Abstract
How common polymorphisms in noncoding genome regions can regulate cellular function remains largely unknown. Here we show that cardiac fibrosis, mimicked using a hydrogel with controllable stiffness, affects the regulation of the phenotypes of human cardiomyocytes by a portion of the long noncoding RNA ANRIL, the gene of which is located in the disease-associated 9p21 locus. In a physiological environment, cultured cardiomyocytes derived from induced pluripotent stem cells obtained from patients who are homozygous for cardiovascular-risk alleles (R/R cardiomyocytes) or from healthy individuals who are homozygous for nonrisk alleles contracted synchronously, independently of genotype. After hydrogel stiffening to mimic fibrosis, only the R/R cardiomyocytes exhibited asynchronous contractions. These effects were associated with increased expression of the short ANRIL isoform in R/R cardiomyocytes, which induced a c-Jun N-terminal kinase (JNK) phosphorylation-based mechanism that impaired gap junctions (particularly, loss of connexin-43 expression) following stiffening. Deletion of the risk locus or treatment with a JNK antagonist was sufficient to maintain gap junctions and prevent asynchronous contraction of cardiomyocytes. Our findings suggest that mechanical changes in the microenvironment of cardiomyocytes can activate the regulation of their function by noncoding loci.
Cardiovascular disease is caused by numerous factors; however, a considerable portion of the risk is caused by genetic factors. More than 106 disease-associated single-nucleotide polymorphisms (SNPs) in the human genome have been identified1. The vast majority of risk-associated SNPs cluster in noncoding regions, complicating our understanding of their function2. As a model locus in which to study the mechanisms of noncoding variants, we investigated the 9p21.3 locus, which has a robust association with a range of diseases, including coronary artery disease (CAD). Despite approximately 21% of the population being homozygous for the risk haplotype3, the impact of the most common haplotypes on cellular function is unclear. The 9p21 locus itself is flanked by the cyclin-dependent kinase inhibitor 2A (CDKN2A) and 2B (CDKN2B) genes and intersects part of a long noncoding RNA known as ANRIL (CDKN2B-AS1)3–5. Analyses of an orthologous knockout mouse, in which parts of chromosome 4 were knocked out, found reduced CDKN2A and CDKN2B expression in cardiac tissue6,7, but such findings are difficult to interpret because of the limited sequence conservation of ANRIL in animals other than primates8. Moreover, expression of ANRIL9 and its alternative splicing may be context-specific, complicating our understanding of the mechanisms of associated risks10,11.
Given the challenges with interpreting the results obtained in previous studies, new methods to create human induced pluripotent stem cells (iPS cells) and to assess their function in context-specific assays could offer an alternative approach12. Patient-specific iPS cells generate the opportunity to examine how specific risk haplotypes affect function, but given substantial genomic variation and the probable presence of additional cardiac risk regulators outside of 9p212,13, we have previously adopted a genome-editing strategy14 to isogenically assess function. To assess relevant changes in the function of cardiomyocytes, it is important to recapitulate niche stresses. For example, myocardial remodelling associated with CAD and subsequent myocardial infarction15 could affect how SNPs regulate the genome, which could affect cardiomyocyte function and lead to a higher prevalence of arrhythmic cardiac death observed in homozygous haplotypes13,16,17; conversely culture on substrates with a stiffness outside of a myocardial range or of constant stiffness would not recapitulate dynamic changes in the niche. Recently developed biomaterials address this with two-stage crosslinking; methacrylated hyaluronic acid (MeHA; Fig. 1a) is initially crosslinked into a soft matrix before in situ dynamic stiffening in the presence of cells18,19 (Fig. 1b–e). The combination of these two technologies serves as a unique system to investigate how the risk haplotype alters function in response to mechanical stress, such as stiffening induced by fibrosis. We find that patient-derived cardiomyocytes, independent of genotype, function normally on static soft matrices, but that on a dynamically stiffened matrix, only cardiomyocytes from homozygous carriers with the risk haplotype contract out of sync with each other, resulting from of a loss of gap junctions through short isoform-specific ANRIL-mediated suppression of the CDKN2A gene and activation of JNK phosphorylation. More generally, our findings indicate that disease modelling with iPS cells may require context-appropriate mechanical stresses and that responses can be induced to identify signalling mechanisms as complex as the regulation of the cardiac transcriptome by noncoding RNAs.
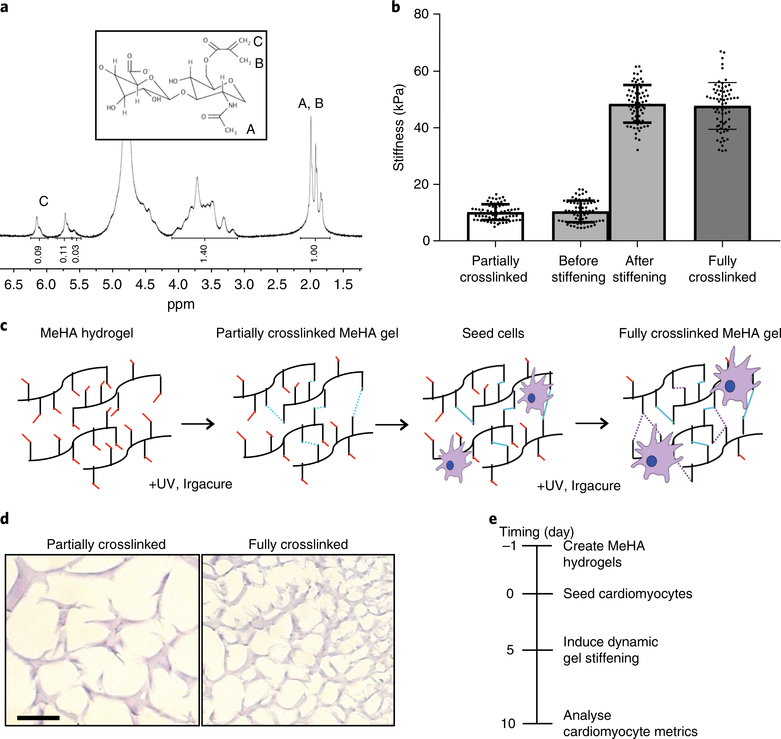
Fig. 1 |. MeHA synthesis and schematic of dynamic stiffening.
a, NMR spectrum of 50 kDa hyaluronic acid after methacrylate functionalization (the degree of methacrylation was around 40%). Inset, the MeHA structure. b, Plot of atomic-force-microscopy-measured stiffness for hydrogels of ‘partially crosslinked’ (10 kPa or ‘physiological’), ‘stiffened’ (a hydrogel originally crosslinked to 10 kPa before additionally stiffened to 50 kPa) and ‘stiff’ (50 kPa) MeHA (n = 70 measurements). Data are mean ± s.d. with individual points. c, Schematic illustrating cell seeding on soft MeHA substrates followed by sequential, in situ dynamic stiffening. d, MeHA hydrogels stained with haemotoxylin and eosin to visualize crosslinking. Scale bar, 10 μm. e, Timeline indicating cell seeding, dynamic stiffening induction and analysis.
Results
Cardiomyocytes with risk variants exhibit asynchronous contractions with dynamic stiffening.
iPS cells from patients homozygous for the risk alleles at rs1333049, rs2383207 and rs10757278 (labelled as R/R) as well as two healthy individuals homozygous for the nonrisk alleles (labelled as N/N; Supplementary Fig. 1a) were obtained from previous, related studies14. Cardiomyocyte differentiation efficiency, analysed by dual expression of cardiac troponin T and smooth muscle actin, was around 85% across multiple patient lines and independent of risk haplotype (Supplementary Fig. 1b); mRNA expression of myosin light chain 2a and 2v and myosin heavy chain 6 and 7 further indicates similar cardiac maturation across patient lines and haplotypes (Supplementary Fig. 1c). Conversely, markers of ectoderm, endoderm, mesoderm and vascular cells were minimally expressed except for the mesenchymal marker THY1, although its expression was still similar across patient lines and haplotypes (Supplementary Fig. 1d).
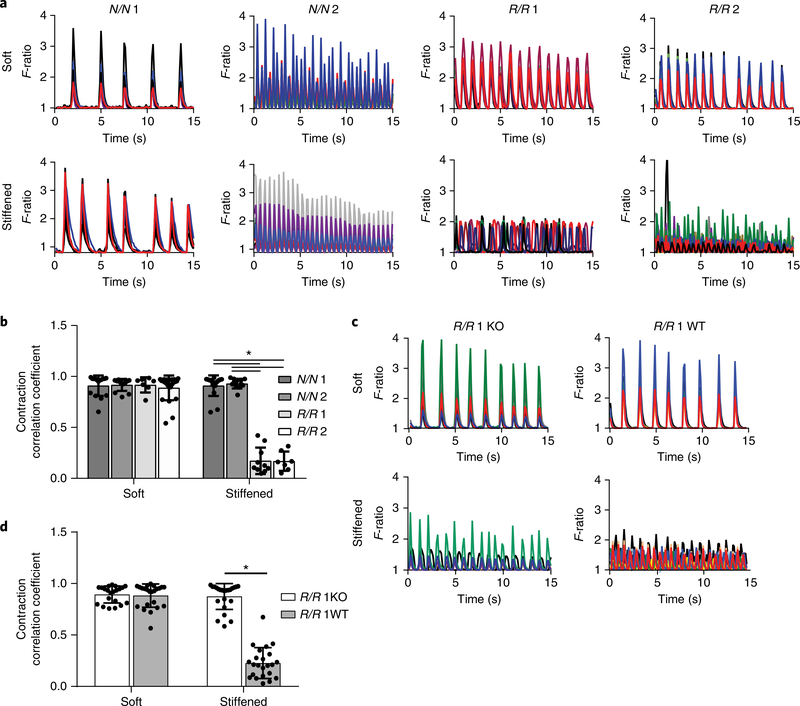
Because the risk haplotype did not affect maturation, we hypothesized that an environmental cue, such as matrix stiffness, which increases fivefold during wall remodelling15, could mimic the mechanical stress that is present during an infarct and induce altered behaviour in R/R cardiomyocytes. When cultured in softer conditions15,20–22 (for example, at 10 kPa (Fig. 1b)), calcium transients in cardiomyocytes within each line were highly coordinated, indicating synchronous excitation–contraction coupling (Fig. 2a,b and Supplementary Videos 1, 2). When cultures were dynamically stiffened15,23,24 (for example, up to 50 kPa ( Fig. 1e)), only cardiomyocytes derived from patients with the risk haplotype exhibited asynchronous contractions, as determined by lower correlation coefficients, compared to N/N cells that lacked the haplotype (Fig. 2a,b and Supplementary Videos 3, 4).
Fig. 2 |. Asynchronous calcium flux in R/R iPS cell-derived cardiomyocytes after dynamic stiffening.
a, Representative spontaneous Ca2+ transients plotted as the fluorescence intensity F-ratio over a 10-s time interval are shown for R/R and N/N cardiomyocytes cultured on soft and stiffened MeHA substrates. The different colours represent transients from different cells. Experiments were performed three independent times. b, Contraction correlation coefficient of N/N and R/R cardiomyocytes. *P < 0.0001 for all comparisons; one-way analysis of variance (ANOVA) followed by Tukey’s multiple comparison test. N/N 1, soft, n = 16 videos, stiffened, n = 17 videos; N/N 2, soft, n = 13 videos, stiffened, n = 12 videos; R/R 1, soft, n = 7 videos, stiffened, n = 10 videos; R/R 2, soft, n = 23 videos, stiffened, n = 7 videos. Data are mean ± s.d. with individual points. c, Representative spontaneous Ca2+ transients were plotted as the fluorescence intensity F-ratio over a 10-s time interval for R/R wild-type (WT) and knockout (KO) cardiomyocytes cultured on soft and stiffened MeHA substrates. The different colours represent transients from different cells. Experiments were performed three independent times. d, Contraction correlation coefficient. *P < 0.0001; two-tailed Student’s t-test. R/R 1 knockout, soft, n = 21 videos, stiffened, n = 23 videos; R/R 1 wild-type, soft, n = 23 videos, stiffened, n = 23 videos. Data are mean ± s.d. with individual points.
To control for genomic variability outside of 9p21, we obtained isogenic lines from which the R/R locus was deleted (R/R knockout; Supplementary Fig. 2a), as well as edited lines in which the locus was not removed (R/R wild-type)14. It should be noted that the deleted region overlaps a portion of both ANRIL and the 9p21 locus, but it is not a complete deletion of either. Haplotype editing did not affect pluripotency (Supplementary Fig. 2b) or lineage commitment, as 85% of cells were cardiac troponin T-positive and smooth muscle actin-positive after differentiation into cardiomyocytes (Supplementary Fig. 2c). Deletion in cardiomyocytes was confirmed through the presence or absence of long ANRIL isoforms, which are contained within the 9p21 locus (Supplementary Fig. 2d). Similar expression of cardiac and noncardiac markers was observed compared to previous lines (Supplementary Fig. 2e,f). R/R wild-type cardiomyocytes contracted asynchronously only on dynamically stiffened substrates, indicating that the editing process did not alter cardiomyocyte function. However, haplotype editing restored synchronous contractions in R/R knockout cardiomyocytes, suggesting that the risk haplotype is responsible for transient synchronization (Fig. 2c,d and Supplementary Videos 5,6).
Although changes in patient-matched calcium transient metrics before and after stiffening were slightly different in R/R cells versus N/N cells, no statistically significant differences were observed for calcium transient frequency, calcium transient amplitude or calcium flux per contraction in R/R knockout versus R/R wild-type cells (Supplementary Fig. 3a–c). These data suggest that Ca2+ handling itself is independent of the risk haplotype. In addition, similar differences were observed on static stiff substrates of 50 kPa (Supplementary Fig. 3d–f), suggesting that dynamic stiffening did not account for the majority of differences in calcium transient metrics. Finally, for both dynamically stiffened and static stiff matrices, we did not observe substantial peak-to-peak or amplitude variability in the calcium transients (Supplementary Fig. 3g–j).
To ensure that the asynchronicity observed in R/R cells was caused by dynamic stiffening and not by the stiffening process, calcium flux or inherent stiffness of the hydrogels, a number of control experiments were performed. Given that the stiffening process utilizes an ultraviolet-light-based polymerization method of free radicals, we assessed whether damage from ultraviolet light contributed to asynchronicity in R/R cells. First, we assessed the effect of exposure to ultraviolet light on cardiomyocytes and found little accumulation of phosphorylated p53 after a 2-min exposure to ultraviolet light, which is the time needed for the substrate to stiffen from the softer 10 to stiffer 50 kPa; although there was significant expression after 15 min (Supplementary Fig. 4a), which is more than sevenfold longer than required from the dynamics studied here. Conversely, we did not find that exposure to ultraviolet light affected connexin-43 localization at gap junctions (Supplementary Fig. 4b). Next, we found that R/R wild-type cardiomyocytes that were cultured on static soft polyacrylamide hydrogels and exposed to ultraviolet light and free radicals for 2 min from ultraviolet-light-activated Irgacure—both of which are required for MeHA hydrogel crosslinking—maintained synchronicity and had similar calcium flux dynamics (Supplementary Fig. 4c–f). Given that polyacrylamide hydrogels are incapable of additional stiffening, these data suggest that dynamic stiffening induced the observed R/R phenotype rather than the crosslinking mechanism. To further rule out that a loss of cell adhesion caused impaired conduction, we assessed cell density before and after stiffening and again no statistically significant differences were observed across patient lines (Supplementary Fig. 4g). To ensure that cardiomyocyte asynchronicity was not due to intrinsic defects in calcium flux, R/R wild-type cardiomyocytes on dynamically stiffened substrates were externally paced. Significantly higher correlation coefficients were present in paced versus unpaced cells (Supplementary Fig. 5a,b), suggesting that asynchronicity is the result of impaired signal propagation, not internal flux. Additional comparisons between cardiomyocytes with and without the R/R haplotype on substrates that were always stiff (50 kPa) indicated that both types of cardiomyocyte mainly showed synchronous contractions (Supplementary Fig. 6a,b). Together, these data suggest that the onset of mechanical stress, rather than a pathologically stiff condition itself, greatly exacerbates the phenotype. Lastly, given that some studies have demonstrated that the stiffness in a healthy human heart can be 20 kPa15, we cultured R/R knockout and wild-type cells on 20 kPa substrates to ensure that it did not induce the pathological results that were observed when substrates were stiffened to 50 kPa. We observed minimal differences in calcium transient metrics (Supplementary Fig. 3k–o) and no decrease in synchronicity (Supplementary Fig. 6c).
Gap junction remodelling results from stiffening.
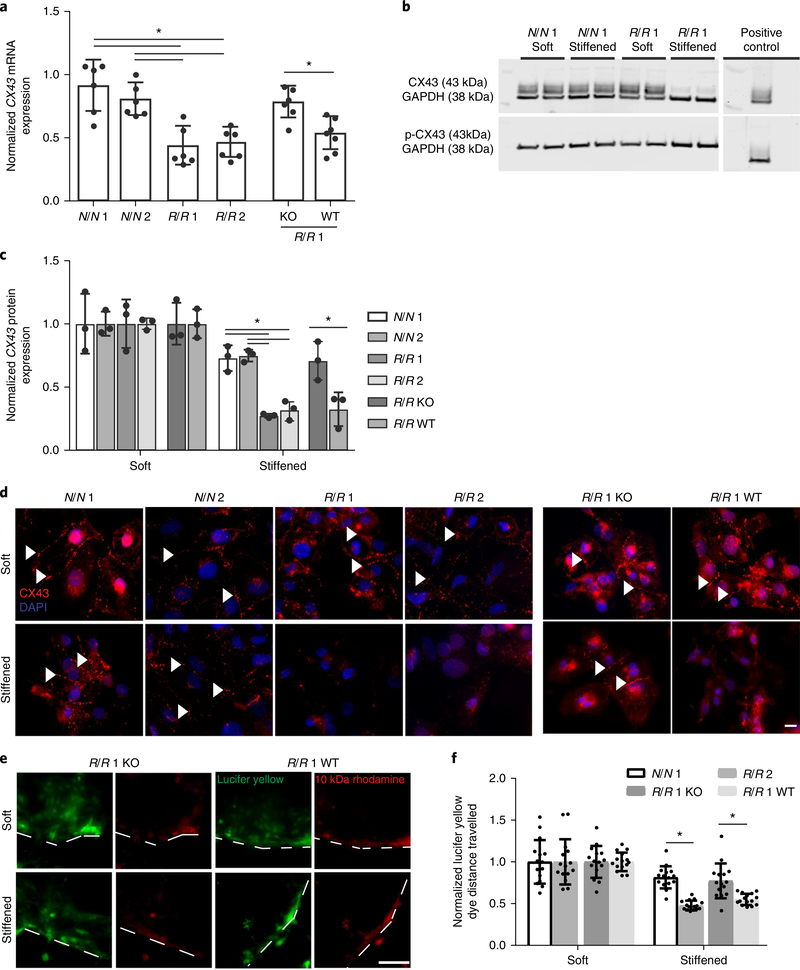
Gap junctions primarily regulate electrical conduction between cardiomyocytes. Therefore, we determined whether alterations in expression or localization of connexin-43, the most predominant gap junction protein found in ventricular cardiomyocytes25, caused asynchronicity in R/R cardiomyocytes when cultures were dynamically stiffened. Dynamic stiffening resulted in reduced mRNA expression in R/R cardiomyocytes (Fig. 3a) but not in R/R cardiomyocytes cultured on static stiff 50 kPa substrates (Supplementary Fig. 7a), again suggesting that the phenotype is induced by stiffening. Consistent with this finding, dynamic stiffening resulted in reduced protein expression of connexin-43 but not destabilization, which is commonly associated with Ser368 phosphorylation (Fig. 3b,c). To further illustrate this, connexin-43 localization was analysed; all genotypes formed functional gap junctions on static soft substrates, whereas dynamic stiffening resulted in reduced connexin-43-containing punctae only in R/R cardiomyocytes (Fig. 3d). All cardiomyocytes on static substrates of 20 and 50 kPa exhibited appropriate connexin-43 expression (Supplementary Fig. 7b,c), suggesting that decreased connexin-43 is unique to risk haplotype cardiomyocytes in the dynamically stiffened conditions and consistent with an increased propensity for arrhythmias. In addition to electrical conduction, gap junctions link the cytoplasms of adjacent cardiomyocytes; if connexin expression is perturbed, functional deficits should exist in cytoplasmic exchange. Indeed, when cardiomyocytes were scraped, cells at the leading edge were loaded with the dye lucifer yellow and 10 kDa rhodamine dextran, and after dynamic stiffening, R/R cardiomyocytes exhibited reduced transfer of lucifer yellow versus cardiomyocytes from other genotypes; dextran, which has a larger size, did not propagate beyond the first row of cells (Fig. 3e,f). These data again suggest that connexins were perturbed preferentially by dynamic stiffening and exclusively in the presence of the risk haplotype.
Fig. 3 |. Gap junction remodelling in R/R iPS cell-derived cardiomyocytes after dynamic stiffening.
a, Expression of connexin-43 mRNA (CX43 also known as GJA1). Values for stiffened substrates were normalized to those of soft substrates. *P = 0.002 for N/N 1 versus R/R 1 and *P = 0.004 for N/N 1 versus R/R 2, *P = 0.0027 for N/N 2 versus R/R 1 and *P = 0.0053 for N/N 2 versus R/R 2; one-way ANOVA followed by Tukey’s multiple comparison test. *P = 0.0054 for R/R 1 knockout versus R/R 1 wild-type; two-tailed Student’s t-test. N/N 1, n = 6 samples; N/N 2, n = 6 samples; R/R 1, n = 6 samples; R/R 2, n = 6 samples; R/R 1 knockout, n = 6 samples; R/R 1 wild-type, n = 7 samples. Data are mean ± s.d. with individual points. b, Blots showing phosphorylated and total connexin-43 expression for the indicated patient lines. c, Quantification of phosphorylated to total connexin-43. Values were normalized to GAPDH expression as a loading control (n = 3 blots). Human umbilical vein endothelial cells (HUVECs) were used as a positive control. *P = 0.002 for N/N 1 versus R/R 1 and *P = 0.003 for N/N 1 versus R/R 2, *P = 0.0015 for N/N 2 versus R/R 1 and *P = 0.003 for N/N 2 versus R/R 2; one-way ANOVA followed by Tukey’s multiple comparison test. *P = 0.032 for R/R 1 knockout versus R/R 1 wild-type; two-tailed Student’s t-test. Data are mean ± s.d. with individual points. d, Immunofluorescence images are shown for connexin-43 (red) and 4,6-diamidino-2-phenylindole (DAPI; blue) for the indicated iPS cell-derived cardiomyocyte patient types and bioreactor conditions. Arrowheads indicate regions of assembled connexin-43. Scale bar, 10 μm. Experiments were performed three independent times. e, Representative images demonstrating the transfer of lucifer yellow dye for the indicated iPS cell-derived cardiomyocyte patient types and bioreactor conditions. Rhodamine dextran images and dashed lines are included to indicate where the cut was made. Scale bar, 100 μm. Experiments were performed three independent times. f, Quantification of the distance the dye travelled (n = 15 cuts) was normalized to the distance travelled on soft substrates. *P = 0.0002 for N/N 1 versus R/R 1 and *P = 0.0011 for R/R 1 knockout versus R/R 1 wild-type; two-tailed Student’s t-test. Data are mean ± s.d. with individual points.
Loss of connexin localization could also impair contractility and disturb myofibril organization as seen in cardiomyopathies and heart failure26–28. Therefore, we assessed sarcomere organization of the Z-band protein α-actinin and found that only R/R cardiomyocytes cultured on dynamically stiffened substrates lacked assembled sarcomeres (Supplementary Fig. 8a); data were substantiated by unbiased fast Fourier transform analyses29,30 of patient-matched organizational differences before and after stiffening as well as static 20 and 50 kPa substrates (Supplementary Fig. 8b–d). Despite global differences in organization of the sarcomeres, the sarcomere assembly process was not impaired as striations, when present, were of mature length (Supplementary Fig. 8e–g), suggesting that function and not differentiation depended on the presence of the risk haplotype.
Specific ANRIL isoforms regulate JNK activation to induce cardiomyocyte asynchronicity.
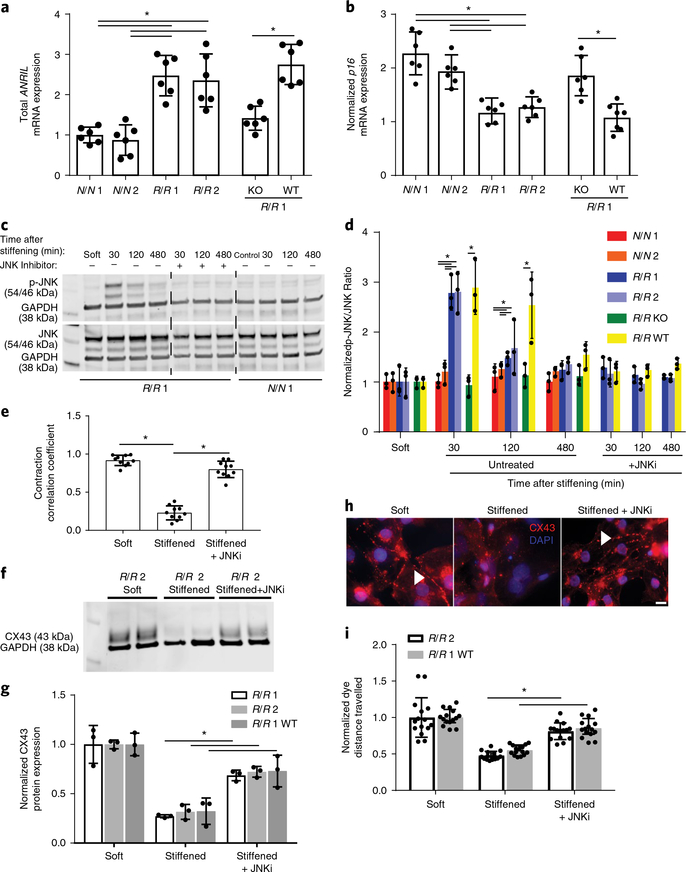
We next determined how the risk haplotype alters expression of the noncoding RNA ANRIL to induce remodelling of the gap junctions. We first examined ANRIL RNA expression, which can be expressed in short or long variants (which include exons 18–19)31. Using primers that would measure total ANRIL (exons 5–6), we found higher expression in R/R cardiomyocytes compared to patient cells either with a normal haplotype or those edited to lack the haplotype (Fig. 4a). In addition, we observed increased expression of the short ANRIL isoform in R/R cardiomyocytes compared to N/N and R/R knockout cardiomyocytes (Supplementary Fig. 9a) but no difference in the expression of the long ANRIL isoform in R/R cardiomyocytes compared to N/N cardiomyocytes (Supplementary Fig. 2d), indicating that effects are mediated by the short isoforms. Despite intrinsic differences in expression as a function of haplotype, ANRIL expression did not change with either dynamic stiffening or static 50 kPa stiffness (Supplementary Fig. 9b–g). Given that ANRIL expression suppresses activity of adjacent regulators of the cell cycle32–34, we next measured the expression of p16, a protein that is encoded by CDKN2A and is associated with cellular stress responses35. Expression of the p16 mRNA transcript was significantly increased in response to dynamic stiffening in N/N and R/R knockout cardiomyocytes compared to R/R cardiomyocytes (Fig. 4b); these data suggest that cellular stress induced by dynamic stiffening is modulated by p16 in the absence of excessive ANRIL to prevent downstream deleterious remodelling. Higher p16 expression appears to be specific to the mechanical stress induced by dynamic stiffening to compensate and maintain cardiomyocyte junctions as higher basal stiffness does not induce a similar response (Supplementary Fig. 10).
Fig. 4 |. JNK-mediated dysfunction in R/R cardiomyocytes in response to a stiffened hydrogel.
a, Expression levels of the mRNA transcrips of total ANRIL using primers directed against exons 5–6. Data are normalized to N/N patient 1. *P = 0.001 for N/N 1 versus R/R 1 and *P = 0.003 for N/N 1 versus R/R 2, *P = 0.0008 for N/N 2 versus R/R 1 and *P = 0.001 for N/N 2 versus R/R 2; one-way ANOVA followed by Tukey’s multiple comparison test. *P = 0.0004 for R/R 1 knockout versus R/R 1 wild-type; two-tailed Student’s t-test. n = 6 samples. Data are mean ± s.d. with individual points. b, Expression levels of p16 mRNA transcript for p16. Stiffened values were normalized to those for cells grown on soft substrates. *P = 0.001 for N/N 1 versus R/R 1 and *P = 0.001 for N/N 1 versus R/R 2, *P = 0.004 for N/N 2 versus R/R 1 and *P = 0.019 for N/N 2 versus R/R 2; one-way ANOVA followed by Tukey’s multiple comparison test. *P = 0.002 for R/R 1 knockout versus R/R 1 wild-type; two-tailed Student’s t-test. N/N 1, n = 6 samples; N/N 2, n = 6 samples; R/R 1, n = 6 samples; R/R 2, n = 6 samples; R/R 1 knockout, n = 6 samples; R/R 1 wild-type, n = 7 samples. Data are mean ± s.d. with individual points. c, Blots showing phosphorylated JNK and total JNK expression for the indicated patient line (bottom). Lanes are shown as indicated for the presence of the JNK inhibitor SP600125 and time course after stiffening. d, Quantification of the ratio of phosphorylated JNK to total JNK expression normalized to GAPDH. Values were normalized to GAPDH expression as a loading control (n = 3 blots). Comparisons between isogenic patient lines were intentionally separated from nonisogenic lines for the time course without inhibitor for clarity. *P = 0.0004 for N/N 1 versus R/R 1 and *P = 0.0004 for N/N 1 versus R/R 2, *P = 0.0009 for N/N 2 versus R/R 1 and *P = 0.0008 for N/N 2 versus R/R 2; one-way ANOVA followed by Tukey’s multiple comparison test at 30 min. *P = 0.0042 for R/R 1 knockout versus R/R 1 wild-type at 30 min; two-tailed Student’s t-test. *P = 0.0321 for N/N 1 versus R/R 1 and *P = 0.0283 for N/N 1 versus R/R 2, *P = 0.0417 for N/N 2 versus R/R 1 and *P = 0.0397 for N/N 2 versus R/R 2; one-way ANOVA followed by Tukey’s multiple comparison test at 120 min. *P = 0.026 for R/R 1 knockout versus R/R 1 wild-type at 120 min; two-tailed Student’s t-test. Mean ± s.d. with individual points. e, Contraction correlation coefficients for R/R cardiomyocytes on soft substrates, stiffened substrates and stiffened substrates treated with the JNK inhibitor (JNKi) SP600125 were plotted (n = 10 videos). *P < 0.0001 for soft versus stiffened and *P = 0.0006 for stiffened versus stiffened treated with JNKi; one-way ANOVA followed by Tukey’s multiple comparison test. Data are mean ± s.d. with individual points. f, Blots showing phosphorylated and total connexin-43 expression for the indicated patient line. g, Quantification of the ratio of phosphorylated connexin-43 to total connexin-43 expression normalized to GAPDH. Values were normalized to GAPDH expression as a loading control (n = 3 blots). *P = 0.0033 for R/R 1 stiffened versus stiffened treated with JNKi, *P = 0.0024 for R/R 2 stiffened versus stiffened treated with JNKi, *P = 0.029 for R/R 1 wild-type stiffened versus stiffened treated with JNKi; two-tailed Student’s t-test. Data are mean ± s.d. with individual points. h, Immunoflurorescence images of connexin-43 (red) and DAPI (blue) for the conditions described above. Scale bar, 10 μm. Experiments were performed three independent times. i, Quantification of the distance the dye travelled (n = 15 cuts). *P < 0.0001 for R/R 2 stiffened versus stiffened treated with JNKi and *P < 0.0001 for R/R 1 wild-type stiffened versus stiffened treated with JNKi; two-tailed Student’s t-test. Data are mean ± s.d. with individual points.
To better understand how changes in ANRIL and p16 expression connect to reduced expression of connexin-43, we performed a phospho-kinase screen that measures 43 different kinase activities. A reduction dependent on dynamic stiffening was observed in the activity of extracellular signal-regulated kinase (ERK) that appeared to be independent of the risk haplotype. In addition, marked upregulation was observed in JNK phosphorylation (p-JNK) as well as the possible downstream targets cyclic AMP-responsive element-binding protein (CREB) and heat-shock protein 27 (HSP27) in R/R cardiomyocytes compared to N/N or R/R knockout cardiomyocytes after dynamic stiffening (Supplementary Fig. 11). To validate this finding, the expression of phosphorylated and total JNK was measured in cardiomyocytes cultured on static soft and dynamically stiffened hydrogels. Expression of p-JNK was significantly increased in R/R cardiomyocytes immediately after dynamic stiffening compared to N/N and R/R knockout cardiomyocytes, in which the expression was suppressed (Fig. 4c,d). We further modulated JNK phosphorylation to determine its impacts on cardiomyocyte function when cultures were dynamically stiffened. In the presence of the p-JNK antagonist SP600125 and after dynamic stiffening, JNK phosphorylation was inhibited (Fig. 4c,d), which restored coordination of calcium transients in adjacent R/R cardiomyocytes despite a dynamically stiffened matrix (Fig. 4e); inhibition also improved α-actinin localization to Z-bands despite dynamic stiffening, but it did not change calcium dynamics (Supplementary Fig. 12a–e). Chronic inhibition improved expression of connexin-43 and altered localization (Fig. 4f–h). Furthermore, inhibition resulted in improved function of gap junctions, as measured by increased dye transfer compared to the dynamically stiffened condition (Fig. 4i).
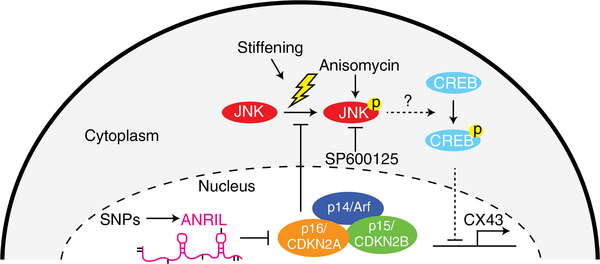
Although this proposed mechanism depends on ANRIL-mediated p-JNK activation, we next sought to determine whether JNK activation alone, without the risk haplotype, could activate downstream components of the pathway. R/R knockout and wild-type cardiomyocytes were cultured on static soft hydrogels and treated with the p-JNK agonist anisomycin in lieu of dynamic stiffening. Anisomycin-treated R/R wild-type cardiomyocytes lost connexin-43 and α-actinin localization consistent with R/R cardiomyocytes cultured on dynamically stiffened hydrogels (Supplementary Fig. 13a–c). Such pathway perturbations together suggest that ANRIL-mediated silencing of p16 enables cardiomyocytes to become sensitive to the detrimental effects of changes in JNK phosphorylation, which include blocking the expression of connexin-43 in the presence of stress. Previous studies have demonstrated that p16 prevents JNK phosphorylation in response to stress35, and p-JNK, in turn, reduces expression of gap junctions in cardiomyocytes and contributes to arrhythmias36–38, potentially through CREB39. Therefore, these data and literature together suggest a complex pathway (Fig. 5), in which the risk haplotype increases expression of the short ANRIL isoform to block p16 inhibition of JNK phosphorylation and CREB-mediated transcriptional suppression of connexin-43.
Fig. 5 |. Proposed mechanism for the regulation of 9p21 in cardiomyocytes.
In R/R cardiomyocytes, increased levels of ANRIL silences CDKN2A and, therefore, in response to stress—for example, stiffening—JNK phosphorylates and activates downstream targets that downregulate connexin-43. In cardiomyocytes without the risk haplotype, there is insufficient ANRIL to silence p16, so stress-mediated JNK phosphorylation is blocked and connexin-43 expression is maintained. Short dashed lines indicate portions of the pathway inferred from the literature35–39. p14/Arf, p14 is also known as Arf; p15/CDKN2B, p15 is also known as CDKN2B; p16/CDKN2A, p16 is also known as CDKN2A.
Discussion
Association studies frequently identify variants in noncoding regions that correlate with disease, but that have mechanisms that are difficult to study. This could be because of linkage disequilibrium40, overlapping risk regulators2,13, divergent animal models6–8 or a requirement for a covariant stressor. As illustrated here, a genome-editing strategy14 to isogenically assess variants in patient-specific iPS cell-derived cardiomyocytes along with their culture on a MeHA hydrogel to mimic remodelling associated with disease15 affected how the risk haplotype regulates the genome, impairing the coordinated contraction of cardiomyocytes in a manner that potentially mirrors disease13,16,17. Although our approach specifically examined the 9p21 locus, the vast majority of loci remain undetermined2 and may be noncoding. Therefore, our data are supportive of a general approach in which suitable biological and cell systems are developed to explore mechanism(s) in vitro and under pathologically appropriate mechanical stress when in vivo assays are impractical or impossible. Of course this approach depends on the success of genetic strategies to appropriately isolate variants, on the affected cell type and may also depend on the maturation state of the iPS cell-derived lineage, which—although it did not have fully mature cardiomyocytes—was similar across genotypes and consistent with previous methods30,41.
The presence of risk variants directly caused asynchronous contractions and altered connexin-43 expression in R/R cardiomyocytes only after dynamic stiffening was induced; this phenotype was abolished when the risk haplotype was deleted isogenically. Increased susceptibility to mechanical stress can be explained by altered JNK activation in the R/R lines: JNK phosphorylation was higher in R/R cardiomyocytes compared to N/N or R/R knockout cardiomyocytes only after dynamic stiffening and could be reversed by p-JNK inhibition. These findings demonstrate stress-induced activation caused by the expression of a noncoding RNA in cardiomyocytes using a biomaterial system.
Although the effects of the risk haplotype or dynamic substrate stiffening on cardiomyocytes has not been clear to date, stress-mediated loss of connexin-43 in cardiomyocytes has been noted both in vitro and in vivo. Rabbit cardiac trabeculae subjected to load-induced hypertrophy in vitro showed downregulation of connexin-43 expression compared to unloaded conditions28, which support previous studies that reported decreased connexin-43 expression in ischaemic, hypertrophic and failing human left ventricles42. In addition, cyclic strain resulted in reduced expression of connexin-43 mRNA (which is encoded by GJA1) and increased heterogeneity in sarcomeric patterns in stem cell-derived cardiomyoctes from patients with dilated cardiomyopathy compared to controls43, suggesting a similar susceptibility to mechanical stress in other stem cell models albeit owing to different mechanisms.
Pathways identified in cardiomyocytes using this strategy are not probably not exclusive to cardiomyocytes. Vascular smooth muscle cells that contain the risk haplotype also exhibit altered p15 and p16 expression44,45, even in primary cells. Global deletion of the orthologous 9p21 region in mice resulted in altered p15 and p16 expression, increased proliferation and decreased senescence in vascular smooth muscle cells, but it failed to increase the development of atherogenic lesions, making the mechanisms by which pathogenesis occurs unclear7. However, as we noted, there is limited sequence conservation of ANRIL in orthologous sequences8, large differences in expression9 and considerably differences in alternative splice variants10,11. Thus, a human-centric approach using genome-edited iPS cells such as those described elsewhere14 may further clarify similarities in cell cycle regulation between cell types. In humans, p16 suppresses JNK phosphorylation35, which is activated in response to environmental stress and which downregulates connexin-43 in cardiomyocytes36–38,46. Hits from the phospho-screen, such as CREB, which has also been shown to transcriptionally silence connexin-4339, and HSP27, a cardioprotective and apoptotic antagonist47, further describe the regulation of arrhythmic contraction without inducing cell death in a human model.
We describe this mechanism in cardiomyocytes—despite caveats that have been raised about the maturity of iPS cell-derived cardiomyocytes—however, the 9p21 locus contributes to a multitude of diseases that are characterized by pathological mechanical and oxidative stress10. Little is known about how stress influences phenotypic manifestation of the 9p21 risk allele in either endothelial cells or vascular smooth muscle cells. Altered interferon-γ-mediated inflammatory signalling in human umbilical vein endothelial cells with 9p21 SNPs contributed to CAD40, but others have shown that interferon-γ acted independently of 9p21 risk variants32. However, given the complexities of ANRIL expression and splicing, broad generalizations between cell types, models and mechanisms are not possible at this time, but given the approach used here, we are confident that additional data will clarify how interactions between stress and noncoding loci more generally alter disease-causing mechanisms.
In summary, our data indicate that the presence of the risk haplotype downregulates connexin-43 expression and localization through JNK activation in response to mechanical stress. These findings, coupled with the loss of sarcomeric organization, suggest a possible mechanism that contributes to asynchronous contraction. However, additional studies in primary tissues are needed to further validate this hypothesis.
Methods
An expanded Methods section is included in the Supplementary Information, with additional details on iPS cell maintenance and cardiomyocyte differentiation, flow cytometry analysis, atomic force microscopy measurements, immunofluorescence assays and analysis, scrape-loading dye transfer assay, quantitative PCR, phospho-kinase array assays, western blot analysis, JNK signalling analysis and electrical stimulation of cardiomyocytes.
MeHA synthesis.
MeHA was synthesized as previously described18,19. In brief, 50 kDa sodium hyaluronate (Lifecore Biomedical, HA40K) was dissolved in deionized water at 1% w/v overnight and reacted with methacrylate anhydride (Sigma-Aldrich, 276685, 6 ml methacrylate anhydride per 1 g sodium hyaluronate) for 8 h on ice at pH 8, followed by overnight incubation at 4 °C, and further reacted with methacrylate anhydride (3 ml methacrylate anhydride per 1 g sodium hyaluronate) at pH 8 on ice for 4 h. The monomer solution was then dialysed (Spectrum Labs, 132655) against deionized water at 4 °C for 3 days followed by lyophilization for another 3 days. The lyophilized MeHA was analysed by 1H magnetic resonance (NMR) to determine the methacrylate substitution ratio by normalization of the peaks at a chemical shift of 6 ppm, representing the methacrylate group, by peaks between 3 and 4 ppm, which represent native HA. Substitution ratios were computed from triplicate NMR analyses (Fig. 1a).
MeHA substrate fabrication.
MeHA was dissolved at 3% in 0.2 M triethanolamine (Sigma-Aldrich, T58300) and phosphate-buffered saline (PBS) solution. Irgacure 2959 (Sigma-Aldrich, 410896), the ultraviolet-light-activated free radical donor, was initially dissolved at 10% in 100% ethanol and then diluted to 0.02% in the MeHA solution. Next, 10 μl of the hydrogel solution was sandwiched between a 12-mm diameter methacrylated glass coverslip to permit hydrogel binding and a nonadherent dichlorodimethylsilane (Acros Organics, AC11331)-activated glass slide to achieve easy detachment and photopolymerized using a transilluminator (4 mW cm−2, UVP) emitting 350 nm wavelength ultraviolet light. Methacrylation of glass coverslips was achieved by first oxidizing the surface through ultraviolet light-ozone exposure (BioForce Nanosciences) followed by functionalization with 20 mM 3-(trimethoxysilyl)propyl methacrylate (Sigma-Aldrich, 440159) in ethanol. Proteins for cell attachment were added by mixing 20 mM 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (ProteoChem, c1100), 50 mM N-hydroxysuccinimide (Alfa Aesar, A10312) and 150 μg ml−1 type I rat tail collagen (Corning, 354236) dissolved in PBS. The collagen-crosslinking solution was added to the hydrogel and incubated overnight at 37 °C.
Seeding cardiomyocytes onto MeHA substrates and in situ dynamic stiffening.
On day 30, cardiomyocytes were dissociated using 0.25% trypsin-EDTA at 37 °C and resuspended in RPMI medium, 20% fetal bovine serum (Gemini Bio Products, 900–108) and 5 μM Y27632 (StemGent, 04–0012). Then, 200,000 cardiomyocytes were seeded on each MeHA hydrogels. Two days later, the medium was changed to RPMI medium supplemented with vitamin B27. On day 5, MeHA hydrogels were stiffened as previously described18,19. Irgacure 2959 was added at 0.05% v/v to cell medium. Following a 30-min incubation without ultraviolet-light exposure, hydrogels were exposed to 350 nm wavelength ultraviolet light for 2 min to generate a stiff 50 kPa hydrogel, washed two times with sterile PBS to remove unreacted Irgacure 2959 and resuspended in medium. It should be noted that the same cell population was used for static soft, dynamically stiffened and static stiff substrates. To produce the dynamically stiffened substrates, only a subset of soft substrates were dynamically stiffened on day 5, ensuring that cells from the same differentiation were seeded on hydrogels of the same composition before the subset were stiffened; static soft and static stiff substrate still underwent the same brief ultraviolet-light treatment in the absence of the photoactivated radical donor. The medium was changed again on day 7 before experiments were carried out on day 10.
To ensure altered cellular behaviour was not caused by ultraviolet-light exposure, cells on soft and stiff substrates were also exposed to ultraviolet light for the same duration as cells on stiffened substrates. In addition, cells were seeded on 10 kPa polyacrylamide hydrogels that are incapable of stiffening and exposed to the stiffening process. Coverslips were methacrylated as described above. A polymer solution containing 10%/0.1% acrylamide/bis-acrylamide (Fisher), 0.5% acrylic acid, 1% v/v of 10% ammonium persulfate (Fisher) and 0.1% v/v of N,N,N′,N′ -tetramethylethylenediamine (VWR International) was prepared. Then, 10 μl of polymerizing hydrogel solution was sandwiched between a functionalized coverslip and a dichlorodimethylsilane-treated glass slide and was allowed to polymerize for 15 min. Collagen-crosslinking solution was added overnight and cells were seeded on hydrogels as described above. On day 5, substrates were incubated with Irgacure 2959 and exposed to ultraviolet light as described above. Calcium metrics were then analysed on day 10 to ensure that there was no difference in behaviour as a result of exposure to free radicals.
Calcium-handling assay.
Calcium imaging was performed by adding 0.5 μM Fluo-4 AM (Thermo Fisher, F-14201) directly into the cell medium for 15 min. Videos were captured using a 20× objective on a Nikon Eclipse TI fluorescence microscope outfitted with a LiveCell imager with Metamorph 7.6 software. Acquisition rate was 25 frames per second. Then, 10 cells from each video were randomly selected and the mean fluorescence intensities were recorded using Fiji and plotted for a minimum of 5 beats. Calcium tracings were then analysed using custom-written code in MATLAB to determine correlation coefficient, mean peak area, mean peak amplitude, transient frequency, peak-to-peak irregularity and amplitude irregularity. The contraction correlation coefficient was determined by computing the Pearson’s correlation coefficients of the fluorescence intensities of each cell to one another within a single video to create a large matrix of correlations. The correlation coefficient for each video was then averaged to obtain a final correlation coefficient value. Mean peak area was calculated by integrating the area under the fluorescence intensities and dividing by the number of peaks. Mean peak amplitude was calculated as the maximum fluorescence peak and transient frequency was defined as the number of peaks. Transient frequency was calculated as the number of calcium transients. Peak-to-peak irregularity was calculated by dividing the s.d. of the time between peaks by the mean time. Amplitude irregularity was calculated by dividing the s.d. of the peaks by the mean as previously described12. To determine the effect of stiffening on each cell line, all calcium values for dynamically stiffened or static stiff substrates at day 10 were determined from cardiomyocytes plated from the same differentiation and normalized to the average of the values on static soft substrates for each cell line from day 10. The raw data and absolute numbers related to the data in the figures are provided in the Supplementary Data.
Statistical analysis.
All experiments were performed using cells from three distinct differentiations and at least five distinct MeHA hydrogels. Bar graphs are represented as mean ± s.d. Box and whisker graphs show the median as a line, box edges are the 25% and 75% quartiles and whiskers extend to the maximum and minimum points. Statistically significant differences among two groups were tested with two-tailed Student’s t-tests. Statistically significant differences among more than two groups were analysed using a one-way ANOVA followed by Tukey’s multiple comparison test. Statistical analyses were performed using Graphpad Prism software, with the threshold for significance set at P < 0.05. Comparisons between isogenic patient lines were intentionally separated from comparisons between nonisogenic lines for clarity. Nonisogenic lines were analysed using an ANOVA and isogenic lines were analysed using a Student’s t-test. All data represent biological triplicates with the number of technical replicates (n) indicated where appropriate.
Supplementary Material
Acknowledgements
We thank M. Ondeck, C. Happe, K. Vincent, X. Ma and A. McCulloch for technical support and helpful discussions regarding cardiomyocyte differentiation, functional assays and pacing; F. Urnov for design of the TALE nucleases. National Institutes of Health (NIH) grants R01AG045428 (A.J.E.), UL1RR025774 and U01HL107436 (Scripps Translational Science Institute, E.J.T. and K.K.B.), U54GM114833 (A.T.) and F32HL126406 (J.K.P.) supported this work. NIH grants T32HL105373 (E.J.T.) and T32AR060712 (A.K.) and the ARCS/Roche Foundation Scholar Award Program in the Life Science (A.K.) provided trainee support. National Science Foundation grant 1463689 (A.J.E.) and the graduate fellowship program (A.K.) also provided support. UC San Diego Frontiers of Innovation Scholars Program award 2-U1041 also provided trainee support (S.K.T.).
Footnotes
Competing interests
The authors declare no competing interests.
Ethical compliance. We complied with all ethical regulations. Informed consent from the human participants was obtained under a study approved by the Scripps IRB (11–5676). Cells were transferred and maintained under a study approved by the UCSD IRB (141315).
Reporting Summary. Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Code availability
All custom written code in this study is available at https://github.com/englea52/Englerlab. It contains code for the calcium handling analysis, correlation coefficient analysis and sarcomere analysis.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its Supplementary Information. Source data for the figures are available from the corresponding author upon reasonable request.
Additional information
Supplementary information is available for this paper at https://doi.org/10.1038/s41551-018-0344-5.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.The 1000 Genomes Project Consortium. A global reference for human genetic variation. Nature 526, 68–74 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schaub MA, Boyle AP, Kundaje A, Batzoglou S & Snyder M Linking disease associations with regulatory information in the human genome. Genome Res. 22, 1748–1759 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Helgadottir A et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science 316, 1491–1493 (2007). [DOI] [PubMed] [Google Scholar]
- 4.McPherson R Chromosome 9p21.3 locus for coronary artery disease: how little we know. J. Am. Coll. Cardiol 62, 1382–1383 (2013). [DOI] [PubMed] [Google Scholar]
- 5.Roberts R & Stewart AFR 9p21 and the genetic revolution for coronary artery disease. Clin. Chem 58, 104–112 (2012). [DOI] [PubMed] [Google Scholar]
- 6.Kim JB et al. Effect of 9p21.3 coronary artery disease locus neighboring genes on atherosclerosis in mice. Circulation 126, 1896–1906 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Visel A et al. Targeted deletion of the 9p21 non-coding coronary artery disease risk interval in mice. Nature 464, 409–412 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarinova O et al. Functional analysis of the chromosome 9p21.3 coronary artery disease risk locus. Arterioscler. Thromb. Vasc. Biol 29, 1671–1677 (2009). [DOI] [PubMed] [Google Scholar]
- 9.Holdt LM & Teupser D Recent studies of the human chromosome 9p21 locus, which is associated with atherosclerosis in human populations. Arterioscler. Thromb. Vasc. Biol 32, 196–206 (2012). [DOI] [PubMed] [Google Scholar]
- 10.Congrains A, Kamide K, Ohishi M & Rakugi H ANRIL: molecular mechanisms and implications in human health. Int. J. Mol. Sci 14, 1278–1292 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Holdt LM et al. Alu elements in ANRIL non-coding RNA at chromosome 9p21 modulate atherogenic cell functions through trans-regulation of gene networks. PLoS Genet. 9, e1003588 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sun N et al. Patient-specific induced pluripotent stem cells as a model for familial dilated cardiomyopathy. Sci. Transl. Med 4, 130ra47 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Larson MG et al. Framingham Heart Study 100k project: genome-wide associations for cardiovascular disease outcomes. BMC Med. Genet 8, S5 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lo Sardo V et al. Unveiling the role of the most impactful cardiovascular risk locus through haplotype editing. Cell 175, 1796–1810 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Berry MF et al. Mesenchymal stem cell injection after myocardial infarction improves myocardial compliance. Am. J. Physiol. Heart Circ. Physiol 290, H2196–H2203 (2006). [DOI] [PubMed] [Google Scholar]
- 16.Lahtinen AM et al. Common genetic variants associated with sudden cardiac death: the FinSCDgen study. PLoS ONE 7, e41675 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newton-Cheh C et al. A common variant at 9p21 is associated with sudden and arrhythmic cardiac death. Circulation 120, 2062–2068 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ondeck MG & Engler AJ Mechanical characterization of a dynamic and tunable methacrylated hyaluronic acid hydrogel. J. Biomech. Eng 138, 021003 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Guvendiren M & Burdick JA Stiffening hydrogels to probe short- and long-term cellular responses to dynamic mechanics. Nat. Commun 3, 792 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Engler AJ et al. Embryonic cardiomyocytes beat best on a matrix with heart-like elasticity: scar-like rigidity inhibits beating. J. Cell Sci 121, 3794–3802 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Young JL & Engler AJ Hydrogels with time-dependent material properties enhance cardiomyocyte differentiation in vitro. Biomaterials 32, 1002–1009 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacot JG, McCulloch AD & Omens JH Substrate stiffness affects the functional maturation of neonatal rat ventricular myocytes. Biophys. J 95, 3479–3487 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fomovsky GM & Holmes JW Evolution of scar structure, mechanics, and ventricular function after myocardial infarction in the rat. Am. J. Physiol. Heart Circ. Physiol 298, H221–H228 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sun Y & Weber KT Infarct scar: a dynamic tissue. Cardiovasc. Res 46, 250–256 (2000). [DOI] [PubMed] [Google Scholar]
- 25.Stauffer BL, Sobus R & Sucharov CC Sex differences in cardiomyocyte connexin43 expression. J. Cardiovasc. Pharmacol 58, 32–39 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kostin S Zonula occludens-1 and connexin 43 expression in the failing human heart. J. Cell. Mol. Med 11, 892–895 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harvey PA & Leinwand LA The cell biology of disease: cellular mechanisms of cardiomyopathy. J. Cell Biol 194, 355–365 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bupha-Intr T, Haizlip KM & Janssen PML Temporal changes in expression of connexin 43 after load-induced hypertrophy in vitro. Am. J. Physiol. Heart Circ. Physiol 296, H806–H814 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monteiro da Rocha A et al. Deficient cMyBP-C protein expression during cardiomyocyte differentiation underlies human hypertrophic cardiomyopathy cellular phenotypes in disease specific human ES cell derived cardiomyocytes. J. Mol. Cell. Cardiol 99, 197–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hinson JT et al. Titin mutations in iPS cells define sarcomere insufficiency as a cause of dilated cardiomyopathy. Science 349, 982–986 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Folkersen L et al. Relationship between CAD risk genotype in the chromosome 9p21 locus and gene expression. Identification of eight new ANRIL splice variants. PLoS ONE 4, e7677 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Almontashiri NAM et al. Interferon-γ activates expression of p15 and p16 regardless of 9p21.3 coronary artery disease risk genotype. J. Am. Coll. Cardiol 61, 143–147 (2013). [DOI] [PubMed] [Google Scholar]
- 33.Roberts R et al. Identifying genes for coronary artery disease: an idea whose time has come. Can. J. Cardiol 23, 7A–15A (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samani NJ & Schunkert H Chromosome 9p21 and cardiovascular disease: the story unfolds. Circ. Cardiovasc. Genet 1, 81–84 (2008). [DOI] [PubMed] [Google Scholar]
- 35.Choi BY et al. The tumor suppressor p16INK4a prevents cell transformation through inhibition of c-Jun phosphorylation and AP-1 activity. Nat. Struct. Mol. Biol 12, 699–707 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Yan J et al. c-Jun N-terminal kinase activation contributes to reduced connexin43 and development of atrial arrhythmias. Cardiovasc. Res 97, 589–597 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warn-Cramer BJ, Cottrell GT, Burt JM & Lau AF Regulation of connexin-43 gap junctional intercellular communication by mitogen-activated protein kinase. J. Biol. Chem 273, 9188–9196 (1998). [DOI] [PubMed] [Google Scholar]
- 38.Petrich BG et al. Targeted activation of c-Jun N-terminal kinase in vivo induces restrictive cardiomyopathy and conduction defects. J. Biol. Chem. 279, 15330–15338 (2004). [DOI] [PubMed] [Google Scholar]
- 39.Zhang X et al. AMPK suppresses connexin43 expression in the bladder and ameliorates voiding dysfunction in cyclophosphamide-induced mouse cystitis. Sci. Rep 6, 19708 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Harismendy O et al. 9p21 DNA variants associated with coronary artery disease impair interferon-γ signaling response. Nature 470, 264–268 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lian X et al. Directed cardiomyocyte differentiation from human pluripotent stem cells by modulating Wnt/Î2-catenin signaling under fully defined conditions. Nat. Protocols 8, 162–175 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Severs NJ, Bruce AF, Dupont E & Rothery S Remodelling of gap junctions and connexin expression in diseased myocardium. Cardiovasc. Res 80, 9–19 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chun YW et al. Differential responses of induced pluripotent stem cell-derived cardiomyocytes to anisotropic strain depends on disease status. J. Biomech 48, 3890–3896 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Almontashiri NAM et al. 9p21.3 coronary artery disease risk variants disrupt TEAD transcription factor-dependent transforming growht factor β regulation of p16 expression in human aortic smooth muscle cells.Circulation 132, 1969–1978 (2015). [DOI] [PubMed] [Google Scholar]
- 45.Motterle A et al. Functional analyses of coronary artery disease associated variation on chromosome 9p21 in vascular smooth muscle cells. Hum. Mol. Genet 21, 4021–4029 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Petrich BG et al. c-Jun N-terminal kinase activation mediates downregulation of connexin43 in cardiomyocytes. Circ. Res 91, 640–647 (2002). [DOI] [PubMed] [Google Scholar]
- 47.Clements RT, Feng J, Cordeiro B, Bianchi C & Sellke FW p38 MAPK-dependent small HSP27 and α B-crystallin phosphorylation in regulation of myocardial function following cardioplegic arrest. Am. J. Physiol. Heart Circ. Physiol 300, H1669–H1677 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.