Abstract
Some of the most unique and compelling survival strategies in the natural world are fixed in isolated species1. To date, molecular insight into these ancient adaptations has been limited, as classic experimental genetics has focused on interfertile individuals in populations2. Here we use a new mapping approach, which screens mutants in a sterile interspecific hybrid, to identify eight housekeeping genes that underlie the growth advantage of Saccharomyces cerevisiae over its distant relative S. paradoxus at high temperature. Pro-thermotolerance alleles at these mapped loci were required for the adaptive trait in S. cerevisiae and sufficient for its partial reconstruction in S. paradoxus. The emerging picture is one in which S. cerevisiae improved the heat resistance of multiple components of the fundamental growth machinery in response to selective pressure. Our study lays the groundwork for the mapping of genotype to phenotype in clades of sister species across Eukarya.
Geneticists since Mendel have sought to understand how and why wild individuals differ. Studies toward this end routinely test for a relationship between genotype and phenotype via linkage or association2. These familiar approaches, though powerful in many contexts, have an important drawback—they can only be applied to interfertile members of the same species. This rules out any case in which an innovation in form or function evolved long ago and is now fixed in a reproductively isolated population.
As organisms undergo selection over long timescales, their traits may be refined by processes quite different from those that happen early in adaptation3,4. We know little about these mechanisms in the wild, expressly because when the resulting lineages become reproductively incompatible, classic statistical-genetic methods cannot be used to analyze them1. To date, the field has advanced largely on the strength of candidate-based studies that implicate a single variant gene in an interspecific trait5,6, with the complete genetic architecture often remaining unknown. Against the backdrop of a few specialized introgression7–10 and molecular-evolution11 techniques available in the field, dissection of complex trait differences between species has remained a key challenge.
Here we develop a new genetic mapping strategy, based on the reciprocal hemizygosity test12,13, and use it to identify the determinants of a difference in high-temperature growth between isolated Saccharomyces yeast species. We validate the contributions of the mapped loci to the thermotolerance trait, and we investigate their evolutionary history.
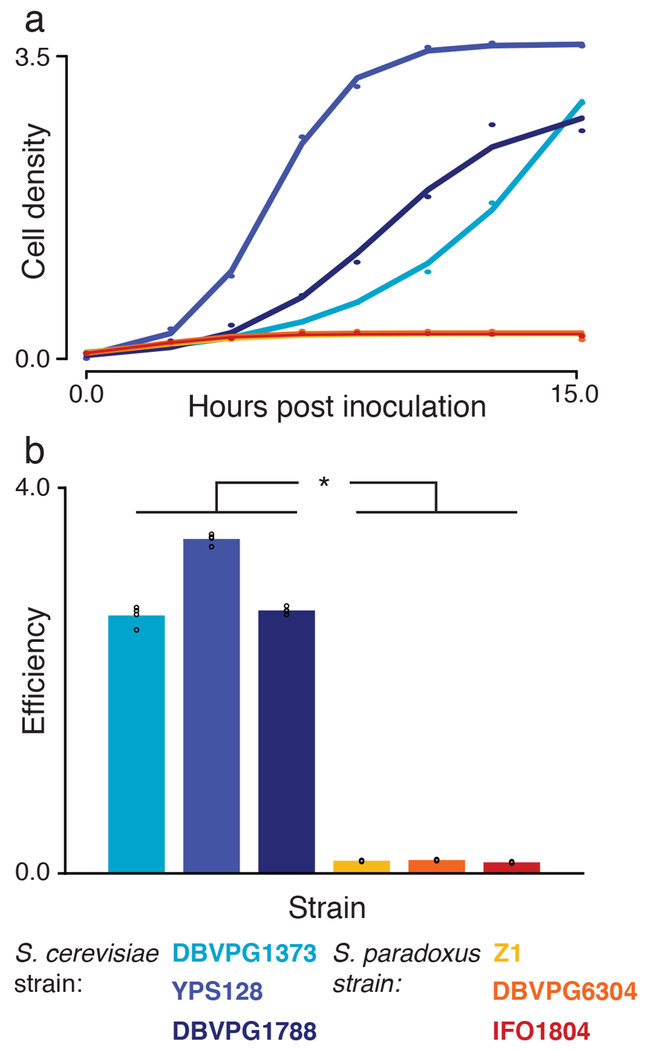
At high temperature, the yeast Saccharomyces cerevisiae grows qualitatively better than other members of its clade14–16, including its closest relative, S. paradoxus, from which it diverged ~5 million years ago17. In culture at 39°C, S. cerevisiae doubled faster than S. paradoxus and accumulated more biomass over a timecourse, a compound trait that we call thermotolerance. The magnitude of differences in thermotolerance between species far exceeded that of strain variation within each species (Figure 1), whereas no such effect was detectable at 28°C (Supplementary Figure 1). The failure by S. paradoxus to grow to high density at 39°C was, at least in part, a product of reduced survival relative to that of S. cerevisiae, as cells of the former were largely unable to form colonies after heat treatment (Supplementary Figure 2). In microscopy experiments, S. paradoxus cells were almost uniformly visible as large-budded dyads after 24 hours at 39°C (Supplementary Figure 3), suggestive of defects late in the cell cycle as a proximal cause of death18. No such tendency was apparent in S. cerevisiae at high temperature, or in either species at 28°C (Supplementary Figure 3).
Figure 1. S. cerevisiae grows better at high temperature than S. paradoxus.
a, Each point reports cell density (OD600) of the indicated wild isolate of S. cerevisiae (blue) or S. paradoxus (orange) over a timecourse of growth at 39°C. Each solid line shows a logistic population growth model fit to the respective cell density measurements. b, Each bar reports mean efficiency (n = 4 cultures) of the indicated strain at 39°C, defined as the difference between cell density at 24 hours of growth and that at the time of inoculation. *, p = 3.78×10−11, two-sample, two-tailed t-test; individual measurements are reported as circles.
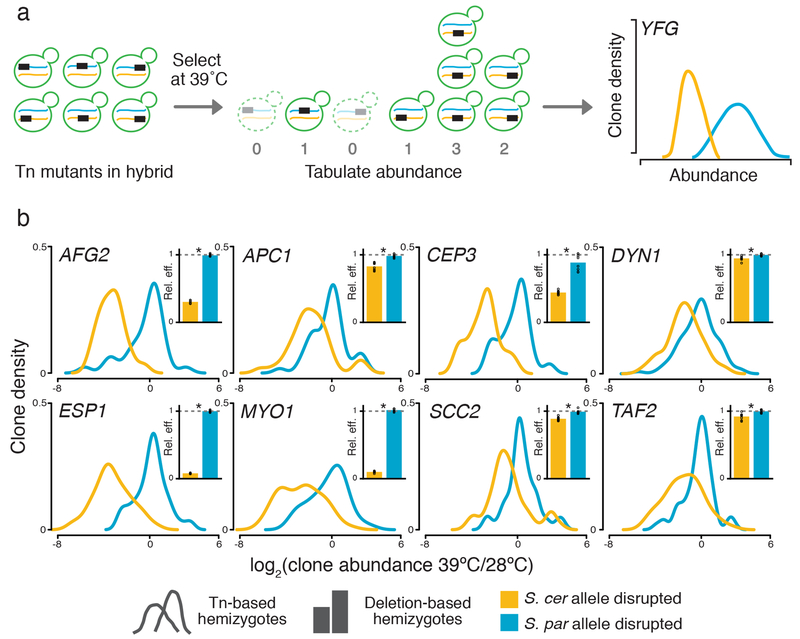
We set out to dissect the genetic basis of S. cerevisiae thermotolerance, using a genomic implementation of the reciprocal hemizygote test12,13 (Figure 2a). For this purpose, we first mated S. cerevisiae strain DBVPG1373, a soil isolate from the Netherlands, with S. paradoxus strain Z1, an English oak tree isolate. The resulting sterile hybrid had a thermotolerance phenotype between those of its purebred parents (Supplementary Figure 4). In this hybrid background we generated hemizygote mutants using a plasmid-borne, selectable PiggyBac transposon system19. We cultured the pool of mutants in bulk for ~7 generations at 39°C and, separately, at 28°C. From cells in each culture we sequenced transposon insertion locations20 as a readout of the genotypes and abundance of mutant hemizygote clones present in the selected sample. In these sequencing data, at each of 4888 genes we detected transposon mutant clones in both species’ alleles in the hybrid (Supplementary Figure 5), with transposon insertions distributed in a largely unbiased manner across the genome (Supplementary Figure 6). For a given gene, we tabulated the abundances of mutants whose transposon insertion fell in the S. cerevisiae allele of the hybrid, after high-temperature selection relative to the 28°C control, and we compared them to the abundance distribution of mutants in the S. paradoxus allele (Figure 2a). Any difference in abundance between these reciprocal hemizygote cohorts can be ascribed to variants between the retained alleles at the respective locus; we refer to the comparison as reciprocal hemizygosity analysis via sequencing (RH-seq). Integrating this approach with a quality-control pipeline (Supplementary Figure 5), in a survey of 3416 high-coverage genes we identified 8 top-scoring hits (false discovery rate 0.01; Figure 2b). At each such locus, disruption of the S. cerevisiae allele in the hybrid was associated with low clone abundance after selection at 39°C relative to 28°C (Figure 2b), reflecting a requirement for the S. cerevisiae allele for thermotolerance. All of the genes mapped by RH-seq were annotated as housekeeping factors: ESP1, DYN1, MYO1, CEP3, APC1, and SCC2 function in chromosome segregation and cytokinesis, and AFG2 and TAF2 in transcription/translation.
Figure 2. Mapping thermotolerance by RH-seq.
a, A transposon (Tn, rectangle) disrupts the allele from S. cerevisiae (blue) or S. paradoxus (orange) of a gene (YFG) in an interspecific hybrid (green). Clones lacking the pro-thermotolerance allele grow poorly at 39°C (dashed outlines), as measured by sequencing and reported in smoothed histograms (traces, colored to indicate the species’ allele that is not disrupted). b, Each panel reports results from one RH-seq hit locus. In the main figure of a given panel, the x-axis reports the log2 of abundance, measured by RH-seq after selection at 39°C, of a clone harboring a transposon insertion in the indicated species’ allele, relative to the analogous quantity for that clone from selection at 28°C. The y-axis reports the proportion of all clones bearing insertions in the indicated allele that exhibited the abundance ratio on the x, as a kernel density estimate. In insets, each bar reports the relative efficiency, calculated as the mean growth efficiency at 39°C (n = 8-12 cultures) of the indicated targeted-deletion hemizygote measured in liquid culture assays, normalized to the analogous quantity for the wild-type hybrid. *, p ≤ 0.002, two-sample, one-tailed t-test; individual measurements are reported as circles. See Supplementary Table 1 for exact p-values and sample numbers.
To evaluate the role in thermotolerance of genes that emerged from RH-seq, we first sought to verify that growth differences between hemizygotes at a given locus were the consequence of allelic variation, and not an artifact of our genomic approach. Toward this end, at each RH-seq hit gene we engineered hemizygotes by targeted deletion of each species’ allele in turn in the hybrid. In growth assays, the strain lacking the S. cerevisiae allele at each gene grew poorly at high temperature (Figure 2b), with little impact at 28°C (Supplementary Figure 7), as inferred from RH-seq. Likewise, at each locus, the S. paradoxus allele made no contribution to the phenotype of the hybrid, since deleting it had no effect (Figure 2B and Supplementary Figure 7). Locus effect sizes from this single-gene validation paradigm largely paralleled the estimates from RH-seq (R2 = 0.74). We conclude that RH-seq hits represent bona fide determinants of thermotolerance in the hybrid.
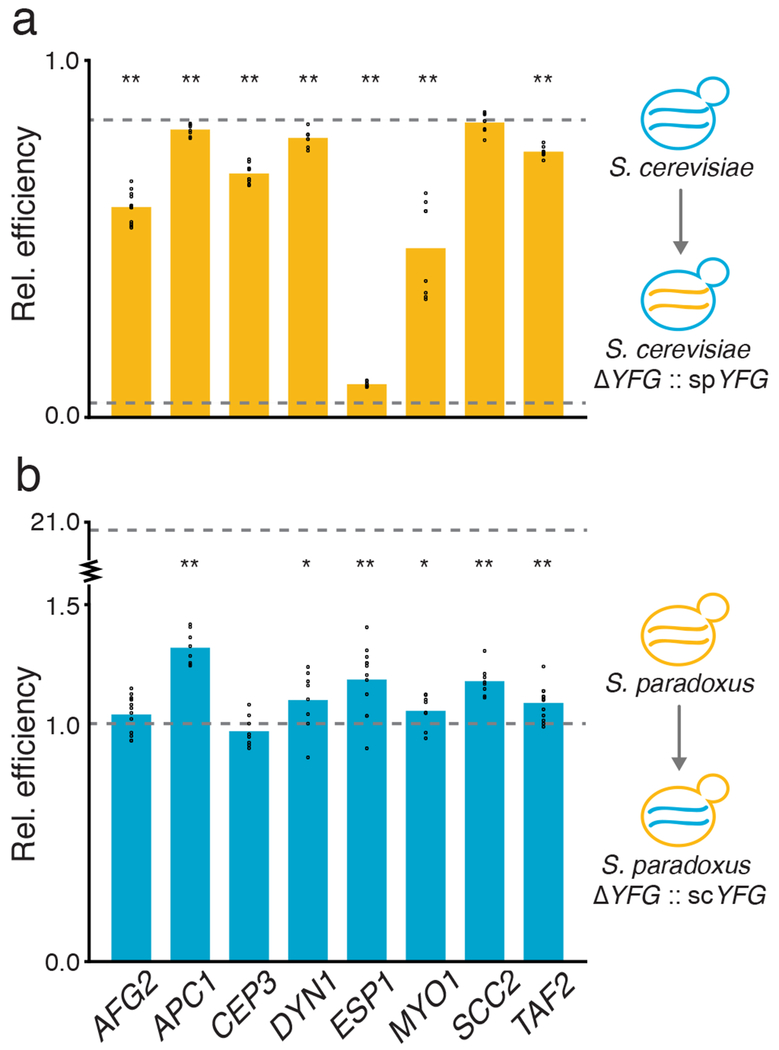
We expected that variation at our RH-seq hits, though mapped by virtue of their impact in the hybrid, could also explain thermotolerance differences between purebred species. As a test of this notion, for each mapped gene in turn, we replaced the two copies of the endogenous allele in each purebred diploid with the allele from the other species. Growth assays of these transgenics established the S. cerevisiae allele of each locus as necessary or sufficient for biomass accumulation at 39°C, or both: thermotolerance in the S. cerevisiae background was compromised by S. paradoxus alleles at 7 of the 8 genes and, in S. paradoxus, improved by S. cerevisiae alleles at 6 of 8 loci (Figure 3). Allele replacements had little impact on growth at 28°C (Supplementary Figure 8). These trends mirrored the direction of locus effects from hemizygotes in the hybrid, though the magnitudes were often different. Most salient were the small effect sizes in S. paradoxus relative to other backgrounds, indicative of strong epistasis in this poorly-performing species (Supplementary Figure 9). Thus, the loci mapped by RH-seq in an interspecies hybrid contribute causally to thermotolerance in purebreds, with effect sizes that depend on the context in which they are interrogated.
Figure 3. S. cerevisiae thermotolerance alleles are necessary and sufficient for growth at high temperature.
a, Each bar reports mean growth efficiency at 39°C, measured in liquid culture assays (n = 8-12 cultures), of an S. cerevisiae strain harboring the S. paradoxus allele at the indicated RH-seq hit locus, relative to the analogous quantity for wild-type S. cerevisiae. b, Data are as in a, except that each bar reports results from a S. paradoxus strain harboring the S. cerevisiae allele at the indicated locus, normalized to wild-type S. paradoxus. In a given panel, the top and bottom dotted lines report the relative efficiency of wild-type S. cerevisiae and S. paradoxus, respectively. *, p ≤ 0.036; **, p ≤ 0.001, one-sample, one-tailed t-test; individual measurements are reported as circles. See Supplementary Table 1 for exact p-values and sample numbers.
Avid growth at high temperature is a defining characteristic of S. cerevisiae as a species, relative to other Saccharomycetes (refs. 14–16 and Figure 1). In principle, the loci mapped by RH-seq could be unique to the genetic architecture of thermotolerance in our focal S. cerevisiae strain, DBVPG1373, or be part of a mechanism common to many S. cerevisiae isolates. In support of the latter model, transgenesis experiments showed that a diverse panel of S. cerevisiae isolates all harbored alleles conferring modest but significant growth benefits at high temperature, and alleles from multiple S. paradoxus isolates were deleterious (Supplementary Figure 10a-b). We detected no such impact at 28°C (Supplementary Figure 10a-b). Similarly, we found elevated sequence divergence from S. paradoxus to be a shared feature of S. cerevisiae strains at the loci mapped by RH-seq (using the absolute divergence measure Dxy; Supplementary Figure 10c). These findings indicate that the S. cerevisiae population accumulated divergent, pro-thermotolerance alleles at appreciable density in the loci mapped by RH-seq, consistent with a role in the trait for these genes across the species. And in the yeast phylogeny, RH-seq hit genes were distinguished by accelerated evolution along the branch leading to S. cerevisiae, as expected if the ancestral program has been conserved among the other species in the clade (Supplementary Figure 10c).
In this work, we have developed the RH-seq method for genome-wide mapping of natural trait variation, and we have used it to elucidate the genetics of thermotolerance in reproductively isolated yeasts. Growth at high temperature is likely a derived character in S. cerevisiae14–16, and the mechanism by which evolution built the trait, after the split from S. paradoxus, has remained unknown. In pursuing the genetics of this putative ancient adaptation, we complement studies of younger, intra-specific variants that erode thermotolerance, in the few S. cerevisiae isolates that have lost the trait relatively recently12,21. We have sought to shed light on more ancient evolutionary events by considering S. paradoxus as a representative of the ancestral state, to which thermotolerant S. cerevisiae can be compared.
Using this approach, we have mapped eight loci at which S. cerevisiae alleles are necessary and sufficient for thermotolerance. As our RH-seq scan did not attain complete genomic coverage, the hits we did find likely represent a lower bound on the complexity of the architecture of the trait. Six of the RH-seq hit genes are essential for growth in standard conditions22, and all eight contribute to fundamental growth processes. ESP1, DYN1, CEP3, APC1, MYO1, and SCC2 mediate mitotic spindle assembly, chromatid cohesion and separation, cytokinesis, and mitotic exit; AFG2 regulates the release of maturation factors from the ribosome; and TAF2 encodes a TFIID subunit. In each case, our growth experiments in the interspecific hybrid have shown that the S. paradoxus allele acts as a hypomorph at high temperature. Our work leaves unanswered exactly how heat-treated S. paradoxus dies in the absence of these functions, though the cells’ large-budded morphology strongly suggests regulated arrest or stochastic failure late in the cell cycle. That said, given that some but not all RH-seq hit loci have roles in mitosis, it is likely only one of the choke points at which S. paradoxus alleles are a liability at high temperature. Assuming that these heat-sensitive alleles also littered the genome of the common ancestor with S. cerevisiae, thermotolerance would have evolved along the S. cerevisiae lineage by resolving each of them, boosting the heat resistance of many housekeeping processes. Such a mechanism would dovetail with the recent finding that, across species, the limiting temperature for cell growth correlates with the melting temperatures of a subset of essential proteins23.
These insights into the evolution of a complex yeast trait serve as a proof of concept for RH-seq. To date, the reciprocal hemizygosity test has led to landmark discoveries in a candidate-gene framework, confirming the effects of variation at a given locus identified by other means12,13. Schemes to scale up the test have generated a genome’s worth of hemizygotes from deletion-strain purebreds, which tend to harbor secondary mutations that come through screens as false positives24,25. As such, a key advantage of RH-seq is that we carry out mutagenesis in the hybrid, which ensures coverage of essential genes and obviates the use of mutation-prone null genotypes. Furthermore, any secondary mutations that do arise in a given hemizygote clone, e.g. during a long competition in the condition of interest, would not have a strong influence on RH-seq mapping, because deep mutagenesis generates many independent clones per gene that are analyzed together. One important caveat of RH-seq, as in single-gene reciprocal hemizygote tests, is the assumption that no epistasis unique to the hybrid will mask the effects of loci underlying a trait difference of interest between the parents. In our case study, the genetic architecture of thermotolerance in the hybrid did bear out as relevant for the purebreds, albeit with locus effect sizes that varied across the backgrounds. More dramatic discrepancies may be particularly likely when the hybrid has a heterotic (i.e. extreme) phenotype and is a poor model for the genetics of the parents26. The choice of a non-heterotic hybrid in which to pursue RH-seq would be analogous to classical linkage mapping in a cross whose progeny have, on average, phenotypes that are intermediate between those of the parents.
In fact, although we have focused here on ancient divergence, the RH-seq method would be just as applicable to individuals within a species, as a high-resolution alternative to linkage analysis. We thus anticipate that RH-seq will accelerate the mapping of genotype to phenotype in many systems, whether the parents of a cross are closely related or members of a species complex that have been locally adapting for millions of years.
Online methods
Strains
Strains used in this study are listed in Supplementary Table 2. Homozygous diploid strains of S. cerevisiae and S. paradoxus used as parents of the interspecific hybrid, and as the backgrounds for allele-swap experiments, were homothallic DBVPG1373 and Z1, respectively. In the case of the hybrid parents, each strain was rendered homozgyous null for URA3 via homologous recombination with a HYGMX cassette, then sporulated; a given mated spore from a dissected tetrad was grown up into a diploid that was homozygous null at URA3 and tested for the presence of both genomes by PCR with species-specific primers.
piggyBac transposon machinery
For untargeted, genome-scale construction of reciprocal hemizygotes in the S. cerevisiae x S. paradoxus hybrid, we adapted methods for piggyBac transposon mutagenesis19 to develop a system in which the transposon machinery was borne on a selectable and counter-selectable plasmid lacking a centromere. We constructed this plasmid (final identifier pJR487) in three steps. In step I we cloned the piggyBac transposase enzyme gene driven by the S. cerevisiae TDH3 promoter (from plasmid p3E1.2, a gift from Malcolm Fraser, Notre Dame) into plasmid pJED104 (which contains URA3, an ARS, and the CEN6 locus, and was a gift from John Dueber, UC Berkeley). For this cloning, the amplification used a forward and reverse primer containing a BamHI and XhoI site, respectively, that upon restriction digest yielded sticky ends for ligation to recipient BamHI and XhoI sites in digested pJED104. We used the resulting plasmid as input into step II, removal of the CEN6 sequence: we first amplified the entire plasmid with primers that initiated outside of CEN6 and were directed away from it, and contained reciprocally complementary NheI sites; sticky ends of the linear PCR product were then ligated together for re-circularization. We used the resulting plasmid as input into step III, the cloning in of a construct comprised of the KANMX cassette flanked by long terminal arms (328bp and 361bp) from the piggyBac transposon. We first amplified KANMX from pUG627 and each transposon arm from p3E1.2, using primers that contained overlapping sequence on the fragment ends that would ultimately be the interior of the construct, and XbaI sites on the fragment ends that would ultimately be the 5’ and 3’-most ends of the construct. We stitched the three fragments together by overlap extension PCR, digested the resulting construct and the plasmid from step II with XbaI, and annealed sticky ends of the two to yield the final pJR487 plasmid.
Untargeted hemizygote construction via transposon mutagenesis
For mutagenesis, pJR487 was gigaprepped using a column kit (Zymo Research) to generate ~11 mg plasmid. To prepare for transformation, JR507 (the S. cerevisiae DBVPG1373 x S. paradoxus Z1 hybrid) was streaked from a −80°C freezer stock onto a yeast peptone dextrose (YPD, 1% yeast extract [BD], 2% yeast peptone [BD], 2% D-glucose [Sigma]) agar plate and incubated for 2 days at 26°C. A single colony was inoculated into 100 mL YPD and shaken at 28°C, 200rpm for ~24 hours. The next day, we transferred cells from this pre-culture, and YPD, to each of four 1 L flasks at the volumes required to attain an optical density at 600 nm (OD600) of 0.2 in 500 mL each. We cultured each for 6 hours at 28°C with shaking at 200rpm. Two of these cultures were combined into 1 L of culture and two into a separate 1 L, and each such culture was subjected to transformation (for a total of two transformations) as follows. The 1 L was split into twenty 50-mL conical tubes. Each aliquot was centrifuged and washed with water and then with 0.1 M lithium acetate (LiOAc, Sigma) mixed with 1X Tris-EDTA buffer (10 mM Tris-HCl and 1.0 mM EDTA); after spin-down, to each tube was added a solution of 0.269 mg of pJR487 mixed 5:1 by volume with salmon sperm DNA (Invitrogen), and then to each was added 3 mL of 39.52% polyethylene glycol, 0.12M LiOAc and 1.2X Tris-EDTA buffer (12 mM Tris-HCl and 1.2 mM EDTA). Tubes were rested for 10 minutes at room temperature, then heat-shocked in a water bath at 39°C for 26 minutes. Cells from all 20 tubes were then combined. We transferred cells from this post-transformation culture, and YPD, to each of three 1 L flasks at the volumes required to attain an OD600 of ~0.35-4 in 500 mL. Each such culture was recovered by shaking at 28°C and 200 rpm for 2 hours. G418 (Geneticin, Gibco) was added to each at a concentration of 300 µg/mL to select for those cells which had taken up the plasmid, and cultures were incubated with 200 rpm shaking at 28°C for two days until each reached an OD600 of ~2.3. All six such selected cultures across the two transformations were combined. We transferred cells from this combined culture, and YPD + G418 (300 ug/mL), to each of two 1 L flasks at the volumes required to attain an OD600 of 0.2 in 500 mL each. We cultured each flask at 28°C and 200 rpm shaking overnight until reaching an OD600 of 2.18 and combined the two cultures again to yield one culture. To cure transformants of the pJR487 URA+ plasmid, we spun down a volume of this master culture and resuspended in water with the volume required to attain a cell density of 1.85 OD600 units/mL. 12 mL of this resuspension were plated (1 mL per 24.1cm x 24.1cm plate) onto plates containing complete synthetic media with 5-fluoroorotic acid (5-FOA) [0.2% drop-out amino acid mix without uracil or yeast nitrogen base (YNB) (US Biological), 0.005% uracil (Sigma), 2% D-glucose (Sigma), 0.67% YNB without amino acids (Difco), 0.075% 5-FOA (Zymo Research)]. After incubation at 28°C to enable colony growth, colonies were scraped off all 12 plates and combined into water at the volume required to attain 40 OD600 units per 900 µL, yielding the final transposon mutant hemizygote pool. This was aliquoted into 1 mL volumes with 10% DMSO and frozen at −80°C.
Thermotolerance phenotyping via selection of the hemizygote pool
One aliquot of the pool of transposon mutant hemizygotes in the JR507 S. cerevisiae DBVPG1373 x S. paradoxus Z1 hybrid background was thawed and inoculated into 150 mL of YPD in a 250 mL flask, and cultured for 7.25 hours at 28°C, with shaking at 200 rpm. We used this timepoint as time zero of our thermotolerance experiment, and took four aliquots of 6.43 mL (7 OD units) as technical replicates for sequencing of transposon insertion positions (see below). 9.19 mL of the remaining culture was back-diluted to an OD600 of 0.02 in a total of 500 mL YPD in each of six 2L glass flasks for cultures that we call selections; three were grown at 28°C and three at 39°C (shaking at 200 rpm) until an OD600 of 1.9-2.12 was reached, corresponding to about 6.5 doublings in each case. Four cell pellets of 7 OD600 units each were harvested from each of these biological replicate flasks, for sequencing as technical replicates (see below). In total, 28 pellets were subjected to sequencing: 4 technical replicates from the time-zero culture; 3 biological replicates, 4 technical replicates each, from the 28°C selection; and 3 biological replicates, 4 technical replicates each, from the 39°C selection (Supplementary Table 3).
Tn-seq library construction
To determine the abundance of transposon mutant hemizygote clones after selection, we first sequenced transposon (Tn) insertions as follows. Each cell pellet from a time zero or selection sample (see above) was thawed on ice, and its genomic DNA (gDNA) was harvested with the ZR Fungal/Bacterial DNA MiniPrep kit (Zymo Research). gDNA was resuspended in DNA elution buffer (Zymo) pre-warmed to 65°C and its concentration was quantified using a Qubit 3.0 fluorometer. Illumina Tn-seq library construction was as described28. Briefly, gDNA was sonicated and ligated with common adapters, and for each fragment deriving from a Tn insertion in the genome, a sequence containing a portion of the transposon and a portion of its genomic context (the Tn-genome junction) was amplified using one primer homologous to a region in the transposon, and another primer homologous to a region in the adapter. See Supplementary Table 4 for the transposon specific primer (“forward primer”), where N’s are random nucleotides, and the indexed adapter-specific primer (“reverse primer”), where the six N’s are a unique index used for multiplexing multiple libraries onto the same Hiseq sequencing lane. Amplification used Jumpstart polymerase (Sigma) and the following cycling protocol: 94°C-2 min, [94°C-30 sec, 65°C-20 sec, 72°C-30 sec] X 25, 72°C-10 min. Sequencing of single-end reads of 150 bp was done over eight lanes on a HiSeq 2500 at the Joint Genome Institute (Walnut Creek, CA). Reads sequenced per library are reported in Supplementary Table 3.
Tn-seq read-mapping and data analysis
For analysis of data from the sequencing of Tn insertion sites in pools of hemizygotes, we first searched each read for a string corresponding to the last 20 base pairs of the left arm of the piggyBac transposon sequence, allowing up to two mismatches. For each Tn-containing read, we then identified the genomic location of the sequence immediately downstream of the Tn insertion site, which we call the genomic context of the insertion, by mapping with BLAT (minimum sequence identity = 95, tile size = 12) against a hybrid reference genome made by concatenating amended S. cerevisiae DBVPG1373 and S. paradoxus Z1 genomes (see below). These genomic-context sequence fragments were of variable length; any case in which the sequence was shorter than 50 base pairs was eliminated from further analysis, as was any case in which a genomic-context sequence mapped to more than one location in the hybrid reference. The resulting data set thus comprised reads containing genomic-context sequences specifically mapping to a single location in either S. cerevisiae DBVPG1373 or S. paradoxus Z1, which we call usable reads. For a given library, given a cohort of usable reads whose genomic-context sequence mapped to the same genomic location, we inferred that these reads originated from clones of a single mutant with the Tn inserted at the respective site, which we call an insertion. In cases where the genomic-context sequences from reads in a given library mapped to positions within 3 bases of each other, we inferred that these all originated from the same mutant genotype and combined them, assigning to them the position corresponding to the single location to which the most reads mapped among those combined. For a given insertion thus defined, we considered the number of associated reads ninsert as a measure proportional to the abundance of the insertion clone in the cell pellet whose gDNA was sequenced. To enable comparison of these abundances across samples, we tabulated the total number of usable reads npellet from each cell pellet, took the average of this quantity across pellets, <npellet>, and multiplied each ninsert by <npellet>/ npellet to yield ainsert, the final normalized estimate of the abundance of the insertion clone in the respective pellet. For any insertions that were not detected in a given pellet’s library (ninsert = 0) but detectable in another library of the data set, we assigned ninsert = 1.
We evaluated, from the mapped genomic-context sequence of each insertion, whether it fell into a gene according to the S. cerevisiae and S. paradoxus genome annotations17,29, and we retained for further analysis only those insertions that fell into genes that were syntenic in the two species. For each such insertion, for each biological replicate corresponding to a selection culture (at 28°C or 39°C), we averaged the normalized abundances ainsert across technical replicates, yielding a single abundance estimate <ainsert>technical for the biological replicate. We then calculated the mean of the latter quantities across all biological replicates of the selection, to yield a final abundance estimate for the insertion in this selection, <ainsert>total. Likewise, for each insertion and selection experiment we calculated CVinsert,total, the coefficient of variation of <ainsert>technical values across biological replicates.
To use Tn-seq data in reciprocal hemizygosity tests, we considered for analysis only genes annotated with the same (orthologous) gene name in the S. cerevisiae and S. paradoxus reference genomes. For each insertion, we divided the <ainsert>total value from the 39°C selection by the analogous quantity from the 28°C selection and took the log2 of this ratio, which we consider to reflect thermotolerance as measured by RH-seq. For each gene in turn, we used a two-tailed Mann-Whitney U test to compare thermotolerance measured by RH-seq between the set of insertions falling into the S. cerevisiae alleles of the gene, against the analogous quantity from the set of insertions falling into the S. paradoxus allele of the gene, and we corrected for multiple testing using the Benjamini-Hochberg method.
We tabulated the number of inserts and genes used as input into the reciprocal hemizygote test, and the number of top-scoring genes emerging from these tests, under each of a range of possible thresholds for coverage and measurement noise parameter values (Supplementary Figure 5). We used in the final analysis the parameter-value set yielding the most extensive coverage and the most high-significance hits: this corresponded to insertions whose abundances had, in the data from at least one of the two selections (at 28°C or 39°C), CVinsert,total ≤ 1.5 and <ainsert>total ≥ 1.1, and genes for which this high-confidence insertion data set contained at least 5 insertions in each species’ allele. This final data set comprised 110,678 high-quality insertions (Supplementary Table 5) in 3416 genes (Supplementary Table 6). We used this complement of data in all display items of this paper with the following exception. To evaluate post facto the reproducibility across replicates of RH-seq measurements on genes called as hits, we first randomly paired each biological replicate at 39°C with one at 28°C; then, from the sequencing data from each pair in turn, we identified insertions whose abundances had <ainsert>total ≥ 1.1 in at least one of the two temperatures for the respective replicate, and genes for which we had at least 5 such insertions in each species’ allele. From these data, for a given RH-seq hit gene we tabulated single-replicate estimates of the abundances of hemizygotes harboring insertions in each species’ homolog, for Columns G-L of Supplementary Table 6.
Amended reference genome construction
We generated reference genomes for S. cerevisiae strain DBVPG1373 and S. paradoxus strain Z1 as follows. Raw genome sequencing reads for each strain were downloaded from the SGRP2 database (see URLs). Reads were aligned using bowtie230 with default options; DBVPG1373 reads were aligned to version R64.2.1 of the reference sequence of the S. cerevisiae type strain S288C (Genbank Assembly Accession GCA_000146045.2), and Z1 reads were aligned to the S. paradoxus strain CBS432 reference sequence31. Single nucleotide variants (SNPs) were called using a pipeline of samtools32, bcftools and bgzip, and were filtered for a quality score (QUAL) of >20 and a combined depth (DP) of >5 and either <65 (S. cerevisiae) or <255 (S. paradoxus). We then amended each reference genome with the respective filtered SNPs: we replaced the S288C allele with that of DBVPG1373 at each filtered SNP using bcftools’ consensus command with default options (42,983 base pairs total), and amendment of the CBS432 sequence was carried out analogously using Z1 alleles (15,126 base pairs total).
Sequence analysis
Dxy analysis.
To evaluate whether sequence divergence from S. paradoxus at RH-seq hit loci was a shared feature of S. cerevisiae isolates, we used the Dxy statistic33, the average number of pairwise differences between S. cerevisiae strains and S. paradoxus, normalized for gene length, as follows. We downloaded S. cerevisiae genomic sequences from the following sources: YJM978, UWOPS83-787, Y55, UWOPS05-217.3, 273614N, YS9, BC187, YPS128, DBVPG6765, YJM975, L1374, DBVPG1106, K11, SK1, 378604X, YJM981, UWOPS87-2421, DBVPG1373, NCYC3601, YPS606, Y12, UWOPS05-227.2, and YS2 from the Yeast Resource Center (see URLs); Sigma1278b, ZTW1, T7, and YJM789 from the Saccharomyces Genome Database (see URLs); and RM11 from NCBI (accession PRJNA13674). For each strain, we extracted the coding sequence of each gene in turn, and we downloaded the S. paradoxus reference sequence for each orthologous coding region from 17. Sequences were aligned using MAFFT34 with default settings. Alignments that did not contain a start and stop codon, or those that contained gaps at greater than 40% of sites were considered poor quality and discarded. We tabulated Dxy for each gene. To evaluate whether the eight RH-seq hit genes were enriched for elevated Dxy, we first tabulated <Dxy>true, the mean value across the eight RH-seq hit genes. We then sampled eight random genes from the set of 3416 genes tested by RH-seq; to account for biases associated with lower rates of divergence among essential genes, the resampled set contained six essential genes and two non-essential genes, mirroring the breakdown of essentiality among the RH-seq hits. Across this random sample we tabulated the mean Dxy, <Dxy>resample. We repeated the resampling 5000 times and used as an empirical p-value the proportion of resamples at which <Dxy>resample ≤ <Dxy>true.
Phylogenetic analysis.
We downloaded orthologous protein coding regions for the type strains of S. cerevisiae, S. paradoxus, and an outgroup, S. mikatae, from 17. For each gene for which ortholog sequences were available in all three species, we aligned the sequences with PRANK35 utilizing the “-codon” option for codon alignment. These alignments were used as input into the codeml module of PAML36, which was run assuming no molecular clock and allowing omega values to vary for each branch in the phylogeny. From the resulting inferences, we tabulated the branch length on the S. cerevisiae lineage for each gene. To evaluate whether sequence divergence of the eight RH-seq hit genes showed signatures of rapid evolution along the S. cerevisiae lineage, we used the resampling test detailed above.
Supplementary Material
Acknowledgements
The authors thank F. AlZaben, A. Flury, G. Geiselman, J. Hong, J. Kim, M. Maurer, and L. Oltrogge for technical assistance, D. Savage for his generosity with microscopy resources, and B. Blackman, S. Coradetti, A. Flamholz, V. Guacci, D. Koshland, C. Nelson, and A. Sasikumar for discussions; we also thank J. Dueber (Department of Bioengineering, UC Berkeley) for the PiggyBac plasmid. This work was supported by R01 GM120430-A1 and by Community Sequencing Project 1460 to RBB at the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility. The work conducted by the latter was supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231.
Footnotes
URLs
SGRP2 Database - ftp://ftp.sanger.ac.uk/pub/users/dmc/yeast/SGRP2/input/strains
Yeast Resource Center - http://www.yeastrc.org/g2p/home.do
Saccharomyces Genome Database - http://www.yeastgenome.org/
RH-seq data analysis scripts - https://github.com/weiss19/rh-seq
Accession Codes RH-seq data are deposited in the Sequence Read Archive (SRA) under the accession SRP156210.
Competing Interests
The authors do not declare any competing financial interests.
Supplementary Information is available in the online version of the paper.
Code availability
Custom Python and R scripts used for RH-seq data analysis are available on Github (see URLs).
For strain construction, growth assays, microscopy and locus effect size methods, see Supplementary Note. Detailed information on experimental design and reagents can be found in the Life Sciences Reporting Summary.
Data availability
RH-seq data are deposited in the Sequence Read Archive (SRA) under the accession SRP156210.
Main text references
- 1.Allen Orr H The genetics of species differences. Trends Ecol Evol 16, 343–350 (2001). [DOI] [PubMed] [Google Scholar]
- 2.Flint J & Mott R Finding the molecular basis of quantitative traits: successes and pitfalls. Nat Rev Genet 2, 437–445, doi: 10.1038/35076585 (2001). [DOI] [PubMed] [Google Scholar]
- 3.Good BH, McDonald MJ, Barrick JE, Lenski RE & Desai MM The dynamics of molecular evolution over 60,000 generations. Nature, doi: 10.1038/nature24287 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Savolainen O, Lascoux M & Merila J Ecological genomics of local adaptation. Nat Rev Genet 14, 807–820, doi: 10.1038/nrg3522 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Nadeau NJ & Jiggins CD A golden age for evolutionary genetics? Genomic studies of adaptation in natural populations. Trends Genet 26, 484–492, doi: 10.1016/j.tig.2010.08.004 (2010). [DOI] [PubMed] [Google Scholar]
- 6.Wray GA Genomics and the Evolution of Phenotypic Traits. Annual Review of Ecology, Evolution, and Systematics 44, 51–72 (2013). [Google Scholar]
- 7.Masly JP & Presgraves DC High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLoS biology 5, e243, doi: 10.1371/journal.pbio.0050243 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greig D A screen for recessive speciation genes expressed in the gametes of F1 hybrid yeast. PLoS genetics 3, e21, doi: 10.1371/journal.pgen.0030021 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Eshed Y & Zamir D An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141, 1147–1162 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lazzarano S et al. Genetic mapping of species differences via in vitro crosses in mouse embryonic stem cells. Proc Natl Acad Sci U S A 115, 3680–3685, doi: 10.1073/pnas.1717474115 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Roop JI, Chang KC & Brem RB Polygenic evolution of a sugar specialization trade-off in yeast. Nature 530, 336–339, doi: 10.1038/nature16938 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Steinmetz LM et al. Dissecting the architecture of a quantitative trait locus in yeast. Nature 416, 326–330, doi: 10.1038/416326a (2002). [DOI] [PubMed] [Google Scholar]
- 13.Stern DL Identification of loci that cause phenotypic variation in diverse species with the reciprocal hemizygosity test. Trends Genet 30, 547–554, doi: 10.1016/j.tig.2014.09.006 (2014). [DOI] [PubMed] [Google Scholar]
- 14.Goncalves P, Valerio E, Correia C, de Almeida JM & Sampaio JP Evidence for divergent evolution of growth temperature preference in sympatric Saccharomyces species. PLoS One 6, e20739, doi: 10.1371/journal.pone.0020739 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Salvado Z et al. Temperature adaptation markedly determines evolution within the genus Saccharomyces. Appl Environ Microbiol 77, 2292–2302, doi: 10.1128/AEM.01861-10 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sweeney JY, Kuehne HA & Sniegowski PD Sympatric natural Saccharomyces cerevisiae and S. paradoxus populations have different thermal growth profiles. FEMS Yeast Res 4, 521–525 (2004). [DOI] [PubMed] [Google Scholar]
- 17.Scannell DR et al. The Awesome Power of Yeast Evolutionary Genetics: New Genome Sequences and Strain Resources for the Saccharomyces sensu stricto Genus. G3 (Bethesda) 1, 11–25, doi: 10.1534/g3.111.000273 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hartwell LH Saccharomyces cerevisiae cell cycle. Bacteriol Rev 38, 164–198 (1974). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitra R, Fain-Thornton J & Craig NL piggyBac can bypass DNA synthesis during cut and paste transposition. EMBO J 27, 1097–1109, doi: 10.1038/emboj.2008.41 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Opijnen T, Lazinski DW & Camilli A Genome-Wide Fitness and Genetic Interactions Determined by Tn-seq, a High-Throughput Massively Parallel Sequencing Method for Microorganisms. Curr Protoc Mol Biol 106, 7 16 11–24, doi: 10.1002/0471142727.mb0716s106 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Parts L et al. Revealing the genetic structure of a trait by sequencing a population under selection. Genome Res 21, 1131–1138, doi: 10.1101/gr.116731.110 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winzeler EA et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science 285, 901–906 (1999). [DOI] [PubMed] [Google Scholar]
- 23.Leuenberger P et al. Cell-wide analysis of protein thermal unfolding reveals determinants of thermostability. Science 355, doi: 10.1126/science.aai7825 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Wilkening S et al. An evaluation of high-throughput approaches to QTL mapping in Saccharomyces cerevisiae. Genetics 196, 853–865, doi: 10.1534/genetics.113.160291 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neurological Disorders: Public Health Challenges. 232 (World Health Organization, 2006). [Google Scholar]
- 26.Sinha H, Nicholson BP, Steinmetz LM & McCusker JH Complex genetic interactions in a quantitative trait locus. PLoS Genet 2, e13, doi: 10.1371/journal.pgen.0020013 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
Methods-only references
- 27.Guldener U, Heck S, Fielder T, Beinhauer J & Hegemann JH A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res 24, 2519–2524 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wetmore KM et al. Rapid quantification of mutant fitness in diverse bacteria by sequencing randomly bar-coded transposons. MBio 6, e00306–00315, doi: 10.1128/mBio.00306-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skelly DA et al. Integrative phenomics reveals insight into the structure of phenotypic diversity in budding yeast. Genome Res 23, 1496–1504, doi: 10.1101/gr.155762.113 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Langmead B & Salzberg SL Fast gapped-read alignment with Bowtie 2. Nat Methods 9, 357–359, doi: 10.1038/nmeth.1923 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liti G et al. Population genomics of domestic and wild yeasts. Nature 458, 337–341, doi: 10.1038/nature07743 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Li H et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079, doi: 10.1093/bioinformatics/btp352 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nei M Molecular Evolutionary Genetics. (Columbia University Press, 1987). [Google Scholar]
- 34.Katoh K & Standley DM MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30, 772–780, doi: 10.1093/molbev/mst010 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loytynoja A & Goldman N webPRANK: a phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinformatics 11, 579, doi: 10.1186/1471-2105-11-579 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang Z PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24, 1586–1591, doi: 10.1093/molbev/msm088 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.