software revealed enrichment in several pathways and biological processes (p < 0.05).
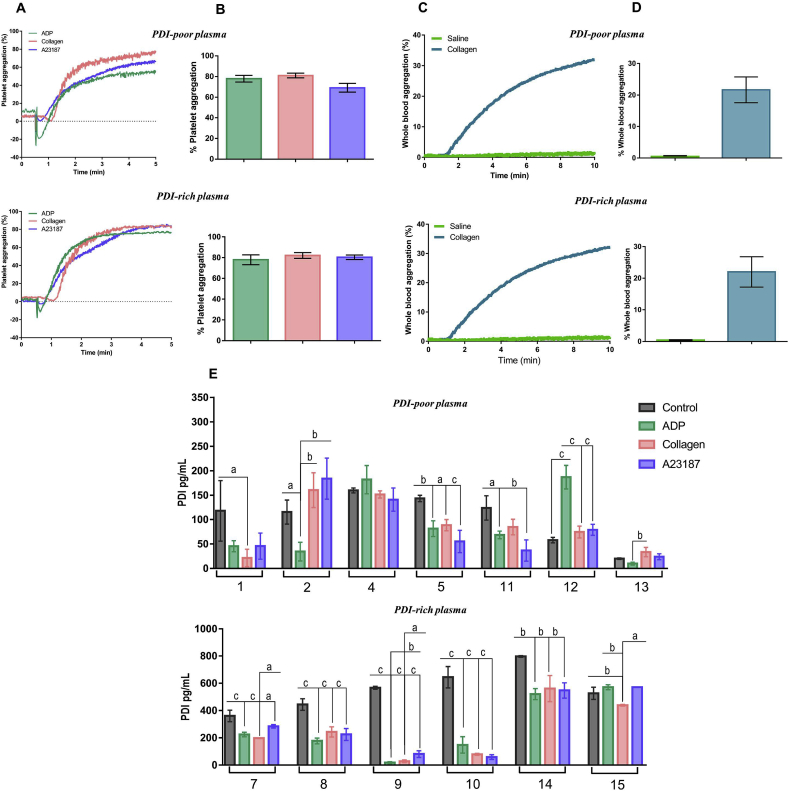
3.5. PDI level-associated protein signatures did not correlate with platelet aggregation patterns
We next interrogated whether the observed differences in PDI plasma levels associate with different platelet function profiles. Platelet aggregation assays were performed in platelet-rich plasma collected from individuals with PDI-poor vs. PDI-rich plasma stimulated for 5 min with ADP (10 μM), collagen (5 μg/mL) or calcium ionophore A23187 (20.5 μM). Platelet aggregation responses to each of these agonists showed no differences between PDI-poor vs. PDI-rich plasma, all of them having on average ca.80% extent of aggregation (Fig. 4A and B). Similar results were observed for whole-blood platelet aggregation (Fig. 4C and D). Since PDI is secreted after platelet activation, we next investigated soluble PDI levels after platelet aggregation. Interestingly, PDI levels in platelet-poor plasma were decreased post-aggregation in most samples regardless of the agonist, with few exceptions (Fig. 4E). Both effects were unrelated to circulating PDI levels or degree of platelet activation.
Fig. 4.
Platelet aggregation in individuals with PDI-poor or PDI-rich plasma. Experiments were performed in platelet-rich plasma (PRP) by light transmission and whole blood by impedance (in both cases 2–3x108 platelets mL−1, 37 °C). Platelets were stimulated with agonists and tracings were recorded for 5 min for PRP and 10 min for whole blood. A. Representative platelet aggregation tracings induced by 10 μM ADP, 5 μg/mL collagen and 20.5 μM A23187. B. Maximal extents of aggregation from (A). No difference was detected among groups. One-way ANOVA followed by Tukey's post test. C. Representative whole blood aggregation tracings induced by 25 μM collagen. D. Maximal extents of aggregation from (C). No difference was detected between PDI-poor and PDI-rich plasma groups for stimulated aggregation. One-way ANOVA followed by Tukey's post test. E. Soluble PDI was measured by ELISA in supernatants of resting (control) or activated PRP as described in Materials and methods. Numbers represent different individuals. All data represent mean ± SEM from 6 to 7 independent experiments. ap<0.05; bp < 0.005; cp < 0.0005 (One-way ANOVA followed by Tukey's post test, for each individual samples).
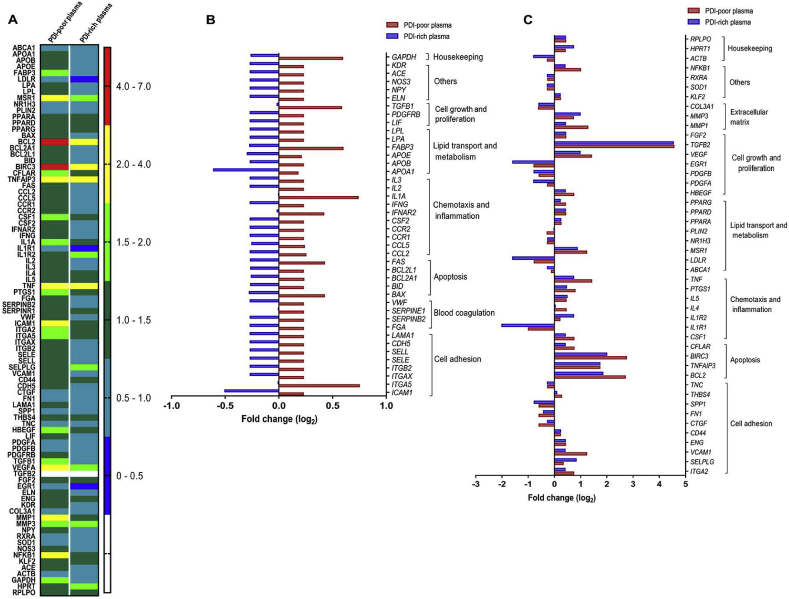
3.6. Distinct PDI levels in plasma recapitulate contrasting endothelial gene expression and secretome patterns
Since EC can secrete PDI [10,49] and pecPDI modulates several EC effects, we assessed effects of incubation of PDI-poor or PDI-rich plasma in cultured EC. Cultured human umbilical vein EC (HUVEC) were incubated for 24 h in medium with standard FBS (5%) or in medium in which FBS was replaced with 5% plasma collected from individuals with low or high PDI levels, and the patterns of EC gene expression and protein secretion were assessed. Gene expression changes were analyzed by PCR array covering 84 prespecified genes (Suppl.Table S3) involved in blood coagulation, cell adhesion, lipid transport/metabolism, chemotaxis/inflammation, cell growth/proliferation and apoptosis. Plasma-exposed EC exhibited a fibroblast-like rather than their normal cobblestone-like phenotype (Suppl.Figs. S5–A), consistent with EC activation, i.e., switch from baseline to a state primed for thrombosis, proliferation and vasoconstriction [50,51]. While shape changes were indistinguishable following incubation with PDI-poor or PDI-rich plasma, EC gene expression responses were significantly different for each type of plasma vs. FBS-controls, suggesting distinct EC activation patterns (Fig. 5A). Overall, 48% of genes were simultaneously upregulated with PDI-poor and downregulated with PDI-rich plasma (Fig. 5B). Main examples include integrins involved in leukocyte-EC adhesion: integrin ITGAL/ITGB2 (ITGB2), αXβ2 (ITGAX), intracellular adhesion molecule 1 (ICAM1), selectins E (SELE) and L (SELL), as well as coagulation-related genes such as von Willebrand factor (VWF), fibrinogen α-chain (FGA) and fibrinolysis inhibitors such as plasminogen-activator inhibitors 1(SERPINE1) and 2(SERPINB2). Identical patterns occurred for 10/17 inflammatory/chemotaxis genes and 6/14 lipid transport/metabolism genes, with the other showing diverse behaviors (Fig. 5B and C). In contrast, there was no clear pattern for growth/proliferation genes (Fig. 5B and C). Among 9 apoptosis-related genes, 5 were upregulated with PDI-poor and downregulated with PDI-rich plasma (FAS, BCL2L1, BCL2A1, BID, BAX), while 4 were upregulated in both cases. Anti-apoptotic genes BCL2 and BIRC3, which also regulate inflammatory signaling and immunity [52,53], underwent the highest fold-changes vs. controls, accentuated with PDI-poor plasma (Fig. 5B and C). However, most cells remained viable after 24 h with PDI-poor or PDI-rich plasmas, similarly to FBS-incubated controls (Suppl.Figs. S5–A). Thus, EC incubation with PDI-poor or PDI-rich plasma recapitulated a gene expression pattern reminiscent of corresponding plasma protein signatures.
Fig. 5.
Effects of incubation with PDI-poor or PDI-rich plasma on endothelial cell expression of several genes covering different biological processes. EC were incubated with 5% PDI-poor plasma, PDI-rich plasma or FBS for 24 h and the expression levels of pre-specified 84 genes were analyzed using RT2 Profile PCR array. A. Heat map of relative expressions of transcripts. B. Transcript levels are shown as Log2 fold-changes for genes which were simultaneously upregulated with PDI-poor and downregulated with PDI-rich plasma. C. Transcript levels are shown as Log2 fold-changes for genes with distinct behavior in response to incubation with PDI-poor or PDI-rich plasma. All expression values were normalized for the expression of β-2-microglobulin. All data represent mean values from 3 independent experiments.
To further pursue this connection, we analyzed the EC secretome profile in preparations run in parallel with those used for gene expression studies. After 24 h-incubation with PDI-poor or PDI-rich plasma, EC cultures were carefully washed from plasma-supplemented medium and incubated for an additional 8 h-period with serum-free media, after which the conditioned medium was collected and submitted to shotgun mass spectrometry-based proteomic analysis. We identified 873 proteins across all conditions: comparison between proteins upregulated with PDI-poor or PDI-rich plasma vs. FBS controls allowed identification of 26 and 60 proteins, respectively (Suppl.Tables S4–5). Consistent with Fig. 3 results in whole plasma, EC secretome showed differential protein expression with PDI-poor vs. PDI-rich plasma (Fig. 6), although differences were more subtle. Coagulation-related proteins, for example, shared some degree of protein identities between PDI-poor and PDI-rich plasma, such as fibrinogen chains and histidine-rich glycoprotein. Also, both conditions associated with proteins involved in immuno-inflammation and antigen-dependent immune response. However, more detailed analysis of proteins involved in these and other pathways/processes uncovered relevant differences in gene ontology profile (Suppl.Figs. S4–C). With PDI-rich plasma, EC secretome preferentially associated with protein folding/unfolded protein response (e.g., BiP) and transport pathways (e.g. transthyretin, serotransferrin), while PDI-poor plasma preferentially associated with proteolysis processes and immune regulation (Fig. 6, Suppl.Figs. S4–D). Of note, PDI, ERp57 (PDIA3) and ERp72 (PDIA4), were preferentially secreted following incubation with PDI-rich but not PDI-poor plasma vs. FBS (Suppl.Figs. S5–B and Suppl.Table S5). Importantly, there was no evidence of cell lysis, since β-actin, a validated EC damage indicator in this assay [10], was undetectable in conditioned medium (Suppl.Figs. S5–B).
Fig. 6.
Secretome proteome and enrichment analysis of endothelial cells in response to incubation with PDI-poor or PDI-rich plasma. EC were incubated with 5% PDI-poor plasma, PDI-rich plasma or FBS-control for 24 h and conditioned medium was generated after 8 h in serum-free media for secretome analysis, as described in Materials and methods. Proteome analysis through shotgun mass spectrometry profiling disclosed subsets of upregulated proteins differentially expressed between EC exposed to PDI-poor or PDI-rich plasma vs. FBS-control. These subsets were significantly enriched in several pathways, biological processes and gene ontology processes (p < 0.05), as analyzed by Metacore software. All data represent results from 3 independent experiments.
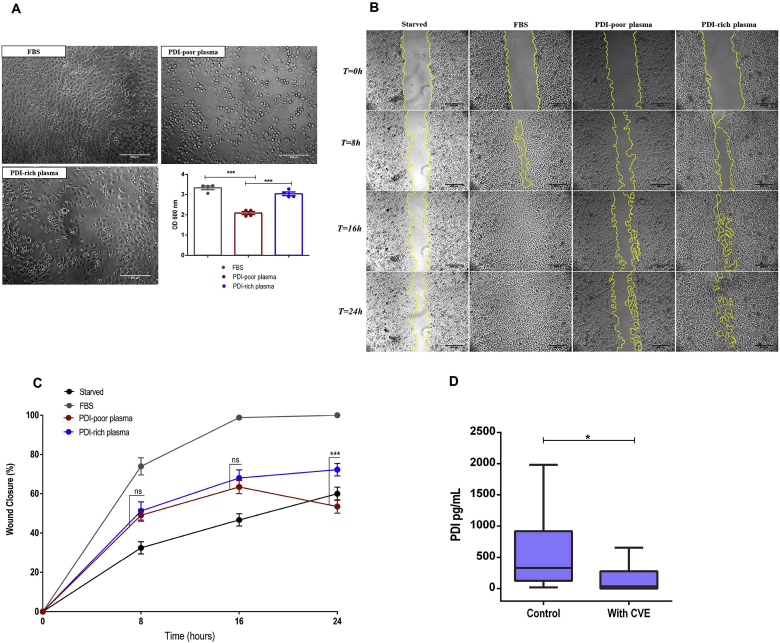
3.7. Functional correlates of distinct PDI-associated protein signatures in EC
Having shown correlation of specific protein signatures and associated EC phenotypes with plasma PDI levels, we inquired whether the distinct observed plasma profiles correlate with functional EC outcomes. For that, we first assessed EC adhesion. Incubation with PDI-poor plasma impaired EC attachment to fibronectin by ca.30% vs. FBS-control (p < 0.001) and was ca.20% lower than with PDI-rich plasma (p < 0.05), while the later condition was not different vs. FBS-control (Fig. 7A). In fact, total plasma proteomics (Fig. 3) showed many adhesion/cell matrix pathways to be preferentially related to PDI-rich plasma. Analogous experiments with EC attachment to collagen-1 showed no difference among exposures to FBS or plasmas (Suppl.Figs. S5–C).
Fig. 7.
Functional effects of PDI-poor or PDI-rich plasma incubation in endothelial cell adhesion and migration. A. HUVEC were pre-incubated with 5% PDI-poor plasma, PDI-rich plasma or FBS for 24 h, seeded (4 × 104 cells/mL) in 96-well plates coated with fibronectin (5 μg/mL) and allowed to adhere for 1 h at 37 °C. After washing, adherent cells were stained with crystal violet, solubilized by SDS and absorbance was measured at 600 nm. Scale bar: 500 μM. Objective: 40x. Data represent mean ± SEM from 5 independent experiments. ***p < 0.001 (One-way ANOVA followed by Tukey's post test) B. HUVEC were seeded at 2 × 105 cells/mL in 24-well plates. After culturing for 48 h, a wound was created by scratching with a sterile pipette tip. Cells were cultured for an additional 24 h in medium as follows: without FBS (starved), with 5% of FBS-control, PDI-poor plasma or PDI-rich plasma. Photomicrographs representing migration into the denuded area at 0 h, 8 h, 16 h and 24 h. The zone between two yellow lines indicates the area occupied by the initial wound. Scale bar: 500 μM. Objective: 5x. C. Percent wound surface coverage by migrating HUVEC showed in (B). Data represent mean ± SEM from 4 independent experiments. ***p < 0.0001 (Two-way ANOVA followed by Tukey's post test). For clarity, statistics are shown only for EC incubated with PDI-poor vs. PDI-rich plasma. D. Plasma PDI concentration measured by ELISA, in a population with cardiovascular events (n = 95, see Table 2 for details) vs. healthy controls (n = 35). Data represent median plus lower and upper quartiles. *p < 0.05 (Mann-Whitney t-test).
We also investigated effects of plasma exposure in EC migration. EC monolayers incubated as described above with FBS, PDI-poor or PDI-rich plasma for 24 h underwent wound-healing assays. FBS-exposed EC showed the highest migration rates (Fig. 7B and C) and completely covered the wounded area after 24 h. In contrast, wound repair was impaired vs. FBS-control to a greater extent following incubation with PDI-poor plasma than with PDI-rich plasma (to 53% and 72% wound area coverage, respectively, p < 0.001) (Fig. 7C). Interestingly, both PDI-poor and PDI-rich plasma yielded analogous migration rates up to ca.16 h of incubation, coincident with 63–65% wound recovery. However, after this stage, EC incubated with PDI-rich plasma continued to migrate to reach 72% covered area at 24 h, while EC incubated with PDI-poor plasma continued to migrate, but mainly in the reverse direction, again re-expanding the wounded area, which had 53% coverage after 24 h (Fig. 7B and C, Suppl.Videos S1–4). Incubation of starved EC with recombinant PDI concentrations up to levels close to those of PDI-rich plasma did not enhance cell migration (not shown). Overall, these results indicate contrasting functional effects of PDI-rich vs. PDI-poor plasma in EC.
Supplementary video related to this article can be found at https://doi.org/10.1016/j.redox.2019.101142.
The following are the supplementary data related to this article:
Video representing migration into the denuded area after incubation with medium without FBS.
Video representing migration into the denuded area after incubation with fetal bovine serum (FBS)-control.
Video representing migration into the denuded area after incubation with PDI-poor plasma.
Video representing migration into the denuded area after incubation with PDI-rich plasma.
4. Discussion
Our results indicate that plasma PDI is detectable in healthy individuals, depicting high inter-individual but low intra-individual variability. Remarkably, PDI levels behave as reporters of distinct plasma protein signatures, with PDI-rich plasma differentially expressing proteins related to cell differentiation, protein processing and housekeeping functions, among others, while PDI-poor plasma differentially displays a subset of proteins associated with coagulation, inflammatory responses and immunoactivation. Moreover, there is close correlation of PDI levels and corresponding protein signatures with EC function and phenotype, since cultured EC incubated with PDI-poor or PDI-rich plasmas recapitulated gene expression and secretome profiles in line with the observed plasma proteome. Furthermore, such signatures directly translated into functional responses, with PDI-poor plasma promoting impaired EC adhesion to fibronectin and disturbed pattern of organized migration vs. PDI-rich plasma. Essex et al. [28] also showed a detectable plasma PDI pool in healthy donors measured by noncommercial ELISA, however at a 250–1000 ng/mL range (∼4–17 nM), significantly higher than in our assays. Reasons for such distinct levels may likely relate to assay conditions and population characteristics. Also, PDI and ERp57 were previously reported in plasma proteome but not clearly associated with cardiovascular risk [54]. While this is to our knowledge the first report assessing plasma concentrations of a PDI family protein, it is not unlikely that other PDIs can also potentially be present in plasma and display analogous behaviors.
While our results indicate that plasma PDI pool is mainly reduced (Fig. 2D), correlations between PDI redox state and activity deserve further discussion. Oxidized PDI can directly introduce disulfides into its substrates, with reduced PDI as end-product; in the ER PDI is reoxidized by Ero1, GSSG and oxidized Prx4 or Gpx7/8 [3]. Reduced PDI can either reduce substrate thiols (with oxidized PDI as end-product) or promote thiol reshuffling of disulfide bonds, yielding reduced PDI [7,55]. Interestingly, kinetic trapping experiments suggest that different substrates are biased to reduction vs. oxidation by PDI. The first comprises platelet substrates including, among others, platelet factor-V [24], annexin V, ERp57 [23] and plasma substrates such as vitronectin, some complement factors, HRG and thrombospondin-1 [22]. In contrast, cathepsin G, glutaredoxin-1, thioredoxin, GP1b and fibrinogen are biased for PDI-dependent oxidation in platelets [23]. In parallel, although PDI does not seem a mass peroxide sensor as discussed before, it is affected by redox potentials [7]. Accordingly, one might expect that plasma PDI would be primarily oxidized [56], contrary to our results, since redox potential of the major plasma thiol pools, CyS/CySS (−80 ± 9 mV) and GSH/GSSG (−137 ± 9 mV) [1] are more oxidizing than PDI redox potential (−190 ± 10 mV). While mechanisms underlying this apparent paradox are yet unclear, this is not unexpected since most plasma thiol pools are not in equilibrium [1], suggesting possible mechanism(s) for their mutual insulation.
Several mechanisms may regulate plasma PDI levels and account for their variability. Our data (Suppl.Fig.5-B, Suppl.Table S5) showed increased PDI secretion by cultured EC exposed to PDI-rich plasma, thus suggesting that specific plasma factors control PDI externalization from EC. Contrarily, platelet activity was similar in individuals with low vs. high plasma PDI, suggesting that PDI level variability is not explained only by distinct amounts of secretion from platelets, which has been well-documented during thrombus formation [30]. In contrast, increased platelet PDI secretion was reported in patients with hemophilia A [57] and a pH-dependent release from platelets has been suggested as possible source of plasma PDI [28]. Meanwhile, there was decrease in soluble PDI following platelet activation (Fig. 4E), which we attribute to sequestration of platelet-derived PDI by its potential substrates [22,23,58]. In fact, PDI is retained in platelets through binding to surface β3 integrins [56,59], and several PDI substrates have been identified in platelet-rich plasma [21]. A similar effect likely occurred with respect to other circulating or EC-attached PDI substrates, which may account for its plasma level variability. Finally, recent observations suggest that single nucleotide polymorphisms are a substantial source of plasma protein variability [60], an issue deserving further investigation for PDI. Overall levels of pecPDI represent a minor fraction of total intracellular PDI pool, but can increase following forced PDI overexpression [61]. Moreover, it is important to point that our study addressed healthy individuals, while disease processes associated with ongoing vascular injury and inflammation may enhance circulating PDI levels due to cell death/leakage [21].
An important result was the contrasting functional effects of PDI-rich plasma, which supported organized EC migration, vs. PDI-poor plasma, which significantly disrupted the migration pattern. In both cases, such effects are likely related to corresponding plasma protein signatures rather than to PDI levels themselves. This is because the pM/nM PDI concentration ranges contrast with higher concentrations of its many plasma substrates (e.g., HRG, 100–150 μg/mL) [45], other thiol proteins (400–600 μM) and low-molecular weight thiols (7.3–9.0 μM for glutathione and 202–281 μM for cysteine [1]). Thus, it is likely that PDI levels behave as a reporter of the amount and type of bound substrates.
PDI adds to other thiol-proteins with potential as disease biomarkers such as glutathione peroxidase-3, quiescin sulfhydryl oxidase, thioredoxin/thioredoxin reductase, glutaredoxin [1] and S-nitrosoglutathione reductase [62]. However, except for a described association of glutaredoxin-1 with endothelial permeability [63], functional endothelial effects related to such proteins remain unknown, as well as their possible correlation with plasma protein signatures. In fact, to the best of our knowledge, our study is the first to describe different plasma proteome profiles and corresponding EC phenotypes related to vascular (dys)function and thus our data open new ways to understand how plasma biomarkers in general, and specifically redox biomarkers, can interplay with disease processes. A clear limitation of our results is that the type of population and the small number of subjects does not allow inferences about biomarker capabilities of PDI levels. Nevertheless, to get a potential insight into this question, we evaluated PDI levels from plasma samples (n = 95) collected in a previously well-described and validated databank from a population composed of individuals with clinically evident vascular disease [64]. Patients who had documented cardiovascular events, namely myocardial infarction, unstable angina and cerebrovascular accident (further detailed at Table 2) showed significantly lower median circulating PDI levels vs. healthy individuals (Fig. 7D), indicating that lower PDI levels may have a general correlation with enhanced cardiovascular risk in patients with chronic atherosclerotic disease. Again, PDI levels were unrelated to other traditional risk factors (gender, LDL-cholesterol levels, HDL-cholesterol levels, triglycerides, diabetes, body mass index – Suppl. Table S6) except age (which showed an association, p < 0.05). These data are in line with our proteomic and functional studies in healthy individuals, but, again, further studies are needed to assess general applicability of PDI levels as risk biomarkers.
Table 2.
Clinical characteristics of a population from a plasma databank who had cardiovascular events.
| n | 95 |
| Age (years) | 50.4 ± 1.39 |
| Male sex (%) | 40 |
| Total cholesterol, mg/dL | 279.3 ± 7.5 |
| HDL cholesterol | 48.6 ± 2.3 |
| LDL cholesterol, mg/dL | 204.8 ± 7.3 |
| Triglycerides, mg/dL | 156.4 ± 9.2 |
| Medical history (%) | |
| Hypertension | 80 |
| Diabetes | 28 |
| FH | 35 |
| Obesity | 14 |
| Type of CVE (%) | |
| Unstable angina | 74 |
| AMI | 70 |
| CVA | 10 |
| PDI levels, pg/mL | |
| Median | 35 |
| Mean | 285.1 |
| 25% percentile | 0 |
| 75% percentile | 277 |
Data represent mean ± SEM unless specified otherwise. HDL: High-density lipoproteins, LDL: Low-density lipoprotein, FH: familial hypercholesterolemia, CVE: Cardiovascular event, AMI: acute myocardial infarction, CVA: cerebrovascular accident.
Overall, our results reinforce that plasma proteins including PDI are both contributors and accessible reporters of disease-associated conditions [65] and provide evidence that these concepts also extend to healthy individuals. Improved identification of suitable reporters, particularly those related to PDI, can yield insights into disease mechanisms involving endothelium-related plasma protein signatures. This redox-related protein may pave the way to the innovative concept of endothelial-related liquid biopsy.
Declaration of interest
None.
Acknowledgements
We thank Laura Ventura and Carolina Fernandes for technical support with blood collection procedure.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101142.
Funding
This work was supported by the CEPID Redoxoma- FAPESP (Fundação de Amparo à Pesquisa do Estado de Sao Paulo) grant 2013/07937-8, in addition to scholarship grants 2014/20595-1 (to PVSO), 2017/19866-9 (to NTS) and 2018/07511-4 (to TCDB). FAPESP grant 2013/25177-0, Conselho Nacional de Desenvolvimento Científico e Tecnológico–CNPq–Grant 305245/2015-5 and Fundação Butantan (to MLS). CAPES (Coordenação de Aperfeiçoamento de Pessoa de Nível Superior) fellowship (to ATAS). FAPESP grants 2013/08711-3, 2014/10068-4 and 2017/25588-1 (to DM-S). CAPES-PROEX grant 154529 (to SG-R).
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Oliveira P.V.S., Laurindo F.R.M. Implications of plasma thiol redox in disease. Clin. Sci. (Lond.) 2018;132:1257–1280. doi: 10.1042/CS20180157. [DOI] [PubMed] [Google Scholar]
- 2.Ellgaard L., Ruddock L.W. The human protein disulphide isomerase family: substrate interactions and functional properties. EMBO Rep. 2005;6:28–32. doi: 10.1038/sj.embor.7400311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Laurindo F.R., Pescatore L.A., Fernandes DeC. Protein disulfide isomerase in redox cell signaling and homeostasis. Free Radic. Biol. Med. 2012;52:1954–1969. doi: 10.1016/j.freeradbiomed.2012.02.037. [DOI] [PubMed] [Google Scholar]
- 4.Appenzeller-Herzog C., Ellgaard L. The human pdi family: versatility packed into a single fold. Biochim. Biophys. Acta. 2008;1783:535–548. doi: 10.1016/j.bbamcr.2007.11.010. [DOI] [PubMed] [Google Scholar]
- 5.Wang L., Wang X., Wang C.C. Protein disulfide-isomerase, a folding catalyst and a redox-regulated chaperone. Free Radic. Biol. Med. 2015;83:305–313. doi: 10.1016/j.freeradbiomed.2015.02.007. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka L.Y., Araújo H.A., Hironaka G.K., Araujo T.L., Takimura C.K., Rodriguez A.I., Casagrande A.S., Gutierrez P.S., Lemos-Neto P.A., Laurindo F.R. Peri/epicellular protein disulfide isomerase sustains vascular lumen caliber through an anticonstrictive remodeling effect. Hypertension. 2016;67:613–622. doi: 10.1161/HYPERTENSIONAHA.115.06177. [DOI] [PubMed] [Google Scholar]
- 7.Soares Moretti A.I., Martins Laurindo F.R. Protein disulfide isomerases: redox connections in and out of the endoplasmic reticulum. Arch. Biochem. Biophys. 2017;617:106–119. doi: 10.1016/j.abb.2016.11.007. [DOI] [PubMed] [Google Scholar]
- 8.Bekendam R.H., Flaumenhaft R. Inhibition of protein disulfide isomerase in thrombosis. Basic Clin. Pharmacol. Toxicol. 2016;119(Suppl 3):42–48. doi: 10.1111/bcpt.12573. [DOI] [PubMed] [Google Scholar]
- 9.Zhou J., Wu Y., Wang L., Rauova L., Hayes V.M., Poncz M., Essex D.W. The c-terminal cghc motif of protein disulfide isomerase supports thrombosis. J. Clin. Invest. 2015;125:4391–4406. doi: 10.1172/JCI80319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Araujo T.L.S., Zeidler J.D., Oliveira P.V.S., Dias M.H., Armelin H.A., Laurindo F.R.M. Protein disulfide isomerase externalization in endothelial cells follows classical and unconventional routes. Free Radic. Biol. Med. 2017;103:199–208. doi: 10.1016/j.freeradbiomed.2016.12.021. [DOI] [PubMed] [Google Scholar]
- 11.Jasuja R., Passam F.H., Kennedy D.R., Kim S.H., van Hessem L., Lin L., Bowley S.R., Joshi S.S., Dilks J.R., Furie B., Furie B.C., Flaumenhaft R. Protein disulfide isomerase inhibitors constitute a new class of antithrombotic agents. J. Clin. Invest. 2012;122:2104–2113. doi: 10.1172/JCI61228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sharda A., Kim S.H., Jasuja R., Gopal S., Flaumenhaft R., Furie B.C., Furie B. Defective pdi release from platelets and endothelial cells impairs thrombus formation in hermansky-pudlak syndrome. Blood. 2015;125:1633–1642. doi: 10.1182/blood-2014-08-597419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crescente M., Pluthero F.G., Li L., Lo R.W., Walsh T.G., Schenk M.P., Holbrook L.M., Louriero S., Ali M.S., Vaiyapuri S., Falet H., Jones I.M., Poole A.W., Kahr W.H., Gibbins J.M. Intracellular trafficking, localization, and mobilization of platelet-borne thiol isomerases. Arterioscler. Thromb. Vasc. Biol. 2016;36:1164–1173. doi: 10.1161/ATVBAHA.116.307461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bekendam R.H., Bendapudi P.K., Lin L., Nag P.P., Pu J., Kennedy D.R., Feldenzer A., Chiu J., Cook K.M., Furie B., Huang M., Hogg P.J., Flaumenhaft R. A substrate-driven allosteric switch that enhances pdi catalytic activity. Nat. Commun. 2016;7:12579. doi: 10.1038/ncomms12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu S., Butkevich A.N., Yamada R., Zhou Y., Debnath B., Duncan R., Zandi E., Petasis N.A., Neamati N. Discovery of an orally active small-molecule irreversible inhibitor of protein disulfide isomerase for ovarian cancer treatment. Proc. Natl. Acad. Sci. U. S. A. 2012;109:16348–16353. doi: 10.1073/pnas.1205226109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xu S., Sankar S., Neamati N. Protein disulfide isomerase: a promising target for cancer therapy. Drug Discov. Today. 2014;19:222–240. doi: 10.1016/j.drudis.2013.10.017. [DOI] [PubMed] [Google Scholar]
- 17.Stopa J.D., Zwicker J.I. The intersection of protein disulfide isomerase and cancer associated thrombosis. Thromb. Res. 2018;164(Suppl 1):S130–S135. doi: 10.1016/j.thromres.2018.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Essex D.W., Wu Y. Multiple protein disulfide isomerases support thrombosis. Curr. Opin. Hematol. 2018;25:395–402. doi: 10.1097/MOH.0000000000000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Essex D.W. Redox control of platelet function. Antioxidants Redox Signal. 2009;11:1191–1225. doi: 10.1089/ars.2008.2322. [DOI] [PubMed] [Google Scholar]
- 20.Mor-Cohen R. Disulfide bonds as regulators of integrin function in thrombosis and hemostasis. Antioxidants Redox Signal. 2016;24:16–31. doi: 10.1089/ars.2014.6149. [DOI] [PubMed] [Google Scholar]
- 21.Sharda A., Furie B. Regulatory role of thiol isomerases in thrombus formation. Expert Rev. Hematol. 2018;11:437–448. doi: 10.1080/17474086.2018.1452612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bowley S.R., Fang C., Merrill-Skoloff G., Furie B.C., Furie B. Protein disulfide isomerase secretion following vascular injury initiates a regulatory pathway for thrombus formation. Nat. Commun. 2017;8:14151. doi: 10.1038/ncomms14151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stopa J.D., Baker K.M., Grover S.P., Flaumenhaft R., Furie B. Kinetic-based trapping by intervening sequence variants of the active sites of protein-disulfide isomerase identifies platelet protein substrates. J. Biol. Chem. 2017;292:9063–9074. doi: 10.1074/jbc.M116.771832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stopa J.D., Neuberg D., Puligandla M., Furie B., Flaumenhaft R., Zwicker J.I. Protein disulfide isomerase inhibition blocks thrombin generation in humans by interfering with platelet factor v activation. JCI Insight. 2017;2 doi: 10.1172/jci.insight.89373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peixoto Á., Geyer R.R., Iqbal A., Truzzi D.R., Soares Moretti A.I., Laurindo F.R.M., Augusto O. Peroxynitrite preferentially oxidizes the dithiol redox motifs of protein-disulfide isomerase. J. Biol. Chem. 2018;293:1450–1465. doi: 10.1074/jbc.M117.807016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J., Mayfield S.P. Protein disulfide isomerase as a regulator of chloroplast translational activation. Science. 1997;278:1954–1957. doi: 10.1126/science.278.5345.1954. [DOI] [PubMed] [Google Scholar]
- 27.Fernandes D.C., Manoel A.H., Wosniak J., Laurindo F.R. Protein disulfide isomerase overexpression in vascular smooth muscle cells induces spontaneous preemptive nadph oxidase activation and nox1 mrna expression: effects of nitrosothiol exposure. Arch. Biochem. Biophys. 2009;484:197–204. doi: 10.1016/j.abb.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 28.Essex D.W., Miller A., Swiatkowska M., Feinman R.D. Protein disulfide isomerase catalyzes the formation of disulfide-linked complexes of vitronectin with thrombin-antithrombin. Biochemistry. 1999;38:10398–10405. doi: 10.1021/bi990694s. [DOI] [PubMed] [Google Scholar]
- 29.Chen K., Lin Y., Detwiler T.C. Protein disulfide isomerase activity is released by activated platelets. Blood. 1992;79:2226–2228. [PubMed] [Google Scholar]
- 30.Chen K., Detwiler T.C., Essex D.W. Characterization of protein disulphide isomerase released from activated platelets. Br. J. Haematol. 1995;90:425–431. doi: 10.1111/j.1365-2141.1995.tb05169.x. [DOI] [PubMed] [Google Scholar]
- 31.Banfi C., Brioschi M., Wait R., Begum S., Gianazza E., Pirillo A., Mussoni L., Tremoli E. Proteome of endothelial cell-derived procoagulant microparticles. Proteomics. 2005;5:4443–4455. doi: 10.1002/pmic.200402017. [DOI] [PubMed] [Google Scholar]
- 32.Raturi A., Miersch S., Hudson J.W., Mutus B. Platelet microparticle-associated protein disulfide isomerase promotes platelet aggregation and inactivates insulin. Biochim. Biophys. Acta. 2008;1778:2790–2796. doi: 10.1016/j.bbamem.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 33.Fan G.Q., Qin R.R., Li Y.H., Song D.J., Chen T.S., Zhang W., Zhong M., Zhang Y., Xing Y.Q., Wang Z.H. Endothelial cells microparticle-associated protein disulfide isomerase promotes platelet activation in metabolic syndrome. Oncotarget. 2016;7:83231–83240. doi: 10.18632/oncotarget.13081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iqbal A., Paviani V., Moretti A.I., Laurindo F.R., Augusto O. Oxidation, inactivation and aggregation of protein disulfide isomerase promoted by the bicarbonate-dependent peroxidase activity of human superoxide dismutase. Arch. Biochem. Biophys. 2014;557:72–81. doi: 10.1016/j.abb.2014.06.013. [DOI] [PubMed] [Google Scholar]
- 35.Raturi A., Mutus B. Characterization of redox state and reductase activity of protein disulfide isomerase under different redox environments using a sensitive fluorescent assay. Free Radic. Biol. Med. 2007;43:62–70. doi: 10.1016/j.freeradbiomed.2007.03.025. [DOI] [PubMed] [Google Scholar]
- 36.Montano S.J., Lu J., Gustafsson T.N., Holmgren A. Activity assays of mammalian thioredoxin and thioredoxin reductase: fluorescent disulfide substrates, mechanisms, and use with tissue samples. Anal. Biochem. 2014;449:139–146. doi: 10.1016/j.ab.2013.12.025. [DOI] [PubMed] [Google Scholar]
- 37.Thery C., Amigorena S., Raposo G., Clayton A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006;30(1) doi: 10.1002/0471143030.cb0322s30. (Chapter 3):Unit 3 22. [DOI] [PubMed] [Google Scholar]
- 38.Giannakopoulos B., Gao L., Qi M., Wong J.W., Yu D.M., Vlachoyiannopoulos P.G., Moutsopoulos H.M., Atsumi T., Koike T., Hogg P., Qi J.C., Krilis S.A. Factor xi is a substrate for oxidoreductases: enhanced activation of reduced fxi and its role in antiphospholipid syndrome thrombosis. J. Autoimmun. 2012;39:121–129. doi: 10.1016/j.jaut.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 39.Moretti A.I.S., Pavanelli J.C., Nolasco P., Leisegang M.S., Tanaka L.Y., Fernandes C.G., Wosniak J., Jr., Kajihara D., Dias M.H., Fernandes D.C., Jo H., Tran N.V., Ebersberger I., Brandes R.P., Bonatto D., Laurindo F.R.M. Conserved gene microsynteny unveils functional interaction between protein disulfide isomerase and rho guanine-dissociation inhibitor families. Sci. Rep. 2017;7:17262. doi: 10.1038/s41598-017-16947-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moraes M.S., Costa P.E., Batista W.L., Paschoalin T., Curcio M.F., Borges R.E., Taha M.O., Fonseca F.V., Stern A., Monteiro H.P. Endothelium-derived nitric oxide (no) activates the no-epidermal growth factor receptor-mediated signaling pathway in bradykinin-stimulated angiogenesis. Arch. Biochem. Biophys. 2014;558:14–27. doi: 10.1016/j.abb.2014.06.011. [DOI] [PubMed] [Google Scholar]
- 41.Garcia S., Silva-Costa L.C., Reis-de-Oliveira G., Guest P.C., Baldasso P.A., Cassoli J.S., Martins-de-Souza D. Identifying biomarker candidates in the blood plasma or serum proteome. Adv. Exp. Med. Biol. 2017;974:193–203. doi: 10.1007/978-3-319-52479-5_15. [DOI] [PubMed] [Google Scholar]
- 42.Tang S., Morgan K.G., Parker C., Ware J.A. Requirement for protein kinase c theta for cell cycle progression and formation of actin stress fibers and filopodia in vascular endothelial cells. J. Biol. Chem. 1997;272:28704–28711. doi: 10.1074/jbc.272.45.28704. [DOI] [PubMed] [Google Scholar]
- 43.Lin L., Gopal S., Sharda A., Passam F., Bowley S.R., Stopa J., Xue G., Yuan C., Furie B.C., Flaumenhaft R., Huang M., Furie B. Quercetin-3-rutinoside inhibits protein disulfide isomerase by binding to its b'x domain. J. Biol. Chem. 2015;290:23543–23552. doi: 10.1074/jbc.M115.666180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prado G.N., Romero J.R., Rivera A. Endothelin-1 receptor antagonists regulate cell surface-associated protein disulfide isomerase in sickle cell disease. FASEB J. 2013;27:4619–4629. doi: 10.1096/fj.13-228577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jones A.L., Hulett M.D., Parish C.R. Histidine-rich glycoprotein: a novel adaptor protein in plasma that modulates the immune, vascular and coagulation systems. Immunol. Cell Biol. 2005;83:106–118. doi: 10.1111/j.1440-1711.2005.01320.x. [DOI] [PubMed] [Google Scholar]
- 46.Tsuchida-Straeten N., Ensslen S., Schäfer C., Wöltje M., Denecke B., Moser M., Gräber S., Wakabayashi S., Koide T., Jahnen-Dechent W. Enhanced blood coagulation and fibrinolysis in mice lacking histidine-rich glycoprotein (hrg) J. Thromb. Haemostasis. 2005;3:865–872. doi: 10.1111/j.1538-7836.2005.01238.x. [DOI] [PubMed] [Google Scholar]
- 47.Druhan L.J., Lance A., Li S., Price A.E., Emerson J.T., Baxter S.A., Gerber J.M., Avalos B.R. Leucine rich α-2 glycoprotein: a novel neutrophil granule protein and modulator of myelopoiesis. PLoS One. 2017;12 doi: 10.1371/journal.pone.0170261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Frey E.A., Miller D.S., Jahr T.G., Sundan A., Bazil V., Espevik T., Finlay B.B., Wright S.D. Soluble cd14 participates in the response of cells to lipopolysaccharide. J. Exp. Med. 1992;176:1665–1671. doi: 10.1084/jem.176.6.1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jasuja R., Furie B., Furie B.C. Endothelium-derived but not platelet-derived protein disulfide isomerase is required for thrombus formation in vivo. Blood. 2010;116:4665–4674. doi: 10.1182/blood-2010-04-278184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sumpio B.E., Riley J.T., Dardik A. Cells in focus: endothelial cell. Int. J. Biochem. Cell Biol. 2002;34:1508–1512. doi: 10.1016/s1357-2725(02)00075-4. [DOI] [PubMed] [Google Scholar]
- 51.Pober J.S., Sessa W.C. Evolving functions of endothelial cells in inflammation. Nat. Rev. Immunol. 2007;7:803–815. doi: 10.1038/nri2171. [DOI] [PubMed] [Google Scholar]
- 52.Badrichani A.Z., Stroka D.M., Bilbao G., Curiel D.T., Bach F.H., Ferran C. Bcl-2 and bcl-xl serve an anti-inflammatory function in endothelial cells through inhibition of nf-kappab. J. Clin. Invest. 1999;103:543–553. doi: 10.1172/JCI2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Damgaard R.B., Gyrd-Hansen M. Inhibitor of apoptosis (iap) proteins in regulation of inflammation and innate immunity. Discov. Med. 2011;11:221–231. [PubMed] [Google Scholar]
- 54.Ganz P., Heidecker B., Hveem K., Jonasson C., Kato S., Segal M.R., Sterling D.G., Williams S.A. Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. J. Am. Med. Assoc. 2016;315:2532–2541. doi: 10.1001/jama.2016.5951. [DOI] [PubMed] [Google Scholar]
- 55.Hudson D.A., Gannon S.A., Thorpe C. Oxidative protein folding: from thiol-disulfide exchange reactions to the redox poise of the endoplasmic reticulum. Free Radic. Biol. Med. 2015;80:171–182. doi: 10.1016/j.freeradbiomed.2014.07.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Flaumenhaft R., Furie B. Vascular thiol isomerases. Blood. 2016;128:893–901. doi: 10.1182/blood-2016-04-636456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Voigtlaender M., Holstein K., Spath B., Bokemeyer C., Langer F. Expression and release of platelet protein disulphide isomerase in patients with haemophilia a. Haemophilia. 2016;22:e537–e544. doi: 10.1111/hae.13074. [DOI] [PubMed] [Google Scholar]
- 58.Flaumenhaft R. Advances in vascular thiol isomerase function. Curr. Opin. Hematol. 2017;24:439–445. doi: 10.1097/MOH.0000000000000362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cho J., Kennedy D.R., Lin L., Huang M., Merrill-Skoloff G., Furie B.C., Furie B. Protein disulfide isomerase capture during thrombus formation in vivo depends on the presence of β3 integrins. Blood. 2012;120:647–655. doi: 10.1182/blood-2011-08-372532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Joshi A., Mayr M. In aptamers they trust. Circulation. 2018;138:2482–2485. doi: 10.1161/CIRCULATIONAHA.118.036823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Araujo T.L.S., Fernandes C.G., Laurindo F.R.M. Golgi-independent routes support protein disulfide isomerase externalization in vascular smooth muscle cells. Redox. Biol. 2017;12:1004–1010. doi: 10.1016/j.redox.2017.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hayashida K., Bagchi A., Miyazaki Y., Hirai S., Seth D., Silverman M.G., Rezoagli E., Marutani E., Mori N., Magliocca A., Liu X., Berra L., Hindle A.G., Donnino M.W., Malhotra R., Bradley M.O., Stamler J.S., Ichinose F. Improvement in outcomes after cardiac arrest and resuscitation by inhibition of s-nitrosoglutathione reductase. Circulation. 2018;139(6) doi: 10.1161/CIRCULATIONAHA.117.032488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Han J., Weisbrod R.M., Shao D., Watanabe Y., Yin X., Bachschmid M.M., Seta F., Janssen-Heininger Y.M.W., Matsui R., Zang M., Hamburg N.M., Cohen R.A. The redox mechanism for vascular barrier dysfunction associated with metabolic disorders: glutathionylation of rac1 in endothelial cells. Redox. Biol. 2016;9:306–319. doi: 10.1016/j.redox.2016.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jannes C.E., Santos R.D., de Souza Silva P.R., Turolla L., Gagliardi A.C., Marsiglia J.D., Chacra A.P., Miname M.H., Rocha V.Z., Filho W.S., Krieger J.E., Pereira A.C. Familial hypercholesterolemia in Brazil: cascade screening program, clinical and genetic aspects. Atherosclerosis. 2015;238:101–107. doi: 10.1016/j.atherosclerosis.2014.11.009. [DOI] [PubMed] [Google Scholar]
- 65.Vegvari A., Welinder C., Lindberg H., Fehniger T.E., Marko-Varga G. Biobank resources for future patient care: developments, principles and concepts. J. Clin. Bioinf. 2011;1:24. doi: 10.1186/2043-9113-1-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cassoli J S., Brandão-Teles C., Santana A G., Souza G H.M.F., Martins-de-Souza D. Ion mobility-enhanced data-independent acquisitions enable a deep proteomic landscape of oligodendrocytes. Proteomics. 2017;17(21) doi: 10.1002/pmic.201700209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Video representing migration into the denuded area after incubation with medium without FBS.
Video representing migration into the denuded area after incubation with fetal bovine serum (FBS)-control.
Video representing migration into the denuded area after incubation with PDI-poor plasma.
Video representing migration into the denuded area after incubation with PDI-rich plasma.