Abstract
Poisson-Boltzmann equation (PBE)-based continuum electrostatics models have been widely used in modeling electrostatic interactions in biochemical processes, particularly in estimating protein-ligand binding affinities. Fast convergence of PBE solvers is crucial in binding affinity computations as numerous snapshots need to be processed. Efforts have been reported to develop PBE solvers on graphics processing units (GPUs) for efficient modeling of biomolecules, though only relatively simple successive over-relaxation and conjugate gradient methods were implemented. However, neither convergence nor scaling properties of the two methods are optimal for large biomolecules. On the other hand, geometric multigrid (MG) has been shown to be an optimal solver on CPUs, though no MG was reported for biomolecular applications on GPUs. This is not a surprise as it is a more complex method and depends on simpler but limited iterative methods such as Gauss-Seidel in its core relaxation procedure. The robustness and efficiency of MG on GPUs are also unclear. Here we present an implementation and a thorough analysis of MG on GPUs. Our analysis shows that robustness is a more pronounced issue than efficiency for both MG and other tested solvers when the single precision is used for complex biomolecules. We further show how to balance robustness and efficiency by utilizing MG’s overall efficiency and conjugate gradient’s robustness, pointing to a hybrid GPU solver with a good balance of efficiency and accuracy. The new PBE solver will significantly improve the computational throughput for a range of biomolecular applications on the GPU platforms.
Graphical Abstract
1. Introduction
Poisson-Boltzmann equation (PBE)-based continuum electrostatics modeling has gained wide acceptance in biomolecular applications, given the crucial roles played by electrostatic interactions in biochemical processes such as protein-protein and protein-ligand recognitions.1–18 For example, PBE-based models have been widely used in the binding affinity computations, such as in the Molecular Mechanics Poisson-Boltzmann Surface Area (MMPBSA) method.19–24 Because of the extremely complex geometries of biomolecules, it is vital to increase the accuracy and efficiency of PBE-based models for nontrivial applications in biomolecular simulations.25–33
For biomolecular applications, it is impossible to solve the PBE analytically. Instead, we rely on numerical solutions. The most widely adopted numerical method is the finite-difference method,34–46 where finite-difference grids are used to discretize the space and build up a set of linear/nonlinear equations. Other traditional numerical schemes include the finite-element method,47–55 the box method,40 the boundary element method,56–71 and the boundary integral method.72, 73 Different from these direct numerical methods, indirect methods such as a free energy functional variational formulation have also been explored to solve PBE,12, 74 where the solution is obtained by searching for the minimum in the functional space. Among these approaches, FDM solvers for the PBE have been incorporated in several programs such as DelPhi,34, 36, 43, 75, 76 APBS,38, 40, 77 UHBD,35, 37 CHARMM/PBEQ,36, 42 and Amber/PBSA.30, 44–46, 78, 79 Numerical PBE schemes have been applied to the prediction of pKa values,80–85 solvation free energies,86–91 and binding free energies,92–98 and the analysis of protein folding and biosynthesis,99–111 and the design of new functional proteins.112, 113
As the computational studies shift to larger and more complex biomolecular systems, both program efficiency and convergence rate become more challenging to address on conventional CPU-based computing platforms. These challenges are more pronounced when incorporating the PBE in typical biomolecular applications such as post processing binding free energy calculations and modeling amino acid mutation effects114, 115 that involve tens of thousands of snapshots. An interesting direction to address these challenges is to explore alternative hardware, such as the graphics processing units (GPUs), which have gradually been employed in a range of computational chemistry problems with impressive speedup, for example in MD simulations116–118 and ab initio quantum mechanical (QM) calculations.119–121 Several publications have also reported to use GPUs to accelerate linear PBE solutions for biomolecular systems with impressive speedup.122–124 However, different from MD or QM simulations, various PBE solvers behave with markedly different efficiency on CPUs.45, 46 This is an important issue to consider when porting PBE calculations to GPUs.
To date only relatively simple PBE solvers were implemented on GPUs, including the successive over-relaxation (SOR) method,122, 123 the conjugate gradient methods (CG),124 and the direct-sum boundary integral method with algebraic GMRES.125 However, our prior analysis of various PBE solvers have shown that the convergence rate of many simple methods is not optimal on CPUs for large systems or tight convergence criteria even if they are simple to implement.45, 46 Specifically for SOR, there are two disadvantages. Firstly, a parallel (i.e. red-black) SOR has to be used on GPUs. However, the convergence rate of the red-black SOR is slower than SOR due to its color-ordered updating approach. Secondly, for most consumer-level GPU cards, single precision operations are widely supported with high efficiency. Double precision operations are at a significant disadvantage. Unfortunately, use of single precision further reduces the convergence rate of the red-black SOR. CG, as one of the best known Krylov subspace methods, has shown to be more stable but its scaling was not optimal for large biomolecular systems.124
Multigrid methods are known to be optimal for solving many linear and nonlinear systems on CPUs. Typically there are two classes of multigrid methods: geometric multigrid and algebraic multigrid.126 Geometric multigrid requires prior physical/mathematical knowledge of the underlying discretization and grid hierarchy, whereas algebraic multigrid only requires the coefficient matrix. Previous studies have shown that geometric multigrid is an optimal PBE solver for biomolecular applications.38, 45, 46, 127, 128 Algebraic multigrid can be used both as a direct PBE solver and as a preconditioner for a CG PBE solver.124, 129 However, its performance was not promising for biomolecules as our previous testing has shown.45, 124 Algebraic multigrid130, 131 and specialized multigrid132–134 were also ported to GPUs on the Nvidia platform for various elliptic partial differential equation problems.
To date, there has been no report of geometric multigrid (MG) as a PBE solver on GPUs. This is not a surprise, as there are many implementation issues, particularly for applications to large and complex biomolecules. What further complicates the issue is the use of iterative methods such as SOR or Gauss-Seidel in its core relaxation procedure, as these simple methods are limited in the widely used single-precision mode as reviewed above. In addition, the robustness and efficiency of MG on GPUs are unclear. Therefore, a thorough analysis of MG on GPUs is a necessary step to realize marked overall efficiency improvement.
In the following, we present an implementation and a thorough analysis of the MG PBE solver for complex biomolecules on GPUs based on the Nvidia Compute Unified Device Architecture (CUDA). Our results show the robustness is a more pronounced issue than efficiency among the tested GPU PBE solvers in the widely used single precision mode. We further show how to balance robustness and efficiency in MG solver for complex biomolecular applications.
2. Methods
2.1. Poisson-Boltzmann Equation
In Poisson-Boltzmann continuum solvent models, both solute and solvent molecules are represented as continua: the solute molecule is treated as a low-dielectric cavity embedded in a high-dielectric continuum representing the solvent molecules collectively. For an electrolyte solution, a Boltzmann term describing the salt effect can be used in the Poisson equation, leading to the well-known Poisson-Boltzmann equation (PBE)
| (1) |
where ε is the dielectric constant, u is the electrostatic potential, ρ is the charge density, and λ is a masking function for the Stern layer. All variables are functions of position vector r. In the salt related term, ni is the number density of ion type i in the bulk electrolyte, qi is the charge of ion type i, k is the Boltzmann constant, and T is the temperature. Obviously the PBE is a non-linear partial differential equation. When qiu(r)/kT is small, the PBE can be linearized as
| (2) |
Only numerical solutions of equation (1) or (2) can be obtained for biomolecules. In this development the equations are discretized with a simple finite-volume scheme. The linearized PBE, for example, can be written at each grid point as follows
| (3) |
where h is the grid spacing, i, j, and k are grid indexes along x, y, and z axes, respectively. εi(i, j, k) is the dielectric constant between grid points (i, j, k) and (i+1, j, k). εj(i, j, k) and εk(i, j, k) are defined similarly. κ2 absorbs all related coefficients in the Boltzmann term. q(i, j, k) is the total solute charge within a cubic volume h3 centered at (i, j, k). The linear system can then be denoted symbolically as
| (4) |
where A is the left-hand-side coefficient matrix and f is the right-hand-side constant in equation (3).
To solve equation (4), a number of solvers have been developed for biomolecular applications, such as successive over-relaxation,135 conjugate gradient,135 (modified) incomplete Cholesky conjugate gradient ((M)ICCG),136–139 geometric multigrid,140 and algebraic multigrid.141 All solvers proceed from an initial guess of u(i, j, k) to approach the solution iteratively. In this manuscript, we present an implementation of the geometric multigrid method on the CUDA GPU platform and a detailed analysis of its performance along with other GPU-friendly methods.
2.2. Geometric Multigrid Method
Geometric multigrid (MG) methods are highly efficient techniques to solve linear equations or nonlinear equations with an algorithm complexity of O(N) for a system of N grid points.142 In the following the basic ideas of MG are first introduced before discussion of their GPU-specific issues.
In MG, a multigrid hierarchy is constructed by partitioning the system into coarse and fine grid levels. Spectral analysis of some of the basic iterative methods such as Gauss-Seidel shows that they are very good at eliminating both high-frequency or oscillatory components of the error on fine grids, and low-frequency or smooth components of the error on coarse grids.142 Therefore, the multigrid hierarchy can be utilized to solve the linear system by using the same basic iterative methods as smoothers on every level. The coarse grid points form a coarse level, and an interpolation operator, defined as a weighted sum of the coarse grid points, is used to interpolate a coarse level solution to a fine level. A restriction operator, usually taken as the transpose of the interpolation operator, is used to restrict a fine-level solution to a coarse level.143, 144 Worth noting is that in our implementation, the restriction and interpolation operators were implemented as harmonic averaging as proposed by Holst et al.38
There are basic and nested strategies for the MG methods, leading to the V-Cycle and F-Cycle algorithms, respectively. By combining relaxation, restriction and interpolation operators, a V-Cycle scheme can be constructed as shown in Figure 1(a), and a recursive V-Cycle scheme is provided in Algorithm 1. The linear restriction and interpolation operators are denoted as and , respectively, below.
Figure 1.
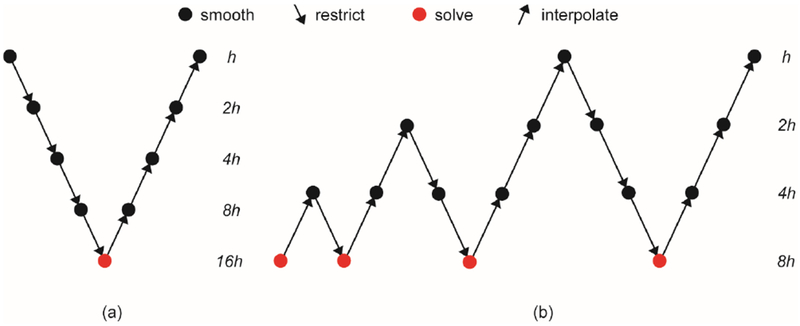
Illustrations of (a) V-Cycle and (b) F-Cycle.
Algorithm 1. MG V-Cycle (Recursive):
Relax ν1 times on Ahuh=fh with a given initial guess νh.
-
If Ωh = coarsest grid, then go to step 4.
Else Correct .
Relax ν2 times on Ahuh=fh with a given initial guess νh.
The idea of using coarsest-grid solutions to obtain improved initial guesses for the finest-grid solutions is called nested iteration, as relaxation on the coarsest grid is not expensive. The algorithm that joins the nested iteration with the V-Cycle leads to the full multigrid V-Cycle, or F-Cycle for short. An F-Cycle scheme is illustrated in the Figure 1(b) and a recursive F-Cycle algorithm is shown in Algorithm 2. In this study, we implemented both V-Cycle and combined F-V-Cycle MG solvers on GPUs and analyzed their performance.
Algorithm 2. MG F-Cycle (Recursive):
-
If Ωh=coarsest grid, set νh ← 0 and go to step 3.
Else Correct .
Solve νh ← Vh (νh, fh) with ν0 cycles.
2.3. Red-Black Gauss-Seidel/SOR Relaxations
As reviewed above, a core routine of the MG method is to relax all grids using an iterative relaxer. There are many basic iterative methods such as Gauss-Seidel and SOR that can be utilized.
The basic idea of SOR is that components of a new approximated solution are used as soon as they are computed.142 To solve the linear equation (4) Au = f, we first introduce a decomposition of A, such as A = D − L − U, where D is the diagonal of A, U and L are the negative upper and lower triangular parts of A, respectively. Equation (4) can then be rewritten as
where ω is an over-relaxation factor. Note that when ω = 1, SOR reduces to Gauss-Seidel. This representation corresponds to solving the jth equation for uj and using latest approximations for components 1, 2, …, j−1.
Defining the Gauss–Seidel/SOR iteration matrix as
we can express the method as the following given an approximate solution v,
The order in which the components of v are updated can be ascending or descending.
Another effective alternative is to update all the even components first and then update all the odd components next. This strategy leads to the red-black Gauss-Seidel/SOR method,145 as illustrated in Figure 2. Notice that the red points only depend on the black points and vice versa. This leads to some significant advantage for parallel computation, as shown in several MPI implementations.75, 146 Here in our MG implementation, the red-black Gauss-Seidel is used as the relaxer at every level except the coarsest level where the red-black SOR is used.
Figure 2.
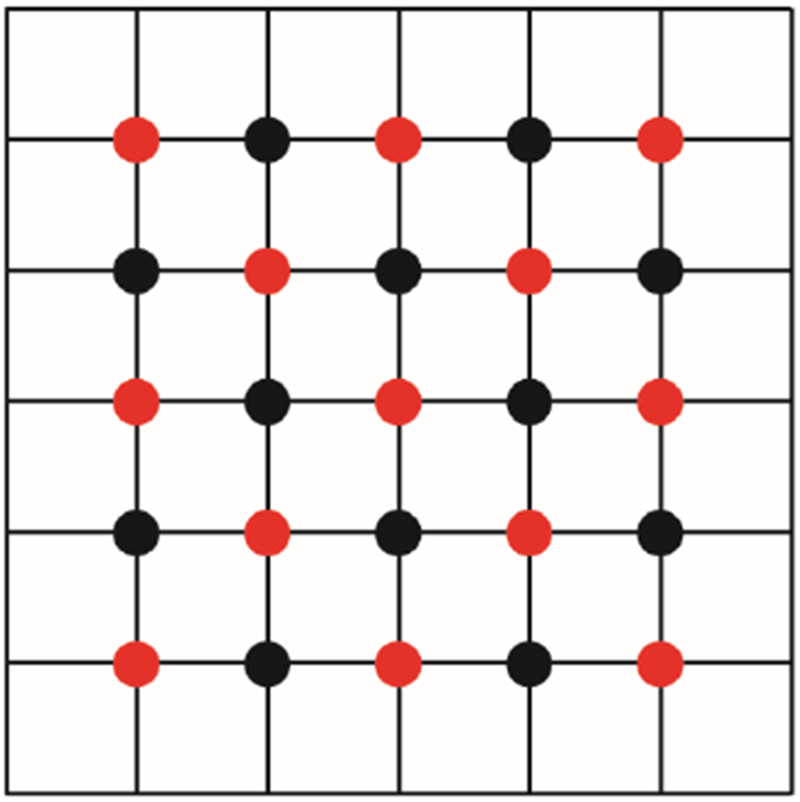
Illustration of a 2-D red-black grid.
2.4. CUDA Unified Memory and Array Optimization
Unified Memory was introduced in CUDA 6. It provides a new programming framework that allows GPU applications to use a single pointer in both CPU functions and GPU kernels. This markedly simplifies memory management. Later CUDA 8 and the Pascal architecture significantly improves the Unified Memory functionalities by introducing 49-bit virtual addressing and on-demand page migration. The large 49-bit virtual addresses are sufficient to enable GPUs to access the entire system memory plus the memory of all GPUs in the system. The Page Migration engine allows GPU threads to fault on non-resident memory accesses, so that the system can migrate pages from anywhere in the system to the GPUs memory on-demand for efficient processing.147
However, page faults and migration can be expensive. To avoid the overhead, data prefetching and usage hint functionalities may be used. In our MG implementations on Pascal architectures, we utilized CUDA Unified Memory and optimized memory access with cudaMemAdvise and cudaMemPrefetchAsync application programming interfaces. On pre-Pascal architectures, we used zero-copy memory to avoid data migrations.134
Similar to CPU implementation of FDPB solvers,44 the matrix arrays were stored in the matrix-free style with padded zeros at grid boundaries so that fast stencil access can be achieved and the conditional branch was avoided in all GPU kernels. When the MG solver is combined with the Jacobi-PCG solver implemented with the CUSP library,148 the matrix-free arrays were transformed into the diagonal matrix format first, which is suited for PBE seven-banded coefficient matrix computation, before feeding into later phase iterations.
2.5. Computational Details
All CUDA PBE solvers were implemented in both single and double precisions in the PBSA program45, 46, 78, 79, 149–161 of the Amber/AmberTools 18 packages.162, 163 The double precision was implemented for the robustness analysis only, and the single precision was used throughout the study unless otherwise specified. The largest 144 biomolecular structures from the Amber PBSA benchmark suite were used in our test.45 The number of atoms in these molecules range from 4,240 to 37,421. Their geometries are quite different, requiring at least 1 million grid points with the default setup. The atomic charges were assigned to those of Cornell et al164 and the atomic radii to the modified Bondi radius set.
All testing runs were performed with the following conditions unless specified otherwise. The convergence criteria of 10−4 and 10−6 were used for low- and high-precision applications, respectively, unless specified otherwise. The grid spacing of 0.5 Ǻ was used. The ratio of the grid dimension over the solute dimension (the fillratio keyword in Amber) was set to 1.5. No electrostatic focusing was applied. The potential values on all grid points were initialized to zero. The dielectric constants were set to 1 and 80 for solute and solvent, respectively. The weighted harmonic average of the solute and solvent dielectric constants was used as the boundary dielectric constants. Therefore, the symmetric and positive-definite coefficient matrices were obtained and suitable for all tested linear solvers. Finally, the conductor boundary condition was used to minimize the setup cost of boundary conditions. All other parameters were set as default in the PBSA module in the Amber/AmberTools 18 package.162
All testing was conducted on a dedicated compute node with two NVIDIA TITAN Xp GPU cards and one Intel Xeon E5-1620 v3 CPU and 16GB main memory. Our time measurements for each solver include all execution time of the core routine code, i.e. time elapsed on device (GPU) and on host (CPU) and also for transferring data between the device and the host.
3. Results and Discussion
In the following we first investigated the robustness of the GPU solvers with a diverse set of large and complex biomolecules and analyzed the cause of observed failures with the MG solver as the focus. Based on the analysis, we proposed to balance algorithm robustness and computational efficiency with a hybrid MG-PCG iteration scheme on GPUs. This is followed by further optimization of the MG iteration. Finally the performance of various GPU solvers is presented, along with a sanity check to confirm the numerical accuracy of the new implementation. We end this section with a discussion of implementation details and potential future improvements.
3.1. Robustness of GPU Solvers
We first tested three representative GPU solvers, SOR, Jacobi-PCG, and MG, in the single-precision modes on a set of 144 large biomolecules. The testing statistics are summarized in Table 1. With a convergence criterion 10−4 (or looser), all solvers were found to converge in all test cases. Once the convergence criterion is tightened to 10−5, SOR fails to converge with all test cases and MG fails to converge with five test cases. Interestingly Jacobi-PCG stands out by passing all tests. With the tightest tested convergence criterion of 10−6, even MG fails almost all except one case. Once again Jacobi-PCG passes all tests.
Table 1.
Number of failures in the test of different GPU solvers with a convergence criterion of 10−4, 10−5, and 10−6, respectively. The maximum allowed iteration steps is 3,000. S/D denotes the single/double precision mode. A total of 144 cases were used.
| Solver | SOR(S) | Jacobi-PCG(S) | MG(S) | SOR(D) | Jacobi-PCG(D) | MG(D) |
|---|---|---|---|---|---|---|
| 10−4 | 0 | 0 | 0 | 0 | 0 | 0 |
| 10−5 | 144 | 0 | 5 | 27 | 0 | 0 |
| 10−6 | 144 | 0 | 143 | 65 | 0 | 0 |
As a comparison all tests were rerun in the double precision mode. Under this condition, SOR still fails to converge in 27 cases at the criterion of 10−5 and 65 cases at that of 10−6. On the other hand, both Jacobi-PCG and MG are able to converge in all cases in all tested convergence criteria (Table 1). It is clear that MG has a better convergence behavior than SOR, but worse than Jacobi-PCG in the single-precision mode, apparently due to its dependence on SOR/Gauss-Seidel as the relaxers.
Inspection of the failed MG test cases shows that these failures are not due to their extra-large system sizes but due to their high net charges. Here we use protein 1TZY to illustrate the common characteristics of all failures. The protein contains 152 positive residues and 68 negative residues among a total of 755 residues and 12,321 atoms; and is discretized onto a finite-difference grid of 16,393,727 points. The residual L2-norm of the finest grid was observed to decrease from the initial 4.27×103 to 7.97×10−2 in eight V-Cycles, then halted around 7.30×10−2 in the following 144 V-Cycles until hitting the maximum relaxation iteration cycles allowed. Specifically, the iteration cycles needed at each level increased from the default setting of ten cycles to the maximum of 1,000 cycles, as shown in the Figure 3. In contrast, MG in the double precision mode was able to converge with a final L2-norm of 3.98×10−2 within seven V-Cycles.
Figure 3.
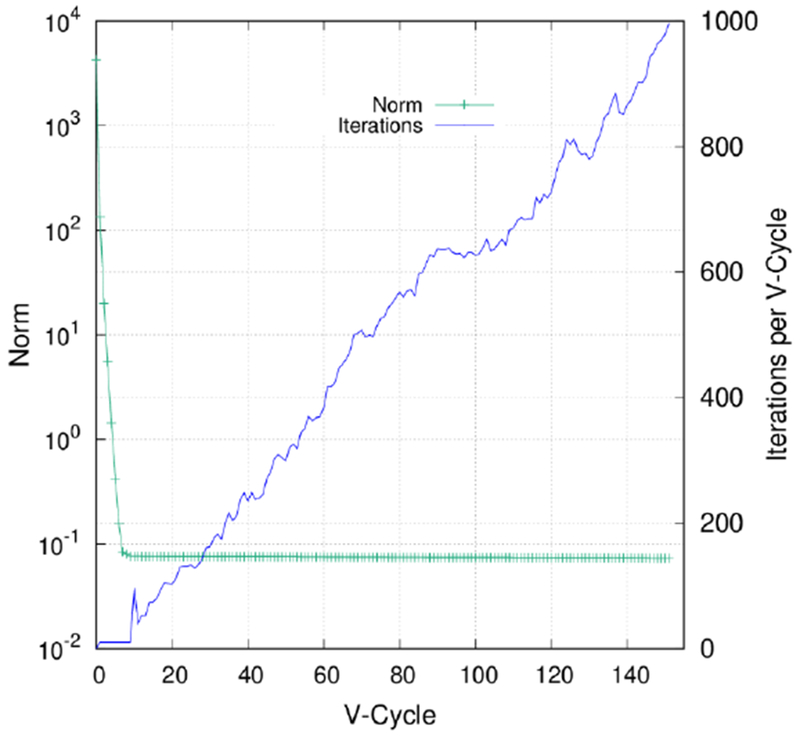
Decreasing L2-norm of the finest grid and increasing iteration cycles needed per V-Cycle of 1TZY. The single precision was used.
We also investigated whether the use of the red-black SOR/Gauss-Seidel instead of the original SOR/Gauss-Seidel was causing the failures by MG. Our test shows that MG with the original Gauss-Seidel also fails in the single precision, with the L2-norm oscillating around 6.50×10−2 after nine V-Cycles. Considering the error-smoothing nature of the MG algorithm, we can conclude that in charge-rich systems, the single-precision SOR/Gauss-Seidel iterations result in too much numerical noise that is too hard to reduce. Worth noting is that at the coarsest level, where the systems are always small, the single-precision SOR/Gauss-Seidel can be used to solve the linear system in exactly ten iterations. Thus, the difficulty faced in the single-precision MG runs is in the relaxation phase at the fine grid levels, but not in the final solution phase at the coarsest grid level.
3.2. Hybrid GPU Solver
Since MG solver is much faster than other GPU solvers if it can converge (most 10−5 cases as shown above) and Jaocbi-PCG is more robust with tighter convergence criteria, we introduced the Jacobi-PCG solver as a backup to address the MG solver’s convergence issue. For the same test case of 1TZY as shown in Figure 4, Jacobi-PCG was able to reduce the L2-norm to 5.43×10−3 in 259 iterations after MG V-Cycles fail to reduce the L2-norm further. Interestingly oscillation in L2-norm occurs at the switching point between the two solvers, but the L2-norm decreases thereafter. This is reasonable since the MG and CG solvers follow very different ideas (iterative vs. Krylov subspace) and thus approach the solution through different paths.
Figure 4.
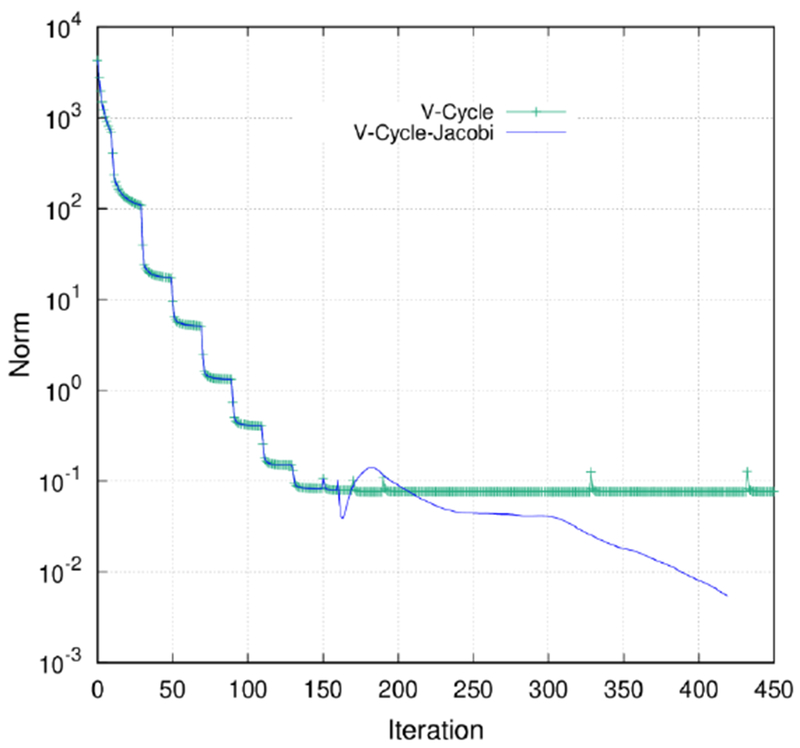
L2-norm of the finest grid versus number of iterations (either Gauss-Seidel iterations or PCG iterations) for 1TZY. Green is for V-Cycle solver. Blue is for V-Cycle-Jacobi-PCG hybrid solver.
Next we investigated the proper switching point between the two solvers. By monitoring the instant un-convergence rate (ruc) in the MG iteration, which is defined as the ratio of the L2-norm of the current step to that of the previous step (ruc = curent_norm/previous_norm), the solver scheduler smoothly switches from MG to Jacobi-PCG when ruc is larger than a preset cutoff. Our analysis shows that V-Cycles generally stop to converge after ruc becomes larger than 0.95, but different cutoff value may lead to different overall performance. To find the optimal cutoff, we analyzed the performance of the hybrid MG-Jacobi solver for twelve randomly selected test cases versus the ruc cutoff at the tight convergence criterion of 10−6. As shown in Figure 5, the cutoff value of 0.9 is a reasonable choice for most test cases.
Figure 5.
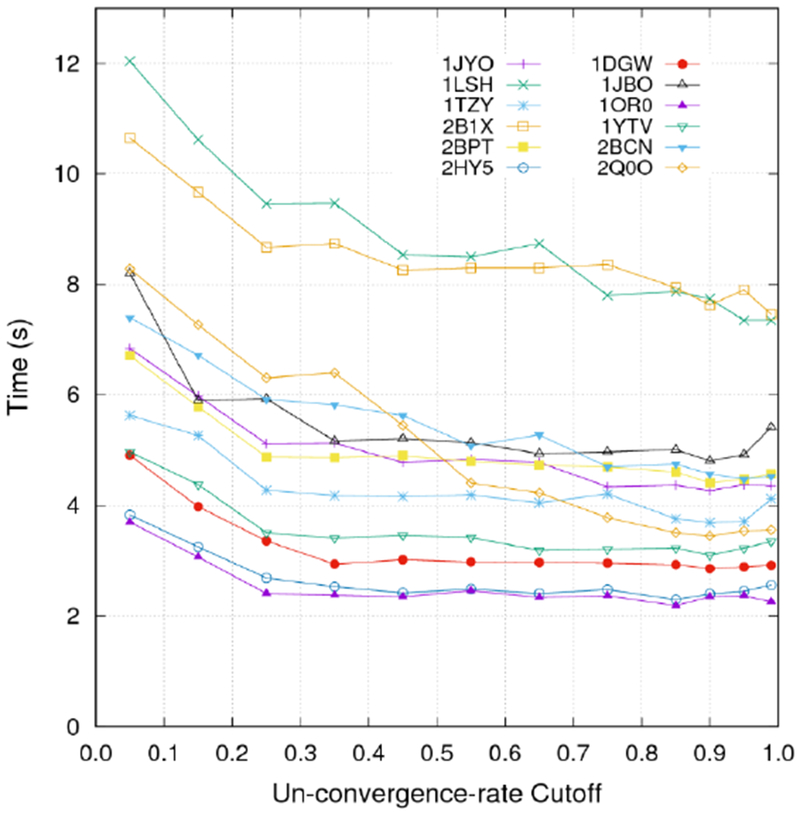
Performances of the hybrid MG-Jacobi-PCG solver for twelve randomly selected test cases versus the cutoff used for ruc. The convergence criterion was set to 10−6.
To illustrate the performance of the hybrid GPU solver, a detailed timing analysis is provided in Table 2 for the five test cases previously failed in the MG (single precision) runs. Worth noting is that the performance of the hybrid GPU solver is still better than the GPU/Jacobi-PCG solver, and the speedup factor is ~2.0. More thorough tests with all 144 protein systems show that the overall speedup ratio of the hybrid MG over Jacobi-PCG can be higher as ~5.0 as shown in Section 3.4.
Table 2.
Timing (in seconds) of the five test cases that previously failed in MG (single precision) runs by GPU/Jacobi-PCG and GPU/MG-Jacobi-PCG at the convergence criterion of 10−6.
| Protein | Ngrid | Jacobi-PCG | MG-Jacobi |
|---|---|---|---|
| 1TZY | 16393727 | 7.87 | 3.71 |
| 1JYO | 19433295 | 8.61 | 4.31 |
| 2BPT | 18727455 | 8.97 | 4.46 |
| 1LSH | 29984175 | 16.54 | 8.08 |
| 2B1X | 27740559 | 15.23 | 7.98 |
3.3. Efficiency Tuning of MG on GPUs
The V-Cycle used in the MG solver usually starts with zero initial potentials. However, it would be beneficial if a better initial guess than the default zero solution is used. The straightforward way is to solve the system on the coarsest grid first and interpolate the solution back to the finest level as the initial guess. To realize this idea, we embedded one F-Cycle at the beginning of the MG cycle to provide a better initial guess for the subsequent V-Cycles. As shown in Figure 6, improvement from 0.8% to 28% was observed compared to the V-Cycle-only MG at least at the tested condition on GPUs. Here the convergence criterion was set to 10−5 to make the problems more challenging while still without many convergence failures. Our analysis shows that one embedded F-Cycle on average saves three or four V-Cycles later.
Figure 6.
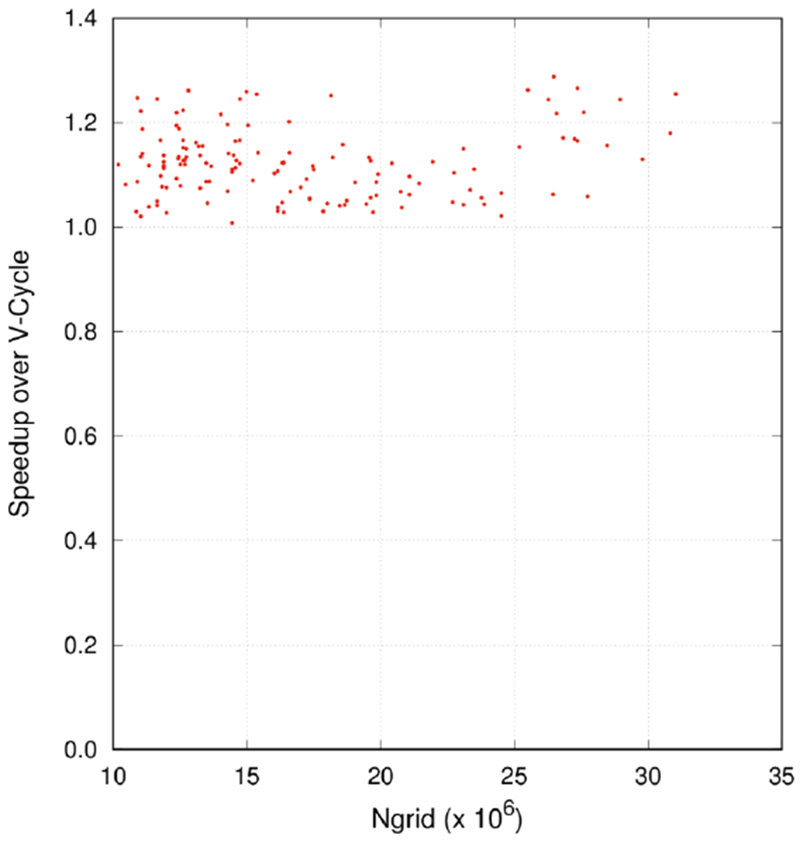
Speedup of 1F-V-Cycle over V-Cycle for the protein test set. The convergence criterion was set to 10−5.
Interestingly, however, using multiple F-Cycles as iterative engine deteriorates the convergence of MG (data not shown). This is reasonable considering that in one F-Cycle (see Figure 1(b)) a better initial guess is already obtained and used for the largest V-Cycle (the final cycle inside F-Cycle) to smooth errors. The solution obtained here should be fed directly into a subsequent V-Cycle to continue smoothing. If, however, another F-Cycle is applied subsequently, the solution would be restricted all way down to provide an initial guess for the coarsest level. This cancels the contribution of the first F-Cycle since on the coarsest level the system is small enough to be easily solved with any initial guess.
3.4. Efficiency Comparison of Various CUDA Solvers
As discussed in Methods, the red-black Gauss-Seidel was used as the smoother and the red-black SOR as the coarsest-level solver in the GPU MG cycles. Thus the performance gain of the GPU MG over the CPU MG can never be higher than that of the GPU SOR over the CPU SOR. Fortunately, it is good to note that the SOR can be implemented with extremely high efficiency as shown in Figure 7: a very high speedup ratios ranging from ~87 to ~109 can be obtained if the problem can be solved with the tested single precision condition. This is consistent with a previous development (Luty and Walker, Personal communication).
Figure 7.
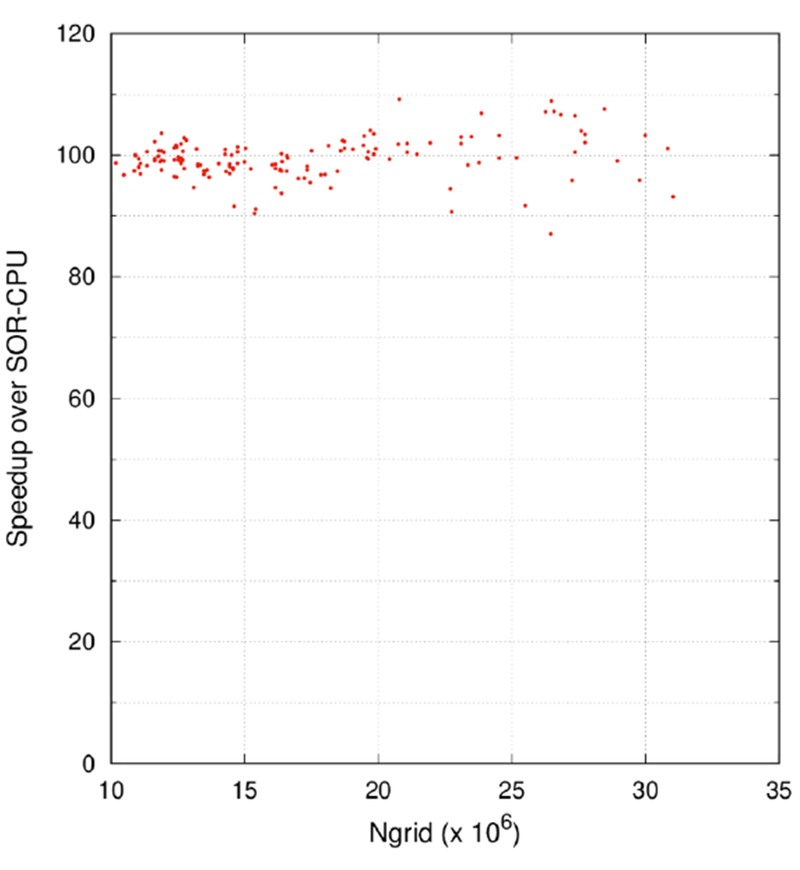
SOR speedup of GPU runs over CPU runs for the protein test set. The convergence criterion was set to 10−4, with which no failure was observed. Here the CPU version is the single-thread version without the red-black labeling.
Next a natural question is how much faster MG can achieve over the already very highly efficient SOR solver on GPUs. As a reference, we also compared the CG-based GPU (Jacobi-PCG) solver.124 Here eight representative proteins that cover from ~12 to ~30 million grid points were selected to measure their solver time with 10−4 and 10−6 convergence criteria. Table 3 shows that the hybrid MG solver overall performs the best among the three solvers with Jacobi-PCG as the second and SOR as the third. With the convergence criterion of 10−4 where SOR can converge, the speedup ratios of the hybrid MG and Jacobi-PCG solvers over SOR are ~3.0 to ~9.0 and ~0.9 to ~1.9, respectively. With the convergence criterion of 10−6 where SOR and pure MG mostly cannot converge, the speedup ratios of the hybrid MG solver over the Jacobi-PCG solver are ~1.9 to ~2.7.
Table 3.
Average time (in seconds) used by GPU solvers for eight selected proteins. The measurement is the time elapsed both on device (GPU) and on host (CPU) and on transferring data between the device and the host. The criteria of 10−4 and 10−6 were used.
| Protein | Ngrid | GPU | |||
|---|---|---|---|---|---|
| SOR | Jacobi-PCG | MG | |||
| Convergence 10−4 | 1OR0 | 12615615 | 3.44 | 3.94 | 0.92 |
| 3EHU | 12737983 | 4.16 | 4.27 | 1.05 | |
| 1YTV | 15039999 | 4.22 | 4.31 | 1.19 | |
| 1DGW | 15368463 | 3.74 | 4.13 | 1.15 | |
| 2BCN | 19625007 | 6.91 | 5.97 | 1.34 | |
| 2Q0O | 21077567 | 6.81 | 6.01 | 1.33 | |
| 1JBO | 25502607 | 14.61 | 7.66 | 1.86 | |
| 1E6Y | 29788591 | 12.80 | 10.01 | 1.86 | |
| Convergence 10−6 | 1OR0 | 12615615 | - | 5.88 | 2.47 |
| 3EHU | 12737983 | - | 5.78 | 2.71 | |
| 1YTV | 15039999 | - | 6.23 | 3.14 | |
| 1DGW | 15368463 | - | 6.21 | 3.05 | |
| 2BCN | 19625007 | - | 8.91 | 4.75 | |
| 2Q0O | 21077567 | - | 10.04 | 3.78 | |
| 1JBO | 25502607 | - | 11.44 | 4.84 | |
| 1E6Y | 29788591 | - | 15.57 | 6.91 | |
Finally, it is interesting to see how the three tested GPU solvers scale with system sizes. Figure 8 shows the overall performances of the three GPU solvers on all 144 test cases. With the convergence criterion of 10−4 (Figure 8(a)), speedup ratios of MG over SOR solvers are from ~2.4 to ~10.4. Clearly the ratios depend on system sizes, but the scaling is roughly linear. This shows the advantage of the MG solver on larger systems, consistent with the original MG development on CPUs as reported in the literature.38, 45 Finally, with the convergence criterion of 10−6 (Figure 8(b)), the hybrid MG performs better than the Jacobi PCG at speedup ratios of ~2.0 to ~5.0, clearly showing the added benefit of incorporating the Jacobi PCG into the hybrid MG solver.
Figure 8.
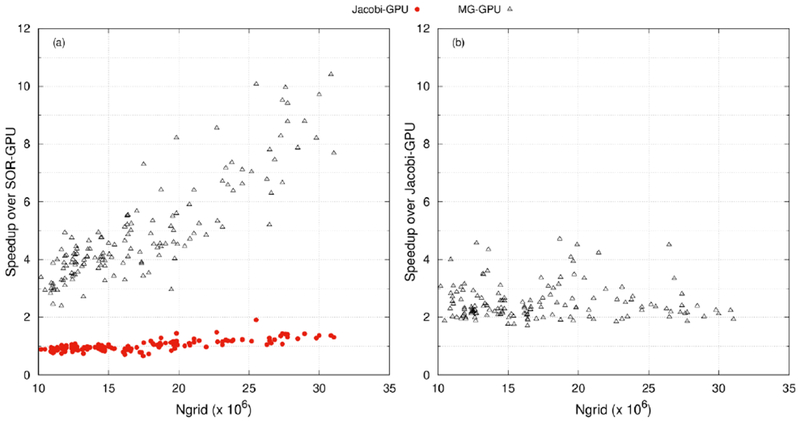
Comparison between GPU solvers SOR, Jacobi-PCG, and MG for the protein test set with (a) SOR-GPU and (b) Jacobi-GPU as the reference, respectively. The convergence criteria were set to 10−4 for (a) and 10−6 for (b), respectively.
3.5. Accuracy of CUDA MG Implementation
Given the default use of single precision on GPUs for optimal efficiency, it is important to confirm that our MG GPU implementation can achieve consistent numerical results with its CPU counterpart. Specifically, the electrostatic solvation energies by both solvers were compared for the large set of biomolecules. As shown in Figure 9, the energies between GPU and CPU implementations correlate quite well with both 10−4 and 10−6 convergence criteria. The final fitting slopes are 1.00016 and 0.999997, respectively, and the asymptotic standard errors are 0.0024% and 0.000025%, respectively. The maximum relative energy errors between the two implementations are 8.76×10−4 and 6.27 ×10−6, respectively, which are consistent with the convergence criteria chosen.
Figure 9.
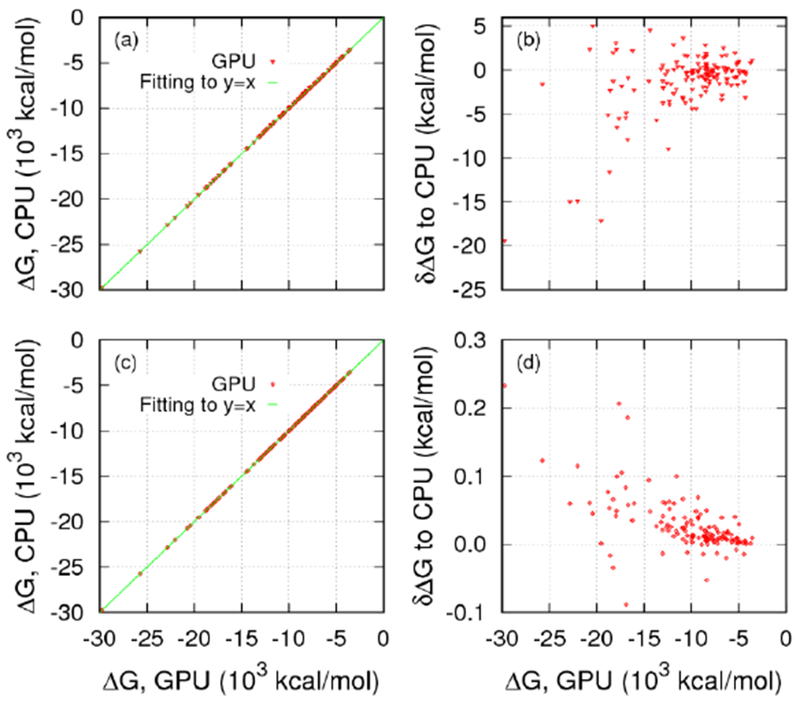
Correlations (a), (c) and differences (b), (d) of electrostatic solvation energies by the MG solver on GPU and on CPU for the protein test set. The convergence criterion was set to 10−4 in (a) and (b) and 10−6 in (c) and (d). The linear regression slopes are 1.00016 and 0.999997, respectively. The asymptotic standard errors are 0.0024% and 0.000025% for the 10−4 and 10−6 criteria, respectively.
3.6. Memory Considerations
Memory usage is crucial for GPU implementations since memory is often limited on most consumer-grade GPUs. In the MG implementation, the typical GPU memory usage is about 75 × Ngrid bytes, where Ngrid is the number of grid points when discretizing the system with the finite difference method. If the MG-Jacobi-PCG hybrid solver is involved in the computation with tighter convergence criteria, the typical GPU memory usage is about 135 × Ngrid bytes. Our analysis of the MG solver showed that NVIDIA Titan Xp cards, which have 12 GB GPU memory, are sufficient to successfully run all our 144 stress tests until host memory hit the limit first. On the older NVIDIA GTX 980 Ti cards with ∼ 6 GB GPU memory, the MG implementation is able to successfully complete calculations with ~ 75.0 million grid points given sufficient host memory. Worth noting is that for extremely large grids, for example those with at least one billion grid points, the MG implementation generally requires about 70 GB memory, which is far beyond the available memory on most consumer-grade GPU cards.
3.7. Future Directions
In the MG cycles, the computation bottleneck is the red-black Gauss-Seidel/SOR relaxation, so that it would be most beneficial to gain further optimization on those operations. In the color-labeled storage patterns, the matrix elements of the same color are accessed nonconsecutively by parallel threads, which causes performance loss. A possible strategy to improve locality and coalescing of memory accesses is to split the matrix storage into two separate red/black groups.165 In this way, bandwidth utilization can be improved by accessing array elements continuously.
Another direction for further optimization is to utilize the on-chip fast shared memory that is private for each thread block, its latency is two orders of magnitude lower than that of the global memory. However, shared memory is more complicated to use and needs careful management. For example, data prefetching is required and comes with a cost when moving data from the global memory to the shared memory of a thread block. In addition, unfavorable overhead may also result from accessing overlapping data between neighboring thread blocks since the shared memory of a thread block is not visible outside the thread block.
Finally, only the linear system solver, i.e. the multi-grid setup and the solution (both F- and V-cycles) have been implemented on GPUs in this study, while the discretization of PBE is still carried out on the host CPU. Porting the entire PBSA program onto GPUs would clearly accelerate the overall efficiency of the program. The remaining bottlenecks in the program are the molecular surface determination and the reaction field energy calculation (as it requires looping over all pairwise terms between surface charges and atomic charges). These two steps dominate roughly 96% of the remaining CPU time, which we expect to be reduced by at least 2/3 after being ported to GPUs. The additional development and other fine-grained optimizations are in progress in our lab. We expect our full GPU implementation of both PBSA and MMPBSA programs to be available along with the release of Amber 2019.
4. Conclusions
In this study, we implemented a PBE MG solver to harvest the computing powers on GPU platforms, and investigated the robustness and efficiency of multiple GPU solvers using a large set of realistic biomolecules. Our analysis shows that the tested GPU solvers have different convergence behaviors when a tight acceptance criterion ≥ 10−5 is used in the single precision mode. SOR was found to be the worst in this regard while Jacobi-PCG the best, and MG in the middle. Failures in MG were found not due to large system sizes but significant numbers of charged residues. It is clear that the use of single precision in the Gauss-Seidel relaxation introduces too much numerical noise that is too hard to reduce. This is in contrast to its much-better convergence behavior when the tests were conducted in the double-precision mode. We therefore developed a hybrid MG/PCG solver to utilize the advantages of high efficiency of MG and robustness of Jacobi-PCG for the widely used single-precision mode on GPUs.
The MG solver was further improved by embedding one F-Cycle before the initiation of V-cycles to provide a better initial guess. Our analysis shows that the strategy on average saves three or four V-Cycles. After incorporating this and other improvements, we further compared the efficiency of three representative GPU solvers. Our analysis shows that GPU/SOR speedup over CPU/SOR can be very impressive with ratios ~100 if the problem can be solved with the tested single precision condition. MG solver overall performs best among the three solvers with Jacobi-PCG as the second and SOR as the third when they all can converge. With the tight convergence criterion of 10−6 where SOR and pure MG mostly cannot converge, the speedup ratios of the hybrid MG solver over Jacobi-PCG are ~2.0 to ~5.0.
Finally, the implementation of the MG solver was validated by comparing electrostatic solvation energies computed on both GPU and CPU. The energies between the two sets correlate quite well with both loose and tight convergence criteria. The maximum relative energy errors between the two are consistent with the convergence criteria chosen. Future directions to improve the PBE calculations were also discussed. The new developments, together with other fine-grained optimizations in progress as implemented in the latest Amber package, will greatly benefit a wide range of biomolecular applications, such as those in MMPBSA binding affinity simulations on the GPU platforms.
5. Acknowledgements
This work was supported by National Institutes of Health/NIGMS (GM093040 & GM079383). We thank Prof. Tai-Sung Lee for useful discussion on CUDA kernel optimization.
6. References
- 1.Davis ME; McCammon JA, Electrostatics in Biomolecular Structure and Dynamics. Chem. Rev. (Washington, DC, U. S.) 1990, 90, 509–521. [Google Scholar]
- 2.Honig B; Sharp K; Yang AS, Macroscopic Models of Aqueous-Solutions - Biological and Chemical Applications. J. Phys. Chem 1993, 97, 1101–1109. [Google Scholar]
- 3.Honig B; Nicholls A, Classical Electrostatics in Biology and Chemistry. Science 1995, 268, 1144–1149. [DOI] [PubMed] [Google Scholar]
- 4.Beglov D; Roux B, Solvation of Complex Molecules in a Polar Liquid: An Integral Equation Theory. J. Chem. Phys 1996, 104, 8678–8689. [Google Scholar]
- 5.Cramer CJ; Truhlar DG, Implicit Solvation Models: Equilibria, Structure, Spectra, and Dynamics. Chem. Rev. (Washington, DC, U. S.) 1999, 99, 2161–2200. [DOI] [PubMed] [Google Scholar]
- 6.Bashford D; Case DA, Generalized Born Models of Macromolecular Solvation Effects. Annu. Rev. Phys. Chem 2000, 51, 129–152. [DOI] [PubMed] [Google Scholar]
- 7.Baker NA, Improving Implicit Solvent Simulations: A Poisson-Centric View. Curr. Opin. Struct. Biol 2005, 15, 137–143. [DOI] [PubMed] [Google Scholar]
- 8.Chen JH; Im WP; Brooks CL, Balancing Solvation and Intramolecular Interactions: Toward a Consistent Generalized Born Force Field. J. Am. Chem. Soc 2006, 128, 3728–3736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Feig M; Chocholousova J; Tanizaki S, Extending the Horizon: Towards the Efficient Modeling of Large Biomolecular Complexes in Atomic Detail. Theor. Chem. Acc 2006, 116, 194–205. [Google Scholar]
- 10.Koehl P, Electrostatics Calculations: Latest Methodological Advances. Curr. Opin. Struct. Biol 2006, 16, 142–151. [DOI] [PubMed] [Google Scholar]
- 11.Im W; Chen J; Brooks CL Peptide and Protein Folding and Conformational Equilibria: Theoretical Treatment of Electrostatics and Hydrogen Bonding with Implicit Solvent Models In Adv. Protein Chem; Academic Press: 2005; Vol. 72, pp 173–198. [DOI] [PubMed] [Google Scholar]
- 12.Lu BZ; Zhou YC; Holst MJ; McCammon JA, Recent Progress in Numerical Methods for the Poisson-Boltzmann Equation in Biophysical Applications. Commun. Comput. Phys 2008, 3, 973–1009. [Google Scholar]
- 13.Wang J; Tan CH; Tan YH; Lu Q; Luo R, Poisson-Boltzmann Solvents in Molecular Dynamics Simulations. Commun. Comput. Phys 2008, 3, 1010–1031. [Google Scholar]
- 14.Altman MD; Bardhan JP; White JK; Tidor B, Accurate Solution of Multi-Region Continuum Biomolecule Electrostatic Problems Using the Linearized Poisson-Boltzmann Equation with Curved Boundary Elements. J. Comput. Chem 2009, 30, 132–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cai Q; Wang J; Hsieh M-J; Ye X; Luo R Chapter Six - Poisson–Boltzmann Implicit Solvation Models In Annu. Rep. Comput. Chem, Ralph AW, Ed.; Elsevier: 2012; Vol. Volume 8, pp 149–162. [Google Scholar]
- 16.Xiao L; Wang C; Luo R, Recent Progress in Adapting Poisson–Boltzmann Methods to Molecular Simulations. J. Theor. Comput. Chem 2014, 13, 1430001. [Google Scholar]
- 17.Botello-Smith WM; Cai Q; Luo R, Biological Applications of Classical Electrostatics Methods. J. Theor. Comput. Chem 2014, 13, 1440008. [Google Scholar]
- 18.Wang C; Greene D; Xiao L; Qi R; Luo R, Recent Developments and Applications of the Mmpbsa Method. Front. Mol. Biosci 2017, 4, 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kollman PA; Massova I; Reyes C; Kuhn B; Huo S; Chong L; Lee M; Lee T; Duan Y; Wang W; Donini O; Cieplak P; Srinivasan J; Case DA; Cheatham TE 3rd, Calculating Structures and Free Energies of Complex Molecules: Combining Molecular Mechanics and Continuum Models. Acc. Chem. Res 2000, 33, 889–97. [DOI] [PubMed] [Google Scholar]
- 20.Srinivasan J; Cheatham TE; Cieplak P; Kollman PA; Case DA, Continuum Solvent Studies of the Stability of DNA, Rna, and Phosphoramidate−DNA Helices. J. Am. Chem. Soc 1998, 120, 9401–9409. [Google Scholar]
- 21.Gohlke H; Case DA, Converging Free Energy Estimates: Mm-Pb(Gb)Sa Studies on the Protein-Protein Complex Ras-Raf. J. Comput. Chem 2004, 25, 238–250. [DOI] [PubMed] [Google Scholar]
- 22.Yang T; Wu JC; Yan C; Wang Y; Luo R; Gonzales MB; Dalby KN; Ren P, Virtual Screening Using Molecular Simulations. Proteins 2011, 79, 1940–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller BR; McGee TD; Swails JM; Homeyer N; Gohlke H; Roitberg AE, Mmpbsa.Py: An Efficient Program for End-State Free Energy Calculations. J. Chem. Theory Comput 2012, 8, 3314–3321. [DOI] [PubMed] [Google Scholar]
- 24.Wang CH; Nguyen PH; Pham K; Huynh D; Le TBN; Wang HL; Ren PY; Luo R, Calculating Protein-Ligand Binding Affinities with Mmpbsa: Method and Error Analysis. J. Comput. Chem 2016, 37, 2436–2446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Warwicker J; Watson HC, Calculation of the Electric-Potential in the Active-Site Cleft Due to Alpha-Helix Dipoles. J. Mol. Biol 1982, 157, 671–679. [DOI] [PubMed] [Google Scholar]
- 26.Bashford D; Karplus M, Pkas of Ionizable Groups in Proteins - Atomic Detail from a Continuum Electrostatic Model. Biochemistry 1990, 29, 10219–10225. [DOI] [PubMed] [Google Scholar]
- 27.Jeancharles A; Nicholls A; Sharp K; Honig B; Tempczyk A; Hendrickson TF; Still WC, Electrostatic Contributions to Solvation Energies - Comparison of Free-Energy Perturbation and Continuum Calculations. J. Am. Chem. Soc 1991, 113, 1454–1455. [Google Scholar]
- 28.Gilson MK, Theory of Electrostatic Interactions in Macromolecules. Curr. Opin. Struct. Biol 1995, 5, 216–223. [DOI] [PubMed] [Google Scholar]
- 29.Edinger SR; Cortis C; Shenkin PS; Friesner RA, Solvation Free Energies of Peptides: Comparison of Approximate Continuum Solvation Models with Accurate Solution of the Poisson-Boltzmann Equation. J. Phys. Chem. B 1997, 101, 1190–1197. [Google Scholar]
- 30.Lu Q; Luo R, A Poisson-Boltzmann Dynamics Method with Nonperiodic Boundary Condition. J. Chem. Phys 2003, 119, 11035–11047. [Google Scholar]
- 31.Luo R; Moult J; Gilson MK, Dielectric Screening Treatment of Electrostatic Solvation. J. Phys. Chem. B 1997, 101, 11226–11236. [Google Scholar]
- 32.Wang J; Tan C; Chanco E; Luo R, Quantitative Analysis of Poisson-Boltzmann Implicit Solvent in Molecular Dynamics. Phys. Chem. Chem. Phys 2010, 12, 1194–1202. [DOI] [PubMed] [Google Scholar]
- 33.Hsieh MJ; Luo R, Exploring a Coarse-Grained Distributive Strategy for Finite-Difference Poisson-Boltzmann Calculations. J. Mol. Model 2011, 17, 1985–1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Klapper I; Hagstrom R; Fine R; Sharp K; Honig B, Focusing of Electric Fields in the Active Site of Copper-Zinc Superoxide Dismutase Effects of Ionic Strength and Amino Acid Modification. Proteins: Struct., Funct., Genet 1986, 1, 47–59. [DOI] [PubMed] [Google Scholar]
- 35.Davis ME; McCammon JA, Solving the Finite-Difference Linearized Poisson-Boltzmann Equation - a Comparison of Relaxation and Conjugate-Gradient Methods. J. Comput. Chem 1989, 10, 386–391. [Google Scholar]
- 36.Nicholls A; Honig B, A Rapid Finite-Difference Algorithm, Utilizing Successive over-Relaxation to Solve the Poisson-Boltzmann Equation. J. Comput. Chem 1991, 12, 435–445. [Google Scholar]
- 37.Luty BA; Davis ME; McCammon JA, Solving the Finite-Difference Nonlinear Poisson-Boltzmann Equation. J. Comput. Chem 1992, 13, 1114–1118. [Google Scholar]
- 38.Holst M; Saied F, Multigrid Solution of the Poisson-Boltzmann Equation. J. Comput. Chem 1993, 14, 105–113. [Google Scholar]
- 39.Forsten KE; Kozack RE; Lauffenburger DA; Subramaniam S, Numerical-Solution of the Nonlinear Poisson-Boltzmann Equation for a Membrane-Electrolyte System. J. Phys. Chem 1994, 98, 5580–5586. [Google Scholar]
- 40.Holst MJ; Saied F, Numerical-Solution of the Nonlinear Poisson-Boltzmann Equation - Developing More Robust and Efficient Methods. J. Comput. Chem 1995, 16, 337–364. [Google Scholar]
- 41.Bashford D An Object-Oriented Programming Suite for Electrostatic Effects in Biological Molecules an Experience Report on the Mead Project. Berlin, Heidelberg, 1997; Springer Berlin Heidelberg: Berlin, Heidelberg, 1997; pp 233–240. [Google Scholar]
- 42.Im W; Beglov D; Roux B, Continuum Solvation Model: Computation of Electrostatic Forces from Numerical Solutions to the Poisson-Boltzmann Equation. Comput. Phys. Commun 1998, 111, 59–75. [Google Scholar]
- 43.Rocchia W; Alexov E; Honig B, Extending the Applicability of the Nonlinear Poisson-Boltzmann Equation: Multiple Dielectric Constants and Multivalent Ions. J. Phys. Chem. B 2001, 105, 6507–6514. [Google Scholar]
- 44.Luo R; David L; Gilson MK, Accelerated Poisson-Boltzmann Calculations for Static and Dynamic Systems. J. Comput. Chem 2002, 23, 1244–1253. [DOI] [PubMed] [Google Scholar]
- 45.Wang J; Luo R, Assessment of Linear Finite-Difference Poisson-Boltzmann Solvers. J. Comput. Chem 2010, 31, 1689–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cai Q; Hsieh M-J; Wang J; Luo R, Performance of Nonlinear Finite-Difference Poisson-Boltzmann Solvers. J. Chem. Theory Comput 2010, 6, 203–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cortis CM; Friesner RA, Numerical Solution of the Poisson-Boltzmann Equation Using Tetrahedral Finite-Element Meshes. J. Comput. Chem 1997, 18, 1591–1608. [Google Scholar]
- 48.Holst M; Baker N; Wang F, Adaptive Multilevel Finite Element Solution of the Poisson-Boltzmann Equation I. Algorithms and Examples. J. Comput. Chem 2000, 21, 1319–1342. [Google Scholar]
- 49.Baker N; Holst M; Wang F, Adaptive Multilevel Finite Element Solution of the Poisson-Boltzmann Equation Ii. Refinement at Solvent-Accessible Surfaces in Biomolecular Systems. J. Comput. Chem 2000, 21, 1343–1352. [Google Scholar]
- 50.Shestakov AI; Milovich JL; Noy A, Solution of the Nonlinear Poisson-Boltzmann Equation Using Pseudo-Transient Continuation and the Finite Element Method. J. Colloid Interface Sci 2002, 247, 62–79. [DOI] [PubMed] [Google Scholar]
- 51.Chen L; Holst MJ; Xu JC, The Finite Element Approximation of the Nonlinear Poisson-Boltzmann Equation. SIAM J. Numer. Anal 2007, 45, 2298–2320. [Google Scholar]
- 52.Xie D; Zhou S, A New Minimization Protocol for Solving Nonlinear Poisson–Boltzmann Mortar Finite Element Equation. Bit. Numer. Math 2007, 47, 853–871. [Google Scholar]
- 53.Lu B; Holst MJ; McCammon JA; Zhou YC, Poisson-Nernst-Planck Equations for Simulating Biomolecular Diffusion-Reaction Processes I: Finite Element Solutions. J. Comput. Phys 2010, 229, 6979–6994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bond SD; Chaudhry JH; Cyr EC; Olson LN, A First-Order System Least-Squares Finite Element Method for the Poisson-Boltzmann Equation. J. Comput. Chem 2010, 31, 1625–1635. [DOI] [PubMed] [Google Scholar]
- 55.M. CC; A. FR, Numerical Solution of the Poisson–Boltzmann Equation Using Tetrahedral Finite‐Element Meshes. J. Comput. Chem 1997, 18, 1591–1608. [Google Scholar]
- 56.Miertus S; Scrocco E; Tomasi J, Electrostatic Interaction of a Solute with a Continuum - a Direct Utilization of Abinitio Molecular Potentials for the Prevision of Solvent Effects. Chem. Phys 1981, 55, 117–129. [Google Scholar]
- 57.Hoshi H; Sakurai M; Inoue Y; Chujo R, Medium Effects on the Molecular Electronic-Structure .1. The Formulation of a Theory for the Estimation of a Molecular Electronic-Structure Surrounded by an Anisotropic Medium. J. Chem. Phys 1987, 87, 1107–1115. [Google Scholar]
- 58.Zauhar RJ; Morgan RS, The Rigorous Computation of the Molecular Electric-Potential. J. Comput. Chem 1988, 9, 171–187. [Google Scholar]
- 59.Rashin AA, Hydration Phenomena, Classical Electrostatics, and the Boundary Element Method. J. Phys. Chem 1990, 94, 1725–1733. [Google Scholar]
- 60.Yoon BJ; Lenhoff AM, A Boundary Element Method for Molecular Electrostatics with Electrolyte Effects. J. Comput. Chem 1990, 11, 1080–1086. [Google Scholar]
- 61.Juffer AH; Botta EFF; Vankeulen BAM; Vanderploeg A; Berendsen HJC, The Electric-Potential of a Macromolecule in a Solvent - a Fundamental Approach. J. Comput. Phys 1991, 97, 144–171. [Google Scholar]
- 62.Zhou HX, Boundary-Element Solution of Macromolecular Electrostatics - Interaction Energy between 2 Proteins. Biophys. J 1993, 65, 955–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Bharadwaj R; Windemuth A; Sridharan S; Honig B; Nicholls A, The Fast Multipole Boundary-Element Method for Molecular Electrostatics - an Optimal Approach for Large Systems. J. Comput. Chem 1995, 16, 898–913. [Google Scholar]
- 64.Purisima EO; Nilar SH, A Simple yet Accurate Boundary-Element Method for Continuum Dielectric Calculations. J. Comput. Chem 1995, 16, 681–689. [Google Scholar]
- 65.Liang J; Subramaniam S, Computation of Molecular Electrostatics with Boundary Element Methods. Biophys. J 1997, 73, 1830–1841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vorobjev YN; Scheraga HA, A Fast Adaptive Multigrid Boundary Element Method for Macromolecular Electrostatic Computations in a Solvent. J. Comput. Chem 1997, 18, 569–583. [Google Scholar]
- 67.Totrov M; Abagyan R, Rapid Boundary Element Solvation Electrostatics Calculations in Folding Simulations: Successful Folding of a 23-Residue Peptide. Biopolymers 2001, 60, 124–133. [DOI] [PubMed] [Google Scholar]
- 68.Boschitsch AH; Fenley MO; Zhou HX, Fast Boundary Element Method for the Linear Poisson-Boltzmann Equation. J. Phys. Chem. B 2002, 106, 2741–2754. [Google Scholar]
- 69.Lu BZ; Cheng XL; Huang JF; McCammon JA, Order N Algorithm for Computation of Electrostatic Interactions in Biomolecular Systems. Proc. Natl. Acad. Sci. U. S. A 2006, 103, 19314–19319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lu B; Cheng X; Huang J; McCammon JA, An Adaptive Fast Multipole Boundary Element Method for Poisson-Boltzmann Electrostatics. J. Chem. Theory Comput 2009, 5, 1692–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bajaj C; Chen S-C; Rand A, An Efficient Higher-Order Fast Multipole Boundary Element Solution for Poisson-Boltzmann-Based Molecular Electrostatics. SIAM J. Sci. Comput 2011, 33, 826–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lu B; Cheng X; Huang J; McCammon JA, Afmpb: An Adaptive Fast Multipole Poisson–Boltzmann Solver for Calculating Electrostatics in Biomolecular Systems. Comput. Phys. Commun 2010, 181, 1150–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Geng W; Krasny R, A Treecode-Accelerated Boundary Integral Poisson–Boltzmann Solver for Electrostatics of Solvated Biomolecules. J. Comput. Phys 2013, 247, 62–78. [Google Scholar]
- 74.Baptista M; Schmitz R; Dünweg B, Simple and Robust Solver for the Poisson-Boltzmann Equation. Phys. Rev. E 2009, 80, 016705. [DOI] [PubMed] [Google Scholar]
- 75.Li C; Li L; Zhang J; Alexov E, Highly Efficient and Exact Method for Parallelization of Grid-Based Algorithms and Its Implementation in Delphi. J. Comput. Chem 2012, 33, 1960–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li L; Li C; Sarkar S; Zhang J; Witham S; Zhang Z; Wang L; Smith N; Petukh M; Alexov E, Delphi: A Comprehensive Suite for Delphi Software and Associated Resources. BMC Biophys 2012, 5, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Baker NA; Sept D; Joseph S; Holst MJ; McCammon JA, Electrostatics of Nanosystems: Application to Microtubules and the Ribosome. Proc. Natl. Acad. Sci 2001, 98, 10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cai Q; Wang J; Zhao H-K; Luo R, On Removal of Charge Singularity in Poisson-Boltzmann Equation. J. Chem. Phys 2009, 130. [DOI] [PubMed] [Google Scholar]
- 79.Wang J; Cai Q; Li Z-L; Zhao H-K; Luo R, Achieving Energy Conservation in Poisson-Boltzmann Molecular Dynamics: Accuracy and Precision with Finite-Difference Algorithms. Chem. Phys. Lett 2009, 468, 112–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Luo R; Head MS; Moult J; Gilson MK, Pk(a) Shifts in Small Molecules and Hiv Protease: Electrostatics and Conformation. J. Am. Chem. Soc 1998, 120, 6138–6146. [Google Scholar]
- 81.Georgescu RE; Alexov EG; Gunner MR, Combining Conformational Flexibility and Continuum Electrostatics for Calculating Pk(a)S in Proteins. Biophys. J 2002, 83, 1731–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nielsen JE; McCammon JA, On the Evaluation and Optimization of Protein X-Ray Structures for Pka Calculations. Protein Sci. 2003, 12, 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Warwicker J, Improved Pk(a) Calculations through Flexibility Based Sampling of a Water-Dominated Interaction Scheme. Protein Sci. 2004, 13, 2793–2805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tang CL; Alexov E; Pyle AM; Honig B, Calculation of Pk(a)S in Rna: On the Structural Origins and Functional Roles of Protonated Nucleotides. J. Mol. Biol 2007, 366, 1475–1496. [DOI] [PubMed] [Google Scholar]
- 85.Wang L; Li L; Alexov E, Pka Predictions for Proteins, Rnas, and Dnas with the Gaussian Dielectric Function Using Delphi Pka. Proteins 2015, 83, 2186–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tan C; Yang L; Luo R, How Well Does Poisson-Boltzmann Implicit Solvent Agree with Explicit Solvent? A Quantitative Analysis. J. Phys. Chem. B 2006, 110, 18680–18687. [DOI] [PubMed] [Google Scholar]
- 87.Nicholls A; Mobley DL; Guthrie JP; Chodera JD; Bayly CI; Cooper MD; Pande VS, Predicting Small-Molecule Solvation Free Energies: An Informal Blind Test for Computational Chemistry. J. Med. Chem 2008, 51, 769–779. [DOI] [PubMed] [Google Scholar]
- 88.Shivakumar D; Deng YQ; Roux B, Computations of Absolute Solvation Free Energies of Small Molecules Using Explicit and Implicit Solvent Model. J. Chem. Theory Comput 2009, 5, 919–930. [DOI] [PubMed] [Google Scholar]
- 89.Brieg M; Setzler J; Albert S; Wenzel W, Generalized Born Implicit Solvent Models for Small Molecule Hydration Free Energies. Phys. Chem. Chem. Phys 2017, 19, 1677–1685. [DOI] [PubMed] [Google Scholar]
- 90.Wang C; Ren P; Luo R, Ionic Solution: What Goes Right and Wrong with Continuum Solvation Modeling. J. Phys. Chem. B 2017, 121, 11169–11179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tan C; Tan Y-H; Luo R, Implicit Nonpolar Solvent Models. J. Phys. Chem. B 2007, 111, 12263–12274. [DOI] [PubMed] [Google Scholar]
- 92.Swanson JMJ; Henchman RH; McCammon JA, Revisiting Free Energy Calculations: A Theoretical Connection to Mm/Pbsa and Direct Calculation of the Association Free Energy. Biophys. J 2004, 86, 67–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bertonati C; Honig B; Alexov E, Poisson-Boltzmann Calculations of Nonspecific Salt Effects on Protein-Protein Binding Free Energies. Biophys. J 2007, 92, 1891–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Brice AR; Dominy BN, Analyzing the Robustness of the Mm/Pbsa Free Energy Calculation Method: Application to DNA Conformational Transitions. J. Comput. Chem 2011, 32, 1431–1440. [DOI] [PubMed] [Google Scholar]
- 95.David L; Luo R; Head MS; Gilson MK, Computational Study of Kni-272, a Potent Inhibitor of Hiv-1 Protease: On the Mechanism of Preorganization. J. Phys. Chem. B 1999, 103, 1031–1044. [Google Scholar]
- 96.Luo R; Head MS; Given JA; Gilson MK, Nucleic Acid Base-Pairing and N-Methylacetamide Self-Association in Chloroform: Affinity and Conformation. Biophys. Chem 1999, 78, 183–193. [DOI] [PubMed] [Google Scholar]
- 97.Luo R; Gilson MK, Synthetic Adenine Receptors: Direct Calculation of Binding Affinity and Entropy. J. Am. Chem. Soc 2000, 122, 2934–2937. [Google Scholar]
- 98.Luo R; Gilson HSR; Potter MJ; Gilson MK, The Physical Basis of Nucleic Acid Base Stacking in Water. Biophys. J 2001, 80, 140–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hsieh MJ; Luo R, Physical Scoring Function Based on Amber Force Field and Poisson-Boltzmann Implicit Solvent for Protein Structure Prediction. Proteins 2004, 56, 475–86. [DOI] [PubMed] [Google Scholar]
- 100.Wen EZ; Luo R, Interplay of Secondary Structures and Side-Chain Contacts in the Denatured State of Bba1. J. Chem. Phys 2004, 121, 2412–2421. [DOI] [PubMed] [Google Scholar]
- 101.Wen EZ; Hsieh MJ; Kollman PA; Luo R, Enhanced Ab Initio Protein Folding Simulations in Poisson-Boltzmann Molecular Dynamics with Self-Guiding Forces. J. Mol. Graphics Modell 2004, 22, 415–424. [DOI] [PubMed] [Google Scholar]
- 102.Lwin TZ; Luo R, Overcoming Entropic Barrier with Coupled Sampling at Dual Resolutions. J. Chem. Phys 2005, 123. [DOI] [PubMed] [Google Scholar]
- 103.Lwin TZ; Zhou RH; Luo R, Is Poisson-Boltzmann Theory Insufficient for Protein Folding Simulations? J. Chem. Phys 2006, 124. [DOI] [PubMed] [Google Scholar]
- 104.Lwin TZ; Luo R, Force Field Influences in Beta-Hairpin Folding Simulations. Protein Sci. 2006, 15, 2642–2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tan Y-H; Luo R, Protein Stability Prediction: A Poisson-Boltzmann Approach. J. Phys. Chem. B 2008, 112, 1875–1883. [DOI] [PubMed] [Google Scholar]
- 106.Korman TP; Tan Y-H; Wong J; Luo R; Tsai S-C, Inhibition Kinetics and Emodin Cocrystal Structure of a Type Ii Polyketide Ketoreductase. Biochemistry 2008, 47, 1837–1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Tan Y; Luo R, Structural and Functional Implications of P53 Missense Cancer Mutations. BMC Biophys. 2009, 2, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Barajas Jesus F.; Phelan Ryan M.; Schaub Andrew J.; Kliewer Jaclyn T.; Kelly Peter J.; Jackson David R.; Luo R; Keasling Jay D.; Tsai S-C, Comprehensive Structural and Biochemical Analysis of the Terminal Myxalamid Reductase Domain for the Engineered Production of Primary Alcohols. Chem. Biol 2015, 22, 1018–1029. [DOI] [PubMed] [Google Scholar]
- 109.Jackson DR; Tu SS; Nguyen M; Barajas JF; Schaub AJ; Krug D; Pistorius D; Luo R; Müller R; Tsai S-C, Structural Insights into Anthranilate Priming During Type Ii Polyketide Biosynthesis. ACS Chem. Biol 2016, 11, 95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Qian T; Wo J; Zhang Y; Song Q; Feng G; Luo R; Lin S; Wu G; Chen HF, Crystal Structure of Stna for the Biosynthesis of Antitumor Drug Streptonigrin Reveals a Unique Substrate Binding Mode. Sci. Rep 2017, 7, 40254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ellis BD; Milligan JC; White AR; Duong V; Altman PX; Mohammed LY; Crump MP; Crosby J; Luo R; Vanderwal CD; Tsai S-C, An Oxetane-Based Polyketide Surrogate to Probe Substrate Binding in a Polyketide Synthase. J. Am. Chem. Soc 2018, 140, 4961–4964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Marshall SA; Vizcarra CL; Mayo SL, One- and Two-Body Decomposable Poisson-Boltzmann Methods for Protein Design Calculations. Protein Sci. 2005, 14, 1293–1304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Greene D; Po T; Pan J; Tabibian T; Luo R, Computational Analysis for the Rational Design of Anti-Amyloid Beta (Abeta) Antibodies. J. Phys. Chem. B 2018, 122, 4521–4536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Petukh M; Li M; Alexov E, Predicting Binding Free Energy Change Caused by Point Mutations with Knowledge-Modified Mm/Pbsa Method. PLoS Comput. Biol 2015, 11, e1004276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Getov I; Petukh M; Alexov E, Saafec: Predicting the Effect of Single Point Mutations on Protein Folding Free Energy Using a Knowledge-Modified Mm/Pbsa Approach. Int. J. Mol. Sci 2016, 17, 512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Götz AW; Williamson MJ; Xu D; Poole D; Le Grand S; Walker RC, Routine Microsecond Molecular Dynamics Simulations with Amber on Gpus. 1. Generalized Born. J. Chem. Theory Comput 2012, 8, 1542–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Páll S; Hess B, A Flexible Algorithm for Calculating Pair Interactions on Simd Architectures. Comput. Phys. Commun 2013, 184, 2641–2650. [Google Scholar]
- 118.Salomon-Ferrer R; Götz AW; Poole D; Le Grand S; Walker RC, Routine Microsecond Molecular Dynamics Simulations with Amber on Gpus. 2. Explicit Solvent Particle Mesh Ewald. J. Chem. Theory Comput 2013, 9, 3878–3888. [DOI] [PubMed] [Google Scholar]
- 119.Ufimtsev IS; Martínez TJ, Quantum Chemistry on Graphical Processing Units. 1. Strategies for Two-Electron Integral Evaluation. J. Chem. Theory Comput 2008, 4, 222–231. [DOI] [PubMed] [Google Scholar]
- 120.Asadchev A; Gordon MS, New Multithreaded Hybrid Cpu/Gpu Approach to Hartree–Fock. J. Chem. Theory Comput 2012, 8, 4166–4176. [DOI] [PubMed] [Google Scholar]
- 121.Titov AV; Ufimtsev IS; Luehr N; Martinez TJ, Generating Efficient Quantum Chemistry Codes for Novel Architectures. J. Chem. Theory Comput 2013, 9, 213–221. [DOI] [PubMed] [Google Scholar]
- 122.Colmenares J; Ortiz J; Rocchia W, Gpu Linear and Non-Linear Poisson–Boltzmann Solver Module for Delphi. Bioinformatics 2014, 30, 569–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Colmenares J; Galizia A; Ortiz J; Clematis A; Rocchia W, A Combined Mpi-Cuda Parallel Solution of Linear and Nonlinear Poisson-Boltzmann Equation. BioMed Res. Int 2014, 2014, 560987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Qi R; Botello-Smith WM; Luo R, Acceleration of Linear Finite-Difference Poisson–Boltzmann Methods on Graphics Processing Units. J. Chem. Theory Comput 2017, 13, 3378–3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Geng W; Jacob F, A Gpu-Accelerated Direct-Sum Boundary Integral Poisson–Boltzmann Solver. Comput. Phys. Commun 2013, 184, 1490–1496. [Google Scholar]
- 126.Stuben K, Algebraic Multigrid (Amg) - Experiences and Comparisons. Appl. Math. Comput 1983, 13, 419–451. [Google Scholar]
- 127.Oberoi H; Allewell NM, Multigrid Solution of the Nonlinear Poisson-Boltzmann Equation and Calculation of Titration Curves. Biophys. J 1993, 65, 48–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Michael H; E. KR; Faisal S; Shankar S, Treatment of Electrostatic Effects in Proteins: Multigrid‐Based Newton Iterative Method for Solution of the Full Nonlinear Poisson–Boltzmann Equation. Proteins: Struct., Funct., Bioinf 1994, 18, 231–245. [DOI] [PubMed] [Google Scholar]
- 129.Haase G; Liebmann M; Douglas CC; Plank G A Parallel Algebraic Multigrid Solver on Graphics Processing Units. Berlin, Heidelberg, 2010; Springer Berlin Heidelberg: Berlin, Heidelberg, 2010; pp 38–47. [Google Scholar]
- 130.Naumov M; Arsaev M; Castonguay P; Cohen J; Demouth J; Eaton J; Layton S; Markovskiy N; Sakharnykh N; Strzodka R Amgx: Scalability and Performance on Massively Parallel Platforms In SIAM workshop on exascale applied mathematics challenges and opportunities. SIAM, 2014; 2014. [Google Scholar]
- 131.Liu H; Yang B; Chen Z, Accelerating Algebraic Multigrid Solvers on Nvidia Gpus. Comput. Math Appl 2015, 70, 1162–1181. [Google Scholar]
- 132.Bolz J; Farmer I; Grinspun E; Schroder P, In ACM SIGGRAPH 2003 Papers; ACM: San Diego, California, 2003, pp 917–924. [Google Scholar]
- 133.Stroia I; Itu L; Niţă C; Lazăr L; Suciu C Gpu Accelerated Geometric Multigrid Method: Performance Comparison on Recent Nvidia Architectures. In 2015 19th International Conference on System Theory, Control and Computing (ICSTCC), 14–16 Oct. 2015, 2015; 2015; pp 175–179. [Google Scholar]
- 134.Layton S; Sakharnykh N; Clark K, Gpu Implementation of Hpgmg-Fv. HPGMG BoF, Supercomputing 2015. [Google Scholar]
- 135.Press WH; Teukolsky SA; Vetterling WT; Flannery BP, Numerical Recipes : The Art of Scientific Computing. Cambridge University Press: Cambridge [Cambridgeshire]; New York, 1986. [Google Scholar]
- 136.Eisenstat SC, Efficient Implementation of a Class of Preconditioned Conjugate-Gradient Methods. SIAM J. Sci. Comput 1981, 2, 1–4. [Google Scholar]
- 137.Meijerink JA; Vandervorst HA, Iterative Solution Method for Linear-Systems of Which Coefficient Matrix Is a Symmetric M-Matrix. Math. Comput 1977, 31, 148–162. [Google Scholar]
- 138.Gustafsson I, A Class of First Order Factorization Methods. Bit. Numer. Math 1978, 18, 142–156. [Google Scholar]
- 139.Meijerink JA; Vandervorst HA, Guidelines for the Usage of Incomplete Decompositions in Solving Sets of Linear-Equations as They Occur in Practical Problems. J. Comput. Phys 1981, 44, 134–155. [Google Scholar]
- 140.Alcouffe RE; Brandt A; Dendy JE; Painter JW, The Multi-Grid Method for the Diffusion Equation with Strongly Discontinuous Coefficients. SIAM J. Sci. Comput 1981, 2, 430–454. [Google Scholar]
- 141.Ruge JW; K., S. Algebraic Multigrid In Multigrid Methods, McCormick SF, Ed.; SIAM: Philadelphia, 1987; Vol. 3, Chapter 4, pp 73–130. [Google Scholar]
- 142.Briggs WL; McCormick SF, A Multigrid Tutorial. Siam: 2000; Vol. 72. [Google Scholar]
- 143.Stuben K, A Review of Algebraic Multigrid. J. Comput. Appl. Math 2001, 128, 281–309. [Google Scholar]
- 144.Gandham R; Esler K; Zhang Y, A Gpu Accelerated Aggregation Algebraic Multigrid Method. Comput. Math Appl 2014, 68, 1151–1160. [Google Scholar]
- 145.Jun Z, Acceleration of Five-Point Red-Black Gauss-Seidel in Multigrid for Poisson Equation. Appl. Math. Comput 1996, 80, 73–93. [Google Scholar]
- 146.Li C; Petukh M; Li L; Alexov E, Continuous Development of Schemes for Parallel Computing of the Electrostatics in Biological Systems: Implementation in Delphi. J. Comput. Chem 2013, 34, 1949–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Harris M, Unified Memory in Cuda 6. GTC On-Demand, NVIDIA 2013. [Google Scholar]
- 148.Nvidia Nvidia Cusp Library. https://developer.nvidia.com/cusp (October 1st, 2016),
- 149.Ye X; Cai Q; Yang W; Luo R, Roles of Boundary Conditions in DNA Simulations: Analysis of Ion Distributions with the Finite-Difference Poisson-Boltzmann Method. Biophys. J 2009, 97, 554–562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Ye X; Wang J; Luo R, A Revised Density Function for Molecular Surface Calculation in Continuum Solvent Models. J. Chem. Theory Comput 2010, 6, 1157–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Cai Q; Ye X; Wang J; Luo R, Dielectric Boundary Force in Numerical Poisson-Boltzmann Methods: Theory and Numerical Strategies. Chem. Phys. Lett 2011, 514, 368–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cai Q; Ye X; Wang J; Luo R, On-the-Fly Numerical Surface Integration for Finite-Difference Poisson-Boltzmann Methods. J. Chem. Theory Comput 2011, 7, 3608–3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cai Q; Ye X; Luo R, Dielectric Pressure in Continuum Electrostatic Solvation of Biomolecules. Phys. Chem. Chem. Phys 2012, 14, 15917–15925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Wang J; Cai Q; Xiang Y; Luo R, Reducing Grid Dependence in Finite-Difference Poisson-Boltzmann Calculations. J. Chem. Theory Comput 2012, 8, 2741–2751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Wang C; Wang J; Cai Q; Li ZL; Zhao H; Luo R, Exploring High Accuracy Poisson-Boltzmann Methods for Biomolecular Simulations. Comput. Theor. Chem 2013, 1024, 34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Xiao L; Cai Q; Ye X; Wang J; Luo R, Electrostatic Forces in the Poisson-Boltzmann Systems. J. Chem. Phys 2013, 139, 094106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Liu X; Wang C; Wang J; Li Z; Zhao H; Luo R, Exploring a Charge-Central Strategy in the Solution of Poisson’s Equation for Biomolecular Applications. Phys. Chem. Chem. Phys 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Botello-Smith WM; Liu X; Cai Q; Li Z; Zhao H; Luo R, Numerical Poisson-Boltzmann Model for Continuum Membrane Systems. Chem. Phys. Lett 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Botello-Smith WM; Luo R, Applications of Mmpbsa to Membrane Proteins I: Efficient Numerical Solutions of Periodic Poisson–Boltzmann Equation. J. Chem. Inf. Model 2015, 55, 2187–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Greene DA; Botello-Smith WM; Follmer A; Xiao L; Lambros E; Luo R, Modeling Membrane Protein–Ligand Binding Interactions: The Human Purinergic Platelet Receptor. J. Phys. Chem. B 2016, 120, 12293–12304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Xiao L; Diao J; Greene DA; Wang J; Luo R, A Continuum Poisson–Boltzmann Model for Membrane Channel Proteins. J. Chem. Theory Comput 2017, 13, 3398–3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Case DA; Brozell SR; Cerutti DS; Cheatham TE III; Cruzeiro VWD; Darden TA; Duke RE; Ghoreishi D; Gohlke H; Goetz AW; Greene D; Harris R; Homeyer N; Izadi S; Kovalenko A; Lee TS; LeGrand S; Li P; Lin C; Liu J; Luchko T; Luo R; Mermelstein DJ; Merz KM; Miao Y; Monard G; Nguyen H; Omelyan I; Onufriev A; Pan F; Qi R; Roe DR; Roitberg A; Sagui C; Schott-Verdugo S; Shen J; Simmerling CL; Swails J; Walker RC; Wang J; Wei H; Wolf RM; Wu X; Xiao L; York DM; Kollman PA, In; University of California, San Francisco., 2018. [Google Scholar]
- 163.Lee T-S; Cerutti DS; Mermelstein D; Lin C; LeGrand S; Giese TJ; Roitberg A; Case DA; Walker RC; York DM, Gpu-Accelerated Molecular Dynamics and Free Energy Methods in Amber18: Performance Enhancements and New Features. J. Chem. Inf. Model 2018, 58, 2043–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Cornell WD; Cieplak P; Bayly CI; Gould IR; Merz KM; Ferguson DM; Spellmeyer DC; Fox T; Caldwell JW; Kollman PA, A 2nd Generation Force-Field for the Simulation of Proteins, Nucleic-Acids, and Organic-Molecules. J. Am. Chem. Soc 1995, 117, 5179–5197. [Google Scholar]
- 165.Konstantinidis E; Cotronis Y, In Proceedings of the 9th international conference on Parallel Processing and Applied Mathematics - Volume Part I; Springer-Verlag: Torun, Poland, 2012, pp 589–598. [Google Scholar]


